Abstract
Phytoplasmas are unculturable, phytopathogenic bacteria that cause economic losses worldwide. As unculturable micro-organisms, phytoplasma taxonomy has been based on the use of the 16S rRNA-encoding gene to establish 16Sr groups and subgroups based on the restriction fragment length polymorphism (RFLP) pattern resulting from the digestion of amplicon (in vitro) or sequence (in silico) with seventeen restriction enzymes. Problems such as heterogeneity of the ribosomal operon and the inability to differentiate closely related phytoplasma strains has motivated the search for additional markers capable of providing finer differentiation of phytoplasma strains. In this study we developed and validated a scheme to classify phytoplasmas based on the use of cpn60 universal target (cpn60 UT) sequences. Ninety-six cpn60 UT sequences from strains belonging to 19 16Sr subgroups were subjected to in silico RFLP using pDRAW32 software, resulting in 25 distinctive RFLP profiles. Based on these results we delineated cpn60 UT groups and subgroups, and established a threshold similarity coefficient for groups and subgroups classifying all the strains analysed in this study. The nucleotide identity among the reference strains, the correspondence between in vitro and in silico RFLP, and the phylogenetic relationships of phytoplasma strains based on cpn60 UT sequences are also discussed.
Keywords: Phytoplasma, Diversity, differentiation, chaperonin 60, taxonomy
Phytoplasmas, first known as mycoplasma-like organisms (Doi et al., 1967), are wall-less, insect-vectored bacteria that cause disease in more than a thousand different plant hosts, affecting weedy, ornamental and crop plants worldwide (Harrison et al., 2014; Pérez-López et al., 2016a). With a small, A-T rich, and distinctively organized genome, phytoplasmas are a well-defined clade inside the class Mollicutes, derived from an Acholeplasma-like ancestor (Zhao et al., 2014, 2015).
Phytoplasmas have not been successfully isolated in axenic cultures, so traditional taxonomic characteristics are difficult to measure and phytoplasma taxonomy remains under the classification criteria specified for uncultured micro-organisms (Murray & Stackebrandt, 1995). In 2004, the International Committee of Systematic Bacteriology Subcommittee for the Taxonomy of Mollicutes, the International Research Program for Comparative Mycoplasmology (IRPCM), proposed the provisional genus ‘Candidatus Phytoplasma’ (IRPCM, 2004). This classification is based on the similarity of 16S rRNA gene sequences supported by phylogenetic analysis, and using this strategy, 38 ‘Candidatus Phytoplasma’ species have been formally described to date (Davis et al., 2013; Harrison et al., 2014; IRPCM, 2004; Nejat et al., 2013). Classification of phytoplasmas is further supported by the 16S rRNA gene through the use of restriction fragment length polymorphism (RFLP) of the 16S rRNA F2nR2 fragment with a set of seventeen endonucleases (Lee et al., 1993, 1998). This approach identifies at least 30 groups of phytoplasmas, designated 16SrI-16SrXXX, with each group containing subgroups designated by letters (Harrison et al., 2014; Pérez-López et al., 2016a; Zhao et al., 2009). The validation of a computer simulated (in silico) RFLP as an alternative to the actual (in vitro) RFLP, along with the development of the interactive online phytoplasma classification tool iPhyClassifier, increased the accuracy of phytoplasma classification based on 16S rRNA gene sequences (Wei et al., 2007, 2008; Zhao et al., 2009).
The use of other genes as part of the scheme of identification and classification of phytoplasmas has been broadly suggested, mainly because closely related strains are not well resolved using the 16S rRNA-encoding gene alone. The 16S–23S rRNA intergenic spacer, 23S rRNA region, rp (ribosomal protein) operon, tuf, rplV (rpl22)–rpsC (rps3), secY, map, uvrB–degV, nusA, secA, and rpoB genes have been used to identify and characterize phytoplasmas (Arnaud et al., 2007; Botti et al., 2003; Hodgetts et al., 2008; Lee et al., 2006; Marcone et al., 2000; Shao et al., 2006; Streten & Gibb, 2005; Valiunas et al., 2013). All these genes have been used to achieve a finer differentiation of phytoplasmas belonging to different species and/or RFLP groups. Another gene used to improve the resolution of phytoplasmas classification is the groEL gene, also known as chaperonin 60 (cpn60) (Dumonceaux et al., 2014; Mitrović et al., 2011,2015). All the genes mentioned above have also been used to differentiate other bacterial species. Lactic acid bacteria have been differentiated and identified using RFLP analysis of rpoB (Claisse et al., 2007), 16S rRNA/16S-23S rRNA intergenic spacer region (Ruiz et al., 2000), and tuf (Park et al., 2012). Moreover, partial cpn60 gene sequences (500 to 550 bp), have been useful to identify novel species such as Lactobacillus selangorensis (Haakensen et al., 2011), Sphingobacterium detergens (Marqués et al., 2012);Methylobacterium gnaphalii (Tani et al., 2012), and Prevotella jejuni (Hedberg et al., 2013), among many others. The cpn60 universal target (cpn60 UT) (Goh et al., 1996), is a fragment of approximately 550 bp that has been extensively used in the study of microbial communities (Town et al., 2014), and suggested as a molecular barcode for the domain Bacteria (Links et al., 2012). While not all Mollicutes encode Cpn60 within their genomes (Clark & Tillier, 2010), genes encoding Cpn60 have been found in all complete phytoplasma genomes reported to date and have been detected in many different phytoplasma subgroups (Andersen et al., 2013; Bai et al., 2006; Kube et al., 2008; Oshima et al., 2004; Tran-Nguyen et al., 2008). However, draft genomes for phytoplasma strains from the 16SrIII group suggest that this subgroup may lack this gene (Saccardo et al., 2012), which would limit the utility of cpn60-based classification tools for this subgroup. Nevertheless, the recent development of methods to access cpn60 UT sequences from phytoplasmas (Dumonceaux et al., 2014), has enabled the use of these sequences to develop diagnostic methods, and facilitates phytoplasma characterization based on polymorphisms detected among the different phytoplasma groups and subgroups (Dumonceaux et al., 2014; Pérez-López et al., 2016b). This primer cocktail has been shown to amplify the cpn60 UT from a diverse array of phytoplasmas (sharing as little as 61 % identity at the nucleotide level) from the major groups of phytoplasmas (Chung et al., 2013; Dumonceaux et al., 2014), although it is acknowledged that this amplification strategy may need to be modified as new sequences accrue, particularly from genomic sequencing efforts. Moreover, nested PCR is possible using previously reported primer sets that span the cpn60 UT of various phytoplasma groups (Kakizawa et al., 2006; Mitrović et al., 2011).
In this study, following the strategy previously used in the phytoplasma classification scheme based on the 16S rRNA gene, we suggest a complementary, coherent system to classify phytoplasmas based on RFLP analysis of cpn60 UT sequences with seven endonucleases. This new classification scheme, besides being phylogenetically valid, allowed a finer differentiation of phytoplasma strains inside the same 16Sr RFLP subgroups, with the identification of cpn60 UT groups and subgroups.
cpn60 UT sequences differentiate phytoplasma clade and subclades
One hundred and thirty-three cpn60 UT sequences were retrieved from the cpnDB (Hill et al., 2004) and NCBI nucleotide sequence databases. Fifty-five cpn60 UT sequences from phytoplasma, along with three sequences belonging to Acholeplasmas, three from Mycoplasmas, one from Clostridia, 19 from Bacillales, six from Lactobacillales, 34 sequences from walled Gram-negative bacterial taxa (Rhizobiales, Enterobacteriaceae, Sphingomonadales, among others), and one sequence from Cyanobacteria used as outgroup, were aligned with clustal x version 1.63b (Thompson et al., 1997) and trimmed to the 552 bp corresponding to the cpn60 UT sequences defined for phytoplasmas (Dumonceaux et al., 2014). A phylogenetic tree was reconstructed by the neighbour-joining method, using the tree-bisection-and-regrafting (TBR) algorithm available in mega6 software package (Tamura et al., 2013), and was bootstrapped 1000 times. We chose neighbour-joining because this method selects pairs of taxa that decrease the overall length of the tree, and because it is computationally less intensive than other methods of calculating phylogeny (Gascuel & Steel, 2006).
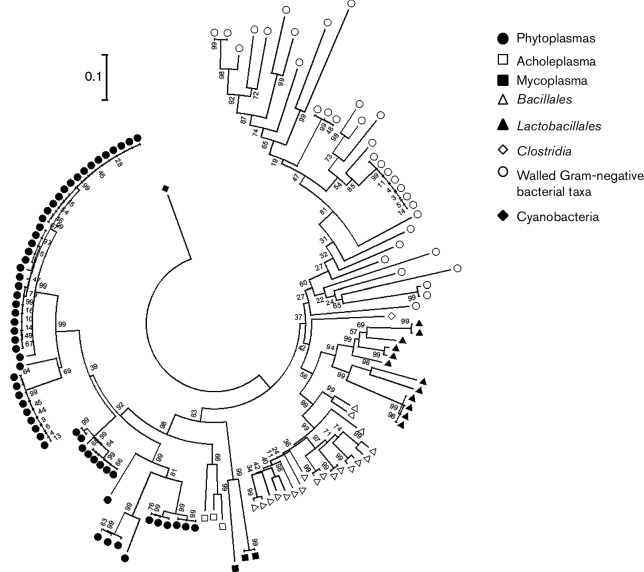
The phylogenetic tree obtained (Fig. 1) showed a clear delineation of the phytoplasma clade, with a differentiation of the three major phytoplasma subclades previously described (Chung et al., 2013; Hogenhout et al., 2008; Zhao et al., 2010, 2014). Similar results were obtained by calculating the tree using the maximum-likelihood method (Yang, 2007) (data not shown). The tree topology corresponded with the topology previously obtained by Wei and colleagues in 2007 using 16S rRNA gene sequences (Wei et al., 2007). This result confirms the ability of cpn60 UT sequences to identify phytoplasmas through cladistics analysis, as previously suggested (Dumonceaux et al., 2014; Pérez-López et al., 2016b).
Fig. 1.
Phylogenetic tree reconstructed using the neighbour-joining method of the cpn60 UT sequences of 163 micro-organisms within the domain Bacteria. We included 9 sequences from phytoplasma, 3 from Acholeplasmas, 3 from Mycoplasmas, 1 from Clostridia, 19 from Bacillales, 6 from Lactobacillales, and 34 from walled Gram-negative bacterial taxa (Rhizobiales, Enterobacteriaceae, Sphingomonadales). The cpn60 UT sequence from Cyanobacteria was used as outgroup. The phylogenetic tree was bootstrapped 1000 times to achieve reliability. Bar, 1 substitution in 10 positions.
To identify a phytoplasma-specific ‘signature’ sequence, corresponding to that reported for the 16S rRNA-encoding gene (IRPCM, 2004), we analysed the sequences shown in Fig. 1 using sigoligo, software that can identify signature sequences (Zahariev et al., 2009). This analysis revealed that the first ~60 nucleotides of the cpn60 UT differentiated phytoplasma sequences from other cpn60 UT sequences (data not shown). Aligning nucleotides 1–58 of all phytoplasma sequences and displaying them using Weblogo (Crooks et al., 2004) suggested a possible phytoplasma-specific signature sequence (5′-GCWAYHNTWTTRGCDCAAARWATVATTCAWMRGGDTTYRAWKYDRTWRAYDYWGGDG-3′; Fig. S1, available in the online Supplementary Material) that yielded only phytoplasma sequences by fasta alignment at cpnDB (Hill et al., 2004) (data not shown). Furthermore, translation of this nucleotide sequence revealed a putative, less degenerate amino acid sequence that similarly functioned as a signature sequence for phytoplasmas: [A(T/V)(V/L)LAQ(S/K/N)MI(H/R/Q)(R/K)GF(D/K)(A/F)(I/V)(D/N)(A/S/L)G; Fig. S1]. Like the nucleotide sequence, this amino acid sequence from randomly selected phytoplasmas yielded only phytoplasma sequences by blastp at cpnDB among the first 100 hits (data not shown).
Differentiating phytoplasmas based on cpn60 UT sequences
So far, phytoplasma cpn60 sequences have been reported from members of the groups 16SrI, 16SrII, 16SrV, 16SrVII, 16SrIX, 16SrX, 16SrXII, 16SrXIII and 16SrXIV (Dumonceaux et al., 2014; Pérez-López et al., 2016b). Altogether, after trimming the cpn60 UT sequence from the five completely sequenced phytoplasma genomes (Andersen et al., 2013; Bai et al., 2006; Kube et al., 2008; Oshima et al., 2013; Tran-Nguyen et al., 2008), from the draft genome belonging to the group 16SrII-A, strain PnWB (Chung et al., 2013) and 16SrIX-B strain SA213 (Quaglino et al., 2015), from the cpn60 sequences reported by Mitrović et al. (2011) for members of the group 16SrI, and members of the group 16SrXIV (Mitrović et al., 2015), from the 3.6 kb DNA fragments obtained by Kakizawa et al. (2006), and the sequences previously obtained by our group, we had 96 cpn60 UT sequences in this study.
The highest cpn60 UT sequence diversity was observed in members of the group 16SrI, with sequences from the subgroups 16SrI- A, B, C, E, F, and P subgroups represented. We also had a cpn60 UT sequence from more than one subgroup inside the 16Sr groups IX, X, XII and XIV. The description of the strains used and the 16Sr and suggested cpn60-based classifications are contained in Table S1.
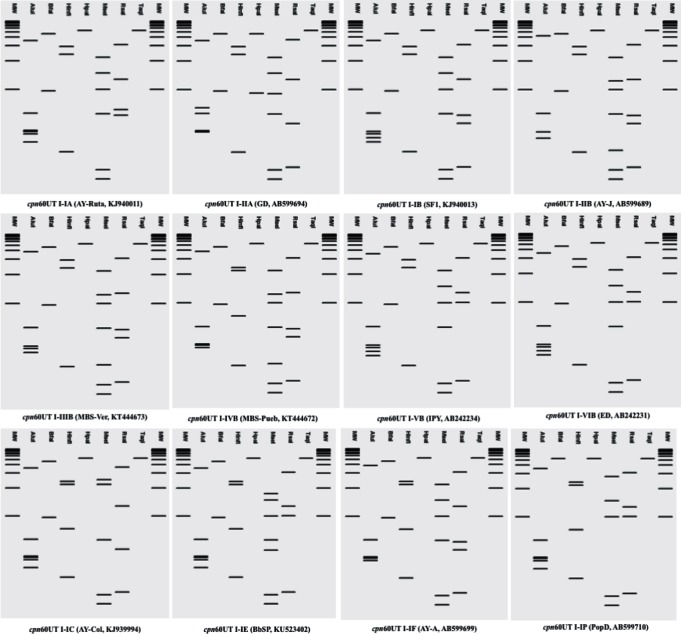
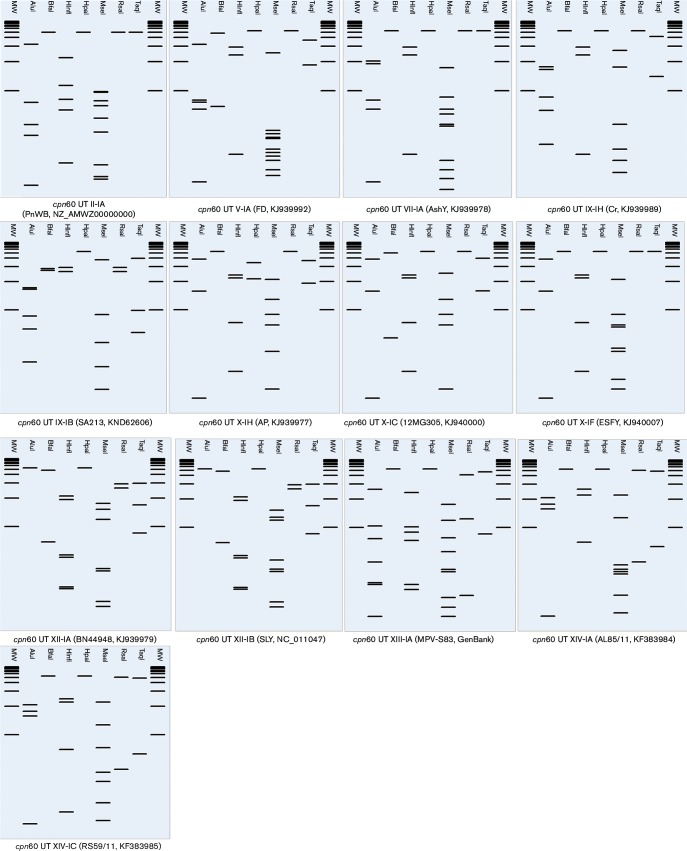
Since the development of the first coherent scheme to differentiate phytoplasmas, the use of RFLP has contributed to an understanding of phytoplasma diversity and has been used to differentiate strains that are phylogenetically closely related. This strategy has been used not only with the 16S rRNA gene, but also with rp (ribosomal protein) operon (Lee et al., 1998), secA (Hodgetts et al., 2008), cpn60 (Mitrović et al., 2011), and recently with rpoB (Valiunas et al., 2013). Following the strategies previously described, and taking into account the restriction sites present in the 552 bp corresponding to cpn60 UT in phytoplasmas, we found seven endonucleases capable of differentiating phytoplasma strains. All the cpn60 UT sequences used in this study were subjected to in silico RFLP with endonucleases AluI, BfaI, HinfI, HpaI, MseI, RsaI and TaqI using pDRAW32 software (AcaClone Software, http://www.acaclone.com). After comparing the RFLP patterns obtained for each strain, we detected 25 different RFLP patterns from 19 16Sr subgroups, which points to the increased diversity observed using cpn60 UT as an additional marker to differentiate phytoplasmas. The highest diversity was detected inside the 16SrI group. We detected two cpn60 RFLP profiles among the strain members of the 16SrI-A subgroup and six distinctive RFLP profiles within the members of the 16SrI-B subgroup, while for the rest of the subgroups we detected only one cpn60 RFLP pattern for each corresponding 16Sr subgroup. The virtual 4 % agarose gel electrophoresis patterns observed for each of the 25 reference strains detected in this study are presented in Figs 2 and3.
Fig. 2.
Distinctive RFLP patterns obtained with pDRAW32 from in silico digestion of cpn60 UT sequence from the 12 representative cpn60 UT subgroups within the group cpn60 UT I. In the computer-simulated digestions, the full set of seven enzymes AluI, BfaI, HinfI, HpaI, MseI, RsaI and TaqI were used. Lanes labelled MW represent Invitrogen 1 kb plus ladder.
Fig. 3.
Distinctive RFLP pattern obtained with pDRAW32 from in silico digestion of cpn60 UT sequence from the 12 representative cpn60 UT subgroups within the groups cpn60 UT II, cpn60 UT V, cpn60 UT VII, cpn60 UT IX, cpn60 UT X, cpn60 UT XII, cpn60 UT XIII and cpn60 UT XIV. In the computer-simulated digestions, the set of seven enzymes AluI, BfaI, HinfI, HpaI, MseI, RsaI and TaqI were used. Lanes labelled MW represent Invitrogen 1 kb plus ladder.
Based on the RFLP patterns observed, we separated the strains into cpn60 UT-based subgroups. To maintain consistency with the established nomenclature based on the 16S rRNA-encoding gene, we named the strains from group 16SrI as cpn60 UT I, 16SrII as cpn60 UT II, and so on. To name subgroups, for example the 16SrI-B, which had until now six different RFLP patterns among strains, we named the cpn60 UT subgroups as cpn60 UT I-IB, cpn60 UT I-IIB, cpn60 UT I-IIIB, (…), cpn60 UT I-VIB. All 96 strains analysed in this study were reclassified based on their cpn60 UT RFLP patterns (Table S1).
To establish the threshold similarity coefficient to delineate new cpn60 UT groups and subgroups, we calculated the similarity coefficients (F) among the 25 reference strains with unique RFLP patterns. We used the formula F=2Nxy / (Nx+Ny) (Nei & Li, 1979), where Nx and Ny are the number of bands resulting from the digestion of cpn60 UT with the seven endonucleases for strain x and strain y, respectively, and Nxy is the number of bands common to both strains. The number of bands generated by digesting the reference cpn60 UT sequences with each of the seven endonucleases used in this study is shown in Table 1.
Table 1. Number of bands produced by RFLP analysis of cpn60 UT sequences from reference phytoplasma strains of cpn60 UT groups.
| No. of bands generated | ||||||||
|---|---|---|---|---|---|---|---|---|
| cpn60 UT group | Strain | AluI | BfaI | HinfI | HpaI | MseI | RsaI | TaqI |
| cpn60 UT I | ||||||||
| cpn60 UT I-IA | AY-Ruta | 6 | 2 | 3 | 1 | 6 | 4 | 1 |
| cpn60 UT I-IIA | GD | 5 | 2 | 3 | 2 | 7 | 4 | 1 |
| cpn60 UT I-IB | SF1 | 7 | 2 | 3 | 1 | 6 | 5 | 1 |
| cpn60 UT I-IIB | AY-J | 5 | 2 | 3 | 1 | 7 | 5 | 1 |
| cpn60 UT I-IIIB | MBS-Ver | 6 | 2 | 3 | 1 | 7 | 5 | 1 |
| cpn60 UT I-IVB | MBS-Pueb | 5 | 2 | 4 | 1 | 7 | 5 | 1 |
| cpn60 UT I-VB | IPY | 7 | 2 | 3 | 1 | 6 | 4 | 1 |
| cpn60 UT I-VIB | ED | 6 | 2 | 3 | 1 | 6 | 4 | 1 |
| cpn60 UT I-IC | AY-Col | 6 | 2 | 4 | 1 | 5 | 4 | 1 |
| cpn60 UT I-IE | BbSP | 6 | 2 | 4 | 1 | 7 | 4 | 1 |
| cpn60 UT I-IF | AY-A | 5 | 2 | 4 | 1 | 6 | 5 | 1 |
| cpn60 UT I-IP | PopD | 6 | 1 | 4 | 1 | 5 | 4 | 1 |
| cpn60 UT II | ||||||||
| cpn60 UT II-IA | PnWB | 6 | 1 | 5 | 1 | 9 | 1 | 1 |
| cpn60 UT V | ||||||||
| cpn60 UT V-IA | FD | 5 | 2 | 3 | 1 | 14 | 1 | 3 |
| cpn60 UT VII | ||||||||
| cpn60 UT VII-IA | AshY | 5 | 1 | 3 | 1 | 12 | 1 | 1 |
| cpn60 UT IX | ||||||||
| cpn60 UT IX-IH | Cr | 6 | 1 | 3 | 1 | 6 | 1 | 2 |
| cpn60 UT IX-IB | SA213 | 6 | 2 | 3 | 1 | 5 | 2 | 3 |
| cpn60 UT X | ||||||||
| cpn60 UT X-IA | AP | 3 | 1 | 4 | 2 | 6 | 1 | 2 |
| cpn60 UT X-IC | 12MG305 | 3 | 2 | 4 | 1 | 5 | 1 | 2 |
| cpn60 UT X-IF | ESFY | 3 | 1 | 4 | 1 | 8 | 1 | 1 |
| cpn60 UT XII | ||||||||
| cpn60 UT XII-IA | BN44948 | 1 | 2 | 6 | 1 | 7 | 2 | 3 |
| cpn60 UT XII-IB | AT | 1 | 2 | 6 | 1 | 8 | 2 | 3 |
| cpn60 UT XIII | ||||||||
| cpn60 UT XIII-IA | MPV-S83 | 7 | 1 | 6 | 1 | 9 | 3 | 2 |
| cpn60 UT XIV | ||||||||
| cpn60 UT XIV-IA | AL85/11 | 4 | 1 | 3 | 1 | 10 | 2 | 2 |
| cpn60 UT XIV-IC | RS59/11 | 4 | 1 | 4 | 1 | 9 | 2 | 2 |
The similarity coefficients among the 25 reference strains are shown in Table 2. We found that the F value between strains from the same cpn60 UT group varied from 0.97 to 0.62, while F values lower than 0.62 belonged to strains classified in a different cpn60 UT group (Table 2). Based on these results we confirmed the presence of two cpn60 UT subgroups inside the cpn60 UT I-A group (cpn60 UT I-IA and cpn60 UT I-IIA), and six subgroups inside the cpn60 UT I-B group (cpn60 UT I-IB to cpn60 UT I-VIB). We suggest 0.97 as the threshold similarity coefficient to delineate new subgroups based on the use of the seven endonucleases previously mentioned, while 0.60 can be considered as the threshold similarity coefficient to delineate new groups. The threshold to delineate new cpn60 UT subgroups (0.97), corresponds with the threshold to delineate new 16S rRNA gene subgroups (Wei et al., 2007).
Table 2. Similarity coefficients obtained from RFLP analysis of cpn60 UT sequences from reference phytoplasma strains.
| cpn60 UT classification | Strains | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | cpn60 UT I-IA | AY-Ruta | 1.00 | |||||||||||||||||||||||
| 2 | cpn60 UT I-IIA | GD | 0.83 | 1.00 | ||||||||||||||||||||||
| 3 | cpn60 UT I-IB | SF1 | 0.95 | 0.84 | 1.00 | |||||||||||||||||||||
| 4 | cpn60 UT I-IIB | AY-J | 0.86 | 0.81 | 0.92 | 1.00 | ||||||||||||||||||||
| 5 | cpn60 UT I-IIIB | MBS-Ver | 0.87 | 0.79 | 0.95 | 0.97 | 1.00 | |||||||||||||||||||
| 6 | cpn60 UT I-IVB | MBS-Pueb | 0.89 | 0.79 | 0.90 | 0.92 | 0.95 | 1.00 | ||||||||||||||||||
| 7 | cpn60 UT I-VB | IPY | 0.89 | 0.81 | 0.97 | 0.89 | 0.92 | 0.89 | 1.00 | |||||||||||||||||
| 8 | cpn60 UT I-VIB | ED | 0.94 | 0.83 | 0.95 | 0.86 | 0.89 | 0.92 | 0.97 | 1.00 | ||||||||||||||||
| 9 | cpn60 UT I-IC | AY-Col | 0.89 | 0.83 | 0.89 | 0.81 | 0.84 | 0.86 | 0.86 | 0.89 | 1.00 | |||||||||||||||
| 10 | cpn60 UT I-IE | BbSP | 0.89 | 0.79 | 0.90 | 0.82 | 0.85 | 0.87 | 0.92 | 0.95 | 0.85 | 1.00 | ||||||||||||||
| 11 | cpn60 UT I-IF | AY-A | 0.92 | 0.81 | 0.92 | 0.89 | 0.92 | 0.95 | 0.89 | 0.92 | 0.92 | 0.92 | 1.00 | |||||||||||||
| 12 | cpn60 UT I-IP | PopD | 0.83 | 0.72 | 0.84 | 0.76 | 0.79 | 0.81 | 0.86 | 0.89 | 0.83 | 0.89 | 0.81 | 1.00 | ||||||||||||
| 13 | cpn60 UT II-IA | PnWB | 0.15 | 0.10 | 0.15 | 0.15 | 0.15 | 0.15 | 0.15 | 0.15 | 0.15 | 0.15 | 0.15 | 0.21 | 1.00 | |||||||||||
| 14 | cpn60 UT IX-IA | Cr | 0.41 | 0.29 | 0.39 | 0.29 | 0.33 | 0.34 | 0.40 | 0.41 | 0.41 | 0.39 | 0.34 | 0.47 | 0.22 | 1.00 | ||||||||||
| 15 | cpn60 UT V-IA | FD | 0.29 | 0.19 | 0.27 | 0.23 | 0.27 | 0.28 | 0.28 | 0.29 | 0.29 | 0.27 | 0.28 | 0.24 | 0.13 | 0.35 | 1.00 | |||||||||
| 16 | cpn60 UT VII-IA | AshY | 0.26 | 0.15 | 0.24 | 0.20 | 0.24 | 0.30 | 0.25 | 0.26 | 0.26 | 0.29 | 0.25 | 0.31 | 0.24 | 0.38 | 0.58 | 1.00 | ||||||||
| 17 | cpn60 UT X-IA | AP | 0.31 | 0.31 | 0.29 | 0.30 | 0.29 | 0.30 | 0.30 | 0.31 | 0.38 | 0.35 | 0.36 | 0.38 | 0.17 | 0.40 | 0.42 | 0.34 | 1.00 | |||||||
| 18 | cpn60 UT X-IC | 12MG305 | 0.39 | 0.32 | 0.36 | 0.38 | 0.36 | 0.38 | 0.38 | 0.39 | 0.45 | 0.42 | 0.44 | 0.39 | 0.18 | 0.41 | 0.38 | 0.35 | 0.74 | 1.00 | ||||||
| 19 | cpn60 UT X-IF | ESFY | 0.41 | 0.35 | 0.39 | 0.40 | 0.39 | 0.40 | 0.40 | 0.41 | 0.47 | 0.44 | 0.46 | 0.47 | 0.27 | 0.44 | 0.45 | 0.43 | 0.80 | 0.76 | 1.00 | |||||
| 20 | cpn60 UT XII-IA | BN44948 | 0.29 | 0.23 | 0.27 | 0.28 | 0.27 | 0.28 | 0.28 | 0.29 | 0.29 | 0.27 | 0.28 | 0.29 | 0.11 | 0.36 | 0.29 | 0.21 | 0.32 | 0.33 | 0.36 | 1.00 | ||||
| 21 | cpn60 UT XII-IB | SYL | 0.28 | 0.22 | 0.26 | 0.27 | 0.26 | 0.27 | 0.27 | 0.28 | 0.28 | 0.26 | 0.27 | 0.28 | 0.10 | 0.35 | 0.29 | 0.26 | 0.31 | 0.32 | 0.35 | 0.97 | 1.00 | |||
| 22 | cpn60 UT XIII-IA | MPV-S83 | 0.38 | 0.38 | 0.40 | 0.42 | 0.41 | 0.42 | 0.42 | 0.43 | 0.48 | 0.45 | 0.47 | 0.48 | 0.13 | 0.35 | 0.33 | 0.27 | 0.47 | 0.38 | 0.50 | 0.49 | 0.48 | 1.00 | ||
| 23 | cpn60 UT XIV-IA | AL85/11 | 0.17 | 0.11 | 0.16 | 0.16 | 0.16 | 0.16 | 0.16 | 0.17 | 0.17 | 0.16 | 0.16 | 0.22 | 0.21 | 0.35 | 0.24 | 0.26 | 0.38 | 0.32 | 0.41 | 0.29 | 0.28 | 0.33 | 1.00 | |
| 24 | cpn60 UT XIV-IC | RS59/11 | 0.17 | 0.11 | 0.16 | 0.16 | 0.16 | 0.16 | 0.16 | 0.17 | 0.16 | 0.16 | 0.16 | 0.22 | 0.21 | 0.35 | 0.19 | 0.26 | 0.31 | 0.32 | 0.35 | 0.23 | 0.22 | 0.29 | 0.94 | 1.00 |
Subgroup cpn60 UT I-IA is represented by Brassica spp. phytoplasma strain AY-Ruta (GenBank accession no. KJ940011), and cpn60 UT I-IIA is represented by Grey dogwood stunt phytoplasma strain GD (GenBank accession no. AB599694). The subgroup cpn60 UT I-IB is represented by Linum usitatissimum phytoplasma strain SF1 (GenBank accession no. KJ940013); cpn60 UT I-IIB is represented by Aster yellow phytoplasma strain AY-J (GenBank accession no. AB599689); cpn60 UT I-IIIB and cpn60 UT I-IVB are represented by Maize bushy stunt phytoplasma, strains MBS-Ver (GenBank accession no. KT444673) and MBS-Pueb (GenBank accession no. KT444672), respectively. Subgroup cpn60 UT I-VB is represented by Iceland poppy yellows phytoplasma strain IPY (GenBank accession no. AB242234), and the subgroup cpn60 UT I-VIB is represented by Eggplant dwarf phytoplasma strain ED (GenBank accession no. AB242231). Subgroup cpn60 UT I-IC is represented by Aster Yellow phytoplasma strain AY-Col (GenBank accession no. KJ939994); cpn60 UT I-IE is represented by Blueberry stunt phytoplasma strain BbSP (GenBank accession no. KU523402); cpn60 UT I-IF is represented by Apricot chlorotic leafroll phytoplasma strain AY-A (GenBank accession no. AB599699); and cpn60 UT I-IP represented by Populus decline phytoplasma strain PopD (GenBank accession no. AB599710).
Inside the groups cpn60 UT II, V, VII, and XIII, we only had strains from one subgroup, so we were not able to detect more than one RFLP pattern. The subgroup cpn60 UT II-IA is represented by Peanut witches’-broom phytoplasma strain PnWB (GenBank accession no. NZ_AMWZ00000000); subgroup cpn60 UT V-IA is represented by the Flavescence doree phytoplasma strain FD (GenBank accession no. KJ939992); the subgroup cpn60 UT VII-IA, on the other hand, is represented by Ash Yellow phytoplasma strain AshY (GenBank accession no. KJ939978). Subgroup cpn60 UT IX-IH and cpn60 UT IX-IB are represented by Catharanthus roseus phoenicium phytoplasma strain Cr (GenBank accession no. KJ939989) and Almond witches’-broom strain SA213 (GenBank accession no. KND62606), respectively. Inside the group cpn60 UT X, we were able to differentiate members of the subgroups cpn60 UT X-IA, represented by Apple proliferation phytoplasma (GenBank accession no. KJ939977), members of the subgroup cpn60 UT X-IC represented by Pear decline phytoplasma strain 12MG305 (GenBank accession no. KJ940000), and members of the subgroup cpn60 UT X-IF represented by the European stone fruit phytoplasma strain ESFY (GenBank accession no. KJ940007). Inside the group cpn60 UT XII we identified two subgroups, subgroup cpn60 UT XII-IA, represented by Bois noir phytoplasma strain BN44948 (GenBank accession no. KJ939979), and subgroup cpn60 UT XII-IB, represented by Strawberry lethal yellow strain AT (GenBank accession no. NC_011047). Subgroup cpn60 UT XIII-IA was represented by Mexican periwinkle virescence strain MPV-S83 (GenBank accession no. KT444668). Finally, we identified two subgroups inside the group cpn60 UT XIV, subgroup cpn60 UT XIV-IA, represented by Bermuda white leaf phytoplasma strain AL85/11 (GenBank accession no. KF383984), and subgroup cpn60 UT XIV-IC represented by Bermuda white leaf phytoplasma strain RS59/11 (GenBank accession no. KF383985).
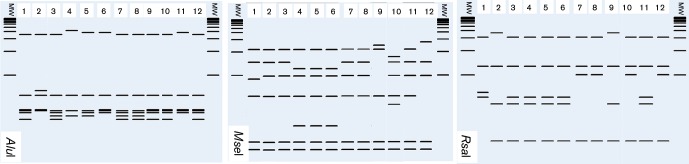
Analysing the RFLP patterns for each group, we identified enzymes capable of differentiating cpn60 UT-subgroups. Subgroups from the group cpn60 UT I can be differentiated through the use of AluI, MseI and RsaI (Fig. 4). Subgroups from group cpn60 UT X can be differentiated using endonucleases HpaI, MseI and TaqI (Fig. 5). Subgroups included in group cpn60 UT XII can be differentiated only by the pattern generated by MseI (Fig. 6), while subgroups within cpn60 UT XIV can be differentiated by HinfI and MseI (Fig. 7). The in vitro RFLP profile from strains within the group cpn60 UT IX differed with six of the seven endonucleases (not shown). Moreover, we observed correspondence between the in silico and in vitro RFLP for 12 phytoplasma strains representing the three major phylogenetic subclades into which phytoplasmas are grouped [(Dumonceaux et al., 2014); not shown].
Fig. 4.
Key restriction enzymes to differentiate strains belonging to the subgroups within the group cpn60 UT I. Lanes 1 and 2 represent subgroups cpn60 UT I-IA and cpn60 UT I-IIA, respectively. Lanes 3 to 8 represent strains cpn60 UT I-IB to cpn60 UT I-VIB, respectively. Lanes 9, 10, 11, and 12 represent strains cpn60 UT I-IC, cpn60 UT I-IE, cpn60 UT I-IF, and cpn60 UT I-IP, respectively. Lanes labelled MW represent Invitrogen 1 kb plus ladder.
Fig. 5.
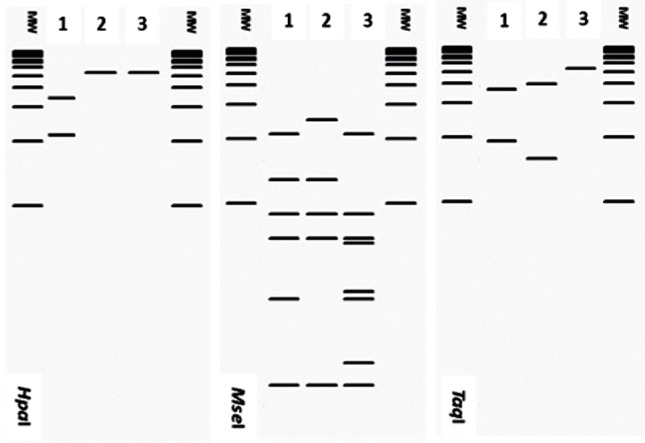
Key restriction enzymes to differentiate strains belonging to the subgroups within the group cpn60 UT X. Lanes 1, 2, and 3 represent subgroups cpn60 UT X-IA, cpn60 UT X-IC, and cpn60 UT X-IF, respectively. Lanes labelled MW represent Invitrogen 1 kb plus ladder.
Fig. 6.

Key restriction enzymes to differentiate strains belonging to the subgroups within the group cpn60 UT XII. Lanes 1 and 2 represent subgroups cpn60 UT XII-IA and cpn60 UT XII-IB, respectively. Lanes labelled MW represent Invitrogen 1 kb plus ladder.
Fig. 7.
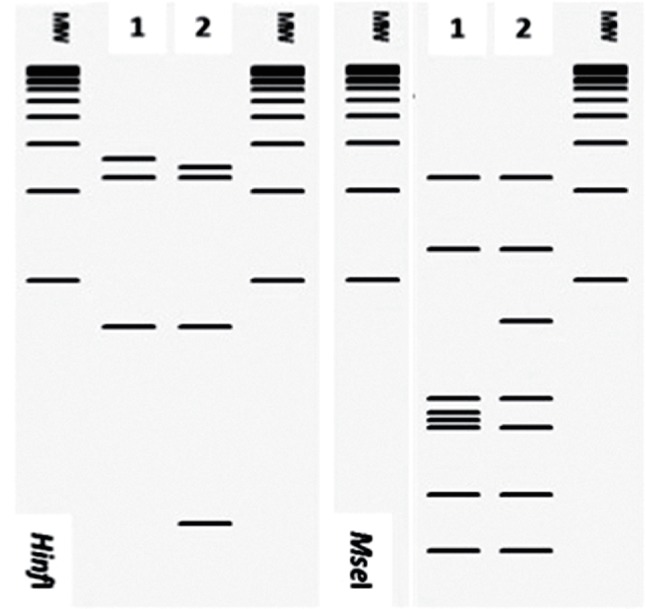
Key restriction enzymes to differentiate strains belonging to the subgroups within the group cpn60 UT XIV. Lanes 1 and 2 represent subgroups cpn60 UT XIV-IA and cpn60 UT XIV-IC, respectively. Lanes labelled MW represent Invitrogen 1 kb plus ladder.
After aligning the 25 cpn60 UT reference strains we detected 92–99 % nucleotide sequence identity among cpn60 UT subgroups within the same group, while the sequence identities between groups was 61–84 %. The variability shown by cpn60 UT sequences was higher compared to the 16Sr RNA gene and other genes previously used as phytoplasma markers. cpn60 UT sequences could differentiate closely related phytoplasma strains more precisely. We observed the same trend between similarity coefficient (Table 2), and nucleotide similarity (Table 3).
Table 3. Nucleotide similarity obtained from the alignment of cpn60 UT sequences from reference phytoplasma strains.
| cpn60 UT classification | Strains | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | cpn60 UT I-IA | AY-Ruta | 1 | |||||||||||||||||||||||
| 2 | cpn60 UT I-IIA | GD | 0.98 | 1 | ||||||||||||||||||||||
| 3 | cpn60 UT I-IB | SF1 | 0.97 | 0.97 | 1 | |||||||||||||||||||||
| 4 | cpn60 UT I-IIB | AY-J | 0.97 | 0.97 | 0.99 | 1 | ||||||||||||||||||||
| 5 | cpn60 UT I-IIIB | MBS-Ver | 0.97 | 0.97 | 0.99 | 0.99 | 1 | |||||||||||||||||||
| 6 | cpn60 UT I-IVB | MBS-Pueb | 0.97 | 0.97 | 0.99 | 0.99 | 0.99 | 1 | ||||||||||||||||||
| 7 | cpn60 UT I-VB | IPY | 0.97 | 0.97 | 0.99 | 0.99 | 0.99 | 0.99 | 1 | |||||||||||||||||
| 8 | cpn60 UT I-VIB | ED | 0.97 | 0.97 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 1 | ||||||||||||||||
| 9 | cpn60 UT I-IC | AY-Col | 0.98 | 0.98 | 0.97 | 0.97 | 0.97 | 0.97 | 0.97 | 0.97 | 1 | |||||||||||||||
| 10 | cpn60 UT I-IE | BbSP | 0.97 | 0.97 | 0.97 | 0.96 | 0.97 | 0.97 | 0.97 | 0.97 | 0.98 | 1 | ||||||||||||||
| 11 | cpn60 UT I-IF | AY-A | 0.97 | 0.97 | 0.97 | 0.96 | 0.97 | 0.97 | 0.97 | 0.97 | 0.98 | 0.98 | 1 | |||||||||||||
| 12 | cpn60 UT I-IP | PopD | 0.94 | 0.94 | 0.94 | 0.93 | 0.94 | 0.94 | 0.94 | 0.94 | 0.94 | 0.94 | 0.94 | 1 | ||||||||||||
| 13 | cpn60 UT II-IA | PnWB | 0.65 | 0.65 | 0.65 | 0.65 | 0.65 | 0.65 | 0.65 | 0.65 | 0.65 | 0.65 | 0.66 | 0.65 | 1 | |||||||||||
| 14 | cpn60 UT IX-IA | Cr | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.71 | 0.7 | 0.69 | 0.66 | 1 | ||||||||||
| 15 | cpn60 UT V-IA | FD | 0.66 | 0.66 | 0.65 | 0.65 | 0.65 | 0.65 | 0.65 | 0.65 | 0.66 | 0.66 | 0.66 | 0.66 | 0.64 | 0.69 | 1 | |||||||||
| 16 | cpn60 UT VII-IA | AshY | 0.62 | 0.63 | 0.62 | 0.62 | 0.62 | 0.62 | 0.62 | 0.62 | 0.63 | 0.63 | 0.63 | 0.63 | 0.63 | 0.7 | 0.83 | 1 | ||||||||
| 17 | cpn60 UT X-IA | AP | 0.76 | 0.76 | 0.76 | 0.76 | 0.76 | 0.76 | 0.76 | 0.76 | 0.76 | 0.76 | 0.76 | 0.75 | 0.64 | 0.74 | 0.71 | 0.69 | 1 | |||||||
| 18 | cpn60 UT X-IC | 12MG305 | 0.75 | 0.76 | 0.76 | 0.76 | 0.76 | 0.76 | 0.76 | 0.76 | 0.76 | 0.75 | 0.76 | 0.76 | 0.64 | 0.73 | 0.7 | 0.69 | 0.94 | 1 | ||||||
| 19 | cpn60 UT X-IF | ESFY | 0.77 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 | 0.77 | 0.65 | 0.74 | 0.71 | 0.69 | 0.95 | 0.95 | 1 | |||||
| 20 | cpn60 UT XII-IA | BN44948 | 0.8 | 0.79 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.79 | 0.79 | 0.81 | 0.63 | 0.69 | 0.61 | 0.61 | 0.74 | 0.74 | 0.75 | 1 | ||||
| 21 | cpn60 UT XII-IB | SYL | 0.8 | 0.79 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.8 | 0.79 | 0.79 | 0.81 | 0.63 | 0.68 | 0.61 | 0.61 | 0.74 | 0.74 | 0.75 | 0.99 | 1 | |||
| 22 | cpn60 UT XIII-IA | MPV-S83 | 0.82 | 0.82 | 0.82 | 0.82 | 0.82 | 0.82 | 0.82 | 0.82 | 0.83 | 0.82 | 0.82 | 0.84 | 0.64 | 0.71 | 0.65 | 0.64 | 0.76 | 0.78 | 0.78 | 0.81 | 0.81 | 1 | ||
| 23 | cpn60 UT XIV-IA | AL85/11 | 0.69 | 0.69 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.69 | 0.7 | 0.7 | 0.7 | 0.67 | 0.75 | 0.72 | 0.71 | 0.76 | 0.76 | 0.76 | 0.67 | 0.67 | 0.69 | 1 | |
| 24 | cpn60 UT XIV-IC | RS59/11 | 0.68 | 0.69 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | 0.69 | 0.7 | 0.69 | 0.69 | 0.67 | 0.75 | 0.71 | 0.7 | 0.75 | 0.75 | 0.76 | 0.67 | 0.67 | 0.68 | 0.96 | 1 |
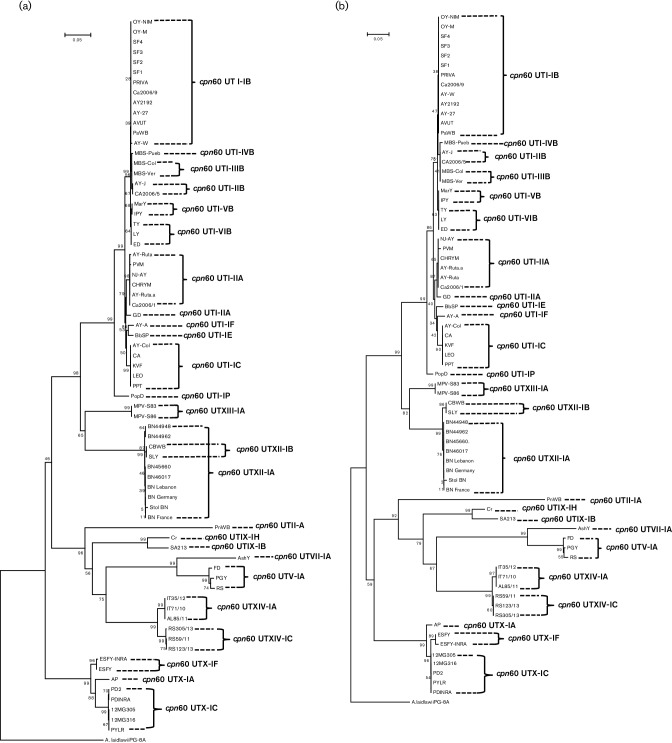
Phylogenetic analysis of cpn60 UT sequences of all the groups and subgroups identified in this study was performed using the neighbour-joining method, using the tree-bisection-and-regrafting (TBR) algorithm available in the mega6 software package (Tamura et al., 2013), with bootstrapping 1000 times for nucleotide (Fig. 8a) and amino acid (Fig. 8b) sequences. Both phylogenetic trees showed distinction between the cpn60 UT groups and subgroups, supporting the results obtained through the RFLP analysis, the calculation of F value and the nucleotide identity among the reference strains. Phylogenetic analysis of cpn60 UT sequences showed a better resolution of the subgroup B, identified inside the group cpn60 UT I (Fig. 8a), while the phylogenetic tree using the amino acid sequences allowed a better resolution of the subgroups identified within the group cpn60 UT XII (Fig. 8b).
Fig. 8.
Phylogenetic tree reconstructed through the neighbuor-joining method of the cpn60 UT nucleotide (a) and amino acid (b) sequences of phytoplasma strains from the cpn60 groups and subgroups described in this study. Strain descriptions and GenBank accession numbers are shown in Table S1. Acholeplasma laidlawii was used as outgroup. Trees were bootstrapped 1000 times to achieve reliability. Bar, 5 substitutions in 100 positions.
The present study confirms previously published work (Dumonceaux et al., 2014; Mitrović et al., 2011, 2015) showing the capability of cpn60 UT sequences to act as an additional marker to differentiate phytoplasmas. Strains that are closely related based on 16S rRNA gene sequence classification were differentiated as members of new subgroups, contributing to a better identification of the strains. Previous studies mentioned a high nucleotide similarity between the cpn60-encoding genes amplified from members of the 16SrI-B subgroup(Kakizawa et al., 2006), but with the increased number of the strains characterized in this study, we showed that the nucleotide variability is higher among strains from the same 16Sr subgroup than was thought.
Protein-encoding genes are known to provide a better strain resolution compared to rRNA-encoding genes (Zeigler, 2003). Unlike the 16S rRNA gene, cpn60 is present in a single copy in the phytoplasma genome, which obviates the taxonomic complications related with the occasional presence of heterogeneous ribosomal operons (Wei et al., 2007; Zhao et al., 2009). The identification of distinct phytoplasma strains is very important to vector studies, epidemiological research and development of management strategies. The classification scheme we describe herein provides a supplementary tool to the existing classification scheme based on the 16S rRNA-based F2nR2 locus. If certain subgroups of phytoplasma are confirmed to lack a gene encoding Cpn60, then this classification scheme will not apply to these groups. However, it has been noted that Mollicutes lacking cpn60 do not tend to invade cells (Clark & Tillier, 2010), so phytoplasmas that do not encode this gene would constitute exceptions among the Mollicutes. Nevertheless, including cpn60 UT among the additional markers used to characterize phytoplasma strains will improve the understanding of phytoplasmas. This study, supported by the cpnDB (Hill et al., 2004), could be the first step in the development of interactive online tools capable of classifying phytoplasmas based on an unknown cpn60 UT sequence amplified from phytoplasmas.
Acknowledgements
This work was supported by the Genomic Research and Development Initiative for the shared priority project on quarantine and invasive species. E. P.-L. thanks CONACYT for PhD scholarship (CVU: 517835) and the Government of Canada for internship at Agriculture and Agri-food Canada-Saskatoon Research Centre.
Supplementary Data
References
- Andersen M. T., Liefting L. W., Havukkala I., Beever R. E.(2013). Comparison of the complete genome sequence of two closely related isolates of ‘Candidatus Phytoplasma australiense’ reveals genome plasticity. BMC Genomics 14529. 10.1186/1471-2164-14-529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud G., Malembic-Maher S., Salar P., Bonnet P., Maixner M., Marcone C., Boudon-Padieu E., Foissac X.(2007). Multilocus sequence typing confirms the close genetic interrelatedness of three distinct flavescence dorée phytoplasma strain clusters and group 16SrV phytoplasmas infecting grapevine and alder in Europe. Appl Environ Microbiol 734001–4010. 10.1128/AEM.02323-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Zhang J., Ewing A., Miller S. A., Jancso Radek A., Shevchenko D. V., Tsukerman K., Walunas T., Lapidus A., et al. (2006). Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J Bacteriol 1883682–3696. 10.1128/JB.188.10.3682-3696.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botti S., Bertaccini A.(2003). Variability and functional role of chromosomal sequences in 16SrI-B subgroup phytoplasmas including aster yellows and related strains. J Appl Microbiol 94103–110. 10.1046/j.1365-2672.2003.01809.x [DOI] [PubMed] [Google Scholar]
- Chung W. C., Chen L. L., Lo W. S., Lin C. P., Kuo C. H.(2013). Comparative analysis of the peanut witches'-broom phytoplasma genome reveals horizontal transfer of potential mobile units and effectors. PLoS One 8e62770. 10.1371/journal.pone.0062770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claisse O., Renouf V., Lonvaud-Funel A.(2007). Differentiation of wine lactic acid bacteria species based on RFLP analysis of a partial sequence of rpoB gene. J Microbiol Methods 69387–390. 10.1016/j.mimet.2007.01.004 [DOI] [PubMed] [Google Scholar]
- Clark G. W., Tillier E. R.(2010). Loss and gain of GroEL in the Mollicutes. Biochem Cell Biol 88185–194. 10.1139/o09-157 [DOI] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J. M., Brenner S. E.(2004). WebLogo: a sequence logo generator. Genome Res 141188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. E., Zhao Y., Dally E. L., Lee I. M., Jomantiene R., Douglas S. M.(2013). ‘Candidatus Phytoplasma pruni’, a novel taxon associated with X-disease of stone fruits, Prunus spp.: multilocus characterization based on 16S rRNA, secY, and ribosomal protein genes. Int J Syst Evol Microbiol 63766–776. 10.1099/ijs.0.041202-0 [DOI] [PubMed] [Google Scholar]
- Doi Y., Teranaka M., Yora K., Asuyama H.(1967). Mycoplasma- or PLT group-like microorganisms found in the phloem elements of plants infected with mulberry dwarf, potato witches' broom, aster yellows, or paulownia witches' broom. Ann Phytopathol Japan 33259–266. 10.3186/jjphytopath.33.259 [DOI] [Google Scholar]
- Dumonceaux T. J., Green M., Hammond C., Perez E., Olivier C.(2014). Molecular diagnostic tools for detection and differentiation of phytoplasmas based on chaperonin-60 reveal differences in host plant infection patterns. PLoS One 9e116039. 10.1371/journal.pone.0116039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascuel O., Steel M.(2006). Neighbor-joining revealed. Mol Biol Evol 231997–2000. 10.1093/molbev/msl072 [DOI] [PubMed] [Google Scholar]
- Goh S. H., Potter S., Wood J. O., Hemmingsen S. M., Reynolds R. P., Chow A. W.(1996). HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J Clin Microbiol 34818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haakensen M., Pittet V., Ziola B.(2011). Reclassification of Paralactobacillus selangorensis Leisner et al. 2000 as Lactobacillus selangorensis comb. nov. Int J Syst Evol Microbiol 612979–2983. 10.1099/ijs.0.027755-0 [DOI] [PubMed] [Google Scholar]
- Harrison N. A., Davis R. E., Oropeza C., Helmick E. E., Narváez M., Eden-Green S., Dollet M., Dickinson M.(2014). ‘Candidatus Phytoplasma palmicola’, associated with a lethal yellowing-type disease of coconut (Cocos nucifera L.) in Mozambique. Int J Syst Evol Microbiol 641890–1899. 10.1099/ijs.0.060053-0 [DOI] [PubMed] [Google Scholar]
- Hedberg M. E., Israelsson A., Moore E. R., Svensson-Stadler L., Wai S. N., Pietz G., Sandström O., Hernell O., Hammarström M. L., Hammarström S.(2013). Prevotella jejuni sp. nov., isolated from the small intestine of a child with coeliac disease. Int J Syst Evol Microbiol 634218–4223. 10.1099/ijs.0.052647-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. E., Penny S. L., Crowell K. G., Goh S. H., Hemmingsen S. M.(2004). cpnDB: a chaperonin sequence database. Genome Res 141669–1675. 10.1101/gr.2649204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgetts J., Boonham N., Mumford R., Harrison N., Dickinson M.(2008). Phytoplasma phylogenetics based on analysis of secA and 23S rRNA gene sequences for improved resolution of candidate species of ‘Candidatus Phytoplasma’. Int J Syst Evol Microbiol 581826–1837. 10.1099/ijs.0.65668-0 [DOI] [PubMed] [Google Scholar]
- Hogenhout S. A., Oshima K., Ammar el-D., Kakizawa S., Kingdom H. N., Namba S.(2008). Phytoplasmas: bacteria that manipulate plants and insects. Mol Plant Pathol 9403–423. 10.1111/j.1364-3703.2008.00472.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- IRPCM Phytoplasma/Spiroplasma Working Team-Phytoplasma Taxonomy Group (2004).‘Candidatus Phytoplasma’, a taxon for the wall-less,non-helical prokaryotes that colonize plant phloem and insects. Int J Syst Evol Microbiol 541243–1255. 10.1099/ijs.0.02854-0 [DOI] [PubMed] [Google Scholar]
- Kakizawa S., Oshima K., Jung H. Y., Suzuki S., Nishigawa H., Arashida R., Miyata S., Ugaki M., Kishino H., Namba S.(2006). Positive selection acting on a surface membrane protein of the plant-pathogenic phytoplasmas. J Bacteriol 1883424–3428. 10.1128/JB.188.9.3424-3428.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kube M., Schneider B., Kuhl H., Dandekar T., Heitmann K., Migdoll A. M., Reinhardt R., Seemüller E.(2008). The linear chromosome of the plant-pathogenic mycoplasma ‘Candidatus Phytoplasma mali’. BMC Genomics 9306. 10.1186/1471-2164-9-306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.-M., Hammond R. W., Davis R. E., Gundersen D. E.(1993). Universal amplification and analysis of pathogen 16S rDNA for classification and identification of mycoplasmalike organisms. Phytopathol 83834–842. 10.1094/Phyto-83-834 [DOI] [Google Scholar]
- Lee I. M., Gundersen-Rindal D. E., Davis R. E., Bartoszyk I. M.(1998). Revised classification scheme of phytoplasmas based on RFLP analyses of 16S rRNA and ribosomal protein gene sequences. Int J Sys Bacteriol 481153–1169. 10.1099/00207713-48-4-1153 [DOI] [Google Scholar]
- Lee I. M., Zhao Y., Bottner K. D.(2006). SecY gene sequence analysis for finer differentiation of diverse strains in the aster yellows phytoplasma group. Mol Cell Probes 2087–91. 10.1016/j.mcp.2005.10.001 [DOI] [PubMed] [Google Scholar]
- Links M. G., Dumonceaux T. J., Hemmingsen S. M., Hill J. E.(2012). The chaperonin-60 universal target is a barcode for bacteria that enables de novo assembly of metagenomic sequence data. PLoS One 7e49755. 10.1371/journal.pone.0049755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcone C., Lee I. M., Davis R. E., Ragozzino A., Seemüller E.(2000). Classification of aster yellows-group phytoplasmas based on combined analyses of rRNA and tuf gene sequences. Int J Syst Evol Microbiol 501703–1713. 10.1099/00207713-50-5-1703 [DOI] [PubMed] [Google Scholar]
- Marqués A. M., Burgos-Díaz C., Aranda F. J., Teruel J. A., Manresa À., Ortiz A., Farfán M.(2012). Sphingobacterium detergens sp. nov., a surfactant-producing bacterium isolated from soil. Int J Syst Evol Microbiol 623036–3041. 10.1099/ijs.0.036707-0 [DOI] [PubMed] [Google Scholar]
- Mitrović J., Kakizawa S., Duduk B., Oshima K., Namba S., Bertaccini A.(2011). The groEL gene as an additional marker for finer differentiation of ‘Candidatus Phytoplasma asteris’-related strains. Ann Appl Biol 15941–48. 10.1111/j.1744-7348.2011.00472.x [DOI] [Google Scholar]
- Mitrović J., Smiljković M., Seemüller E., Reinhardt R., Hüttel B., Büttner C., Bertaccini A., Kube M., Duduk B.(2015). Differentiation of ‘Candidatus Phytoplasma cynodontis’ based on 16S rRNA and groEL genes and identification of a new subgroup 16SrXIV-C. Plant Dis 991578–1583. 10.1094/PDIS-01-15-0061-RE [DOI] [PubMed] [Google Scholar]
- Murray R. G., Stackebrandt E.(1995). Taxonomic note: implementation of the provisional status Candidatus for incompletely described procaryotes. Int J Syst Bacteriol 45186–187. 10.1099/00207713-45-1-186 [DOI] [PubMed] [Google Scholar]
- Nei M., Li W. H.(1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A 765269–5273. 10.1073/pnas.76.10.5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejat N., Vadamalai G., Davis R. E., Harrison N. A., Sijam K., Dickinson M., Abdullah S. N., Zhao Y.(2013). ‘Candidatus Phytoplasma malaysianum’, a novel taxon associated with virescence and phyllody of Madagascar periwinkle (Catharanthus roseus). Int J Syst Evol Microbiol 63540–548. 10.1099/ijs.0.041467-0 [DOI] [PubMed] [Google Scholar]
- Oshima K., Kakizawa S., Nishigawa H., Jung H. Y., Wei W., Suzuki S., Arashida R., Nakata D., Miyata S., et al. (2004). Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat Genet 3627–29. 10.1038/ng1277 [DOI] [PubMed] [Google Scholar]
- Oshima K., Maejima K., Namba S.(2013). Genomic and evolutionary aspects of phytoplasmas. Front Microbiol 4230. 10.3389/fmicb.2013.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-H., Jung J.-H., Seo D.-H., Lee H.-L., Kim G.-W., Park S.-Y., Shin W.-C., Hong S., Park C.-S.(2012). Differentiation of lactic acid bacteria based on RFLP analysis of the tuf gene. Food Science and Biotechnology 21911–915. 10.1007/s10068-012-0119-9 [DOI] [Google Scholar]
- Pérez-López E., Luna-Rodríguez M., Olivier C. Y., Dumonceaux T. J.(2016a). The underestimated diversity of phytoplasmas in Latin America. Int J Syst Evol Microbiol 66492–513. 10.1099/ijsem.0.000726 [DOI] [PubMed] [Google Scholar]
- Pérez-López E., Olivier C. Y., Luna-Rodríguez M., Rodríguez Y., Iglesias L. G., Castro-Luna A., Adame-García J., Dumonceaux T. J.(2016b). Maize bushy stunt phytoplasma affects native corn at high elevations in Southeast Mexico. Eur J Plant Pathol 145963–971. 10.1007/s10658-016-0883-0 [DOI] [Google Scholar]
- Quaglino F., Kube M., Jawhari M., Abou-Jawdah Y., Siewert C., Choueiri E., Sobh H., Casati P., Tedeschi R., et al. (2015). ‘Candidatus Phytoplasma phoenicium’ associated with almond witches'-broom disease: from draft genome to genetic diversity among strain populations. BMC Microbiol 15148. 10.1186/s12866-015-0487-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A., Poblet M., Mas A., Guillamón J. M.(2000). Identification of acetic acid bacteria by RFLP of PCR-amplified 16S rDNA and 16S-23S rDNA intergenic spacer. Int J Syst Evol Microbiol 501981–1987. 10.1099/00207713-50-6-1981 [DOI] [PubMed] [Google Scholar]
- Saccardo F., Martini M., Palmano S., Ermacora P., Scortichini M., Loi N., Firrao G.(2012). Genome drafts of four phytoplasma strains of the ribosomal group 16SrIII. Microbiology 1582805–2814. 10.1099/mic.0.061432-0 [DOI] [PubMed] [Google Scholar]
- Shao J. Y., Jomantiene R., Dally E. L., Zhao Y., Lee I.-M., Nuss D. L., Davis R. E.(2006). Phylogeny and characterization of phytoplasmal NusA and use of the nusA gene in detection of group 16SrI strains. J Plant Pathol 88193–201. [Google Scholar]
- Streten C., Gibb K. S.(2005). Genetic variation in Candidatus Phytoplasma australiense. Plant Pathology 548–14. 10.1111/j.1365-3059.2005.01113.x [DOI] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.(2013). mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 302725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani A., Sahin N., Kimbara K.(2012). Methylobacterium gnaphalii sp. nov., isolated from leaves of Gnaphalium spicatum. Int J Syst Evol Microbiol 622602–2607. 10.1099/ijs.0.037713-0 [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G.(1997). The clustal_x windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 254876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town J., Annand H., Pratt D., Dumonceaux T., Fonstad T.(2014). Microbial community composition is consistent across anaerobic digesters processing wheat-based fuel ethanol waste streams. Bioresour Technol 157127–133. 10.1016/j.biortech.2014.01.074 [DOI] [PubMed] [Google Scholar]
- Tran-Nguyen L. T., Kube M., Schneider B., Reinhardt R., Gibb K. S.(2008). Comparative genome analysis of ‘Candidatus Phytoplasma australiense’ (subgroup tuf-Australia I; rp-A) and ‘Ca. Phytoplasma asteris’ strains OY-M and AY-WB. J Bacteriol 1903979–3991. 10.1128/JB.01301-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiunas D., Jomantiene R., Davis R. E.(2013). Evaluation of the DNA-dependent RNA polymerase β-subunit gene (rpoB) for phytoplasma classification and phylogeny. Int J Syst Evol Microbiol 633904–3914. 10.1099/ijs.0.051912-0 [DOI] [PubMed] [Google Scholar]
- Wei W., Davis R. E., Lee I. M., Zhao Y.(2007). Computer-simulated RFLP analysis of 16S rRNA genes: identification of ten new phytoplasma groups. Int J Syst Evol Microbiol 571855–1867. 10.1099/ijs.0.65000-0 [DOI] [PubMed] [Google Scholar]
- Wei W., Lee I. M., Davis R. E., Suo X., Zhao Y.(2008). Automated RFLP pattern comparison and similarity coefficient calculation for rapid delineation of new and distinct phytoplasma 16Sr subgroup lineages. Int J Syst Evol Microbiol 582368–2377. 10.1099/ijs.0.65868-0 [DOI] [PubMed] [Google Scholar]
- Yang Z.(2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 241586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zahariev M., Dahl V., Chen W., Lévesque C. A.(2009). Efficient algorithms for the discovery of DNA oligonucleotide barcodes from sequence databases. Mol Ecol Resour 958–64. 10.1111/j.1755-0998.2009.02651.x [DOI] [PubMed] [Google Scholar]
- Zeigler D. R.(2003). Gene sequences useful for predicting relatedness of whole genomes in bacteria. Int J Syst Evol Microbiol 531893–1900. 10.1099/ijs.0.02713-0 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Wei W., Lee I. M., Shao J., Suo X., Davis R. E.(2009). Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the peach X-disease phytoplasma group (16SrIII). Int J Syst Evol Microbiol 592582–2593. 10.1099/ijs.0.010249-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wei W., Davis R. E., Lee I.-M.(2010). Recent advances in 16S rRNA gene-based phytoplasma differentiation, classification and taxonomy. Phytoplasmas: Genomes, Plant Hosts and Vector, 64–92. Edited by Weintraub P., Jones P.Wallingford, UK: CABI Publishing. [Google Scholar]
- Zhao Y., Davis R. E., Wei W., Shao J., Jomantiene R.(2014). Phytoplasma genomes: evolution through mutually complementary mechanisms, gene loss and horizontal acquisition.. Genomics of Plant-Associated Bacteria, 235–271. Edited by Gross D. C.Berlin Heidelberg: Springer-Verlag. [Google Scholar]
- Zhao Y., Davis R. E., Wei W., Lee I. M.(2015). Should ‘Candidatus Phytoplasma’ be retained within the order Acholeplasmatales? Int J Syst Evol Microbiol 651075–1082. 10.1099/ijs.0.000050 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.