Abstract
TRAF6 (tumor necrosis factor receptor-associated factor 6) is a RING (really interesting new gene) domain ubiquitin (Ub) ligase that mediates the activation of protein kinases, such as transforming growth factor β-activated kinase (TAK1) and IκB kinase (IKK), by catalyzing the formation of a unique polyubiquitin chain linked through Lys-63 of Ub. Here, we present evidence that TIFA (TRAF-interacting protein with a forkhead-associated domain, also known as T2BP) activates IKK by promoting the oligomerization and Ub ligase activity of TRAF6. We show that recombinant TIFA protein, but not TRAF6-binding-defective mutant, can activate IKK in crude cytosolic extracts. Furthermore, TIFA activates IKK in an in vitro reconstitution system consisting of purified proteins, including TRAF6, the TAK1 kinase complex, and Ub-conjugating enzyme complex Ubc13–Uev1A. Interestingly, a fraction of recombinant TIFA protein exists as high-molecular-weight oligomers, and only these oligomeric forms of TIFA can activate IKK. Importantly, TIFA induces the oligomerization and polyubiquitination of TRAF6, which leads to the activation of TAK1 and IKK through a proteasome-independent mechanism.
TRAFs [tumor necrosis factor (TNF) receptor (TNFR)-associated factors] are a family of proteins that participate in signal transduction of TNFR and IL-1 receptor (IL-1R)/toll-like receptor (TLR) superfamilies (1–5). A wide range of biological functions, such as inflammatory response, immunity, and bone metabolism, rely on the proper functions of TRAF proteins to activate transcription factors, including NF-κB, c-JUN, and activating transcription factor 2 (ATF-2). Seven members of the TRAF family have been identified recently in mammals. All TRAF proteins except TRAF7 contain a highly conserved TRAF/MATH (meprin and TRAF-homology) domain at the C terminus, which mediates homotypic and heterotypic TRAF–TRAF interactions, as well as their association with members of the TNFR and IL-1R–TLR receptor superfamilies. Two additional functional domains, the zinc finger domain and the RING domain, are located at the N terminus of TRAF2–TRAF7.
The RING domain, which uses a unique cross-brace arrangement to chelate zinc, is important for downstream signaling by TRAF proteins (1, 6, 7). Many RING domain proteins have been shown recently to function as ubiquitin (Ub) ligases (E3) that mediate polyubiquitination of target proteins, which are subsequently degraded by the 26S proteasome (8). Unlike most E3s, the primary function of TRAF2 and TRAF6 is not to target protein degradation but to activate downstream kinase cascades (9). These TRAF proteins function in conjunction with the Ub-conjugating enzyme (E2) complex Ubc13–Uev1A (also known as Mms2) to catalyze the formation of a unique polyubiquitin chain linked through Lys-63 (K63) of Ub (10). This K63-linked polyubiquitination has a signaling function in initiating a protein kinase cascade by activating the transforming growth factor β-activated kinase (TAK1) complex (7). TAK1 phosphorylates and activates the IκB kinase (IKK) complex, which contains the catalytic subunits IKKα and IKKβ as well as an essential regulatory subunit NF-κB essential modulator (NEMO). IKK subsequently phosphorylates the NF-κB inhibitor IκB and targets this inhibitor for ubiquitination by the SCF (Skp1–Cullin-1 F box)–βTrCP E3 complex. Ubiquitinated IκBis then degraded by the proteasome, thus allowing NF-κB to enter the nucleus to regulate gene expression.
TIFA [TRAF-interacting protein with a forkhead-associated (FHA) domain] was identified as a TRAF6-interacting protein in a yeast two-hybrid screen (11). The same molecule also was isolated from an independent mammalian two-hybrid screen as a TRAF2-binding protein (12). In addition to the FHA domain, which is known to bind phosphothreonine and phosphoserine (13), TIFA contains a consensus TRAF6-binding motif (14). TIFA activates NF-κB and c-JUN N-terminal kinase (JNK), possibly by linking TRAF6 to IL-1R-associated kinase (IRAK-1) in the IL-1 pathway (11), but the biochemical mechanism underlying this activation is unknown. In this article, we demonstrate that TIFA activates IKK in an in vitro reconstitution system consisting of purified proteins, including TAK1, TRAF6, and other ubiquitination enzymes. Furthermore, we show that TIFA induces the oligomerization and polyubiquitination of TRAF6, which in turn activates TAK1 and IKK.
Materials and Methods
Plasmids and Proteins. Human TIFA cDNA was cloned by PCR and then subcloned into pET14b (Novagen) for expression in Escherichia coli. His6-tagged proteins were purified by using nickel columns (Qiagen, Valencia, CA) according to the manufacturer's instructions. TIFA mutants were generated by using the QuikChange kit (Stratagene) and verified by automatic DNA sequencing. The ORFs of wild-type and mutant TIFA were subcloned into the mammalian expression vector pEF-FLAG-IRES-Puro. cDNA and proteins of E1, TRIKA1 (E2, Ubc13–Uev1A), TRIKA2 (TAK1–TAB1–TAB2), TRAF6 (E3), and IKK have been described (7, 10).
Gel-Filtration Chromatography. Wild-type or mutant TIFA (1 μg) was added to 20 μl of Ubc13 eluate (6.4 mg/ml, containing endogenous TRAF6; see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). After 1 h of incubation at 4°C, the mixture was applied to Superdex 200 (2.4-ml bed volume) by using the smart system (Pharmacia), and fractions (100 μl) were collected for analysis by immunoblotting with anti-TRAF6 antibody (D10).
Glycerol-Gradient Ultracentrifugation. Wild-type or mutant TIFA (5 μg) was incubated with 50 μl of Ubc13 eluate as described above, and the mixtures were then applied to a 2-ml 10–50% glycerol gradient in 50 mM Tris·HCl, pH 7.5/150 mM NaCl. After centrifugation at 250,000 × g for 3 h in a TLS-55 rotor (Beckman Coulter) at 4°C, fractions (200 μl) were analyzed by immunoblotting with an antibody against penta-His (detecting His6-TIFA) or TRAF6.
In Vitro Assay for IKK Activation. To measure the activation of IKK by TIFA in vitro, Jurkat or HeLa cytosolic extracts (10 mg/ml in 20 mM Tris·HCl, pH 7.5/150 mM NaCl/0.5 mM DTT/0.5 mM PMSF/0.1% Nonidet P-40) were incubated with recombinant TIFA protein (50 ng) and 35S-labeled IκBα in a 10-μl reaction mixture containing an ATP-regenerating system (10). The reaction mixture was incubated at 30°C for 1 h, followed by SDS/PAGE and imaging with a PhosphorImager (Molecular Dynamics). To identify the factors required for the activation of IKK by TIFA, the same IKK assay was used except that the cell extracts were replaced by Ubc13-depleted HeLa S100, Ubc13–Uev1A (1 μM), and column fractions.
Ubiquitination Assay. To detect the Ub ligase activity of TRAF6, reaction mixtures containing E1 (0.1 μM), Ubc13–Uev1A (0.4 μM), TRAF6 (≈0.1 μM), Ub (60 μM), and ATP (2 mM) were incubated in the presence or absence of TIFA at 30°C for 1 h. The reaction products were then resolved by SDS/PAGE and detected by immunoblotting with an antibody against TRAF6 (D10) or Ub (P4D1). See Supporting Materials and Methods for additional information.
Results
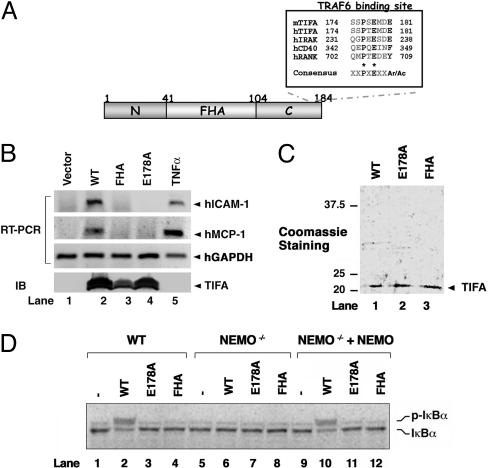
TIFA Activates IKK in a Cell-Free System. TIFA is a 20-kDa protein with an FHA domain and a TRAF6-binding site (Fig. 1A). Overexpression of TIFA leads to activation of IKK and NF-κB in a manner that depends on the FHA domain and TRAF6-binding site (11). For example, a point mutation in TIFA, E178A, abolishes its binding to TRAF6 as well as its ability to activate the expression of an NF-κB reporter (11). To determine whether overexpression of TIFA can induce the expression of endogenous NF-κB target genes, we transfected expression vectors encoding wild-type or mutant TIFA into human embryonic kidney (HEK) 293 cells, and then used RT-PCR to examine the expression of two NF-κB target genes, hICAM-1 (human intercellular adhesion molecule 1) and hMCP-1 (human mono-cyte chemoattractant protein 1). As shown in Fig. 1B, overexpression of wild-type TIFA induced the expression of hICAM-1 and hMCP-1 without affecting the constitutive expression of GAPDH. In contrast, the FHA-domain mutant (G50E/S66A) and TRAF6-binding-defective mutant (E178A) of TIFA failed to activate any of the NF-κB target genes. These results indicate that overexpression of TIFA is sufficient to activate the expression of endogenous NF-κB target genes.
Fig. 1.
TIFA activates IKK in a cell-free system. (A) Diagram of the primary structure of TIFA and alignment of the TRAF6-binding site. Ar/Ac represents an aromatic or acidic amino acid. (B) Overexpression of TIFA is sufficient to induce the expression of endogenous NF-κB target genes. Plasmids encoding Flag-tagged wild-type or mutant TIFA were transfected into HEK 293 cells. At 48 h after transfection, the expression of hICAM-1, hMCP-1, and human GAPDH (hGAPDH) RNA was analyzed by RT-PCR, and the expression of TIFA proteins was examined by immunoblotting with the Flag antibody. As a positive control, HEK 293 cells were stimulated with TNFα (10 ng/ml). WT, wild type; E178A, TRAF6-binding-defective mutant; FHA, FHA-domain mutant G50E/S66A. (C) Expression and purification of recombinant TIFA. His-tag TIFA and mutants were expressed in E. coli and purified with nickel nitrilotriacetic acid (Ni-NTA) resin. (D) Phosphorylation of IκBα in crude cell extracts. In vitro translated, 35S-labeled IκBα was used as the substrate in the reactions that contain protein extracts from wild-type Jurkat cells (lanes 1–4) or NEMO-deficient Jurkat cells (lanes 5–12), TIFA or its mutants, and ATP. In lanes 9–12, recombinant NEMO was added to the NEMO-deficient extracts to restore IKK activation. p-IκBα, phosphorylated IκBα.
To identify the factors that link TIFA to IKK and NF-κB activation, we developed an in vitro kinase assay that measures IKK activation after the addition of TIFA protein to crude cell extracts. Recombinant His6-tagged wild-type FHA-domain mutant (G50E/S66A) and TRAF6-binding-defective mutant (E178A) of TIFA were expressed in E. coli and then purified by using nickel columns (Fig. 1C). Addition of wild-type TIFA, but not any of the mutants, to Jurkat T cell extracts led to the phosphorylation of 35S-labeled IκBα, as indicated by the electrophoretic mobility shift (Fig. 1D). Furthermore, TIFA-dependent activation of IKK in the cell-free system depends on an intact IKK complex because the extracts prepared from NEMO-deficient Jurkat cells (15) failed to phosphorylate IκBα in response to TIFA. Addition of recombinant NEMO protein to the NEMO-deficient extracts restored TIFA-dependent phosphorylation of IκBα. Therefore, the in vitro TIFA-inducible system shown here recapitulates the functional domain requirement of TIFA and the requirement of NEMO for IKK activation.
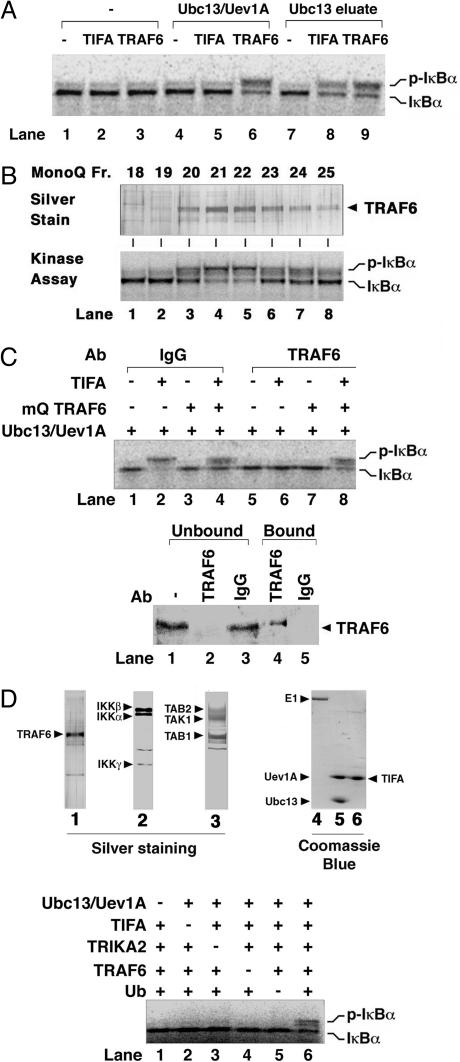
Ubc13–Uev1A and TRAF6 Link TIFA to IKK Activation. It has been shown that TIFA interacts with TRAF2 (12) and TRAF6 (11) and that activation of IKK by TRAF2 and TRAF6 requires the Ub-conjugating enzyme complex Ubc13–Uev1A (10). To determine whether TIFA-induced activation of IKK requires Ubc13–Uev1A, we carried out an in vitro IKK-activation assay by using HeLa cytosolic extracts (S100), which are depleted of Uev1A by Sepharose beads coated with Ubc13. As shown in Fig. 2A, TIFA was unable to activate IKK in Uev1A-depleted extracts (lane 2). Even when recombinant Ubc13 and Uev1A proteins were added to the reaction, TIFA failed to activate IKK (lane 5), whereas recombinant TRAF6 was able to restore IKK activation (lane 6). To test whether the Ubc13 column retained another factor that is required for IKK activation by TIFA, we eluted proteins bound to the column with 0.5 M NaCl and used the eluate (designated as Ubc13 eluate) in the IKK assay. Indeed, the Ubc13 eluate restored IKK activation by TIFA (lane 8), indicating that another Ubc13-associated factor distinct from Ubc13–Uev1A is required for TIFA activation of IKK. In a separate series of experiments, we further purified the IKK stimulatory factor from the Ubc13 eluate and identified this factor as TRAF6, which mediates IKK activation by BCL10 and MALT1 (two signaling proteins of the T cell receptor pathway) (16). To determine whether TRAF6 also mediates TIFA-dependent IKK activation, we tested MonoQ fractions containing purified TRAF6 for their ability to activate IKK in the presence of TIFA. As shown in Fig. 2B, the TIFA-dependent IKK stimulatory activity (Lower) copurified very well with TRAF6 protein (Upper; the identity of the TRAF6 protein was confirmed by mass spectrometry and immunoblotting; ref. 16). To confirm that TRAF6 was required for IKK activation by TIFA in the extracts, we immunodepleted TRAF6 from the extracts and examined the resulting effect on IKK activation (Fig. 2C). Depletion of TRAF6 from HeLa S100 abolished its ability to support TIFA-dependent IKK activation (Fig. 2C, lane 6), whereas the control extracts retained full IKK activation potential (lane 2). Addition of purified TRAF6 (Mono-Q fraction 21) to the TRAF6-depleted extracts restored IKK activation in response to TIFA (lane 8). These results indicate that endogenous TRAF6 is required for IKK activation by TIFA in vitro, whereas recombinant TRAF6 overexpressed and purified from insect cells is able to activate IKK independently of TIFA or other upstream regulators. We have previously shown that a fraction of recombinant TRAF6 protein forms oligomers that activate IKK constitutively, thus explaining the difference in the regulation of endogenous versus recombinant TRAF6 (16).
Fig. 2.
TIFA activates IKK through Ubc13–Uev1A and TRAF6. (A) Activation of IKK by TIFA requires Ubc13–Uev1A and another Ubc13-associated factor. HeLa S100 was incubated with Ubc13–Sepharose to remove endogenous Uev1A and other proteins that bound to the resin. The supernatant was used as “Ubc13-depleted extract” for IKK assays in the presence of recombinant TRAF6 (lanes 3, 6, and 9) or TIFA (lanes 2, 5, and 8). In lanes 4–6, recombinant Ubc13–Uev1A protein was added to the reaction, whereas in lanes 7–9, protein that bound to the Ubc13 column was eluted (Ubc13 eluate) and added to the assays. (B) Identification of TRAF6 as a Ubc13-interacting protein that is required for IKK activation by TIFA. Endogenous TRAF6 protein was purified as described in ref. 16, and fractions from the last MonoQ step were analyzed by silver staining (Upper) or IKK kinase assays (Lower). (C) TRAF6 is required for IKK activation by TIFA in vitro. HeLa S100 was subjected to immunoprecipitation with an antibody against TRAF6 (lanes 5–8) or a control IgG (lanes 1–4). The supernatants were then analyzed by immunoblotting with a TRAF6 antibody (Lower) or by IKK assays (Upper) in the presence of Ubc13–Uev1A, TIFA, and purified endogenous TRAF6, as indicated. (D) (Upper) Silver or Coomassie blue staining of purified proteins. TRAF6 (native, purified from HeLa S100; lane 1), IKK (native, purified from HeLa S100; lane 2), TRIKA2 (recombinant, purified from Sf9; lane 3), E1 (recombinant, purified from Sf9; lane 4), Ubc13–Uev1A (recombinant, purified from E. coli; lane 5), and TIFA (recombinant, purified from E. coli; lane 6). (Lower) Reconstitution of TIFA-induced IKK pathway in vitro. Purified native IKK complex and TRAF6 were incubated with recombinant proteins of TIFA, Ubc13–Uev1A, Ub, and TAK1–TAB1–TAB2 complex (TRIKA2) as indicated. The reconstitution reaction mixture also contained ATP, E1, and 35S-labeled IκBα.
Reconstitution of IKK Activation by TIFA in Vitro. We next attempted to reconstitute TIFA-dependent IKK activation in vitro by using purified proteins (Fig. 4D Top). The reconstitution reactions contain E1, Ubc13–Uev1A (E2), TRAF6 (E3), Ub, TRIKA2 (TAK1–TAB1–TAB2 complex expressed and purified from baculovirus-infected insect cells), purified endogenous IKK complex, TIFA, 35S-labeled IκBα (purified from in vitro translation system), and ATP (see Materials and Methods). As shown in Fig. 2D (Lower), the presence of all of these components in the reaction was required to reconstitute IKK activation by TIFA (compare lane 6 with other lanes). These results demonstrate that TIFA activates IKK through TRAF6 and TAK1 complex in a ubiquitination-dependent manner.
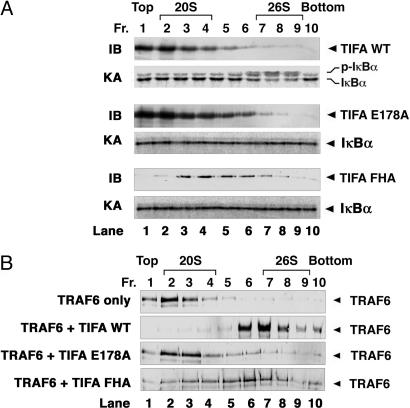
Fig. 4.
TIFA induces TRAF6 oligomerization and IKK activation. (A) Oligomerized TIFA activates IKK in vitro. Recombinant wild-type or mutant (E178A or FHA) His6-TIFA proteins were fractionated by glycerol-gradient ultracentrifugation (10–50%). Fractions were collected (percentage of glycerol increases from top to bottom) and analyzed by immunoblotting with a penta-His-specific antibody (IB) or by IKK kinase assays (KA). (B) TIFA induces the oligomerization of TRAF6. Ubc13 eluate enriched in native TRAF6 (labeled as TRAF6) was incubated alone or with TIFA proteins as indicated at 4°C for 1 h. The mixtures were fractionated by glycerol-gradient ultracentrifugation as described above. Fractions were collected and analyzed by immunoblotting with a TRAF6 antibody.
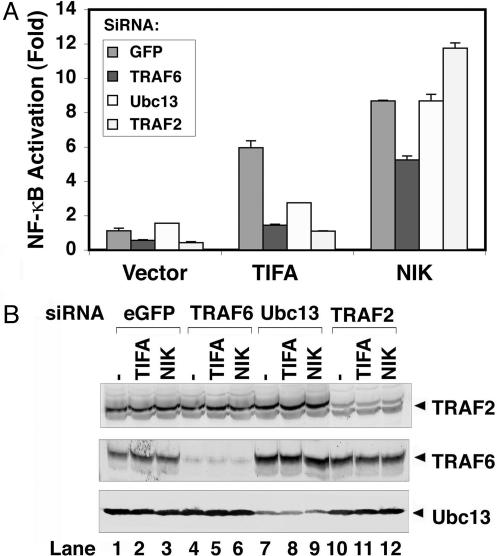
NF-κB Activation by TIFA Requires Ubc13, TRAF2, and TRAF6. To determine whether Ubc13, TRAF2, and TRAF6 are required for TIFA-mediated NF-κB activation in living cells, we used RNA interference to silence the expression of these genes in HEK 293 cells transfected with TIFA expression vector and a luciferase reporter gene under the control of three tandem repeats of NF-κB-binding sites (Fig. 3A). Small interfering RNA (siRNA) oligonucleotides targeting Ubc13, TRAF6, or TRAF2 decreased the expression of the corresponding proteins (Fig. 3B). Overexpression of TIFA activated the NF-κB-luciferase reporter in cells transfected with GFP siRNA (as a control; Fig. 3A). In contrast, siRNA targeting Ubc13, TRAF6, or TRAF2 strongly inhibited TIFA-mediated NF-κB activation. As a control, none of the siRNA oligonucleotides significantly affected NF-κB activation by NF-κB-inducing kinase (NIK), which is likely to activate NF-κB by means of direct phosphorylation of IKKα (17). Together, these results suggest that Ubc13, TRAF6, and TRAF2 are required for TIFA-mediated activation of NF-κB. The requirement for both TRAF6 and TRAF2 in TIFA signaling is reminiscent of the CD40 signaling pathway in which genetic ablation of either TRAF6 or TRAF2 impaired NF-κB activation (18–21). Thus, TRAF2 and TRAF6 have nonoverlapping functions in some signaling pathways (see Discussion).
Fig. 3.
Ubc13, TRAF6, and TRAF2 are required for NF-κB activation by TIFA. Double-stranded siRNA oligomers corresponding to the sequence of GFP (control), Ubc13, TRAF2, or TRAF6 were transfected into HEK 293 cells. After 2 consecutive days of transfection of siRNA, cells were transfected with a NF-κB luciferase reporter with the expression constructs encoding Flag-tagged TIFA, NF-κB-interacting kinase (NIK), or vector alone. (A) The luciferase activity was measured and normalized for transfection efficiency, as determined by co-transfection of a constitutively expressed β-galactosidase reporter. (B) The efficiency of RNA interference was verified by immunoblotting.
TIFA Forms Oligomers to Activate IKK. Previous studies have suggested that TRAF proteins are regulated by oligomerization, which may in turn be controlled by some upstream signaling molecules (7, 16). To determine whether TIFA forms oligomers, we analyzed the molecular sizes of purified TIFA protein by glycerol-gradient ultracentrifugation. Whereas most of the TIFA protein was present in fractions containing a low concentration of glycerol (top of the gradient), a very small amount of TIFA formed oligomers or aggregates that migrated toward the bottom of the gradient where the concentration of glycerol was higher (Fig. 4A). Interestingly, only these oligomeric forms of TIFA, whose apparent molecular size was similar to that of the 26S proteasome, were able to activate IKK. The TIFA mutant defective in TRAF6 binding, E178A, had a similar distribution pattern to that of wild-type TIFA in the glycerol gradient but was unable to activate IKK. Mutations in the FHA domain caused a shift in the distribution of TIFA toward the middle of the gradient, but none of the fractions containing TIFA–FHA mutant was able to activate IKK.
TIFA Promotes the Oligomerization of TRAF6. TIFA was identified as a TRAF6-binding protein. Thus, one possible mechanism by which TIFA oligomers activate IKK is to induce the oligomerization of TRAF6. To test this possibility, we incubated a partially purified TRAF6 fraction with wild-type or mutant TIFA protein and then determined the molecular distribution of TRAF6 by glycerol-gradient ultracentrifugation. In the absence of TIFA, most of TRAF6 was present in low-molecular-weight fractions (Fig. 4B). Significantly, when wild-type TIFA was added, TRAF6 migrated to the high-molecular-weight fractions. The TIFA mutant that does not bind to TRAF6 was also unable to induce TRAF6 oligomerization, whereas the FHA mutant of TIFA, which still binds to TRAF6, caused TRAF6 to sediment in the glycerol gradient in a pattern that is similar to that of TIFA–FHA mutant itself (Fig. 4A). These results suggest that TIFA binds to TRAF6 and induces TRAF6 oligomerization. It is interesting to note that although most of TIFA was present as low-molecular-weight species (Fig. 4A, Top), only the high-molecular-weight forms of TIFA cosedimented with TRAF6 (Fig. 4B), suggesting that oligomerization of TIFA greatly enhances its ability to bind to TRAF6.
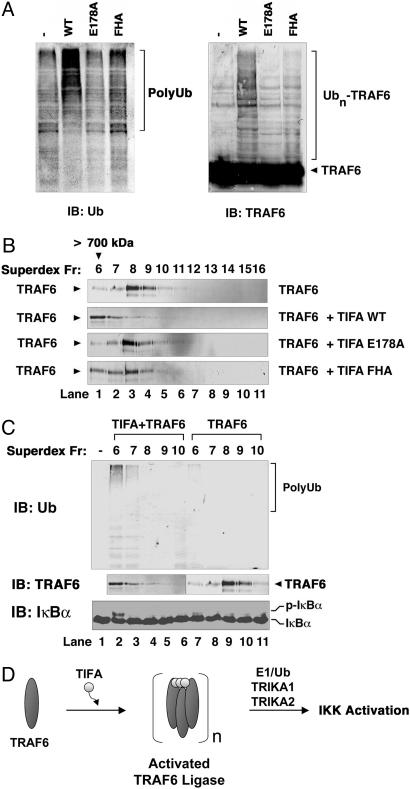
TIFA Enhances the Ub Ligase Activity of TRAF6. TRAF6 is a RING domain Ub ligase (E3), which, in conjunction with Ubc13–Uev1A, catalyzes K63-linked polyubiquitination that is required for IKK activation (10). We have previously shown that the Ub ligase activity of TRAF6 can be enhanced by forced dimerization of an artificial dimerization domain (7) or by oligomerization induced by BCL10 and MALT1 (16). To determine whether TIFA enhanced the Ub ligase activity of endogenous TRAF6, we performed in vitro ubiquitination assay in the presence of E1, Ubc13–Uev1A, purified endogenous TRAF6, Ub, and ATP (Fig. 5A). Addition of wild-type, but not mutant, TIFA into the reaction significantly enhanced the synthesis of polyubiquitin chains as well as the polyubiquitination of TRAF6. To determine whether the enhanced Ub ligase activity of TRAF6 was due to TIFA-induced oligomerization, we incubated TRAF6 with wild-type or mutant TIFA proteins and then separated the mixtures by gel-filtration chromatography (Superdex 200). As shown in Fig. 5B, addition of wild-type, but not mutant, TIFA protein led to oligomerization of TRAF6, which eluted from the gel filtration column in the void volume (>700 kDa). Importantly, these high-molecular-weight forms of TRAF6 displayed greatly increased Ub ligase activity as compared with TRAF6 of lower-molecular-weight species, despite a similar amount of TRAF6 protein in these fractions (Fig. 5C Top and Middle). Moreover, only the fraction with significantly higher Ub ligase activity of TRAF6 (Fig. 5C, lane 2, fraction 6) was able to activate IKK (Fig. 5C Bottom). This activity was not due to the presence of TIFA because we had removed His6-TIFA from the fraction by using a nickel-affinity column (data not shown). Together, these results indicate that TIFA activates IKK by inducing the oligomerization and ubiquitination of TRAF6 (Fig. 5D).
Fig. 5.
Oligomerization of TRAF6 by TIFA activates its Ub ligase activity. (A) TIFA promotes polyubiquitination of TRAF6. Recombinant wild-type or mutant TIFA proteins were incubated with TRAF6 in a reaction mixture containing E1, Ubc13–Uev1A (E2), Ub, and ATP. After incubation at 30°C for 1 h, reaction products were analyzed by immunoblotting with an antibody against Ub (Left) or TRAF6 (Right). (B) TIFA induces oligomerization of TRAF6. TIFA was incubated with TRAF6 as described in the legend of Fig. 4B, and the mixture was fractionated by gel-filtration chromatography on Superdex 200. Fractions from the column were analyzed by immunoblotting with a TRAF6 antibody. (C) The Ub ligase and IKK-stimulatory activities of TRAF6 are induced by TIFA-mediated oligomerization. (Top) The Superdex 200 fractions shown in B were depleted of His6-TIFA by using nickel-affinity resin and then incubated in a reaction mixture containing E1, Ubc13–Uev1A, Ub, and ATP. Polyubiquitin chain synthesis was analyzed by immunoblotting with a Ub antibody. (Bottom) Aliquots containing TRAF6 of different molecular sizes from the Superdex 200 column were also incubated with HeLa S100 to determine the IKK-stimulatory activity of TRAF6 fractions. After incubation at 30°C for1hinthe presence of ATP, phosphorylation of endogenous IκBα was analyzed by immunoblotting with an IκBα-specific antibody. (D) A biochemical model of IKK activation by TIFA. TIFA forms oligomers upon stimulation of cells with some NF-κB agonists. The oligomerized forms of TIFA bind to TRAF6 and promote TRAF6 oligomerization. As a result, the TRAF6 Ub ligase is activated to catalyze K63-linked polyubiquitination in conjunction with the Ubc13–Uev1A E2 complex (TRIKA1). Ubiquitinated TRAF6 is recruited to the TRIKA2 complex, which contains the Ub receptor TAB2 and the protein kinase TAK1. TAK1 is then activated to phosphorylate IKKβ at key serine residues within the activation loop, thereby activating IKK.
Discussion
In this article, we have investigated the biochemical mechanism of IKK activation by TIFA. We show that TIFA activates IKK in crude cell extracts in a manner that depends on an intact FHA domain and a TRAF6-binding motif. By using this in vitro activation system, we have identified Ubc13–Uev1A and TRAF6 as the E2 and E3, respectively, that mediate IKK activation by TIFA through a Ub-dependent mechanism. Importantly, we have succeeded in the reconstitution of IKK activation by TIFA in vitro by using purified proteins including the TAK1 kinase complex, TRAF6 and other ubiquitination enzymes. Furthermore, we have provided evidence that TIFA forms oligomers that activate IKK by inducing the oligomerization and polyubiquitination of TRAF6.
Our data showed that the FHA domain and TRAF6-binding site of TIFA are required for IKK activation in vitro. A single point mutation in the TRAF6-binding site of TIFA abolished its ability to induce TRAF6 oligomerization and ubiquitination, indicating that the direct binding between TIFA and TRAF6 is essential for its function. However, the biochemical function of FHA domain in TIFA is less clear. Interestingly, it has been shown that mutations of TIFA in the FHA domain cause this mutant protein to form a pentamer or hexamer, whereas the wild-type TIFA appears to be a trimer (11). In this study, we have found that a small fraction of TIFA forms high-molecular-weight oligomers, and that only these large oligomers are capable of inducing TRAF6 oligomerization and activating IKK. Although the FHA-domain mutation causes TIFA to form intermediate-sized multimers, these multimers fail to activate IKK or induce TRAF6 oligomerization. Thus, it appears that the activity of TRAF6 as a RING domain Ub ligase and an IKK activator depends on its highly oligomerized forms.
More than 70 members of RING domain proteins have been observed in discrete subcellular structures that can be visualized by immunostaining and confocal microscopy (22). A good example is provided by the breast tumor suppressor BRCA1, which forms discrete subnuclear foci (23). Purified RING domain of BRCA1 self-assembles in vitro into supramolecular assemblies, which have higher Ub ligase activity than do the tetrameric or dimeric forms of BRCA1. A cancer-predisposing mutation (C64G) in BRCA1 abolishes the self-assembly and Ub ligase activity of the protein in vivo and in vitro. The Ub ligase activity of this BRCA1 mutant can be recovered partially by forced oligomerization. These studies, together with our finding that the higher-molecular-weight forms of TRAF6 have higher Ub ligase activity (Fig. 5), suggest that RING domain proteins may facilitate polyubiquitination by coupling multiple Ub thio-ester-bound E2 to each other when the RING domains are assembled into supramolecular structures. In support of this model, multiple gold-stained UbcH5C molecules and nanogold-stained Ub chains have been found to colocalize with the BRCA1/BARD1 supramolecular structures by electron microscopy (23).
In addition to binding to TRAF6, TIFA has also been found to interact with TRAF2 (12), although the exact TRAF2-binding site has not been mapped. In keeping with the role of TRAF2 in TIFA signaling, we have found that RNA interference of TRAF2 blocked NF-κB activation by TIFA. However, our in vitro studies did not reveal a requirement of TRAF2 in IKK activation by TIFA. Unlike TRAF6, TRAF2 does not bind to the Ubc13 column (data not shown). In addition, TRAF2 is not required to reconstitute IKK activation by TIFA in vitro, although it remains to be determined whether TRAF2 could enhance IKK activation in the reconstitution system. Although TRAF2 and TRAF6 appear to be involved separately in the TNFα and IL-1 pathways, respectively, both are required for CD40 signaling (18–21). In addition, TRAF2 and TRAF6 also have redundant functions in T cell receptor signaling (16). Thus, the roles of TRAF2 and TRAF6 depend on the context of specific stimuli, and it is possible that these two proteins cooperate in certain signaling pathways, some of which may involve TIFA. However, the physiological functions of TIFA remain to be determined by further genetic studies.
Our studies on TIFA have uncovered a mechanism of TRAF6 Ub ligase activation that involves inducible oligomerization. TRAF6 then functions with Ubc13–Uev1A to catalyze the synthesis of K63-linked polyubiquitin chains that are conjugated to target proteins, including NEMO and TRAF6 (7, 10, 16). Polyubiquitination of TRAF6 then leads to the activation of IKK through the TAK1–TAB1–TAB2 kinase complex (7). Recently, we found that TAB2 and its homologue TAB3 bind preferentially to K63-linked polyubiquitin chains through a conserved novel zinc finger (NZF) domain, which is essential for TAK1 and IKK activation (24). Thus, polyubiquitination of TRAF6 likely facilitates its interaction with the TAB2–TAK1 complex, thereby leading to TAK1 activation. TAK1 then phosphorylates IKKβ at two conserved serine residues in the activation loop, resulting in IKK activation. Such an oligomerization→ubiquitination→phosphorylation cascade has also been shown to operate in T lymphocytes, in which antigenic stimulation of T cell receptors leads to the recruitment and clustering of essential signaling proteins, including CARMA1, BCL10, and MALT1 (16, 25). Together, these studies suggest that different signaling pathways may use distinct adaptor proteins, such as MALT1 and TIFA, as the endogenous “oligomerizers” to activate Ub ligases such as TRAF6, which then initiate protein kinase cascades that are important for stress responses, inflammation, and immunity.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01-GM63692, Welch Foundation Grant I1389, and American Cancer Society Grant RSG0219501TBE. Z.J.C. is a Leukemia and Lymphoma Society Scholar and a Burroughs Wellcome Fund Investigator in Pathogenesis of Infectious Diseases.
Author contributions: C.-K.E., L.S., and Z.J.C. designed research; C.-K.E. and L.S. performed research; C.-K.E., L.S., and J.-I.I. contributed new reagents/analytical tools; C.-K.E., L.S., and Z.J.C. analyzed data; and C.-K.E. and Z.J.C. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IKK, IκB kinase; TNF, tumor necrosis factor; TNFR, TNF receptor; IL-1R; IL-1 receptor; Ub, ubiquitin; FHA, forkhead-associated domain; TAK1, transforming growth factor β-activated kinase; NEMO, NF-κB essential modulator; siRNA, small interfering RNA; HEK, human embryonic kidney.
References
- 1.Rothe, M., Wong, S. C., Henzel, W. J. & Goeddel, D. V. (1994) Cell 78, 681-692. [DOI] [PubMed] [Google Scholar]
- 2.Cao, Z., Xiong, J., Takeuchi, M., Kurama, T. & Goeddel, D. V. (1996) Nature 383, 443-446. [DOI] [PubMed] [Google Scholar]
- 3.Ishida, T., Mizushima, S., Azuma, S., Kobayashi, N., Tojo, T., Suzuki, K., Aizawa, S., Watanabe, T., Mosialos, G., Kieff, E., et al. (1996) J. Biol. Chem. 271, 28745-28748. [DOI] [PubMed] [Google Scholar]
- 4.Inoue, J., Ishida, T., Tsukamoto, N., Kobayashi, N., Naito, A., Azuma, S. & Yamamoto, T. (2000) Exp. Cell Res. 254, 14-24. [DOI] [PubMed] [Google Scholar]
- 5.Wu, H. & Arron, J. R. (2003) BioEssays 25, 1096-1105. [DOI] [PubMed] [Google Scholar]
- 6.Baud, V., Liu, Z. G., Bennett, B., Suzuki, N., Xia, Y. & Karin, M. (1999) Genes Dev. 13, 1297-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang, C., Deng, L., Hong, M., Akkaraju, G. R., Inoue, J. & Chen, Z. J. (2001) Nature 412, 346-351. [DOI] [PubMed] [Google Scholar]
- 8.Pickart, C. M. (2004) Cell 116, 181-190. [DOI] [PubMed] [Google Scholar]
- 9.Sun, L. & Chen, Z. J. (2004) Curr. Opin. Cell Biol. 16, 119-126. [DOI] [PubMed] [Google Scholar]
- 10.Deng, L., Wang, C., Spencer, E., Yang, L., Braun, A., You, J., Slaughter, C., Pickart, C. & Chen, Z. J. (2000) Cell 103, 351-361. [DOI] [PubMed] [Google Scholar]
- 11.Takatsuna, H., Kato, H., Gohda, J., Akiyama, T., Moriya, A., Okamoto, Y., Yamagata, Y., Otsuka, M., Umezawa, K., Semba, K. & Inoue, J. (2003) J. Biol. Chem. 278, 12144-12150. [DOI] [PubMed] [Google Scholar]
- 12.Kanamori, M., Suzuki, H., Saito, R., Muramatsu, M. & Hayashizaki, Y. (2002) Biochem. Biophys. Res. Commun. 290, 1108-1113. [DOI] [PubMed] [Google Scholar]
- 13.Li, J., Lee, G. I., Van Doren, S. R. & Walker, J. C. (2000) J. Cell Sci. 113, 4143-4149. [DOI] [PubMed] [Google Scholar]
- 14.Ye, H., Arron, J. R., Lamothe, B., Cirilli, M., Kobayashi, T., Shevde, N. K., Segal, D., Dzivenu, O. K., Vologodskaia, M., Yim, M., et al. (2002) Nature 418, 443-447. [DOI] [PubMed] [Google Scholar]
- 15.Harhaj, E. W., Good, L., Xiao, G., Uhlik, M., Cvijic, M. E., Rivera-Walsh, I. & Sun, S. C. (2000) Oncogene 19, 1448-1456. [DOI] [PubMed] [Google Scholar]
- 16.Sun, L., Deng, L., Ea, C. K., Xia, Z. P. & Chen, Z. J. (2004) Mol. Cell 14, 289-301. [DOI] [PubMed] [Google Scholar]
- 17.Ling, L., Cao, Z. & Goeddel, D. V. (1998) Proc. Natl. Acad. Sci. USA 95, 3792-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hostager, B. S., Haxhinasto, S. A., Rowland, S. L. & Bishop, G. A. (2003) J. Biol. Chem. 278, 45382-45390. [DOI] [PubMed] [Google Scholar]
- 19.Lomaga, M. A., Yeh, W. C., Sarosi, I., Duncan, G. S., Furlonger, C., Ho, A., Morony, S., Capparelli, C., Van, G., Kaufman, S., et al. (1999) Genes Dev. 13, 1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen, L. T., Duncan, G. S., Mirtsos, C., Ng, M., Speiser, D. E., Shahinian, A., Marino, M. W., Mak, T. W., Ohashi, P. S. & Yeh, W. C. (1999) Immunity 11, 379-389. [DOI] [PubMed] [Google Scholar]
- 21.Naito, A., Azuma, S., Tanaka, S., Miyazaki, T., Takaki, S., Takatsu, K., Nakao, K., Nakamura, K., Katsuki, M., Yamamoto, T. & Inoue, J. (1999) Genes Cells 4, 353-362. [DOI] [PubMed] [Google Scholar]
- 22.Kentsis, A. & Borden, K. L. (2000) Curr. Protein Pept. Sci. 1, 49-73. [DOI] [PubMed] [Google Scholar]
- 23.Kentsis, A., Gordon, R. E. & Borden, K. L. (2002) Proc. Natl. Acad. Sci. USA 99, 15404-15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanayama, A., Seth, R. B., Sun, L., Ea, C. K., Hong, M., Shaito, A., Chiu, Y. H., Deng, L. & Chen, Z. J. (2004) Mol. Cell 15, 535-548. [DOI] [PubMed] [Google Scholar]
- 25.Thome, M. & Tschopp, J. (2003) Trends Immunol. 24, 419-424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.