Abstract
The “BH3-only” proapoptotic BCL-2 family members initiate the intrinsic apoptotic pathway. A small interfering RNA knockdown of BIM confirms this BH3-only member is important for the cytokine-mediated homeostasis of hematopoietic cells. We show here that the phosphorylation status of BIM controls its proapoptotic activity. IL-3, a hematopoietic survival factor, induces extracellular signal-regulated kinase/mitogen-activated protein kinase-mediated phosphorylation of BIM on three serine sites (S55, S65, and S100). After IL-3 withdrawal, only nonphosphorylated BIM interacts with the multidomain proapoptotic effector BAX. Phosphorylation of BIM on exposure of cells to IL-3 dramatically reduces the BIM/BAX interaction. A nonphosphorylatable BIM molecule (S55A, S65A, and S100A) demonstrates enhanced interaction with BAX and enhanced proapoptotic activity. Thus, ERK/mitogen-activated protein kinase-dependent phosphorylation of BIM in response to survival factor regulates BIM/BAX interaction and the pro-death activity of BIM.
Keywords: apoptosis, BCL-2 family, mitogen-activated protein kinase, BH3-only
Growth factors and cytokines signal survival through their cognate receptors, activating signaling pathways often composed of protein kinase cascades. On the other hand, inadequate growth factor initiates a signaling cascade that leads to cell death (1–4). Both pro- and antiapoptotic BCL-2 family members lie downstream of these proximal signals. The “multidomain” BCL-2 members either suppress (e.g., BCL-2 and BCL-XL) or promote apoptosis (e.g., BAX and BAK), whereas the “BH3-only” subfamily members identified to date (e.g., BAD, BID, and BIM) function to promote cell death. It has become clear that BH3-only proteins are upstream initiators of apoptosis. BIM was identified as a BH3-only protein that induces apoptosis and can be antagonized by antiapoptotic BCL-2 family members. Studies in BIM-deficient mice/cells indicate important roles for BIM in hematopoietic cell homeostasis (1, 5, 6). The activity of BIM can be modulated at the transcriptional and posttranslational levels. Transcriptional control of BIM involves the Jun N-terminal kinase (JNK) in neurons (7, 8), and phosphatidylinositol 3-OH-kinase (PI3-K) and Ras-extracellular signal-regulated kinase (ERK) pathways in hematopoietic, neuronal, and epithelial cells (9–12). Posttranslational regulation of BIM activity has been proposed through sequestration to a microtubular dynein motor complex by interaction with dynein light chain/LC8 (13). Recently, it has been reported that phosphorylation and ubiquitynation of BIM can regulate its protein level (14–17). In the present study, we show that IL-3 induces ERK/mitogen-activated protein kinase (MAPK)-mediated phosphorylation of BIM on three serine sites (S55, S65, and S100) and that ERK-dependent phosphorylation of BIM regulates BIM/BAX interaction and the proapoptotic activity of BIM.
Materials and Methods
Plasmid Construction. pSuper-BIM was constructed by inserting the sequences of mouse BIM (GenBank accession no. AF032459, nucleotide 486-506) into psuper.retro (Oligoengine, Seattle) according to the manufacturer's protocol. BIM extra long (BIMEL) cDNA was cloned from FL5.12 total RNA by RT-PCR. The cDNA was cloned into pGEX-4T1 for GST fusion protein or pCDNA3 for hemagglutinin (HA)-tagged protein. Potential phosphorylation sites were changed by PCR based site-directed mutagenesis by using QuikChange kit (Stratagene). The integrity was verified by sequencing. The GFP-fused with constitutively active MAPK kinase (MEK) plasmid was kindly provided by S. Grant (Virginia Commonwealth University).
Transfection. Transfection was performed by electroporation by using a Bio-Rad electroporator. The cells were suspended in Iscove's Modified Dulbecco's Medium (Invitrogen) (4 × 106/400 μl) with 10 μg of DNA, and electroporation was done in 0.4-cm cuvette with 300 V, 500 μF.
Chemicals and Abs. U0126, LY294002, and SB203580 were purchased from Sigma. SP600125 was purchased from Calbiochem. An Ab for HA was purchased from Covance, Princeton, NJ. Abs were purchased as follows: BIM (202000) and VDAC1 from Calbiochem; BAX (N-20), α-tubulin, and phospho-JNK from Santa Cruz Biotechnology; and phospho-ERK, phospho-AKT, and phospho-p38 from Cell Signaling Technology.
Generation of Phosphorylation Site-Specific BIM Abs. To generate anti-BIM pS65 and pS100 antigens, phosphopeptides (CHGSPQGPLAPPApSPGPFA for pS65 and GYFSFDTDRpSPAPMSC for pS100) were synthesized at Tufts University Core Facility. Rabbits were immunized at Covance, Princeton, NJ. The Abs were affinity-purified by a protein A-Sepharose column followed by one round of negative selection against nonphosphorylated peptide and one round of positive selection against phosphorylated peptide. An Ab for phospho-S55 was purchased from Upstate Biotechnology, Lake Placid, NY.
Western Blot Analysis and Immunoprecipitation. Whole-cell lysates were prepared with lysis buffer {20 mM Tris (pH 7.4)/137 mM NaCl/1 mM DTT/1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate/20 mM NaF/10 mM β-glycerophosphate, and a protease inhibitor mixture (Sigma)}. Equal amounts of proteins were loaded on SDS/PAGE, transferred to a nitrocellulose membrane, and analyzed by immunoblotting. For immunoprecipitation, equal amounts of protein were precleared with protein A/G beads (Pierce) and incubated with the appropriate Abs on ice for 2 h. Then the Ab complexes were captured with protein A/G beads at 4°C for 1 h. After washing three times with the same lysis buffer, the beads were resuspended in the sample buffer and separated by SDS/PAGE.
Subcellular Fractionation. The fractionation was performed as described in ref. 18.
In Vitro Kinase Assay. The assay was performed with 5 μCi (1 Ci = 37 GBq) of [γ-32P]ATP and 50 μM cold ATP in a buffer of 50 mM Tris (pH 7.4), 10 mM MgCl2, 1 mM DTT, 1 unit purified ERK2 (Cell Signaling Technology), and 10 μg of each GST fusion protein. The reaction mixtures were incubated at 30°C for 20 min and terminated by the addition of the SDS sample loading buffer. Proteins were resolved on an SDS/PAGE and visualized by autoradiography.
Cell Viability Assay. Cell death was quantitated by Annexin-VFITC (Becton Dickinson) staining according to the manufacturer's protocol, followed by flow cytometric analysis by using FACScan (Becton Dickinson). In Fig. 4, cells were incubated with PI to identify nonviable cells and analyzed by FACS, gating GFP-positive transfected cells.
Fig. 4.
Phosphorylation of BIM by constitutive active MEK counters IL-3 withdrawal death. (A) FL5.12 BCL-2/BIM WT or BCL-2/BIM S55AS65AS100A cells were transfected with either GFP (control) or GFP-constitutively active MEK1 (CA-MEK) plasmids, respectively. Total cell extracts prepared at 24 h after IL-3 deprivation were subjected to Western blots with anti-phospho-S65 BIM and anti-HA (for total BIM and CA-MEK, respectively). (B) Transfected cells from above were deprived of IL-3 for 24 h, then stained with PI to identify nonviable cells, and analyzed by FACS, gating on GFP-positive transfected cells. Percentage protection relative to controls was calculated as 100 × [1 - (percentage apoptosis of cells transfected with CA-MEK plasmid/percentage apoptosis of cells transfected with control plasmid)]. Results are the mean ± SD of two independent experiments.
Results
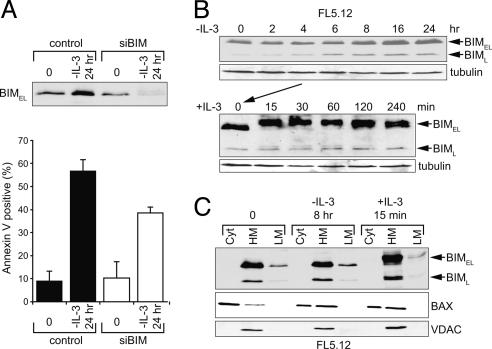
We initiated a study of the regulation of BIM by growth/survival factors by observing whether BIM plays a role in the IL-3-dependent mouse pro-B cell line, FL5.12. We introduced a small interfering RNA, pSuper-BIM construct to knockdown expression of endogenous BIM. The expression of BIMEL is strongly inhibited by small interfering BIM at 24 h after IL-3 deprivation (Fig. 1A). Cell survival after IL-3 withdrawal is also enhanced by small interfering BIM, ≈35%, as compared to control cells (Fig. 1 A). These data indicate that BIM is a proapoptotic factor after IL-3 deprivation. BIM-independent pathways also contribute to IL-3 withdrawal death. Substantial BIMEL and a lesser amount of BIM long (BIML) protein is present in FL5.12 cells cultured in IL-3, but neither BIM short (BIMS) nor other isoforms were detected. BIM protein levels remained unchanged for 8 h after IL-3 withdrawal. After 16–24 h of IL-3 deprivation, only a modest ≈2-fold increase in BIMEL and BIML was noted (Fig. 1B). Consequently, we searched for evidence of a protein modification. IL-3 was added back after 4 h of deprivation, which induced a prominent shift in the electrophoretic mobility of both BIMEL and BIML on SDS/PAGE (Fig. 1 B and C). Similar observations were also made in BAF3, another mouse IL-3-dependent cell line, in TF-1, a human granulocyte macrophage-colony stimulating factor-dependent erythroleukemia cell line, and in mouse embryonic fibroblasts when IGF-1 was used as a survival factor (data not shown). Total levels of BIM protein did not vary substantially throughout the time course, but nearly all of the protein was modified, suggesting that for IL-3-mediated cell survival, a posttranslational regulation might be the critical event for BIM. In contrast, in other cell types, such as epithelial cells (15, 16, 19) or osteoclasts (17), BIM protein levels are regulated by ubiquitin-dependent degradation. This suggests cell-type specific mechanisms of BIM regulation.
Fig. 1.
The expression and distribution of BIM are under the control of IL-3. (A) Small interfering RNA knockdown of BIM expression protects from apoptosis induced by IL-3 deprivation. (Upper) IL-3 was withdrawn from FL5.12 cells transfected with either pSuper-BIM (small interfering BIM) or pSuper (control). Total cell extracts were subjected to Western blot with anti-BIM. (Lower) Cell viabilities were determined at the indicated time points by Annexin V-PI staining. Values represent the mean ± SD of three experiments. (B) IL-3 was withdrawn from FL5.12 cells for the indicated times. Total cell extracts were subjected to Western blots with anti-BIM. Arrows indicate the position of BIM isoforms, BIMEL and BIML, respectively (Upper). IL-3 was withdrawn from FL5.12 cells for 4 h, followed by readdition of IL-3 for the indicated times (Lower). (C) FL5.12 cells cultured in IL-3 (time 0), IL-3 deprived for 8 h (-IL-3), and deprived cells reexposed to IL-3 for 15 min (+IL-3) are shown. Cells were fractionated into cytosol (Cyt), heavy membrane (HM), and light membrane (LM) fractions. Western blots were performed with anti-BIM, anti-BAX, and anti-VDAC1.
We next assessed BIM localization after 8 h of IL-3 deprivation (-IL-3, 8 h) and after IL-3 readdition (+IL-3, 15 min), as compared to cells maintained in IL-3 (t = 0) (Fig. 1C). FL5.12 cells were subcellularly fractionated into the soluble S100 cytosolic fraction, the heavy membrane fraction (enriched for mitochondria), and the light membrane (enriched for endoplasmic reticulum). Under all conditions, both BIMEL and BIML forms localized predominantly to the mitochondrial fraction, to a lesser extent to the endoplasmic reticulum fraction, and were undetectable in the cytosolic fraction. After reexposure to IL-3, a prominent band-shift in the migration of both BIMEL and BIML was observed on SDS/PAGE in both heavy membrane and light membrane fractions. The subcelluar distribution of BIM did not vary substantially in the presence or absence of IL-3. In contrast, BAX, which is localized to the cytosol or loosely attached to mitochondrial membranes in the presence of IL-3, translocates to the mitochondria after IL-3 withdrawal (18, 20). Because FL5.12 cells are dependent on IL-3 to suppress apoptosis, the above data suggest that the function of BIM at the mitochondria might be regulated by posttranslational modification(s).
Growth factor-dependent survival of cells is mediated in part through kinase signaling pathways, including ERK and PI3-K. We first examined the effects of specific kinase inhibitors on the IL-3-induced mobility shift of BIM. The mobility shift of BIM on SDS/PAGE was strongly inhibited by MEK1/2 inhibitors (U0126 and PD98059), but not by a PI3-K inhibitor (LY294002) (Fig. 2A, and data not shown). These data suggest the MEKERK pathway appears to be required for BIM phosphorylation. This effect has also been noted in PC12 neuronal cells cultured in nerve growth factor (11) and epithelial cells cultured in epidermal growth factor (16). The proposed phosphorylation of BIM might prove to be a common event in growth factor mediated survival. Treatment of FL5.12 cells with MEK1/2 inhibitors partially inhibited IL-3-mediated cell survival (≈30%), providing a physiological correlation with the apparent inhibition of phosphorylation (data not shown).
Fig. 2.
ERK/MAPK is a prominent kinase for BIM phosphorylation. (A) IL-3 was withdrawn from FL5.12 BCL-2/HA-BIM WT cells for 4 h (-IL-3), followed by readdition of IL-3 for 15 min (+IL-3). Kinase inhibitors [MEK inhibitor, U0126 (10 μM), and PI3-K inhibitor, LY294002 (10 μM)] were added at the time of IL-3 starvation. Total cell extracts were subjected to Western blots with anti-HA (for detection of BIM), anti-phospho-ERK, anti-phospho-AKT, and anti-phospho-JNK. (B) Schematic representation of major BIM isoforms and the positions of the phosphorylation sites identified in this study. The position of BH3 domain is underlined. (C) ERK2 in vitro kinase assay was performed with GST-ELK1 (positive control), GST-BAD WT and GST-BAD S112AS136A (negative controls), and GST-BIM as substrates. Arrow indicates phosphorylated BIM protein. (D) HA-tagged BIM WT or indicated mutants were transiently transfected into FL5.12 BCL-2 cells. After transfection (24 h), IL-3 was withdrawn for 4 h (-IL-3), followed by readdition of IL-3 for 15 min (+IL-3). Total cell extracts were subjected to Western blot with anti-HA. Arrows indicate the position of BIMEL and BIML, respectively. (E) IL-3 was withdrawn from FL5.12 BCL-2/BIM WT cells for4h(-IL-3), followed by readdition of IL-3 for 15 min (+IL-3). Kinase inhibitors [MEK inhibitor, U0126 (10 μM), PI3-K inhibitor, LY294002 (10 μM), JNK inhibitor SP600125 (10 μM), and p38 inhibitor SB203580 (10 μM)] were added at the time of IL-3 starvation. Total cell extracts were subjected to Western blots with anti-HA (for detection of BIM), anti-phospho-ERK, anti-phospho-JNK, and anti-phospho-p38 Abs. BIM site specific phosphorylations were determined by phospho-S55, phospho-S65, and phospho-S100 specific BIM Abs, respectively.
The primary amino acid sequence of BIMEL contains several consensus phosphorylation sites for ERK (Fig. 2B). An in vitro kinase assay with purified ERK2 and recombinant BIMEL protein demonstrated that BIMEL is a direct substrate for ERK (Fig. 2C). To identify BIM phosphorylation sites, selected sites were mutated to Ala. We chose six sites (amino acid residues 55, 65, 73, 100, 112, and 114 of mouse BIM) conforming to the ERK consensus sequence S/T-P or P-X-S/T-P. When the three serines, S55, S65, and S100, were all mutated to Ala, the mobility shift induced by IL-3 was eliminated (Fig. 2D), indicating that these three serines are major phosphorylation residues. These residues are also conserved in human as well as rat BIM, suggesting that this regulation will prove conserved. Conversely, substitutions of S73, T112, or S114 did not affect the mobility of BIM (data not shown). The S100 mutation eliminated the mobility shift of BIML (Fig. 2D), suggesting that other potential ERK phosphorylation sites, i.e., T112 and S114, are not phosphorylated under these experimental conditions. We then generated or obtained phosphorylation-specific BIM Abs to sites S55, S65, and S100 to confirm the importance of these sites and monitor their phosphorylation status. The specificity of each Ab was confirmed by Ala-substituted BIM mutants (data not shown). All three phosphor-serine Abs demonstrate site specific phosphorylation of BIM after IL-3 exposure. The dependence of all three sites of phosphorylation on the ERK pathway was confirmed by using the MEK inhibitor, U0126 (Fig. 2E). A PI3-K inhibitor, LY294002, did not inhibit any of the sites of BIM phosphorylation. Moreover, a JNK inhibitor, SP600125, and a p38-MAPK inhibitor, SB203580, did not affect the phosphorylation, indicating that ERK is the major kinase involved in IL-3 induced BIM phosphorylation (Fig. 2E).
To determine whether the phosphorylation of BIM regulates its proapoptotic activity, we examined the phosphorylation site-specific mutants of BIM. BIM was originally identified as a BH3-only protein that interacts with BCL-2. The balance between pro- and antiapoptotic BCL-2 family members is critical for regulating cell death. Therefore, several independent sets of FL5.12 BCL-2 stable clones expressing comparable levels of WT or mutant BIM were generated. The nonphosphorylatable BIMEL mutant (i.e., BIM S55AS65AS100A) proved to be a stronger death-agonist after IL-3 deprivation than BIM WT (Fig. 3). Of note, we have successfully maintained clones expressing nonphosphorylatable BIM in IL-3, indicating that BIM-independent pathways are capable of sustaining the survival of these cells. It has been reported that S65 (S69, human) of BIM can be phosphorylated by ERK (14, 15). We observed that the S65A substitution most affected the SDS/PAGE based mobility shift of BIMEL (Fig. 2D). However, in addition to S65, we found that S55 and S100 are also phosphorylated by ERK after IL-3 stimulation. Moreover, a BIM mutant in which two serines are substituted (S65A and S100A) did not demonstrate any enhancement of apoptotic activity in our system (data not shown), indicating that the dephosphorylation of all three serines may be required for the full killing activity of BIM.
Fig. 3.
Nonphosphorylatable BIM (S55A, S65A, and S100A) enhances IL-3 withdrawal death. Independent clones of FL5.12 BCL-2 cells expressing comparable levels of WT or a serine-substituted BIMEL mutant (BIM S55AS65AS100A) were deprived of IL-3, and cell death was quantitated by Annexin-V-PI staining, 24 h after deprivation. Results are the mean ± SD of three independent experiments. We analyzed four independent clones and the results are reproducible.
We next examined whether constitutively active MEK1 would result in phosphorylation of BIM and affect apoptosis after IL-3 withdrawal. Constitutively active MEK1 resulted in S65 phosphorylation (Fig. 4A) and partially countered apoptosis after IL-3 withdrawal (Fig. 4B). To assess whether the phosphorylation of BIM contributed to MEK1-ERK mediated survival, the inhibition of apoptosis by constitutively active MEK1 was compared in FL5.12 cells expressing BIM WT vs. the BIM S55AS65AS100A mutant. Approximately 30% of the MEK1-mediated protective effect was eliminated in cells with the nonphosphorylatable BIM mutant (Fig. 4B). This result indicates that MEK1-ERK cell survival is mediated in part by the phosphorylation of BIM. The residual effect of MEK1-ERK on the survival of cells with the nonphosphorylatable BIM suggests the possibility that ERK may target additional apoptotic molecules.
What are the molecular mechanisms by which phosphorylation could regulate BIM activity? It has been proposed that BIM may interact not only with antiapoptotic BCL-2 family members, such as BCL-2 (21) and MCL-1 (22), but also with proapoptotic members, such as BAX, to directly activate the apoptotic cascades (23–25). Therefore, we addressed whether the phosphorylation status of BIM affected interactions with BAX, BCL-2, or MCL-1. Mitochondria-enriched fractions were prepared from FL5.12 BCL-2/HA-BIM WT cells in the presence of IL-3 (time 0), after IL-3 deprivation (-IL-3, 8 h), or after re-addition of IL-3 (+IL-3, 15 min) (Fig. 5A). The amount of BIM, and also BCL-2, at the mitochondria was relatively unchanged by the various conditions (Fig. 5A). However, the phosphorylation of BIM at S65, which was undetectable after IL-3 withdrawal, was noted in the presence of IL-3, induced most robustly by the readdition of IL-3 (Fig. 5A). As expected, levels of BAX at the mitochondria increased after IL-3 withdrawal and did not change substantially after IL-3 readdition (Fig. 5A). Using these same extracts, we examined BIM/BAX, BIM/BCL-2, and BIM/MCL-1 interactions by coimmunoprecipitation experiments. Irrespective of the phosphorylation status of BIM, similar amounts of BCL-2, as well as MCL-1 were coimmunoprecipitated with HA-BIM WT (Fig. 5B and data not shown). In contrast, HA-BIM WT coimmunoprecipitated with BAX at mitochondria only after IL-3 deprivation when BIM is dephosphorylated (Fig. 5B). In long-term culture with IL-3 (time 0), substantial amounts of BIM are not phosphorylated but the amount of BAX at the mitochondria in the presence of IL-3 is minimal and perhaps inadequate to accurately assess an interaction with BAX. After deprivation of IL-3 for 8 h, BIM is dephosphorylated, BAX has translocated to the mitochondria, and BIM now coprecipitates with BAX. In contrast, after reexposure to IL-3 for 15 min, BIM is rapidly phosphorylated and phosphorylated BIM has lost its ability to interact with BAX (Fig. 5B). When the mitochondria-enriched fractions from FL5.12 BCL-2/H A-BIM S55AS65AS100A cells were analyzed by the same procedure, the immunoprecipitation of nonphosphorylatable BIM resulted in the coprecipitation of substantial amounts of BAX as well as BCL-2 in either the presence or absence of IL-3 (Fig. 5C). These data strongly suggest that only dephosphorylated BIM can interact with BAX, and that IL-3 induced phosphorylation of BIM prevents its interaction with BAX.
Fig. 5.
BIM/BAX interaction is altered by BIM phosphorylation. (A) FL5.12 BCL-2/HA-BIM WT cells were cultured in IL-3 (time 0), deprived of IL-3 for 8 h (-IL-3), or followed by the readdition of IL-3 for 15 min (+IL-3). Mitochondria-enriched heavy membrane fractions were subjected to Western blots with anti-BAX, anti-BCL-2, anti-HA (total BIM), and anti-phospho-S65 BIM. (B) The above fractions were immunoprecipitated with anti-BAX or anti-HA, respectively. The immunoprecipitates were analyzed by Western blots by using anti-HA or anti-BCL-2 to examine the interaction of BIM/BAX or BIM/BCL-2, respectively. (C) FL5.12 BCL-2/HA-BIM S55AS65AS100A cells were deprived of IL-3 for 8 h (-IL-3) or followed by readdition of IL-3 for 15 min (+IL-3). The mitochondria-enriched heavy membrane fractions were prepared, and Western blots (Upper) and immunoprecipitations (Lower) were performed.
Discussion
The combination of small interfering RNA loss-of-function, signal transduction, and detailed phosphorylation studies indicates that the regulated phosphorylation of BIM is a substantial component of cytokine-dependent survival signaling. In the presence of survival factor (e.g., IL-3), most of BAX is in the cytosol whereas most of BIM is at the mitochondria, perhaps helping to ensure their lack of interaction. Moreover, exposure to IL-3 results in a MEK-ERK-dependent phosphorylation of BIM that precludes its interaction with the “multidomain” death effector BAX. Preventing this interaction would be predicted to interfere with the capacity of the “activator” BH3-only protein BIM to trigger the oligomerization of BAX and subsequent apoptosis (25). In contrast, after IL-3 withdrawal, BIM is dephosphorylated and can interact with BAX, which has translocated to the mitochondria where it serves as a critical gateway to apoptosis (26). Even in IL-3-starved cells, if IL-3 is readded before they have passed the point of no return, the phosphorylation of BIM would prevent its interaction with BAX that has already translocated to the mitochondria. Of note, BIM total protein levels are stable up to 4 h after IL-3 readdition (Fig. 1B). It has been shown that BIM phosphorylation can promote the degradation of BIM through the proteasome pathway (15, 19). However, in this IL-3-dependent cell, substantial amounts of BIM protein persist after its phosphorylation. Perhaps phosphorylated BIM, which cannot interact with BAX, can also be degraded, providing two mechanisms whereby cytokine driven phosphorylation of BIM eliminates its proapoptotic effect. Although BIM primarily interacts with the BCL-2 family members, it has been proposed that the interaction of BIM with dynein light chain/LC8 also regulates the induction of apoptosis (13). In this IL-3-dependent hematopoietic cell system, BIM was predominantly localized to mitochondria and did not dramatically change localization upon cytokine withdrawal. In addition, the capacity of BIM to interact with LC8 did not change as the phosphorylation status of BIM changed, as assessed by coimmunoprecipitation experiments (data not shown) (27). On the other hand, phosphorylation of BIML at T112 (mouse) by JNK has been noted to result in the release of dynein light chain after UV irradiation of fibroblasts (28). Thus, although several MAPK family consensus phosphorylation sites exist in BIM, the specific kinase responsible for BIM phosphorylation appears to be specified by selected signals.
Conclusions
Activation of the ERK/MAPK pathway by survival factors inhibits the intrinsic apoptotic pathway. However, it was not entirely clear whether this pathway is interrupted principally upstream or downstream of the mitochondria. It has been recently reported that the activity of caspase-9 can be inhibited through phosphorylation by ERK (29), whereas here BH3-only BIM, which operates upstream to the essential BAX, BAK mitochondrial gateway (26), is clearly a target. Therefore, multiple targets of ERK/MAPK phosphorylation may coordinately inhibit cell death. As one example, MAPK activation plays a role in T cell development (3, 30), and a BIM-deficient mouse indicates BIM plays a role in negative selection, suggesting the role of BIM phosphorylation should be examined in T cell development. Regulation of BIM by ERK may also prove important in the developmental control of cell death and suppression of apoptosis in cancer. The extent of BIM phosphorylation should be determined in tumor cells, which often have a constitutively active ERK/MAPK pathway, to determine whether BIM's phosphorylation status influences the cytotoxic response to cancer therapeutics.
Acknowledgments
We thank S. Grant for reagents; P. Dent, S. Grant, J. Opferman, and O. Takeuchi for discussion and comments on the manuscript; and E. Smith for manuscript preparation. This work was supported in part by the Jeffress Memorial Trust, the American Society of Hematology Scholar Award (to H.H.), and a National Institutes of Health grant (to S.J.K.).
Author contributions: H.H. and S.J.K. designed research; H.H., B.Q., A.R.-V., and S.J.K. performed research; H.H., B.Q., A.R.-V., and S.J.K. analyzed data; B.Q. and A.R.-V. contributed new reagents/analytic tools; and H.H. and S.J.K. wrote the paper.
Abbreviations: ERK, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase; JNK, Jun N-terminal kinase; MEK, MAPK kinase; PI3-K, phosphatidylinositol 3-OH-kinase; BIML or EL, BIM long or extra long; HA, hemagglutinin.
References
- 1.Bouillet, P., Purton, J. F., Godfrey, D. I., Zhang, L. C., Coultas, L., Puthalakath, H., Pellegrini, M., Cory, S., Adams, J. M. & Strasser, A. (2002) Nature 415, 922-926. [DOI] [PubMed] [Google Scholar]
- 2.Khaled, A. R. & Durum, S. K. (2002) Nat. Rev. Immunol. 2, 817-830. [DOI] [PubMed] [Google Scholar]
- 3.Sohn, S. J., Rajpal, A. & Winoto, A. (2003) Curr. Opin. Immunol. 15, 209-216. [DOI] [PubMed] [Google Scholar]
- 4.Marrack, P. & Kappler, J. (2004) Annu. Rev. Immunol. 22, 765-787. [DOI] [PubMed] [Google Scholar]
- 5.Bouillet, P., Metcalf, D., Huang, D. C., Tarlinton, D. M., Kay, T. W., Kontgen, F., Adams, J. M. & Strasser, A. (1999) Science 286, 1735-1738. [DOI] [PubMed] [Google Scholar]
- 6.Hildeman, D. A., Zhu, Y., Mitchell, T. C., Bouillet, P., Strasser, A., Kappler, J. & Marrack, P. (2002) Immunity 16, 759-767. [DOI] [PubMed] [Google Scholar]
- 7.Putcha, G. V., Moulder, K. L., Golden, J. P., Bouillet, P., Adams, J. A., Strasser, A. & Johnson, E. M. (2001) Neuron 29, 615-628. [DOI] [PubMed] [Google Scholar]
- 8.Whitfield, J., Neame, S. J., Paquet, L., Bernard, O. & Ham, J. (2001) Neuron 29, 629-643. [DOI] [PubMed] [Google Scholar]
- 9.Shinjyo, T., Kuribara, R., Inukai, T., Hosoi, H., Kinoshita, T., Miyajima, A., Houghton, P. J., Look, A. T., Ozawa, K. & Inaba, T. (2001) Mol. Cell. Biol. 21, 854-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dijkers, P. F., Birkenkamp, K. U., Lam, E. W., Thomas, N. S., Lammers, J. W., Koenderman, L. & Coffer, P. J. (2002) J. Cell Biol. 156, 531-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biswas, S. C. & Greene, L. A. (2002) J. Biol. Chem. 277, 49511-49516. [DOI] [PubMed] [Google Scholar]
- 12.Reginato, M. J., Mills, K. R., Paulus, J. K., Lynch, D. K., Sgroi, D. C., Debnath, J., Muthuswamy, S. K. & Brugge, J. S. (2003) Nat. Cell Biol. 5, 733-740. [DOI] [PubMed] [Google Scholar]
- 13.Puthalakath, H., Huang, D. C., O'Reilly, L. A., King, S. M. & Strasser, A. (1999) Mol. Cell 3, 287-296. [DOI] [PubMed] [Google Scholar]
- 14.Ley, R., Ewings, K. E., Hadfield, K., Howes, E., Balmanno, K. & Cook, S. J. (2004) J. Biol. Chem. 279, 8837-8847. [DOI] [PubMed] [Google Scholar]
- 15.Luciano, F., Jacquel, A., Colosetti, P., Herrant, M., Cagnol, S., Pages, G. & Auberger, P. (2003) Oncogene 22, 6785-6793. [DOI] [PubMed] [Google Scholar]
- 16.Marani, M., Hancock, D., Lopes, R., Tenev, T., Downward, J. & Lemoine, N. R. (2004) Oncogene 23, 2431-2441. [DOI] [PubMed] [Google Scholar]
- 17.Akiyama, T., Bouillet, P., Miyazaki, T., Kadono, Y., Chikuda, H., Chung, U. I., Fukuda, A., Hikita, A., Seto, H., Okada, T., et al. (2003) EMBO J. 22, 6653-6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross, A., Jockel, J., Wei, M. C. & Korsmeyer, S. J. (1998) EMBO J. 17, 3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ley, R., Balmanno, K., Hadfield, K., Weston, C. & Cook, S. J. (2003) J. Biol. Chem. 278, 18811-18816. [DOI] [PubMed] [Google Scholar]
- 20.Wolter, K. G., Hsu, Y. T., Smith, C. L., Nechushtan, A., Xi, X. G. & Youle, R. J. (1997) J. Cell Biol. 139, 1281-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Connor, L., Strasser, A., O'Reilly, L. A., Hausmann, G., Adams, J. M., Cory, S. & Huang, D. C. (1998) EMBO J. 17, 384-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Opferman, J. T., Letai, A., Beard, C., Sorcinelli, M. D., Ong, C. C. & Korsmeyer, S. J. (2003) Nature 426, 671-676. [DOI] [PubMed] [Google Scholar]
- 23.Marani, M., Tenev, T., Hancock, D., Downward, J. & Lemoine, N. R. (2002) Mol. Cell. Biol. 22, 3577-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, X., Dai, S., Zhu, Y., Marrack, P. & Kappler, J. W. (2003) Immunity 19, 341-352. [DOI] [PubMed] [Google Scholar]
- 25.Letai, A., Bassik, M. C., Walensky, L. D., Sorcinelli, M. D., Weiler, S. & Korsmeyer, S. J. (2002) Cancer Cell 2, 183-192. [DOI] [PubMed] [Google Scholar]
- 26.Wei, M. C., Zong, W. X., Cheng, E. H., Lindsten, T., Panoutsakopoulou, V., Ross, A. J., Roth, K. A., MacGregor, G. R., Thompson, C. B. & Korsmeyer, S. J. (2001) Science 292, 727-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seward, R. J., von Haller, P. D., Aebersold, R. & Huber, B. T. (2003) Mol. Immunol. 39, 983-993. [DOI] [PubMed] [Google Scholar]
- 28.Lei, K. & Davis, R. J. (2003) Proc. Natl. Acad. Sci. USA 100, 2432-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allan, L. A., Morrice, N., Brady, S., Magee, G., Pathak, S. & Clarke, P. R. (2003) Nat. Cell Biol. 5, 647-654. [DOI] [PubMed] [Google Scholar]
- 30.Werlen, G., Hausmann, B., Naeher, D. & Palmer, E. (2003) Science 299, 1859-1863. [DOI] [PubMed] [Google Scholar]