ABSTRACT
Viral inclusion bodies (IBs), or replication factories, are unique structures generated by viral proteins together with some cellular proteins as a platform for efficient viral replication, but little is known about the mechanism underlying IB formation and fusion. Our previous study demonstrated that the interaction between the nucleoprotein (N) and phosphoprotein (P) of human parainfluenza virus type 3 (HPIV3), an enveloped virus with great medical impact, can form IBs. In this study, we found that small IBs can fuse with each other to form large IBs that enhance viral replication. Furthermore, we found that acetylated α-tubulin interacts with the N-P complex and colocalizes with IBs of HPIV3 but does not interact with the N-P complex of human respiratory syncytial virus or vesicular stomatitis virus and does not colocalize with IBs of human respiratory syncytial virus. Most importantly, enhancement of α-tubulin acetylation using the pharmacological inhibitor trichostatin A (TSA), RNA interference (RNAi) knockdown of the deacetylase enzymes histone deacetylase 6 (HDAC6) and sirtuin 2 (SIRT2), or expression of α-tubulin acetyltransferase 1 (α-TAT1) resulted in the fusion of small IBs into large IBs and effective viral replication. In contrast, suppression of acetylation of α-tubulin by overexpressing HDAC6 and SIRT2 profoundly inhibited the fusion of small IBs and viral replication. Our findings offer previously unidentified mechanistic insights into the regulation of viral IB fusion by acetylated α-tubulin, which is critical for viral replication.
IMPORTANCE Inclusion bodies (IBs) are unique structures generated by viral proteins and some cellular proteins as a platform for efficient viral replication. Human parainfluenza virus type 3 (HPIV3) is a nonsegmented single-stranded RNA virus that mainly causes lower respiratory tract disease in infants and young children. However, no vaccines or antiviral drugs for HPIV3 are available. Therefore, understanding virus-host interactions and developing new antiviral strategies are increasingly important. Acetylation on lysine (K) 40 of α-tubulin is an evolutionarily conserved modification and plays an important role in many cellular processes, but its role in viral IB dynamics has not been fully explored. To our knowledge, our findings are the first to show that acetylated α-tubulin enhances viral replication by regulating HPIV3 IB fusion.
KEYWORDS: inclusion bodies, human parainfluenza virus type 3, acetylated α-tubulin
INTRODUCTION
Human parainfluenza virus type 3 (HPIV3) is a nonsegmented single-stranded RNA virus (NSV) that belongs to the Paramyxoviridae family and mainly causes lower respiratory tract disease in infants and young children (1). However, no vaccines are available for HPIV3, and therefore, a deeper understanding of the replication of HPIV3 and its interaction with its host cell is needed to facilitate studies of viral pathogenesis and the development of novel therapeutic approaches.
The HPIV3 genome is encapsidated by the nucleoprotein (N) to form the N-RNA complex, which serves as a template to interact with the RNA-dependent RNA polymerase complex consisting of a large protein (L) and a phosphoprotein (P) cofactor. N-RNA association with RNA-dependent RNA polymerase forms an active ribonucleoprotein complex that is necessary for transcription and replication to generate six monocistronic mRNAs encoding six structural proteins and an antigenome intermediate.
Like the Ns of most NSVs, N of HPIV3 consists of a highly conserved N-terminal core (Ncore) and a hypervariable C-terminal tail (Ntail). Ncore is required for N self-assembly and RNA binding to form the N-RNA complex (2), and Ntail is mainly responsible for the binding of the N-RNA complex to P (3). P also contains two domains, an N-terminal domain and a C-terminal domain. The N terminus of P chaperones N0 (free of RNA) and forms the N0-P complex to prevent the N aggregation and nonspecific binding with cellular RNAs (4) and direct N encapsidation of nascent genomic RNA during replication (5, 6). The main N0 binding site has been localized to the extreme N-terminal 40 amino acids (aa) of P of HPIV3 (7). The C terminus of P is responsible for the oligomerization of P, and an oligomerization domain was mapped between aa 423 and 457 of P of HPIV3 (8).
Viral inclusion bodies (IBs), or viral replication factories, which are accumulated aggregates of viral proteins, are commonly generated in a variety of animal viruses, such as DNA viruses (e.g., herpesviruses and poxviruses) and several RNA virus families (e.g., togaviruses, reoviruses, flaviviruses, coronaviruses, and bunyaviruses) (9, 32). Similarly, some NSVs, such as human respiratory syncytial virus (hRSV), rabies virus, human metapneumovirus, and HPIV3, can also form IBs, but the mechanisms and the details of the IB formation are poorly understood (10–13). Our recent study showed that the association of N with P of HPIV3 is the minimal requirement for the formation of IBs, which contain viral RNA, N, P, and polymerase in HPIV3-infected cells (13). Furthermore, Hoenen et al. showed that Ebola virus IBs are highly dynamic structures that fuse together to form larger IBs (14). Therefore, a better understanding of the dynamics of IB fusion in infected cells would undoubtedly provide novel insight into the life cycle of RNA viruses. However, the lack of a method for viral IB purification has hampered the study of IB composition, and the mechanisms involved in the fusion of most viral IBs have not been described yet. In this report, we present evidence that IBs are dynamic structures in the process of viral infection; small IBs move to each other for fusion and become large while decreasing in number. Most importantly, we further demonstrate that the N-P complex interacts with acetylated α-tubulin, IBs colocalize with acetylated α-tubulin, and acetylated α-tubulin promotes the fusion of small IBs into large IBs, thereby enhancing viral RNA synthesis.
RESULTS
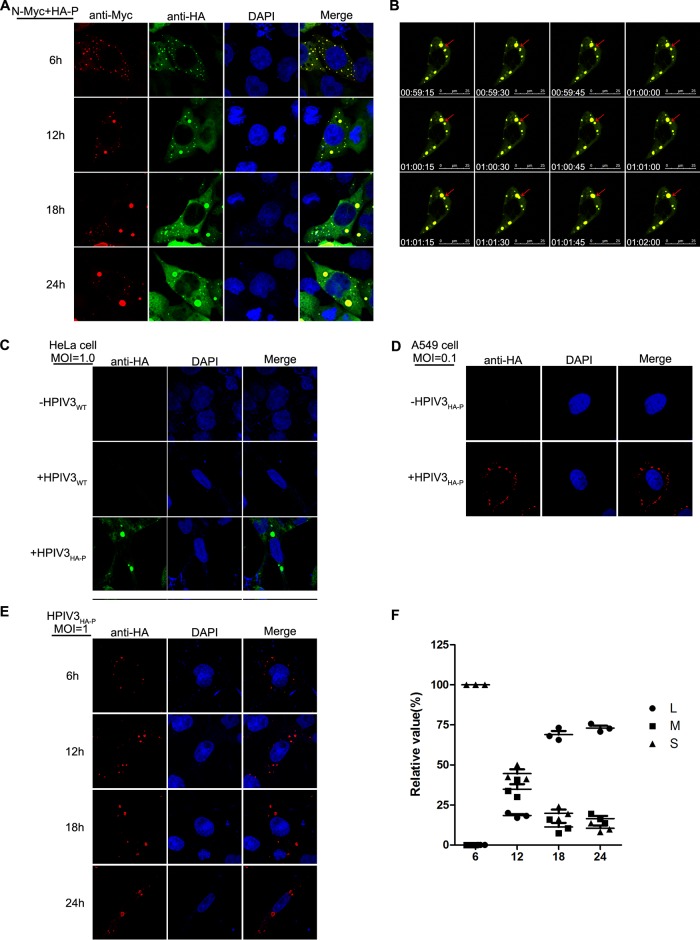
The appearance of large IBs is a kinetic fusion process derived from small IBs.
Our previous work demonstrated that the HPIV3 N association with P is the minimal requirement for the formation of cytoplasmic IBs (13), but the mechanisms of IBs formation are still not clear. To better analyze the IB formation process, we performed time course studies in which HeLa cells were fixed at 6-h intervals for 24 h after coexpression of N and P. Many small IBs with a mean cross-sectional area of less than 0.8 μm2 appeared at 6 h posttransfection and then gradually became larger, with a mean cross-sectional area between 3 μm2 and 8 μm2 from12 h to 18 h posttransfection. Finally, at 24 h posttransfection, the size of the IBs came to the peak, with individual IBs having cross-sectional areas of greater than 10 μm2 while decreasing in number (Fig. 1A), indicating that the large IBs derived from the fusion of small IBs. To confirm this, we visualized the expression of green fluorescent protein (GFP)-N and mCherry-P using live-cell microscopy with a finer temporal resolution of 15 s per picture at 12 h posttransfection and found that the small IBs were mobile and fused with each other to form the large IBs (Fig. 1B; Movie S1 in the supplemental material).
FIG 1.
Morphogenesis of IB fusion. (A) Time course analysis of IB fusion. HeLa cells transfected with plasmids encoding N and P were fixed at 6 h, 12 h, 18 h, and 24 h and subjected to immunofluorescence (IF) staining with antibodies against Myc and HA and appropriate fluorescent secondary antibodies. (B) Kinetic fusion process of small IBs fusing into large IBs. Cells expressing GFP-N and mCherry-P were visualized by live-cell imaging at 12 h posttransfection, and the fusion event is marked with a red arrow. (C and D) HPIV3HA-P forms IBs in HeLa and A549 cells. HeLa cells and A549 cells grown on coverslips were infected with HPIV3HA-P at MOIs of 1.0 and 0.1. At 24 h postinfection, cells were fixed and prepared for IF assay. Anti-HA antibody was used for visualizing the IBs. (E and F) Time course analysis of IB fusion in HPIV3HA-P-infected cells. HeLa cells infected with HPIV3HA-P at an MOI of 1.0 were fixed at different time points and evaluated via IF microscopy. The cross-sectional areas of IBs at different time points were measured. Large, medium, and small IBs were quantified at different time points in HPIV3HA-P-infected cells, and at least 50 cells were included for each group. The graph shows the quantifications of large, medium, and small IBs relative to IBs of all sizes at different time points. Values are means ± standard deviations (SD) from three experiments.
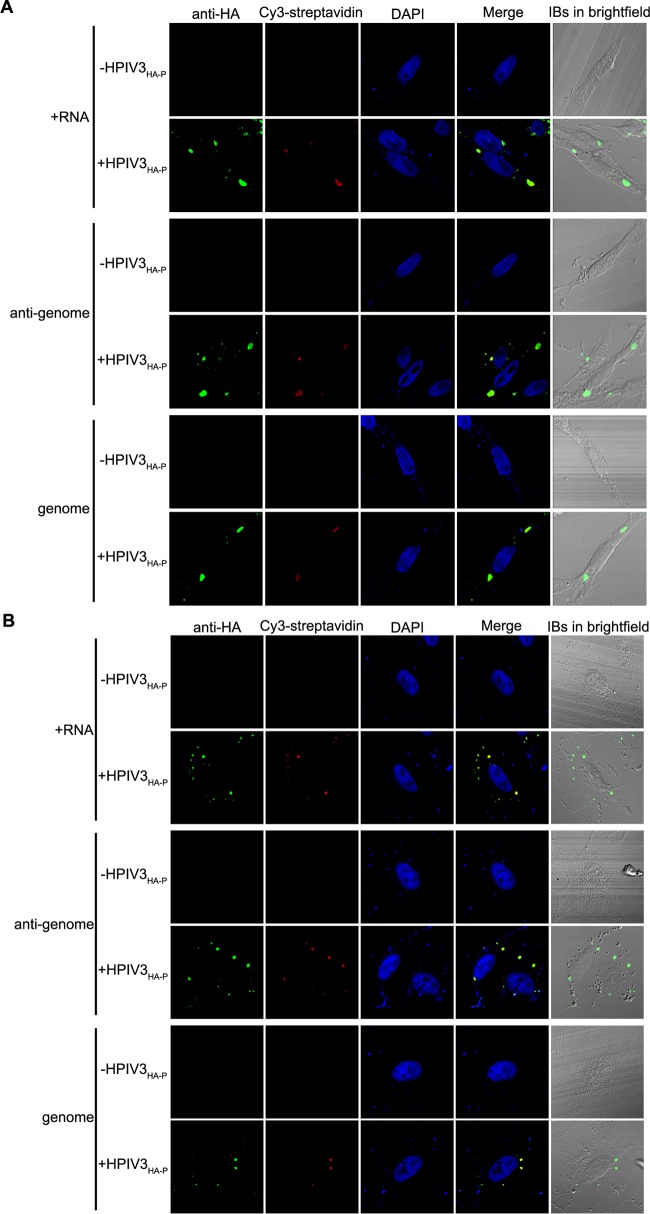
To determine whether IB formation and fusion also occur in HPIV3-infected cells, we rescued a recombinant HPIV3 with a hemagglutinin (HA) tag fused to the N terminus of P (HPIV3HA-P) and found that IBs also appeared in HPIV3HA-P-infected HeLa and A549 cells (Fig. 1C and D). In accordance with the results obtained by coexpression of N and P, small IBs were observed at the early stage of infection and gradually became larger at 12 h, 18 h, and 24 h postinfection (Fig. 1E). The percentages of large (with a mean cross-sectional area larger than 8 μm2), medium (with a mean cross-sectional area between 3 μm2 to 8 μm2), and small (with a mean cross-sectional area less than 3 μm2) IBs at different time points were evaluated and presented as a scatter plot (Fig. 1F), which showed that the percentages of large IBs were highest at 24 h postinfection, indicating that the large IBs indeed derived from the fusion of small IBs in HPIV3-infected cells. Furthermore, viral plus-strand RNAs, antigenomic RNA, and genomic RNA were all observed to localize in IBs in HPIV3-infected HeLa and A549 cells via the use of biotinylated oligonucleotide probes (Fig. 2), indicating that IBs are indeed viral RNA synthesis sites.
FIG 2.
Viral RNAs accumulate in viral IBs in HPIV3HA-P-infected HeLa cells and A549 cells. HeLa cells (A) and A549 cells (B) infected with HPIV3HA-P (MOI = 1) for 24 h postinfection were fixed and subjected to FISH analyses as described in Materials and Methods. Biotinylated oligonucleotides were used to detect N mRNA, antigenomic RNA, and genomic RNA, and anti-HA antibody was used to visualize IBs in HPIV3HA-P-infected cells.
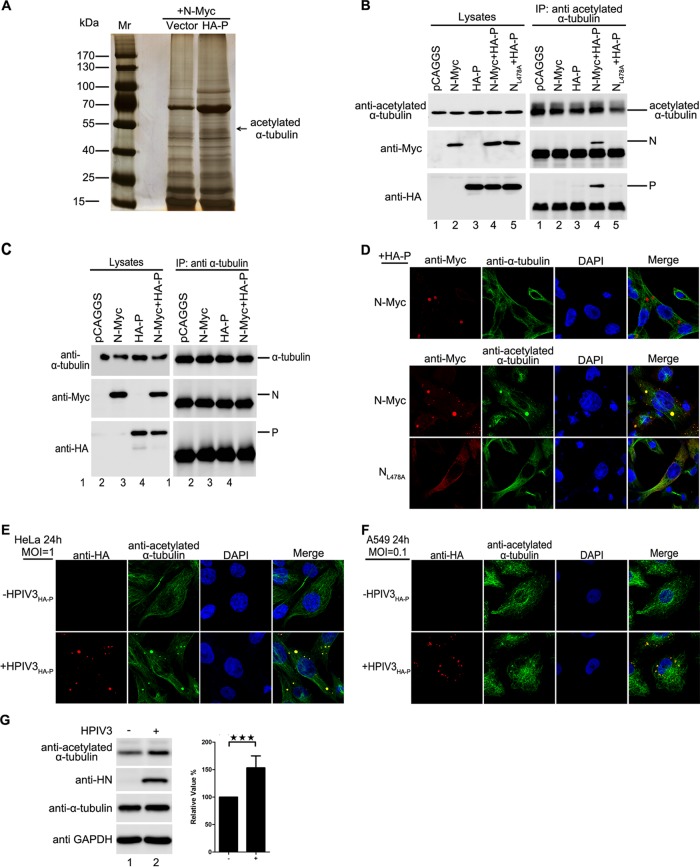
Acetylated α-tubulin interacts with the N-P complex.
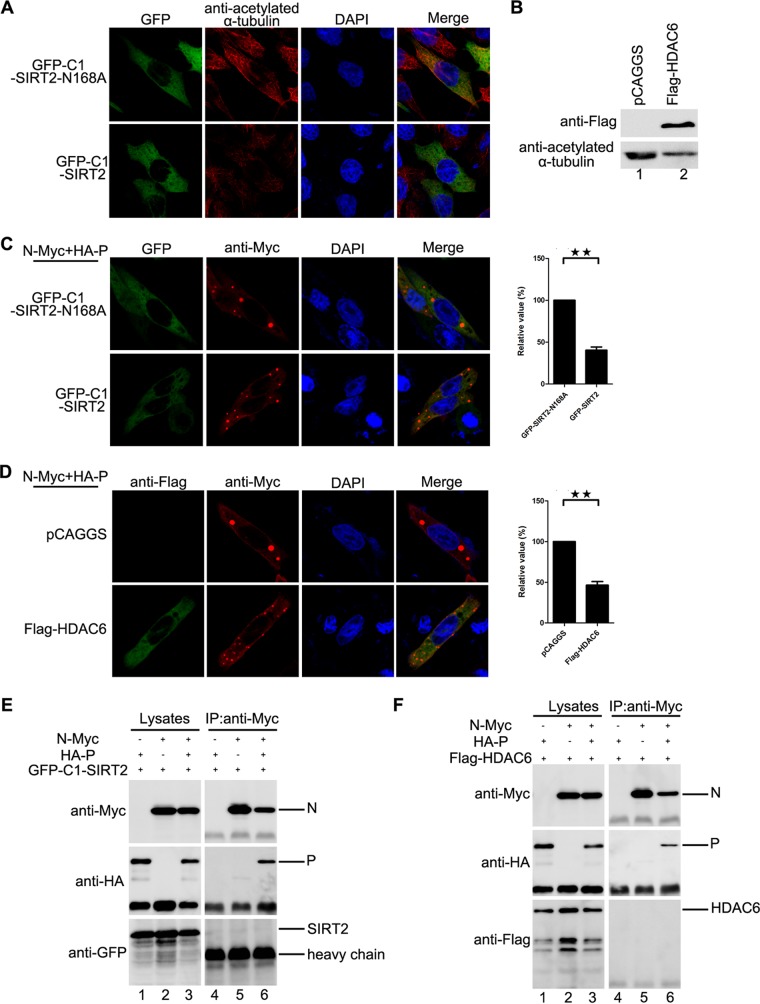
Since we concluded that large IBs were derived from the fusion of small IBs (Fig. 1), we sought to determine which cellular factor(s) regulates the fusion of IBs and how it does so. For this purpose, we transiently coexpressed N and P of HPIV3 in HEK293T cells and used the N-P complex as bait proteins to perform immunoprecipitation (IP). Eluted proteins were separately fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained via silver staining, followed by mass spectrometry (MS). Finally, we found that acetylated α-tubulin, but not α-tubulin, interacted with the N-P complex (Fig. 3A). Further co-IP analysis confirmed that acetylated α-tubulin interacted only with the N-P complex and not with N or P alone (Fig. 3B, lanes 2 to 4). In addition, acetylated α-tubulin interacted with neither NL478A nor P when NL478A and P were coexpressed (Fig. 3B, lane 5), since NL478A-RNA lost the ability to interact with P (13). Moreover, we confirmed that α-tubulin cannot interact with N or P alone or with the N-P complex (Fig. 3C). In addition, we found that acetylated α-tubulin, but not α-tubulin, colocalized with IBs when N and P were coexpressed, and coexpression of NL478A and P did not change the distribution of acetylated α-tubulin (Fig. 3D). Similarly, acetylated α-tubulin colocalized with IBs in HPIV3HA-P-infected HeLa and A549 cells (Fig. 3E and F). Meanwhile, we also found that the HPIV3 infection significantly enhanced the expression of acetylated α-tubulin (Fig. 3G). Taken together, our results show that acetylated α-tubulin interacts with the N-P complex and colocalizes with viral IBs.
FIG 3.
Acetylated α-tubulin interacts with the N-P complex and colocalizes with IBs. (A) Acetylated α-tubulin interaction with the N-P complex identified by IP/MS. HEK293T cells were transfected with plasmids encoding P and N. At 48 h posttransfection, the cells were harvested and subjected to co-IP, followed by silver staining, and the bands of interest were isolated and analyzed by MS. (B) Confirmation of the interaction of acetylated α-tubulin and the N-P complex by co-IP. HEK293T cells were transfected with plasmids encoding P and N individually or jointly. At 48 h posttransfection, the cells were harvested and subjected to co-IP with anti-acetylated α-tubulin antibody. (C) There is no interaction between α-tubulin and the N-P complex. HEK293T cells were transfected with plasmids encoding P and N individually or jointly. At 48 h posttransfection, the cells were harvested and subjected to co-IP assay. IP was performed with anti-α-tubulin antibody. (D) Acetylated α-tubulin colocalizes with IBs. HeLa cells were transfected with plasmids encoding P and N or NL478A. At 24 h posttransfection, cells were fixed and stained with antibody against Myc to visualize the IBs and with antibody against tubulin/acetylated α-tubulin to visualize α-tubulin/acetylated α-tubulin. Images were analyzed via IF microscopy. (E and F) Acetylated α-tubulin colocalizes with IBs in HPIV3HA-P-infected cells. HeLa cells and A549 cells were infected with HPIV3HA-P at an MOI of 1.0 or mock infected. At 24 h postinfection, cells were fixed with HA-P to visualize acetylated α-tubulin. Images were analyzed via IF microscopy. (G) HPIV3 infection increases acetylated α-tubulin expression. HeLa cells were infected with HPIV3 at an MOI of 1. At 12 h postinfection, cells were harvested and the lysates were analyzed via Western blotting. The increased amount of acetylated tubulin was measured and is shown in the graph. Values are means ± SD from three experiments. Student's t test: ***, P < 0.001.
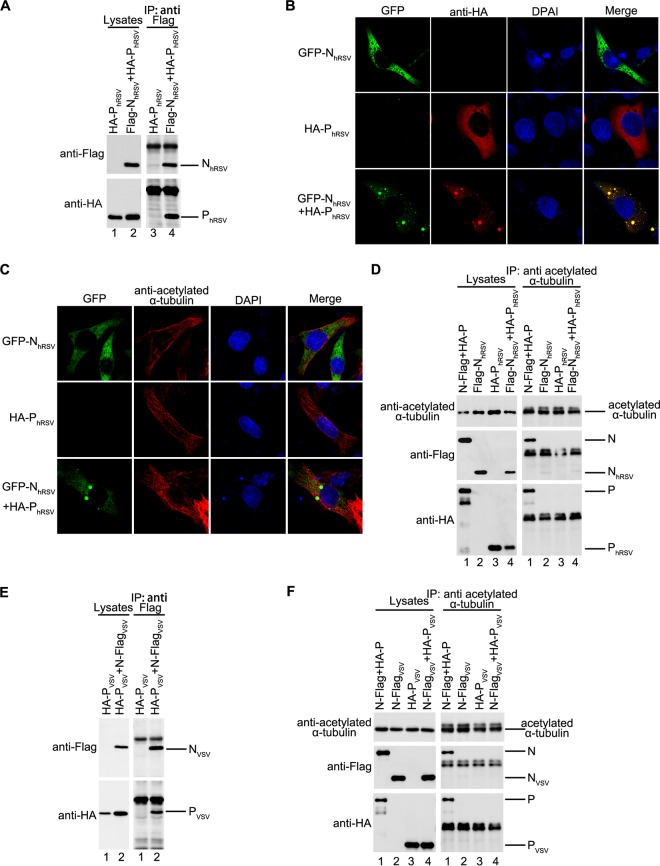
Interaction of acetylated α-tubulin with N-P complex is specific to HPIV3.
Next, we sought to determine whether the interaction of the N-P complex with acetylated α-tubulin is a unique mechanism of HPIV3 or a general feature of NSVs. Therefore, we repeated the immunofluorescence and co-IP assay under the condition of coexpression of N and P of human respiratory syncytial virus (hRSV) or vesicular stomatitis virus (VSV). Although N and P of both hRSV and VSV formed an N-P complex (Fig. 4A and E), acetylated α-tubulin interacted with the N-P complex of neither hRSV nor VSV (Fig. 4D and F). Furthermore, we confirmed that coexpression of N and P of hRSV can form IBs, as reported previously (15) (Fig. 4B). In addition, IB fusion in hRSV occurred as it did in HPIV3 (see Movie S2 in the supplemental material), but acetylated α-tubulin did not colocalize with IBs of hRSV (Fig. 4C). These results show that the association of acetylated α-tubulin with the N-P complex and colocalization with IBs were specific to HPIV3.
FIG 4.
Interaction between acetylated α-tubulin and the N-P complex is unique for HPIV3. (A) Interaction between N and P of RSV. HEK293T cells were transfected with plasmids encoding NRSV and PRSV individually or jointly. At 48 h posttransfection, cells were harvested and subjected to co-IP assay. The IP was performed using anti-Flag polyclonal antibody. (B) Visualization of the N-P complex of RSV. HeLa cells were transfected with plasmids encoding HA-PhRSV and GFP-NhRSV individually or jointly. At 24 h posttransfection, cells were fixed and stained with antibody against HA to visualize P. Images were analyzed by fluorescence microscopy. (C) Acetylated α-tubulin does not colocalize with the N-P complex of RSV. HeLa cells were treated as described for panel B, and immunofluorescence staining with acetylated α-tubulin antibody and Flag antibody was performed. (D) Acetylated α-tubulin interacts with the N-P complex of HPIV3 but not with that of RSV. HEK293T cells were transfected with plasmids encoding Flag-NRSV and HA-PRSV individually or jointly with the HA-P and N-Flag of HPIV3 as a positive control. At 48 h posttransfection, the cells were harvested and subjected to co-IP assay. IP was performed with anti-acetylated α-tubulin antibody. (E) Interaction between N and P of VSV. HEK293T cells were transfected with plasmids encoding N and P of VSV individually or jointly. At 48 h posttransfection, cells were harvested and subjected to co-IP assay. The IP was performed using anti-Flag polyclonal antibody. (F) Acetylated α-tubulin does not interact with the N or P of VSV. HEK293T cells were transfected with plasmids encoding NVSV and PVSV individually or jointly. N and P of HPIV3 were used as a positive control. At 48 h posttransfection, the cells were harvested and subjected to co-IP assay. IP was performed with anti-acetylated α-tubulin antibody.
Acetylated α-tubulin is required for the IB fusion of HPIV3 and enhances viral RNA synthesis.
Having established that acetylated α-tubulin associates with the N-P complex and colocalizes with IBs, we sought to determine whether acetylated α-tubulin regulates the fusion of small IB into large, mature IBs through its interaction with the N-P complex and thus facilitates viral RNA synthesis. For this purpose, first we expressed two deacetylases of α-tubulin, sirtuin 2 (SIRT2) and histone deacetylase 6 (HDAC6), and found that their expression indeed reduced the level of acetylated α-tubulin, but a catalytically inactive mutant of SIRT2, SIRT2-N168A, had no effect on the level of acetylated α-tubulin (Fig. 5A and B). Consistent with levels of acetylated α-tubulin, compared to the expression of SIRT2-N168A, the expression of SIRT2 dramatically decreased the number of large IBs (Fig. 5C). Similarly, the expression of HDAC6 dramatically decreased the number of large IBs (Fig. 5D), indicating that acetylated α-tubulin may be required for the fusion of small IBs into large IBs. Furthermore, the expression of SIRT2 dramatically reduced the fusion of small IBs into large IBs, but the expression of SIRT2-N168A did not (see Movies S3 and S4 in the supplemental material). To rule out the possibility that apart from reducing α-tubulin acetylation, SIRT2 and HDAC6 might also inhibit the fusion of IBs via interacting with the N-P complex directly, we also performed co-IP to examine the interaction of SIRT2 or HDAC6 with the N-P complex. Neither SIRT2 nor HDAC6 could be coimmunoprecipitated by the N-P complex (Fig. 5E and F).
FIG 5.
Acetylated α-tubulin is required for the fusion of IBs of HPIV3. (A) Effect of SIRT2 expression on the level of acetylated α-tubulin. HeLa cells transfected with plasmids encoding GFP-SIRT2-N168A or GFP-SIRT2 were fixed and stained with antibody against acetylated α-tubulin at 24 h posttransfection. Images were analyzed by fluorescence microscopy. (B) Effect of HDAC6 expression on the level of acetylated α-tubulin. HeLa cells transfected with plasmids encoding empty vector or HDAC6 were harvested at 24 h posttransfection, and the acetylated α-tubulin was measured by Western blotting. (C) Effects of SIRT2 expression on the fusion of IBs. HeLa cells transfected with plasmids encoding N, P, GFP-SIRT2-N168A, or GFP-SIRT2 were fixed and stained with antibody against Myc to visualize the IBs at 24 h posttransfection. Images were analyzed via IF microscopy. IB formation was categorized as described for Fig. 1F. The percentage of large IBs in the cells expressing GFP-SIRT2-N168A or GFP-SIRT2 is shown in the graph. Values are means ± SD from three experiments. Student's t test: **, P < 0.01. (D) Effects of HDAC6 expression on the fusion of IBs. HeLa cells transfected with plasmids encoding N, P, empty vector, or HDAC6 were stained with antibody against Myc to visualize the IBs and with antibody against Flag to visualize HDAC6 at 24 h posttransfection. Images were analyzed by IF microscopy. The percentage of large IBs in the cells expressing empty vector or Flag-HDAC6 is shown in the graph. Values are means ± SD from three experiments. Student's t test: **, P < 0.01. (E and F) Neither GFP-SIRT2 nor HDAC6 interacts with the N-P complex in a co-IP assay. HEK293T cells were transfected with plasmids encoding P, N, GFP-SIRT2, or HDAC6. At 48 h posttransfection, the cells were harvested and subjected to co-IP assay as described in Materials and Methods. The IP was performed using anti-Myc antibody and anti-HA antibody, and anti-Myc, anti-GFP antibody, and anti-Flag antibody were used for Western bolts.
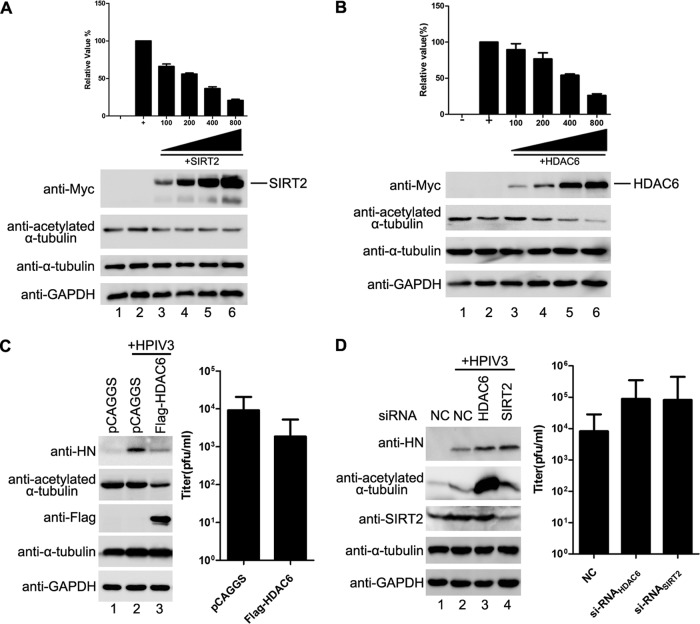
Next, we sought to determine whether acetylated α-tubulin is required for viral RNA synthesis. We first performed an HPIV3 minigenome assay in the presence of Myc-SIRT2 or -HDAC6, and the results showed that the rate of RNA synthesis was lower in SIRT2- or HDAC6-expressing cells than in cells expressing empty vectors, with a significant dose-dependent effect (Fig. 6A and B). In addition, the expression of HDAC6 also reduced viral hemagglutinin-neuraminidase (HN) protein expression (Fig. 6C, left panel) and viral titer (Fig. 6C, right panel) in HPIV3-infected cells.
FIG 6.
Acetylated α-tubulin is required for viral RNA synthesis by HPIV3. (A) Effect of SIRT2 expression on RNA synthesis of the HPIV3 minigenome. Plasmids encoding N, P, L, minigenome, and SIRT2 (100, 200, 400, and 800 ng) were transfected into vTF7-3-infected HeLa cells. Relative luciferase activity and protein expression were analyzed according to the manufacturer's instructions. Values are means ± SD from three experiments. (B) Effects of HDAC6 expression on RNA synthesis of the HPIV3 minigenome. vTF7-3-infected HeLa cells were transfected with plasmids encoding N, P, L, minigenome, and HDAC6 (100, 200, 400, and 800 ng). Relative luciferase activity and protein expression were detected as described for panel A. (C) Effects of HDAC6 expression on RNA synthesis in HPIV3-infected cells. HeLa cells were transfected with empty vector or plasmid encoding HDAC6 for 24 h. Cells were then infected with HPIV3 at an MOI of 1.0. At 24 h postinfection, cells were harvested and analyzed for viral HN expression via Western blotting (left) and virions production (right). (D) Effects of HDAC6 and SIRT2 depletion on RNA synthesis in HPIV3-infected cells. Negative-control siRNA-, siHDAC6-, and siSIRT2-transfected HeLa cells were infected with HPIV3 (MOI = 1). At 24 h postinfection, cell lysates were analyzed for expression of HN, HDAC6, and SIRT2 via Western blotting (left) and the medium were analyzed for production of virions (right).
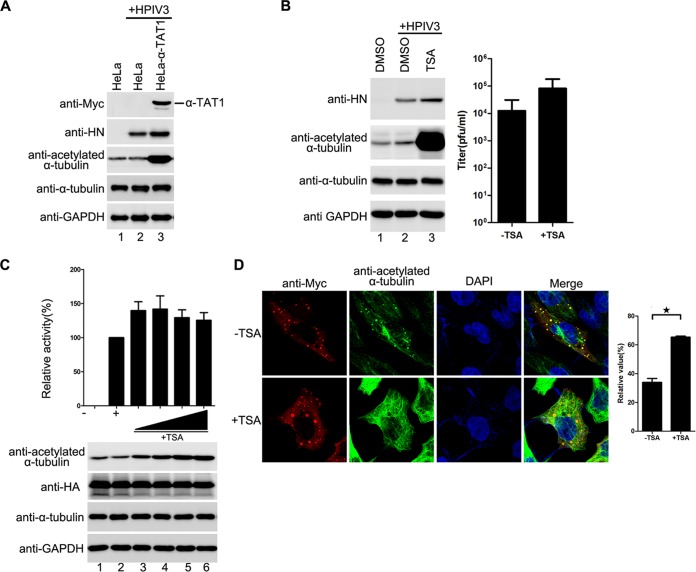
Having confirmed that exogenous expression of SIRT2 and HDAC6 reduced viral RNA synthesis, we sought to determine whether depletion of endogenous SIRT2 or HDAC6 would have the opposite effect. Thus, we knocked down SIRT2 or HDAC6 by RNA interference (RNAi) in HPIV3-infected cells, which resulted in elevated levels of α-tubulin acetylation and increased viral HN protein synthesis (Fig. 6D, left panel) and viral titer (Fig. 6D, left panel). To exclude possibilities by which SIRT2 or HDAC6 could mediate viral RNA synthesis other than by regulating the deacetylation of α-tubulin and to confirm that acetylated α-tubulin indeed contributes to viral RNA synthesis, we also established an α-tubulin acetyltransferase 1 (α-TAT1) stably expressed HeLa cell line, as α-TAT1 is considered to be a major α-tubulin acetyltransferase in mammalian cells. Consistent with depletion of HDAC6 and SIRT2, acetylated α-tubulin and viral HN protein levels were higher in stably α-TAT1-expressing cells than in normal HeLa cells (Fig. 7A).
FIG 7.
An increased level of acetylated α-tubulin promotes the fusion of IBs and viral RNA synthesis by HPIV3. (A) Effects of stable expression of α-TAT1 on RNA synthesis in HPIV3-infected cells. Normal HeLa cells or HeLa cells stably expressing α-TAT1 were infected with HPIV3 at an MOI of 1.0. At 24 h postinfection, viral HN was measured via Western blotting. The data shown represent three independent experiments. (B) Effects of TSA treatment on RNA synthesis in HPIV3-infected cells. HeLa cells treated with 2 μM TSA or mock treated for 12 h were infected with HPIV3 at an MOI of 1. At 12 h postinfection, cells were harvested, and viral HN was measured via Western blotting. In addition, the medium was analyzed for production of virions (right). (C) Effect of TSA treatment on RNA synthesis in the HPIV3 minigenome. Plasmids encoding N, P, L, and the minigenome were transfected into vTF7-3-infected HeLa cells. At 5 h posttransfection, the transfection medium was replaced with 30, 60, 120, or 240 nM TSA or mock treated. At 24 h postinfection, relative luciferase activity and protein expression were detected as described for Fig. 6A. Values are means ± SD from three experiments. (D) Effects of TSA treatment on the fusion of IBs. HeLa cells were transfected with plasmids encoding N and P. At 5 h posttransfection, the transfection medium was replaced with 60 nM TSA or mock treated. At 5 h posttransfection, cells were fixed and stained with antibody against Myc to visualize the IBs and with antibody against acetylated α-tubulin to visualize acetylated α-tubulin. Images were analyzed via IF microscopy. IB formation was categorized as described for Fig. 1F. Values are means ± SD from three experiments. Student's t test: *, P < 0.05.
Similarly, when cells were treated with trichostatin A (TSA), an inhibitor of the HDAC family, the acetylated α-tubulin and viral HN protein levels (Fig. 7B, left panel), viral titer (Fig. 7B, right panel), and RNA synthesis level were higher in TSA-treated cells than in mock-treated cells (Fig. 7C). Accordingly, IBs were much larger in TSA-treated cells than in mock-treated cells, in a dose-dependent manner (Fig. 7D).
Acetylated α-tubulin is dispensable for the fusion of IBs and viral RNA synthesis of RSV.
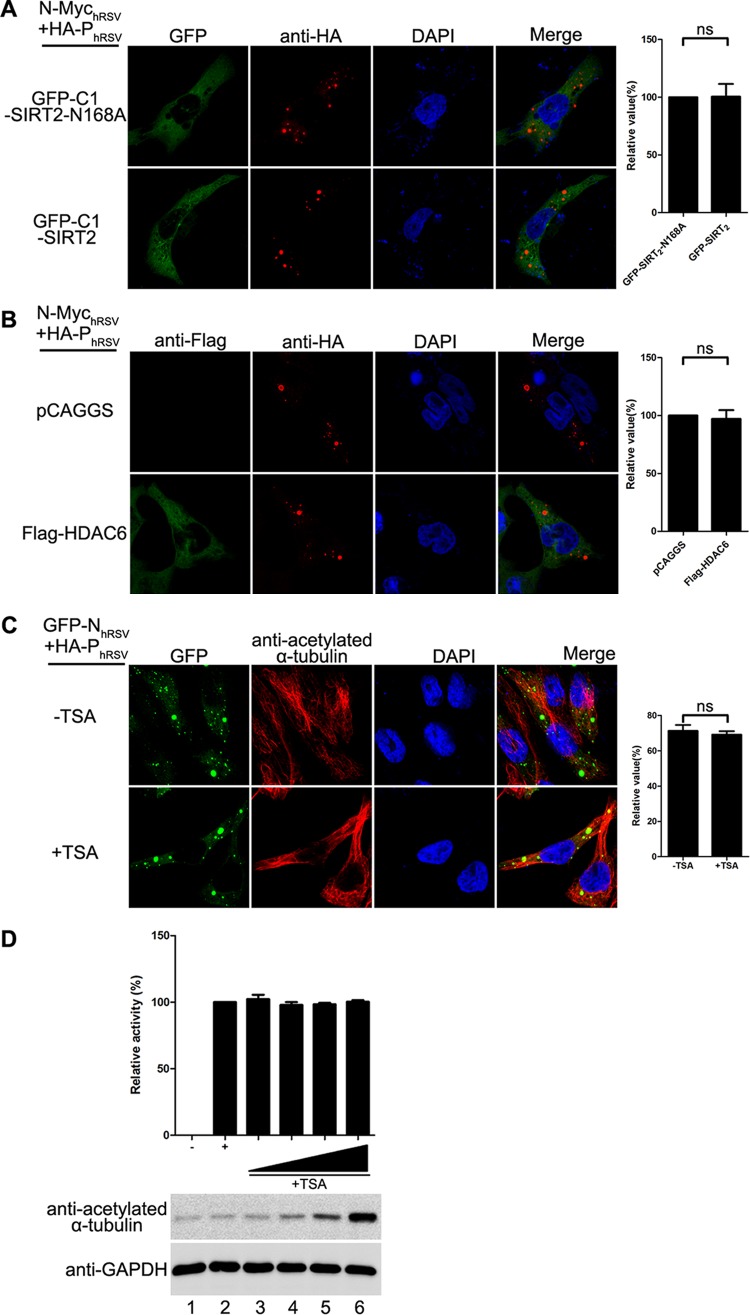
Next, we repeated the same examination for RSV IB formation and RNA synthesis. Although the IBs of RSV also underwent fusion, the reduction of acetylated α-tubulin caused by SIRT2 and HDAC6 had no effect on the fusion of the RSV IBs (Fig. 8A and B), suggesting that reliance on acetylated α-tubulin for the fusion of IBs was specific to HPIV3. Meanwhile, the increased level of acetylated α-tubulin induced by TSA treatment had no effect on the size of IBs and RNA synthesis of RSV (Fig. 8C and D). Taken together, our results demonstrate that it is a specific feature of HPIV3 that acetylated α-tubulin is required for the fusion of IBs and RNA synthesis.
FIG 8.
Acetylated α-tubulin is dispensable for the fusion of IBs and viral RNA synthesis of RSV. (A and B) Effect of SIRT2 (A) or HDAC6 (B) expression on the IB fusion of RSV. HeLa cells transfected with plasmids encoding HA-PhRSV, N-MychRSV GFP-SIRT2-N168A, GFP-SIRT2, Flag-HDAC6 or vector were fixed and stained with antibody against HA and Flag at 24 h posttransfection. Images were analyzed by fluorescence microscopy. Values are means ± SD from three experiments. Student's t test: ns, nonsignificant. (C) Effects of TSA on the IB fusion of RSV. HeLa cells were transfected with plasmids encoding GFP-N or P of RSV. At 5 h posttransfection, the transfection medium was replaced with 60 nM TSA or mock treated. At 24 h posttransfection, cells were fixed and stained with antibody against Myc to visualize the IBs. Images were analyzed by fluorescence microscopy. IB fusion was categorized as described for Fig. 1F. Values are means ± SD from three experiments. Student's t test: ns, nonsignificant. (D) Effects of TSA on RNA synthesis by RSV. BSR-T7 cells infected with vTF7-3 were transfected with plasmids encoding N, P, L, M2-1, and the minigenome of RSV. At 24 h posttransfection, the medium was replaced with 0.25, 0.5, 1.0, or 2.0 μM TSA or mock treated. At 48 h posttransfection, cells were harvested, and the relative CAT activity was detected according to the manufacturer's instructions. Values are mean ± SD from three experiments.
DISCUSSION
Our previous study showed that N-P interaction is the minimal requirement for the formation of IBs and that IBs contain viral RNA, N, P, and polymerase in HPIV3-infected cells (13). Here, we further demonstrated that large IBs derive from small IBs via a kinetic fusion process, which is critical for viral RNA synthesis. Therefore, we conclude that some cellular proteins may regulate IB fusion and viral RNA synthesis via interaction with the N-P complex of HPIV3. Indeed, we found that acetylated α-tubulin interacted with the N-P complex, IBs colocalized with acetylated α-tubulin, and viral RNAs were colocalized in IBs. Our findings provide evidence that acetylated α-tubulin regulates the fusion of IBs via interaction with the N-P complex and enhances viral RNA synthesis. Suppression of acetylation of α-tubulin by overexpressing deacetylases HDAC6 and SIRT2 profoundly inhibited the fusion of small IBs and viral RNA synthesis. Conversely, increasing acetylation of α-tubulin using the pharmacological inhibitor TSA or RNAi knockdown of HDAC6 and SIRT2 or by expressing α-TAT1 resulted in the fusion of small IBs into large IBs and effectively promoted viral RNA synthesis. In conclusion, our results reveal that fusion of HPIV3 IBs regulated by acetylated α-tubulin is required for viral RNA synthesis and suggest that acetylated α-tubulin may be involved in viral RNA synthesis by acting as a scaffolding protein, forming a platform that may be critical in N-P complex interaction with other cellular proteins.
We found that the acetylated tubulin modulation of N-P-induced IB formation may be specific to HPIV3: (i) acetylated α-tubulin, not α-tubulin, interacts with the N-P complex and colocalizes with IBs of HPIV3, (ii) acetylated α-tubulin was unable to interact with the N-P complexes of hRSV and VSV and did not colocalize with IBs of hRSV, and (iii) variations in the degree of α-tubulin acetylation had no effect on the RNA synthesis of hRSV. All these data suggest that paramyxoviruses may use different cellular elements inside or outside the cytoskeleton for replication, which supports the following findings from previous studies: (i) genome transcription of hRSV requires actin and can be further enhanced by an actin-modulatory protein, profilin (16), and (ii) no colocalization of actin or tubulin and individual measles virus (MeV) proteins was observed, although actin is packaged within the infectious particles of both MeV and Sendai virus (17).
Acetylated microtubules are generally thought to be more stable than unmodified microtubules, but no direct evidence has shown that acetylation modification of microtubules directly influences tubulin polymerization or depolymerization kinetics in vitro (18). In addition, no clear differences in microtubule structure or tubulin conformation have been found between electron cryomicroscopy reconstructions of maximally deacetylated or acetylated microtubules (19), and the levels at which kinesin-1 binds to acetylated microtubules and deacetylated microtubules are similar (20, 21). These findings require further investigation into the mechanism by which acetylation of α-tubulin enhances the fusion of IBs of HPIV3 via interaction with the N-P complex. It is possible that the N-P complex recognizes modified αK40 and gains access to the luminal surface of microtubules and then mediates N-P complex interaction with cellular proteins that bind directly to the lumen of the microtubules, where these cellular proteins assist IB fusion for effective viral RNA synthesis. Although the identity of these cellular proteins is unknown, the presence of luminal particles has been shown (22). In addition, as acetylated α-tubulin associates predominantly with the membrane by directly interacting with the transport protein Na+,K+-ATPase (23), it is also possible that acetylated α-tubulin pulls the N-P complex into the intracellular membranes, which provides a platform for efficient IB fusion and subsequent RNA synthesis. This scenario resembles findings of a recent study that surprisingly showed that reovirus, a nonenveloped, double-stranded RNA virus, forms IBs that recruit cell membranes for viral replication (24). Additional experiments will be needed to test these possibilities further.
Without doubt, acetylation modification indeed endows microtubules with new features, and several reports have shown the roles of acetylated microtubules in virus infection: herpesvirus and adenovirus induce microtubule acetylation to facilitate viral entry (25, 26); influenza A virus infection enhances the levels of acetylated α-tubulin to facilitate the transport of viral components (27); and HIV-1 induces the acetylation of microtubules to promote infection, and the overexpression or depletion of HDAC6 inhibits or enhances HIV-1 infection, respectively (28, 29). However, the direct role of acetylation modification in virus infection remains unclear.
Here we identify an entirely new and unique role for the acetylation of α-tubulin as a regulator of RNA synthesis through its ability to enhance the fusion of IBs of HPIV3. Our future research will focus on identifying cellular proteins which are involved in acetylated α-tubulin mediated fusion of viral IBs.
MATERIALS AND METHODS
Cell cultures and virus.
HeLa, A549, HEK293T, and LLC-MK2 cells (China Center for Type Culture Collection [CCTCC]) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco) containing 8% fetal bovine serum (FBS) (Gibco) at 37°C. 293T-T7 cells (derived from 293T cells expressing T7 polymerase in this study) and HeLa-αTAT1 cells (derived from HeLa cells expressing α-TAT1 in this study) were maintained in DMEM containing 8% FBS and 1 μg/ml puromycin. BSR-T7 cells (Harbin Veterinary Research Institute, CAAS) were maintained in DMEM containing 8% FBS and 1 μg/ml G418. HPIV3 (NIH 47885) and HPIV3HA-P were amplified in LLC-MK2 cells as described previously (13). Recombinant vaccinia virus (vTF7-3) expressing T7 RNA polymerase was amplified in HeLa cells by inoculation at a multiplicity of infection (MOI) of 0.1.
Plasmid constructs.
The plasmids pOCUS-HPIV3, pGEM4-N, pGEM4-P, and pGEM4-L, plasmids encoding the HPIV3 minigenome, HA-tagged P of HPIV3, Flag-tagged N, Myc-tagged N, and its mutant NL478A of HPIV3, and HA-tagged P and Flag-tagged N of VSV have been described previously (13, 30). The construct encoding P of HPIV3 with an mCherry epitope tag at its N terminus was generated by PCR-based cloning techniques and cloned into pCAGGS. The constructs encoding N of HPIV3 with a GFP epitope were generated by PCR-based cloning techniques and cloned into pCAGGS. cDNAs encoding P with an HA tag, N with a Flag and GFP tag, L protein, and M2-1 were amplified from the RSV-infected cells via reverse transcription-PCR and cloned into pCAGGS, pBS, pGADT7, and pGBKT7, respectively. Plasmid encoding HDAC6 was amplified from HeLa cells via reverse transcription-PCR and cloned into pCAGGS and pGBKT7, respectively. GFP-SIRT2 and GFP-SIRT2-N168A were kindly provided by E. M. Verdin (Gladstone Institute of Cardiovascular Disease, University of California). The cDNA of HDAC6 or SIRT2 was amplified by PCR using plasmids encoding Flag-HDAC6 and GFP-SIRT2 as templates, and PCR products were cloned into pGBKT7. The cDNA of α-TAT1 was amplified by PCR with plasmids pEF5B-FRT-GFP-αTAT1 (purchased from Addgene, plasmid 27099) as a template, and PCR products were cloned into pCAGGS and pHAGE-CMV-MCS-IRES, respectively. Plasmids carrying the HPIV3 genome with an HA tag fused to the N terminus of the P gene were constructed as described previously (13). All constructs were verified by DNA sequencing.
In vivo minigenome assay of HPIV3.
The in vivo minigenome assay was performed as described by Hoffman and Banerjee (31) with minor modifications. Briefly, HeLa cell monolayers grown on 12-well plates were infected with vTF7-3 at 37°C for 1 h. The plasmids pcDNA3.0-N (250 ng), pGADT7-P (62.5 ng), and pGEM4-L (100 ng), and the plasmid carrying the minigenome (50 ng) of HPIV3 were transfected with increasing amounts of glutathione S-transferase (GST), or GST-PN40, HA-M, pGBDT7-HDAC6, or pGBDT7-SIRT2 (100 ng, 200 ng, 400 ng, and 800 ng) by using Lipofectamine 2000 (Invitrogen). Total amounts of plasmid were always kept constant. At 5 h posttransfection, the transfection medium was replaced by DMEM contains 4% FBS. At 24 h posttransfection, cells were lysed in 150 μl of lysis buffer, from which 20-μl aliquots were used to determine luciferase activity according to the manufacturer's instructions (luciferase assay kit; Roche). All assays were repeated at least three times for accuracy.
In vivo minigenome assay of RSV.
BSR-T7 cell monolayers grown on 6-well plates were infected with vTF7-3 at 37°C for 1 h. The plasmids pGBDT7-N (1.0 μg), pGADT7-P (1.0 μg), pBS-L (0.25 μg), pBS-minigenome-CAT (1.0 μg), and pGBDT7-M2-1 (1.5 μg), encoding N, P, L, M2-1, and RSV minigenome, respectively, were transfected with Lipofectamine 2000 (Invitrogen). At 5 h posttransfection, the transfection medium was replaced by DMEM containing 4% FBS. At 24 h posttransfection, the medium was replaced by DMEM containing 4% FBS with 0.25, 0.5, 1.0, or 2.0 μM TSA or mock treated. At 48 h posttransfection, cells were harvested, and the cell lysates were subjected to a chloramphenicol acetyltransferase (CAT) enzyme-linked immunosorbent assay (ELISA) for detection of the relative CAT activity as described by Chen et al. (30). All assays were repeated at least three times for accuracy.
In vivo co-IP.
Appropriate plasmids were transfected into 293T cells via calcium phosphate transfection. At 48 h posttransfection, the cells were harvested and prepared as described by Zhang et al. (13). The cell lysate was precleaned with protein G-Sepharose 4 Fast Flow medium. Antibody-protein complexes were purified with protein G beads and washed with washing buffer (5% [wt/vol] sucrose, 5 mM Tris-HCl [pH 7.4], 5 mM EDTA [pH 8.0], 500 mM NaCl, and 1% [vol/vol] Triton X-100) three times. After boiling in SDS sample buffer, the proteins were separated via SDS-PAGE and processed for immunoblotting.
Silver staining.
HA-P and/or N-Myc was transfected into 293T cells in 10-cm dishes. At 48 h posttransfection, cells were harvested and prepared for coimmunoprecipitation as described above. The proteins were subjected to SDS-PAGE, and then the gel was fixed in 50% methanol (MeOH)–10% acetic acid for 10 min, followed by washing in double-distilled water (ddH2O) for 30 min. The gel was then incubated with dithiothreitol (DTT) (32.5 μl of 0.1 M DTT in 100 ml ddH2O) for 10 min, rinsed with ddH2O 2 times, and submerged in 0.1% silver nitrate for 10 min. The gel was then rinsed briefly with ddH2O, the developer was added quickly. and the gel was rocked in the developer until bands appeared. The reaction was stopped with 2.3 M citric acid, and the gel was rocked in ddH2O.
Immunofluorescence analysis.
HeLa cells were fixed with 4% paraformaldehyde for 20 min at room temperature and then permeabilized with 0.2% Triton X-100 for 20 min at room temperature. After blocking with 3% bovine serum albumin in phosphate-buffered saline, cells were immunostained with appropriate primary antibodies and incubated for 1 h. The cells were then washed with 1% bovine serum albumin and incubated with goat anti-mouse immunoglobulin G-fluorescein or goat anti-rabbit immunoglobulin G-rhodamine secondary antibody for 1 h. Nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole). Images were acquired with an Olympus confocal FV1000 microscope. For live-cell imaging, HeLa cells were seeded into 20-mm dishes 24 h prior to transfection and then transfected with GFP-N and mCherry-P of HPIV3 or GFP-N and HA-P of RSV with or without GFP-SIRT2N168A or GFP-SIRT2. At 12 h posttransfection, cells incubated in a 37°C and 5% CO2 environment were visualized on a Leica fluorescence microscope using a 63× oil immersion objective with a temporal resolution of 15 s per picture.
FISH.
Fluorescence in situ hybridization (FISH) was performed by using 3′-biotinylated oligonucleotides complementary to specific sequences of viral RNA. The probe used to detect plus-strand RNA hybridizes the antigenome and N mRNA. The probe used to detect the antigenome was designed in order to avoid the detection of the mRNA, whereas the probe used to detect the genome was a mixture of two probes designed to hybridize to the leader RNA and N gene of genome. The sequences TCTCAAGGGAAAAGAACTGGAAGATCAGTTACAGAAATTAGG and ATGTTGAGCCTATTTGATACATTTAATGCACGTAGGCAAGAA hybridized to the genome and the N gene of genome, respectively, and were used to detect the genome of HPIV3. The sequences TTACATGTTTTTCTTACTTTTTGTCTATCTTTAGGCTTAATG and TTCTTGCCTACGTGCATTAAATGTATCAAATAGGCTCAACAT hybridized to the antigenome and N mRNA, respectively, and were used to detect the antigenome and plus-strand RNA.
The FISH experiments were performed as described by Lahaye et al. (11). Briefly, HPIV3HA-P-infected cells in 24-well plates grown on coverslips were fixed with 2% formaldehyde for 10 min and permeabilized with 0.1% Triton X-100 for 10 min at room temperature. After treatment with 200 U of DNase I/ml for 30 min at 37°C, cells were prehybridized in hybridization buffer (50% formamide, 10% dextran sulfate, 4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) for 30 min at 37°C before being incubated with 200 μl of hybridization mix containing 200 ng of biotinylated probe, 200 μg of herring testis DNA, and 140 μl of hybridization buffer. Coverslips were then put in a humidified hybridization chamber at 37°C overnight and washed three times for 5 min with 2× SSC and then three times with 0.5× SSC. Cells were then processed for immunofluorescence staining. The P protein was stained with a mouse anti-HA monoclonal antibody (MAb) (Sigma; 1:2,000), followed by incubation with goat anti-mouse IgG-fluorescein (Thermo; 1:200) secondary antibody for 2 h at room temperature. Viral biotinylated RNA was detected by incubation with streptavidin conjugated to cyanin 3 (Cy3).
RNAi.
Negative-control small interfering RNAs (siRNAs) and siRNAs targeted SIRT2 and HDAC6 were transfected into HeLa cells grown on 6-well plates using Lipofectamine 2000 (Invitrogen). At 5 h posttransfection, the transfection medium was replaced by DMEM containing 8% FBS. At 24 h posttransfection, the medium was replaced and cells were infected by HPIV3 at an MOI of 1. At 48 h posttransfection, cells were harvested and lysed. Cell lysates were subjected to Western blotting for detection of the viral protein. The duplex nucleotides of siRNA specific for the mRNAs of HDAC6 and SIRT2 and a control siRNA were purchased from Genepharma, Shanghai, China. The target sequences for SIRT2 and HDAC6 were CTAAGCTGGATGAAAGAGAA and GGTAATGGAACTCAGCACA, respectively.
α-TAT1 stable expression cell line.
293T cells in 10-cm dishes were transfected with pHAGE-CMV-MCS-IRES-α-TAT1, psPAX2, and pMD2G. At 48 h posttransfection, the medium was harvested and filtered with a 0.22-μm filter. The clarified supernatant was used to infect HeLa cells monolayers in 6-well plates, and then cells that stably express α-TAT1 were selected by addition of puromycin at a final concentration of 1 μg/ml.
Transfection and recovery of recombinant HPIV3.
293-T7 cells in 6-well plates, grown to 40% confluence, were transfected with pGEM4-N (400 ng), pGEM4-P (400 ng), pGEM4-L (200 ng), and pOCUS-HPIV3HA-P (4 g) via calcium phosphate transfection at 37°C. Four days later, the cells were frozen, thawed, scraped, and centrifuged at 5,000 rpm at 4°C for 5 min. The clarified supernatant was collected and layered onto LLC-MK2 cell monolayers for amplification for 4 days. The LLC-MK2 cells then were harvested, and viral titers were determined. A single recovered recombinant HPIV3 isolated by being picked up as an agar plug during the titer determination was used to infect fresh LLC-MK2 cells for amplification.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (grants 31630086, 81471939, and 81271816).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01802-16.
REFERENCES
- 1.Collins PL, Chanock RM, McIntosh K. 1996. Parainfluenza viruses, p 1205–1241. In Knipe DM, Howley PM (ed), Fields virology. Lippincott-Raven Publications, Philadelphia, PA. [Google Scholar]
- 2.Myers TM, Pieters A, Moyer SA. 1997. A highly conserved region of the Sendai virus nucleocapsid protein contributes to the NP-NP binding domain. Virology 229:322–335. doi: 10.1006/viro.1996.8429. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Glendening C, Linke H, Parks CL, Brooks C, Udem SA, Oglesbee M. 2002. Identification and characterization of a regulatory domain on the carboxyl terminus of the measles virus nucleocapsid protein. J Virol 76:8737–8746. doi: 10.1128/JVI.76.17.8737-8746.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curran J, Marq JB, Kolakofsky D. 1995. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J Virol 69:849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howard M, Wertz G. 1989. Vesicular stomatitis virus RNA replication: a role for the NS protein. J Gen Virol 70:2683–2694. doi: 10.1099/0022-1317-70-10-2683. [DOI] [PubMed] [Google Scholar]
- 6.Chen M, Ogino T, Banerjee AK. 2007. Interaction of vesicular stomatitis virus P and N proteins: identification of two overlapping domains at the N terminus of P that are involved in N0-P complex formation and encapsidation of viral genome RNA. J Virol 81:13478–13485. doi: 10.1128/JVI.01244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De BP, Hoffman MA, Choudhary S, Huntley CC, Banerjee AK. 2000. Role of NH(2)- and COOH-terminal domains of the P protein of human parainfluenza virus type 3 in transcription and replication. J Virol 74:5886–5895. doi: 10.1128/JVI.74.13.5886-5895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhary SK, Malur AG, Huo Y, De BP, Banerjee AK. 2002. Characterization of the oligomerization domain of the phosphoprotein of human parainfluenza virus type 3. Virology 302:373–382. doi: 10.1006/viro.2002.1668. [DOI] [PubMed] [Google Scholar]
- 9.Netherton C, Moffat K, Brooks E, Wileman T. 2007. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv Virus Res 70:101–182. doi: 10.1016/S0065-3527(07)70004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia J, Garcia-Barreno B, Vivo A, Melero JA. 1993. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 195:243–247. doi: 10.1006/viro.1993.1366. [DOI] [PubMed] [Google Scholar]
- 11.Lahaye X, Vidy A, Pomier C, Obiang L, Harper F, Gaudin Y, Blondel D. 2009. Functional characterization of Negri bodies (NBs) in rabies virus-infected cells: evidence that NBs are sites of viral transcription and replication. J Virol 83:7948–7958. doi: 10.1128/JVI.00554-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derdowski A, Peters TR, Glover N, Qian R, Utley TJ, Burnett A, Williams JV, Spearman P, Crowe JE Jr. 2008. Human metapneumovirus nucleoprotein and phosphoprotein interact and provide the minimal requirements for inclusion body formation. J Gen Virol 89:2698–2708. doi: 10.1099/vir.0.2008/004051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang S, Chen L, Zhang G, Yan Q, Yang X, Ding B, Tang Q, Sun S, Hu Z, Chen M. 2013. An amino acid of human parainfluenza virus type 3 nucleoprotein is critical for template function and cytoplasmic inclusion body formation. J Virol 87:12457–12470. doi: 10.1128/JVI.01565-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoenen T, Shabman RS, Groseth A, Herwig A, Weber M, Schudt G, Dolnik O, Basler CF, Becker S, Feldmann H. 2012. Inclusion bodies are a site of ebolavirus replication. J Virol 86:11779–11788. doi: 10.1128/JVI.01525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lifland AW, Jung J, Alonas E, Zurla C, Crowe JE Jr, Santangelo PJ. 2012. Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. J Virol 86:8245–8258. doi: 10.1128/JVI.00215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burke E, Mahoney NM, Almo SC, Barik S. 2000. Profilin is required for optimal actin-dependent transcription of respiratory syncytial virus genome RNA. J Virol 74:669–675. doi: 10.1128/JVI.74.2.669-675.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyrrell DL, Norrby E. 1978. Structural polypeptides of measles virus. J Gen Virol 39:219–229. doi: 10.1099/0022-1317-39-2-219. [DOI] [PubMed] [Google Scholar]
- 18.Maruta H, Greer K, Rosenbaum JL. 1986. The acetylation of alpha-tubulin and its relationship to the assembly and disassembly of microtubules. J Cell Biol 103:571–579. doi: 10.1083/jcb.103.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howes SC, Alushin GM, Shida T, Nachury MV, Nogales E. 2014. Effects of tubulin acetylation and tubulin acetyltransferase binding on microtubule structure. Mol Biol Cell 25:257–266. doi: 10.1091/mbc.E13-07-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soppina V, Herbstman JF, Skiniotis G, Verhey KJ. 2012. Luminal localization of alpha-tubulin K40 acetylation by cryo-EM analysis of fab-labeled microtubules. PLoS One 7:e48204. doi: 10.1371/journal.pone.0048204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter WJ, Beranek V, Fischermeier E, Diez S. 2012. Tubulin acetylation alone does not affect kinesin-1 velocity and run length in vitro. PLoS One 7:e42218. doi: 10.1371/journal.pone.0042218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouchet-Marquis C, Zuber B, Glynn AM, Eltsov M, Grabenbauer M, Goldie KN, Thomas D, Frangakis AS, Dubochet J, Chretien D. 2007. Visualization of cell microtubules in their native state. Biol Cell 99:45–53. doi: 10.1042/BC20060081. [DOI] [PubMed] [Google Scholar]
- 23.Casale CH, Previtali G, Barra HS. 2003. Involvement of acetylated tubulin in the regulation of Na+,K+-ATPase activity in cultured astrocytes. FEBS Lett 534:115–118. doi: 10.1016/S0014-5793(02)03802-4. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez de Castro I, Zamora PF, Ooms L, Fernandez JJ, Lai CM, Mainou BA, Dermody TS, Risco C. 2014. Reovirus forms neo-organelles for progeny particle assembly within reorganized cell membranes. mBio 5:e00931-13. doi: 10.1128/mBio.00931-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naranatt PP, Krishnan HH, Smith MS, Chandran B. 2005. Kaposi's sarcoma-associated herpesvirus modulates microtubule dynamics via RhoA-GTP-diaphanous 2 signaling and utilizes the dynein motors to deliver its DNA to the nucleus. J Virol 79:1191–1206. doi: 10.1128/JVI.79.2.1191-1206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warren JC, Rutkowski A, Cassimeris L. 2006. Infection with replication-deficient adenovirus induces changes in the dynamic instability of host cell microtubules. Mol Biol Cell 17:3557–3568. doi: 10.1091/mbc.E05-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Husain M, Harrod KS. 2011. Enhanced acetylation of alpha-tubulin in influenza A virus infected epithelial cells. FEBS Lett 585:128–132. doi: 10.1016/j.febslet.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 28.Valenzuela-Fernandez A, Alvarez S, Gordon-Alonso M, Barrero M, Ursa A, Cabrero JR, Fernandez G, Naranjo-Suarez S, Yanez-Mo M, Serrador JM, Munoz-Fernandez MA, Sanchez-Madrid F. 2005. Histone deacetylase 6 regulates human immunodeficiency virus type 1 infection. Mol Biol Cell 16:5445–5454. doi: 10.1091/mbc.E05-04-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabo Y, Walsh D, Barry DS, Tinaztepe S, de Los Santos K, Goff SP, Gundersen GG, Naghavi MH. 2013. HIV-1 induces the formation of stable microtubules to enhance early infection. Cell Host Microbe 14:535–546. doi: 10.1016/j.chom.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Zhang S, Banerjee AK, Chen M. 2013. N-terminal phosphorylation of phosphoprotein of vesicular stomatitis virus is required for preventing nucleoprotein from binding to cellular RNAs and for functional template formation. J Virol 87:3177–3186. doi: 10.1128/JVI.02761-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffman MA, Banerjee AK. 2000. Precise mapping of the replication and transcription promoters of human parainfluenza virus type 3. Virology 269:201–211. doi: 10.1006/viro.2000.0223. [DOI] [PubMed] [Google Scholar]
- 32.Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol Cell 97:147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.