Summary
Retrograde signals from the plastid regulate photosynthesis‐associated nuclear genes and are essential to successful chloroplast biogenesis. One model is that a positive haem‐related signal promotes photosynthetic gene expression in a pathway that is abolished by the herbicide norflurazon. Far‐red light (FR) pretreatment and transfer to white light also results in plastid damage and loss of photosynthetic gene expression. Here, we investigated whether norflurazon and FR pretreatment affect the same retrograde signal.
We used transcriptome analysis and real‐time reverse transcription−polymerase chain reaction (RT‐PCR) to analyse the effects of these treatments on nuclear gene expression in various Arabidopsis (Arabidopsis thaliana) retrograde signalling mutants.
Results showed that the two treatments inhibited largely different nuclear gene sets, suggesting that they affected different retrograde signals. Moreover, FR pretreatment resulted in singlet oxygen (1O2) production and a rapid inhibition of photosynthetic gene expression. This inhibition was partially blocked in the executer1executer2 mutant, which is impaired in 1O2 signalling.
Our data support a new model in which a 1O2 retrograde signal, generated by chlorophyll precursors, inhibits expression of key photosynthetic and chlorophyll synthesis genes to prevent photo‐oxidative damage during de‐etiolation. Such a signal would provide a counterbalance to the positive haem‐related signal to fine tune regulation of chloroplast biogenesis.
Keywords: chlorophyll synthesis, chloroplast development, photosynthesis, regulation of gene expression, retrograde signalling, singlet oxygen (1O2), tetrapyrroles
Introduction
Communication between the nucleus and plastids (most notably the chloroplasts) is crucial for plant cell function. The nucleus maintains control over most aspects of chloroplast development and function (Jarvis & López‐Juez, 2013), but it has been recognized for over three decades that chloroplasts also exert a retrograde influence on nuclear gene expression (Bradbeer et al., 1979). Many signalling molecules have been implicated in plastid‐to‐nucleus communication (Kleine et al., 2009; Pfannschmidt, 2010; Chi et al., 2013; Chan et al., 2016), with the best characterized operating in mature plants in response to a range of stresses (Estavillo et al., 2011; Xiao et al., 2012). Reactive oxygen species (ROS) have also been shown to be important, with chloroplast‐derived superoxide, hydrogen peroxide (H2O2) and singlet oxygen (1O2) all able to regulate nuclear gene expression (Galvez‐Valdivieso & Mullineaux, 2010). In particular, extensive characterization of the fluorescent (flu) mutant of Arabidopsis (Meskauskiene et al., 2001) has revealed an important role for chloroplast‐derived 1O2 in mediating stress acclimation and cell death responses (Kim et al., 2012; Kim & Apel, 2013). In this experimental system, 1O2 is generated by photo‐excitation of the chlorophyll precursor protochlorophyllide (Pchlide), which accumulates in dark‐grown flu seedlings (op den Camp et al., 2003). The nature of this 1O2 signalling pathway is unknown, but, as 1O2 signalling has a short half‐life, signals would need to originate within the chloroplast. Some possible components have been identified, the most prominent of which are EXECUTER1 (EX1; Wagner et al., 2004) and EX2 (Lee et al., 2007). These related, chloroplast‐localized proteins are both required for flu‐mediated induction of 1O2‐regulated genes (Lee et al., 2007). Recently, Woodson et al. (2015) also identified a protoporphyrin IX‐induced, 1O2‐signalling pathway leading to ubiquitin‐mediated degradation of damaged chloroplasts that may be important in stress adaptation.
In contrast to signals involved in environmental stress responses, signals mediating retrograde signalling in seedlings during chloroplast biogenesis have proved elusive. Many of the early studies on retrograde signalling demonstrated a catastrophic loss of nuclear gene expression either in mutant seedlings lacking functional chloroplasts (Harpster et al., 1984; Hess et al., 1994) or in wild‐type (WT) seedlings subjected to chemical treatments that disrupt chloroplast function (Mayfield & Taylor, 1984; Oelmüller et al., 1986). The most commonly used treatment, the herbicide norflurazon (NF), inhibits carotenoid synthesis, causing plastid‐specific photo‐oxidative damage and resulting in a severe reduction in expression of photosynthetic genes, exemplified by LIGHT HARVESTING CHLOROPHYLL A/B BINDING PROTEIN 1.2 (LHCB1.2) encoding a light‐harvesting chlorophyll‐binding protein (Strand et al., 2003; Koussevitzky et al., 2007; Moulin et al., 2008; Aluru et al., 2009). What we know about this biogenic retrograde signal has come mostly from the identification of genomes uncoupled (gun) mutants in Arabidopsis that retain partial LHCB1.2 expression in NF‐bleached seedlings (Susek et al., 1993; Mochizuki et al., 2001; Larkin et al., 2003; Koussevitzky et al., 2007; Woodson et al., 2011). Of the original five gun mutants identified, the gun2, gun3, gun4 and gun5 mutations are all in genes involved in tetrapyrrole synthesis (Mochizuki et al., 2001; Larkin et al., 2003), with the gun5 mutation residing in the H subunit of magnesium (Mg) ‐chelatase (CHLH) and resulting in reduced synthesis of Mg‐porphyrins (Mochizuki et al., 2001). GUN1 is a chloroplast‐localized pentatricopeptide‐repeat protein that is predicted to have nucleotide‐binding activity (Koussevitzky et al., 2007) and, in contrast to gun2‐gun5, gun1 can also rescue nuclear gene expression under other conditions affecting chloroplast development, such as treatment with lincomycin, an inhibitor of plastid translation (Gray et al., 2003; Koussevitzky et al., 2007). Initial analysis of the tetrapyrrole‐related gun mutants led to the hypothesis that the tetrapyrrole Mg‐protoporphyrin IX, a chlorophyll biosynthesis intermediate, is a mobile retrograde signal (Strand et al., 2003). This was not supported by further biochemical and genetic studies (Mochizuki et al., 2008; Moulin et al., 2008) and instead a new model has been put forward in which a ferrochelatase1 (FC1)‐dependent, haem‐related signal acts positively to promote expression of nuclear photosynthesis genes (Woodson et al., 2011). However, a role for Mg‐protoporphyrin as an inhibitory plastid signal continues to be proposed (e.g. Kindgren et al., 2012).
A strong inhibition of nuclear gene expression is also observed following a pretreatment of Arabidopsis seedlings with far‐red light (FR) before transfer to white light (WL) (McCormac & Terry, 2002, 2004). Under FR, the phytochrome A photoreceptor (phyA) induces expression of nuclear‐encoded chloroplast proteins, but as FR cannot be utilized by the light‐dependent chlorophyll synthesis enzyme protochlorophyllide oxidoreductase (POR), chloroplast development is stalled (Barnes et al., 1996). Instead, accumulation of Pchlide and depletion of the POR proteins, which bind and buffer photosensitive Pchlide, result in severe photo‐oxidative damage to chloroplasts (Sperling et al., 1997; McCormac & Terry, 2004) and inhibition of nuclear gene expression (McCormac & Terry, 2002, 2004). Here, we tested whether NF and FR pretreatments target the same retrograde signal by measuring their impact on global gene expression. Our analysis shows that not only are the response profiles different, but the FR pretreatment identifies a previously undescribed pathway in which 1O2 mediates the inhibition of photosynthesis‐related nuclear genes. This novel inhibitory retrograde signalling pathway would provide a counterbalance to a positive haem‐related signal driving chloroplast biogenesis during seedling development.
Materials and Methods
Plant material and accessions
The WT Arabidopsis (Arabidopsis thaliana L.) line used in this study was Columbia (Col‐0). The single mutants gun1 and gun5 and the gun1gun5 double mutant have been previously described (Vinti et al., 2000; Mochizuki et al., 2001), as has the phyA mutant (in the Col‐0 background) (see McCormac & Terry, 2002). The ex1, ex2 and ex1ex2 double mutants have been previously described (Wagner et al., 2004; Lee et al., 2007). Standard growth conditions (including growth medium and light sources) were as described previously (McCormac & Terry, 2002). Arabidopsis Genome Initiative accessions for genes mentioned in this study are given in Supporting Information Table S1.
RNA extraction
Total RNA extraction was carried out as previously described (McCormac et al., 2001), but with the addition of a further purification step using the Qiagen RNeasy kit according to the manufacturer's instructions. Total RNA samples for reverse transcription−polymerase chain reaction (RT‐PCR) analysis were treated using the method described by Manning (1991) for the removal of polysaccharides. Polysaccharides were precipitated using 0.1 volumes of 1 M sodium acetate (NaOAc), pH 4.5, and 0.4 volumes of ethylene glycol monobutyl ether (2‐BE). The sample was incubated on ice for 30 min and centrifuged at 20 000 g for 10 min, and RNA precipitation from the supernatant was achieved by adding a further 0.6 volumes (with respect to the original RNA sample) 2‐BE, incubation for 30 min on ice and centrifugation at 20 000 g for 10 min. The pellet was washed consecutively with 40 mM NaOAc (pH 4.5) : 2‐BE (1 : 1), 70% ethanol (v/v) and 100% ethanol and air dried.
Microarray analysis
For the FR pretreatment experiment, WT and phyA seedlings were grown on 1 × Murashige and Skoog (MS) salts without sucrose for 1 d in dark (D) followed by 2 d of continuous FR (or maintained for 2 d in D) before transfer to continuous WL for 1 d. Under these conditions, WT seedlings retain RNA and membrane integrity (McCormac & Terry, 2004). However, to ensure a full block‐of‐greening response in seedlings depleted of Pchlide, gun1gun5 seedlings were grown for 3 d in FR before growth for 1 d in WL, with gun1gun5 control seedlings receiving 3 d of D before 1 d of WL. For NF treatment, WT and gun1gun5 seedlings were grown for 3 d in D followed by 3 d in WL on a medium containing 1 × MS salts and 1.5% (w/v) sucrose with or without 5 μM NF. RNA samples for each treatment were extracted from two fully independent experiments that were analysed separately (with the exception of phyA samples, which had one replicate). Microarrays were produced by the GARNet facility (University of Nottingham, Nottingham, UK) using 22K Affymetrix (Santa Clara, CA, USA) ATH1 Arabidopsis chips. Full microarray data sets are deposited in the National Center for Biotechnology Information (NCBI) GEO database (http://www.ncbi.nlm.nih.gov/gds; FR = GSE6169; NF = GSE5726). Analysis of the normalized data was conducted using Microsoft Excel and normalized signal data were filtered for a positive ‘transcript‐present’ score in both replicates of the WT control or treatment samples. Genes inhibited following an FR pretreatment were identified according to a consistent (i.e. both replicates) signal fold‐ratio of FR : D ≤ 0.5. Rescue of gene expression after an FR pretreatment in gun1gun5 and phyA mutants compared with WT was calculated as the mutant treatment : control ratio divided by the WT treatment : control ratio, using a cut‐off of 1.5‐fold. Genes inhibited by NF were identified according to a consistent signal fold‐ratio of NF : control ≤ 0.5. A criterion of ≥ 1.5‐fold increase in NF‐treated gun1gun5 seedlings compared with WT NF‐treated samples was used to identify gun1gun5 rescued genes. For both NF and FR arrays, induced genes were identified according to a signal fold‐ratio of treatment : control ≥ 2.0 in both replicates. All further analysis of the microarray data, including comparisons with other microarray data sets, was performed in Microsoft Excel. Heat maps were generated using Multiexperiment Viewer (mev v.4.8.1; Saeed et al., 2003).
Real‐time RT‐PCR
For direct comparison with microarray data using real‐time RT‐PCR, WT and gun1gun5 seedlings were grown in the presence of NF or received an FR pretreatment, along with the respective controls, under the same conditions as described in the previous section for microarray analysis. In addition, the NF experiment was also carried out in the absence of sucrose. cDNA synthesis and real‐time PCR were carried out as described by McCormac & Terry (2004) and primer pairs are given in Table S1. Transcript abundance was calculated relative to 18S rRNA within each sample. The real‐time RT‐PCR data for each treatment are expressed relative to the respective WT control samples and the signal values for the corresponding array data were normalized accordingly. For time‐course analyses, WT, gun1 and gun5 seedlings were grown for 1 d in D followed by 2 d in FR or kept for 3 d in D (± 5 μM NF) on medium without sucrose. All seedlings were transferred to WL at t = 0 and total RNA samples extracted at the times indicated. Transcript abundance was calculated relative to 18S rRNA within each sample. For comparison of WT and the ex mutants, seedlings were grown for 1 d or 2 d in D followed by either 2 d in FR or 2 d in D (controls), and then transferred to WL for 24 h. Transcript abundance was calculated using real‐time RT‐PCR relative to ACTIN DEPOLYMERIZING FACTOR 2 (ADF2) within each sample and confirmed using a second reference gene, YELLOW LEAF SPECIFIC GENE 8 (YLS8). As shown in Fig. S1, all three reference genes give an equivalent response for the protocols used in this study.
DanePy fluorescence quenching
Seedlings were grown with or without a 2‐d FR pretreatment or in D for 2 d with or without 5 μM NF and infiltrated with 50 mM KPO4 (pH 7.2) (1% v/v ethanol) containing 200 μM DanePy (a gift from Kalman Hideg, University of Pécs, Hungary) using a plastic syringe as described by Hideg et al. (2002). Twenty seedlings per treatment were infiltrated with 2 ml of solution and incubated under WL for 5 h. Fluorescence spectra of samples (excitation 330 nm) were measured using an F‐2000 spectrophotometer (Hitachi, Tokyo, Japan) and values were recorded for the emission maxima (532 nm); seedlings were removed from the solution before measurement.
Imaging Singlet Oxygen Sensor Green fluorescence
Seedlings were grown for 2 d in the dark, followed by a 3‐d FR pretreatment. At 150 min before the end of the third day in FR, seedlings were immersed in a solution of 10 μM Singlet Oxygen Sensor Green (SOSG; ThermoFisher, Waltham, MA, USA) for 2 h in FR, then gently blotted dry and returned to their growth environment for 30 min. Seedlings were transferred to WL and excised cotyledons were imaged with fluorescence microscopy using a Zeiss Axioplan2 microscope (excitation = 470/40 nm; dichroic = 495 nm (LP); emission = 525/50 nm) with an integration time of 100 ms. Control seedlings either remained in the dark for 5 d or were imaged after FR without SOSG treatment to account for background fluorescence from the plant tissue. For the first time‐point (0 h, before transfer to WL), slides were prepared in the dark under a dim green safelight and maintained in the dark before imaging by wrapping in foil. All images were acquired using the same objective lens (×10), and intensity histograms were kept constant for all images shown. The SOSG signal for each sample was determined in Imagej (NIH, Bethesda, MD, USA) by assessing the signal averaged over the area of one cotyledon. Each data point represents the mean SOSG signal of three cotyledons from seedlings assayed in independent biological replicates. The same microscope settings were used to acquire all images.
Pigment analysis
Chlorophyll and Pchlide was assayed for 20 seedlings as described in Stephenson & Terry (2008) and Stephenson et al. (2009), respectively.
Results
NF and FR pretreatments target different retrograde signals
To test whether the retrograde signal after an FR pretreatment was the same as that after an NF treatment, we compared gene expression profiles using the 22K Affymetrix ATH1 Arabidopsis microarray in WT (Col‐0) and in gun1gun5 double mutant seedlings in which GUN signalling is blocked (Fig. 1a). For both data sets there was strong correlation between replicates that was confirmed in correlation plots of all microarray data (Fig. S2). In WT seedlings treated with NF, there was a two‐fold down‐regulation of 761 genes (Fig. 1b; Table S2), which represents c. 3% of the genes present on the array. Comparison with other data sets for NF treatment showed a large overlap, with 228 of the 704 NF down‐regulated genes identified by Aluru et al. (2009) and 491 of the 1140 genes identified by Koussevitzky et al. (2007) represented in this gene cohort, even though these studies were performed in more mature plants and under different experimental conditions. When WT seedlings were grown for 2 d in FR before transfer to WL for 24 h, 442 genes were identified as two‐fold inhibited (Fig. 1b; Table S3) and, as expected, this response showed an almost complete rescue in the phyA mutant (Table S3). In total, 1140 different genes showed a two‐fold inhibition of expression in response to either NF or FR pretreatment, but just 63 (6%) were common to both (Fig. 1b; Table S4). This strongly suggests that the retrograde signalling pathways initiated by the two treatments are essentially distinct. Nevertheless, inhibition of both gene cohorts was mitigated in the gun1gun5 mutant, with 154 (35%) rescued in gun1gun5 (defined as a 1.5‐fold increase in expression compared with WT) following an FR pretreatment, and 326 (43%) genes rescued after NF treatment (Fig. 1a,b; Tables S2, S3, S5). Evidence that the two treatments target different retrograde signals is also provided by the finding of differences in the predicted intracellular targeting of the proteins encoded by the retrograde‐regulated gene sets (Table S6), and the limited overlap of the gene groups induced by NF and FR pretreatment (Fig. 1a,b; Tables S7, S8, S9). FR pretreatment resulted in a two‐fold induction of 263 genes compared with D‐treated controls of which 192 (73%) were rescued (no induction after FR pretreatment) by the gun1gun5 mutations (Fig. 1a,b; Table S7). The induction of gene expression was also blocked by the phyA mutation (Table S7). NF treatment resulted in a two‐fold induction of 367 genes and just 43 of these were rescued in the gun1gun5 mutant (Fig. 1a,b; Table S8). Again, the overlap between the two WT gene sets was low, with only 37 (6%) genes induced by both treatments (Fig. 1b; Table S9), further supporting the conclusion that these retrograde responses are distinct.
Figure 1.
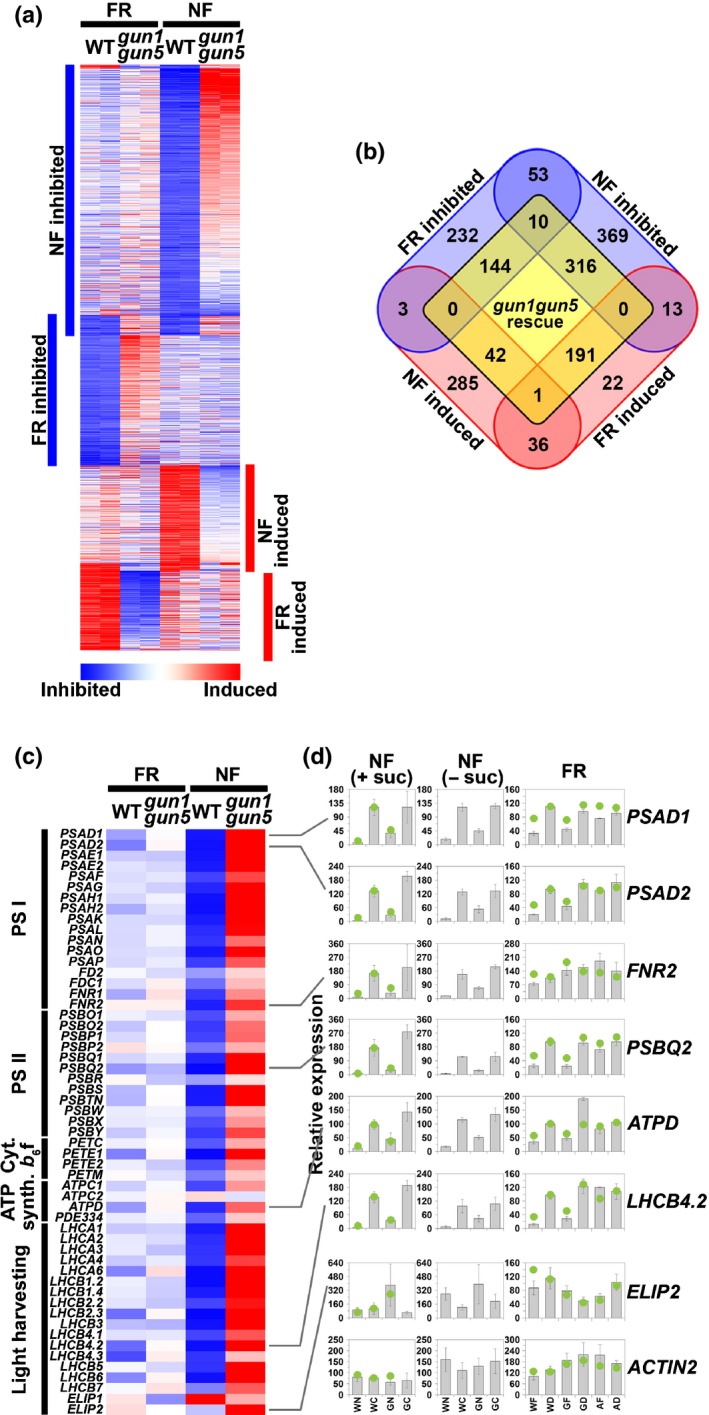
Analysis of the transcriptional response under white light of wild‐type (WT; Columbia (Col‐0)) and gun1gun5 Arabidopsis thaliana seedlings treated with either 5 μM norflurazon (NF) or a far‐red light (FR) pretreatment. (a) Heatmap depicting a gene cluster analysis of microarray data for nuclear genes inhibited (blue) or induced (red) at least two‐fold by either NF or FR pretreatment in WT in two independent biological replicates. For the FR gun1gun5 column, expression is shown as the difference between treatment and controls in gun1gun5 relative to the difference in WT. For the NF gun1gun5 column, expression is shown as the difference between NF‐treated WT and gun1gun5. (b) Venn diagram demonstrating the number of genes inhibited (blue) or induced (red) at least two‐fold by NF or an FR pretreatment, and the number of genes rescued 1.5‐fold in gun1gun5 (yellow). (c) Heat map depicting microarray analysis of inhibited (blue) or induced (red) photosynthesis‐related genes grouped by photosynthetic complex. Ratios are the mean of two independent experiments. Columns are represented as in (b). (d) Real‐time reverse transcription−polymerase chain reaction (RT‐PCR) analysis of representative photosynthesis‐related genes. For the NF experiments, WT and gun1gun5 seedlings were grown in the presence (WN and GN) or absence (WC and GC) of 5 μM NF with and without 1.5% (w/v) sucrose. For the FR pretreatment experiments, WT, gun1gun5 or phytochrome A (phyA) mutant seedlings were grown with a pretreatment of FR (WF, GF and AF, respectively) or kept in darkness (WD, GD and AD, respectively). Data shown are the mean ± SE (n = 3 (NF) or n = 7 (FR) independent experiments) with array data represented by green dots, normalized to the WD (FR) or WC (NF) real‐time RT‐PCR values. gun, genomes uncoupled;PSA, photosystem I (PSI) subunit;FD, FERREDOXIN;FNR, ferredoxin:NADP(H) oxidoreductase; PSB,PSII subunit; PET, cytochrome b 6 f subunit;ATPD,ATP synthase subunit;PDE, PIGMENT DEFECTIVE;LHC, LIGHT HARVESTING CHLOROPHYLL A/B BINDING PROTEIN; ELIP, EARLY LIGHT‐INDUCIBLE PROTEIN.
The different impacts of NF and an FR pretreatment on nuclear gene expression were most clearly apparent when examining genes encoding components of the photosynthetic light reactions (Fig. 1c,d). Of the 55 nuclear‐encoded, photosynthesis‐related genes on the microarray, 47 were more than two‐fold inhibited in NF‐treated seedlings (of which 42 showed a > 1.5‐fold rescue by gun1gun5), but only nine were inhibited > 1.5‐fold following an FR pretreatment (Fig. 1c). However, this set of responsive genes encoded at least one member of each of the major photosynthetic complexes (i.e. photosystem I (PSI), PSII, cytochrome b 6 f and ATP synthase), as well as a representative of each of the LHCA and LHCB gene families (Fig. 1c). This relationship was confirmed by real‐time PCR analysis of seven photosynthesis‐related genes (Fig. 1d). As the NF experiment was carried out in the presence of sucrose to follow standard protocols, the real‐time PCR experiments were also performed in the absence of sucrose, as sucrose has been shown to be an important regulator of photosynthetic gene expression (Hanson & Smeekens, 2009). The data in Fig. 1(d) demonstrate that the real‐time PCR analysis was consistent with the respective expression profiles determined from the array. In addition, the presence or absence of sucrose was not found to significantly influence the qualitative response to NF.
Tetrapyrroles have been strongly implicated as signalling molecules in plastid‐to‐nucleus signalling (Strand et al., 2003; Woodson et al., 2011; Terry & Smith, 2013), and we also examined the impact of an FR pretreatment on the expression of tetrapyrrole synthesis genes in the microarray data set and by real‐time PCR (Fig. 2). We previously demonstrated that NF treatment resulted in a severe and global knockdown in the expression of chlorophyll synthesis genes (Moulin et al., 2008). In contrast to the situation on NF, FR pretreatment had a selective effect on tetrapyrrole synthesis, with only a few genes showing an inhibitory response (Fig. 2a). These included HEMA1, encoding glutamyl‐tRNA reductase, CHLH, GUN4 and CHLOROPHYLL A OXYGENASE (CAO), which correspond to a small cohort of key regulatory genes in the pathway (Matsumoto et al., 2004; Stephenson & Terry, 2008), and FC2, which has also previously been shown to be regulated by light and NF (Singh et al., 2002; Moulin et al., 2008). To confirm these results, we undertook real‐time PCR on 12 genes of the tetrapyrrole biosynthesis pathway (Fig. 2b). In general, expression was in close agreement with the microarray data and, in particular, the down‐regulation of HEMA1, CHLH, GUN4, CAO and FC2 after an FR pretreatment was confirmed. In addition, the FR pretreatment also induced the expression of some tetrapyrrole biosynthesis genes (Fig. 2a) including GLUTAMYL tRNA SYNTHETASE, encoding glutamyl‐tRNA synthetase, HEMA2, PROTOPORPHYRINOGEN OXIDASE 2 (PPO2) and FC1, all of which are associated with nonphotosynthetic haem synthesis. These genes were also induced by NF (Moulin et al., 2008), suggesting that haem synthesis for hemoproteins required in response to oxidative stress is protected following both treatments.
Figure 2.
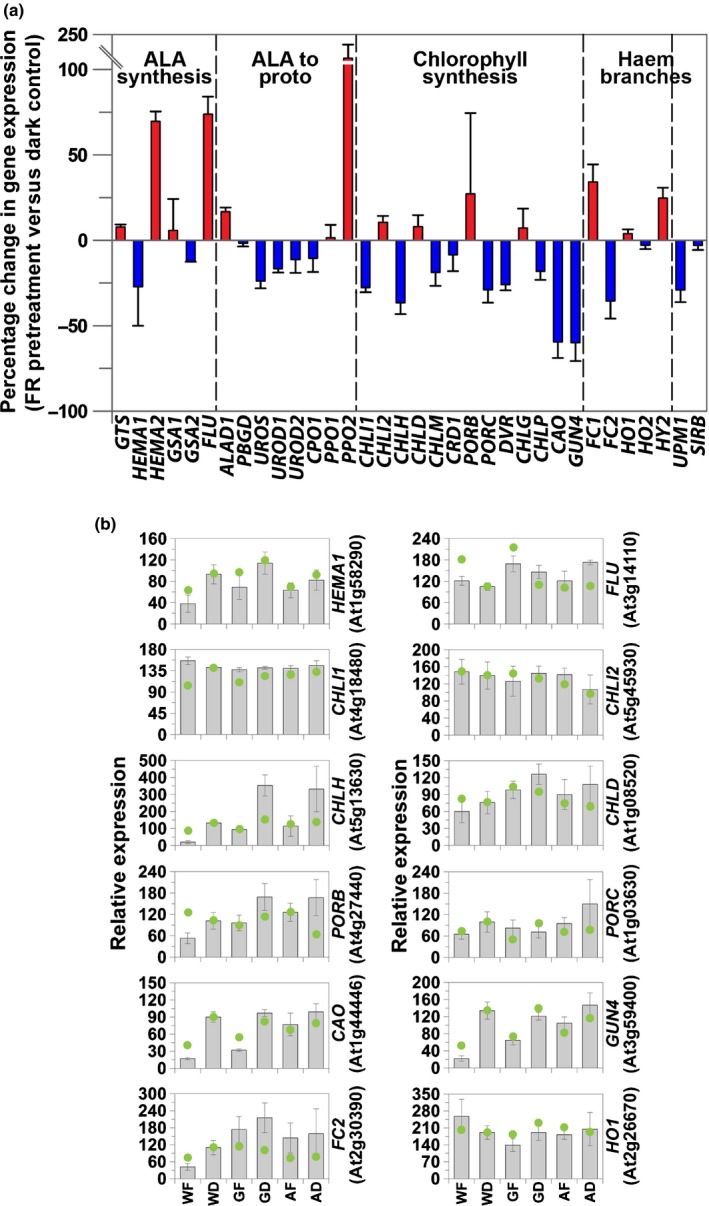
The effect of a far‐red light (FR) pretreatment on expression of Arabidopsis thaliana genes encoding enzymes involved in the tetrapyrrole pathway. (a) Microarray analysis of tetrapyrrole pathway genes in wild‐type (WT) seedlings in white light after an FR pretreatment compared with a dark (D) pretreated control, with induced genes represented by red bars and inhibited genes by blue bars. Data shown are the mean and range of two independent experiments. (b) Real‐time reverse transcription−polymerase chain reaction (RT‐PCR) analysis of tetrapyrrole synthesis genes. WT, gun1gun5 or phytochrome A (phyA) seedlings were grown with a pretreatment of FR (WF, GF and AF, respectively) or D (WD, GD and AD, respectively). Data shown are the mean ± SE (n = 7 independent experiments) and array data are represented by green dots, normalized to the WD real‐time RT‐PCR values. ALA, aminolevulinic acid; gun, genomes uncoupled;GTS, GLUTAMYL TRNA SYNTHETASE;HEMA, glutamyl tRNA reductase;GSA, GLUTAMATE‐1‐SEMIALDEHYDE 2,1‐AMINOMUTASE;FLU, FLUORESCENT IN BLUE LIGHT;ALAD,ALA DEHYDRATASE;PBGD, PORPHOBILINOGEN DEAMINASE;UROS, UROPORPHYRINOGEN III SYNTHASE;UROD, UROPORPHYRINOGEN III DECARBOXYLASE;CPO, COPROPORPHYRINOGEN III OXIDASE;PPO, PROTOPORPHYRINOGEN IX OXIDASE;CHLI, magnesium chelatase subunit I;CHLH, magnesium chelatase subunit H;CHLD, magnesium chelatase subunit D;CHLM, S‐adenosyl‐L‐methionine:magnesium protoporphyrinogen IX methyltransferase;CRD, COPPER RESPONSE DEFECT;POR, PROTOCHLOROPHYLLIDE OXIDOREDUCTASE;DVR, DIVINYL REDUCTASE;CHLG, chlorophyll synthase;CHLP, geranylgeranyl pyrophosphate reductase;CAO, CHLOROPHYLL A OXYGENASE;FC, FERROCHELATASE;HO, HAEM OXYGENASE;HY, ELONGATED HYPOCOTYL;UPM, UROPORPHYRINOGEN III METHYLTRANSFERASE;SIR, SIROHYDROCHLORIN FERROCHLEATASE.
1O2 is implicated as the retrograde signal after an FR pretreatment
An FR pretreatment has been shown to lead to an increase in Pchlide (Sperling et al., 1997; McCormac & Terry, 2002) and we hypothesized that the signal leading to the retrograde regulation described in this study might be similar to the signal resulting in the induction of 1O2‐responsive genes in the flu mutant of Arabidopsis, which also accumulates high concentrations of Pchlide (Meskauskiene et al., 2001; op den Camp et al., 2003). As shown in Fig. 3(a), an FR pretreatment did indeed result in the induction of known 1O2‐responsive genes (op den Camp et al., 2003; Danon et al., 2005; Lee et al., 2007; Kim & Apel, 2013). We also compared our array data after an FR pretreatment to gene expression profiles for the two other well‐characterized 1O2 signalling systems: the flu mutant (op den Camp et al., 2003) and the chlorophyll b‐less chlorina1 mutant (Ramel et al., 2013). In both these cases, 1O2‐regulated transcriptomes were determined using plants at the rosette stage and using different time‐points. Nevertheless, there was good overlap of our data with both experimental systems. For example, out of the 70 genes induced specifically by 1O2 (op den Camp et al., 2003), 40 were also induced to some degree in both replicates of the FR pretreatment array. Also, of the 263 genes up‐regulated after an FR pretreatment, 157 were more than two‐fold induced in flu after 2 h, with 130 out of 442 down‐regulated genes also down‐regulated two‐fold in flu (op den Camp et al., 2003). Similarly, 47 of the 263 genes induced by an FR pretreatment were also induced in chlorina1, with 80 of the 442 inhibited genes also down‐regulated in chlorina1 (Ramel et al., 2013).
Figure 3.
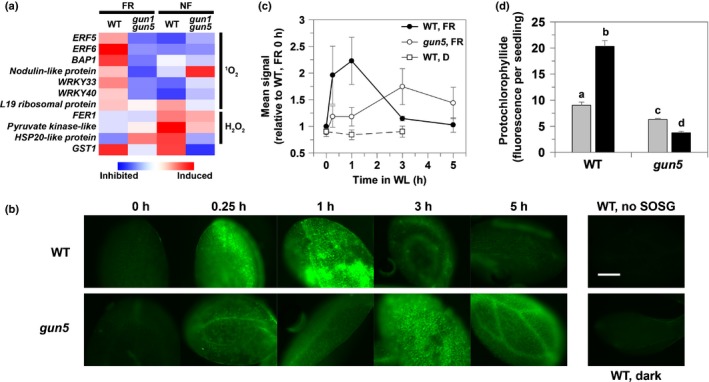
Evidence for the involvement of singlet oxygen (1O2) in the response to a far‐red light (FR) pretreatment. (a) Heat map depicting microarray analysis of reactive oxygen species marker genes responsive to 1O2 or hydrogen peroxide (H2O2) in wild‐type (WT) Arabidopsis thaliana seedlings after norflurazon (NF) or an FR pretreatment. Ratios are the mean of two independent experiments. GLUTATHIONE S‐TRANSFERASE 1 (GST1) is an example of a general stress‐responsive gene. (b) Singlet Oxygen Sensor Green (SOSG) staining to detect singlet oxygen generation on transfer of FR‐treated seedlings to white light (WL). Time‐points represent the number of hours after transfer to WL. Controls were FR‐pretreated WT seedlings after 1 h in WL without SOSG (‘WT, no SOSG’), and WT seedlings that did not receive the FR pretreatment, but were maintained in the dark for 3 d (‘WT, dark’). Images shown are representative of three independent biological replicates of WT and genomes uncoupled 5 (gun5) seedlings, and were taken using the same microscope settings and were produced from the same look‐up table; bar, 200 μm. (c) Image intensity analysis for three independent biological replicates of the SOSG staining experiment outlined in (b). Data shown are mean ± SE. (d) Protochlorophyllide content of WT and gun5 seedlings grown in the dark (grey bars) or after an FR pretreatment (black bars) corresponding to the 0 h time‐point in (b). Data shown are the mean + SE (n = 3 independent experiments), with different letters denoting significant differences between group means (P < 0.05; Student's t‐test). ERF, ETHYLENE RESPONSE FACTOR; BAP, BON ASSOCIATION PROTEIN; WRKY, WRKY DNA‐BINDING PROTEIN;FER, FERRETIN;HSP, HEAT SHOCK PROTEIN;GST, GLUTATHIONE S‐TRANSFERASE.
To confirm whether 1O2 was produced in WL after transfer from FR, we measured 1O2 production using Singlet Oxygen Sensor Green (SOSG; Flors et al., 2006). Fluorescence was rapidly and strongly induced in WT seedlings after transfer to WL from FR, with a fluorescence signal clearly detectable after 15 min and a maximum signal by 1 h (Fig. 3b,c). No induction of 1O2 was observed in the first 1 h after transfer from dark to WL (Fig. 3b,c). By contrast, the gun5 mutant, which contains severely reduced Pchlide after an FR treatment (Fig. 3d), showed a much attenuated response with a shallower peak of fluorescence that was also seen far later than in WT seedlings (Fig. 3b,c). The reason for some 1O2 production in the gun5 mutant when Pchlide concentrations were low is not clear. One possibility is that the gun5 mutation, which leads to a decrease in Mg‐chelatase activity, results in an accumulation of the Mg‐chelatase substrate, protoporphyrin IX, which is also a photosensitizer. This could result in some 1O2 production under longer WL periods as the flux through the tetrapyrrole pathway increases. Consistent with our SOSG results, the dansyl‐based ROS sensor, DanePy, which is specifically quenched by 1O2 (Hideg et al., 2002), also showed fluorescence quenching after an FR pretreatment in WT seedlings, but not in a phyA mutant (Fig. S3). NF treatment might also be expected to produce 1O2 as a result of photo‐excitation of chlorophyll in the absence of carotenoids, as has been observed for light‐grown seedlings treated with NF (Kim & Apel, 2013). However, seedlings treated with NF from germination do not show a 1O2 response (Kim & Apel, 2013) and no evidence for 1O2 production was observed here (Figs 3a, S3). By contrast, FR pretreatment did not induce H2O2‐specific transcripts (op den Camp et al., 2003), and these were instead elevated after NF treatment (Fig. 3a).
To investigate how rapidly changes in nuclear gene expression could be observed after an FR pretreatment, we conducted a time course expression profile over 3 h for seven ROS‐responsive genes (Fig. 4a) and six photosynthesis‐related genes (Fig. 4b). The microarray data were obtained with the gun1gun5 double mutant and therefore to break this response down further we conducted this experiment using the monogenic gun1 and gun5 mutants. Upon transfer to WL, the FR‐pretreated WT seedlings displayed a strong and rapid up‐regulation of two 1O2‐responsive genes, BON ASSOCIATION PROTEIN 1 (BAP1) and nodulin‐like protein (Fig. 4a). This induction was abolished in the gun5 mutant, but was more rapid in the gun1 mutant than in WT (Fig. 4b), consistent with the more severe effect of an FR pretreatment on the gun1 mutant (McCormac & Terry, 2004). Although some 1O2 production was observed in gun5 (Fig. 3b,c), it was only apparent after 3 h, which may have been too late to induce gene expression in this assay. Control (3 d in the dark before transfer to WL without NF) and NF‐treated seedlings of all lines showed little induction of ROS‐responsive genes over this time course (Fig. 4a). Photosynthesis‐related genes were induced after transfer from the dark to WL. However, in parallel to the rapid induction of 1O2‐induced genes, expression of photosynthesis‐related genes was strongly inhibited in FR‐pretreated seedlings, with differences observed from control samples after just 30 min in WL in some cases (Figs 4b, S4). Furthermore, the most sensitive transcripts, GUN4 and PHOTOSYSTEM II SUBUNIT Q2 (PSBQ2), were depleted within 0.5 h in WL to below the levels seen at the time of initial transfer from FR. The rapid response in gene expression was consistent with the induction of 1O2 observed within 15 min in the SOSG assay (Fig. 3b,c). Again, as seen for induction of 1O2‐responsive genes, gun1 mutant seedlings showed an exacerbated inhibitory response to an FR pretreatment over the first 30 min, although gun1 seedlings had higher levels of expression on transfer to WL (as noted previously; McCormac & Terry, 2004), while the gun5 mutant completely rescued the early WL response of GUN4, GLUTAMATE‐1‐SEMIALDEHYDE 2,1‐AMINOMUTASE 2 (GSA2) and HEMA1 from inhibition by an FR pretreatment and partially rescued all other genes (Figs 4b, S4). Again, this rescue was consistent with the attenuated production of 1O2 as shown by SOSG (Fig. 3c). We previously showed the effect of the gun1gun5 mutations on gene expression after an FR pretreatment and 24 h in WL (Figs 1, 2). To enable a direct comparison with the gun5 single mutant, we also analysed expression in gun5 at this time‐point (Fig. S5). Under these conditions, the gun5 mutant was able to rescue expression to a similar degree to the gun1gun5 double mutant.
Figure 4.
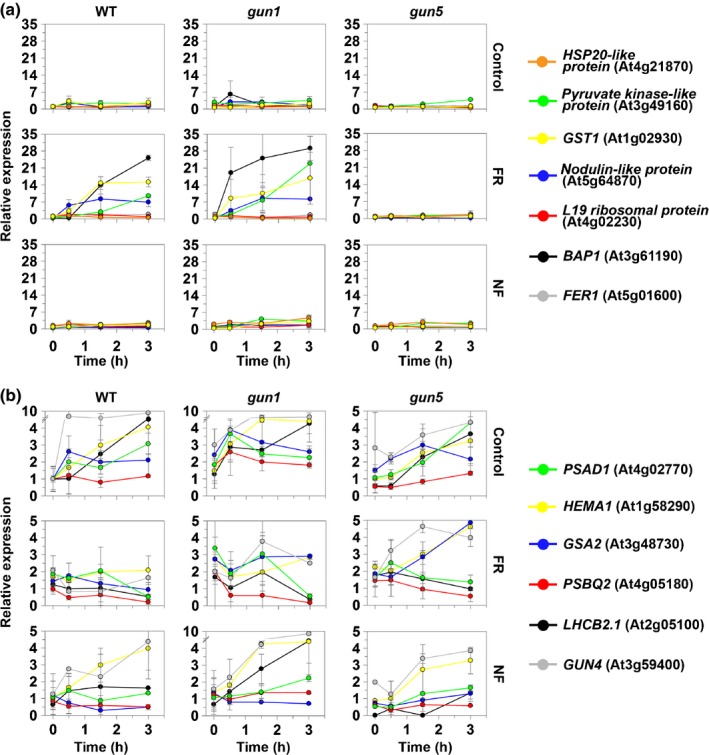
Time‐course of changes in nuclear gene expression in response to norflurazon (NF) and a far‐red light (FR) pretreatment measured by real‐time reverse transcription−polymerase chain reaction (RT‐PCR). Wild‐type (WT), genomes uncoupled 1 (gun1) and gun5 Arabidopsis thaliana seedlings were grown for 3 d in the dark (D) in the presence (NF) or absence (control) of NF or grown for 1 d in D followed by a 2‐d FR pretreatment (FR). All samples were then transferred to white light for 3 h. (a) Reactive oxygen species marker genes. (b) Photosynthesis‐/tetrapyrrole‐associated genes. Data shown are the mean ± SE (n = 3 independent experiments), normalized to the WT control (t = 0) value for each transcript series. HSP, HEAT SHOCK PROTEIN;GST, GLUTATHIONE S‐TRANSFERASE;BAP, BON ASSOCIATION PROTEIN;FER, FERRETIN;PSA, photosystem I (PSI) subunit; HEMA, glutamyl tRNA reductase; GSA, GLUTAMATE‐1‐SEMIALDEHYDE 2,1 AMINOMUTASE;PSB,PSII SUBUNIT;LHCB, LIGHT HARVESTING CHLOROPHYLL A/B BINDING PROTEIN.
Changes in photosynthetic gene expression after NF treatment were less pronounced than after an FR pretreatment and showed partial rescue in the gun1 mutant, but not in gun5, over this 3‐h period (Fig. 4b). The two retrograde signals can therefore be further distinguished by the relative impact of the gun1 and gun5 mutations on the responses.
Retrograde signalling after an FR pretreatment is partially dependent on EXECUTER proteins
To examine further the hypothesis that retrograde signalling after an FR pretreatment is dependent on 1O2, we examined the effect of the 1O2 signalling mutants ex1 (Wagner et al., 2004) and ex2 (Lee et al., 2007) on photosynthetic gene expression. Using our standard conditions of 1 d in the dark before the 2‐d FR treatment, the ex2 mutant showed a partial rescue of greening and this was substantially increased in the ex1ex2 double mutant (Fig. 5a,b). Rescue was not a result of a reduction in Pchlide concentrations (Fig. S6). Under these conditions, expression of HEMA1, GUN4 and LHCB2.1 was significantly higher in the ex2 single mutant and the ex1ex2 double mutant, with expression restored to c. 30–50% in the latter (Fig. 5d). Interestingly, when the dark period was extended to 2 d, we still saw a strong rescue of nuclear gene expression in the ex1ex2 double mutant, but in this case partial rescue was observed in ex1 and not ex2 (Figs 5c,e, S6). A greater role for EX1 compared with EX2 was previously observed for the rescue of gene expression in the flu mutant (Lee et al., 2007). Analysis of expression data for EX1 and EX2 following germination showed that EX2 is initially elevated compared with EX1, with EX1 expression induced later in development (Fig. 5f). This is consistent with the observed earlier role for EX2.
Figure 5.
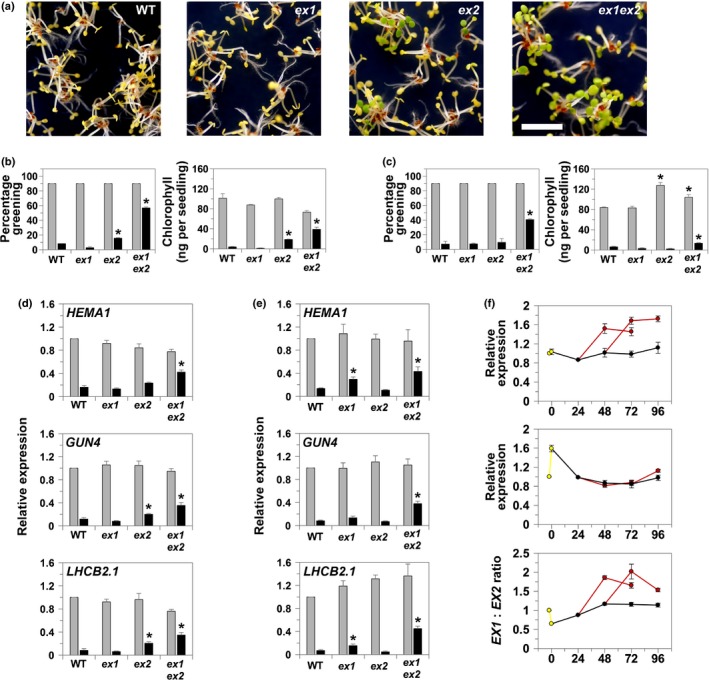
The white light (WL) response of executer (ex) mutants to a far‐red light (FR) pretreatment. (a) Representative WL phenotype of ex1, ex2 and ex1ex2 Arabidopsis thaliana mutant seedlings after an FR pretreatment following 1 d in the dark (D); bar, 5 mm. (b, c) Percentage greening and total chlorophyll content of ex1, ex2 and ex1ex2 seedlings after an FR (black bars) or dark control (grey bars) pretreatment following an initial incubation in D for (b) 1 d or (c) 2 d. (d, e) Real‐time reverse transcription−polymerase chain reaction (RT‐PCR) analysis of HEMA1 encoding glutamyl tRNA reductase, GENOMES UNCOUPLED 4 (GUN4) and LIGHT HARVESTING CHLOROPHYLL A/B BINDING PROTEIN 2.1 (LHCB2.1) expression in ex1, ex2 and ex1ex2 seedlings in WL after an FR (black bars) or D control (grey bars) pretreatment following an initial incubation in D for (d) 1 d or (e) 2 d. For (b–e), data shown are mean + SE (n = 4 independent experiments). (f) Quantitative RT‐PCR analysis of EX1 and EX2 expression throughout the two different growth regimes used to assess the response of ex mutants to an FR pretreatment, with the EX1:EX2 expression ratio also given. Data shown are the mean ± SE (n = 3 independent experiments). Line colours correspond to light conditions (yellow, WL; black, D; red, FR). Asterisks denote a significant increase compared to WT (P < 0.05; Student's t‐test).
Discussion
NF and FR pretreatments affect different chloroplast‐to‐nucleus signals
The primary biogenic retrograde signal affected by NF treatment is proposed to be a positive signal that is dependent on FC1 enzyme activity in the chloroplast (Woodson et al., 2011). Our data suggest that the signal generated by an FR pretreatment is different from the NF signal based on the distinctiveness of the overall gene expression profiles affected by the two treatments, and the relative impact of the gun1 and gun5 mutations on the signals. We did, however, observe a small, common set of inhibited genes that showed a significant enrichment for genes encoding predicted chloroplast‐targeted proteins. This included many genes required for the synthesis of the photosynthetic complexes, as well as genes critical for chloroplast development such as GOLDEN2‐LIKE 2 (GLK2; Waters et al., 2009). Some overlap in regulation is not surprising as any informational signal affecting chloroplast biogenesis is likely to converge on a few key regulatory genes. The alternative scenario that the common gene set is responding to a unique signal generated under both conditions, with the regulation of condition‐specific genes under the control of separate, additional signals, is far less likely. In this regard, the FR pretreatment resulted in the selective inhibition of just a few chlorophyll synthesis genes, including HEMA1, GUN4, CHLH and CAO, which have previously been identified as key regulatory genes in the pathway (Matsumoto et al., 2004; Stephenson & Terry, 2008). These results are therefore consistent with the FR pretreatment initiating a targeted and specific down‐regulation of chlorophyll synthesis under these conditions rather than a general inhibition of all the pathway components as seen after NF treatment (Moulin et al., 2008). Detailed analysis of gene expression for the four complexes of the photosynthetic light reactions showed a similar pattern. NF treatment caused a strong down‐regulation of almost all photosynthetic genes, while an FR pretreatment only affected a few in each photosystem and just one for ATP synthase (ATPD encoding the δ subunit) and the cytochrome b 6 f complex (PLASTOCYANIN 1 (PETE1)). It is tempting to speculate that the genes specifically regulated by an FR pretreatment also reflect key regulatory targets for each photosynthetic complex, as seen for the tetrapyrrole pathway.
The role of 1O2 in plastid‐to‐nucleus communication
Previous work has unequivocally demonstrated that flu mutant seedlings generate a 1O2 signal on transfer to WL (op den Camp et al., 2003; Kim et al., 2012), resulting in a severe response leading to seedling death (Danon et al., 2005; Kim et al., 2012), and attention has focused on the role of 1O2 signalling in stress (Ramel et al., 2013; Zhang et al., 2014). Our experimental design, in which light‐regulated photosynthetic genes are induced during the FR treatment, has now allowed us to reveal a previously undiscovered role for 1O2 signalling as a regulatory retrograde signal during chloroplast biogenesis. By contrast, other studies on 1O2 signalling have generally not been conducted during the biogenic phase of chloroplast development. The proposed 1O2 signal works rapidly to inhibit photosynthetic gene expression within 30 min and, in response to moderate increases in chlorophyll precursors that might occur in nature (as compared with the severe conditions of a flu mutation or FR pretreatment), would produce an acclimatory response that would serve to modulate chlorophyll synthesis to achieve an optimal synthesis rate under challenging environmental conditions. Under more severe conditions, 1O2 production results in chloroplast degradation via a ubiquitin‐mediated pathway (Woodson et al., 2015) and ultimately cell death (Danon et al., 2005; Kim et al., 2012).
One study that is potentially similar to ours investigated the phytochrome regulatory mutants phytochrome interacting factor 1 (pif1) and pif3. These mutants show elevated Pchlide as the PIF1 and PIF3 proteins are required to repress chloroplast development in darkness (Huq et al., 2004; Shin et al., 2009; Stephenson et al., 2009). In fact, dark‐grown pif mutants behave in a similar way to the situation we observe after an FR pretreatment, with activation of phytochrome responses, but insufficient light for photoconversion of Pchlide. Consistent with this, pif1 and pif3 mutants also produce 1O2 on transfer to WL (Chen et al., 2013), with significant overlap of the gene sets regulated by the two treatments: 137 of the 263 genes induced by an FR pretreatment were also induced in pif1 seedlings and 115 of the 442 genes inhibited by an FR pretreatment were also inhibited in pif1. Interestingly, the rice mutant faded green leaf, which lacks PROTOCHLOROPHYLLIDE OXIDOREDUCTASE B, also accumulates 1O2 in light‐grown plants and showed a strong down‐regulation of photosynthesis‐related genes including HEMA1, CHLH, CAO and LHCB1 (Sakuraba et al., 2013).
Our data therefore support a new model in which two tetrapyrrole‐related signals regulate photosynthesis‐related nuclear genes (Fig. 6). NF treatment inhibits the synthesis of a specific FC1‐dependent haem pool that would normally promote photosynthesis‐related nuclear gene expression (Woodson et al., 2011), most probably by permitting normal light induction of these genes (Ruckle et al., 2007; Martin et al., 2016). This signal measures the general requirement for plastid proteins as a function of the number and developmental status of the plastids and has a broad effect on the expression of nuclear photosynthetic genes. However, under conditions in which tetrapyrrole synthesis is elevated and synthesis of potentially damaging chlorophyll intermediates might compromise seedling survival, there is a rapid down‐regulation of selected key regulatory genes to prevent overaccumulation of tetrapyrroles and repress chloroplast development. Such conditions might include severe shade (i.e. similar conditions to those used in this study), nutritional deficiencies such as low metal availability, or the presence of contaminants in the soil that alter tetrapyrrole flux. Our data suggest that this inhibitory signal is a 1O2‐mediated signal generated by direct excitation of free chlorophyll intermediates (Terry & Smith, 2013). Although the signal analysed in this study is primarily generated by Pchlide overaccumulation, in principle any porphyrin (or chlorin) could generate such a signal (Redmond & Gamlin, 1999), including Mg‐protoporphyrin IX, and this observation may reconcile some discrepancies in the literature (Strand et al., 2003; Zhang et al., 2011; Kindgren et al., 2012). The current study has focused on the situation during chloroplast biogenesis in which there is a large increase in flux through the tetrapyrrole pathway that brings new dangers to a developing seedling. In the future, it will be interesting to test whether such a signalling pathway could operate in mature plants. At this later developmental stage, the major source of 1O2 is the excitation of chlorophyll molecules in the light‐harvesting antenna complexes and the photosystem II (PSII) reaction centres (Hideg et al., 1998; Triantaphylidès & Havaux, 2009). Overexcitation of these complexes would result in increased 1O2 production and photoinhibition, and an inhibition of the tetrapyrrole pathway via 1O2 signalling could form part of an integrated response to this problem. It is also noteworthy that the PSII genes PSBQ2 and PSAD1 were both rapidly inhibited in this study, suggesting that regulation of photosystem components is an important function of this signalling pathway. Such a pathway would serve as part of the operational chloroplast signalling network conveying the impact of the environment on chloroplast status to the rest of the cell (Pogson et al., 2008).
Figure 6.
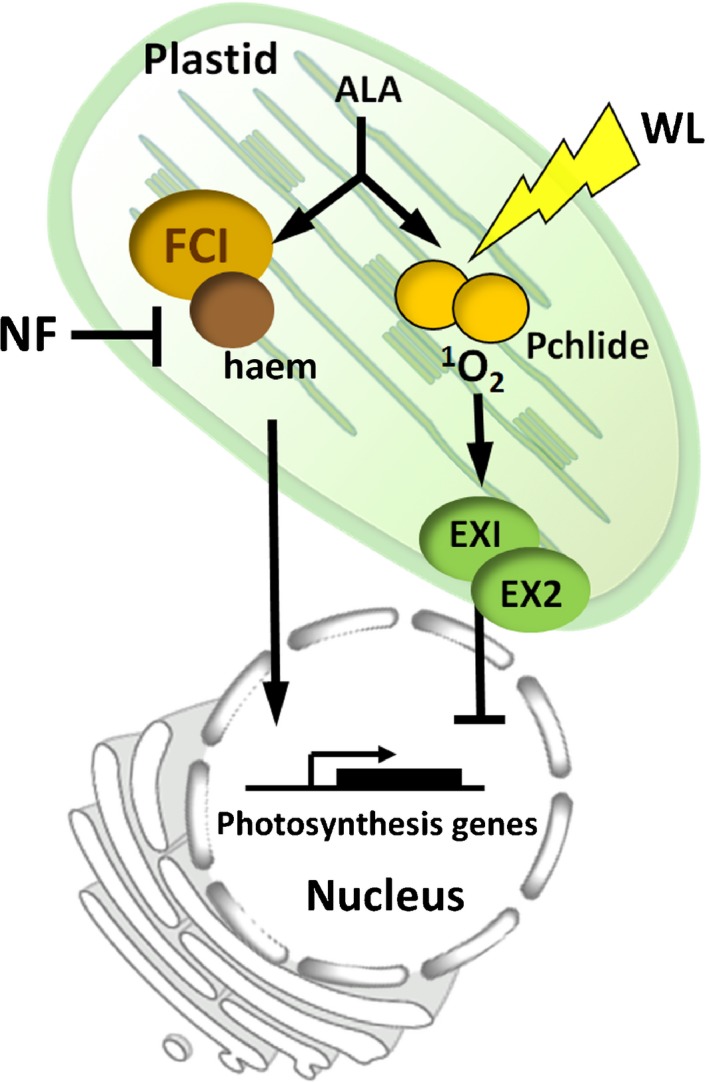
A model for tetrapyrrole regulation of nuclear gene expression. Both norflurazon (NF) and a far‐red light (FR) pretreatment result in inhibition of nuclear gene expression by activating different signalling pathways. NF treatment inhibits a ferrochelatase1‐dependent haem‐related positive signal that promotes photosynthetic gene expression (Woodson et al., 2011). After an FR pretreatment, there is an accumulation of protochlorophyllide, which generates a singlet oxygen (1O2) signal on transfer to white light (WL). This results in the activation of 1O2 marker genes and an inhibition of specific photosynthesis‐related genes in a signalling pathway that is partially dependent on EXECUTER1 (EX1) and EX2. ALA, aminolevulinic acid.
In our study, retrograde regulation of photosynthetic gene expression by 1O2 signalling was only partially mediated by EX1 and EX2, suggesting that other pathways may also be involved. One possibility is the carotenoid oxidation product β‐cyclocitral (Ramel et al., 2012), which functions independently of the EX proteins and mediates inhibition of some photosynthetic genes, such as GUN4, CAO and FC2, that are the most repressed following an FR pretreatment (Ramel et al., 2013). There are also a number of other possibilities for signalling molecules, including dihydroactinidiolide, another secondary metabolite of β‐carotene, which is EX‐independent (Shumbe et al., 2014), and products of EX‐dependent enzymatic lipid peroxidation (Przybyla et al., 2008). The zinc finger protein METHYLENE BLUE SENSITIVITY1 has also been proposed to play a role in 1O2 signalling (Shao et al., 2013). Understanding the relationship between these different 1O2 signalling pathways will be key to elucidating 1O2‐mediated retrograde signalling of photosynthetic gene expression in the future.
In summary, our data identify the primary consequence of an FR pretreatment as the production of 1O2, which leads to the inhibition of expression of nuclear‐encoded chloroplast proteins via a chloroplast‐generated signal that is distinct from that observed after NF treatment. The flu, chlorina and fc2 mutants have all proved invaluable for studying the cellular consequences of 1O2 production (Ramel et al., 2013; Zhang et al., 2014; Woodson et al., 2015) and we believe that an FR pretreatment may prove to be equally useful for investigating 1O2 responses as it allows the controlled and noninvasive induction of chloroplast‐localized 1O2 in the absence of any requirement for a specific mutant background. This should permit further dissection of the acclimatory and stress‐responsive roles attributed to 1O2 signalling in plants.
Author contributions
M.T.P. and A.C.M. designed and performed experiments, analysed data and contributed to writing the manuscript. A.G.S. analysed data and contributed to writing the manuscript. M.J.T. designed experiments, analysed data and wrote the manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Changes in GUN4 and HEMA1 expression in response to a far‐red light pretreatment and a norflurazon treatment assessed with different real‐time RT‐PCR reference genes.
Fig. S2 Correlation plots of raw expression values from the microarray data set.
Fig. S3 Detection of singlet oxygen production via quenching of DanePy fluorescence after a far‐red light pretreatment and a norflurazon treatment.
Fig. S4 Time‐course of changes in photosynthetic gene expression in response to a far‐red pretreatment measured by real‐time RT‐PCR.
Fig. S5 Rescue of nuclear gene expression in gun5 after a far‐red pretreatment.
Fig. S6 Response of ex mutants to a far‐red pretreatment.
Table S1 List of genes referred to in this paper with real‐time PCR primer sequences given (where used)
Table S2 List of 761 genes inhibited at least two‐fold in WL in WT after NF treatment
Table S3 List of 442 genes inhibited at least two‐fold in WL in WT after FR pretreatment
Table S4 List of 63 genes inhibited at least two‐fold in WT by both FR and NF treatments
Table S5 gun1gun5 rescue of genes differentially expressed in WT
Table S6 Predicted localization of protein products of differentially expressed genes identified through microarray analysis
Table S7 List of 263 genes induced at least two‐fold in WL in WT after an FR pretreatment
Table S8 List of 367 genes induced at least two‐fold in WL in WT after NF treatment
Table S9 List of 37 genes induced at least two‐fold in WT by both FR and NF treatments
Acknowledgements
We thank Kálmán Hideg (University of Pécs, Hungary) for the generous gift of DanePy, and Éva Hideg (Institute of Plant Biology, Biological Research Center, Hungary) for advice on the assay. The gun mutants were gifts from Enrique López‐Juez (Royal Holloway College, UK) and Joanne Chory (Salk Institute for Biological Studies, USA). The ex mutants were a gift from Klaus Apel (Boyce Thompson Institute for Plant Research, USA). We thank GARNet (NASC, Nottingham, UK) for the microarray service. Thanks also to Clark Lagarias (UC Davis, USA) for critical reading of an early version of this manuscript. This work was funded by BBSRC grants 51/P17214 and BB/J018139/1 to M.J.T. and BB/J018694/1 to A.G.S.
References
- Aluru MR, Zola J, Foudree A, Rodermel SR. 2009. Chloroplast photooxidation‐induced transcriptome reprogramming in Arabidopsis immutans white leaf sectors. Plant Physiology 150: 904–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes SA, Nishizawa NK, Quaggio RB, Whitelam GC, Chua N‐H. 1996. Far‐red light blocks greening of Arabidopsis seedlings via a phytochrome A‐mediated change in plastid development. Plant Cell 8: 601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbeer JW, Atkinson YE, BörnerT Hagemann R. 1979. Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid‐synthesized RNA. Nature 279: 816–817. [Google Scholar]
- op den Camp RGL, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Göbel C, Feussner I et al 2003. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ. 2016. Learning the languages of the chloroplast: retrograde signaling and beyond. Annual Review of Plant Biology 67: 25–53. [DOI] [PubMed] [Google Scholar]
- Chen D, Xu G, Tang W, Jing Y, Ji Q, Fei Z, Lin R. 2013. Antagonistic basic helix‐loop‐helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis . Plant Cell 25: 1657–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Sun X, Zhang L. 2013. Intracellular signaling from plastid to nucleus. Annual Review of Plant Biology 64: 559–582. [DOI] [PubMed] [Google Scholar]
- Danon A, Miersch O, Felix G, op den Camp RG, Apel K. 2005. Concurrent activation of cell death‐regulating signaling pathways by singlet oxygen in Arabidopsis thaliana . Plant Journal 41: 68–80. [DOI] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H et al 2011. Evidence for a SAL1‐PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis . Plant Cell 23: 3992–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flors C, Fryer MJ, Waring J, Reeder B, Bechtold U, Mullineaux PM, Nonell S, Wilson MT, Baker NR. 2006. Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, Singlet Oxygen Sensor Green® . Journal of Experimental Botany 57: 1725–1734. [DOI] [PubMed] [Google Scholar]
- Galvez‐Valdivieso G, Mullineaux PM. 2010. The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiologia Plantarum 138: 430‐439. [DOI] [PubMed] [Google Scholar]
- Gray JC, Sullivan JA, Wang J‐H, Jerome CA, Maclean D. 2003. Coordination of plastid and nuclear gene expression. Philosophical Transactions of the Royal Society B 358: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J, Smeekens S. 2009. Sugar perception and signaling – an update. Current Opinion in Plant Biology 12: 562–567. [DOI] [PubMed] [Google Scholar]
- Harpster MH, Mayfield SP, Taylor WC. 1984. Effects of pigment‐deficient mutants on the accumulation of photosynthetic proteins in maize. Plant Molecular Biology 3: 59–71. [DOI] [PubMed] [Google Scholar]
- Hess WR, Muller A, Nagy F, Borner T. 1994. Ribosome deficient plastids affect transcription of light‐induced nuclear genes: genetic evidence for a plastid‐derived signal. Molecular and General Genetics 242: 305–312. [DOI] [PubMed] [Google Scholar]
- Hideg E, Barta C, Kálai T, Vass I, Hideg K, Asada K. 2002. Detection of singlet oxygen and superoxide with fluorescent sensors in leaves under stress by photoinhibition or UV Radiation. Plant and Cell Physiology 43: 1154–1164. [DOI] [PubMed] [Google Scholar]
- Hideg E, Kálai T, Hideg K, Vass I. 1998. Photoinhibition of photosynthesis in vivo results in singlet oxygen production detection via nitroxide‐induced fluorescence quenching in broad bean leaves. Biochemistry 37: 11405–11411. [DOI] [PubMed] [Google Scholar]
- Huq E, Al‐Sady B, Hudson M, Kim C, Apel K, Quail PH. 2004. Phytochrome‐interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941. [DOI] [PubMed] [Google Scholar]
- Jarvis P, López‐Juez E. 2013. Biogenesis and homeostasis of chloroplasts and other plastids. Nature Reviews Molecular Cell Biology 14: 787–802. [DOI] [PubMed] [Google Scholar]
- Kim C, Apel K. 2013. 1O2‐mediated and EXECUTER‐dependent retrograde plastid‐to‐nucleus signaling in Norflurazon‐treated seedlings of Arabidopsis thaliana . Molecular Plant 6: 1580–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Meskauskiene R, Zhang S, Lee KP, Ashok ML, Blajecka K, Herrfurth C, Feussner I, Apel K. 2012. Chloroplasts of Arabidopsis are the source and a primary target of a plant‐specific programmed cell death signaling pathway. Plant Cell 24: 3026–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P, Noren L, Lopez JDB, Shaikhali J, Strand A. 2012. Interplay between HEAT SHOCK PROTEIN 90 and HY5 controls PhANG expression in response to the GUN5 plastid signal. Molecular Plant 5: 901–913. [DOI] [PubMed] [Google Scholar]
- Kleine T, Voigt C, Leister D. 2009. Plastid signalling to the nucleus: messengers still lost in the mists? Trends in Genetics 25: 185–190. [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto‐Martins G, Surpin M, Lim J, Mittler R, Chory J. 2007. Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719. [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J. 2003. GUN4, a regulator of chlorophyll synthesis and intracellular signalling. Science 299: 902–906. [DOI] [PubMed] [Google Scholar]
- Lee KP, Kim C, Landgraf F, Apel K. 2007. EXECUTER1‐ and EXECUTER2‐dependent transfer of stress‐related signals from the plastid to the nucleus of Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 104: 10270–10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K. 1991. Isolation of nucleic‐acids from plants by differential solvent precipitation. Analytical Biochemistry 195: 45–50. [DOI] [PubMed] [Google Scholar]
- Martin G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E. 2016. Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light‐induced transcriptional network. Nature Communications 7: 11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto F, Obayashi T, Sasaki‐Sekimoto Y, Ohta H, Takamiya K‐I, Masuda T. 2004. Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini‐array system. Plant Physiology 135: 2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield SP, Taylor WC. 1984. Carotenoid‐deficient maize seedlings fail to accumulate light‐harvesting chlorophyll a/b binding protein (LHCP) mRNA. European Journal of Biochemistry 144: 79–84. [DOI] [PubMed] [Google Scholar]
- McCormac AC, Fischer A, Kumar AM, Söll D, Terry MJ. 2001. Regulation of HEMA1 expression by phytochrome and a plastid signal during de‐etiolation in Arabidopsis thaliana . Plant Journal 25: 549–561. [DOI] [PubMed] [Google Scholar]
- McCormac AC, Terry MJ. 2002. Loss of nuclear gene expression during the phytochrome A‐mediated far‐red block of greening response. Plant Physiology 130: 402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac AC, Terry MJ. 2004. The nuclear genes Lhcb and HEMA1 are differentially sensitive to plastid signals and suggest distinct roles for the GUN1 and GUN5 plastid‐signalling pathways during de‐etiolation. Plant Journal 40: 672–685. [DOI] [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K. 2001. FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 98: 12826–12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. 2001. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg‐chelatase H subunit in plastid‐to‐nucleus signal transduction. Proceedings of the National Academy of Sciences, USA 98: 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A. 2008. The steady‐state level of Mg‐protoporphyrin IX is not a determinant of plastid‐to‐nucleus signaling in Arabidopsis . Proceedings of the National Academy of Sciences, USA 105: 15184–15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin M, McCormac AC, Terry MJ, Smith AG. 2008. Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg‐protoporphyrin IX accumulation. Proceedings of the National Academy of Sciences, USA 105: 15178–15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelmüller R, Levitan I, Bergfeld R, Rajasekhar V, Mohr H. 1986. Expression of nuclear genes as affected by treatments acting on the plastids. Planta 168: 482–492. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T. 2010. Plastidial retrograde signalling – a true ‘‘plastid factor’’ or just metabolite signatures? Trends in Plant Science 15: 427–435. [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Förster B, Small ID. 2008. Plastid signalling to the nucleus and beyond. Trends in Plant Science 13: 602–609. [DOI] [PubMed] [Google Scholar]
- Przybyla D, Göbel C, Imboden A, Hamberg M, Feussner I, Apel K. 2008. Enzymatic, but not non‐enzymatic, 1O2‐mediated peroxidation of polyunsaturated fatty acids forms part of the EXECUTER1‐dependent stress response program in the flu mutant of Arabidopsis thaliana . Plant Journal 54: 236–248. [DOI] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou‐Taconnat L, Triantaphylides C, Havaux M. 2012. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proceedings of the National Academy of Sciences, USA 109: 5535–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F, Ksas B, Akkari E, Mialoundama AS, Monnet F, Krieger‐Liszkay A, Ravanat J‐L, Mueller MJ, Bouvier F, Havaux M. 2013. Light‐induced acclimation of the Arabidopsis chlorina1 mutant to singlet oxygen. Plant Cell 25: 1445–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond RW, Gamlin JN. 1999. A compilation of singlet oxygen yields from biologically relevant molecules. Photochemistry and Photobiology 70: 391–475. [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM. 2007. Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M et al 2003. TM4: a free, open‐source system for microarray data management and analysis. BioTechniques 34: 374–378. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Rahman ML, Cho SH, Kim YS, Koh HJ, Yoo SC, Paek NC. 2013. The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant Journal 74: 122–133. [DOI] [PubMed] [Google Scholar]
- Shao N, Duan GY, Bock R. 2013. A mediator of singlet oxygen responses in Chlamydomonas reinhardtii and Arabidopsis identified by a luciferase‐based genetic screen in algal cells. Plant Cell 25: 4209–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee C‐H, Lee D, Choi G. 2009. Phytochromes promote seedling light responses by inhibiting four negatively‐acting phytochrome‐interacting factors. Proceedings of the National Academy of Sciences, USA 106: 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumbe L, Bott R, Havaux M. 2014. Dihydroactinidiolide, a high light‐induced β‐carotene derivative that can regulate gene expression and photoacclimation in Arabidopsis . Molecular Plant 7: 1248–1251. [DOI] [PubMed] [Google Scholar]
- Singh DP, Cornah JE, Hadingham S, Smith AG. 2002. Expression analysis of the two ferrochelatase genes in Arabidopsis in different tissues and under stress conditions reveals their different roles in haem biosynthesis. Plant Molecular Biology 50: 773–788. [DOI] [PubMed] [Google Scholar]
- Sperling U, van Cleve B, Frick G, Apel K, Armstrong GA. 1997. Overexpression of light‐dependent PORA or PORB in plants depleted of endogenous POR by far‐red light enhances seedlings survival in white light and protects against photooxidative damage. Plant Journal 12: 649–658. [DOI] [PubMed] [Google Scholar]
- Stephenson PG, Fankhauser C, Terry MJ. 2009. PIF3 is a repressor of chloroplast development. Proceedings of the National Academy of Sciences, USA 106: 7654–7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson PG, Terry MJ. 2008. Light signalling pathways regulating the Mg‐chelatase branchpoint of chlorophyll synthesis during de‐etiolation in Arabidopsis thaliana . Photochemical and Photobiological Sciences 7: 1243–1252. [DOI] [PubMed] [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J. 2003. Chloroplast to nucleus communication triggered by accumulation of Mg‐protoporphyrin IX. Nature 421: 79–83. [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J. 1993. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799. [DOI] [PubMed] [Google Scholar]
- Terry MJ, Smith AG. 2013. A model for tetrapyrrole synthesis as the primary mechanism for plastid‐to‐nucleus signaling during chloroplast biogenesis. Frontiers in Plant Science 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantaphylidès C, Havaux M. 2009. Singlet oxygen in plants: production, detoxification and signalling. Trends in Plant Science 14: 219–228. [DOI] [PubMed] [Google Scholar]
- Vinti G, Hills A, Campbell S, Bowyer JR, Mochizuki N, Chory J, Lopez‐Juez E. 2000. Interactions between hy1 and gun mutants of Arabidopsis, and their implications for plastid/nuclear signalling. Plant Journal 24: 883–894. [DOI] [PubMed] [Google Scholar]
- Wagner D, Przybyla D, op den Camp R, Kim C, Landgraf F, Lee KP, Würsch M, Laloi C, Nater M, Hideg E et al 2004. The genetic basis of singlet oxygen‐induced stress responses of Arabidopsis thaliana . Science 306: 1183–1185. [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. 2009. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Joens MS, Sinson AB, Gilkerson J, Salomé PA, Weigel D, Fitzpatrick JA, Chory J. 2015. Ubiquitin facilitates a quality‐control pathway that removes damaged chloroplasts. Science 350: 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez‐Ruiz JM, Chory J. 2011. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Current Biology 21: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EEK, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K. 2012. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress‐response genes. Cell 149: 1525–1535. [DOI] [PubMed] [Google Scholar]
- Zhang S, Apel K, Kim C. 2014. Singlet oxygen‐mediated and EXECUTER‐dependent signalling and acclimation of Arabidopsis thaliana exposed to light stress. Philosophical Transactions of the Royal Society B 369: 20130227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z‐W, Yuan S, Feng H, Xu F, Cheng J, Shang J, Zhang D‐W, Lin H‐H. 2011. Transient accumulation of Mg‐protoporphyrin IX regulates expression of PhANGs – new evidence for the signaling role of tetrapyrroles in mature Arabidopsis plants. Journal of Plant Physiology 168: 714–721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Changes in GUN4 and HEMA1 expression in response to a far‐red light pretreatment and a norflurazon treatment assessed with different real‐time RT‐PCR reference genes.
Fig. S2 Correlation plots of raw expression values from the microarray data set.
Fig. S3 Detection of singlet oxygen production via quenching of DanePy fluorescence after a far‐red light pretreatment and a norflurazon treatment.
Fig. S4 Time‐course of changes in photosynthetic gene expression in response to a far‐red pretreatment measured by real‐time RT‐PCR.
Fig. S5 Rescue of nuclear gene expression in gun5 after a far‐red pretreatment.
Fig. S6 Response of ex mutants to a far‐red pretreatment.
Table S1 List of genes referred to in this paper with real‐time PCR primer sequences given (where used)
Table S2 List of 761 genes inhibited at least two‐fold in WL in WT after NF treatment
Table S3 List of 442 genes inhibited at least two‐fold in WL in WT after FR pretreatment
Table S4 List of 63 genes inhibited at least two‐fold in WT by both FR and NF treatments
Table S5 gun1gun5 rescue of genes differentially expressed in WT
Table S6 Predicted localization of protein products of differentially expressed genes identified through microarray analysis
Table S7 List of 263 genes induced at least two‐fold in WL in WT after an FR pretreatment
Table S8 List of 367 genes induced at least two‐fold in WL in WT after NF treatment
Table S9 List of 37 genes induced at least two‐fold in WT by both FR and NF treatments


