ABSTRACT
Despite success in viral inhibition and CD4 T cell recovery by highly active antiretroviral treatment (HAART), HIV-1 is still not curable due to the persistence of the HIV-1 reservoir during treatment. One patient with acute myeloid leukemia who received allogeneic hematopoietic stem cell transplantation from a homozygous CCR5 Δ32 donor has had no detectable viremia for 9 years after HAART cessation. This case has inspired a field of HIV-1 cure research focusing on engineering HIV-1 resistance in permissive cells. Here, we employed a glycosylphosphatidylinositol (GPI)-scFv X5 approach to confer resistance of human primary CD4 T cells to HIV-1. We showed that primary CD4 T cells expressing GPI-scFv X5 were resistant to CCR5 (R5)-, CXCR4 (X4)-, and dual-tropic HIV-1 and had a survival advantage compared to control cells ex vivo. In a hu-PBL mouse study, GPI-scFv X5-transduced CD4 T cells were selected in peripheral blood and lymphoid tissues upon HIV-1 infection. Finally, GPI-scFv X5-transduced CD4 T cells, after being cotransfused with HIV-infected cells, showed significantly reduced viral loads and viral RNA copy numbers relative to CD4 cells in hu-PBL mice compared to mice with GPI-scFv AB65-transduced CD4 T cells. We conclude that GPI-scFv X5-modified CD4 T cells could potentially be used as a genetic intervention against both R5- and X4-tropic HIV-1 infections.
IMPORTANCE Blocking of HIV-1 entry is one of most promising approaches for therapy. Genetic disruption of the HIV-1 coreceptor CCR5 by nucleases in T cells is under 2 clinical trials and leads to reduced viremia in patients. However, the emergence of viruses using the CXCR4 coreceptor is a concern for therapies applying single-coreceptor disruption. Here, we report that HIV-1-permissive CD4 T cells engineered with GPI-scFv X5 are resistant to R5-, X4-, or dual-tropic virus infection ex vivo. In a preclinical study using hu-PBL mice, we show that CD4 T cells were protected and that GPI-scFv X5-transduced cells were selected in HIV-1-infected animals. Moreover, we show that GPI-scFv X5-transduced CD4 T cells exerted a negative effect on virus replication in vivo. We conclude that GPI-scFv X5-modified CD4 T cells could potentially be used as a genetic intervention against both R5- and X4-tropic HIV-1 infections.
KEYWORDS: CD4 protection, GPI-anchored scFv, HIV-1, gene therapy, humanized mice
INTRODUCTION
Although new infections by human immunodeficiency virus type 1 (HIV-1) have decreased 35% since 2000, as of 2014, there are still approximately 36.9 million people living with HIV-1 (59). The implementation of highly active antiretroviral treatment (HAART) has dramatically reduced morbidity and mortality associated with AIDS and prolonged the life expectancy of individuals infected with HIV-1 (1). Nonetheless, the cessation of HAART results in a rapid rebound of viral replication and ensuing CD4 depletion due to the persistence of HIV-1-infected reservoir cells. As a result, patients need lifelong adherence to antiretroviral drugs to control HIV-1 infection (2). The appearance of drug resistance (3, 4) as well as severe adverse side effects (5) led the scientific community to explore curative approaches that control viral replication in the absence of HAART and restore immune function. Cure of HIV-1 infection remains the major challenge to global public health.
The only “functional cure” case so far is known as the “Berlin patient,” who received transplantation of hematopoietic stem cells from a CCR5 Δ32 homozygous donor prior to interruption of antiretroviral therapy. After HAART interruption, HIV-1 in lymphoid tissues of this patient has remained undetectable for 9 years, giving rise to the hope for a cure of HIV-1 infection (6). CCR5 (R5) is the major coreceptor required for HIV-1 entry (7). Individuals with a naturally occurring homozygous mutant CCR5 gene (CCR5 Δ32) express truncated dysfunctional CCR5, resulting in resistance to HIV-1 infection without adverse health effects (8, 9). One of the underlying benefits of CCR5 disruption in the Berlin patient is the inhibition of new infection by R5-tropic HIV-1 strains. This case indicates that enabling HIV-1 target cells to resist virus entry can prevent viral infection and restore functional immune cells or even the immune system.
To date, many approaches have been tested to modify autologous HIV-1-susceptible cells to prevent virus entry. As a choice of top priority, profound efforts have been made to knock down or knock out CCR5 expression, including the use of intrabodies (10), RNA interference (RNAi) (11–13), transcription activator-like nucleases (TALENs) (14, 15), zinc finger nucleases (ZFNs) (16–21), and clustered regularly interspaced short palindromic repeat (CRISPR)-CAS9 nucleases (22, 23). Preclinical evaluation of CCR5 disruption by ZFNs has been tested in a humanized mouse model. Mice engrafted with gene-modified cells displayed reduced viremia, selection of CCR5-disrupted cells, and protection of CD4 T cells in peripheral blood and lymphoid organs when challenged with R5-tropic virus (16, 19). A more recent clinical trial using CCR5-disrupted autologous CD4 T cell transplantation showed a relative survival advantage during treatment interruption and improved trafficking to rectal mucosa of modified cells (24), suggesting the safety, persistence, and resistance to infection of gene-modified CD4 T cells in infected patients. However, at a late stage of infection in patients, viruses using the other coreceptor, CXCR4 (X4), emerge, which is correlated with rapid disease progression (25, 26). The shift of HIV-1 tropism to CXCR4 was also reported for humanized mice transplanted with CCR5-disrupted hematopoietic stem cells as well as one patient who received an adoptive transfer of stem cells with the CCR5 Δ32 mutation (27). Approaches to block CXCR4 expression were also developed (21, 28–30), but CXCR4 disruption alone exhibited only partial protection upon X4-tropic virus infection (28). However, simultaneous editing of CCR5 and CXCR4 conferred robust protection against CD4 loss in humanized mice infected with R5- and X4-tropic viruses (31).
An alternative approach to protect HIV target cells from both R5- and X4-tropic HIV-1 strains utilizes a membrane-bound C-peptide entry inhibitor (maC46), which is derived from the C-terminal heptad repeat 2 (HR2) region of HIV-1 Env gp41 (32, 33). Cells expressing mC46 alone (32) or mC46 combined with other antivirus factors (34, 35) were resistant to both R5-tropic and X4-tropic virus infections in humanized mice and were positively selected in pigtail macaques infected with a dual-tropic simian-human immunodeficiency virus (SHIV) strain (36).
Previously, we demonstrated that an anti-HIV-1 single-chain fragment variant (scFv) derived from human anti-HIV Env antibody X5, when expressed on the cell surface via lipid rafts of the plasma membrane through a glycosylphosphatidylinositol (GPI) anchor, exhibits extremely potent and broad neutralization activity against diverse HIV-1 strains (37). CD4 T cell lines expressing GPI-anchored scFv X5 (GPI-scFv X5) inhibit a broad range of R5-, X4-, and dual-tropic strains as well as quasispecies infection. In addition, GPI-scFv X5 also blocked the transfer of viral particles by dendritic cells to CD4 T cells (W. Wang, C. Ye, and P. Zhou, unpublished data). These results suggest the great potential of GPI-scFv X5 as an alternative approach for the engineering of cell resistance to HIV-1 infection. Hence, we carried out a proof-of-concept study to test the feasibility of this approach. We interrogated the ability of GPI-scFv X5 to protect human primary CD4 T cells upon HIV-1 infection and designed a preclinical evaluation of this strategy in vivo using the hu-PBL NOD/Rag1−/−/IL-2rγ−/− (NRG) mouse model. Lentiviral vectors (lentivectors) encoding GPI-scFv X5 or AB65 (anti-influenza virus hemagglutinin [HA] control scFv vector) were generated to modify primary CD4 T cells. We show that transduction of primary CD4 T cells with GPI-scFv X5, but not GPI-scFv AB65, conferred robust protection of CD4 cells, resulted in a survival advantage, and exerted a negative effect on HIV-1 replication during infection with R5- or X4-tropic strains both ex vivo and in vivo. Thus, we developed an easy and feasible approach using GPI-scFv X5 to protect CD4 T cells from R5- and X4-tropic virus infections ex vivo and in vivo.
RESULTS
Human primary CD4 T cells expressing GPI-scFv AB65 or X5 show comparable proliferation capacities.
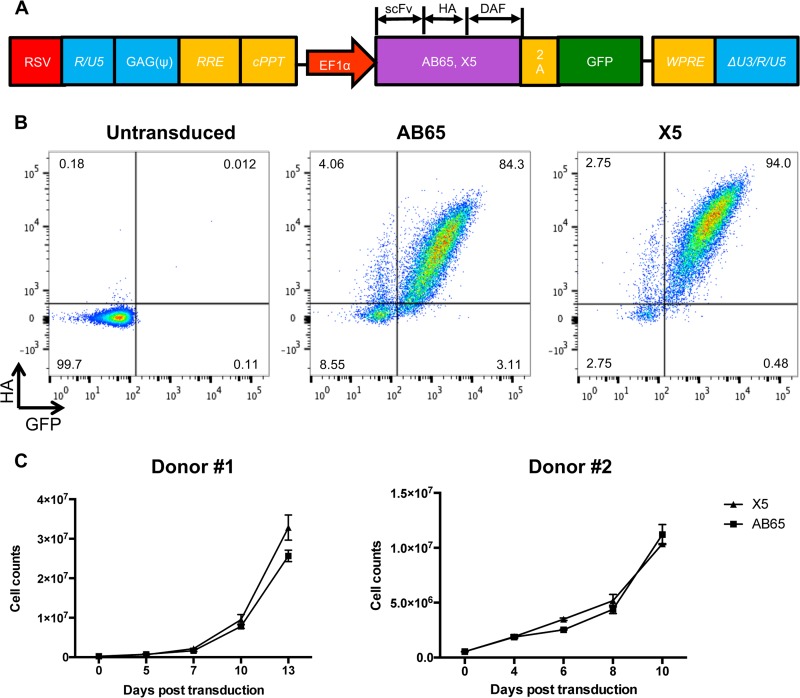
To investigate whether GPI-scFv X5 could confer protection from HIV-1 infection in primary CD4 T cells, we generated lentiviral vector constructs encoding GPI-scFv X5 or AB65. The latter was derived from an anti-influenza virus HA antibody and used here as a control. Sequences expressing GPI-scFv along with an HA tag at the C terminus were linked to a green fluorescent protein (GFP)-encoding sequence via an internal 2A protein splicing signal. Expression of GPI-scFv and GFP was under the control of the EF1α promoter (Fig. 1A). Figure 1B shows representative transduction efficiencies at a multiplicity of infection (MOI) of 5. GPI-scFv X5 or AB65 was highly expressed on CD4 T cells, with 94% and 84.3% GFP-HA-double-positive cells, respectively. High transduction efficiencies were obtained with cells isolated from different donors (data not shown). To test whether GPI-scFv X5- or AB65-transduced cells differed in cell proliferation, 2.5 × 105 transduced cells were expanded and maintained in culture medium containing 100 to 300 IU/ml human interleukin-2 (hIL-2). Figure 1C shows that cells transduced with GPI-scFv X5 had a proliferation rate similar to that of cells transduced with GPI-scFv AB65, and after a 10-day period of postransduction, the numbers of cells expanded 40-fold. We conclude that GPI-scFv X5- and AB65-transduced primary CD4 T cells had comparable proliferation capacities.
FIG 1.
Human primary CD4 T cells expressing GPI-scFv AB65 or X5 show comparable levels of proliferation. (A) Schematic diagram of the lentiviral vector expressing GPI-scFv. Sequences encoding GPI-scFv AB65 or X5 with an HA tag at the C terminus are linked to GFP-encoding sequences via a 2A signal. The GPI signal is derived from the C-terminal 34 amino acids of delay-accelerating factor (DAF). The expression cassette is driven by the EF1α promoter in the pRRL lentiviral vector. RSV, respiratory syncytial virus; RRE, Rev response element; cPPT, central polypurine track; WPRE, woodchuck hepatitits virus posttranscription regulatory element. (B) Expression of GPI-scFv AB65 or X5 in primary CD4 T cells. CD4 T cells were isolated and activated with anti-CD3/anti-CD28 beads in the presence of IL-2 for 24 h. Cells were then transduced with the indicated lentiviral vectors. FACS plots show the expression of GPI-scFv at 5 days postransduction. Representative data from one of five experiments are shown. GFP is on the x axis, and HA is on the y axis. Mock, untransduced cells; X5, cells transduced with a lentivirus encoding GPI-scFv X5; AB65, cells transduced with a lentivirus encoding GPI-scFv AB65. (C) Growth curve of CD4 T cells after transduction. Data are from two independent experiments with 2 donors. Error bars represent the SD of data from biological duplicates of each experiments.
HIV-1 resistance and survival advantage of GPI-scFv X5-transduced human primary CD4 T cells ex vivo.
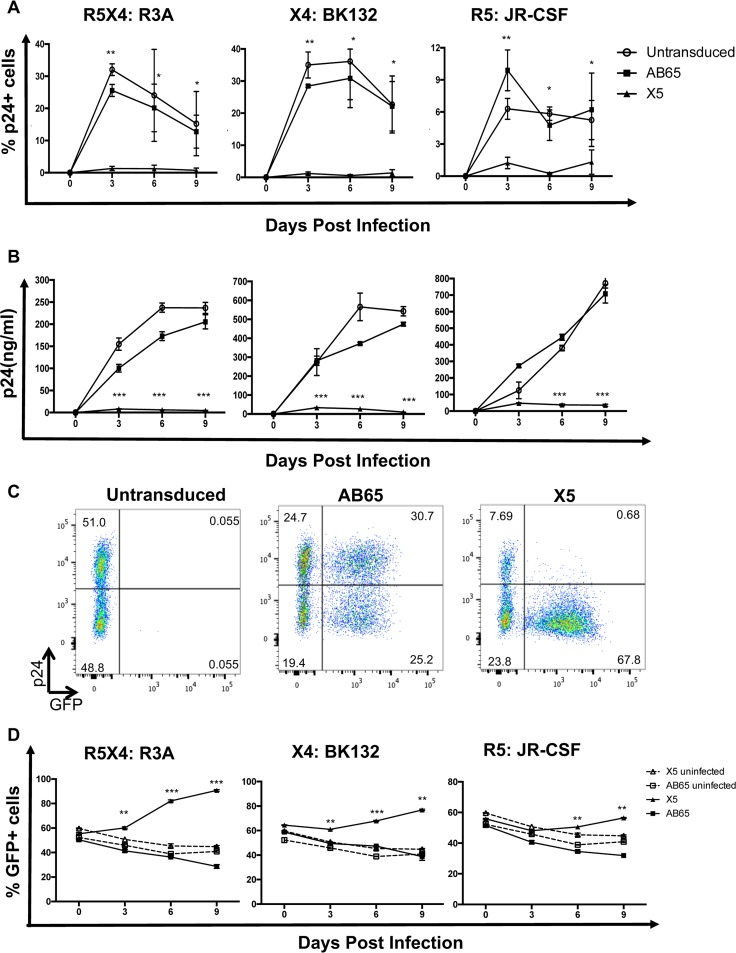
To test whether GPI-scFv X5 protects transduced primary CD4 T cells from HIV-1 infection, untransduced or GPI-scFv X5- or AB65-transduced cells were infected with HIV R3A (dual tropic), JR-CSF (R5 tropic), or BK132 (X4 tropic) at an MOI of 0.01 (100 ng p24 per million cells in the presence of 8 ng/ml Polybrene). After infection, cells were cultured for 9 days. Every 3 days, cells and culture supernatants were collected, HIV-1 replication was measured by intracellular p24 staining, and the p24 level in culture supernatants was determined. As expected, untransduced and GPI-scFv AB65-transduced cells had robust HIV-1 replication after infections by all three viruses (Fig. 2A and B). In contrast, only 3% of GPI-scFv X5-transduced cells were infected by R3A, BK132, or JR-CSF (Fig. 2A). On day 9 postinfection, p24 levels in culture supernatants of GPI-scFv X5-transduced cells were 40- to 70-fold lower for R3A, 6- to 10-fold lower for BK132, and 30- to 50-fold lower for JR-CSF infection than those in untransduced or GPI-scFv AB65-transduced control cells (Fig. 2B).
FIG 2.
HIV resistance and survival advantage of GPI-scFv X5-transduced human primary CD4 T cells ex vivo. (A) Percentage of p24-positive cells over thecourse of infection. The intracellular p24 level was monitored 3, 6, and 9 days after HIV-1 infection. Results from three independent experiments with 3 donors are summarized. (B) HIV-1 replication was monitored by a p24 ELISA 3, 6, and 9 days after infection. Data are results from one representative experiment of three. Error bars represent the SD of data from triplicate p24 ELISAs. (C) Intracellular staining of the p24 Gag protein post-HIV-1 infection. Mock-transduced or GPI-scFv-transduced cells were cocultured with cells infected with R3A, BK132, or JR-CSF. Cells were permeabilized and stained with anti-p24 antibody. GFP is on the x axis, and p24 is on the y axis. Representative data show intracellular p24 levels after BK132 infection. (D) Percentage of GFP+/HA+ cells during coculture of infected or uninfected CD4 T cells with the indicated transduced cells. Dashed lines represent transduced cells cocultured with uninfected CD4 T cells. Solid lines represent transduced cells cocultured with infected CD4 T cells. Error bars represent data from biological duplicates from one experiment. P values represent the differences between the AB65 and X5 groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To investigate whether GPI-scFv X5-transduced cells resist to cell-to-cell spread, infected or uninfected primary CD4 T cells were cocultured with untransduced or GPI-scFv AB65- or X5-transduced cells. Both GFP-positive (GFP+) and GFP-negative GPI-scFv AB65-transduced cells were infected, whereas only GFP-negative GPI-scFv X5-transduced cells were infected by HIV-1 BK132 (Fig. 2C) as well as JR-CSF and R3A (data not shown). These data show that GPI-scFv X5-transduced CD4 T cells are resistant to not only free virus infection but also cell-to-cell spread of HIV-1.
To test whether GPI-scFv X5-transduced cells should be protected and have a survival advantage, we measured the percentage of scFv-transduced cells in the coculture system. When GPI-scFv X5-transduced cells were cocultured with infected CD4 cells, the percentage of GPI-scFv-positive cells increased over the course of coculture. However, GPI-scFv X5-transduced cells cocultured with uninfected cells or GPI-scFv AB65-transduced cells cocultured with infected or uninfected cells had no selection of scFv-positive cells. As a result, the percentages of GFP+ GPI-scFv X5-transduced cells cocultured with infected CD4 cells were significantly higher than those of GPI-scFv AB65-transduced cells cocultured with infected CD4 cells (P < 0.01 or 0.001) (Fig. 2D). Of note, the strength of selection was relevant to the pathogenesis of virus strains. GPI-scFv X5-transduced cells were rapidly selected by highly pathogenic strains R3A and BK132, while selection was relatively slow for lowly pathogenic strain JRCSF. Taken together, GPI-scFv X5-transduced primary CD4 T cells are resistant to both cell-free and cell-associated HIV-1 infections.
Equal engraftment efficiencies of GPI-scFv X5- and AB65-transduced human CD4 T cells among all hu-PBL mice.
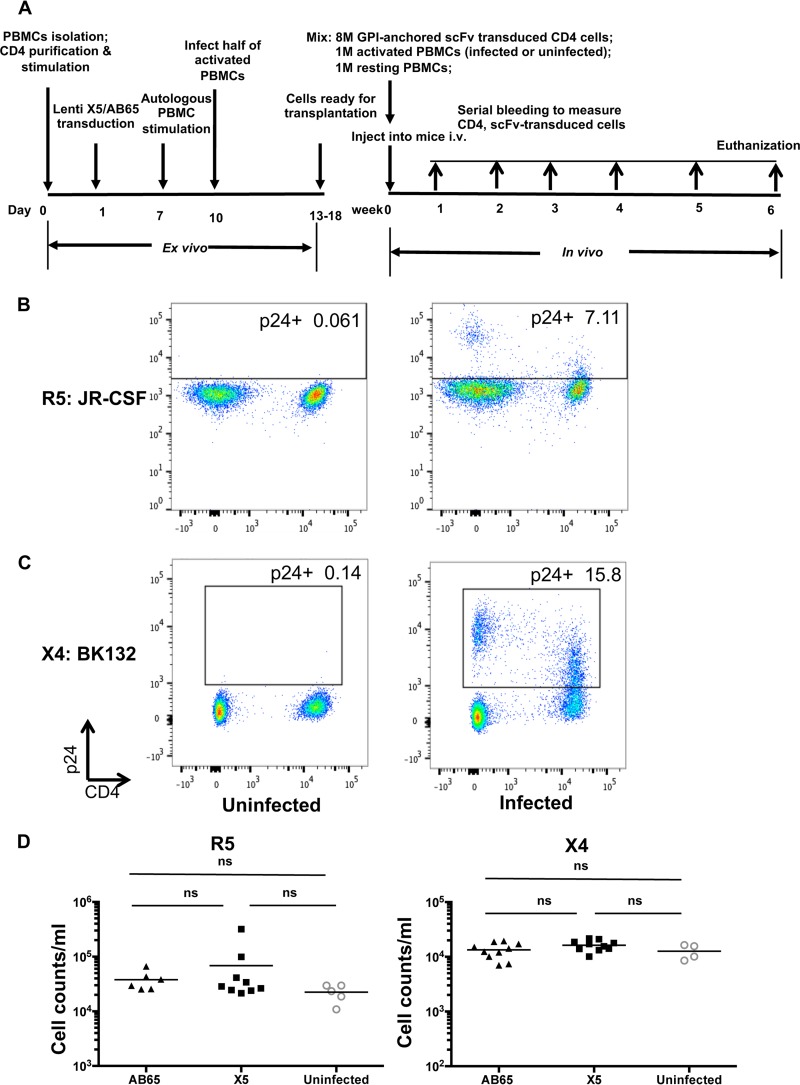
To explore the therapeutic potential of GPI-scFv X5-transduced human CD4 T cells, we used a hu-PBL mouse model with an experimental procedure similar to the one reported previously by Perez et al. (19). In this procedure, human CD4 T cells transduced with GPI-scFv X5 or AB65 were mixed with HIV-1-infected or uninfected peripheral blood mononuclear cells (PBMCs). Cell mixtures along with 1 × 106 resting PBMCs (to enhance engraftment) were cotransfused into NRG mice (Fig. 3A). This approach resembles “therapeutic” conditions since modified T cells were cotransfused with infected cells. In vivo protective activity against HIV-1 infection was tested in three independent experiments by using cells from different donors. In these experiments, mice were randomly assigned to four treatment groups, defined as (i) X5/uninfected, comprising mice receiving X5-transduced CD4 cells and uninfected activated PBMCs; (ii) X5/R5 or X5/X4, composed of mice receiving X5-transduced CD4 cells or HIV-infected activated PBMCs, respectively; (iii) AB65/uninfected, comprising mice receiving AB65-transduced CD4 cells and uninfected activated PBMCs; and (iv) AB65/R5 or AB65/X4, comprising mice receiving AB65-transduced CD4 cells and R5- or X4-tropic HIV-infected activated PBMCs, respectively. Infection of activated PBMCs was confirmed prior to injection (Fig. 3B and C). Peripheral blood was collected on the indicated days after infusion and analyzed by flow cytometry. Results for mice in the X5/uninfected and AB65/uninfected groups were combined and defined as the uninfected group. Figure 3B shows that at 1 week posttransfusion, mice in the X5/R5, AB65/R5, and uninfected groups or mice in the other three groups, the X5/X4, AB65/X4, and uninfected groups, exhibited comparable engraftment, as measured by human CD45 cell counts in peripheral blood.
FIG 3.
Equal engraftment efficiencies among all hu-PBL mice. (A) Experimental outline of the generation of GPI-scFv X5- or AB65-transduced CD4 T cells and R5- or X4-tropic HIV-1-infected PBMCs and cotransfusion of transduced CD4 T cells or infected or uninfected PBMCs along with resting PBMCs into NRG mice. CD4 cells were isolated, stimulated, and transduced. Activated PBMCs from the same donor were infected with BK132 or JR-CSF and mixed with GPI-scFv AB65- or X5-transduced CD4 T cells. Mixtures of uninfected activated PBMCs and transduced CD4 T cells were used as uninfected controls. Additionally, 1 × 106 resting PBMCs were added to mixed cells, as previously reported (19). The mixtures of cells were injected into NRG mice intravenously (i.v.). Mice were bled every week to monitor numbers of CD4 cells and GFP-positive cells. At 6 weeks posttransplantation, mice were necropsied, and spleen and bone marrow were collected for analysis. 8M, 8 million. (B and C) Infection of activated PBMCs prior to infusion. PBMCs were stimulated with anti-CD3/anti-CD28 beads. Cells were infected with JR-CSF (B) or BK132 (C) at an MOI of 0.01 or with 100 ng p24 per million cells. Infection was determined by intracellular staining of HIV-1 Gag p24. (D) Engraftment efficiency of human cells at 1 week posttransplantation. Human CD45 cell counts were measured (uninfected/R5, n = 5; X5/R5, n = 9; AB65/R5, n = 6; uninfected/X4, n = 4; X5/X4, n = 10; AB65/X4, n = 10). (Left) Engraftment of infused cells in mice infected with JR-CSF. (Right) Engraftment of infused cells in mice infected with BK132. Mean values are shown for each group. ns, not significant.
Protection of CD4 T cells in peripheral blood by GPI-scFv X5 during HIV-1 infection in vivo.
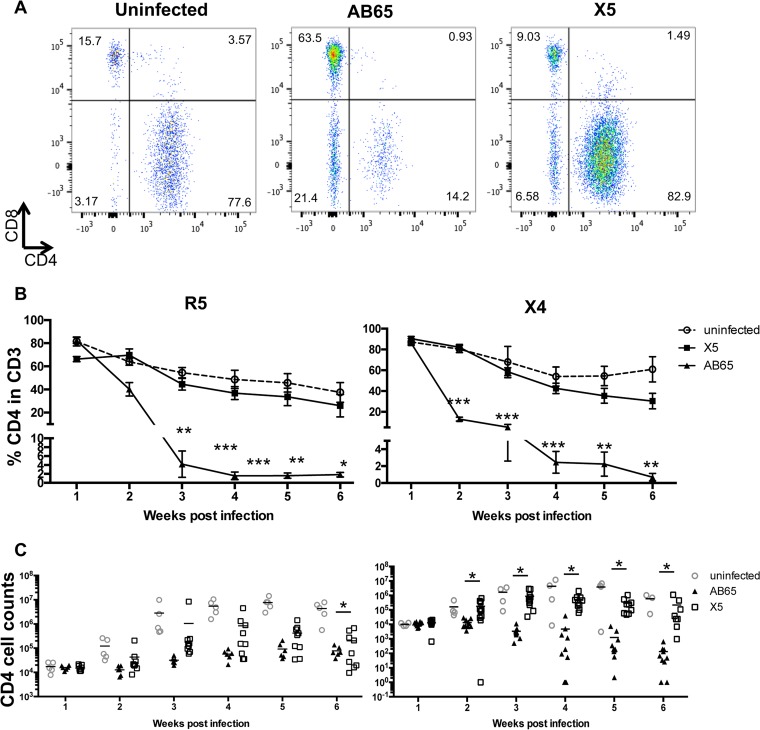
CD4 cell depletion caused by HIV-1 infection in patients is one of the major syndromes of AIDS. To test whether GPI-scFv X5-transduced CD4 T cells are protected in the presence of HIV-1 infection in vivo, after transfusion, the percentages and numbers of CD4 T cells were monitored weekly. Figure 4A shows the gating of CD4 and CD8 T cells in the human CD3 T cell population of representative hu-PBL mice from each of the three groups 2 weeks after infection with X4-tropic virus. After cotransfusion with R5-tropic virus-infected cells, the average percentages of CD4 T cells in the CD3 cell population of mice in the uninfected or AB65/R5 group were about 80% at 1 week postinfection, while in mice in the X5/R5 group, the percentage was about 65%. The average percentages for mice in the AB65/R5 group decreased to below 5% at 3 weeks postinfection (wpi) and were maintained at about 2% until the end of the experiment. In contrast, the average percentages of CD4 cells in the CD3 cell population of mice in the X5/R5 group were significantly higher than those in mice in the AB65/R5 group from the second week after infection (P < 0.05) but were similar to those in mice in the uninfected group. The difference in the percentages of CD4 cells between the X5/R5 and AB65/R5 groups reached a maximum of 23-fold (Fig. 4B, left). Moreover, on average, the CD4 counts in peripheral blood of mice in the X5/R5 group were 0.5 to 1 log unit higher than those in mice in the AB65/R5 group from 2 weeks postinfection, but the differences were not statistically significant, except for those at 6 weeks postinfection (Fig. 4C, left).
FIG 4.
Protection of CD4 T cells in peripheral blood in vivo. (A) Gating of CD4 and CD8 T cells in the human CD3 T cell population of representative hu-PBL mice from each of the three groups 2 weeks after X4-tropic virus infection. (B) Percentage of human CD4 cells in human CD3 cells in blood during infection. Percentages of CD4 cells were determined by flow cytometry. (C) CD4 cell counts during HIV-1 infection. Left panels show results from JR-CSF infection; right panels show results from BK132 infection. *, P < 0.05; **, P < 0.01; ***, P < 0.001. P values represent the differences between the AB65/R5 and X5/R5 groups and between the AB65/X4 and X5/X4 groups.
Similarly, after cotransfusion with X4-tropic virus-infected cells, average percentages of CD4 T cells dropped to 5% for mice in the AB65/X4 group, while for mice in the X5/X4 group, average percentages of CD4 T cells were similar to those for mice in the uninfected group and significantly higher than those for mice in the AB65/X4 group. The maximum difference in the percentages of CD4 T cells between the AB65/X4 and X5/X4 groups reached 43-fold (Fig. 4B, right). Surprisingly, on average, the CD4 counts for mice in the X5/X4 group were 1 to 3 log units higher than those for mice in the AB65/X4 group from 2 weeks postinfection, and the differences were statistically significant (P < 0.05) (Fig. 4C, right). This is likely due to the highly pathogenic activity of the X4-tropic BK132 strain. Thus, we demonstrate that CD4 T cells in hu-PBL mice transfused with GPI-scFv X5-transduced cells are protected during infection by both X4- and R5-tropic viruses.
GPI-scFv X5-transduced CD4 T cells are selected in peripheral blood during HIV-1 infection.
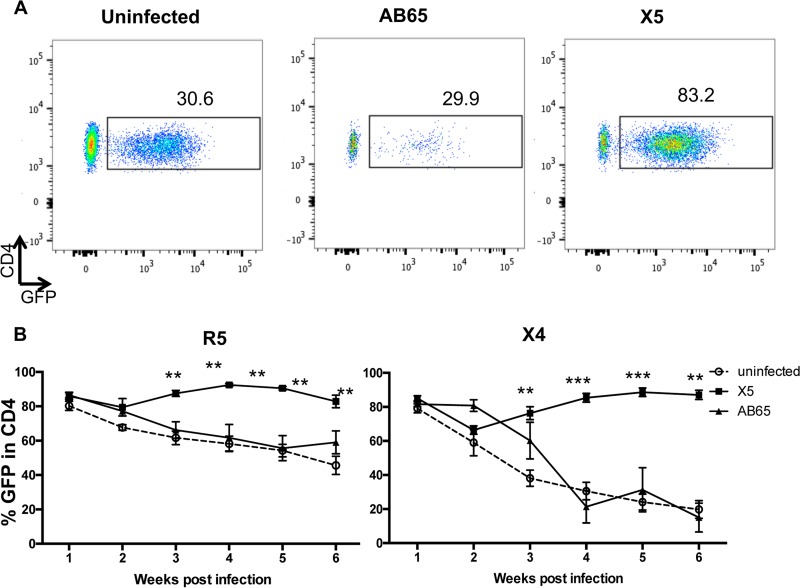
To investigate whether the protection of CD4 cells is due to the selection of GPI-scFv X5-transduced CD4 T cells in vivo, the percentage of GFP+ cells in CD4 cell populations was determined by flow cytometry. Figure 5A shows the gating of CD4+/GFP+ cells in human CD4 T cell populations of representative hu-PBL mice from each of the three groups 4 weeks after X4-tropic virus infection. After cotransfusion with both R5- and X4-tropic virus-infected cells, average percentages of CD4+/GFP+ cells in mice in the X5/X4 and X5/R5 groups dropped slightly at 2 weeks postinfection and then gradually increased and remained at 70 to 90% afterwards (Fig. 5B). In contrast, average percentages of CD4+/GFP+ cells in mice in the uninfected and AB65/R5 groups gradually decreased from 80% to about 50%. The differences in the average percentages of CD4+/GFP+ cells between mice in the X5/R5 and AB65/R5 groups as well as between mice in the X5/R5 and uninfected groups were significant (P < 0.01) (Fig. 5B, left). More significantly, average percentages of CD4+/GFP+ cells in mice in the uninfected and AB65/X4 groups dropped from 80% to about 20%. The differences in the average percentages of CD4+/GFP+ cells between mice in the X5/X4 and AB65/X4 groups as well as between mice in the X5/X4 and uninfected groups were very significant (from P values of <0.01 to P values of <0.001) (Fig. 5B, right). Thus, these results strongly suggest that a selective advantage for GPI-scFv X5-transduced CD4 T cells occurs during both R5- and X4-tropic HIV-1 infections.
FIG 5.
Selection of GPI-scFv X5-transduced cells in peripheral blood in vivo. (A) Gating of CD4+/GFP+ cells in human CD4 T cell populations of representative hu-PBL mice from each of the three groups 4 weeks after X4-tropic virus infection. (B) Percentage of CD4+/GFP+ cells in human CD4 T cells in mouse blood during infection. Percentages of GFP cells were determined by flow cytometry. Left panels show results from JR-CSF infection; right panels show results from BK132 infection. *, P < 0.05; **, P < 0.01; ***, P < 0.001. P values represent the differences between the AB65/R5 and X5/R5 groups and between the AB65/X4 and X5/X4 groups.
GPI-scFv X5-transduced CD4 T cells reduce HIV-1 replication in hu-PBL mice.
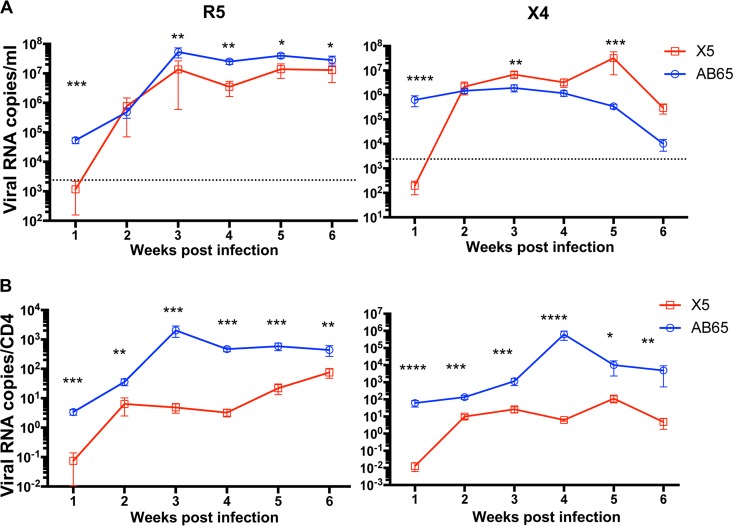
We analyzed replication kinetics in individual mice cotransfused with different cells. One week after cotransfusion with R5-tropic virus-infected cells, the average plasma HIV-1 RNA load was 3,509 copies/ml in X5/R5 mice, while it was 53,771 copies/ml in mice in the AB65/R5 group (P < 0.01). At later time points, plasma viremia increased significantly in both groups. However, the average plasma viral loads for the X5/R5 group were significantly lower than those for the AB65/R5 group (P < 0.05 or P < 0.01) (Fig. 6A, left). One week after cotransfusion with X4-tropic virus-infected cells, the average plasma viral RNA load was 642 copies/ml in mice in the X5/X4 group, while it was 627,274 copies/ml in mice in the AB65/X4 group (P < 0.0001). HIV-1 plasma loads increased significantly in both groups of mice from 2 to 6 wpi. The total plasma viral loads in mice in the X5/X4 group were actually higher than those in mice in the AB65/X4 group (Fig. 6A, right).
FIG 6.
GPI-scFv X5-transduced CD4 T cells reduce blood viral loads in hu-PBL mice. (A) Temporal viral load curve. (B) Temporal normalized numbers of viral RNA copies per CD4 cell in plasma. (Left) Data from JR-CSF infection. (Right) Data from BK132 infection. (Top) Viral loads throughout the experiment. Data represent average viral loads in infected mice. (Bottom) Numbers of viral RNA copies per CD4 cell throughout the experiment. Data represent average numbers of viral RNA copies per CD4 cell. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Because X4-tropic HIV-1 infection led to a dramatic depletion of CD4 T cells, the average CD4 T cell counts in mice in the X5/X4 group were significantly higher than those in mice in the AB65/X4 group (Fig. 4C), suggesting that the more profound CD4 depletion in mice in the AB65/X4 group than in mice in the X5/X4 group may be responsible for the lower viral loads seen in mice in the AB65/X4 group at 1 week posttransfusion. We therefore also calculated the number of HIV-1 RNA copies per CD4 cell. We show that 1 week after cotransfusion with R5-tropic virus-infected cells, the average number of viral RNA copies per CD4 cell in mice in the X5/R5 group was 49 times lower than that in mice in the AB65/R5 group (P < 0.001). During the 6 weeks posttransfusion, the numbers of HIV-1 RNA copies/CD4 cell in mice in the X5/R5 group were significantly lower than those in mice in the AB65/R5 group (P < 0.01 or P < 0.001) (Fig. 6B, left). Similarly, 1 week after cotransfusion with X4-tropic virus-infected cells, the average number of viral RNA copies per CD4 cell in mice in the X5/X4 group was 4,783 times lower than that in mice in the AB65/X4 group (P < 0.0001). Afterwards, the numbers increased in both groups of mice. However, the numbers in mice in the X5/X4 group were still significantly lower than those in mice in the AB65/X4 group (P < 0.05 to P < 0.0001) (Fig. 6B, right). Taken together, we conclude that GPI-scFv X5-transduced CD4 T cells indeed exert a negative effect on viral loads in hu-PBL mice.
GPI-scFv X5-transduced CD4 T cells are selected in lymphoid tissues during HIV-1 infection.
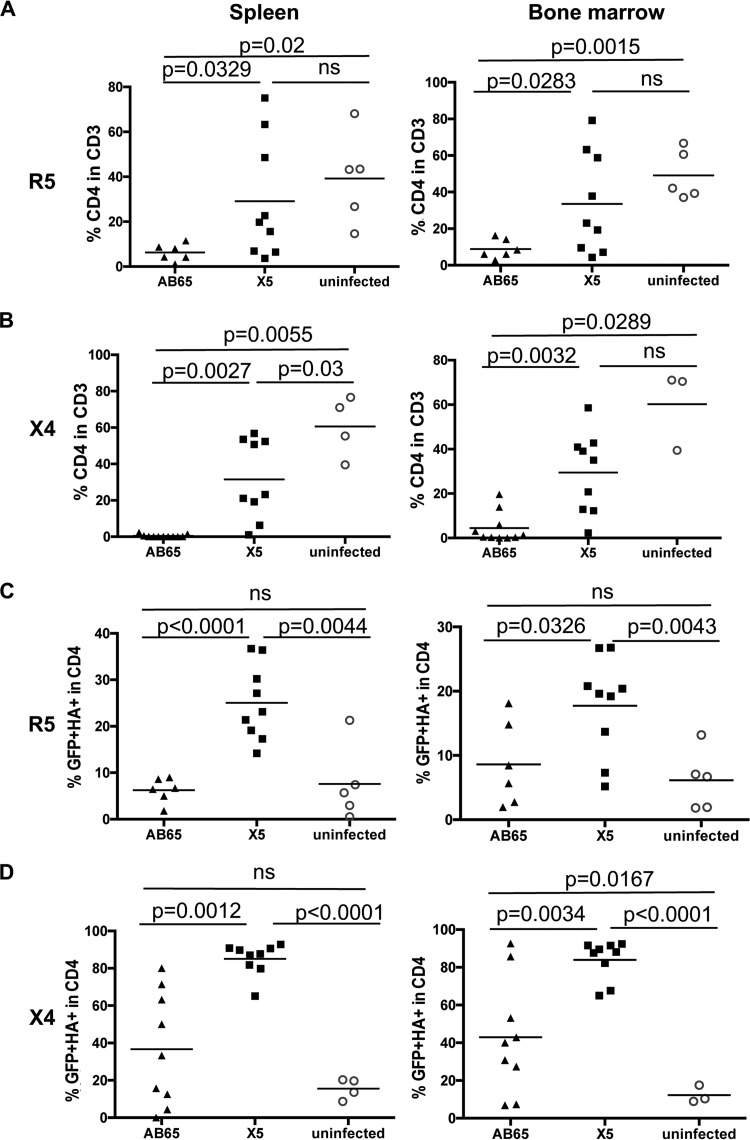
At 6 weeks posttransfusion, mice were necropsied, and splenocytes and bone marrow cells were isolated. The numbers of GPI-scFv X5- or AB65-transduced CD4 T cells as well as HIV-1 replication in lymphoid tissues were measured. Figure 7A shows that consistent with the results observed for peripheral blood, mice in the X5/R5 group, while similar to mice in the uninfected group, had significantly higher average percentages of CD4 T cells in spleen (29.12%) than did mice in the AB65/R5 group (6.27%) (P = 0.0329). Mice in the X5/R5 group, while similar to mice in the uninfected group, also had a significantly higher average percentage of CD4 T cells in bone marrow (33.58%) than did mice in the AB65/R5 group (8.91%) (P = 0.0283).
FIG 7.
Selection of GPI-scFv X5-transduced human CD4 T cells in lymphoid tissues during HIV-1 infection in vivo. Shown are percentages of CD4 cells in CD3 T cells (A and B) and scFv-positive cells in CD4 T cells (C and D) from mouse spleen or bone marrow. (A and C) Data from mice infected with JR-CSF. (B and D) Data from mice infected with BK132. Spleen and bone marrow cells were collected on the day of euthanasia. Cells were stained as described in Materials and Methods. Mice with very low numbers of CD4 T cells were excluded when the counts determined by flow cytometry were below 10. Mean values for each group are shown.
Similarly, after cotransfusion with X4-tropic virus-infected cells, mice in the X5/X4 group, while similar to mice in the uninfected group, had a significantly higher average percentage of CD4 T cells in spleen (31.58%) than did mice in the AB65/X4 group (0.43%) (P = 0.0027). Mice in the X5/X4 group, while similar to mice in the uninfected group, also had a significantly higher average percentage of CD4 T cells in bone marrow (29.41%) than did mice in the AB65/X4 group (4.49%) (P = 0.0032) (Fig. 7B). Thus, the survival advantage for CD4 T cells was also observed in spleens and bone marrow of mice transfused with GPI-scFv X5-transduced CD4 T cells compared with mice transfused with GPI-scFv AB65-transduced CD4 T cells.
Figure 7C shows that, consistent with the results observed for peripheral blood, mice in the X5/R5 group had a significantly higher average percentage of HA+/GFP+ cells in spleen (25.02%) than did mice in the AB65/R5 and uninfected groups (6.24% and 7.57%, respectively) (P < 0.001 and P < 0.01, respectively). Mice in the X5/R5 group also had a significantly higher average percentage of HA+/GFP+ cells in bone marrow (17.75%) than did mice in the AB65/R5 and uninfected groups (8.61% and 6.14%, respectively) (P < 0.05 and P < 0.01, respectively).
Similarly, after cotransfusion with X4-tropic virus-infected cells, mice in the X5/X4 group had a significantly higher average percentage of HA+/GFP+ cells in spleen (85.38%) than did mice in the AB65/X4 and uninfected groups (36.71% and 15.58%, respectively) (P = 0.0012 and P < 0.001, respectively). Mice in the X5/X4 group also had a significantly higher average percentage of HA+/GFP+ cells in bone marrow (83.94%) than did mice in the AB65/X4 and uninfected groups (42.97% and 12.28%, respectively) (P = 0.0034 and P < 0.001, respectively) (Fig. 7D). Thus, these results further demonstrate that GPI-scFv X5 was stably expressed in vivo and that GPI-scFv X5-transduced CD4 T cells were selected in lymphoid tissues during HIV-1 infection.
GPI-scFv X5-transduced CD4 T cells reduce HIV-1 replication in lymphoid tissues in hu-PBL mice.
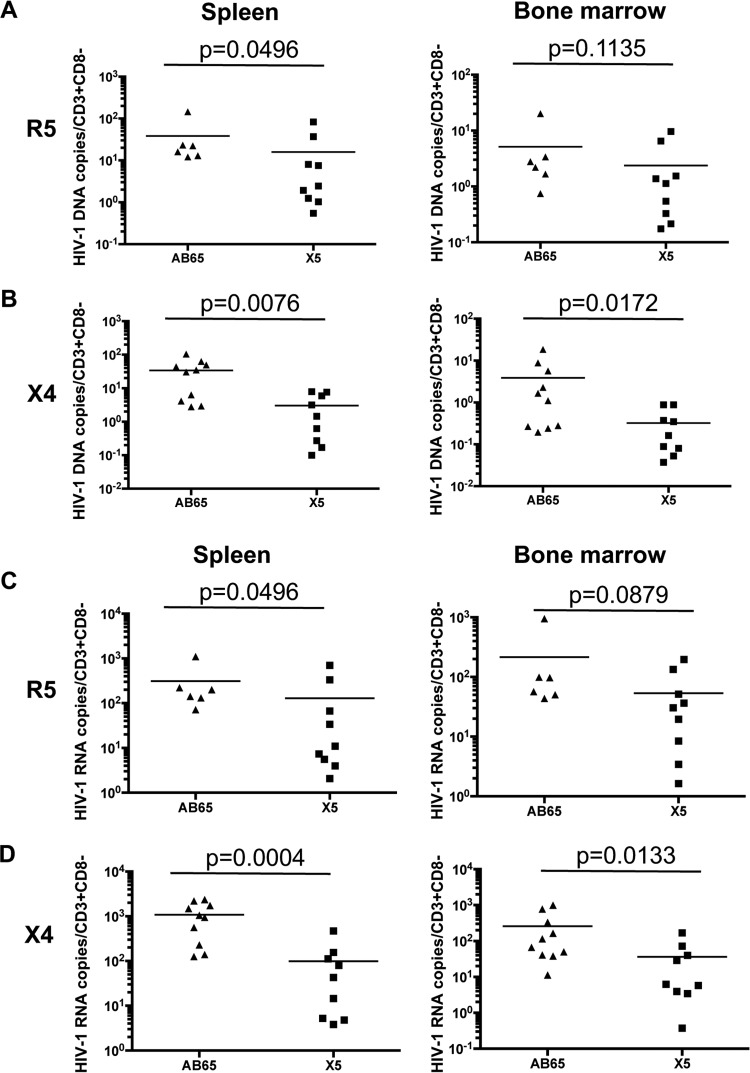
To test the effect of GPI-scFv-transduced CD4 T cells on HIV-1 replication in lymphoid tissues, we measured HIV-1 DNA or RNA copy numbers in splenocytes and bone marrow cells and calculated the number of DNA or RNA copies per CD3+/CD8− cell to include both CD4+ T cells and HIV-infected CD4 T cells with downregulated CD4 expression (38). Figure 8A shows that mice in the X5/R5 group had significantly lower HIV-1 DNA copy numbers per human CD4 T cell in spleens than did mice in the AB65/R5 group (P = 0.0496). Mice in the X5/R5 group also had lower average DNA copy numbers per human CD4 T cell in bone marrow than did mice in the AB65/R5 group (but no statistically significant difference [P = 0.1135]). Similarly, after cotransfusion with X4-tropic virus-infected cells, mice in the X5/X4 group had 11 times fewer HIV-1 DNA copies per human CD4 T cell in spleens than did mice in the AB65/X4 group (P = 0.0076). Mice in the X5/X4 group also had 12 times fewer average HIV-1 DNA copies per CD4 T cell in the bone marrow than did mice in the AB65/X4 group (P = 0.0032) (Fig. 8B).
FIG 8.
Negative effect of GPI-scFv X5-transduced CD4 T cells on viral replication in lymphoid tissues. Shown are normalized HIV-1 DNA (A and B) and RNA (C and D) copy numbers in spleen and bone marrow. HIV-1 DNA or RNA copy numbers were divided by CD3+ CD8− cell counts. (A and C) Data from mice infected with JR-CSF. (B and D) Data from mice infected with BK132. Spleen and bone marrow cells were collected on the day of euthanasia. Due to very low cell recovery, the result for one mouse in the X5/X4 group was excluded. Mean values for each group are shown.
We also measured the HIV-1 RNA levels in lymphoid tissues. Figure 8C shows that mice in the X5/R5 group had significantly lower average HIV-1 RNA copy numbers than did mice in the AB65/R5 group in spleens (P = 0.0496) or in bone marrow (P = 0.0879). Similarly, after cotransfusion with X4-tropic virus-infected cells, mice in the X5/X4 group had 10 times fewer average HIV-1 RNA copies per CD4 T cell than did mice in the AB65/X4 group in spleens (P = 0.0004) or 7 times fewer copies in the bone marrow (P = 0.0133) (Fig. 8D). Taken together, we conclude that GPI-scFv X5-transduced CD4 T cells exert a negative effect on viral replication in lymphoid tissues in hu-PBL mice.
DISCUSSION
In the present study, we employed GPI-scFv X5, which is derived from human anti-HIV-1 monoclonal antibody (MAb) X5, to confer resistance of primary CD4 T cells to HIV-1 infection. We show that GPI-scFv X5-transduced CD4 T cells were resistant to R5-, X4-, and dual-tropic virus infections and displayed a survival advantage in the presence of HIV-1 strains ex vivo. In a preclinical study using a semitherapeutic strategy in hu-PBL mice, we show that after cotransfusion with R5-tropic virus-infected cells, averages of 2- to 15-fold reductions in viral loads as well as 5- to 400-fold reductions in the number of viral RNA copies per CD4 cell were observed for mice in the X5/R5 group compared to those for mice in the AB65/R5 group, and after cotransfusion with X4-tropic virus-infected cells, averages of 13- to 4,000-fold reductions in the number of viral RNA copies per CD4 cell were observed for mice in the X5/X4 group compared to those for mice in the AB65/X4 group throughout the experiments (Fig. 6). Moreover, we show that there was significant protection of CD4 T cells in peripheral blood, spleen, and bone marrow in mice in the X5/R5 and X5/X4 groups compared to mice in the AB65/R5 and AB65/X4 groups (Fig. 4B and C and 7A). In the presence of R5- or X4-tropic virus, GPI-scFv X5-transduced cells were enriched 1.8- or 4.4-fold in peripheral blood, 3.3- or 5.5-fold in spleens, and 2.9- or 6.8-fold in bone marrow, respectively (Fig. 5B and 7B), implying a selective advantage for GPI-scFv X5-transduced cells in vivo. Thus, these results strongly suggest that GPI-scFv X5 protects CD4 T cells from depletion not only by R5-tropic virus but also by X4-tropic virus infection and has a great potential to be a therapeutic candidate for HIV-1 gene therapy.
Extensive studies have been carried out to evaluate the feasibility of entry inhibition. The most promising study utilized a disruption of the CCR5 gene by ZFN. A recent clinical trial using autologous CD4 cells modified by ZFN/CCR5 demonstrated a decrease in the viral load by an average of 1.2 log units from peak viremia during treatment disruption for 4 out of 6 patients (24). However, the possible appearance of X4-tropic virus in the late stages of the disease requires approaches to engineer HIV-1 target cell resistance to X4- or R5X4-tropic virus. Wilen et al. reported partial protection of CXCR4-edited CD4 cells in humanized mice (28). Env sequencing and tropism testing showed that a shift to CCR5 usage by the virus accounted for the depletion of CXCR4-disrupted cells at later time points. It seems that the ablation of both CCR5 and CXCR4 confers full protection against HIV-1 infection for all three tropisms (31, 39). Although gene editing provides a permanent and lifelong inherited phenotype for progeny cells of modified cells, potential off-target effects should be carefully investigated when this strategy is applied to clinical use, especially in light of the high level of homology between the CCR5 and CCR2 genes (40, 41). Additionally, the relatively lower efficiency of genome editing by ZFN or other nucleases limits the amount of biallelic disrupted cells for a given number of cells for infusion. Another drawback with the CCR5 gene knockout strategy is the potential for disease exacerbation. For example, it has been shown that trafficking of CCR5 gene knockout leukocytes to the central nervous system (CNS) after West Nile virus infection is reduced, leading to more severe disease (42, 43). Thus, in the present study, we delivered GPI-scFv X5 at an 80% average transduction efficiency to human primary T cells using a lentiviral vector. Since the safety of gene transfer by lentiviral vectors (44) has been demonstrated in several clinical trials, our approach is able to achieve a large amount of infection-resistant cells by a single low dose (MOI of 5) for transduction.
X5 is a human monoclonal Fab neutralizing antibody selected by the gp120-CD4-CCR5 fusion complex from an antibody phage display library and significantly neutralizes primary HIV-1 isolates (45). It recognizes a CD4-inducible epitope located in the coreceptor-binding site (amino acid residues 417 to 432) of gp120 (46, 47). It was reported previously that the scFv form of X5 is more effective than the Fab fragment or the whole antibody in neutralization (48). It is likely that after gp120-CD4 binding, the available space is not sufficient to accommodate large molecules. Despite the broad range of neutralization activities of X5, we and others showed that free scFv X5 had a relatively lower level of inhibition of virus entry (37, 49). X5 function is great enhanced by the addition of soluble CD4. In this way, GPI-anchored scFv X5 may efficiently capture the epitope exposed after virus attachment to target cells and block virus fusion at an early stage, since CD4 resides in the lipid raft membrane. Although the inhibitory activity of GPI-scFv X5 expressed on human CD4 T cell lines was demonstrated previously by our group, the in vivo function of GPI-scFv X5-transduced primary CD4 T cells addressed in the present study extends these findings to a more clinically relevant system.
A limitation of the present study is that the prolonged therapeutic impact of GPI-scFv X5 has not been addressed. In this proof-of-concept study, we have shown vigorous protection by GPI-scFv X5 in a humanized mouse model of acute HIV infection. We have also shown the persistent expression of GPI-anchored scFv (X5 and control AB65) within lymphoid tissues at the end of in vivo experiments (Fig. 7B). However, since the human T cells transplanted into mice resulted in xenogeneic graft-versus-host activity, we were not able to investigate longer effects of modified cells on HIV infection and immune cell repopulation. Moreover, the effect of GPI-scFv X5 on chronic infection and the influence of ex vivo manipulation on cell proliferation, immune function, persistence, and homing should be studied further. Previous clinical studies described the maintenance of cells modified by gene transfer for up to a decade (50–52), and these cells were detected in rectal mucosa, implying persistence and trafficking to various organs of modified cells. Interestingly, a similar approach using membrane-anchored C-peptide entry inhibitor (mC46)-expressing hematopoietic stem cell transplantation in pigtail macaques showed that modified SHIV-specific CD4 T cells were maintained in mC46-treated animals (36). A more robust T lymphocyte response, including gamma interferon (IFN-γ) production and an enhanced production of antibodies against SHIV, was also observed in animals transfused with mC46-treated cells. Whether these enhanced immunological responses are mC46 specific or beneficial for protected SHIV-1-resistant CD4 cells (or other myeloid cells) should be investigated further. Thus, the next step is to test GPI-scFv X5-transduced autologous CD4 T cells in an SHIV-challenged macaque model.
Because GPI-scFvs contain human VH and VL gene segments and an artificial linker, expression of the scFvs on the cell surface may lead to immune-mediated rejection of cells expressing GPI-scFvs when used in human patients in a clinical setting. However, such immune-mediated rejection did not occur in the studies of mC46 (a membrane-bound version of C46) described above (36, 53). In both an immunocompetent macaque model as well as a phase I human trial, mC46-modified macaque hematopoietic stem cells or mC46-modified autologous human CD4 T cells expanded in the host, without significant host immune responses to mC46. Nonetheless, it is possible that GPI-scFvs may be immunogenic, and it will be important to test their immunogenicity in immunocompetent hosts in vivo. Finally, a single use of GPI-scFv X5 may lead to escape mutants in long-term infections in clinical settings. Therefore, a mixture of GPI-anchored antibody derivatives, such as GPI-scFv X5 and trimeric GPI-HCDR3 PG16 (54), that target different epitopes in HIV-1 Env could be preferable.
MATERIALS AND METHODS
Ethics statement.
All experiments using human blood and all animal experiments were conducted according to guidelines for the housing and care of laboratory animals (60) and in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) (approval 14-100.0) at the University of North Carolina at Chapel Hill. Human blood was purchased from the Gulf Coast Regional Blood Center, Houston, TX.
Lentiviral vector production and titration.
Lentiviral vectors were generated by using transient transfection of HEK293T cells (American Type Culture Collection [ATCC]) with calcium phosphate and the lentivectors pRRL, encoding GPI-anchored scFvs; delta 8.9, encoding Gag/Pol/Rev; and pLP/VSVG; encoding the vesicular stomatitis virus G (VSVG) envelope, as described previously (22). Cell culture supernatants were collected at 48 h posttransfection and concentrated via ultracentrifugation at 25,000 rpm for 2 h at 4°C. Concentrated lentivectors were resuspended in serum-free RPMI 1640 and titrated by the transduction of HEK293T cells. Titers of concentrated lentivectors were calculated by quantification of the number of GFP-positive cells 2 days after transduction.
Human primary CD4 T cell culture and transduction.
Methods for the isolation of human PBMCs were described previously (55), and PBMCs were cryopreserved until use. CD4 T cells were negatively isolated from PBMCs by using an untouched Dynabeads human CD4 T cell kit (catalog number 11352D; Invitrogen) according to the manufacturer's instructions. Isolated CD4 T cells were stimulated with anti-CD3/anti-CD28-coated magnetic beads (catalog number 11131D; Gibco) at a bead-to-cell ratio of 1:1 and maintained in RPMI 1640 (Invitrogen) supplemented with 10% fetal bovine serum (FBS) plus 100 to 300 IU/ml human recombinant IL-2 (NIH AIDS Reagent Program) (here referred to as complete RPMI medium). Twenty-four hours after stimulation, concentrated lentiviral vectors were added to cells at an MOI of 5. Beads were removed at day 5 after stimulation. Transduction efficiency was determined as the percentage of GFP-positive cells determined by using fluorescence-activated cell sorter (FACS) analysis. Transduced T cells were expanded and maintained in complete RPMI.
Flow cytometry.
All staining procedures except intracellular staining were performed on ice in FACS buffer (phosphate-buffered saline [PBS] containing 2% FBS and 2 mM EDTA). Intracellular staining was performed by using a BD Cytofix/Cytoperm kit (catalog number 554714) according to the manufacturer's instructions. The HA tag was detected by staining of cells with rabbit anti-HA antibody (catalog number H6908; Sigma) followed by a secondary Alexa Fluor 647-conjugated anti-rabbit IgG antibody (clone poly4064; BioLegend). Phycoerythrin (PE)-conjugated anti-p24 antibody (clone KC57; Beckman Coulter) was used to detect intracellular HIV-1 Gag proteins. Other antibodies used in ex vivo experiments were (i) PE-CD4 (clone OKT4; BioLegend) and 7-aminoactinomycin D (catalog number A1310; Life Technologies) for cell counting and (ii) Live/Dead yellow dye (catalog number L34959; Life Technologies) and peridinin chlorophyll protein (PerCP)/Cy5.5-CD4 (clone OKT4; BioLegend) for infection staining. Other antibodies used for in vivo experiments were (i) PE-human CD45 (clone HI30; BioLegend), PE/Cy7-CD3 (clone OKT3; BioLegend), and 7-aminoactinomycin D for cell counting and (ii) Live/Dead yellow dye, Pacific Orange-mouse CD45 (reference number MCD4530; Life Technologies), allophycocyanin (APC)/Cy7-human CD45 (clone HI30; BioLegend), Pacific Blue-CD3 (clone UCHT1; BioLegend), PerCP/Cy5.5-CD4 (clone OKT4; BioLegend), and PE/Cy7-CD8 (clone HIT8a; BioLegend) for surface marker staining. Cell counting and analysis were performed by using a Guava easyCyte flow cytometer (Millipore). Other staining samples were run on a BD LSR Fortessa or Canto II instrument. Results were analyzed by using FlowJo 10.0.7 (TreeStar).
Ex vivo HIV-1 challenge.
X4-tropic strain BK132, R5-tropic strain JRCSF, and R5X4-dual-tropic strain R3A were used for ex vivo challenges. R3A and JR-CSF were generated by transfection of HEK293T cells. BK132 was obtained from the NIH AIDS Reagent Program and propagated in activated CD4 T cells. Virus stocks were titrated on MagiX4R5 cells and analyzed by a p24 enzyme-linked immunosorbent assay (ELISA). Human primary CD4 T cells were stimulated and transduced as described above. A total of 1 × 106 stimulated cells were infected with R3A, JR-CSF, or BK132 at an MOI of 0.01 or with 100 ng p24 per million cells in the presence of 8 ng/ml Polybrene via spinning at 1,500 × g for 2 h at 37°C. Cells were then washed with RPMI 1640 twice and resuspended in 1.5 ml complete RPMI medium on a 12-well plate. At days 3, 6, and 9 postinfection, supernatants were collected and stored at −20°C for further p24 detection via an ELISA. Cells were counted after Live/Dead staining and CD4 surface staining. HIV-1 infection was determined by flow cytometry. Cells were stained for the surface HA tag and CD4 marker, followed by intracellular staining of the p24 Gag protein. For the coculture system, 5 × 105 primary CD4 cells were infected with R3A, JR-CSF, or BK132 at a dose of 100 ng p24 per million cells. Cells were then washed with medium and added to uninfected untransduced or GPI-scFv X5- or AB65-transduced cells, respectively. HIV-1 infection was determined by intracellular p24 staining as described above.
In vivo testing of GPI-scFv X5-transduced human CD4 T cells using a hu-PBL mouse model.
A total of three in vivo experiments were carried out. In each experiment, cells from one healthy human donor were used to generate GPI-scFv X5- or AB65-transduced primary CD4 T cells, HIV-1-infected and uninfected PBMCs, and resting PBMCs. NRG mice (Jackson Laboratory) were irradiated as previously described (56). In each experiment, mice at 4 to 6 weeks of age were randomly assigned to 4 groups (n = 2 to 3 for each uninfected group and n = 6 to 10 for each infected group) to control for sex and age effects. CD4 T cells were stimulated, transduced, and expanded as described above. Resting PBMCs were stimulated with CD3/CD28 beads. Beads were removed 3 days after stimulation. Cells were allowed to recover for 2 days and then infected with JR-CSF or BK132 at an MOI of 0.01. On the day of transfusion, 8 × 106 GPI-scFv X5- or AB65-transduced CD4 T cells with 1 × 106 uninfected or infected PBMCs in 200 μl PBS were injected into each mouse via tail vein. Additionally, 1 × 106 autologous resting PBMCs were also injected into each mouse to promote engraftment, using methods similar to those reported previously by Perez et al. (19).
Engraftment efficiency was assessed at day 7 posttransfusion by counting human CD45 cells in mouse blood using a flow cytometer. Blood was bled through the tail vein, and plasma was isolated by centrifugation at 5,000 rpm for 5 min and stored at −80°C for further viral RNA extraction. For cell pellets, red blood cells were lysed by using Ammonium-Chloride-Potassium (ACK) lysis buffer (catalog number A1049201; Gibco) at room temperature for 5 min. Cells were then resuspended in FACS buffer and stained for surface markers, including mCD45, hCD45, CD3, CD4, CD8, and the HA tag, followed by flow cytometry analysis. Cells were counted by using a Guava easyCyte flow cytometer (Millipore) after staining of live/dead cells and human CD45 and CD3.
At 6 weeks posttransfusion, splenocytes and bone marrow cells were isolated following euthanasia. Single-cell suspensions were obtained by passing cells through a 70-μm filter. Surface markers, including mCD45, hCD45, CD3, CD4, CD8, and the HA tag, were analyzed by flow cytometry after staining. Genomic DNA and total RNA were isolated from splenocytes and bone marrow cells by using a DNeasy blood and tissue kit (Qiagen) and an RNeasy Mini Plus kit (Qiagen), respectively.
Measurement of viral loads in plasma samples.
The viral load was determined as previously described (57, 58). Briefly, viral RNA from each mouse and standard plasma (catalog number 3443; NIH AIDS Reagent Program) were extracted (QIAamp viral RNA minikit; Qiagen) from plasma of infected humanized mice and analyzed by reverse transcription-quantitative PCR (RT-qPCR) (TaqMan One-Step RT-qPCR master mix; ABI). The primers and probe used for RT-qPCR were forward primer 5′-CAATGGCAGCAATTTCACCA-3′, reverse primer 5′-ATGCCAAATTCCTGCTTGA-3′, and probe FAM (6-carboxyfluorescein)-CCCACCAACAGGCRGCCTTAACYG-QSY7 (TAMRA [6-carboxymethyltetrarhodamine]).
Measurement of HIV-1 DNA or RNA levels.
Total spleen or bone marrow cells corresponding to 1 × 105 human CD45 cells were lysed. Genomic DNA was extracted according to the manufacturer's instructions (DNeasy blood and tissue kit; Qiagen). A total of 1 × 106 ACH2 cells were serially diluted and mixed with human PBMCs from a healthy donor to obtain a total of 1 × 106 cells. Genomic DNAs from ACH2 cell-PBMC mixtures were used as standard samples. Cell-associated RNA and RNA standards were extracted by using an RNeasy Mini Plus kit (Qiagen) and reverse transcribed into first-strand cDNA (Superscript III reverse transcriptase; Invitrogen). HIV-1 DNA copies and cDNA copies were detected by quantitative PCR (TaqMan Universal master mix; Applied Biosystems) using the primers and probe described above, and copy numbers were calculated according to standard curves.
Statistical analysis.
Statistical analysis was performed by using GraphPad Prim software version 5.0. Averages and standard deviation (SD) were calculated. Unpaired two-tailed t tests with Welch's correction or Mann-Whitney U tests were used to compare data between two groups. All results were considered significant when the P value was <0.05.
ACKNOWLEDGMENTS
The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 BK132 from Nelson Michael and human recombinant IL-2 from Maurice Gately. The HIV RNA quantification standard and ACH2 cells were obtained from Hoffmann-La Roche Inc. We thank Christopher M. Murphy from the Lineberger Comprehensive Cancer Center at the University of North Carolina at Chapel Hill for critical reading of the manuscript. We thank UNC DLAM for support and Liqun Chi and Yaxu Wu from the Lineberger Comprehensive Cancer Center at the University of North Carolina at Chapel Hill for technical support.
This work was supported by research grants from the Chinese National Science Foundation (CNSF) (31170871) and the National Science and Technology Major Project (2014ZX10001-001) and a CNSF-NIH joint grant (81361120406) to P.Z.; U.S. NIH grants AI095097 and R01AI080432 to L.S.; and a China Scholarship Council grant (file 201504910661) to C.Y. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication
REFERENCES
- 1.Yeni PG, Hammer SM, Hirsch MS, Saag MS, Schechter M, Carpenter CC, Fischl MA, Gatell JM, Gazzard BG, Jacobsen DM, Katzenstein DA, Montaner JS, Richman DD, Schooley RT, Thompson MA, Vella S, Volberding PA. 2004. Treatment for adult HIV infection: 2004 recommendations of the International AIDS Society-USA Panel. JAMA 292:251–265. doi: 10.1001/jama.292.2.251. [DOI] [PubMed] [Google Scholar]
- 2.Blankson JN, Persaud D, Siliciano RF. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu Rev Med 53:557–593. doi: 10.1146/annurev.med.53.082901.104024. [DOI] [PubMed] [Google Scholar]
- 3.Eshleman SH, Krogstad P, Jackson JB, Wang YG, Lee S, Wei LJ, Cunningham S, Wantman M, Wiznia A, Johnson G, Nachman S, Palumbo P. 2001. Analysis of human immunodeficiency virus type 1 drug resistance in children receiving nucleoside analogue reverse-transcriptase inhibitors plus nevirapine, nelfinavir, or ritonavir (Pediatric AIDS Clinical Trials Group 377). J Infect Dis 183:1732–1738. doi: 10.1086/320728. [DOI] [PubMed] [Google Scholar]
- 4.Hirsch MS, Brun-Vezinet F, Clotet B, Conway B, Kuritzkes DR, D'Aquila RT, Demeter LM, Hammer SM, Johnson VA, Loveday C, Mellors JW, Jacobsen DM, Richman DD. 2003. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA panel. Clin Infect Dis 37:113–128. doi: 10.1086/375597. [DOI] [PubMed] [Google Scholar]
- 5.Carr A. 2003. Toxicity of antiretroviral therapy and implications for drug development. Nat Rev Drug Discov 2:624–634. doi: 10.1038/nrd1151. [DOI] [PubMed] [Google Scholar]
- 6.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, Schneider T, Hofmann J, Kucherer C, Blau O, Blau IW, Hofmann WK, Thiel E. 2009. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 7.Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 8.Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, MacDonald ME, Stuhlmann H, Koup RA, Landau NR. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367–377. doi: 10.1016/S0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 9.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth RJ, Collman RG, Doms RW, Vassart G, Parmentier M. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 10.Swan CH, Buhler B, Steinberger P, Tschan MP, Barbas CF III, Torbett BE. 2006. T-cell protection and enrichment through lentiviral CCR5 intrabody gene delivery. Gene Ther 13:1480–1492. doi: 10.1038/sj.gt.3302801. [DOI] [PubMed] [Google Scholar]
- 11.DiGiusto DL, Krishnan A, Li L, Li H, Li S, Rao A, Mi S, Yam P, Stinson S, Kalos M, Alvarnas J, Lacey SF, Yee JK, Li M, Couture L, Hsu D, Forman SJ, Rossi JJ, Zaia JA. 2010. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med 2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myburgh R, Ivic S, Pepper MS, Gers-Huber G, Li D, Audige A, Rochat MA, Jaquet V, Regenass S, Manz MG, Salmon P, Krause KH, Speck RF. 2015. Lentivector knockdown of CCR5 in hematopoietic stem and progenitor cells confers functional and persistent HIV-1 resistance in humanized mice. J Virol 89:6761–6772. doi: 10.1128/JVI.00277-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizu S, Ringpis GE, Marsden MD, Cortado RV, Wilhalme HM, Elashoff D, Zack JA, Chen IS, An DS. 2015. RNAi-mediated CCR5 knockdown provides HIV-1 resistance to memory T cells in humanized BLT mice. Mol Ther Nucleic Acids 4:e227. doi: 10.1038/mtna.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T. 2011. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res 39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sather BD, Romano Ibarra GS, Sommer K, Curinga G, Hale M, Khan IF, Singh S, Song Y, Gwiazda K, Sahni J, Jarjour J, Astrakhan A, Wagner TA, Scharenberg AM, Rawlings DJ. 2015. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci Transl Med 7:307ra156. doi: 10.1126/scitranslmed.aac5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, Cannon PM. 2010. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, Krymskaya L, Wang J, Henley J, Rao A, Cao LF, Tran CA, Torres-Coronado M, Gardner A, Gonzalez N, Kim K, Liu PQ, Hofer U, Lopez E, Gregory PD, Liu Q, Holmes MC, Cannon PM, Zaia JA, DiGiusto DL. 2013. Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther 21:1259–1269. doi: 10.1038/mt.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maier DA, Brennan AL, Jiang S, Binder-Scholl GK, Lee G, Plesa G, Zheng Z, Cotte J, Carpenito C, Wood T, Spratt SK, Ando D, Gregory P, Holmes MC, Perez EE, Riley JL, Carroll RG, June CH, Levine BL. 2013. Efficient clinical scale gene modification via zinc finger nuclease-targeted disruption of the HIV co-receptor CCR5. Hum Gene Ther 24:245–258. doi: 10.1089/hum.2012.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, Guschin DY, Rupniewski I, Waite AJ, Carpenito C, Carroll RG, Orange JS, Urnov FD, Rebar EJ, Ando D, Gregory PD, Riley JL, Holmes MC, June CH. 2008. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi G, Choi JG, Bharaj P, Abraham S, Dang Y, Kafri T, Alozie O, Manjunath MN, Shankar P. 2014. CCR5 gene editing of resting CD4(+) T cells by transient ZFN expression from HIV envelope pseudotyped nonintegrating lentivirus confers HIV-1 resistance in humanized mice. Mol Ther Nucleic Acids 3:e198. doi: 10.1038/mtna.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan J, Wang J, Crain K, Fearns C, Kim KA, Hua KL, Gregory PD, Holmes MC, Torbett BE. 2012. Zinc-finger nuclease editing of human cxcr4 promotes HIV-1 CD4(+) T cell resistance and enrichment. Mol Ther 20:849–859. doi: 10.1038/mt.2011.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Ye C, Liu J, Zhang D, Kimata JT, Zhou P. 2014. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS One 9:e115987. doi: 10.1371/journal.pone.0115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang H, Minder P, Park MA, Mesquitta WT, Torbett BE, Slukvin II. 2015. CCR5 disruption in induced pluripotent stem cells using CRISPR/Cas9 provides selective resistance of immune cells to CCR5-tropic HIV-1 virus. Mol Ther Nucleic Acids 4:e268. doi: 10.1038/mtna.2015.42. [DOI] [PubMed] [Google Scholar]
- 24.Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, Holmes MC, Gregory PD, Ando DG, Kalos M, Collman RG, Binder-Scholl G, Plesa G, Hwang WT, Levine BL, June CH. 2014. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med 185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esbjornsson J, Mansson F, Martinez-Arias W, Vincic E, Biague AJ, da Silva ZJ, Fenyo EM, Norrgren H, Medstrand P. 2010. Frequent CXCR4 tropism of HIV-1 subtype A and CRF02_AG during late-stage disease—indication of an evolving epidemic in West Africa. Retrovirology 7:23. doi: 10.1186/1742-4690-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kordelas L, Verheyen J, Beelen DW, Horn PA, Heinold A, Kaiser R, Trenschel R, Schadendorf D, Dittmer U, Esser S, Essen HIV AlloSCT Group. 2014. Shift of HIV tropism in stem-cell transplantation with CCR5 Delta32 mutation. N Engl J Med 371:880–882. doi: 10.1056/NEJMc1405805. [DOI] [PubMed] [Google Scholar]
- 28.Wilen CB, Wang J, Tilton JC, Miller JC, Kim KA, Rebar EJ, Sherrill-Mix SA, Patro SC, Secreto AJ, Jordan AP, Lee G, Kahn J, Aye PP, Bunnell BA, Lackner AA, Hoxie JA, Danet-Desnoyers GA, Bushman FD, Riley JL, Gregory PD, June CH, Holmes MC, Doms RW. 2011. Engineering HIV-resistant human CD4+ T cells with CXCR4-specific zinc-finger nucleases. PLoS Pathog 7:e1002020. doi: 10.1371/journal.ppat.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou P, Chen S, Wang S, Yu X, Chen Y, Jiang M, Zhuang K, Ho W, Hou W, Huang J, Guo D. 2015. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci Rep 5:15577. doi: 10.1038/srep15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson J, Banerjea A, Planelles V, Akkina R. 2003. Potent suppression of HIV type 1 infection by a short hairpin anti-CXCR4 siRNA. AIDS Res Hum Retroviruses 19:699–706. doi: 10.1089/088922203322280928. [DOI] [PubMed] [Google Scholar]
- 31.Didigu CA, Wilen CB, Wang J, Duong J, Secreto AJ, Danet-Desnoyers GA, Riley JL, Gregory PD, June CH, Holmes MC, Doms RW. 2014. Simultaneous zinc-finger nuclease editing of the HIV coreceptors ccr5 and cxcr4 protects CD4+ T cells from HIV-1 infection. Blood 123:61–69. doi: 10.1182/blood-2013-08-521229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez EE, Riley JL, Carroll RG, von Laer D, June CH. 2005. Suppression of HIV-1 infection in primary CD4 T cells transduced with a self-inactivating lentiviral vector encoding a membrane expressed gp41-derived fusion inhibitor. Clin Immunol 115:26–32. doi: 10.1016/j.clim.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 33.Zahn RC, Hermann FG, Kim EY, Rett MD, Wolinsky SM, Johnson RP, Villinger F, von Laer D, Schmitz JE. 2008. Efficient entry inhibition of human and nonhuman primate immunodeficiency virus by cell surface-expressed gp41-derived peptides. Gene Ther 15:1210–1222. doi: 10.1038/gt.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burke BP, Levin BR, Zhang J, Sahakyan A, Boyer J, Carroll MV, Colon JC, Keech N, Rezek V, Bristol G, Eggers E, Cortado R, Boyd MP, Impey H, Shimizu S, Lowe EL, Ringpis GE, Kim SG, Vatakis DN, Breton LR, Bartlett JS, Chen IS, Kitchen SG, An DS, Symonds GP. 2015. Engineering cellular resistance to HIV-1 infection in vivo using a dual therapeutic lentiviral vector. Mol Ther Nucleic Acids 4:e236. doi: 10.1038/mtna.2015.10. [DOI] [PubMed] [Google Scholar]
- 35.Petit NY, Baillou C, Burlion A, Dorgham K, Levacher B, Amiel C, Schneider V, Lemoine FM, Gorochov G, Marodon G. 2016. Gene transfer of two entry inhibitors protects CD4(+) T cell from HIV-1 infection in humanized mice. Gene Ther 23:144–150. doi: 10.1038/gt.2015.101. [DOI] [PubMed] [Google Scholar]
- 36.Younan PM, Polacino P, Kowalski JP, Peterson CW, Maurice NJ, Williams NP, Ho O, Trobridge GD, Von Laer D, Prlic M, Beard BC, DeRosa S, Hu SL, Kiem HP. 2013. Positive selection of mC46-expressing CD4+ T cells and maintenance of virus specific immunity in a primate AIDS model. Blood 122:179–187. doi: 10.1182/blood-2013-01-482224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen M, Arora R, Wang H, Liu L, Kimata JT, Zhou P. 2010. GPI-anchored single chain Fv—an effective way to capture transiently-exposed neutralization epitopes on HIV-1 envelope spike. Retrovirology 7:79. doi: 10.1186/1742-4690-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenberg M, DeTulleo L, Rapoport I, Skowronski J, Kirchhausen T. 1998. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr Biol 8:1239–1242. doi: 10.1016/S0960-9822(07)00518-0. [DOI] [PubMed] [Google Scholar]
- 39.Anderson J, Banerjea A, Akkina R. 2003. Bispecific short hairpin siRNA constructs targeted to CD4, CXCR4, and CCR5 confer HIV-1 resistance. Oligonucleotides 13:303–312. doi: 10.1089/154545703322616989. [DOI] [PubMed] [Google Scholar]
- 40.Pattanayak V, Ramirez CL, Joung JK, Liu DR. 2011. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods 8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cradick TJ, Fine EJ, Antico CJ, Bao G. 2013. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res 41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glass WG, McDermott DH, Lim JK, Lekhong S, Yu SF, Frank WA, Pape J, Cheshier RC, Murphy PM. 2006. CCR5 deficiency increases risk of symptomatic West Nile virus infection. J Exp Med 203:35–40. doi: 10.1084/jem.20051970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durrant DM, Daniels BP, Pasieka T, Dorsey D, Klein RS. 2015. CCR5 limits cortical viral loads during West Nile virus infection of the central nervous system. J Neuroinflammation 12:233. doi: 10.1186/s12974-015-0447-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tebas P, Stein D, Binder-Scholl G, Mukherjee R, Brady T, Rebello T, Humeau L, Kalos M, Papasavvas E, Montaner LJ, Schullery D, Shaheen F, Brennan AL, Zheng Z, Cotte J, Slepushkin V, Veloso E, Mackley A, Hwang WT, Aberra F, Zhan J, Boyer J, Collman RG, Bushman FD, Levine BL, June CH. 2013. Antiviral effects of autologous CD4 T cells genetically modified with a conditionally replicating lentiviral vector expressing long antisense to HIV. Blood 121:1524–1533. doi: 10.1182/blood-2012-07-447250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moulard M, Phogat SK, Shu Y, Labrijn AF, Xiao X, Binley JM, Zhang MY, Sidorov IA, Broder CC, Robinson J, Parren PW, Burton DR, Dimitrov DS. 2002. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc Natl Acad Sci U S A 99:6913–6918. doi: 10.1073/pnas.102562599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darbha R, Phogat S, Labrijn AF, Shu Y, Gu Y, Andrykovitch M, Zhang MY, Pantophlet R, Martin L, Vita C, Burton DR, Dimitrov DS, Ji X. 2004. Crystal structure of the broadly cross-reactive HIV-1-neutralizing Fab X5 and fine mapping of its epitope. Biochemistry 43:1410–1417. doi: 10.1021/bi035323x. [DOI] [PubMed] [Google Scholar]
- 47.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang CC, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol 77:10557–10565. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang MY, Shu Y, Rudolph D, Prabakaran P, Labrijn AF, Zwick MB, Lal RB, Dimitrov DS. 2004. Improved breadth and potency of an HIV-1-neutralizing human single-chain antibody by random mutagenesis and sequential antigen panning. J Mol Biol 335:209–219. doi: 10.1016/j.jmb.2003.09.055. [DOI] [PubMed] [Google Scholar]
- 50.Mitsuyasu RT, Anton PA, Deeks SG, Scadden DT, Connick E, Downs MT, Bakker A, Roberts MR, June CH, Jalali S, Lin AA, Pennathur-Das R, Hege KM. 2000. Prolonged survival and tissue trafficking following adoptive transfer of CD4zeta gene-modified autologous CD4(+) and CD8(+) T cells in human immunodeficiency virus-infected subjects. Blood 96:785–793. [PubMed] [Google Scholar]
- 51.Scholler J, Brady TL, Binder-Scholl G, Hwang WT, Plesa G, Hege KM, Vogel AN, Kalos M, Riley JL, Deeks SG, Mitsuyasu RT, Bernstein WB, Aronson NE, Levine BL, Bushman FD, June CH. 2012. Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci Transl Med 4:132ra53. doi: 10.1126/scitranslmed.3003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macpherson JL, Boyd MP, Arndt AJ, Todd AV, Fanning GC, Ely JA, Elliott F, Knop A, Raponi M, Murray J, Gerlach W, Sun LQ, Penny R, Symonds GP, Carr A, Cooper DA. 2005. Long-term survival and concomitant gene expression of ribozyme-transduced CD4+ T-lymphocytes in HIV-infected patients. J Gene Med 7:552–564. doi: 10.1002/jgm.705. [DOI] [PubMed] [Google Scholar]
- 53.van Lunzen J, Glaunsinger T, Stahmer I, von Baehr V, Baum C, Schilz A, Kuehlcke K, Naundorf S, Martinius H, Hermann F, Giroglou T, Newrzela S, Muller I, Brauer F, Brandenburg G, Alexandrov A, von Laer D. 2007. Transfer of autologous gene-modified T cells in HIV-infected patients with advanced immunodeficiency and drug-resistant virus. Mol Ther 15:1024–1033. [DOI] [PubMed] [Google Scholar]
- 54.Liu L, Wang W, Yang L, Ren H, Kimata JT, Zhou P. 2013. Trimeric glycosylphosphatidylinositol-anchored HCDR3 of broadly neutralizing antibody PG16 is a potent HIV-1 entry inhibitor. J Virol 87:1899–1905. doi: 10.1128/JVI.01038-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reszka-Blanco NJ, Sivaraman V, Zhang L, Su L. 2015. HIV-1 Env and Nef cooperatively contribute to plasmacytoid dendritic cell activation via CD4-dependent mechanisms. J Virol 89:7604–7611. doi: 10.1128/JVI.00695-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li G, Cheng M, Nunoya J, Cheng L, Guo H, Yu H, Liu YJ, Su L, Zhang L. 2014. Plasmacytoid dendritic cells suppress HIV-1 replication but contribute to HIV-1 induced immunopathogenesis in humanized mice. PLoS Pathog 10:e1004291. doi: 10.1371/journal.ppat.1004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhen A, Kamata M, Rezek V, Rick J, Levin B, Kasparian S, Chen IS, Yang OO, Zack JA, Kitchen SG. 2015. HIV-specific immunity derived from chimeric antigen receptor-engineered stem cells. Mol Ther 23:1358–1367. doi: 10.1038/mt.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsao LC, Guo H, Jeffrey J, Hoxie JA, Su L. 2016. CCR5 interaction with HIV-1 Env contributes to Env-induced depletion of CD4 T cells in vitro and in vivo. Retrovirology 13:22. doi: 10.1186/s12977-016-0255-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.UNAIDS, World Health Organization. 2015. AIDS by the numbers, p 3. World Health Organization, Geneva, Switzerland: http://www.unaids.org/sites/default/files/media_asset/AIDS_by_the_numbers_2015_en.pdf. [Google Scholar]
- 60.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]