Abstract
β-Catenin is a central effector of Wnt signaling in embryonic and stem cell development and in tumorigenesis. Here, through a mass spectrometric analysis of a β-catenin protein complex, we identified 12 proteins as putative β-catenin interactors. We show that one of them, 14-3-3ζ, enhances β-catenin-dependent transcription by maintaining a high level of β-catenin protein in the cytoplasm. More importantly, 14-3-3ζ facilitates activation of β-catenin by the survival kinase Akt and colocalizes with activated Akt in intestinal stem cells. We propose that Akt phosphorylates β-catenin, which results in 14-3-3ζ binding and stabilization of β-catenin, and these interactions may be involved in stem cell development.
Keywords: stem cells
Beta-catenin plays a key role in embryonic development and tumorigenesis by controlling the expression of Wnt-responsive genes (1–3). In the absence of Wnt signaling, β-catenin attaches to cadherins and α-catenin at the cell membrane adherens junctions (4); the remaining excess β-catenin forms a complex with scaffolding protein Axin and tumor suppressor adenomatous polyposis coli (APC), where it is phosphorylated by glycogen synthase kinase 3β (GSK-3β) at multiple serine and threonine residues in the amino terminus (5, 6). The phosphorylated β-catenin is marked by the F-box protein β-Trcp for degradation through the ubiquitination-proteasome pathway (7, 8). In response to Wnt signals, Dishevelled protein (Dsh) is recruited to the Axin complex to inhibit GSK-3β, resulting in cytosolic accumulation and subsequent nuclear translocation of β-catenin, where it binds to the transcription factor T cell factor (TCF)/lymphoid enhancer factor and transactivates Wnt target genes. Mutations that stabilize β-catenin either by the gain-of-function mutations at the putative GSK-3 phosphoration sites or the loss-of-function mutations in tumor suppressor proteins APC and axin have been associated with colorectal cancer and hepatocellular carcinoma (9, 10). Overexpression of the constitutively active (mutated) β-catenin in hematopoietic stem cells leads to enhanced self-renewal (11).
Although the two pools of β-catenin in the cell, the membrane-bound form and the soluble form, exert distinct functions, they have also been shown to compete for binding partners such as E-cadherin and APC (12). Alternative Wnt-nonresponsive regulation of β-catenin that involves presenilin and protein kinase A independent of axin/GSK-3 also has been reported (13). Thus, multiple signaling events can impinge on β-catenin, affecting its stability and/or subcellular localization. Although a great deal has been learned from the studies of β-catenin-interacting proteins that affect its activity either positively (Brg-1, CBP, and Pin1) or negatively (ICAT) (14–17), it remains clear that current knowledge of the components of the Wnt/β-catenin pathway is still not sufficient to fully explain all aspects of β-catenin function. For instance, little is known about how the cytoplasmic-nuclear translocation of β-catenin is regulated. Additional β-catenin interactors keep being identified that contribute extra levels of regulation as exemplified by the most recent findings of Pygo and Chibby (18, 19). Identifying more β-catenin interactors will undoubtedly further advance our understanding of the mechanisms of its function and will provide potential new therapeutic targets.
Here, we conducted a global proteomic analysis of a β-catenin protein complex by using coimmunoprecipitation and tandem MS (MS/MS). We identified 12 potential β-catenin interaction partners in the complex and confirmed that one of the proteins, 14-3-3ζ, is a functional β-catenin-interacting protein. 14-3-3ζ enhances β-catenin-dependent transcription by stabilizing the β-catenin protein in the cytoplasm. More importantly, 14-3-3ζ facilitates activation of β-catenin by the survival kinase Akt, and it colocalizes with activated Akt in intestinal stem cells (ISCs). We propose that Akt phosphorylates β-catenin, which leads to 14-3-3ζ binding and stabilization of β-catenin. These findings have important implications in the understanding of stem cell development and tumorigenesis.
Methods
Cell Cultures. 293T and DLD-1 cells were grown in DMEM with 10% FBS. Chinese hamster ovary (CHO) cells were maintained in Ham's F-12 medium supplemented with 10% FBS. HeLa cells were cultured in MEM with 10% FBS. All of the cells were incubated at 37°C and 5% CO2.
Plasmid Constructs and Antibodies. The myc-tagged human β-catenin plasmid was a kind gift from Hans Clevers, University Medical Center, Utrecht, The Netherlands. The FLAG-tagged 14-3-3ζ plasmid was provided by Haian Fu, Emory University, Atlanta. The pTOPFLASH and pFOPFLASH constructs were provided by Randall Moon, University of Washington, Seattle. The constitutively activated and kinase-mutated Akt plasmids were provided by Benyi Li, University of Kansas, Kansas City. All of the antibodies used were purchased from commercial sources as indicated: anti-β-catenin antibody (BD Transduction Laboratories, Lexington, KY), anti-14-3-3ζ anti-body (C-16) (Santa Cruz Biotechnology), anti-phospho-β-catenin (Thr-41/Ser-45) and (Ser-33/37/Thr-41) (Cell Signaling Technology, Beverly, MA), anti-myc mAb (9E10) (Research Diagnostics, Flanders, NJ), and anti-FLAG M2 mAb (Sigma).
Transient Transfection Assays. Transfections were carried out by using calcium phosphate in 6-cm tissue culture dishes or Lipofectamine (Invitrogen) in 6-well plates as described in the manufacturer's protocol. One day after transfection the cells were lyzed for immunoprecipitation or luciferase assays by using the Dual-Luciferase Reporter Assay Systems (Promega).
Immunoprecipitation. One milligram of whole-cell lysate in 500 μl in lysis buffer (20 mM Tris, pH 7.5/150 mM NaCl/1 mM EDTA/1 mM EGTA/1% Triton X-100/2.5 mM sodium pyrophosphate/1mM β-glycerolphosphate, supplemented with fresh 1 mM PMSF and complete protease inhibitor from Roche Applied Science) was precleared with 6 μg of IgG1 and 50 μl of protein A-Sepharose for 2 h, then immunoprecipitated with 12 μg of specific primary antibody and rocked at 4°C overnight. Fifty microliters of protein A-Sepharose was used to pull the pellet. After washing the pellet once in lysis buffer and twice in water, the protein complex was eluted in 100 μl of 5% acetic acid, pH 3, speed-vacuum dried, and resuspended in water.
MS Analyses. Proteins eluted with 5% acetic acid were denatured, reduced, and subjected to tryptic digestion. Resulting peptides were analyzed by microcapillary reverse-phase chromatography-electrospray (ESI) MS by using an LCQ mass spectrometer (ThermoFinnigan, San Jose, CA) equipped with a combination C18 trap ESI-emitter/microcapillary liquid chromatography column design. Mass spectra were acquired by data-dependent ion selection, which was achieved by automatic switching between single MS and MS/MS modes. Proteins were identified from MS/MS spectra by using a database search engine called comet to search against the latest versions of National Cancer Institute human protein databases. The resulting proteins were analyzed by using peptide prophet and interact programs and validated manually.
Results
Identification of β-Catenin-Interacting Proteins by MS/MS. In an effort to detect proteins interacting with β-catenin, we conducted a MS/MS analysis of a β-catenin-containing protein complex, immunoprecipitated from 293T cells overexpressing human β-catenin. The precipitated protein complex was trypsinized and analyzed through microcapillary liquid chromatography MS/MS followed by protein database searching of the generated spectra. Fifteen proteins were identified in the β-catenin complex after subtracting common proteins that were also found in a control sample composed of a mock-transfected control complex isolated under identical conditions. The detection of β-catenin and two of its known interacting partners α-1 catenin and tubulin α-1 (20, 21) in the β-catenin complex validated the experiment. The other 12 proteins are listed in Table 1. They include 14-3-3ζ, transcription factor Btf3, plasminogen activator inhibitor mRNA binding protein 1, 34/67-kDa laminin receptor (P40), 60s acidic ribosomal protein p2, heterogeneous nuclear ribonucleoprotein (hnRNP) a1, small nuclear ribonucleoprotein sm d1, l-lactate dehydrogenase h chain, and several heat shock proteins (Hsp60, 70, 71, and 90). This list of plausible β-catenin-interacting proteins lays the groundwork for testing hypotheses regarding β-catenin function and regulation, from the regulation of basal transcription (Btf3) (22) to telomerase activity (both hnRNP a1 and Hsp90 are components of telomerase) (23). However, stringent biochemical analyses need to be performed for further confirmation of the putative functional roles these proteins may play with β-catenin.
Table 1. List of proteins identified in coimmunoprecipitated β-catenin complex by MS/MS.
| Gene name | Protein description and peptide sequences | No. unique peptides |
|---|---|---|
| SW:CTNB | β-catenin | 15 |
| R.AAMFPETLDEGMQIPSTQFDAAHPTNVQR.L | ||
| K.AAVMVHQLSK.K | ||
| K.AAVSHWQQQSYLDSGIHSGATTTAPSLSGK.G | ||
| K.EAAEAIEAEGATAPLTELLHSR.N | ||
| R.GLNTIPLFVQLLYSPIENIQR.V | ||
| K.HAVVNLINYQDDAELATR.A | ||
| R.HQEAEMAQNAVR.L | ||
| K.LIILASGGPQALVNIMR.T | ||
| K.LLNDEDQVVVNK.A | ||
| K.MLGSPVDSVLFYAITTLHNLLLHQEGAK.M | ||
| R.MSEDKPQDYK.K | ||
| R.TEPMAWNETADLGLDIGAQGEPLGYR.Q | ||
| R.TMQNTNDVETAR.C | ||
| R.TSMGGTQQQFVEGVR.M | ||
| R.SPQMVSAIVR.T | ||
| SW:CTN1 | α-1 catenin (cadherin-associated protein) (α e-catenin) | 2 |
| K.AEVQNLGGELVVSGVDSAMSLIQAAK.N | ||
| R.VLTDAVDDITSIDDFLAVSENHILEDVNK.C | ||
| SW:TBA1 | Tubulin α-1 chain, brain-specific | 3 |
| K.TIGGGDDSFNTFFSETGAGK.H | ||
| K.VGINYQPPTVVPGGDLAK.V | ||
| K.EDAANNYAR.G | ||
| SW:143Z | 14-3-3 protein ζ δ (protein kinase c inhibitor protein-1) (kclp-1) (factor activating exoenzyme s) (fas) | 1 |
| K.GIVDQSQQAYQEAFEISK.K | ||
| SW:BTF3 | Transcription factor btf3 (rna polymerase b transcription factor 3) | 1 |
| K.QLTEMLPSILNQLGADSLTSLR.R | ||
| PIR2:T12456 | Hypothetical protein DKFZp564M2423.1 | 1 |
| R.FDQLFDDESDPFEVLK.A | ||
| SW:CH60 | 60-kDa heat shock protein, mitochondrial precursor (hsp60) | 1 |
| R.ALMLQGVDLLADAVAVTMGPK.G | ||
| SW:HS71 | Heat shock 70-kDa protein 1 (hsp70.1) | 3 |
| R.IINEPTAAAIAYGLDR.T | ||
| K.SINPDEAVAYGAAVQAAILMGDK.S | ||
| R.TTPSYVAFTDTER.L | ||
| SW:HS7C | Heat shock cognate 71-kDa protein | 1 |
| K.NQVAMNPTNTVFDAK.R | ||
| SW:HS9B | Heat shock protein hsp 90-β (hsp 84) (hsp 90) | 1 |
| K.HSQFIGYPITLYLEK.E | ||
| SW:LDHH | L-lactate dehydrogenase h chain (ec 1.1.1.27) (ldh-b) | 3 |
| K.GYTNWAIGLSVADLIESMLK.N | ||
| K.SLADELALVDVLEDK.L | ||
| K.LIAPVAEEEATVPNNK.I | ||
| SW:RLA2 | 60s acidic ribosomal protein p2 | 1 |
| K.NIEDVIAQGIGK.L | ||
| SW:ROA1 | Heterogeneous nuclear ribonucleoprotein a1 (helix-destabilizing protein) (single-strand binding protein) (hnrnp core protein a1) | 1 |
| R.NQGGYGGSSSSSSYGSGR.R | ||
| SW:RSP4 | 40s ribosomal protein sa (p40) (34 67-kDa laminin receptor) (colon carcinoma laminin-binding protein) (nem 1chd4) | 1 |
| K.FAAATGATPIAGR.F | ||
| SW:SMD1 | Small nuclear ribonucleoprotein sm d1 (snrnp core protein d1) (sm-d1) (sm-d autoantigen) | 1 |
| R.YFILPDSLPLDTLLVDVEPK.V |
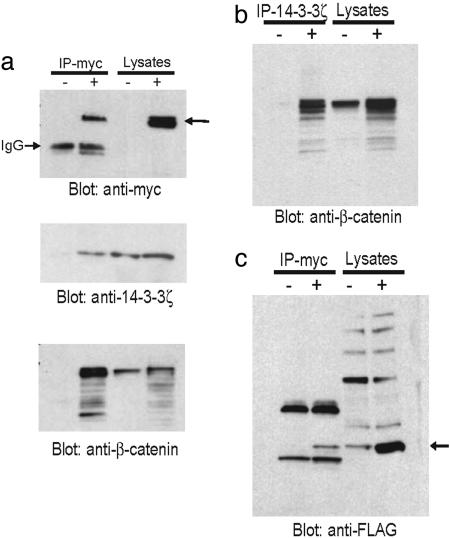
14-3-3ζ Interacts with β-Catenin and Activates β-Catenin-Dependent Promoters. 14-3-3ζ is a member of a family of conserved regulatory proteins that can bind to a plethora of functionally diverse signaling molecules (24). A Western blot using 14-3-3ζ-specific antibody confirms the presence of 14-3-3ζ in the β-catenin complex (Fig. 1a). Reciprocal immunoprecipitation using an anti-14-3-3ζ antibody also precipitates β-catenin (Fig. 1b). Furthermore, cotransfection of myc-tagged β-catenin and FLAG-tagged 14-3-3ζ results in the formation of a complex containing both exogenously introduced proteins (Fig. 1c). Thus, compelling evidence shows that 14-3-3ζ interacts with β-catenin in 293T cells.
Fig. 1.
14-3-3ζ interacts with β-catenin in vivo. 293T cells were transfected with myc-tagged β-catenin plasmid. Twenty-four hours later, the whole-cell lysates were immunoprecipitated (IP) with either anti-myc (a) or anti-14-3-3ζ (b) antibodies and immunoblotted with indicated antibodies. (c) 293T cells were cotransfected with myc-β-catenin and FLAG-14-3-3ζ. The whole-cell lysates were immunoprecipitated (IP) with anti-myc antibody and blotted with anti-FLAG antibody.
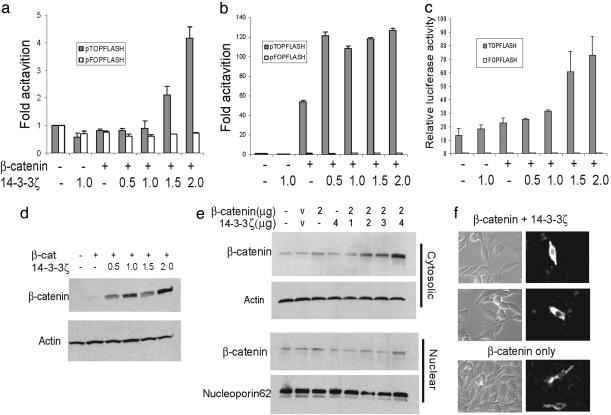
A major function of β-catenin in Wnt signaling pathway is to activate Wnt-responsive genes in conjunction with the transcription factor Tcf. We next tested the functional significance of the interaction between 14-3-3ζ and β-catenin by conducting Tcf reporter assays using pTOPFLASH and its mutant counterpart negative control plasmid pFOPFLASH. 14-3-3ζ by itself shows little effect on the transcription activity of the pTOPFLASH reporter (Fig. 2 a–c). However, when cotransfected with β-catenin, 14-3-3ζ activates the pTOPFLASH reporter in a dose-dependent manner in CHO and DLD1 colorectal adenocarcinoma cells up to 3-fold. A 2-fold activation was observed in 293T cells, and the reporter activity saturated rapidly in the presence of 14-3-3ζ because of the high endogenous level of β-catenin. No significant changes of transcription activity were observed on the mutant reporter pFOPFLASH in any of the three cell lines. These findings clearly demonstrate that the binding of 14-3-3ζ to β-catenin is functionally significant by enhancing the transcription of β-catenin target promoters.
Fig. 2.
14-3-3ζ increases β-catenin-dependent transcription and steady-state level of β-catenin in the cytoplasm. Three different cell lines, CHO (a), 293T (b), and DLD1 colon adenocarcinoma (c) cells, were transfected with pTOPFLASH and pFOPFLASH reporter and increasing amounts of 14-3-3ζ in the absence or presence of β-catenin. The Renilla luciferase reporter pRL-TK was cotransfected as internal control for transfection efficiency. The experiments were repeated in duplicate at least three times. Shown are means ± SD from one representative experiment. Whole-cell lysates from CHO cells transfected with β-catenin and 14-3-3ζ (d) or fractionated cytosolic and nuclear extract (e) were immunoblotted with β-catenin antibody. (f) Transfected CHO cells were fixed in methanol and stained with FITC-conjugated anti-β-catenin antibody.
14-3-3ζ Increases the Steady-State Level of β-Catenin in Cytoplasm. To explore the underlying mechanism for enhanced β-catenin-dependent transcription, we examined whether the protein levels of β-catenin were affected by 14-3-3ζ binding. CHO cells were cotransfected with β-catenin and increasing concentrations of 14-3-3ζ. As shown in Fig. 2d, the levels of β-catenin protein are much higher in the presence of 14-3-3ζ. We further fractionated the transfected CHO cells into either nuclear or nonnuclear (contains both cytosolic and membrane) fractions. Western blots showed that the increase of β-catenin protein occurred mainly in the nonnuclear fractions with only small changes in the nuclear fractions (Fig. 2e). An immunofluorescent staining in transfected CHO cells showed both cytosolic and nuclear distribution of β-catenin. However, the accumulation of β-catenin in the presence of 14-3-3ζ is more prominent in cytosol (Fig. 2f). Thus, the interaction between 14-3-3ζ and β-catenin results in an elevated steady-state level of β-catenin in the cytoplasm.
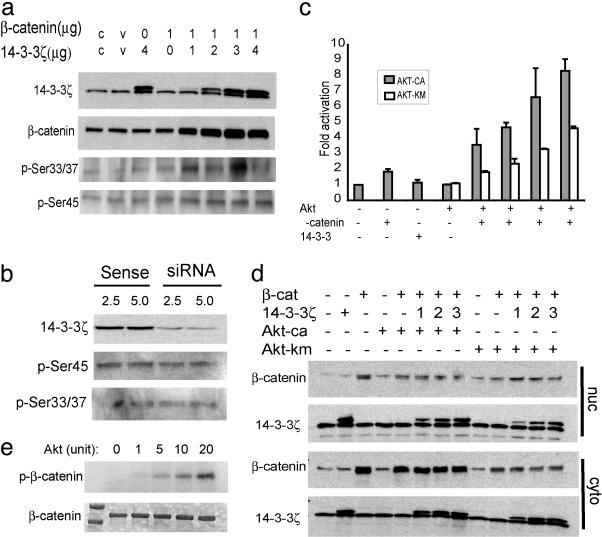
14-3-3 binds mainly to serine/threonine-phosphorylated proteins (24). The amino terminus of β-catenin contains four serine and threonine residues that are sequentially phosphorylated by casein kinase Iα at Ser-45 followed by GSK-3β at Thr-41, Ser-37, and Ser-33 (25, 26). The Ser-33/37 phosphorylation creates recognition sites for β-Trcp, which leads to the ubiquitination and degradation of β-catenin. Using antibodies that specifically recognize either Ser-33/37- or Ser-45-phosphorylated β-catenin, we showed that phosphorylation at these sites was not drastically affected by 14-3-3ζ (Fig. 3a). In addition, mutations of all of these serine and threonine sites do not abolish binding and transactivation by 14-3-3ζ (data not shown). RNA interference knockdown of 14-3-3ζ protein in HeLa cell exerted little effect on β-catenin phosphorylation at Ser-45 and Ser-33/37 (Fig. 3b). These findings suggest that (i) cellular 14-3-3ζ level is in excess of β-catenin and (ii) quite possibly, phosphorylation events outside of the aforementioned GSK-3 phosphorylation sites are needed for the binding of 14-3-3ζ and the enhanced stability and/or activity of β-catenin.
Fig. 3.
14-3-3ζ enhances activation of β-catenin by Akt. (a) Whole-cell lysates from transfected CHO cells were immunoblotted with antibodies that specifically recognize either Ser-33/37- or Ser-45-phosphorylated β-catenin. (b) HeLa cells were transfected with14-3-3ζ-specific small interference RNA (siRNA), and whole-cell lysates were blotted with the indicated antibodies. (c) CHO cells were transfected with pTOPFLASH or pFOPFLASH reporter in the presence of constitutive Akt (AKT-ca) or the kinase domain mutated Akt (Akt-km) together with β-catenin and 14-3-3ζ. (d) CHO cell transfectants were fractionated into either nuclear or cytosolic fractions and blotted with β-catenin and 14-3-3ζ antibodies. (e) Purified human β-catenin protein was incubated with increasing amounts of Akt kinase in the presence of radioactive ATP. The resolved protein gel was exposed to x-ray film (Upper) after staining with Coomassie blue (Lower).
14-3-3ζ Facilitates Activation of β-Catenin by Akt. 14-3-3 molecules are known to bind to an array of cellular signaling molecules. One well characterized function is to cooperate with survival kinase Akt by binding to the Akt-phosphorylated Bcl-2 family member BAD to antagonize its proapoptotic activity (27). A recent study showed that Akt-phosphorylated Ataxin-1 can be bound and stabilized by 14-3-3 (28). These findings prompted us to examine whether Akt is also involved in the regulation of β-catenin by 14-3-3ζ. The Tcf reporter assay using pTOPFLASH in CHO cells showed that Akt by itself does not change the reporter activity very much (Fig. 3c), which is consistent with earlier findings (29). When cotransfected with β-catenin, a 1- to 2-fold activation can be achieved by the constitutively active Akt (Akt-ca) but not Akt with a mutated kinase domain (Akt-km). More importantly, when 14-3-3ζ is included in the transfection, an additional 2- to 3-fold of activation can be achieved in a dose-dependent manner. Although activation in the presence of both constitutively active and mutant Akt can be enhanced by 14-3-3ζ, the kinase mutation does reduce the total contribution of Akt to the activation of the pTOPFLASH promoter by ≈40%. No significant changes were observed on the control reporter pFOPFLASH (data not shown). Consistent with this result, cotransfection of β-catenin with Akt-ca and 14-3-3ζ helps maintain high levels of β-catenin protein in the cytoplasm (but not in the nucleus, Fig. 3d), whereas Akt-km does not help maintain high β-catenin levels even in the presence of 14-3-3ζ (Fig. 3d). Furthermore, more β-catenin protein can be immunoprecipitated from 293T cells cotransfected with Akt-ca more effectively than the mutant Akt (data not shown), suggesting that Akt facilitates the stabilization of β-catenin. Although it has been shown that Akt acts through Dishevelled and GSK-3β to regulate β-catenin (29), our data indicate that Akt may also act directly on β-catenin to facilitate 14-3-3ζ binding. To test this hypothesis, we conducted an in vitro phosphorylation assay using purified β-catenin and Akt proteins. As shown in Fig. 3e, a dose-dependent phosphorylation of β-catenin by Akt kinase was observed. Taken together, these data suggest a model in which Akt phosphorylates β-catenin, resulting in 14-3-3ζ binding and increased transactivation. Of note, even in the absence of Akt kinase activity, 14-3-3ζ still enhances the activation of β-catenin (Fig. 3c). Thus Akt kinase activity is necessary but may not be sufficient to sustain the full activation of β-catenin by 14-3-3ζ.
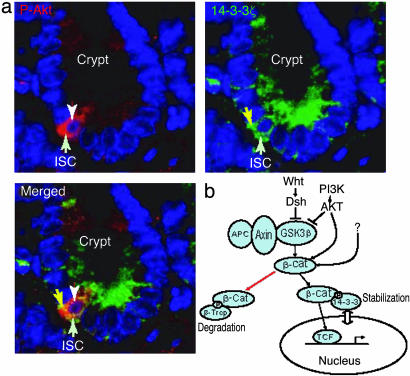
β-Catenin Colocalizes with Activated Akt in ISCs. β-Catenin plays an important role in regulating stem cell self-renewal (11). Using bone morphogenetic protein (BMP) receptor type IA conditioned knockout mice, we showed the development of profuse polyps in the mouse gastrointestinal tract caused by expansion of ISC population resulting from the inactivation of tumor suppressor PTEN and the concomitant activation of phosphatidylinositol 3-kinase (PI3-kinase)/Akt (L.L., unpublished data). Here, we have demonstrated a mechanism for the stablization of β-catenin by the concerted functioning of 14-3-3ζ and Akt. We next ask whether this mode of action is also true in a real stem cell environment. The intestinal crypt-villus from BMP receptor type IA knockout mice was costained with either 14-3-3ζ-specific antibody or antibody that recognized phosphorylated Akt (activated Akt). Although 14-3-3 proteins are known to be broadly expressed, the most abundant expression of 14-3-3ζ along the crypt-villus of small intestine appears to be in the cytoplasm of ISCs [the fourth/fifth cells from the bottom of the crypt (30); Fig. 4a]. A similar expression pattern of phosphorylated Akt was also observed in the ISCs. Thus, a striking colocalization of 14-3-3ζ with activated Akt was found specifically in vivo in the ISCs (Fig. 4a), which is consistent with their functional cooperation.
Fig. 4.
Colocalization of phophorylated Akt with 14-3-3ζ in ISCs. (a) The crypt-villus of intestine from BMP receptor type 1A knockout mouse were stained with antiphosphorylated Akt-specific (red) (Upper Left) or anti-14-3-3ζ-specific (green) (Upper Right) antibodies. Counterstaining with 4′,6-diamidino-2-phenylindole reveals the cell nuclei (blue). (b) A model for the stabilization mechanism of β-catenin by Akt and 14-3-3ζ.
Discussion
We used a MS-based proteomic approach to analyze a β-catenin-containing protein complex. We identified 12 proteins as putative β-catenin interactors, with a particular focus on the interaction between β-catenin and 14-3-3ζ. We found that 14-3-3ζ enhances the activity of β-catenin by using a pTOPFLASH reporter plasmid containing multiple β-catenin/Tcf binding sites. Interestingly, we showed that 14-3-3ζ binding results in increased steady-state levels of β-catenin protein. We further showed that the survival kinase PI3-kinase/Akt activates β-catenin through interactions with14-3-3ζ and the kinase activity of Akt is required for this activation. We propose that Akt phosphorylates β-catenin, which leads to binding of 14-3-3ζ and subsequent stabilization of β-catenin (Fig. 4b). This mechanism can be either Wnt-dependent or independent. Because 14-3-3ζ does not block phosphorylation of β-catenin by GSK-3β (Fig. 3a), we postulate that its binding to β-catenin causes a conformational change of β-catenin, disengaging the binding of β-Trcp to the phosphorylated Ser-33/37, resulting in an increase of cytosolic β-catenin. This model supports previous findings that a number of the Akt-downstream substrates are regulated in a similar manner, i.e., 14-3-3 stabilizes its target proteins after they are primed by Akt phosphorylation. Our results provide biochemical and functional evidence to support the hypothesis that multiple signaling events, including Wnt and PI3-kinase/Akt, impinge on β-catenin and that 14-3-3ζ serves as an essential coordinator for different pathways, and may explain the controversy regarding the regulation of β-catenin by Akt. The absense of P-Akt (phosphorylated, i.e., activated form of Akt), β-catenin, or 14-3-3ζ in the assay system used by other researchers could abrogate the regulation of β-catenin by Akt.
The increase of β-catenin in the cytoplasm appears to be at odds with its transcriptional activity. One explanation is that nuclear accumulation of β-catenin appears to be a transient event, and our analyses using transiently transfected cells may have missed this time window. Alternatively, 14-3-3 proteins may mediate dynamic nucleocytoplasmic transport of their binding ligands (31). Hence 14-3-3ζ may chaperone β-catenin to ensure a balanced nuclear and cytosolic allocation, while at the same time blocking degradation by the axin/APC complex. While this manuscript was in preparation, Lo and colleagues (32) reported that in Caenorhabditis elegans 14-3-3/PAR-5 binds to and mediates nuclear export of TCF/POP1. It is not clear which 14-3-3 isoform is the mammalian orthologue responsible for binding to TCF, because different isoforms of 14-3-3 afford different rates of nucleocytoplasmic transport (33); nonetheless, the critical role of 14-3-3 proteins in the β-catenin/TCF pathway is evident.
14-3-3 proteins mediate a general survival-promoting function, mainly through antagonizing the proapoptotic function of BAD (34). Members of the 14-3-3 family have been shown to be involved in cancers (35). 14-3-3 proteins also play a role in early Xenopus development (36). It has been shown that 14-3-3 proteins enhance nuclear localization of telomerase (37). We show that inactivation of the BMP signal and subsequent activation of PI3-kinase/Akt promotes ISC self-renewal through the activation of β-catenin and telomerase, both of which are self-renewal factors (L.L., unpublished data). We found that 14-3-3ζ interacts with β-catenin and enhances Akt activation of β-catenin. Even more strikingly, we demonstrated that P-Akt and 14-3-3ζ predominantly and specifically coexist in the ISCs of the BMP receptor type IA conditioned knockout mice, where the PI3-kinase/Akt pathway is constitutively activated. Our data provide direct evidence for the involvement of this interaction in stem cell development. These findings have great implications in the understanding of stem cell development and perhaps also tumorigenesis.
Finally, we show that combining the interdisciplinary approaches of MS proteomic technology with biochemical and genetic analysis is a powerful way to elucidate fundamental signal transduction pathways in developmental systems.
Acknowledgments
We thank Dr. Hans Clevers for the myc-tagged human β-catenin plasmid; Dr. Randall Moon for the pTOPFLASH and pFOPFLASH constructs; Dr. Haian Fu for the 14-3-3ζ plasmid; Dr. Benyi Li for the constitutively activated and kinase-mutated Akt plasmids; Drs. Wenqing Xu and Yi Xing for providing purified human β-catenin protein; and Drs. Hui Zhang and Yong Chi for valuable discussions. This work was supported by National Institutes of Health Grants P01DK53074 (to L.H.) and N01-HV-28179 (to R.A.) and the Stowers Institute for Medical Research (L.L.).
Author contributions: Q.T. and L.H. designed research; Q.T., M.C.F., W.A.T., and X.C.H. performed research; L.L. and R.A. contributed new reagents/analytic tools; Q.T. analyzed data; and Q.T., R.A., and L.H. wrote the paper.
Abbreviations: APC, adenomatous polyposis coli; GSK-3β, glycogen synthase kinase 3β; TCF, T cell factor; MS/MS, tandem MS; CHO, Chinese hamster ovary; BMP, bone morphogenetic protein; ISC, intestinal stem cell; PI3-kinase, phosphatidylinositol 3-kinase.
References
- 1.Miller, J. R. & Moon, R. T. (1996) Genes Dev. 10, 2527-2539. [DOI] [PubMed] [Google Scholar]
- 2.Cadigan, K. M. & Nusse, R. (1997) Genes Dev. 11, 3286-3305. [DOI] [PubMed] [Google Scholar]
- 3.Bienz, M. & Clevers, H. (2000) Cell 103, 311-320. [DOI] [PubMed] [Google Scholar]
- 4.Zhurinsky, J., Shtutman, M. & Ben-Ze'ev, A. (2000) J. Cell Sci. 113, 3127-3139. [DOI] [PubMed] [Google Scholar]
- 5.Peifer, M., Pai, L. M. & Casey, M. (1994) Dev. Biol. 166, 543-556. [DOI] [PubMed] [Google Scholar]
- 6.Yost, C., Torres, M., Miller, J. R., Huang, E., Kimelman, D. & Moon, R. T. (1996) Genes Dev. 10, 1443-1454. [DOI] [PubMed] [Google Scholar]
- 7.Aberle, H., Bauer, A., Stappert, J., Kispert, A. & Kemler, R. (1997) EMBO J. 16, 3797-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu, C., Kato, Y., Zhang, Z., Do, V. M., Yankner, B. A. & He, X. (1999) Proc. Natl. Acad. Sci. USA 96, 6273-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morin, P. J., Sparks, A. B., Korinek, V., Barker, N., Clevers, H., Vogelstein, B. & Kinzler, K. W. (1997) Science 275, 1787-1790. [DOI] [PubMed] [Google Scholar]
- 10.Satoh, S., Daigo, Y., Furukawa, Y., Kato, T., Miwa, N., Nishiwaki, T., Kawasoe, T., Ishiguro, H., Fujita, M., Tokino, T., et al. (2000) Nat. Genet. 24, 245-250. [DOI] [PubMed] [Google Scholar]
- 11.Reya, T., Duncan, A.W., Ailles, L., Domen, J., Scherer, D. C., Willert, K., Hintz, L., Nusse, R. & Weissman, I. L. (2003) Nature 423, 409-414. [DOI] [PubMed] [Google Scholar]
- 12.Miller, J. R. & Moon, R. T. (1997) J. Cell Biol. 139, 229-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang, D. E., Soriano, S., Xia, X., Eberhart, C. G., De Strooper, B., Zheng, H. & Koo, E. H. (2002) Cell 110, 751-762. [DOI] [PubMed] [Google Scholar]
- 14.Barker, N., Hurlstone, A., Musisi, H., Miles, A., Bienz, M. & Clevers, H. (2001) EMBO J. 20, 4935-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takemaru, K. I. & Moon, R. T. (2000) J. Cell Biol. 149, 249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryo, A., Nakamura, M., Wulf, G., Liou, Y. C. & Lu, K. P. (2001) Nat. Cell Biol. 3, 793-801. [DOI] [PubMed] [Google Scholar]
- 17.Tago, K., Nakamura, T., Nishita, M., Hyodo, J., Nagai, S., Murata, Y., Adachi, S., Ohwada, S., Morishita, Y., Shibuya, H. & Akiyama, T. (2000) Genes Dev. 14, 1741-1749. [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson, B., Townsley, F., Rosin-Arbesfeld, R., Musisi, H. & Bienz, M. (2002) Nat. Cell Biol. 4, 367-373. [DOI] [PubMed] [Google Scholar]
- 19.Takemaru, K., Yamaguchi, S., Lee, Y. S., Zhang, Y., Carthew, R. W. & Moon, R. T. (2003) Nature 422, 905-909. [DOI] [PubMed] [Google Scholar]
- 20.Knudsen, K. A., Soler, A. P., Johnson, K. R. & Wheelock, M. J. (1995) J. Cell Biol. 130, 67-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufmann, U., Kirsch, J., Irintchev, A., Wernig, A. & Starzinski-Powitz, A. (1999) J. Cell Sci. 112, 55-68. [DOI] [PubMed] [Google Scholar]
- 22.Zheng, X. M., Black, D., Chambon, P. & Egly, J.M. (1990) Nature 344, 556-559. [DOI] [PubMed] [Google Scholar]
- 23.Ford, L. P., Wright, W. E. & Shay, J. W. (2002) Oncogene 21, 580-583. [DOI] [PubMed] [Google Scholar]
- 24.Fu, H., Subramanian, R. R. & Masters, S. C. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 617-647. [DOI] [PubMed] [Google Scholar]
- 25.Liu, C., Li, Y., Semenov, M., Han, C., Baeg, G. H., Tan, Y., Zhang, Z., Lin, X. & He, X. (2002) Cell 108, 837-847. [DOI] [PubMed] [Google Scholar]
- 26.Amit, S., Hatzubai, A., Birman, Y., Andersen, J. S., Ben-Shushan, E., Mann, M., Ben-Neriah, Y. & Alkalay, I. (2002) Genes Dev. 16, 1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Datta, S. R., Dudek, H., Tao, X., Masters, S., Fu, H., Gotoh, Y. & Greenberg, M. E. (1997) Cell 91, 231-241. [DOI] [PubMed] [Google Scholar]
- 28.Chen, H. K., Fernandez-Funez, P., Acevedo, S. F., Lam, Y. C., Kaytor, M. D., Fernandez, M. H., Aitken, A., Skoulakis, E. M., Orr, H. T., Botas, J. & Zoghbi, H. Y. (2003) Cell 113, 457-468. [DOI] [PubMed] [Google Scholar]
- 29.Fukumoto, S., Hsieh, C. M., Maemura, K., Layne, M. D., Yet, S. F., Lee, K. H., Matsui, T., Rosenzweig, A., Taylor, W. G., Rubin, J. S., et al. (2001) J. Biol. Chem. 276, 17479-17483. [DOI] [PubMed] [Google Scholar]
- 30.Booth, C. & Potten, C. S. (2000) J. Clin. Invest. 105, 1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunet, A., Kanai, F., Stehn, J., Xu, J., Sarbassova, D., Frangioni, J. V., Dalal, S. N., DeCaprio, J. A., Greenberg, M. E. & Yaffe, M. B. (2002) J. Cell Biol. 156, 817-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lo, M. C., Gay, F., Odom, R., Shi, Y. & Lin, R. (2004) Cell 117, 95-106. [DOI] [PubMed] [Google Scholar]
- 33.van Hemert, M. J., Niemantsverdriet, M., Schmidt, T., Backendorf, C. & Spaink, H. P. (2004) J. Cell Sci. 117, 1411-1420. [DOI] [PubMed] [Google Scholar]
- 34.Masters, S. C., Subramanian, R. R., Truong, A., Yang, H., Fujii, K., Zhang, H. & Fu, H. (2002) Biochem. Soc. Trans. 30, 360-365. [DOI] [PubMed] [Google Scholar]
- 35.Nakanishi, K., Hashizume, S., Kato, M., Honjoh, T., Setoguchi, Y. & Yasumoto, K. (1997) Hum. Antibodies 8, 189-194. [PubMed] [Google Scholar]
- 36.Wu, C. & Muslin, A. J. (2002) Mech. Dev. 119, 45-54. [DOI] [PubMed] [Google Scholar]
- 37.Seimiya, H., Sawada, H., Muramatsu, Y., Shimizu, M., Ohko, K., Yamane, K. & Tsuruo, T. (2000) EMBO J. 19, 2652-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]