ABSTRACT
HIV-1's Rev protein forms a homo-oligomeric adaptor complex linking viral RNAs to the cellular CRM1/Ran-GTP nuclear export machinery through the activity of Rev's prototypical leucine-rich nuclear export signal (NES). In this study, we used a functional fluorescently tagged Rev fusion protein as a platform to study the effects of modulating Rev NES identity, number, position, or strength on Rev subcellular trafficking, viral RNA nuclear export, and infectious virion production. We found that Rev activity was remarkably tolerant of diverse NES sequences, including supraphysiological NES (SNES) peptides that otherwise arrest CRM1 transport complexes at nuclear pores. Rev's ability to tolerate a SNES was both position and multimerization dependent, an observation consistent with a model wherein Rev self-association acts to transiently mask the NES peptide(s), thereby biasing Rev's trafficking into the nucleus. Combined imaging and functional assays also indicated that NES masking underpins Rev's well-known tendency to accumulate at the nucleolus, as well as Rev's capacity to activate optimal levels of late viral gene expression. We propose that Rev multimerization and NES masking regulates Rev's trafficking to and retention within the nucleus even prior to RNA binding.
IMPORTANCE HIV-1 infects more than 34 million people worldwide causing >1 million deaths per year. Infectious virion production is activated by the essential viral Rev protein that mediates nuclear export of intron-bearing late-stage viral mRNAs. Rev's shuttling into and out of the nucleus is regulated by the antagonistic activities of both a peptide-encoded N-terminal nuclear localization signal and C-terminal nuclear export signal (NES). How Rev and related viral proteins balance strong import and export activities in order to achieve optimal levels of viral gene expression is incompletely understood. We provide evidence that multimerization provides a mechanism by which Rev transiently masks its NES peptide, thereby biasing its trafficking to and retention within the nucleus. Targeted pharmacological disruption of Rev-Rev interactions should perturb multiple Rev activities, both Rev-RNA binding and Rev's trafficking to the nucleus in the first place.
KEYWORDS: CRM1, Gag, RNA trafficking, Rev, exportin-1, human immunodeficiency virus, nuclear export signal, nuclear pore complex, nucleolus, retroviruses
INTRODUCTION
A core challenge to eukaryotic gene expression is ensuring strong but transient interactions between newly transcribed messenger RNAs (mRNAs) in the nucleus and export receptors at nuclear pore complexes (NPCs) (1–3). For spliced mRNAs, posttranscriptional regulatory factors program export receptor recruitment, the formation of export complexes, and subsequent transit through the hydrophobic core of the NPC (4, 5). mRNA dissociation from the NPC is also crucial and is regulated by RNA binding proteins that couple nuclear egress to mRNA turnover, trafficking, and translation machineries in the cytoplasm (6, 7).
Tight regulation of mRNA nucleocytoplasmic transport is also crucial to the replication of many viruses, including retroviruses such as human immunodeficiency virus type 1 (HIV-1). Retroviruses have necessarily evolved to overcome strong cellular blocks to the nuclear export of RNA species bearing introns (8–11). Full-length, intron-retaining retroviral RNAs are transcribed in the nucleus and, upon export to the cytoplasm, serve both as the viral mRNA translated to generate Gag and Gag-Pol structural proteins, as well as the genomic RNA substrate (gRNA) bound by Gag for encapsidation into assembling virions (12–14). To ensure full-length RNA nuclear export (and, in some instances, the export of additional partially spliced viral mRNAs), retroviruses employ cis-acting RNA elements that directly recruit RNA binding proteins that form functional ribonucleoprotein (RNP) transport complexes that facilitate interactions with the NPC (8, 15–17).
HIV-1's Rev response element (RRE), the best-studied example of a cis-acting nuclear export element, hijacks the cellular chromosomal region maintenance 1 (CRM1, also known as exportin-1 or XPO1) nuclear export receptor (18–22) through coordinated interactions with the viral Rev protein. Rev is translated from fully spliced viral mRNAs, trafficked to the nucleus through interactions between its arginine-rich nuclear localization signal (NLS) and importin-β (23, 24), and multimerizes on the RRE as either monomers or dimers (25–31). Rev recruits CRM1 in complex with Ran-GTP to the RRE through the activity of a prototypical leucine-rich nuclear export signal (NES) found in Rev's disordered C-terminal domain (27, 32–34). Rev, CRM1, and Ran-GTP complexes form cooperatively in the nucleus, traverse the nuclear pore, and then disassemble in the cytoplasm when Ran-GTP is hydrolyzed to Ran-GDP (32, 35–37). Rev and CRM1 are thought to then recycle to the nucleus to mediate subsequent rounds of viral RNA nuclear export.
Rev's exploitation of CRM1 using an NES reflects viral mimicry of a conserved, constitutive mechanism for cellular protein nuclear export. Hundreds of NES peptides are proposed for both cellular and viral nuclear substrates (38–43). However, only a handful have been validated or carefully studied. Rev and the cellular protein kinase A inhibitor (PKI) were the first proteins found to encode discrete NES peptides (44, 45), discoveries that facilitated the identification of CRM1 as the receptor responsible for NES recognition (32, 33, 46, 47). CRM1 is a toroid-shaped protein consisting of 21 antiparallel alpha helices known as Huntington, elongation factor 3, protein phosphatase 2A, and TOR1 (HEAT) repeats (48–50). Functional studies and subsequent crystal structures of RanGTP-CRM1-NES complexes revealed that NESs are engaged by CRM1 through a RanGTP-dependent, hydrophobic pocket defined by surface-exposed CRM1 HEAT repeats 11 and 12 (34, 48, 49). Typical NES peptides are 10 to 15 amino acids in length and often conform to a consensus of regularly spaced hydrophobic residues, Φ1x2-3Φ2x2-3Φ3xΦ4, wherein Φ is a hydrophobic residue (often leucine) and x represents any residue (34, 41, 44, 45, 51, 52).
Most NES peptides exhibit weak affinity for CRM1/Ran-GTP (∼μM range), thus favoring efficient release in response to Ran-GTP hydrolysis in the cytoplasm (1, 34). However, some viruses such as the parvovirus minute virus of mice (MVM) or the alphavirus Venezuelan equine encephalitis virus (VEEV) encode proteins bearing “supraphysiological” NES (SNES) peptides that bind CRM1 tightly and arrest cellular trafficking at the NPC with little to no reliance on Ran-GTP (34, 53–55). For MVM, the SNES is located in the capsid protein and promotes export of bulky, intact capsids from the nucleus to the cytoplasm just prior to cell lysis (55). In contrast, a SNES in VEEV's capsid protein arrests CRM1 at the NPC, thought to abrogate nuclear export in order to halt cellular antiviral signaling pathways (53). Equivalent SNES peptides have not been identified for cellular proteins. However, high-affinity or high-avidity interactions with CRM1 are likely crucial to the efficient export of “bulky” nuclear cargos such as rRNA-protein complexes (1, 56, 57).
Other cellular and viral cargoes (e.g., RNP complexes formed by influenza's NS2 protein) promote interactions with CRM1 by encoding two or more discrete NES peptides within the same protein (58–61). Alternatively, proteins such as Rev form oligomeric RNP complexes that present multiples of identical NES peptides to CRM1 or a CRM1 multimer (22, 62–67). Recent structure-function studies from us and others (68, 69), as well as elegant structural work from Frankel and coworkers (27, 70), strongly suggest that Rev multimerization (consisting of 6 to 14 monomers on the RRE) is needed to form a multi-NES complex capable of recruiting at least two molecules of CRM1.
How Rev balances strong nuclear localization and nuclear export signals in space and in time in order to optimize the timing and magnitude of late viral gene expression has only partially been characterized. In the present study, we provide evidence that Rev-Rev interactions serve to mask the NES and thereby promote Rev's accumulation in the nucleus and sufficient access to viral RRE-bearing RNAs. We show that Rev is remarkably tolerant of diverse NES peptides or NES configurations, including SNES peptides predicted to bind to CRM1 even in the absence of Ran-GTP. Rev's capacity to tolerate a SNES was both position and multimerization dependent, suggesting a novel mechanism wherein Rev oligomerization not only regulates export complex formation but also biases Rev's trafficking to and retention in the nucleus.
RESULTS
Rev is robustly tolerant of changes of NES number, position, and identity.
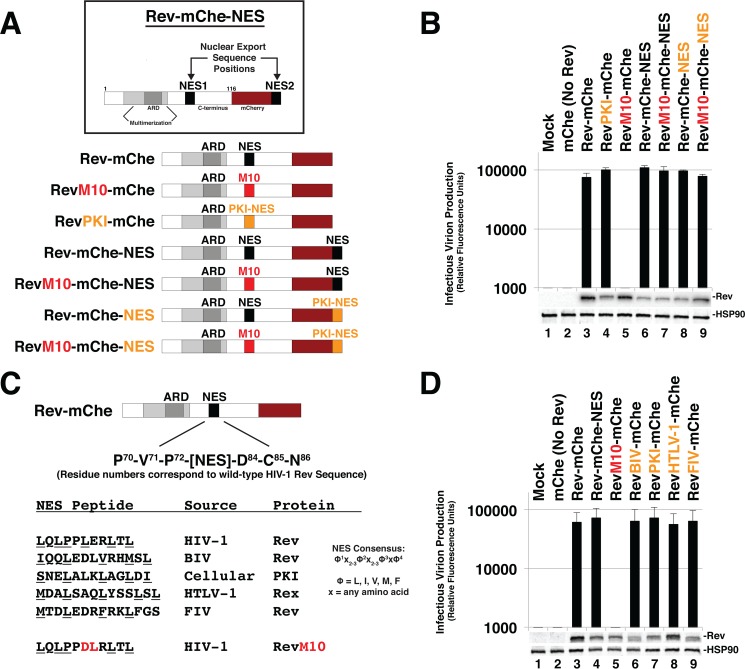
To study the role of NES number and context on HIV-1 mRNA trafficking dynamics and infectious virion production, we coexpressed wild-type or mutated versions of Rev-mCherry (Rev-mChe) fusion proteins (Fig. 1A) in trans with plasmids encoding full-length Rev-minus infectious HIV-1 yellow fluorescent protein (YFP)-encoding reporter viruses (NL4-3/E-R-Rev-/YFP) and vesicular stomatitis virus G glycoprotein (VSV-G) for pseudotyping. As previously described (68), the expression of Rev-mChe or Rev-mChe-NES variants yielded similar levels of infectious virus production from human cells (Fig. 1B, compare lanes 3 and 6). Mutational inactivation of NES1 (RevM10-mChe) completely abrogated virus production (Fig. 1B, lane 5). This phenotype was fully rescued by appending a functional NES (either derived from Rev itself or PKI) to the C terminus of this protein (RevM10-mChe-NES) (Fig. 1B, compare lanes 5 and 7). Thus, the positional context of a functional NES (either in the native NES1 position, residues 73 to 83, or at the C-terminal NES2 position) has little to no bearing on Rev-mChe's capacity to transactivate viral late gene expression, at least when provided in trans and at defined levels of Rev expression.
FIG 1.
Rev is robustly tolerant of changes to NES number, position, or identity. (A) Cartoon indicating Rev-mCherry (Rev-mChe) variants and relevant NES positions or modifications, native position (NES1), altered identity (PKI), inactivated (M10), and C-terminal position (NES2). Rev's arginine-rich domain (ARD, amino acids 34 to 50) encodes the nuclear localization signal (NLS) and RNA-binding activities. Rev's native NES (NES1 position; amino acids 73 to 83) is located within the disordered C-terminal domain (B) Capacity of Rev variants depicted in 1A to trans-complement Rev-minus HIV-1 YFP reporter viruses. 293T cells were transfected with plasmids encoding full-length, NL4-3-derived E-R-Rev-/YFP reporter proviruses, VSV-G, and either mCherry alone (No Rev control) or the indicated Rev-mChe variant. The lane 1 control lacks proviral DNA. Cell lysates and supernatants were harvested at 48 h posttransfection and processed for immunoblotting, and equivalent amounts of supernatant were used to infect target HeLa cells in order to gauge infectious virion production based on YFP fluorescence at 48 h postinfection (viral infectivity assay). Error bars represent standard deviations from the mean for three independent experiments. Rev and HSP90 (loading control) were detected using anti-Rev and anti-HSP90 antisera. (C) Depiction of additional Rev-mChe variants bearing alternative NES sequences. Predicted NES consensus-defining amino acids are underlined. BIV, bovine immunodeficiency virus; PKI, protein kinase A inhibitor; HTLV-1, human T-lymphotropic virus type 1; FIV, feline immunodeficiency virus. (D) Activities of the Rev variants shown in panel C were determined by viral infectivity assay as described for panel B.
To address NES identity, we replaced Rev's native NES (LQLPPLERLTL) with the well-characterized NES peptide (SNELALKLAGLDI) derived from PKI (44, 45). Despite conserved activity in mediating CRM1 binding, the Rev and PKI NES peptides differ in terms of CRM1 binding strength (71–73), are structurally distinct in the context of how they interface with CRM1's NES binding pocket (34), and may be involved in recruiting alternative cellular factors in addition to CRM1 (e.g., the HIV cofactor eIF5A) (74). Despite these differences, replacement of the Rev NES with the PKI NES in the context of Rev-mChe, Rev-mChe-NES or RevM10-mChe-NES yielded wild-type levels of infectious virion production (Fig. 1B, compare lanes 4 and 3, compare lanes 8 and 6, and compare lanes 9 and 5). We also did not observe differences for confirmed NES peptides derived from Rev-equivalent proteins found in other retroviruses, including human T-lymphotropic virus type 1 (HTLV-1), bovine immunodeficiency virus, or feline immunodeficiency virus (Fig. 1C and D). Thus, similar to differential NES quantity or positional context within the protein's structure, Rev's native NES identity had little to no impact on Rev's activity in this assay.
Evidence for position-dependent Rev NES masking.
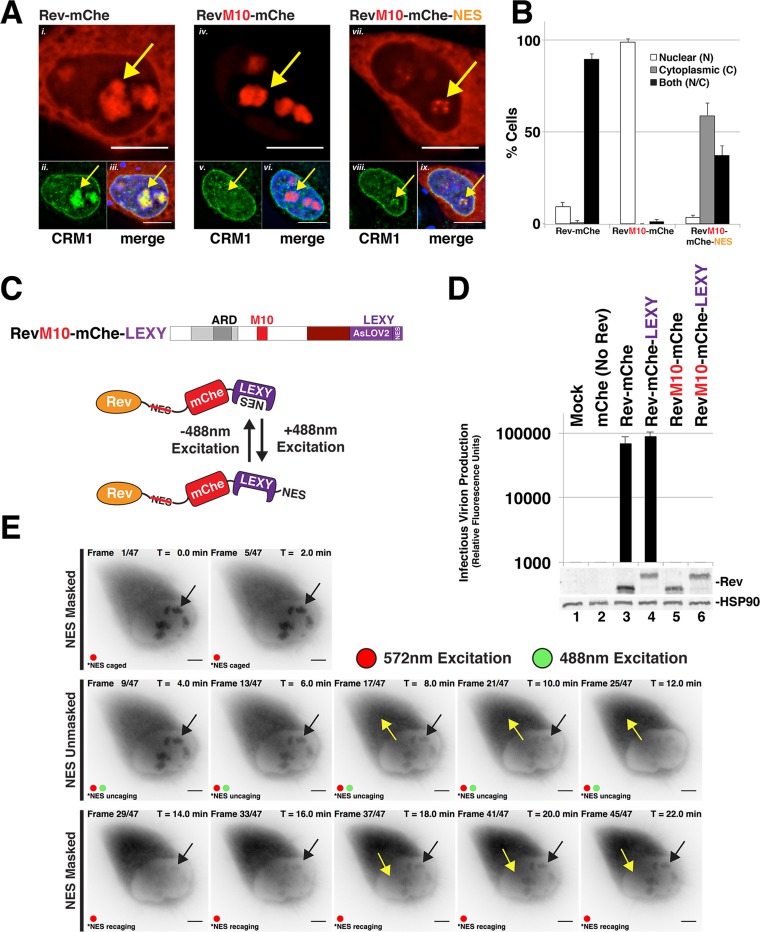
Despite the above nominal effects on infectious virion production, we did observe notable differences to Rev-mChe steady-state subcellular localization when the NES was moved from the native position to the C terminus of the RevM10-mChe fusion protein (Fig. 2A and quantification in 2B) or when replaced with the PKI NES (Fig. 3). As expected, wild-type Rev-mChe accumulated in the cytoplasm and colocalized with CRM1 in the nucleolus in the strong majority of cells (Fig. 2A, panels i to iii, and 2B) while the RevM10-mChe mutant was largely restricted to the nucleolus and did not recruit CRM1 (Fig. 2A, panels iv to vi, and Fig. 2B). Interestingly, the RevM10-mChe-NES variant was predominantly detected in the cytoplasm (Fig. 2A, panels vii to ix, and Fig. 2B) and, when observed in the nucleus, was most frequently in association with small, bright punctae in the periphery of nucleolar structures (as defined by spherical organelles devoid of DAPI [4′,6′-diamidino-2-phenylindole stain]) (Fig. 2A, panels vii to ix).
FIG 2.
Evidence for Rev exhibiting position-dependent NES masking. (A) Context-specific NES effects on Rev's subcellular localization. HeLa cells transfected to express E-R-Rev-/Luc and the indicated Rev-mChe variants were fixed, permeabilized, and DAPI stained 24 h posttransfection. Endogenous CRM1 was detected by indirect immunofluorescence using anti-CRM1 antisera. Yellow arrows highlight nucleolar accumulation of Rev and/or CRM1. Scale bars, 10 μm. (B) RevM10-mChe-NES exhibits less accumulation at or near the nucleolus. Rev-mChe subcellular distribution was quantified in individual cells as primarily nuclear (N), cytoplasmic (C), or equivalent in both compartments (N/C). Error bars represent the standard deviations from the mean for three independent transfections. (C) Depictions of RevM10-mChe-LEXY construct and blue light-regulated NES unmasking using the LEXY regulatory module (75). (D) Control experiment demonstrating that the activity of Rev-mChe-LEXY and RevM10-mChe-LEXY variants is equivalent to Rev-mChe constructs lacking LEXY. Viral infectivity was measured as for Fig. 1B. Error bars represent the standard deviations from the mean for three independent experiments. (E) Image panel shows selected frames from a representative time-lapse fluorescence microscopy experiment capturing mCherry fluorescence from RevM10-mChe-LEXY in HeLa cells. Red circles indicate exposure to 572-nm wavelength light (mCherry acquisition wavelength), and green circles indicate exposure to 488-nm wavelength light (LEXY activation wavelength). Black arrows indicate nucleolar Rev accumulation sites, and yellow arrows indicate direction of Rev transitions over time. Scale bars, 10 μm.
FIG 3.
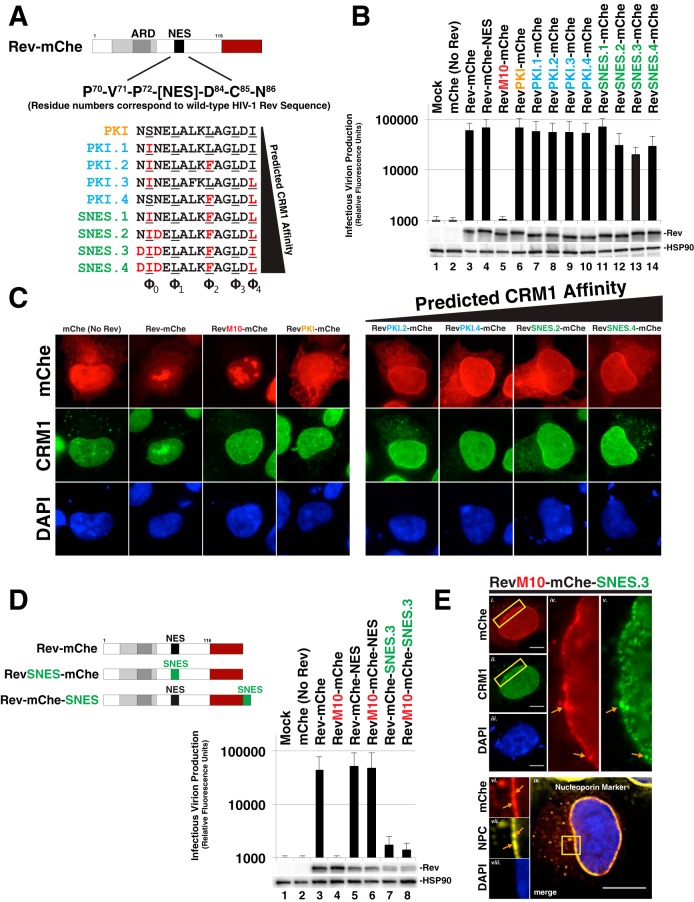
Rev tolerates supraphysiological NES domains in a position-dependent manner. (A) Panel of NES domains predicted to exhibit increasing CRM1 binding strength in the context of Rev-mChe. Wild-type PKI NES is labeled orange (same variant from Fig. 1). PKI NES-derived sequences with predicted increases in CRM1 affinity are labeled blue. PKI NES-derived sequences with predicted supraphysiological CRM1 binding affinity (i.e., bind to CRM1 even in the absence of Ran-GTP; SNESs) are labeled green. Amino acids shown red predicted to confer the increase of CRM1 affinity. (B) Even supraphysiological NESs had only modest effects (∼2-fold decreases) on Rev function in our trans-complementation infectivity assay described for Fig. 1B. Error bars represent the standard deviations from the mean for five independent experiments. (C) Nucleolar localization was decreased when Rev encoded an NES with increased CRM1 affinity. HeLa cells were transfected and prepared as for Fig. 2A. (D) Combining increases to NES strength with changes to NES position potently inhibits viral infectivity. Diagram of relevant Rev NES strength/context variants with our infectivity assay demonstrating >10-fold losses to infectious virion production. Error bars represent the standard deviations from the mean for five independent experiments. (E) Representative indirect immunofluorescence images showing Rev-mChe-SNES colocalizing with CRM1 and nucleoporins at the nuclear membrane. Yellow rectangles indicate regions of interest and orange arrows highlight colocalization. Scale bars, 10 μm.
We hypothesized that the observed differences to Rev's steady-state distribution reflected position-dependent NES exposure (see Fig. 1A). To test this hypothesis, we exploited a cutting-edge optogenetic approach recently developed by Niopek et al. wherein a C-terminal masked NES was designed to be conditionally unmasked under the control of blue light (75). In this system, blue light (480 to 500 nm excitation) destabilizes an Avena sativa phototropin-1 LOV2 core domain (AsLOV2), thus unmasking a rationally designed NES peptide. NES exposure triggers nuclear export through the CRM1 pathway. To control the trafficking of visible Rev, we fused the LEXY domain to the C terminus of Rev-mChe (Rev-mChe-LEXY) and RevM10-mChe (RevM10-mChe-LEXY) (Fig. 2C). We confirmed that the addition of the LEXY domain had no effect on Rev activity relative to the unmodified controls (Fig. 2D, compare lanes 3 and 4) and that, in the absence of blue light, the LEXY domain fully suppressed its masked NES in the context of the RevM10-mChe fusion protein (Fig. 2D, compare lanes 5 and 6).
In single cells, expression of RevM10-mChe-LEXY revealed a high degree of nucleolar accumulation in the absence of blue light, as expected (Fig. 2E). However, we also observed a significant amount of cytoplasmic fluorescence with these constructs relative to RevM10-mChe (compare Fig. 2E to Fig. 2A, panel iv), albeit with no apparent gene transactivation activity as per Fig. 2D. Strikingly, exposure to blue light led to rapid evacuation of RevM10-mChe-LEXY from the nucleolus, with the bulk of the signal moving to the cytoplasm over a time course of ∼10 min (Fig. 2E, “NES unmasked” panels). Subsequent cessation of blue light exposure led to rapid recovery of the nucleolar mCherry signal (Fig. 2E, “NES masked” panels). We attributed these light-dependent effects on Rev localization to AsLOV2-regulation of NES unmasking and remasking. Based on this observation combined with there being a greater tendency of wild-type Rev-mChe to accumulate at the nucleolus at steady-state relative to a condition where the functional NES was moved to the protein's C terminus (RevM10-mChe-NES; Fig. 2A), we reasoned that Rev's general tendency to accumulate in the nucleolus reflects a mechanism of position-dependent NES masking.
Rev tolerates supraphysiological NES peptides in a position-dependent manner.
Considering the remarkable ability of Rev to tolerate diverse NES peptides and configurations, we next tested the effects of progressively strengthening Rev-CRM1 interactions using NES peptides recently characterized by Görlich and coworkers that exhibit substantially increased CRM1 affinity in vitro (depicted in Fig. 3A) (34). Using the PKI NES as a base model, these investigators determined changes to core NES residues that would enhance NES affinity for the CRM1 binding pocket, thus leading to the derivation of several stronger and even supraphysiological NES (SNES) peptides, defined by their capacity to bind to CRM1 even in the absence of Ran-GTP in vitro (34). Remarkably, we found each of these progressively stronger NES peptides in the NES1 position to be functional for infectious virus production in our trans-complementation assay (Fig. 3B). Even the strongest predicted SNES peptides had only modest, up to ∼2-fold inhibitory effects on Rev function relative to the Rev-mChe or RevPKI-mChe controls (Fig. 3B, compare lanes 11 to 14 to lanes 3 and 6). Despite these moderate effects, we did detect noticeably altered CRM1 and Rev-mChe variant colocalization away from the nucleolus, with both proteins predominantly accumulating together at or near the nuclear membrane at steady state (Fig. 3C). Thus, similar to NES position (Fig. 2), stronger Rev-CRM1 interactions affect Rev steady-state subcellular distribution but have relatively little bearing on the capacity of Rev to promote infectious virus production.
Considering that Rev-mChe localization differed from RevM10-mChe-NES (Fig. 2A), we also tested whether SNES activity was position dependent by moving the SNES from the native position (NES1) to the C-terminal position (NES2). Strikingly, this modification almost completely abolished infectious virion production (Fig. 3D, compare lane 3 to lanes 7 and 8), correlating with and even more dramatic accumulation of colocalized Rev-mChe-SNES and CRM1 at or near the nuclear membrane based on visual analysis (Fig. 3E, panels i to v, SNES.3 is shown). Interestingly, the Rev-mChe-SNES protein was not evenly distributed along the nuclear envelope, but enriched near NPCs, as shown by colocalization with a nucleoporin-specific marker (mab414) (Fig. 3E, panels vi to ix). This result was consistent with a prior study by Fornerod and coworkers using Rev-NES fusions as a technique to demonstrate SNES-dependent arrest of CRM1 at the NPC in association with the nucleoporin Nup358 (54). Both Rev-mChe-SNES and RevM10-mChe-SNES constructs yielded similar inhibition of infectious virus production, consistent with the C-terminal position of the dominantly acting SNES causing this block (Fig. 3D, compare lanes 7 and 8). This result revealed that Rev activity can tolerate a SNES peptide, but only when the SNES is in the native position (NES1, see Fig. 1A) located within Rev's disordered carboxy-terminal domain.
A C-terminal SNES blocks Rev's ability to export viral RNA to the cytoplasm.
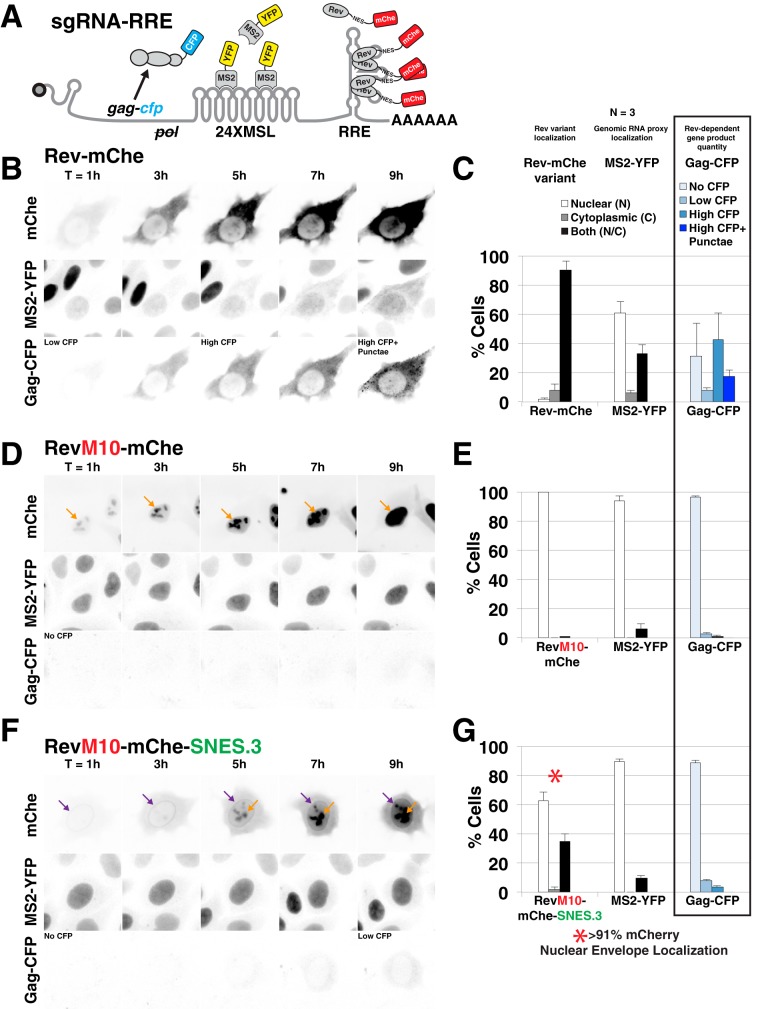
To address the mechanism by which a C-terminal SNES reduced infectious virus production, we used both live cell imaging using YFP-tethered Rev/RRE-dependent viral mRNAs (Fig. 4) and fluorescence in situ hybridization (FISH) (see Fig. 5) to visualize native, unspliced viral transcripts in the presence or absence of our relevant Rev NES variants. We hypothesized that the C-terminal SNES was arresting Rev/CRM1 complexes at the NPC, thereby inducing a roadblock to viral RNA (vRNA) nuclear export. Three-color live cell imaging of vRNA nuclear export was performed using MS2-YFP tagged surrogate, intron-retaining and thus Rev-dependent viral mRNAs encoding Gag-CFP (76) (Fig. 4A). Wild-type Rev-mChe expression led to a progressive transition of vRNAs from the nucleus to the cytoplasm in ∼40% of transfected cells imaged at 24 h posttransfection, a finding consistent with mRNA nuclear export (Fig. 4B, “MS2-YFP” panels, and Fig. 4C). In these cells, Gag-CFP was synthesized coincident with mRNA nuclear export and ultimately formed surface punctae consistent with the onset of virus particle assembly (Fig. 4B, “Gag-CFP” panels, and Fig. 4C). In contrast, viral RNAs (as measured using the MS2-YFP proxy) remained sequestered in the nucleus when coexpressed either with the RevM10-mChe control that does not bind to CRM1 (Fig. 4D and quantification in Fig. 4E) or when coexpressed with Rev-mChe-SNES (Fig. 4F and quantification in Fig. 4G). These observations were consistent with a block to RNA nuclear export. Moreover, the time-resolved imaging revealed that the Rev-mChe-SNES first accumulated at the nuclear envelope prior to amassing at the nucleolus at later time points (Fig. 4F, compare the 3-h and 7-h time points), with nuclear envelope localization observed for >91% of cells measured at 24 posttransfection (Fig. 4G).
FIG 4.
SNES-arrested Rev/CRM1 complexes block Rev's ability to export vRNAs from the nucleus. (A) Diagram of subgenomic, intron-retaining HIV-1 gag-pol mRNA for live-cell imaging. This mRNA encodes 24 copies of MS2 coat protein binding loop (24XMSL) and a CFP-labeled Gag protein (Gag-CFP) for tracking Rev-dependent viral mRNA nuclear export and late viral gene expression (See Materials and Methods and reference 76 for additional information). (B to G) Live cell imaging of Rev-mChe variants, viral mRNA trafficking, and Gag-CFP expression in HeLa cells stably producing MS2-YFP (HeLa.MS2-YFP) over a 9-h interval. Image capture was initiated <1 h posttransfection and fixed at ∼24 h posttransfection for endpoint analysis. (B) Wild-type Rev-mChe supports MS2-YFP translocation from the nucleus to the cytoplasm and Gag-CFP expression consistent with Rev-dependent viral mRNA nuclear export and translation. (C) Endpoint analysis of Rev, viral mRNA labeled by MS2-YFP proxy, and Gag-CFP. Rev and MS2-YFP distribution was quantified in individual cells for fluorescence signal as primarily nuclear (N), cytoplasmic (C), or readily detectable in both compartments (N/C). Gag-CFP was quantified based upon CFP expression level (no CFP, low CFP, or high CFP) and distribution of signal (diffuse or diffuse with punctate). Error bars represent the standard deviations from three independent transfections. A total of >300 cells were scored per condition for all transfection replicates combined. (D and E) RevM10-mChe is restricted to the nucleus (orange arrows) and does not activate viral mRNA export or Gag-CFP expression. (F and G) RevM10-mChe-SNES first accumulates at the nuclear envelope (purple arrows) and at later time points in the nucleolus (orange arrows) but does not trigger detectable viral mRNA export and supports only low levels of Gag-CFP expression. The red asterisk in panel G indicates that RevM10-mChe-SNES localization typically included signal at the nuclear envelope.
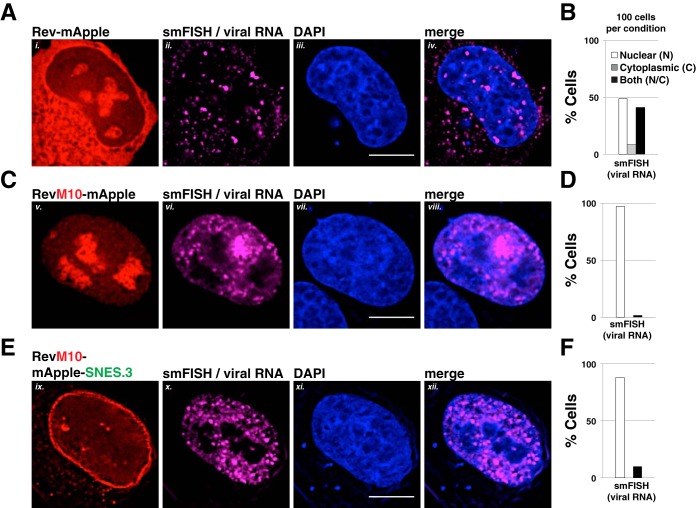
FIG 5.
A C-terminal SNES blocks Rev-dependent viral mRNA export. Direct visualization of unspliced HIV-1 mRNA using fluorescence in situ hybridization (FISH). HeLa cells transfected to express the E-R-Rev-/Luc construct and the indicated Rev variant were processed as for Fig. 2A at 24 h posttransfection. FISH probes targeting the gag-pol reading frame of the E-R-Rev-/Luc construct were used to detect unspliced viral RNA (shown in magenta). Scale bars correspond to 10 μm. (A, C, and E) Representative images of Rev (red), mRNA (FISH, magenta), and nuclear DNA (DAPI, blue) for wild-type Rev-mApple (A), RevM10-mApple (C), and RevM10-mApple-SNES3 (E). (B, D, and F) RNA distribution was quantified in individual cells for RNA localization as primarily nuclear (N), cytoplasmic (C), or readily detectable in both compartments (N/C). A total of 100 cells were scored per condition.
To more clearly elucidate whether Rev-dependent vRNAs were arrested at or near the NPC for Rev bearing a C-terminal SNES, we also performed single-molecule RNA FISH for these three conditions using a dye-conjugated oligonucleotide probe set specific for the gag-pol reading frame of a full-length HIV-1 reporter virus (E-R-Rev-/Luc) (Fig. 5). In this experiment, the Rev variants were tagged with monomeric Apple fluorescent protein (mApple) rather than mCherry in order to avoid spectral overlap with the Quasar 670 dye probe set. When expressed with wild-type Rev-mApple, vRNA was detected in the cytoplasm in ∼50% of cells at 24 h posttransfection, which is consistent with nuclear export (Fig. 5A, panels i to iv, and quantification in Fig. 5B). In contrast, vRNA was restricted to the nucleoplasm in almost 100% of cells in the presence of RevM10-mApple, as expected (Fig. 5C, panels v to viii, and quantification in Fig. 5D). For RevM10-mApple-SNES.3, although much of the Rev variant protein (and, by proxy, CRM1 as indicated in Fig. 3E) was clustered at the nuclear membrane, we observed that the bulk of the viral RNA was distributed throughout the nucleoplasm but excluded from the nucleolus (as defined using the DAPI stain or Rev-mApple accumulation) and rarely at the nuclear envelope, more similar to RevM10 (Fig. 5E, panels ix to xii, and quantification in Fig. 5F). Based on this observation and the fact that the first site of SNES-arrested Rev accumulation was at the nuclear envelope, revealed by live cell imaging (Fig. 4F), we reasoned that the C-terminal SNES traps Rev/CRM1 complexes at the NPC even prior to vRNA/RRE-binding.
Rev's capacity to tolerate a SNES is multimerization dependent.
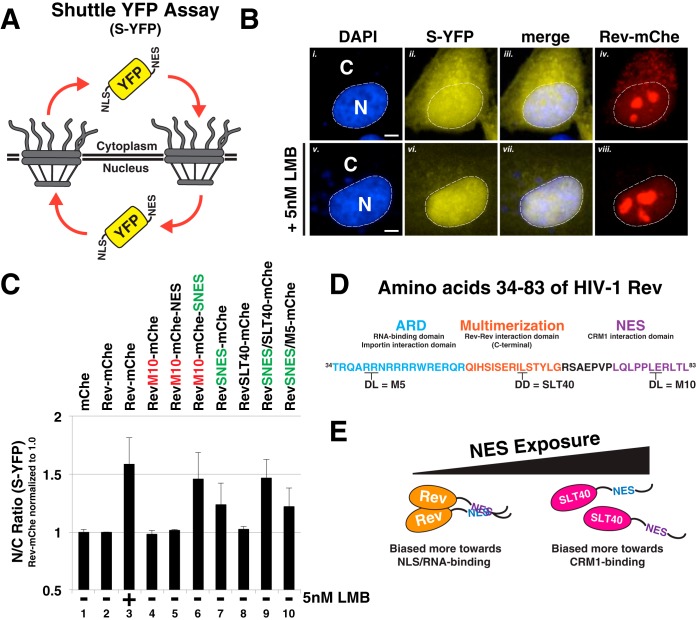
We also tested whether SNES-mediated arrest of Rev trafficking was specific to Rev-dependent vRNA or, instead, reflected a global arrest of CRM1-mediated nuclear export. To this end, we transfected Rev-mChe, RevM10-mChe-NES, and RevM10-mChe-SNES into HeLa cells expressing a “shuttling” YFP (S-YFP) reporter protein modified to bear an N-terminal NLS peptide and a C-terminal NES (PKI) peptide (Fig. 6A and B). Disruption to either nuclear import or export was measured by net per cell changes to bulk S-YFP levels in the nucleus versus the cytoplasm using a computational cell-segmentation strategy (68, 76). The data are presented as a ratio of nuclear to cytoplasmic fluorescence (N/C ratio) normalized to the wild-type Rev-mChe condition (Fig. 6C, condition 2). RevM10-mChe-SNES expression led to increases to the relative nuclear abundance of S-YFP almost identical to when the CRM1 inhibitor leptomycin B (LMB) was added to a concentration of 5 nM (Fig. 6C, compare conditions 3 and 6). This result was indicated a global block to the CRM1 pathway. In contrast, Rev variants encoding wild-type or inactivated (M10) NES peptides, independent of NES position, exhibited N/C ratios similar to that of mChe alone and Rev-mChe controls (Fig. 6C, compare conditions 4 and 5 to condition 2). Interestingly, Rev variants bearing the SNES in the native position (RevSNES-mChe) were not innocuous but exhibited an intermediate phenotype (Fig. 6C, compare conditions 2 and 6 to condition 7). We perceived this result as consistent with Rev's capacity to at least moderately mask the SNES in the NES1 position, thereby maintaining relatively high levels of infectious virion production as shown in Fig. 3B.
FIG 6.
Rev's capacity to mask a SNES is both context and multimerization dependent. (A) Diagram depicting shuttle YFP (S-YFP) reporter assay. This assay employs NLS- and NES-directed nucleocytoplasmic trafficking of S-YFP reporter to determine the disruptive effect of Rev variants encoding different NES configurations. (B) Inhibition of CRM1-dependent export promotes S-YFP nuclear accumulation. The image panel depicts individual transfected HeLa cells producing S-YFP and the indicated Rev variant (the example shown is Rev-mChe) in the absence (panels i to iv) or presence (panels v to viii) of 5 nM leptomycin B (LMB) as a control for CRM1-dependent nuclear export inhibition. Transfected HeLa cells were fixed at 24 h posttransfection and DAPI stained to demarcate nuclear and cytoplasmic compartments (nuclear border labeled by white dotted line). Scale bars, 10 μm. (C) Multimerization-deficient Rev encoding SNES in native context phenocopies C-terminal SNES and LMB treatment. The nuclear-to-cytoplasmic ratio (N/C ratio) of YFP fluorescence was measured from individual cells producing the indicated Rev variants in an S-YFP assay. Cells were transfected to express the indicated Rev variant and viral RNA and processed as for panel B. Nuclear and cytoplasmic YFP fluorescence was measured from YFP- and mCherry-positive HeLa cells and assigned using DAPI as a marker for nuclear-specific fluorescence signal for reference (see Fig. 6B, compare panels i to ii to panels v to vi). Error bars represent the standard deviations from the mean for four independent experiments (>250 cells measured per condition). Automated cell segmentation and fluorescence quantification were performed using KNIME/FIJI. (D) Wild-type HIV-1 Rev sequence (amino acids 34 to 83) indicating functional activities and amino acid substitutions conferring loss of RNA binding (M5), Rev multimerization (SLT40), and CRM1 binding (M10). (E) Model for regulation of multimerization-dependent NES masking. Physical masking of SNES in native NES1 context decreases interaction potential with CRM1, thus promoting the cytoplasmic accumulation of S-YFP, as measured in panel C.
We thought the most parsimonious explanations for position-dependent NES (or SNES) masking activity would be either Rev-Rev multimerization or Rev-RNA binding acting to physically cloak the NES. Rev's nuclear import and RNA binding are both regulated by Rev's N-terminal arginine-rich domain (ARD), with residues flanking the ARD regulating Rev multimerization (depicted in Fig. 6D). We thus compared the capacity of a well-characterized Rev ARD mutant defective in RNA binding (RevM5) (77) and another (RevSLT40) mutant that is multimerization deficient (78) to mask a SNES in our shuttle YFP assay. RevSNES/M5-mChe proteins exhibited an intermediate block to S-YFP nuclear export similar to the RevSNES-mChe variant, in contrast to the RevSNES/SLT40-mChe variant that blocked nuclear export to a more complete extent, similar to the Rev-mChe-SNES and LMB conditions (Fig. 6C, compare lanes 9 and 10 to lanes 6 and 7). Thus, Rev multimerization reduces the impact of a SNES in the NES1 position on global CRM1-dependent nuclear export, consistent with a protective or “masking” NES regulatory feature controlled through Rev-Rev interactions (see Fig. 6E for model).
NES position regulates optimal levels of infectious virion production.
Despite the above observations, the relevance of Rev's NES masking function to natural infection was unclear considering that a SNES is certainly a nonphysiological scenario (HIV's native NES is actually thought to be relatively weak [34, 73]). In addition, we had also observed that moving the NES to what we predicted was an “unmasked” C-terminal position (i.e., RevM10-mChe-NES) affected Rev trafficking (Fig. 2) but was not at all deleterious to viral infectivity in our trans-complementation assay (Fig. 1B). That said, we speculated that levels of Rev expression in these experiments were likely to exceed levels of Rev observed during natural infection, thus prompting us to carry out a careful, head-to-head titration of both Rev-mChe and RevM10-mChe-NES plasmids using 2-fold dilutions and assessing the effects on infectious virus release (Fig. 7). As previously demonstrated (Fig. 1B), we observed no differences to infectious virion production for either Rev variant when expressed at relatively high levels (Fig. 7, compare conditions 5 through 8). In contrast, Rev-mChe activity was relatively robust at low levels of expression (even at ∼100-fold dilution) compared to RevM10-mChe-NES (Fig. 7, compare lanes 8 to 12 to lane 5). Thus, while an identical NES peptide can be functional in either position (NES1 or NES2) of our Rev-mChe fusion protein, the peptides are clearly not equally active at low levels of Rev expression. Combined with our other observations, such a result is consistent with the notion that Rev is most active when able to mask its NES peptide.
FIG 7.
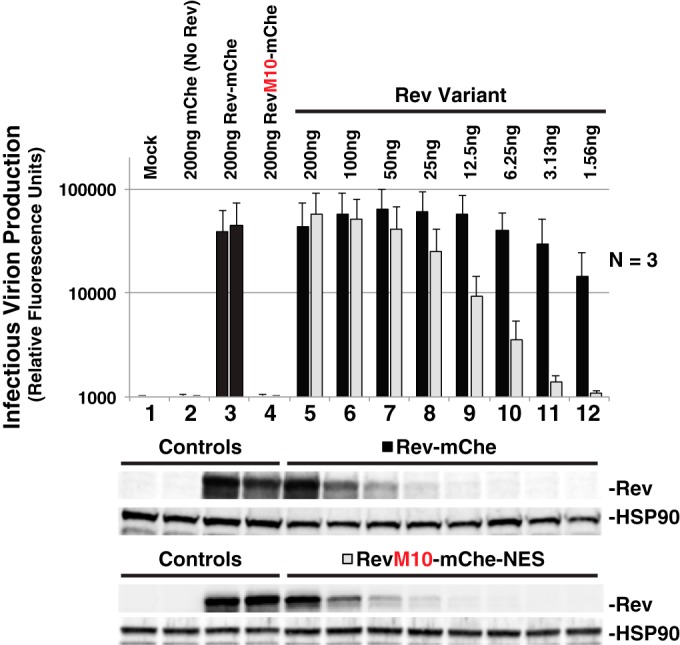
NES context regulates optimal infectious virion production. Titration of Rev-mChe and RevM10-mChe-NES plasmids reveals functional differences between variants in our trans-complementation viral infectivity assay, as described for in Fig. 1B. Error bars represent the standard deviations from the mean for three independent experiments.
DISCUSSION
In this study we provide evidence that the strength and position of Rev's NES plays an important role in regulating Rev trafficking and viral infectivity. Using a functional Rev-mChe fusion protein as a modular platform, we found that Rev activity in human cells is remarkably tolerant of changes to Rev NES peptide sequence (i.e., native or C-terminal positions) and strength (Fig. 1 and 3). Significant recent progress has been made in elucidating unique structural features underpinning the formation of functional RRE/Rev/CRM1 transport complexes (25–27, 29, 29, 30, 70, 79). In contrast, little is yet known regarding the spatiotemporal regulation of these complexes, i.e., how, where, and when they are formed, transited to and through the nuclear pore, and turned over in the cytoplasm in the context of single cells. Nawroth et al. recently provided evidence that Rev's multimerization on the RRE and recruitment of CRM1 occurs cotranscriptionally in the nucleoplasm and not at the nucleolus (80). Consistent with this observation, we recently showed using long-term (>24 h) live cell imaging that Rev/RRE-dependent transcripts typically build up in the nucleoplasm prior to a punctuated, CRM1-dependent en masse nuclear export event (76). Heterologous NES peptides have long been known to support Rev activity either in the context of Rev trafficking or RRE-dependent gene expression (52, 71, 72, 81, 82). Accordingly, and based on the hypothesis that the timing of viral RNA nuclear export is crucial to rates of Gag/Gag-Pol synthesis and genome encapsidation, we anticipated that altering the strength or number of Rev-CRM1 interactions via NES modulation would negatively impact rates of infectious virion production. Thus, we found it striking that even drastic modulations (e.g., SNES peptides that bind very tightly to CRM1) yielded only relatively minor (∼2-fold) impacts on virus output (Fig. 3B).
Regarding the mechanism underpinning Rev's NES tolerance, we propose that under native conditions, Rev is able to mask its NES peptide prior to nuclear entry, engagement of viral RNA, and formation of higher-order Rev/RRE complexes (working model in Fig. 8). We propose this model for three reasons. First, moving the NES from its native position to the C terminus of the Rev-mChe fusion protein caused it to accumulate preferentially in the cytoplasm rather than the nucleolus (Fig. 2A). This phenotype was recapitulated in striking fashion by transiently exposing a C-terminal NES on a RevM10 mutant protein using optogenetics-based control (Fig. 2B). Thus, Rev's tendency to accumulate at the nucleolus at steady state appears to inversely correlate with the degree of NES exposure when outside the native NES1 position. Second, the observation that Rev activity was largely tolerant of SNES peptides predicted to bind CRM1 very tightly (Fig. 3B) serves as additional evidence for Rev NES masking. Moving the SNES to the C-terminal, “unmasked” region of the Rev-mChe fusion protein almost completely abolished infectious virion production (Fig. 3D), a block explained by the disruption of Rev's capacity to export viral RNAs from the nucleus (Fig. 4 and 5). Third, a multimerization-defective mutant of Rev (RevSLT40), but not a multimerization-competent but RNA-binding defective mutant (RevM5), was unable to tolerate a SNES even in the native NES1 position, as measured in our S-YFP shuttling assay (Fig. 6). Thus, Rev self-association with or without RNA binding is likely the source of NES masking (refer to model in Fig. 8).
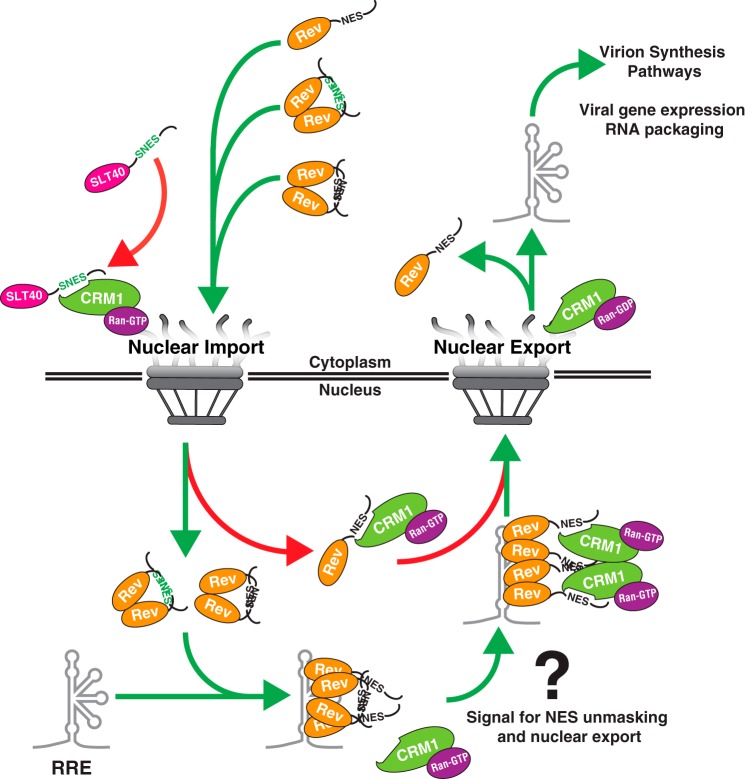
FIG 8.
Working model for multimerization-dependent NES masking in Rev's nucleocytoplasmic trafficking scheme. Our data suggest Rev-Rev interactions promote Rev's nucleolar accumulation by inhibiting CRM1 from accessing Rev's NES prior to and during nuclear import (left NPC, green arrows). An unmasked NES disrupts Rev's capacity to accumulate in the nucleolus by blocking nuclear import (red arrow in cytoplasm, SNES) or driving immediate export of Rev from the nucleus (red arrow in nucleus, NES). In the nucleus, one or more Rev NES peptides are likely exposed through an unknown mechanism during formation of Rev/RNA complexes, thus serving as a signal for the recruitment of CRM1 (right NPC, green arrows). Nuclear export and dissolution of the complex promotes recycling of Rev and downstream events in the HIV-1 life cycle.
Rev is known to self-associate even in the absence of viral RNA (83), so that multimerization in the cytoplasm likely serves as an effective means of masking the NES and thereby biasing Rev's trafficking to the nucleus and accumulation at the nucleolus. NES masking is likely to be a prevalent feature of cell biology (84, 85) and has been shown to regulate the nucleocytoplasmic transport of key transcription factors and signaling molecules (86, 87) through either protein intermolecular interactions (e.g., the oncogene BRCA2 interacting with regulatory protein DSS1) (88) or protein intramolecular interactions (e.g., regulated nuclear retention of the transcription factor NFAT1) (89–91). Conversely, it is worth noting that NLS peptides have also been found to be masked in both cellular and viral protein contexts (85, 91–93). To our knowledge, this study is the first to directly implicate NES masking as a regulatory feature of Rev trafficking and the HIV-1 life cycle and may be relevant to previous observations from Daelemans and coworkers wherein targeted disruption of Rev multimerization caused reduced steady-state levels of Rev in the nucleolus (94, 95). Interestingly, Gu et al. have also provided evidence for cell-type-specific Rev NLS masking regulated by a cellular cofactor MDFIC (also known as the human I-mfa domain-containing protein or HIC) (96). Taken together, the balance of Rev NES and NLS masking within relevant cells is likely to provide HIV-1 with a level of nucleocytoplasmic transport control that extends beyond core cycling in accordance with Ran-GTP turnover.
If important for nuclear accumulation, NES masking would, of course, have to be conditionally reversed in order to promote vRNA nuclear export. Whether Rev's transient or long-term association with the nucleolus has any functional relevance remains undefined (22, 68, 97, 98). However, that Rev-mChe bearing a C-terminal NES localizes poorly to the nucleolus and is less active than wild-type Rev at low levels of expression (Fig. 7) is consistent with a hypothesis wherein the nucleolus plays a role as sink, favoring Rev sequestration over time. We also recently reported that Rev can undergo striking, en masse transitions from the nucleolus to the cytoplasm (68), a phenotype practically identical to the transitions observed in our optogenetics-based NES unmasking experiment (Fig. 2D). Thus, one or more transient cell signaling events in the nucleus may regulate Rev NES unmasking, thereby activating CRM1 binding and subsequent export activity. It is attractive to speculate that unmasking of Rev's NES is regulated by phosphorylation, considering that Rev has long been known to be a phosphoprotein (99, 100) and that there are compelling examples (e.g., NFAT1 NES masking) of cellular proteins wherein differential phosphorylation toggles NES or NLS exposure (90, 101, 102).
In broader terms, we perceive Rev's NES tolerance as further evidence of the remarkable plasticity of the multisubunit RRE/Rev/CRM1 nexus (25, 26, 67, 79). We previously showed that appending a second NES peptide to HIV-1 Rev or doubling the number of RRE sequences per viral RNA transcript was sufficient to overcome a profound block to HIV-1 in murine cells, attributable to a species-specific feature of the murine CRM1 ortholog (mCRM1) (68). This, combined with additional genetic studies from Hoffmann et al. (69) and recent structural work by Booth et al., suggests that multiple NES domains are critical for export of the Rev/RRE complex and needed to recruit at least two molecules of CRM1 (70). Recruitment of multiple CRM1 proteins to a multi-NES complex likely evolved as a robust, modular mechanism by which to gauge, buffer, and ensure efficient export of large, otherwise complex viral ribonucleoprotein assemblies. Indeed, Rev-like proteins and RRE-like response elements are conserved features of all viruses in the Lentiviridae (e.g., HIV-1 and HIV-2) and Deltaretroviridae (e.g., HTLV-1 and HTLV-2) families of retroviruses and are also found in a subset of other retroviruses (103–110). The relevant activities (multimerization, nuclear import, RNA binding, and nuclear export) are thought to be conserved among these proteins, although the sequences and, in some instances, domain organizations are divergent (63, 66, 111). This raises the question of whether both Rev multimerization and NES masking is conserved among these viruses and, if so, what features dictate NES exposure. Moreover, that Rev self-interaction is implicated in multiple stages of viral trafficking (both Rev nuclear import and Rev-RRE binding and export) reemphasizes the potential impact of targeting Rev multimerization as an antiviral strategy.
MATERIALS AND METHODS
Cell lines.
Human HeLa cervical carcinoma and human embryonic kidney 239T cells were obtained from the American Type Culture Collection. All cell lines were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 1% l-glutamine, and 1% penicillin-streptomycin. For all experiments, cells were maintained at 37°C and 5% CO2 in a humidified incubator. The derivation of clonal HeLa cells stably expressing the MS2 bacteriophage coat protein fused to yellow fluorescent protein (HeLa.MS2-YFP, Fig. 4) is described elsewhere (76).
Plasmids.
The pNL4-3 E-R-Rev-/YFP reporter plasmid was generated by replacing the luciferase gene in plasmid pNL4-3 E-R-Rev-/Luc (68, 112, 113) with yfp cDNA using NotI and XhoI restriction sites. The construction of plasmids encoding functional Rev-mCherry (Rev-mChe) and the Rev-mChe-NES, RevM10-mChe, and RevM10-mChe-NES variants have been described elsewhere (68). All Rev-mChe or NES mutants were generated by replacing the wild-type or C-terminal Rev-mChe NES sequence (LQLPPLERLTL) with alternative NES peptide sequences using overlapping PCR and insertion into pcDNA3.1(+) (Invitrogen) using NheI and XhoI cut sites. A subset of Rev-mChe variants were modified to encode the mApple fluorophore instead of mCherry to avoid spectral overlap with the Quasar 670 dye used for RNA FISH (described below). The LEXY domain was derived from plasmid pLexATF-T2A-NLS-LexA-KRAB-mCherry-LEXY (kindly provided by Barbara Di Ventura and Roland Eils; Addgene plasmid 72662) (75) and added to the C terminus of Rev- and RevM10-mChe constructs using BsrGI and XhoI cut sites. Plasmids encoding visible intron-retaining gag-pol mRNAs modified to produce Gag fused to cyan fluorescent protein (Gag-CFP) and bearing 24 copies of the MS2 bacteriophage stem-loop (24XMSL) are described elsewhere (76). The shuttle YFP (S-YFP) reporter plasmid was constructed by fusing yfp cDNA to sequence encoding a modified c-myc N-terminal nuclear localization signal (NLS) (MPAAKRVKLD) and sequence encoding a C-terminal protein kinase A inhibitor NES (NSNELALKLAGLDI). BamHI and XhoI cut sites were used to insert the fused amplicon (NLS-YFP-NES) into pcDNA3.1(+).
Viral infectivity assays and immunoblot analysis.
For viral infectivity assays, producer 293T cells were plated at ∼40% confluence in six-well tissue culture treated dishes 24 h prior to DNA plasmid transfection. Cells were transfected using polyethylenimine (PEI; catalog no. 23966 [Polysciences, Inc.]) with 1,000 ng of E-R-Rev-/YFP, 200 ng of Rev variant, and 100 ng of VSV-G for pseudotyping, followed by culture medium exchange at 4 h posttransfection. Supernatants and producer cell lysates were harvested at 48 h posttransfection. For immunoblotting, producer cell monolayers were washed with phosphate-buffered saline, lysed with radioimmunoprecipitation assay buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate) containing complete protease inhibitor cocktail (Roche). Producer cell lysates were prepared for immunoblot by sonicating, centrifugation for 10 min at 1,000 × g, and boiling in 2× dissociation buffer (62.5 mM Tris-HCl [pH 6.8], 10% glycerol, 2% sodium dodecyl sulfate [SDS], 10% β-mercaptoethanol) at a 1:1 ratio prior SDS-PAGE and transfer to nitrocellulose membranes. Immunoblot analyses were performed as previously described (68, 114) using mouse HIV-1 Rev antiserum (Abcam, ab85529, or Santa Cruz Biosciences, sc-69729) and rabbit anti-HSP90 antiserum (Santa Cruz Biosciences, sc-7947) detected using anti-mouse or anti-rabbit secondary antisera conjugated to either horseradish peroxidase (Pierce) or either of two infrared fluorophores, IRDye680 or IRDye800 (LI-COR Biosciences).
For infectivity measures, fresh supernatants were filtered and used to infect HeLa cells plated 24 h prior at ∼40% confluence in 12-well tissue culture-treated dishes. At 48 h postinfection, target cells were fixed using 4% paraformaldehyde (PFA), permeabilized using 0.2% Triton X-100, and stained with DAPI (4′,6′-diamidino-2-phenylindole4′,6-diamidino-2-phenylindole). YFP and DAPI fluorescence were measured using a Cytation 5 imaging reader (Biotek Instruments, Inc.) operated by Gen5 software (v 2.07) using the following excitation/emission monochromator ranges (wavelengths in nanometers): 490 to 510/520 to 550 (YFP) and 340 to 380/420 to 480 (DAPI). YFP fluorescence was normalized to cell number based on the relative DAPI signal.
Imaging.
All imaging experiments were performed on a Nikon Ti-Eclipse inverted wide-field epifluorescent deconvolution microscope (Nikon Corporation). Images were collected using an Orca-Flash 4.0 C11440 (Hamamatsu Photonics) camera and Nikon NIS Elements software (v 4.20.03) using 20× (N.A. 0.75), 40× (N.A. 0.95), 60× (N.A. 1.40), and 100× (N.A. 1.45) Plan Apo objective lenses and the following excitation/emission filter set ranges (wavelengths in nanometers): 395 to 409/430 to 480 (DAPI), 418 to 442/458 to 482 (CFP), 480 to 500/507 to 543 (GFP), 490 to 510/520 to 550 (YFP), 543 to 567/579 to 631 (mApple), 555 to 589/602 to 662 (mChe), and 630 to 660/669 to 741 (iRFP). For immunofluorescence detection of CRM1 and nucleoporins, cells were plated on glass coverslips or glass-bottom tissue culture dishes and transfected as described above. Cells were fixed at 24 h posttransfection in 4% PFA prior to permeabilization using 0.2% Triton X-100 and incubation with either rabbit anti-CRM1 antiserum (ab24189; Abcam) or mouse anti-nucleoporin antiserum (Mab414; Covance) prior to detection using Alexa Fluor 488 or Alexa Fluor 594 secondary antibodies (Invitrogen).
For live cell imaging, cells were plated at ∼40% confluence in eight-well 1.5H glass-bottom slides (Ibidi) prior to PEI transfection with 100 ng of RRE-sgRNA (pGag-CFP/24xMSL/RRE) plasmid (76) plus 20 to 50 ng of plasmids encoding each Rev-mChe variant. Image capture spanned 24 h starting at approximately 1 h posttransfection. Cells were maintained at 37°C, ∼50% humidity, and 5% CO2 in a Pathology Devices Livecell stage-top incubator (Pathology Devices, Inc.). Single images were acquired every 60 min. Cells and transfections for LEXY imaging were carried out as for the live cell imaging described above, except with image capture spanning only ∼22 min with images acquired every 30 s in three consecutive phases: (i) NES masked state (2 min), (ii) blue light induced unmasked NES state (10 min using 480 to 500/507 to 543 filter sets), and (iii) recovery of the NES masked state (10 min). All movies were postprocessed and analyzed using FIJI/NIH ImageJ2 (115, 116).
Single-molecule FISH.
Stellaris FISH RNA probes (LGC Biosearch Technologies) were custom designed using the Stellaris probe designer in order to recognize NL4-3 gag-pol nucleotides 386 to 4614. A total of 48 individual probes (20 nucleotides in length) conjugated to Quasar 670 dye (SMF-1065-5) were pooled and used for RNA detection. Cells were plated, transfected, and fixed as for fixed-cell analyses ∼24 h posttransfection. Subsequent FISH was performed according to the manufacturer's protocol adapted for eight-well 1.5H glass-bottom slides (Ibidi). We performed 70% ethanol permeabilization and probe hybridization steps with 18- to 24-h incubations. Imaging was done as described above.
ACKNOWLEDGMENTS
This study was supported by a Clinical and Translational Type I pilot grant from the University of Wisconsin Institute for Clinical and Translational Research (NIH/NCATS UL1TR000427) and by NIH grant RO1AI110221A1 to N.M.S. R.T.B. received training support from NIH NRSA award T32 AI078985. G.M.P. is a fellow of the Morgridge Institute for Research and received training support from NIH NRSA award T32 CA 157322.
REFERENCES
- 1.Kutay U, Güttinger S. 2005. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol 15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Wente SR, Rout MP. 2010. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol 2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Güttler T, Görlich D. 2011. Ran-dependent nuclear export mediators: a structural perspective. EMBO J 30:3457–3474. doi: 10.1038/emboj.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Görlich D, Kutay U. 1999. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 5.Siddiqui N, Borden KLB. 2012. mRNA export and cancer. Wiley Interdiscip Rev RNA 3:13–25. doi: 10.1002/wrna.101. [DOI] [PubMed] [Google Scholar]
- 6.Natalizio BJ, Wente SR. 2013. Postage for the messenger: designating routes for nuclear mRNA export. Trends Cell Biol 23:365–373. doi: 10.1016/j.tcb.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strambio-De-Castillia C, Niepel M, Rout MP. 2010. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol 11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 8.Cullen BR. 2003. Nuclear mRNA export: insights from virology. Trends Biochem Sci 28:419–424. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 9.Harris ME, Hope TJ. 2000. RNA export: insights from viral models. Essays Biochem 36:115–127. doi: 10.1042/bse0360115. [DOI] [PubMed] [Google Scholar]
- 10.Le Sage V, Mouland AJ. 2013. Viral subversion of the nuclear pore complex. Viruses 5:2019–2042. doi: 10.3390/v5082019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yarbrough ML, Mata MA, Sakthivel R, Fontoura BMA. 2014. Viral subversion of nucleocytoplasmic trafficking. Traffic Cph Den 15:127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butsch M, Boris-Lawrie K. 2002. Destiny of unspliced retroviral RNA: ribosome and/or virion? J Virol 76:3089–3094. doi: 10.1128/JVI.76.7.3089-3094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson SF, Telesnitsky A. 2010. Retroviral RNA dimerization and packaging: the what, how, when, where, and why. PLoS Pathog 6:e1001007. doi: 10.1371/journal.ppat.1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuzembayeva M, Dilley K, Sardo L, Hu W-S. 2014. Life of psi: how full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 454-455:362–370. doi: 10.1016/j.virol.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cochrane AW, McNally MT, Mouland AJ. 2006. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology 3:18. doi: 10.1186/1742-4690-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammarskjöld ML. 2001. Constitutive transport element-mediated nuclear export. Curr Top Microbiol Immunol 259:77–93. [DOI] [PubMed] [Google Scholar]
- 17.Swanson CM, Malim MH. 2006. Retrovirus RNA trafficking: from chromatin to invasive genomes. Traffic 7:1440–1450. doi: 10.1111/j.1600-0854.2006.00488.x. [DOI] [PubMed] [Google Scholar]
- 18.Daly TJ, Cook KS, Gray GS, Maione TE, Rusche JR. 1989. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature 342:816–819. doi: 10.1038/342816a0. [DOI] [PubMed] [Google Scholar]
- 19.Heaphy S, Dingwall C, Ernberg I, Gait MJ, Green SM, Karn J, Lowe AD, Singh M, Skinner MA. 1990. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell 60:685–693. doi: 10.1016/0092-8674(90)90671-Z. [DOI] [PubMed] [Google Scholar]
- 20.Hope TJ. 1999. The ins and outs of HIV. Rev Arch Biochem Biophys 365:186–191. [DOI] [PubMed] [Google Scholar]
- 21.Malim MH, Tiley LS, McCarn DF, Rusche JR, Hauber J, Cullen BR. 1990. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell 60:675–683. doi: 10.1016/0092-8674(90)90670-A. [DOI] [PubMed] [Google Scholar]
- 22.Pollard VW, Malim MH. 1998. The HIV-1 Rev protein. Annu Rev Microbiol 52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 23.Henderson BR, Percipalle P. 1997. Interactions between HIV rev and nuclear import and export factors: the rev nuclear localisation signal mediates specific binding to human importin-β1. J Mol Biol 274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- 24.Truant R, Cullen BR. 1999. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin β-dependent nuclear localization signals. Mol Cell Biol 19:1210–1217. doi: 10.1128/MCB.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai Y, Tambe A, Zhou K, Doudna JA. 2014. RNA-guided assembly of Rev-RRE nuclear export complexes. eLife 3:e03656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daugherty MD, D'Orso I, Frankel AD. 2008. A solution to limited genomic capacity: using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA. Mol Cell 31:824–834. doi: 10.1016/j.molcel.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daugherty MD, Liu B, Frankel AD. 2010. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat Struct Mol Biol 17:1337–1342. doi: 10.1038/nsmb.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, Grimes JM. 2010. Implications of the HIV-1 Rev dimer structure at 3.2 Å resolution for multimeric binding to the Rev response element. Proc Natl Acad Sci U S A 107:5810–5814. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiMattia MA, Watts NR, Cheng N, Huang R, Heymann JB, Grimes JM, Wingfield PT, Stuart DI, Steven AC. 2016. The structure of HIV-1 Rev filaments suggests a bilateral model for Rev-RRE assembly. Structure 24:1068–1080. doi: 10.1016/j.str.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang X, Wang J, O'Carroll IP, Mitchell M, Zuo X, Wang Y, Yu P, Liu Y, Rausch JW, Dyba MA, Kjems J, Schwieters CD, Seifert S, Winans RE, Watts NR, Stahl SJ, Wingfield PT, Byrd RA, Le Grice SFJ, Rein A, Wang Y- X. 2013. An unusual topological structure of the HIV-1 Rev response element. Cell 155:594–605. doi: 10.1016/j.cell.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain C, Belasco JG. 2001. Structural model for the cooperative assembly of HIV-1 Rev multimers on the RRE as deduced from analysis of assembly-defective mutants. Mol Cell 7:603–614. doi: 10.1016/S1097-2765(01)00207-6. [DOI] [PubMed] [Google Scholar]
- 32.Fornerod M, Ohno M, Yoshida M, Mattaj IW. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051–1060. doi: 10.1016/S0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 33.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 34.Güttler T, Madl T, Neumann P, Deichsel D, Corsini L, Monecke T, Ficner R, Sattler M, Görlich D. 2010. NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat Struct Mol Biol 17:1367–1376. doi: 10.1038/nsmb.1931. [DOI] [PubMed] [Google Scholar]
- 35.Askjaer P, Jensen TH, Nilsson J, Englmeier L, Kjems J. 1998. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J Biol Chem 273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- 36.Cullen BR. 1998. HIV-1 auxiliary proteins: making connections in a dying cell. Cell 93:685–692. doi: 10.1016/S0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 37.Dahlberg JE, Lund E. 1998. Functions of the GTPase Ran in RNA export from the nucleus. Curr Opin Cell Biol 10:400–408. doi: 10.1016/S0955-0674(98)80017-3. [DOI] [PubMed] [Google Scholar]
- 38.la Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S. 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel PEDS 17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 39.Fu S-C, Imai K, Horton P. 2011. Prediction of leucine-rich nuclear export signal containing proteins with NESsential. Nucleic Acids Res 39:e111. doi: 10.1093/nar/gkr493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu S-C, Huang H-C, Horton P, Juan H-F. 2013. ValidNESs: a database of validated leucine-rich nuclear export signals. Nucleic Acids Res 41:D338–D343. doi: 10.1093/nar/gks936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu D, Grishin NV, Chook YM. 2012. NESdb: a database of NES-containing CRM1 cargoes. Mol Biol Cell 23:3673–3676. doi: 10.1091/mbc.E12-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu D, Farmer A, Collett G, Grishin NV, Chook YM. 2012. Sequence and structural analyses of nuclear export signals in the NESdb database. Mol Biol Cell 23:3677–3693. doi: 10.1091/mbc.E12-01-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu D, Marquis K, Pei J, Fu S-C, Cağatay T, Grishin NV, Chook YM. 2015. LocNES: a computational tool for locating classical NESs in CRM1 cargo proteins. Bioinforma Oxf Engl 31:1357–1365. doi: 10.1093/bioinformatics/btu826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischer U, Huber J, Boelens WC, Mattajt LW, Lührmann R. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 45.Wen W, Meinkotht JL, Tsien RY, Taylor SS. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 46.Neville M, Stutz F, Lee L, Davis LI, Rosbash M. 1997. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol CB 7:767–775. doi: 10.1016/S0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 47.Stade K, Ford CS, Guthrie C, Weis K. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041–1050. doi: 10.1016/S0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 48.Dong X, Biswas A, Süel KE, Jackson LK, Martinez R, Gu H, Chook YM. 2009. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature 458:1136–1141. doi: 10.1038/nature07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monecke T, Güttler T, Neumann P, Dickmanns A, Görlich D, Ficner R. 2009. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science 324:1087–1091. doi: 10.1126/science.1173388. [DOI] [PubMed] [Google Scholar]
- 50.Petosa C, Schoehn G, Askjaer P, Bauer U, Moulin M, Steuerwald U, Soler-López M, Baudin F, Mattaj IW, Müller CW. 2004. Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol Cell 16:761–775. doi: 10.1016/j.molcel.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 51.Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR. 1996. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol 16:4207–4214. doi: 10.1128/MCB.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyer BE, Meinkoth JL, Malim MH. 1996. Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev proteins: identification of a family of transferable nuclear export signals. J Virol 70:2350–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Atasheva S, Fish A, Fornerod M, Frolova EI. 2010. Venezuelan equine Encephalitis virus capsid protein forms a tetrameric complex with CRM1 and importin alpha/beta that obstructs nuclear pore complex function. J Virol 84:4158–4171. doi: 10.1128/JVI.02554-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Engelsma D, Bernad R, Calafat J, Fornerod M. 2004. Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J 23:3643–3652. doi: 10.1038/sj.emboj.7600370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engelsma D, Valle N, Fish A, Salomé N, Almendral JM, Fornerod M. 2008. A supraphysiological nuclear export signal is required for parvovirus nuclear export. Mol Biol Cell 19:2544–2552. doi: 10.1091/mbc.E08-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fischer U, Schäuble N, Schütz S, Altvater M, Chang Y, Faza MB, Panse VG. 2015. A non-canonical mechanism for Crm1-export cargo complex assembly. eLife 2015:4. doi: 10.7554/eLife.05745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.West M, Hedges JB, Lo K-Y, Johnson AW. 2007. Novel interaction of the 60S ribosomal subunit export adapter Nmd3 at the nuclear pore complex. J Biol Chem 282:14028–14037. doi: 10.1074/jbc.M700256200. [DOI] [PubMed] [Google Scholar]
- 58.Bhattacharya S, Schindler C. 2003. Regulation of Stat3 nuclear export. J Clin Invest 111:553–559. doi: 10.1172/JCI15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ferrer M, Rodríguez JA, Spierings EA, de Winter JP, Giaccone G, Kruyt FAE. 2005. Identification of multiple nuclear export sequences in Fanconi anemia group A protein that contribute to CRM1-dependent nuclear export. Hum Mol Genet 14:1271–1281. doi: 10.1093/hmg/ddi138. [DOI] [PubMed] [Google Scholar]
- 60.Huang S, Chen J, Chen Q, Wang H, Yao Y, Chen J, Chen Z. 2013. A second CRM1-dependent nuclear export signal in the influenza A virus NS2 protein contributes to the nuclear export of viral ribonucleoproteins. J Virol 87:767–778. doi: 10.1128/JVI.06519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neufeld KL, Nix DA, Bogerd H, Kang Y, Beckerle MC, Cullen BR, White RL. 2000. Adenomatous polyposis coli protein contains two nuclear export signals and shuttles between the nucleus and cytoplasm. Proc Natl Acad Sci U S A 97:12085–12090. doi: 10.1073/pnas.220401797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fernandes J, Jayaraman B, Frankel A. 2012. The HIV-1 Rev response element. RNA Biol 9:6–11. doi: 10.4161/rna.9.1.18178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakano K, Watanabe T. 2012. HTLV-1 Rex: the courier of viral messages making use of the host vehicle. Front Microbiol 3:330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nekhai S, Jeang K-T. 2006. Transcriptional and posttranscriptional regulation of HIV-1 gene expression: role of cellular factors for Tat and Rev. Future Microbiol 1:417–426. doi: 10.2217/17460913.1.4.417. [DOI] [PubMed] [Google Scholar]
- 65.Sherpa C, Rausch JW, Le J, Grice SF, Hammarskjold M-L, Rekosh D. 2015. The HIV-1 Rev response element (RRE) adopts alternative conformations that promote different rates of virus replication. Nucleic Acids Res 43:4676–4686. doi: 10.1093/nar/gkv313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shida H. 2012. Role of nucleocytoplasmic RNA transport during the life cycle of retroviruses. Front Microbiol 3:179. doi: 10.3389/fmicb.2012.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rausch JW, Le Grice SFJ. 2015. HIV Rev assembly on the Rev response element (RRE): a structural perspective. Viruses 7:3053–3075. doi: 10.3390/v7062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aligeti M, Behrens RT, Pocock GM, Schindelin J, Dietz C, Eliceiri KW, Swanson CM, Malim MH, Ahlquist P, Sherer NM. 2014. Cooperativity among Rev-associated nuclear export signals regulates HIV-1 gene expression and is a determinant of virus species tropism. J Virol 88:14207–14221. doi: 10.1128/JVI.01897-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoffmann D, Schwarck D, Banning C, Brenner M, Mariyanna L, Krepstakies M, Schindler M, Millar DP, Hauber J. 2012. Formation of trans-activation competent HIV-1 Rev:RRE complexes requires the recruitment of multiple protein activation domains. PLoS One 7:e38305. doi: 10.1371/journal.pone.0038305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Booth DS, Cheng Y, Frankel AD. 2014. The export receptor Crm1 forms a dimer to promote nuclear export of HIV RNA. eLife 3:e04121. doi: 10.7554/eLife.04121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Corredor AG, Archambault D. 2012. The bovine immunodeficiency virus Rev protein: identification of a novel nuclear import pathway and nuclear export signal among retroviral Rev/Rev-like proteins. J Virol 86:4892–4905. doi: 10.1128/JVI.05132-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Henderson BR, Eleftheriou A. 2000. A comparison of the activity, sequence specificity, and CRM1 dependence of different nuclear export signals. Exp Cell Res 256:213–224. doi: 10.1006/excr.2000.4825. [DOI] [PubMed] [Google Scholar]
- 73.Askjaer P, Bachi A, Wilm M, Bischoff FR, Weeks DL, Ogniewski V, Ohno M, Niehrs C, Kjems J, Mattaj IW, Fornerod M. 1999. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol Cell Biol 19:6276–6285. doi: 10.1128/MCB.19.9.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Elfgang C, Rosorius O, Hofer L, Jaksche H, Hauber J, Bevec D. 1999. Evidence for specific nucleocytoplasmic transport pathways used by leucine-rich nuclear export signals. Proc Natl Acad Sci U S A 96:6229–6234. doi: 10.1073/pnas.96.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Niopek D, Wehler P, Roensch J, Eils R, Di Ventura B. 2016. Optogenetic control of nuclear protein export. Nat Commun 7:10624. doi: 10.1038/ncomms10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pocock GM, Becker JT, Swanson CM, Ahlquist P, Sherer NM. 2016. HIV-1 and M-PMV RNA nuclear export elements program viral genomes for distinct cytoplasmic trafficking behaviors. PLoS Pathog 12:e1005565. doi: 10.1371/journal.ppat.1005565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Malim MH, Cullen BR. 1991. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell 65:241–248. doi: 10.1016/0092-8674(91)90158-U. [DOI] [PubMed] [Google Scholar]
- 78.Thomas SL, Oft M, Jaksche H, Casari G, Heger P, Dobrovnik M, Bevec D, Hauber J. 1998. Functional analysis of the human immunodeficiency virus type 1 Rev protein oligomerization interface. J Virol 72:2935–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jayaraman B, Crosby DC, Homer C, Ribeiro I, Mavor D, Frankel AD. 2014. RNA-directed remodeling of the HIV-1 protein Rev orchestrates assembly of the Rev-Rev response element complex. eLife 3:e04120. doi: 10.7554/eLife.04120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nawroth I, Mueller F, Basyuk E, Beerens N, Rahbek UL, Darzacq X, Bertrand E, Kjems J, Schmidt U. 2014. Stable assembly of HIV-1 export complexes occurs cotranscriptionally. RNA 20:1–8. doi: 10.1261/rna.038182.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fridell RA, Bogerd HP, Cullen BR. 1996. Nuclear export of late HIV-1 mRNAs occurs via a cellular protein export pathway. Proc Natl Acad Sci U S A 93:4421–4424. doi: 10.1073/pnas.93.9.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fridell RA, Fischer U, Lührmann R, Meyer BE, Meinkoth JL, Malim MH, Cullen BR. 1996. Amphibian transcription factor IIIA proteins contain a sequence element functionally equivalent to the nuclear export signal of human immunodeficiency virus type 1. Rev Proc Natl Acad Sci U S A 93:2936–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zapp ML, Hope TJ, Parslow TG, Green MR. 1991. Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: a dual function for an arginine-rich binding motif. Proc Natl Acad Sci U S A 88:7734–7738. doi: 10.1073/pnas.88.17.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kau TR, Way JC, Silver PA. 2004. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer 4:106–117. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- 85.Komeili A, O'Shea EK. 2000. Nuclear transport and transcription. Curr Opin Cell Biol 12:355–360. doi: 10.1016/S0955-0674(00)00100-9. [DOI] [PubMed] [Google Scholar]
- 86.Craig E, Zhang Z-K, Davies KP, Kalpana GV. 2002. A masked NES in INI1/hSNF5 mediates hCRM1-dependent nuclear export: implications for tumorigenesis. EMBO J 21:31–42. doi: 10.1093/emboj/21.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. 1999. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J 18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jeyasekharan AD, Liu Y, Hattori H, Pisupati V, Jonsdottir AB, Rajendra E, Lee M, Sundaramoorthy E, Schlachter S, Kaminski CF, Ofir-Rosenfeld Y, Sato K, Savill J, Ayoub N, Venkitaraman AR. 2013. A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization. Nat Struct Mol Biol 20:1191–1198. doi: 10.1038/nsmb.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hogan PG, Chen L, Nardone J, Rao A. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev 17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 90.Okamura H, Aramburu J, García-Rodríguez C, Viola JP, Raghavan A, Tahiliani M, Zhang X, Qin J, Hogan PG, Rao A. 2000. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell 6:539–550. doi: 10.1016/S1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- 91.Zhu J, McKeon F. 1999. NF-AT activation requires suppression of Crm1-dependent export by calcineurin. Nature 398:256–260. doi: 10.1038/18473. [DOI] [PubMed] [Google Scholar]
- 92.Kumar GR, Shum L, Glaunsinger BA. 2011. Importin α-mediated nuclear import of cytoplasmic poly(A) binding protein occurs as a direct consequence of cytoplasmic mRNA depletion. Mol Cell Biol 31:3113–3125. doi: 10.1128/MCB.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Song J, Nagano-Fujii M, Wang F, Florese R, Fujita T, Ishido S, Hotta H. 2000. Nuclear localization and intramolecular cleavage of N-terminally deleted NS5A protein of hepatitis C virus. Virus Res 69:109–117. doi: 10.1016/S0168-1702(00)00206-9. [DOI] [PubMed] [Google Scholar]
- 94.Vercruysse T, Daelemans D. 2013. HIV-1 Rev multimerization: mechanism and insights. Curr HIV Res 11:623–634. [DOI] [PubMed] [Google Scholar]
- 95.Vercruysse T, Pardon E, Vanstreels E, Steyaert J, Daelemans D. 2010. An intrabody based on a llama single-domain antibody targeting the N-terminal alpha-helical multimerization domain of HIV-1 Rev prevents viral production. J Biol Chem 285:21768–21780. doi: 10.1074/jbc.M110.112490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gu L, Tsuji T, Jarboui MA, Yeo GP, Sheehy N, Hall WW, Gautier VW. 2011. Intermolecular masking of the HIV-1 Rev NLS by the cellular protein HIC: novel insights into the regulation of Rev nuclear import. Retrovirology 8:17. doi: 10.1186/1742-4690-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Michienzi A, Cagnon L, Bahner I, Rossi JJ. 2000. Ribozyme-mediated inhibition of HIV 1 suggests nucleolar trafficking of HIV-1 RNA. Proc Natl Acad Sci U S A 97:8955–8960. doi: 10.1073/pnas.97.16.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wolff H, Hadian K, Ziegler M, Weierich C, Kramer-Hammerle S, Kleinschmidt A, Erfle V, Brack-Werner R. 2006. Analysis of the influence of subcellular localization of the HIV Rev protein on Rev-dependent gene expression by multi-fluorescence live-cell imaging. Exp Cell Res 312:443–456. doi: 10.1016/j.yexcr.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 99.Cochrane AW, Golub E, Volsky D, Ruben S, Rosen CA. 1989. Functional significance of phosphorylation to the human immunodeficiency virus Rev protein. J Virol 63:4438–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hauber J, Bouvier M, Malim MH, Cullen BR. 1988. Phosphorylation of the rev gene product of human immunodeficiency virus type 1. J Virol 62:4801–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Harreman MT, Kline TM, Milford HG, Harben MB, Hodel AE, Corbett AH. 2004. Regulation of nuclear import by phosphorylation adjacent to nuclear localization signals. J Biol Chem 279:20613–20621. doi: 10.1074/jbc.M401720200. [DOI] [PubMed] [Google Scholar]
- 102.Jans DA, Hübner S. 1996. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol Rev 76:651–685. [DOI] [PubMed] [Google Scholar]
- 103.Hofacre A, Nitta T, Fan H. 2009. Jaagsiekte sheep retrovirus encodes a regulatory factor, Rej, required for synthesis of Gag protein. J Virol 83:12483–12498. doi: 10.1128/JVI.01747-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Indik S, Günzburg WH, Salmons B, Rouault F. 2005. A novel, mouse mammary tumor virus encoded protein with Rev-like properties. Virology 337:1–6. doi: 10.1016/j.virol.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 105.Mertz JA, Simper MS, Lozano MM, Payne SM, Dudley JP. 2005. Mouse mammary tumor virus encodes a self-regulatory RNA export protein and is a complex retrovirus. J Virol 79:14737–14747. doi: 10.1128/JVI.79.23.14737-14747.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mertz JA, Lozano MM, Dudley JP. 2009. Rev and Rex proteins of human complex retroviruses function with the MMTV Rem-responsive element. Retrovirology 6:10. doi: 10.1186/1742-4690-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Müllner M, Salmons B, Günzburg WH, Indik S. 2008. Identification of the Rem-responsive element of mouse mammary tumor virus. Nucleic Acids Res 36:6284–6294. doi: 10.1093/nar/gkn608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nitta T, Hofacre A, Hull S, Fan H. 2009. Identification and mutational analysis of a Rej response element in Jaagsiekte sheep retrovirus RNA. J Virol 83:12499–12511. doi: 10.1128/JVI.01754-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang J, Bogerd HP, Peng S, Wiegand H, Truant R, Cullen BR. 1999. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc Natl Acad Sci U S A 96:13404–13408. doi: 10.1073/pnas.96.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang J, Bogerd H, Le SY, Cullen BR. 2000. The human endogenous retrovirus K Rev response element coincides with a predicted RNA folding region. RNA N Y N 6:1551–1564. doi: 10.1017/S135583820000100X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tiley LS, Malim MH, Cullen BR. 1991. Conserved functional organization of the human immunodeficiency virus type 1 and visna virus Rev proteins. J Virol 65:3877–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 113.He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol 69:6705–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sherer NM, Swanson CM, Papaioannou S, Malim MH. 2009. Matrix mediates the functional link between human immunodeficiency virus type 1 RNA nuclear export elements and the assembly competency of Gag in murine cells. J Virol 83:8525–8535. doi: 10.1128/JVI.00699-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. 2015. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol Reprod Dev 82:518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]