ABSTRACT
The alphaviruses Venezuelan equine encephalitis virus (VEEV), eastern equine encephalitis virus (EEEV), and western equine encephalitis virus (WEEV) are arthropod-borne positive-strand RNA viruses that are capable of causing acute and fatal encephalitis in many mammals, including humans. VEEV was weaponized during the Cold War and is recognized as a select agent. Currently, there are no FDA-approved vaccines or therapeutics for these viruses. The spread of VEEV and other members of this family due to climate change-mediated vector range expansion underscores the need for research aimed at developing medical countermeasures. These viruses utilize programmed −1 ribosomal frameshifting (−1 PRF) to synthesize the viral trans-frame (TF) protein, which has previously been shown to be important for neuropathogenesis in the related Sindbis virus. Here, the alphavirus −1 PRF signals were characterized, revealing novel −1 PRF stimulatory structures. −1 PRF attenuation mildly affected the kinetics of VEEV accumulation in cultured cells but strongly inhibited its pathogenesis in an aerosol infection mouse model. Importantly, the decreased viral titers in the brains of mice infected with the mutant virus suggest that the alphavirus TF protein is important for passage through the blood-brain barrier and/or for neuroinvasiveness. These findings suggest a novel approach to the development of safe and effective live attenuated vaccines directed against VEEV and perhaps other closely related −1 PRF-utilizing viruses.
IMPORTANCE Venezuelan equine encephalitis virus (VEEV) is a select agent that has been weaponized. This arthropod-borne positive-strand RNA virus causes acute and fatal encephalitis in many mammals, including humans. There is no vaccine or other approved therapeutic. VEEV and related alphaviruses utilize programmed −1 ribosomal frameshifting (−1 PRF) to synthesize the viral trans-frame (TF) protein, which is important for neuropathogenesis. −1 PRF attenuation strongly inhibited VEEV pathogenesis in mice, and viral replication analyses suggest that the TF protein is critical for neurological disease. These findings suggest a new approach to the development of safe and effective live attenuated vaccines directed against VEEV and other related viruses.
KEYWORDS: ribosomal frameshifting, virus, alphavirus, vaccine, neuropathology
INTRODUCTION
Venezuelan equine encephalitis virus (VEEV) belongs to the family Togaviridae, genus Alphavirus, which is further subdivided into Old World and New World alphaviruses on the basis of their geographical distribution. The Old World alphaviruses include Sindbis virus (SINV), Semliki Forest virus (SFV), and chikungunya virus (CHIKV); these generally cause diseases resulting in fever, rash, and arthritic disease. The New World alphaviruses are categorically encephalitic and include eastern equine encephalitis virus (EEEV) and western equine encephalitis virus (WEEV), in addition to VEEV. VEEV, EEEV, and WEEV infections in humans result in 1%, 50 to 78%, and 3 to 7% mortality rates, respectively (1). Mortality rates in horses are overall more severe at 20 to 80% for VEEV, 70 to 90% for EEEV, and 3 to 50% for WEEV (1). There are no FDA-licensed vaccines or therapeutics for VEEV, EEEV, and WEEV infections, underscoring the need to investigate the molecular biology and pathogenic mechanisms of these pathogens.
The small genomes of RNA viruses limit their coding capacity. In response, they can expand their coding capacity by utilizing alternative splicing (2), RNA editing (3), leaky ribosomal scanning (4), and programmed ribosomal frameshifting (PRF) (4–6). In −1 PRF, the ribosome encounters a cis-acting element, termed the “frameshift signal,” which causes it to stall on the mRNA over a special slippery sequence. The ribosome is then directed to shift 1 nucleotide backward, allowing it to synthesize a protein with an alternative C-terminal peptide sequence (5). Typical −1 PRF signals are composed of three modules arranged in the following 5′-to-3′ direction: a heptameric slippery site, a short spacer, and a strong RNA secondary structural element, often an mRNA pseudoknot (7). Multiple RNA viruses, including West Nile virus, human immunodeficiency virus (HIV), and severe acute respiratory syndrome coronavirus (SARS-CoV), utilize −1 PRF (4, 6, 8).
Alphaviruses were bioinformatically predicted to use −1 PRF to generate the trans-frame (TF) protein, which shares its N-terminal region with the 6K protein (9). Mass spectrometric analyses were used to confirm TF protein production by multiple alphavirus family members, including SFV, SINV, and CHIKV (9, 10). An SINV-based molecular genetics-based study examining the consequences of altering the production, size, or sequence of the TF protein revealed reduced levels of SINV release from both mammalian and mosquito cells, without influencing genomic replication, specific infectivity, or migration of the envelope protein to the cell surface, suggesting that the TF protein was likely involved in viral assembly (10). Moreover, SINV with a mutated TF protein was attenuated in an SINV neuropathogenic mouse model. To date there have been no studies on the role and importance of the TF protein in the New World alphaviruses. The current study characterizes the frameshifting signals in New World alphaviruses (VEEV, EEEV, and WEEV). The results of standard molecular genetics analyses are consistent with the prediction that these viruses utilize −1 PRF to synthesize their TF proteins. VEEV harboring a silent protein-coding mutation that attenuates −1 PRF activity (VEEVPRFm) displayed mildly decreased viral replication kinetics in vitro. Strikingly, however, mice infected with VEEVPRFm showed dramatically increased survival and decreased clinical signs of disease compared to mice infected with wild-type WT VEEV. These data demonstrate that frameshifting is a critical mechanism utilized by alphaviruses to encode the TF protein, which is important for pathogenesis, and suggest −1 PRF attenuation as a general strategy for exploring the rational development of live attenuated vaccines.
RESULTS
Alphavirus-derived sequences promote efficient levels of −1 PRF.
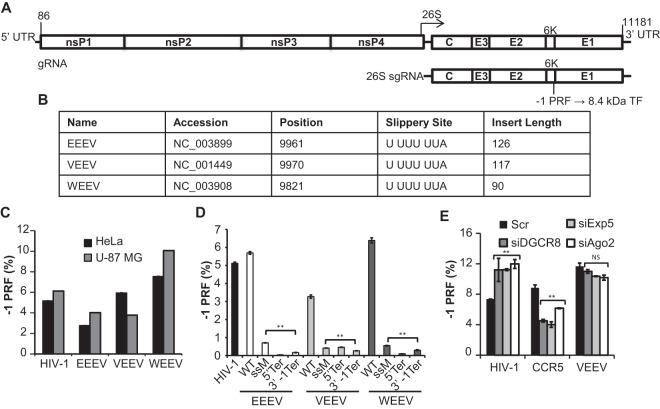
The alphavirus genomes consist of a single positive-strand RNA that harbors two open reading frames (ORFs) (Fig. 1A). The 5′ ORF encodes four nonstructural proteins, while five structural proteins are encoded by the 26S subgenomic RNA. These two ORFs are translated as polyproteins which are proteolytically processed into the mature proteins (reviewed in reference 11). In two Old World alphaviruses, SINV and CHIKV, −1 PRF events in the 6K gene result in production of the 8.4-kDa trans-frame (TF) protein (10). Similar signals are predicted to be located near the 3′ end of the sequence encoding the 6K protein in the New World alphaviruses (12), and the 8.4-kDa TF protein is produced consequent to −1 PRF events. Strategies for cloning the predicted −1 PRF signals from EEEV, VEEV, and WEEV into dual-luciferase reporters (13) were determined by identifying their conserved 3′ slippery sites and in silico RNA folding of 3′ sequences (14). Information pertaining to these clones is shown in Fig. 1B. Figure 1C shows that all of these sequences promoted efficient levels of −1 PRF in both HeLa and U-87 MG cell lines. These findings are in general agreement with prior measurements of VEEV and EEEV frameshifting efficiencies measured by dual-luciferase assays (9). To rule out the possibilities that these sequences harbor internal ribosome entry site activity, cryptic promoters, or cryptic splice sites, a series of mutants harboring slippery site inactivation mutations (ssM), in-frame termination codons prior to the slippery sites (5′ Ter), and −1 frame termination codons 3′ of the virus-derived sequences (3′ −1 Ter) were tested. Dual-luciferase reporter assays ascertained that all of these mutations significantly reduced apparent −1 PRF activity (Fig. 1D), indicating that these sequences encode bona fide −1 PRF signals. A prior study demonstrated that −1 PRF can be naturally regulated in cells by microRNAs (miRNAs) (15). To investigate this possibility, miRNA processing or export was inhibited by knocking down expression of DGCR8, Exportin 5, or Argonaute 2 (Ago2) in U-87 MG cells with small interfering (siRNA), and the effects on VEEV-mediated −1 PRF were subsequently assayed. As positive controls, −1 PRF mediated by the HIV-1 and CCR5 −1 PRF signals was also assayed. Results from these experiments suggest that, unlike HIV-1 and CCR5, VEEV −1 PRF is not regulated by miRNAs in this cell type (Fig. 1E). Similarly, siRNA knockdown of Ago1 in HeLa cells did not significantly affect the −1 PRF promoted by the VEEV, EEEV, or WEEV sequences in HeLa cells (data not shown).
FIG 1.
Identification and monitoring of −1 PRF in the alphaviruses. (A) General schematic of the alphavirus genome and subgenomic transcript. The specific nucleotide numbers are given for the boundaries of the VEEV untranslated regions (UTRs). This family of viruses harbors −1 PRF signals toward the 3′ end of the 6K structural protein-encoding mRNA. The resulting frameshift product is an 8.4-kDa protein called the trans-frame (TF) protein. gRNA, genomic RNA; sgRNA, subgenomic RNA. (B) Accession numbers of the sequences from which the predicted −1 PRF signals were cloned, nucleotide positions of the first base of the predicted slippery sites, the slippery site sequences (the 0 frame is indicated by spaces), and the lengths of inserts (in numbers of nucleotides) initially tested for −1 PRF activities. (C) The predicted −1 PRF signals derived from EEEV, VEEV, and WEEV were cloned into the dual-luciferase reporter plasmid pJD175f, and their ability to promote efficient levels of frameshifting was measured in both HeLa cells and U-87 MG astrocyte cells. The −1 PRF signal from HIV-1 was employed as a positive control. (D) Site-directed mutagenesis was utilized to genetically validate the −1 PRF activities of the EEEV-, VEEV-, and WEEV-derived sequences in U-87 MG cells. WT, wild type; ssM, silent mutations of the slippery sites; 5′ Ter, 0-frame termination codons introduced upstream of the slippery sites; 3′ −1 Ter, termination codons inserted in the −1 reading frame 3′ of the virus-derived sequences. (E) miRNA processing or export was inhibited by siRNA knockdown of DGCR8 (siDGCR8), Exportin 5 (siExp5), or Argonaute 2 (siAgo2) in U-87 MG cells, and the rates of −1 PRF promoted by the indicated sequences were monitored. Control samples were transfected with siRNAs harboring scrambled (Scr) sequences. For assays of −1 PRF, a minimum of three or more biological replicates was performed in triplicate until statistical significance was achieved, as previously described (11). Bars represent standard errors of the means. **, P < 0.01; NS, not significant.
Efficient alphavirus −1 PRF is stimulated by stem-loop mRNA structural elements.
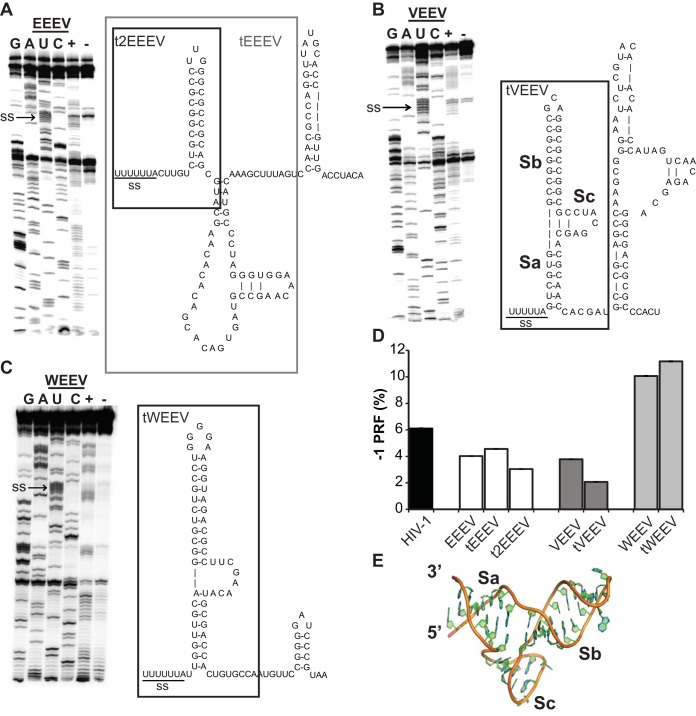
In classic −1 PRF signals, RNA structural elements located immediately 3′ of heptameric slippery sites serve as kinetic traps to stimulate frameshifting by stalling elongating ribosomes over the slippery site (7). Typically, these are mRNA pseudoknot structures, although −1 PRF can be stimulated by simple stem-loop structures as well (16). Selective 2′-hydroxyl acylation and primer extension (SHAPE) (17) was employed to characterize the nature of the EEEV, VEEV, and WEEV −1 PRF stimulating elements (Fig. 2A to C). Analysis of the SHAPE data revealed the presence of tandem stem-loops rather than RNA pseudoknot structures for all three of the viral sequences. Dual-luciferase reporter assays of 3′ truncation mutants (tEEEV, t2EEEV, tVEEV, and tWEEV) revealed that only the slippery site proximal stem-loops are required to promote efficient rates of −1 PRF (Fig. 2D). Guided molecular dynamics simulations of the tVEEV element suggests that this folds into a novel V-shaped structure comprising three stems, labeled Sa, Sb, and Sc (Fig. 2B and E). How this may promote efficient levels of −1 PRF is discussed below.
FIG 2.
The EEEV (A), VEEV (B), and WEEV (C) −1 PRF signals were chemically resolved using selective 2′-hydroxyl acylation and primer extension (SHAPE). RNAs for the −1 PRF signal of each alphavirus were transcribed from their corresponding reporter plasmids, RNA samples were treated with N-methylisatoic anhydride (NMIA), primer extension reactions were carried out, the reaction mixtures were separated through 8% urea-PAGE, and 32P-labeled cDNA products were visualized using a Typhoon phosphorimager (left). (Right) RNA structures deduced from the SHAPE data. The 3′ truncation mutants are indicated in the boxed regions. (D) Identification of the minimal structures required to promote efficient levels of −1 PRF (defined as >1%). The truncated sequences indicated in panels A to C were cloned into pJD175f dual-reporter plasmids and assayed for their ability to promote efficient levels of frameshifting in U-87 MG cells. For assays of −1 PRF, a minimum of three or more biological replicates was performed in triplicate until statistical significance was achieved, as previously described (11). Bars represent standard errors of the means. (E) Predicted three-dimensional structure of the tVEEV −1 PRF stimulatory element.
Ablation of −1 PRF mildly decreases VEEV production in vitro.
Prior studies have suggested that viral −1 PRF signals have evolved to promote frameshifting at very precise rates and that changes in −1 PRF efficiencies have detrimental effects on virus propagation (18–20). To test the importance of −1 PRF on virus propagation in cultured cells, a silent protein-coding change was introduced into the VEEV infectious clones for the TC83 vaccine strain and the highly pathogenic Trinidad Donkey (TrD) strain (21) to create pTC83PRFm and pTrDPRFm, respectively (Fig. 3A). Vero cells were infected with TC83, TC83PRFm, TrD, or TrDPRFm as described in Materials and Methods, and samples were collected for analysis at 3, 6, 9, 18, and 24 h postinfection (hpi). Surprisingly, although statistically significant effects were observed at some of the later time points, ablation of −1 PRF within the TC83 backbone had minimum effects on virus titers and viral RNA accumulation (Fig. 3B and E). Similar results were observed with C6/36 mosquito cells (data not shown). Disruption of the −1 PRF signal within TrD resulted in decreased viral titers (∼1.5 logs) starting at 9 hpi (Fig. 3F). Viral RNA levels were not affected until 18 hpi, which is consistent with a defect in viral assembly (Fig. 3C). Replication analysis was also performed in differentiated AP-7 (dAP-7) rat neuronal cells, which represent a more relevant physiological model of VEEV infection. Viral RNA levels did not differ between TrD and TrDPRFm (Fig. 3D), but a decrease in viral titers was observed with TrDPRFm (Fig. 3G), supporting the notion that the TF protein plays a role in viral assembly. dAP-7 cells had exited the cell cycle, whereas Vero cells were cycling, which may be one reason for the observed differences in viral RNA levels between the two cell types. With several substitutions within E2 and E1 between TrD and TC83 (as noted within Materials and Methods), this may indicate that there is less dependence on the −1 PRF signal for the attenuated TC83 strain in vitro.
FIG 3.
Ablation of −1 PRF decreases VEEV release in vitro. (A) Schematic diagram indicating silent coding nucleotide substitutions ablating the −1 PRF signals in the VEEV TC83 and TrD infectious clones (VEEVPRFm). (B to G) Replication kinetics of TC83 and TC83PRFm in Vero cells (B and E), TrD and TrDPRFm in Vero cells (C and F), and TrD and TrDPRFm in dAP-7 cells (D and G) infected at an MOI of 1. (B to D) Total RNAs were isolated from cells, and the absolute quantities (for TC83-infected cells) and relative quantities (for TrD-infected cells) of VEEV genomic copies were determined by qRT-PCR. For TrD-infected cells, relative fold change values were calculated using the number of genomic copies of the respective virus detected at 3 hpi. Data are plotted as means with standard deviations (n = 9 experiments for TC83-infected cells, n = 6 experiments for TrD-infected Vero cells, n = 6 experiments for TrD-infected dAP-7 cells). (E to G) Medium supernatants were isolated at the indicated times, and infectious viral titers were determined by plaque assay. Data are plotted as means with standard deviations (n = 9 experiments for TC83-infected cells, n = 6 experiments for TrD-infected Vero cells, n = 6 experiments for TrD-infected dAP-7 cells). For all data sets, a multiple unpaired t test analysis with the Holm-Sidak correction was applied to each time point. Bars represent standard deviations. *, P < 0.05.
Ablation of −1 PRF strongly attenuates VEEV pathogenesis.
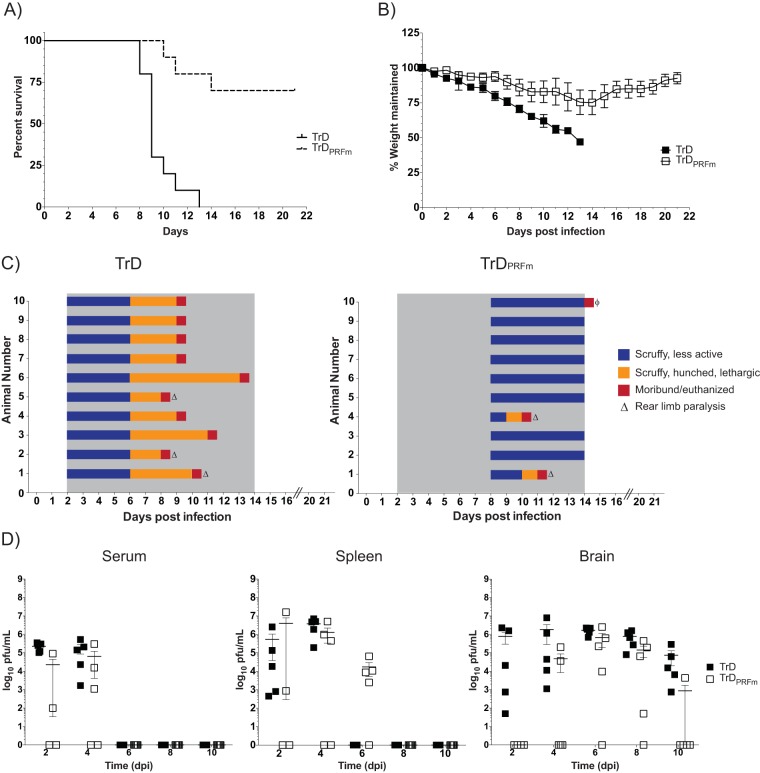
Prior studies in which −1 PRF was ablated in flaviviruses revealed that the NS1′ frameshift PRF product is required for neuroinvasion and replication in both avian and insect hosts (22, 23). Similarly, deletion of the 6K gene reduced the pathogenesis of the Ross River alphavirus in mice (24). To determine the importance of the −1 PRF signal for VEEV pathogenesis, mice were exposed to 1 × 105 PFU/ml of VEEV TrD or VEEV TrDPRFm for 10 min via the aerosol route. Two groups of mice were followed for 21 days in order to assess survival, while others were sacrificed over the course of infection to assess viral kinetics in vivo. Kaplan-Meier survival analysis shows that ablation of −1 PRF had a strong negative effect on VEEV-induced mortality (Fig. 4A). VEEV TrD-infected mice succumbed to infection beginning at 8 days postinfection (dpi), with all mice succumbing by 13 dpi. In contrast, 70% of VEEV TrDPRFm-infected mice survived the infection. Weight loss (Fig. 4B) and clinical symptoms of disease (Fig. 4C) were less severe and delayed in VEEV TrDPRFm-infected mice compared to VEEV TrD-infected mice. In parallel experiments, virus titers in the serum, spleens, and brains of infected mice were monitored every 2 days for 10 days total. In all of the VEEV TrD-infected mice, virus was detected in the blood and spleen early after infection (2 and 4 dpi) and cleared by 6 dpi (Fig. 4D). In contrast, following infection with VEEV TrDPRFm, virus was detectable in the blood and spleen in only 50% of the mice at 2 dpi and in only 60% of the mice at 4 dpi. Virus was also detected in the spleen at 6 dpi in 80% of the VEEV TrDPRFm-infected mice. Plaque assays of brains revealed the presence of high levels of virus in the VEEV TrD-infected mice at all time points tested. In contrast, virus was not detectable in the VEEV TrDPRFm-infected mice until 4 dpi and was cleared in 80% of those mice by 10 dpi. These results indicate that the dissemination of VEEV TrDPRFm is altered in vivo, resulting in less viral replication within the brain and overall decreased pathogenesis.
FIG 4.
Ablation of −1 PRF strongly attenuates VEEV pathogenesis. (A) BALB/c mice were infected with VEEV TrD or VEEV TrDPRFm by aerosol exposure. Animals were monitored for 21 days postchallenge, and survival curves were determined. The data plotted represent those for 10 animals per group. (B) Mice were monitored for weight loss daily over 21 days. The percentage of weight maintained (relative to the starting weight) was determined. The data plotted represent the mean values and standard deviations for 10 animals per group. (C) Mice were also monitored at least daily for clinical symptoms of disease over 21 days. Data are plotted per animal per day. The gray shaded area indicates the time frame when clinical disease was observed in VEEV TrD-infected mice. ϕ, one animal had to be euthanized due to self-mutilation. Necropsy indicated no signs of disease in this mouse. (D) Mice were infected as described in the legend to panel A and were sacrificed at 2, 4, 6, 8, and 10 dpi. Brain, spleen, and serum were harvested. Viral titers were determined by plaque assays. The data plotted represent means and standard errors of the means for five animals per condition. Filled and open squares, VEEV TrD and VEEV TrDPRFm, respectively. Samples without detectable plaques were plotted as 1 PFU/ml.
DISCUSSION
Canonical −1 PRF signals are described as being composed of three elements arranged from 5′ to 3′ in the following order: a heptameric N NNW WWH slippery site, a vaguely defined short spacer segment, and a downstream structural element which is typically a variation of an mRNA pseudoknot (reviewed in reference 25). Examples in which efficient −1 PRF is stimulated by stem-loop structures are rare, but the most well-documented of these is the HIV-1 frameshift signal (26–28). However, while the HIV-1 stem-loop is sufficient to promote efficient −1 PRF, the local genomic RNA secondary structure influences −1 PRF efficiency (29), suggesting that more complex and dynamic interactions between the ribosome and −1 PRF stimulating RNA structural elements are involved. The novel predicted tVEEV structure (Fig. 2E) may provide an explanation for how these structural elements may stimulate −1 PRF. Here, the internal loop results in a structure in which two of the stems (labeled Sb and Sc) abut one another. We suggest that, as this structure enters the ribosomal mRNA entry tunnel, Sb and Sc are compressed toward one another, bringing their phosphodiester backbones in close proximity. At a certain point, the ensuing charge repulsion may drive the decompression or opening up of the structure, resulting in the backward movement of the mRNA relative to the ribosome, i.e., a −1 frameshift. We envision that emerging computational and single-molecule experimental platforms can be applied to test this novel mechanistic model of −1 PRF. Interestingly, a prior study indicated that a VEEV deletion mutant harboring only 32 nucleotides (nt) of sequence (predicted to retain the Sb structure while eliminating the Sa and Sc structural elements) stimulated −1 PRF approximately twice as well as VEEV with the native sequence (9). As discussed in that work, these findings indicate that a diverse array of 3′ RNA structures is capable of promoting efficient levels of −1 PRF.
Programmed −1 ribosomal frameshifting was first discovered in retroviruses, where it directs the synthesis of Gag-Pol polyproteins (30). Subsequent studies using retroviruses (31) and totiviruses (32) demonstrated that changes in −1 PRF efficiency affect virus production. From this, a bioeconomics model emerged in which −1 PRF rates are optimized to maximize virus particle assembly by ensuring the synthesis of the correct stoichiometric ratios of the structural Gag-derived proteins to the enzymes encoded by the Pol open reading frame (ORF) (reviewed in reference 20). These findings engendered interest in targeting −1 PRF for antiviral therapeutics (reviewed in reference 33). In parallel, early studies also examined −1 PRF signals in virus families where they do not occur between ORFs encoding structural and enzymatic ORFs, e.g., in coronaviruses and luteoviruses. The finding that the barley yellow dwarf virus uses −1 PRF as a developmental switch from the initial translation of nonstructural proteins to the translation of proteins involved in viral genome replication represented an expansion of our understanding of the utility of this molecular mechanism (34). Similarly, research in coronaviruses revealed that −1 PRF also serves as a switch, in this case, from expression of immediate early nonstructural proteins that are implicated in modulating the innate immune response to the next developmental step of the viral program, expression of viral replication machinery (reviewed in references 35 and 36). Nonetheless, alteration of −1 PRF efficiency in SARS-CoV severely impacted its infectivity in tissue culture, reinforcing the idea of −1 PRF as an antiviral therapeutic target (19). In flaviviruses, the viral positive-strand RNA genome encodes a single ORF in which the structural proteins are encoded by the 5′ third of the genome and the 3′ two-thirds encodes the nonstructural proteins. In these viruses, the location of the −1 PRF signal in the first nonstructural gene (NS1) has been proposed to ensure the production of large amounts of structural proteins for virus particle assembly and smaller amounts of the nonstructural proteins (6). Interestingly, lower rates of −1 PRF correlate with decreased pathogenicity in West Nile virus (6), and production of the NS1′ frameshift product is critical for neuroinvasiveness in West Nile and Japanese encephalitis viruses (22, 37). In these viruses, the NS1′ protein is thought to be important for virion assembly (22, 38). Additionally, −1 PRF has now been demonstrated to be used to control the expression of a large fraction of cellular genes in eukaryotes by functioning to control mRNA stability (reviewed in reference 39). Thus, we suggest that −1 PRF is an ancient, basic biological regulatory mechanism that has been evolutionarily selected for numerous end uses. On a final note, the observation that −1 PRF in alphaviruses does not appear to be regulated by host cellular miRNAs is not surprising from an evolutionary point of view. Like many RNA viruses, their evolution has followed a generalist strategy favoring the ability to produce acute, high-yield infections in a wide range of host organisms, as opposed to the strategy used by viruses that have coevolved with a single or closely related host species over a long period of time. Thus, HIV-1 with its long coevolutionary history with the great apes would be expected to utilize host-encoded miRNAs, while the alphaviruses would not.
While alphaviruses are related to flaviviruses, alphavirus genomes are arranged such that the nonstructural proteins are located in the 5′ ORF, while the structural protein genes are in a separate 3′ ORF and are expressed from the 26S subgenomic RNA (Fig. 1A). In alphaviruses, production of the 8.4-kDa TF protein may have two consequences. First, because E1 is a structural protein, −1 PRF may play a role in virion assembly by controlling E1 expression levels, and thus, altered E1 production could negatively interfere with virion assembly (10, 40). The data presented in Fig. 3 partially support this model, as ablation of the −1 PRF signal resulted in decreased virion production/release in vitro. This small decrease in virion production may provide just enough of a difference to enable the host to mount an effective immune response, as evidenced by the longer viral residence times of TrDPRFm in the spleen (Fig. 4D). Alternatively, the TF protein itself may have a biological role separate from viral particle assembly. This is supported by the observation that −1 PRF inhibition attenuated VEEV pathogenicity and altered viral spread in mice. Consistent with the information on the flavivirus NS1′ protein in the literature, the observation that the TrDPRFm virus promoted decreased viral titers in the brains of infected mice suggests that the VEEV TF protein may be important for passage through the blood-brain barrier and/or for neuroinvasiveness (10, 22, 37). It is important to note that these two options are not mutually exclusive, in that decreased TF protein expression and the accompanying increase in E1 levels may influence viral assembly/release while at the same time impact an as yet unidentified role of the TF protein. There have been no studies to date examining the interactions between the TF protein and other viral or cellular proteins. Such analyses would lend great insight into the role of the TF protein and will be the focus of future studies. It is also worth noting that the attenuation of TC83 is due to a substitution at position 120 of the E2 glycoprotein (41); thus, it is possible that the differences observed between TC83 with the PRF mutation and TrD may be due to disruption of an interaction between E2 and the TF protein.
The development of VEEV as a biological weapon in the United States and former USSR and a documented history of over 150 cases of serious laboratory infections by VEEV (1) led to it being included as a select agent by the government of the United States of America (http://www.selectagents.gov/SelectAgentsandToxinsList.html). As noted above, the FDA has not approved any vaccines or therapeutics for the equine encephalitis viruses. The attenuated vaccine strain, TC83, was generated in the early 1960s by serially passing VEEV 83 times in guinea pig heart cells (42). TC83 poses a high risk for reversion due to the fact that it harbors only two attenuating mutations (41) and can also be transmitted by mosquito vectors (43). Because of these risks, coupled with its demonstrated ability to cause mild to severe flu-like symptoms in approximately 25% of volunteers and the fact that it promoted seroconversion in only 80% of volunteers (44), TC83 has only limited utility for use in humans, and its use is limited to laboratory personnel and members of the military at risk of contracting an infection with the virus (45). More recent live attenuated vaccine candidates are based on the VEEV TrD infectious clone used in the current study. These include clones with insertion of specific point mutations or a mutation in the PE2 cleavage signal combined with a mutation that rescues E1 gene function. The resulting V3526 strain is safe and immunogenic in nonhuman primates and mice and has a lower risk for mosquito transmission (reviewed in reference 45). The finding in this study that attenuation of −1 PRF strongly attenuated VEEV neuropathogenicity of the virus represents a promising new avenue of inquiry toward the development of safe and effective live attenuated vaccines directed against VEEV and perhaps other −1 PRF-utilizing members of the Togavirus and Flavivirus families. In addition, the novel −1 PRF-stimulating mRNA elements identified here may also serve as targets for small-molecule therapeutics directed against these viruses.
MATERIALS AND METHODS
Computational prediction of viral PRF signals.
The sequences of virus genomes predicted to contain programmed −1 PRF signals were imported from NCBI (http://www.ncbi.nlm.nih.gov) into the J. D. Dinman lab −1 PRF database (http://prfdb.umd.edu) (14). PRFdb search algorithms were used to identify N NNW WWH slippery site sequences in the genome, as well as the alternate polypeptide sequences consequent to a recoding event. Feynman diagrams of the predicted downstream stimulatory structures were generated using the folding algorithms NUPAK (46) and RNAfold (47) as guides for subsequent cloning.
Dual-reporter plasmid construction and bacterial transformation.
All dual-reporter plasmids were adapted from the dual-luciferase read-through control plasmid pLuci (pJD175f) (13). The multiple-cloning site of pJD175f was digested at the SalI and SacI restriction sites using the respective Fast-Digest restriction enzymes (Thermo Fisher). The restriction digestion products were separated via 1% agarose gel electrophoresis, visualized by ethidium bromide staining and UV detection, and isolated by use of a NucleoSpin gel and PCR cleanup kit (Macherey-Nagel). Experimental plasmid inserts containing putative frameshift signals were generated as gBlocks (IDT) using complementary oligonucleotides and cloned into the linearized plasmid backbone by use of an in-fusion HD cloning kit (Clontech Laboratories). Specifically, the following virus-derived sequences were cloned into pJD175f: a 126-nt EEEV-derived sequence beginning at nt 9961, a 117-nt VEEV-derived sequence beginning at nt 9970, and a 90-nt WEEV-derived sequence beginning at nt 9821. Assembly products were transformed into Stellar competent Escherichia coli cells (Clontech Laboratories) and spread onto LB agar plates containing 50 μg/ml carbenicillin. Positive clones were verified by DNA sequencing (Genewiz). Plasmid and primer sequences are available upon request. Additionally, a series of frameshift reporter negative controls based on these three clones were constructed using oligonucleotide-directed site-specific mutagenesis. These consisted of (i) insertion of a 0-frame UAA termination codon immediately 5′ of the slippery sites in each plasmid (denoted 5′ Ter), (ii) silent coding mutagenesis of the U UUU UUA slippery sites to G UUC UUG (named ssM), and (iii) insertion of a UAA termination codon in the −1 frame after the viral sequences (denoted 3′ −1 Ter).
Cell culture.
HeLa (catalog number CCL-2), U-87 MG (catalog number HTB-14), and Vero (catalog number CCL-81) cells were obtained from ATCC (Manassas, VA). Cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 1% l-glutamine and 10% heat-inactivated fetal bovine serum (FBS) and maintained in a humidified 37°C incubator with 5% CO2. Cycling AP-7 rat olfactory bulb neuronal cells were cultured in DMEM supplemented with 1% l-glutamine and 10% FBS at 33°C with 10% CO2 (48). For in vitro differentiation, cycling AP-7 cells were plated in 6-well plates at a seeding density of 2.0 × 105 cells per well. On the next day, the medium was changed to DMEM containing 1% l-glutamine, 1% penicillin-streptomycin, 10% FBS, 2 μg/ml insulin, 40 μM dopamine hydrochloride, and 100 μM ascorbic acid. The cells were maintained at 39°C in 5% CO2 for 5 to 6 days before infection.
Plasmid and siRNA transfection.
HeLa or U-87 MG cells were seeded at 0.6 × 105 cells per well into 24-well plates in 0.5 ml of DMEM enhanced with 1% l-glutamine, 15% FBS, and 1× penicillin-streptomycin at 37°C in 5% CO2. Following a 24-h incubation, control and experimental dual-luciferase reporter plasmids were then transfected into cells using a Fugene HD transfection kit at a 3:1 transfection reagent-to-DNA ratio. siRNAs directed against human DBCR8, Exportin 5, Argonaute 1, Argonaute 2, or scrambled sequences were transfected into U-87 MG or HeLa cells as previously described (15).
Assays of programmed −1 ribosomal frameshifting.
The frameshifting efficiency of the experimental reporter plasmids was assayed as previously described (49) using a dual-luciferase reporter assay system kit (Promega). At 24 h posttransfection, the cell culture medium was aspirated and the cells were rinsed twice with 1× phosphate-buffered saline (PBS) before disruption with 1× passive lysis buffer. Cell lysates were assayed in triplicate in a 96-well plate. Firefly and Renilla luciferase activity was quantified using a GloMax 96 microplate luminometer (Promega).
Chemical modification analyses of −1 PRF promoting RNA structural elements.
mRNAs were structurally assayed using SHAPE (50, 51). DNA templates for mRNA secondary structure analysis were generated by PCR amplification using a DreamTaq DNA polymerase kit (Thermo Fisher). Forward and reverse primers for the Renilla and firefly regions on reporter plasmid pJD175f were used to amplify the inserted PRF sequence and attach a T7 promoter sequence and Kozak sequence at the 5′ end of the amplicons. Amplicons were isolated by agarose gel purification, and in vitro-transcribed RNA was generated using a T7 MEGAscript kit (Life Technologies). In vitro transcripts were purified using a MEGAclear cleanup kit (Life Technologies), and the quality of the RNA transcripts was assessed by agarose gel electrophoresis. Nine picomoles of the PRF RNA templates was denatured at 65°C for 4 min and refolded at 37°C for 20 min in 5× folding buffer (400 mM Tris HCl, pH 8.0, 800 mM NH4Cl, 55 mM magnesium acetate). Probing of flexible bases in the RNA transcripts was conducted through N-methylisatoic anhydride (NMIA) acylation of unprotected 2′-OH groups. Primer extension with γ-32P-radiolabeled probes and reverse transcription were carried out as reported elsewhere (50, 51). cDNA products were separated through an 8% urea gel and visualized on a phosphorimager. The visual clarity of the gel images was adjusted on Adobe Photoshop Lightroom (version 5) software.
Three-dimensional structural modeling of the tVEEV −1 PRF stimulatory element.
All-atom models were generated using the MC-Fold and MC-Sym pipeline programs (52). Initially, the RNA sequence of tVEEV was imported into the MC-Fold program to generate a series of secondary structures. Over 1,000 structures were explored in total, and the top 20 were selected for further consideration on the basis of their energetic scores. Among these, the one whose secondary structure was most consistent with the SHAPE experiments was submitted to MC-Sym for three-dimensional modeling. Two hundred structures were subsequently generated, and these were subjected to energy minimization and solvent refinement, yielding the 15 best models. The highest-scoring model was selected to represent the predicted 3-dimensional structure of tVEEV shown in Fig. 2E.
Introducing −1 PRF mutations into infectious VEEV clones.
Synonymous substitutions to disrupt the −1 PRF signals in the TC83 and TrD genomes were introduced by overlapping PCR extension, using standard techniques. The silent slippery site mutations consisted of the changes T9964G, T9967C, and A9970G to change the U UUU UUA slippery sites to G UUC UUG within the pTC83 (VEEV) and pV3000 (TrD) plasmids. There are six amino acid changes between the TC83 strain and the TrD strain, and all of these lie within the structural coding region: four in E2 (K7N, H85Y, T120R, V192D, T296I) and one in E1 (L161I [41, 53]). Furthermore, the genome of the V3000 clone of TrD utilized for this study also encodes two additional changes within E2 (one previously published, N239I [54], and one unpublished, E323G). All plasmid constructs were verified by restriction enzyme digestion and sequencing. Plasmid and primer sequences are available upon request.
VEEV stocks.
Viral stocks were produced by electroporation of in vitro-transcribed viral RNA generated from either the pTC83 plasmid (41), the pV3000 plasmid (TrD [21]), or the PRF mutant pTC83PRFm and pV3000PRF plasmids. In brief, the viral cDNA was linearized with restriction enzyme and then purified using a MinElute PCR purification kit (Qiagen) according to the manufacturer's directions. Capped RNAs were synthesized using a MEGAscript kit (Invitrogen) with a 2:1 ratio of cap analog [m7G(5′)ppp(5′)G; NEB] to GTP and treated with the DNase I supplied with the kit. RNA was then isolated with an RNeasy minikit with a second DNase I on-column digestion (Qiagen). The RNA integrity and concentration were determined by gel electrophoresis and determination of the absorbance at 260 nm, respectively. In vitro-transcribed viral RNAs were electroporated into BHK-J cells utilizing a 2-mm-gap cuvette (model BTX ECM 630 exponential decay wave electroporator; Harvard Apparatus, Holliston, MA). After trypsinization, cells were washed twice and resuspended in cold Dulbecco's phosphate-buffered saline without Ca2+ and Mg2+ (D-PBS; RNase-free) at 1.25 × 107 cells/ml. An aliquot of the cell suspension (400 μl) was mixed with 1 μg of RNA transcripts, placed into the cuvette, and pulsed once at 860 V, a 25-μF capacitance, and a 950-Ω resistance. Cells were allowed to recover for 5 min at room temperature and resuspended in complete minimal essential medium (MEM; Gibco Invitrogen). Cells from three replicate electroporations were plated in three 75-cm2 culture flasks for virus production. On the next day (∼12 h postelectroporation [hpe]), transfection medium was replaced with fresh MEM. Medium supernatants were harvested at several time points, pooled, and stored at 4°C. After the last collection, supernatants were then filtered (pore size, 0.2 μm), aliquoted, and stored at −80°C. Viral titers were determined by plaque assay on Vero cells.
Analysis of viral kinetics.
VEEV RNA replication and infectious viral titers were determined within Vero cells. Vero cells (seeded in 12-well plates) were infected at a multiplicity of infection (MOI) of 1 for 1 h. After the inoculum was removed, the cells were washed twice with D-PBS and cultured further in complete medium. At 3, 6, 9, 18, and 24 h postinfection (hpi), the supernatants were collected and the cells were washed once with D-PBS and lysed in TRIzol-LS (TC83 viruses) or RLT buffer (RNeasy kit; TrD viruses). Both sample sets were stored at −80°C until they could be further processed. Infectious virus titers were determined by plaque assay on Vero cells. Differentiated AP-7 (dAP-7) cells were infected with either TrD or TrDPRFm at an MOI of 1 and maintained under differentiation conditions throughout the experiment. Supernatants and RNA lysates were harvested at 3, 9, 18, 24, and 48 hpi.
Total cellular RNA was isolated from TRIzol-LS lysates utilizing a Direct-zol RNA miniprep kit (Zymo Research, Irvine, CA) or an RNeasy minikit (Qiagen) according to the manufacturers' directions. RNA quality and concentration were analyzed by gel electrophoresis and determination of the absorbance at 260 nm, respectively. For TrD virus-containing samples, a high-capacity RNA-to-cDNA kit (Thermo Fisher) was used to generate cDNA. Quantification of viral RNA was determined by quantitative reverse transcription-PCR (qRT-PCR; TC83) or quantitative PCR (qPCR; TrD) using a StepOnePlus real-time PCR system (Applied Biosystems). Primer pairs (forward primer, 5′-TCTGACAAGACGTTCCCAATCA-3′; reverse primer, 5′-GAATAACTTCCCTCCGACCACA-3′) and TaqMan probe (5′-6-carboxyfluorescein-TGTTGGAAGGGAAGATAAACGGCTACGC-6-carboxy-N,N,N′,N′-tetramethylrhodamine-3′) for nucleotides 7931 to 8005 of VEEV TC83 were originally described previously (55). The reaction mixtures for TC83 were assembled using an RNA UltraSense one-step quantitative RT-PCR system (Invitrogen), and absolute quantification was calculated on the basis of the threshold cycle (CT) relative to that on the standard curve. For TrD reactions, TaqMan gene expression master mix (Thermo Fisher) was used, and quantification relative to that at the 3-hpi time point was calculated on the basis of the 2−ΔΔCT method using 18S rRNA (56). Undetermined CT values were given a value of 40 for analysis.
Animal experiments.
Six- to 8-week-old female BALB/c mice were obtained from Harlan Laboratories. Groups of 35 mice were infected with VEEV TrD or VEEV TrDPRFm using Biaera's AereoPm system, a whole-body chamber, and a three-jet Collison nebulizer. They were exposed to 1 × 105 PFU/ml of VEEV TrD or VEEV TrDPRFm for 10 min. Hanks' balanced salt solution (HBSS) plus 1% FBS was used for the viral aerosol. Ten animals from each group were observed for survival over the course of 21 days. Five animals from each group were euthanized on days 2, 4, 6, 8, and 10 postinfection to determine the kinetics of disease in the mouse system. Serum, spleen, and brain were collected from each animal. Organs were homogenized using an Omni Bead Ruptor 4 (Omni International) and then centrifuged at 10,000 rpm for 10 min. The supernatants were analyzed by plaque assays to determine viral titers. All VEEV TrD experiments were performed in animal biosafety level 3 (ABSL-3) facilities, in accordance with the National Research Council's Guide for the Care and Use of Laboratory Animals (57) and under George Mason University IACUC protocol number 0331.
Statistical analyses.
Frameshifting efficiencies were calculated and statistical analyses were performed as previously described (11). Frameshifting assays were independently repeated a minimum of three times as technical triplicates. Data were normally distributed, and statistical analyses were conducted using Student's t test. Statistical analysis for viral kinetics was performed using Prism, version 6, software (GraphPad). Multiple unpaired t test analysis with the Holm-Sidak correction of the titer and qRT-PCR data sets was applied with the assumption that all comparisons had the same scatter.
ACKNOWLEDGMENTS
The content of this article does not necessarily reflect the position or the policy of the federal government, and no official endorsement should be inferred.
We thank Pamela Glass, United States Army Medical Research Institute of Infectious Disease, for providing the TrD molecular clone and Ilya Frolov, University of Alabama, for providing the TC83 molecular clone. We also thank Diane Griffin, Johns Hopkins Bloomberg School of Public Health, for providing the dAP-7 rat neuronal cells.
This work was funded through a Defense Threat Reduction Agency grant, HDTRA1-13-1-0005. J.A.K. was supported by NIH institutional training grant 2T32AI051967-06A1.
REFERENCES
- 1.Zacks MA, Paessler S. 2010. Encephalitic alphaviruses. Vet Microbiol 140:281–286. doi: 10.1016/j.vetmic.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leblanc J, Weil J, Beemon K. 2013. Posttranscriptional regulation of retroviral gene expression: primary RNA transcripts play three roles as pre-mRNA, mRNA, and genomic RNA. Wiley Interdiscip Rev RNA 4:567–580. doi: 10.1002/wrna.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanchez A, Trappier SG, Mahy BW, Peters CJ, Nichol ST. 1996. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc Natl Acad Sci U S A 93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Firth AE, Brierley I. 2012. Non-canonical translation in RNA viruses. J Gen Virol 93:1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinman JD. 2006. Programmed ribosomal frameshifting goes beyond viruses: organisms from all three kingdoms use frameshifting to regulate gene expression, perhaps signaling a paradigm shift. Microbe 1:521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moomau C, Musalgaonkar S, Khan YA, Jones JE, Dinman JD. 2016. Structural and functional characterization of programmed ribosomal frameshift signals in West Nile virus strains reveals high structural plasticity among cis-acting RNA elements. J Biol Chem 291:15788–15795. doi: 10.1074/jbc.M116.735613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinman JD. 2012. Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip Rev RNA 3:661–673. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plant EP, Perez-Alvarado GC, Jacobs JL, Mukhopadhyay B, Hennig M, Dinman JD. 2005. A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal. PLoS Biol 3:e172. doi: 10.1371/journal.pbio.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung BY-W, Firth AE, Atkins JF. 2010. Frameshifting in alphaviruses: a diversity of 3′ stimulatory structures. J Mol Biol 397:448–456. doi: 10.1016/j.jmb.2010.01.044. [DOI] [PubMed] [Google Scholar]
- 10.Snyder JE, Kulcsar KA, Schultz KLW, Riley CP, Neary JT, Marr S, Jose J, Griffin DE, Kuhn RJ. 2013. Functional characterization of the alphavirus TF protein. J Virol 87:8511–8523. doi: 10.1128/JVI.00449-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs JL, Dinman JD. 2004. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res 32:e160–e170. doi: 10.1093/nar/gnh157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firth A, Chung B, Fleeton M, Atkins J. 2008. Discovery of frameshifting in alphavirus 6K resolves a 20-year enigma. Virol J 5:108. doi: 10.1186/1743-422X-5-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 14.Belew AT, Hepler NL, Jacobs JL, Dinman JD. 2008. PRFdb: a database of computationally predicted eukaryotic programmed −1 ribosomal frameshift signals. BMC Genomics 9:339. doi: 10.1186/1471-2164-9-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belew AT, Meskauskas A, Musalgaonkar S, Advani VM, Sulima SO, Kasprzak W, Shapiro BA, Dinman JD. 2014. Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway. Nature 512:265–269. doi: 10.1038/nature13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunkle JA, Dunham CM. 2015. Mechanisms of mRNA frame maintenance and its subversion during translation of the genetic code. Biochimie 114:90–96. doi: 10.1016/j.biochi.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mortimer SA, Weeks KM. 2007. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc 129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 18.Hung M, Patel P, Davis S, Green SR. 1998. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 assembly and replication. J Virol 72:4819–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plant EP, Rakauskaite R, Taylor DR, Dinman JD. 2010. Achieving a golden mean: mechanisms by which coronaviruses ensure synthesis of the correct stoichiometric ratios of viral proteins. J Virol 84:4330–4340. doi: 10.1128/JVI.02480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brierley I. 1995. Ribosomal frameshifting on viral RNAs. J Gen Virol 76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 21.Davis NL, Willis LV, Smith JF, Johnston RE. 1989. In vitro synthesis of infectious Venezuelan equine encephalitis virus RNA from a cDNA clone: analysis of a viable deletion mutant. Virology 171:189–204. doi: 10.1016/0042-6822(89)90526-6. [DOI] [PubMed] [Google Scholar]
- 22.Melian EB, Hall-Mendelin S, Du F, Owens N, Bosco-Lauth AM, Nagasaki T, Rudd S, Brault AC, Bowen RA, Hall RA, van den Hurk AF, Khromykh AA. 2014. Programmed ribosomal frameshift alters expression of West Nile virus genes and facilitates virus replication in birds and mosquitoes. PLoS Pathog 10:e1004447. doi: 10.1371/journal.ppat.1004447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melian EB, Hinzman E, Nagasaki T, Firth AE, Wills NM, Nouwens AS, Blitvich BJ, Leung J, Funk A, Atkins JF, Hall R, Khromykh AA. 2010. NS1′ of flaviviruses in the Japanese encephalitis virus serogroup is a product of ribosomal frameshifting and plays a role in viral neuroinvasiveness. J Virol 84:1641–1647. doi: 10.1128/JVI.01979-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor A, Melton JV, Herrero LJ, Thaa B, Karo-Astover L, Gage PW, Nelson MA, Sheng K-C, Lidbury BA, Ewart GD, McInerney GM, Merits A, Mahalingam S. 2016. Effects of an in-frame deletion of the 6k gene locus from the genome of Ross River virus. J Virol 90:4150–4159. doi: 10.1128/JVI.03192-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV. 2016. Ribosomal frameshifting and transcriptional slippage: from genetic steganography and cryptography to adventitious use. Nucleic Acids Res 44:7007–7078. doi: 10.1093/nar/gkw530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. 1988. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 27.Staple DW, Butcher SE. 2003. Solution structure of the HIV-1 frameshift inducing stem-loop RNA. Nucleic Acids Res 31:4326–4331. doi: 10.1093/nar/gkg654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staple DW, Butcher SE. 2005. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J Mol Biol 349:1011–1023. doi: 10.1016/j.jmb.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Miranda P, Becker JT, Benner BE, Blume A, Sherer NM, Butcher SE. 2016. Stability of HIV frameshift site RNA correlates with frameshift efficiency and decreased virus infectivity. J Virol 90:6906–6917. doi: 10.1128/JVI.00149-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacks T. 1990. Translational suppression in gene expression in retroviruses and retrotransposons. Curr Top Microbiol Immunol 157:93–124. [DOI] [PubMed] [Google Scholar]
- 31.Sheju-Shilaga M, Crowe SM, Mak J. 2001. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J Virol 75:1834–1841. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinman JD, Wickner RB. 1992. Ribosomal frameshifting efficiency and Gag/Gag-Pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J Virol 66:3669–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinman JD, Ruiz-Echevarria MJ, Peltz SW. 1998. Translating old drugs into new treatments: identifying compounds that modulate programmed −1 ribosomal frameshifting and function as potential antiviral agents. Trends Biotechnol 16:190–196. doi: 10.1016/S0167-7799(97)01167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barry JK, Miller WA. 2002. A −1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc Natl Acad Sci USA 99:11133–11138. doi: 10.1073/pnas.162223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brierley I, Dos Ramos FJ. 2005. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res 119:23–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plant EP, Dinman JD. 2008. The role of programmed −1 ribosomal frameshifting in coronavirus propagation. Front Biosci 13:4873–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young LB, Melian EB, Khromykh AA. 2013. NS1′ colocalizes with NS1 and can substitute for NS1 in West Nile virus replication. J Virol 87:9384–9390. doi: 10.1128/JVI.01101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winkelmann ER, Widman DG, Suzuki R, Mason PW. 2011. Analyses of mutations selected by passaging a chimeric flavivirus identify mutations that alter infectivity and reveal an interaction between the structural proteins and the nonstructural glycoprotein NS1. Virology 421:96–104. doi: 10.1016/j.virol.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Advani VM, Dinman JD. 2016. Reprogramming the genetic code: the emerging role of ribosomal frameshifting in regulating cellular gene expression. Bioessays 38:21–26. doi: 10.1002/bies.201500131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukhopadhyay S, Zhang W, Gabler S, Chipman PR, Strauss EG, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. 2006. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 14:63–73. doi: 10.1016/j.str.2005.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinney RM, Chang GJ, Tsuchiya KR, Sneider JM, Roehrig JT, Woodward TM, Trent DW. 1993. Attenuation of Venezuelan equine encephalitis virus strain TC-83 is encoded by the 5′-noncoding region and the E2 envelope glycoprotein. J Virol 67:1269–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berge TO, Banks IS, Tigertt WD. 1961. Attenuation of Venezuelan equine encephalomyelitis virus by in vitro cultivation in guinea pig heart cells. Am J Hyg (Lond) 73:209–218. [Google Scholar]
- 43.Pedersen CE, Robinson DM, Cole FE. 1972. Isolation of the vaccine strain of Venezuelan equine encephalomyelitis virus from mosquitoes in Louisiana. Am J Epidemiol 95:490–496. [DOI] [PubMed] [Google Scholar]
- 44.Pittman PR, Makuch RS, Mangiafico JA, Cannon TL, Gibbs PH, Peters CJ. 1996. Long-term duration of detectable neutralizing antibodies after administration of live-attenuated VEE vaccine and following booster vaccination with inactivated VEE vaccine. Vaccine 14:337–343. doi: 10.1016/0264-410X(95)00168-Z. [DOI] [PubMed] [Google Scholar]
- 45.Paessler S, Weaver SC. 2009. Vaccines for Venezuelan equine encephalitis. Vaccine 27(Suppl 4):D80–D85. doi: 10.1016/j.vaccine.2009.07.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. 2011. NUPACK: analysis and design of nucleic acid systems. J Comput Chem 32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 47.Lorenz R, Bernhart SH, zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA package 2.0. Algorithms Mol Biol 6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schultz KLW, Vernon PS, Griffin DE. 2015. Differentiation of neurons restricts arbovirus replication and increases expression of the alpha isoform of IRF-7. J Virol 89:48–60. doi: 10.1128/JVI.02394-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harger JW, Dinman JD. 2003. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA 9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 51.Wilkinson KA, Merino EJ, Weeks KM. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 52.Parisien M, Major F. 2008. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 53.Johnson BJB, Kinney RM, Kost CL, Trent DW. 1986. Molecular determinants of alphavirus neurovirulence: nucleotide and deduced protein sequence changes during attenuation of Venezuelan equine encephalitis virus. J Gen Virol 67:1951–1960. doi: 10.1099/0022-1317-67-9-1951. [DOI] [PubMed] [Google Scholar]
- 54.Bernard KA, Klimstra WB, Johnston RE. 2000. Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology 276:93–103. doi: 10.1006/viro.2000.0546. [DOI] [PubMed] [Google Scholar]
- 55.Julander JG, Skirpstunas R, Siddharthan V, Shafer K, Hoopes JD, Smee DF, Morrey JD. 2008. C3H/HeN mouse model for the evaluation of antiviral agents for the treatment of Venezuelan equine encephalitis virus infection. Antiviral Res 78:230–241. doi: 10.1016/j.antiviral.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 57.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]