Abstract
Background:
The second leading cause of cancer deaths in women is breast cancer. Germline mutations in susceptibility breast cancer gene BRCA1 increase the lifetime risk of breast cancer. Eighty-one large genomic rearrangements (LGRs) have been reported up to date in BRCA1 gene, and evaluation of these rearrangements helps with precise risk assessment in high-risk individuals. In this study, we have investigated LGRs in BRCA1 among Iranian high-risk breast cancer families.
Materials and Methods:
Seventy patients with breast cancer who were identified negative for point mutations or small deletions/insertions of BRCA1 gene were selected. Deletions and duplications of BRCA1 gene were evaluated using multiplex ligation-dependent probe amplification (MLPA).
Results:
Two deletions, deletion of exons 1A/1B-2 and exon 24, were detected in two patients with breast cancer. The former alteration was found in a woman with a strong family history of breast cancer while the latter one was detected in a woman with early onset of breast cancer.
Conclusion:
Although our data confirm that LGRs in BRCA1 comprise a relatively small proportion of mutations in hereditary breast cancer in the Iranian population, MLPA analysis might be considered for screening of LGRs in high-risk individuals. It is worth to note that our results are consistent with previous studies in various Asian and European countries.
Keywords: BRCA1 gene, breast cancer, large genomic rearrangements, multiplex ligation-dependent probe amplification
INTRODUCTION
BRCA1 gene, which is located on the long arm of chromosome 17 (17q21) [Figure 1], spans approximately 81 kb and encodes a protein of 1863 amino acids.[1] BRCA1 is an essential tumor suppressor, suppressing genome instability.[2]
Figure 1.
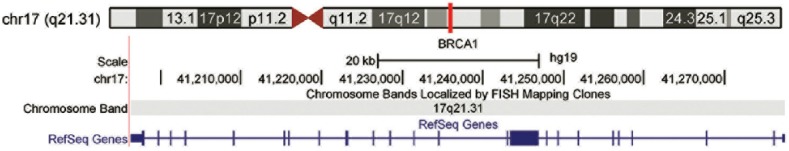
Genomic location of BRCA1 gene on chromosome 17. Genome assembly used to produce (http://genome.ucsc.edu)
Breast cancer is the second leading cause of cancer deaths in women worldwide.[3] BRCA-mutant individuals and families have a notable increased risk of developing breast and ovarian cancers.[4] Five percent of all breast cancers are hereditary, and nearly 16% of hereditary breast cancers are due to germline mutations in BRCA1/2 genes.[5] The screening of BRCA1/2 mutations is a common component of risk evaluation and management of familial breast cancer, bilateral breast cancer, and early-onset breast cancer.[6] Familial breast cancer includes 20–30% of all breast cancer cases. It is also reported that most of the families with <6 breast cancer cases and no ovarian cancer do not carry BRCA1/2 mutations that can be detected by routine sequencing protocols.[7] Various mutations including nonsense mutations, small deletions and insertions, and large genomic rearrangements (LGRs) are reported in BRCA1 gene.[8] Furthermore, these mutations can result in recurrent mutations of other specific genes (such as RB1) that may lead to other cancers.[9] The incidence of LGRs in BRCA1/2 genes is very low,[10] and these LGRs occur in a small percentage (<1%) of patients tested for hereditary breast and ovarian cancers.[11] Furthermore, the prevalence of LGRs varies from 0% (Afrikaner and French-Canadian populations) to 27% (the Dutch population) of all BRCA1 mutations in different populations.[12] The characterization of three founder BRCA1 LGRs, deletion of exon 13, exon 22,[13] and exons 1A/1B-2,[14] in the Dutch population explains high frequency of LGRs (27%).
DNA double-strand break (DSB) is one of the most common DNA repair mechanisms within normal cells. The BRCA-mutated breast cancer cells lack the homologous recombination that is required for error-free DSB repair.[15] Most LGRs detected in BRCA1 result from unequal homologous recombination events involving Alu repeats, comprising 41.5% of the gene, and BRCA1 pseudogene (ψBRCA1), which is located 30 kb upstream of the BRCA1.[16]
The description of multiplex ligation-dependent probe amplification (MLPA) test for detection of LGRs in BRCA1 in 2001 has increased the number of LGRs identified in different populations so that nearly 61 of the 81 reported LGRs are identified by MLPA.[17] Before the development of MLPA, detection of LGRs was mostly carried out by applying different approaches such as Southern blot, long-range PCR, fluorescence in situ hybridization-based methods, and quantitative multiplex PCR of short fragments.
MLPA can be used as the first step for genetic screening of women with family history of breast cancer in populations with high frequency of LGRs. In this study, LGRs in BRCA1 gene using MLPA in seventy patients without detectable point mutation or small deletions/insertions were investigated.
MATERIALS AND METHODS
Patients
The BRCA1 rearrangements analysis was performed on seventy patients with breast cancer that completed the consent form [Table 1]. Our study was approved by the Ethics Committee of Isfahan University of Medical Sciences. The criteria used for selection of patients were as follows: (1) early onset of breast cancer (under 35 years old), (2) two or more cases of breast cancer in the family, (3) bilateral breast cancer, or (4) men with breast cancer. All patients were negative for BRCA1 point mutations or small deletions/insertions. This study was performed in breast cancer research center of Isfahan University of Medical Sciences.
Table 1.
Characteristics of the patients

DNA ISOLATION AND MULTIPLEX LIGATION-DEPENDENT PROBE AMPLIFICATION ANALYSIS
DNA was extracted from blood leukocytes using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The exact DNA concentration was determined using a NanoDrop instrument (Thermo 2000c) after dilution to 50 ng/ml. The BRCA1-MLPA analysis was carried out using the SALSA MLPA test kit P002-C2 (MRC Holland, The Netherland) according to the manufacturer’s instructions. P002-C2 BRCA1 probemix contains probes for each exon of the BRCA1 gene and nine reference probes that are included for normalization purposes. P087 MLPA kit was used for confirmation of the obtained data. This kit is designed to detect deletions/duplications of one or more sequences in the BRCA1 gene in a DNA sample.
In brief, the ligation reaction was performed using 100 ng of target DNA in the following steps: Denaturation at 98°C for 5 min, hybridization using BRCA1-MLPA probemix at 60°C for 16 h, and ligation using Ligase-65 mix at 54°C for 15 min followed by ligase inactivation at 98°C for 5 min. After the ligation step, multiplex polymerase chain reaction was performed using the appropriate PCR primers, dNTPs, SALSA PCR buffer, and SALSA polymerase for thirty cycles (95°C for 30 s, 60°C for 30 s, and 72°C for 60 s) followed by one cycle at 72°C for 20 min. Probe amplification products were analyzed on 3130 capillary sequencer (Applied Biosystems, UK) with a 36 cm capillary array and POP-4™ polymer (Applied Biosystems) by mixing with 0.4 μl of the GeneScan™-500 LIZ™ size standard (Applied Biosystems) and 9 μl of Hi-Di Formamide (Applied Biosystems). The results (size and the peak area) were exported from the DNA sequencer and analyzed by GeneMarker software. It converts data from any sequencing system, offers two normalization and analysis methods. A ratio under 0.7 was taken as a sign of the presence of only one copy of a sequence in the BRCA1 gene.
RESULTS
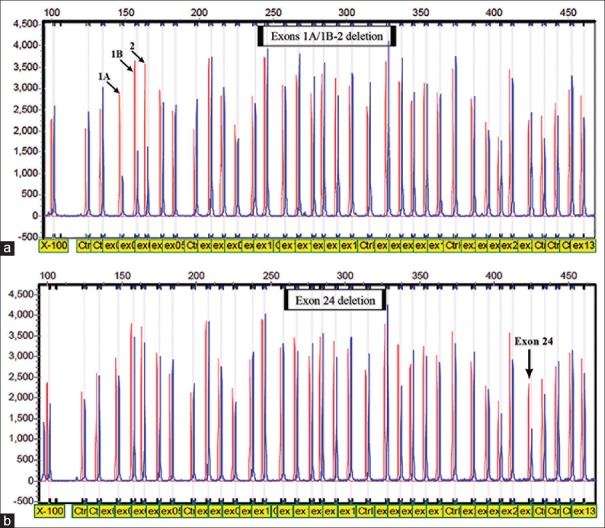
MLPA analysis of genomic DNA for seventy high-risk individuals who were found negative for point mutations or small insertions/deletions in the BRCA1 revealed two different rearrangements [Figure 2]. We detected a deletion of exons 1A/1B-2 in a patient (F307) with a strong family history of breast cancer, with the mother, grandmother, and two other relatives affected with breast cancer. A deletion of exon 24 was found in a woman (F792) with early onset breast cancer (age 30) and with the grandmother affected with breast cancer. The obtained results were confirmed using confirmation MLPA kit (P087).
Figure 2.
Multiplex ligation-dependent probe amplification electropherogram. (a) Analysis of individual carrying deletion of exons 1A/1B-2. (b) Analysis of individual with deletion of exon 24
DISCUSSION
In this study, we carried out MLPA analyses of seventy patients with breast cancer to identify the prevalence and spectrum of LGRs in the BRCA1 gene. Our results indicate that LGRs account for 2.8% of high-risk families without point mutations and small insertion/deletion.
The frequency of LGRs in our study (2.8% of mutation-negative families) was comparable with the results reported in Singapore (3%),[18] Malaysia (8%),[19] and in various European populations.[20,21] However, it is not in agreement with previous study in the Iranian population.[22] Based on the previous study, the frequency of LGRs in the Iranian population was reported 0%. In this study, eight Iranian high-risk breast cancer families without point mutations in BRCA1/2 were investigated by semiquantitative multiplex PCR method. However, in our study, seventy patients negative for point mutations in BRCA1 were analyzed by MLPA analysis. Nevertheless, the comparison between two studies is rather inappropriate because the sample size and the criteria for family selection are largely different.
MLPA analysis of the BRCA1 gene led to the identification of two BRCA1 deletions, exons 1A/1B-2 and exon 24 deletions. To date, six different exons 1A/1B-2 deletions, with different break points, in different populations have been identified.[23] Two different types of exons 1A/1B-2 deletions are considered founder mutations in the Dutch population.[14] Exon 24 deletion is identified in the Greek[24] and German populations. This exon is involved in four more deletions in combination with other exons, exons 21–24 deletion,[25,26] and exons 23–24 deletion.[23,27] Since deletion of exons 1A/1B-2 and exon 24 removes two essential regions for RNA expression, promoter, polyA tail, and 3'-untranslated region, respectively, the possible result of these deletions is loss of RNA transcript.[24,28]
Our data verify that large BRCA1 deletions or duplications comprise a relatively small proportion of mutations in hereditary breast cancer in the Iranian population. Since the identification of large rearrangements in the BRCA1 gene is important for clinical management of individuals affected with breast cancer[29] and for preventive measures for healthy carriers of a familial BRCA1 mutation,[30] MLPA analysis should be considered for screening of LGRs in high-risk individuals.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
AUTHORS’ CONTRIBUTION
MS contributed in conception of the work, conducting the study, revising the draft and agreed for all aspects of the work
EE contributed in conception of the work, conducting the study and agreed for all aspects of the work
EN contributed in conducting the study, writing the draft and agreed for all aspects of the work
AS contributed in drafting and revising the draft and agreed for all aspects of the work
MS contributed in conducting the study and agreed for all aspects of the work
NN contributed in drafting and agreed for all aspects of the work
SF contributed in conception of the work and agreed for all aspects of the work
LD contributed in revising the draft and agreed for all aspects of the work
FM contributed in conception of the work, conducting the study, revising the draft and agreed for all aspects of the work.
Acknowledgments
This work was funded by grant number 187155 from the Deputy for Research, Isfahan University of Medical Sciences, Isfahan, Iran. This study was performed in the Genetics Laboratory of Alzahra University Hospital. We are grateful to patients with breast cancer for their participation in this study.
REFERENCES
- 1.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–82. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 3.Kwong A, Chen J, Shin VY, Ho JC, Law FB, Au CH, et al. The importance of analysis of long-range rearrangement of BRCA1 and BRCA2 in genetic diagnosis of familial breast cancer. Cancer Genet. 2015;208:448–54. doi: 10.1016/j.cancergen.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 4.Rudnicka H, Debniak T, Cybulski C, Huzarski T, Gronwald J, Lubinski J, et al. Large BRCA1 and BRCA2 genomic rearrangements in Polish high-risk breast and ovarian cancer families. Mol Biol Rep. 2013;40:6619–23. doi: 10.1007/s11033-013-2775-0. [DOI] [PubMed] [Google Scholar]
- 5.van der Groep P, van der Wall E, van Diest PJ. Pathology of hereditary breast cancer. Cell Oncol (Dordr) 2011;34:71–88. doi: 10.1007/s13402-011-0010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li YT, Ni D, Yang L, Zhao Q, Ou JH. The prevalence of BRCA1/2 mutations of triple-negative breast cancer patients in Xinjiang multiple ethnic region of China. Eur J Med Res. 2014;19:35. doi: 10.1186/2047-783X-19-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yiannakopoulou E. Etiology of familial breast cancer with undetected BRCA1 and BRCA2 mutations: Clinical implications. Cell Oncol (Dordr) 2014;37:1–8. doi: 10.1007/s13402-013-0158-0. [DOI] [PubMed] [Google Scholar]
- 8.Palma M, Ristori E, Ricevuto E, Giannini G, Gulino A. BRCA1 and BRCA2: The genetic testing and the current management options for mutation carriers. Crit Rev Oncol Hematol. 2006;57:1–23. doi: 10.1016/j.critrevonc.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Jönsson G, Staaf J, Vallon-Christersson J, Ringnér M, Gruvberger-Saal SK, Saal LH, et al. The retinoblastoma gene undergoes rearrangements in BRCA1-deficient basal-like breast cancer. Cancer Res. 2012;72:4028–36. doi: 10.1158/0008-5472.CAN-12-0097. [DOI] [PubMed] [Google Scholar]
- 10.Rouleau E, Jesson B, Briaux A, Nogues C, Chabaud V, Demange L, et al. Rare germline large rearrangements in the BRCA1/2 genes and eight candidate genes in 472 patients with breast cancer predisposition. Breast Cancer Res Treat. 2012;133:1179–90. doi: 10.1007/s10549-012-2009-5. [DOI] [PubMed] [Google Scholar]
- 11.Jackson SA, Davis AA, Li J, Yi N, McCormick SR, Grant C, et al. Characteristics of individuals with breast cancer rearrangements in BRCA1 and BRCA2. Cancer. 2014;120:1557–64. doi: 10.1002/cncr.28577. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Casado Z, Romero I, Fernandez-Serra A, Rubio L, Llopis F, Garcia A, et al. A de novo complete BRCA1 gene deletion identified in a Spanish woman with early bilateral breast cancer. BMC Med Genet. 2011;12:134. doi: 10.1186/1471-2350-12-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrij-Bosch A, Peelen T, van Vliet M, van Eijk R, Olmer R, Drüsedau M, et al. BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat Genet. 1997;17:341–5. doi: 10.1038/ng1197-341. [DOI] [PubMed] [Google Scholar]
- 14.van den Ouweland AM, Dinjens WN, Dorssers LC, van Veghel-Plandsoen MM, Brüggenwirth HT, Withagen-Hermans CJ, et al. Deletion of exons 1a-2 of BRCA1: A rather frequent pathogenic abnormality. Genet Test Mol Biomarkers. 2009;13:399–406. doi: 10.1089/gtmb.2008.0155. [DOI] [PubMed] [Google Scholar]
- 15.Vollebergh MA, Lips EH, Nederlof PM, Wessels LF, Wesseling J, Vd Vijver MJ, et al. Genomic patterns resembling BRCA1-and BRCA2-mutated breast cancers predict benefit of intensified carboplatin-based chemotherapy. Breast Cancer Res. 2014;16:R47. doi: 10.1186/bcr3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith TM, Lee MK, Szabo CI, Jerome N, McEuen M, Taylor M, et al. Complete genomic sequence and analysis of 117 kb of human DNA containing the gene BRCA1. Genome Res. 1996;6:1029–49. doi: 10.1101/gr.6.11.1029. [DOI] [PubMed] [Google Scholar]
- 17.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim YK, Lau PT, Ali AB, Lee SC, Wong JE, Putti TC, et al. Identification of novel BRCA large genomic rearrangements in Singapore Asian breast and ovarian patients with cancer. Clin Genet. 2007;71:331–42. doi: 10.1111/j.1399-0004.2007.00773.x. [DOI] [PubMed] [Google Scholar]
- 19.Kang P, Mariapun S, Phuah SY, Lim LS, Liu J, Yoon SY, et al. Large BRCA1 and BRCA2 genomic rearrangements in Malaysian high risk breast-ovarian cancer families. Breast Cancer Res Treat. 2010;124:579–84. doi: 10.1007/s10549-010-1018-5. [DOI] [PubMed] [Google Scholar]
- 20.Hansen Tv, Jønson L, Albrechtsen A, Andersen MK, Ejlertsen B, Nielsen FC. Large BRCA1 and BRCA2 genomic rearrangements in Danish high risk breast-ovarian cancer families. Breast Cancer Res Treat. 2009;115:315–23. doi: 10.1007/s10549-008-0088-0. [DOI] [PubMed] [Google Scholar]
- 21.Engert S, Wappenschmidt B, Betz B, Kast K, Kutsche M, Hellebrand H, et al. MLPA screening in the BRCA1 gene from 1,506 German hereditary breast cancer cases: Novel deletions, frequent involvement of exon 17, and occurrence in single early-onset cases. Hum Mutat. 2008;29:948–58. doi: 10.1002/humu.20723. [DOI] [PubMed] [Google Scholar]
- 22.Pietschmann A, Mehdipour P, Mehdipour P, Atri M, Hofmann W, Hosseini-Asl SS, et al. Mutation analysis of BRCA1 and BRCA2 genes in Iranian high risk breast cancer families. J Cancer Res Clin Oncol. 2005;131:552–8. doi: 10.1007/s00432-005-0678-8. [DOI] [PubMed] [Google Scholar]
- 23.Sluiter MD, van Rensburg EJ. Large genomic rearrangements of the BRCA1 and BRCA2 genes: Review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res Treat. 2011;125:325–49. doi: 10.1007/s10549-010-0817-z. [DOI] [PubMed] [Google Scholar]
- 24.Armaou S, Konstantopoulou I, Anagnostopoulos T, Razis E, Boukovinas I, Xenidis N, et al. Novel genomic rearrangements in the BRCA1 gene detected in Greek breast/ovarian cancer patients. Eur J Cancer. 2007;43:443–53. doi: 10.1016/j.ejca.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 25.Walsh T, Casadei S, Coats KH, Swisher E, Stray SM, Higgins J, et al. Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA. 2006;295:1379–88. doi: 10.1001/jama.295.12.1379. [DOI] [PubMed] [Google Scholar]
- 26.Ticha I, Kleibl Z, Stribrna J, Kotlas J, Zimovjanova M, Mateju M, et al. Screening for genomic rearrangements in BRCA1 and BRCA2 genes in Czech high-risk breast/ovarian cancer patients: High proportion of population specific alterations in BRCA1 gene. Breast Cancer Res Treat. 2010;124:337–47. doi: 10.1007/s10549-010-0745-y. [DOI] [PubMed] [Google Scholar]
- 27.Buffone A, Capalbo C, Ricevuto E, Sidoni T, Ottini L, Falchetti M, et al. Prevalence of BRCA1 and BRCA2 genomic rearrangements in a cohort of consecutive Italian breast and/or ovarian cancer families. Breast Cancer Res Treat. 2007;106:289–96. doi: 10.1007/s10549-007-9499-6. [DOI] [PubMed] [Google Scholar]
- 28.Suen TC, Goss PE. Transcription of BRCA1 is dependent on the formation of a specific protein-DNA complex on the minimal BRCA1 Bi-directional promoter. J Biol Chem. 1999;274:31297–304. doi: 10.1074/jbc.274.44.31297. [DOI] [PubMed] [Google Scholar]
- 29.Smith KL, Isaacs C. BRCA mutation testing in determining breast cancer therapy. Cancer J. 2011;17:492–9. doi: 10.1097/PPO.0b013e318238f579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calderon-Margalit R, Paltiel O. Prevention of breast cancer in women who carry BRCA1 or BRCA2 mutations: A critical review of the literature. Int J Cancer. 2004;112:357–64. doi: 10.1002/ijc.20429. [DOI] [PubMed] [Google Scholar]