Abstract
Respiratory syncytial virus (RSV) causes substantial morbidity and mortality in young children and the elderly. There are currently no licensed RSV vaccines, and passive prophylaxis with the monoclonal antibody palivizumab is restricted to high-risk infants in part due to its modest efficacy. Although it is widely agreed that an effective RSV vaccine will require the induction of a potent neutralizing antibody response against the RSV fusion (F) glycoprotein, little is known about the specificities and functional activities of RSV F-specific antibodies induced by natural infection. Here, we have comprehensively profiled the human antibody response to RSV F by isolating and characterizing 364 RSV F-specific monoclonal antibodies from the memory B cells of three healthy adult donors. In all donors, the antibody response to RSV F is comprised of a broad diversity of clones that target several antigenic sites. Nearly half of the most potent antibodies target a previously undefined site of vulnerability near the apex of the prefusion conformation of RSV F (preF), providing strong support for the development of RSV vaccine candidates that preserve the membrane-distal hemisphere of the preF protein. Additionally, the antibodies targeting this new site display convergent sequence features, thus providing a future means to rapidly detect the presence of these antibodies in human vaccine samples. Many of the antibodies that bind preF-specific surfaces are over 100 times more potent than palivizumab, and several cross-neutralize human metapneumovirus (HMPV). Taken together, the results have implications for the design and evaluation of RSV vaccine candidates and offer new options for passive prophylaxis.
One Sentence Summary
High-throughput profiling of anti-RSV F antibody repertoires reveals new opportunities for vaccine design and passive therapy.
Introduction
Respiratory syncytial virus (RSV) is the leading cause of infant hospitalization in the United States and accounts for an estimated 64 million infections and 160,000 deaths world-wide each year. However, despite decades of research, the development of a safe and effective vaccine against RSV has remained elusive, highlighting the need for novel strategies that induce protective immune responses. Neutralizing antibodies have been shown to protect against severe RSV disease in humans and animal models, and therefore it is widely agreed that an effective RSV vaccine should induce a robust neutralizing antibody response (1–3).
Similar to other pneumoviruses, RSV expresses two major surface glycoproteins: the fusion protein (F) and the attachment protein (G). Although both have been shown to induce protective neutralizing antibody responses, F is less genetically variable than G, is absolutely required for infection, and is the target for the majority of neutralizing activity in human serum (4–8). RSV F is also the target of the monoclonal antibody palivizumab, which is used to passively protect high-risk infants from severe disease (9). Consequently, the RSV F protein is considered to be a highly attractive target for vaccines and antibody-based therapies.
The mature RSV F glycoprotein initially exists in a metastable prefusion conformation (preF) (10), before undergoing a conformational change that leads to insertion of the hydrophobic fusion peptide into the host-cell membrane. Subsequent refolding of F into a stable, elongated postfusion conformation (postF) (11, 12) results in fusion of the viral and host-cell membranes. Due to its inherent instability, the preF protein has the propensity to prematurely trigger into postF, both in solution and on the viral surface (13). Recently, stabilization of preF has been achieved by protein engineering (14, 15), and stabilized preF has been shown to induce higher titers of neutralizing antibodies than postF in animal models (15).
Despite the importance of neutralizing antibodies in protection against severe RSV disease, our understanding of the human antibody response to RSV has been limited to studies of human sera and a small number of RSV-specific human monoclonal antibodies (16–19). The epitopes recognized by these human antibodies, as well as several murine antibodies, have defined at least four ‘antigenic sites’ on RSV F (1, 10, 16, 18–20) (table S1). Three of these sites—I, II, and IV—are present on both pre- and postF, whereas antigenic site Ø exists exclusively on preF. Additional preF-specific epitopes have been defined by antibodies MPE8 (17) and AM14 (21). Although serum-mapping studies have shown that site Ø-directed antibodies are responsible for a large proportion of the neutralizing antibody response in most individuals (8), there are additional antibody specificities that contribute to serum neutralizing activity that remain to be defined. In addition, it is unknown whether certain antibody sequence features are required for recognition of certain neutralizing sites, as observed for other viral targets (22–25). Finally, understanding the relationship between neutralization potency and epitope specificity will be critical in the selection and design of vaccine antigens that induce potent neutralizing responses.
To address these questions, we isolated an extensive panel of RSV F-specific monoclonal antibodies from the memory B cells of three healthy adult donors and used these antibodies to comprehensively map the antigenic topology of RSV F. The results show that a large proportion of the RSV F-specific human antibody repertoire is comprised of neutralizing antibodies, many of which exhibit remarkable potency. The most potent antibodies target two distinct antigenic sites that are located near the apex of the preF trimer, providing strong support for the development of preF-based vaccine candidates that preserve these antigenic sites. Furthermore, the highly potent neutralizing antibodies described here represent new opportunities for the prevention of severe RSV disease by passive immunoprophylaxis.
Results
Large-scale isolation of RSV F-specific monoclonal antibodies from healthy adult donors
In order to comprehensively profile the human antibody response to RSV F, we aimed to isolate and characterize approximately 100 monoclonal antibodies from the memory B cells of each of three healthy adult donors. Although these donors did not have a documented history of RSV infection, healthy adults are expected to have had multiple RSV infections throughout life (26). We assessed the magnitude of the memory B cell response to RSV F by staining peripheral B cells with a mixture of fluorescently labeled pre- and postfusion RSV F sorting probes (fig. S1) (11, 15). Both proteins were dual-labeled in order to eliminate background due to non-specific fluorochrome binding (27). Flow cytometric analysis revealed that 0.04–0.18% of class-switched (IgG+ and IgA+) peripheral B cells were specific for RSV F (Fig. 1A, fig. S2), which is significantly lower than the percentage of RSV F-specific cells observed after experimental RSV infection, suggesting that these three donors were probably not recently exposed to RSV (28). Index sorting showed that 17–38% of circulating RSV F-specific B cells express IgA, indicating that IgA memory B cells to RSV F are present in peripheral blood (Fig. 1B). Approximately 200 RSV F-specific B cells from each donor sample were single-cell sorted, and antibody variable heavy (VH) and variable light (VL) chain sequences were rescued by single-cell PCR (29). Over 100 cognate heavy and light chain pairs from each donor were subsequently cloned and expressed as full-length IgGs in an engineered strain of Saccharomyces cerevisiae for further characterization (30). Preliminary binding studies showed that approximately 80% of antibodies cloned from RSV F-specific B cells bound to recombinant RSV F proteins (table S2).
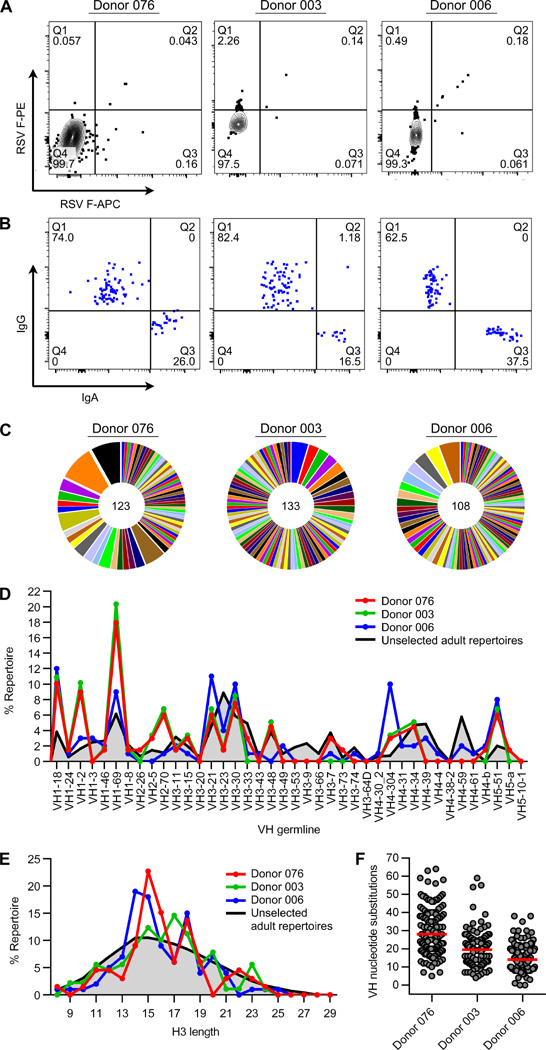
Fig. 1.
Anti-RSV F repertoire cloning. (A) RSV F-specific B cell sorting. FACS plots show RSV F reactivity of IgG+ and IgA+ B cells from three healthy adult donors. B cells in quadrant 2 (Q2) were single cell sorted. (B) Isotype analysis. Index sort plots show the percentage of RSV F-specific B cells that express IgG or IgA. (C) Clonal lineage analysis. Each slice represents one clonal lineage; the size of the slice is proportional to the number of clones in the lineage. The total number of clones is shown in the center of the pie. Lineage numbering can be found in Data file S1. Clonal lineages were assigned based on the following criteria: 1) matching of variable and joining gene segments; 2) identical CDR3 lengths; and 3) >80% homology in CDR3 nucleotide sequences. (D) VH repertoire analysis. VH germline genes were considered to be enriched in RSV repertoires if at least two out of three donors showed >3-fold enrichment over non-RSV-specific repertoires (33). (E) CDRH3 length distribution. (F) Somatic hypermutation in VH (excluding CDRH3). Red bars indicate the average number of nucleotide substitutions. Each clonal lineage is only represented once in (D) and (E). Data for non-RSV reactive IgGs were derived from published sequences obtained by high-throughput sequencing of re-arranged antibody variable gene repertoires from healthy individuals (33).
Sequence analysis of RSV F-specific antibody repertoires
Sequence analysis of the isolated monoclonal antibodies revealed that all three RSV-F specific repertoires were highly diverse, each containing between 70 and 98 unique lineages (Fig. 1C, Data file S1). This result is in stark contrast to the relatively restricted repertoires observed in HIV-infected patients (31) or in healthy donors after influenza vaccination (32). Compared to non-RSV-reactive antibodies (33), the RSV F-specific repertoires were skewed toward certain VH germline genes (VH1–18, VH1–2, VH1–69, VH2–70, VH4–304, and VH5–51) (Fig. 1D). A bias toward VH1–69 has also been observed in anti-HIV-1, anti-influenza, and anti-HCV repertoires (34–36), and recent studies have shown that there is a significant increase in the relative usage of VH1–18, VH1–2, and VH1–69 during acute dengue infection (37). Hence, it appears that these particular germline gene segments may have inherent properties that facilitate recognition of viral envelope proteins. The distribution of heavy chain third complementarity-determining region (CDRH3) lengths in RSV F-specific antibody repertoires was skewed towards lengths of 14–18 amino acids compared to unselected repertoires (Fig 1E). The average level of somatic hypermutation (SHM) varied between the three donor repertoires, ranging between 16 and 30 nucleotide substitutions per VH gene (excluding CDRH3) (Fig. 1F), which is comparable to the average level of SHM observed in anti-influenza antibody repertoires (32, 38) and consistent with the recurrent nature of RSV infection (26). Interestingly, several antibodies contained greater than 50 VH gene nucleotide substitutions, suggesting that multiple rounds of RSV infection can result in antibodies with very high levels of SHM.
A large proportion of antibodies bind exclusively to preF
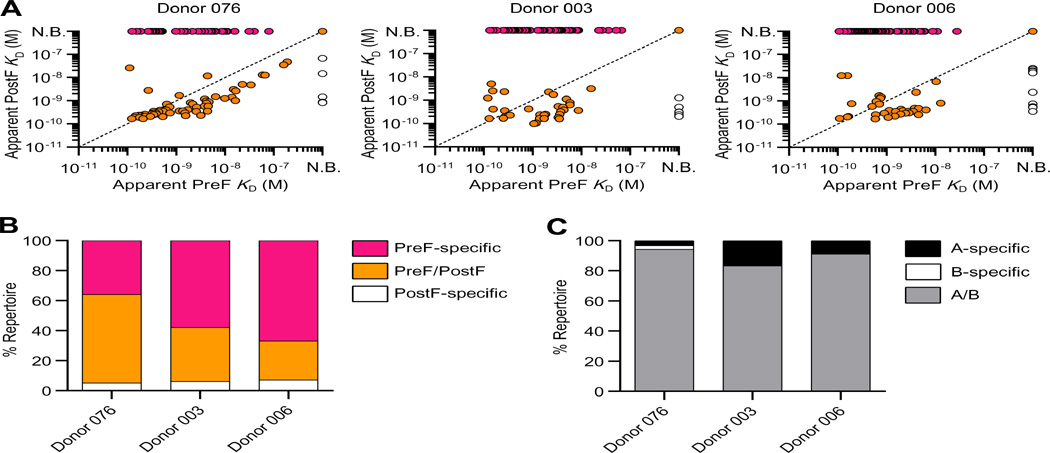
We next measured the apparent binding affinities of the IgGs to furin-cleaved RSV F ectodomains stabilized in the prefusion (DS-Cav1) or postfusion (F ΔFP) conformation using biolayer interferometry (11, 15). In all three donor repertoires, a relatively large proportion of the antibodies (36–67%) bound exclusively to preF (Fig. 2A, B). The vast majority of remaining antibodies bound to both pre- and postF, with only 5–7% of antibodies showing exclusive postF specificity (Fig. 2A, B). The low prevalence of postF-specific antibodies in these donor repertoires is consistent with the observation that less than 10% of anti-RSV F serum-binding activity specifically targets postF (8). However, the majority of cross-reactive antibodies bound with higher apparent affinity to postF (Fig 2A), suggesting that these antibodies were probably elicited by and/or affinity matured against postF in vivo. Hence, the significantly higher proportion of preF- versus postF-specific antibodies is likely due to the higher immunogenicity of the unique surfaces on preF compared to postF, rather than an increased abundance of preF in vivo. Finally, as expected based on the relatively high degree of sequence conservation of RSV F proteins, most of the antibodies bound to F proteins derived from subtypes A and B (Fig. 2C).
Fig. 2.
Antibody preferences for conformational state and subtype of RSV F are similar across three adult donor repertoires. (A) IgG affinities for preF and postF are plotted for each donor. PreF-specific antibodies are colored pink, preF/postF-cross reactive antibodies are orange, and postF-specific antibodies are white. N.B., non-binder (B) Percentage of antibodies within each donor repertoire that are preF-specific, preF/postF-cross reactive or postF-specific. (C) Percentage of antibodies within each donor repertoire that bind specifically to RSV F derived from subtype A (black), subtype B (white), or both subtypes A and B (grey).
Since certain antiviral antibody specificities have been associated with poly- and autoreactivity (39–41), we also tested the RSV antibodies for polyreactivity using a previously described high-throughput assay that correlates with down-stream behaviors such as serum clearance (42, 43). One hundred and seventy-seven clinical antibodies, as well as several broadly neutralizing HIV-1 antibodies, were also included for comparison. In contrast to many previously described HIV-1 broadly neutralizing antibodies, the vast majority of RSV F-specific antibodies lacked substantial polyreactivity in this assay (fig. S3).
RSV F-specific antibodies target six major antigenic sites
To map the epitopes recognized by the RSV F-specific antibodies, we first performed competitive binding experiments using a previously described yeast-based assay (44). Antibodies were initially tested for competition with D25, AM14 and MPE8—three previously described preF-specific antibodies (10, 17, 21)—and motavizumab, an affinity-matured variant of palivizumab that binds to both pre- and postF (10, 11, 45). Non-competing antibodies were then tested for competition with a site IV-directed mAb (101F) (46), a site I-directed antibody (ADI-13390) (table S3), and two high affinity antibodies from the panel (ADI-14443 and ADI-14469) that did not strongly compete with each other or any of the control antibodies (table S3). Each antibody was assigned to a bin based on the results of this competition assay (Data file S1).
In order to increase the resolution of our epitope assignments, we also measured the binding of each antibody to a panel of preF variants using a luminex-based assay (Data file S1). Each variant contained 2–4 mutations clustered together to form a patch on the surface of preF. A total of nine patches that uniformly covered the surface of preF were generated (fig. S4). Deglycosylated preF was also included to identify antibodies targeting glycan-dependent epitopes. Previously characterized antibodies D25, AM14 and motavizumab were used to validate the assay (fig. S4). The combined bin and patch data were then used to assign each antibody to a single antigenic site (Fig. 3A and B), which we defined based on previously determined structures, resistance mutations, and secondary structure of the F protein. Overall, these data show that the large majority of isolated antibodies target six dominant antigenic sites on prefusion RSV F (Ø, I, II, III, IV, and V). Interestingly, only a small proportion of the isolated antibodies had binding profiles similar to that of AM14, suggesting that antibodies targeting this quaternary epitope are not commonly elicited during natural infection. None of the antibodies were sensitive to deglycosylation of F, demonstrating that glycan-dependent antibodies are also rarely elicited by natural RSV infection. Importantly, all three donor repertoires showed highly similar epitope coverage, suggesting that the majority of healthy adults produce antibodies targeting each of these six antigenic sites.
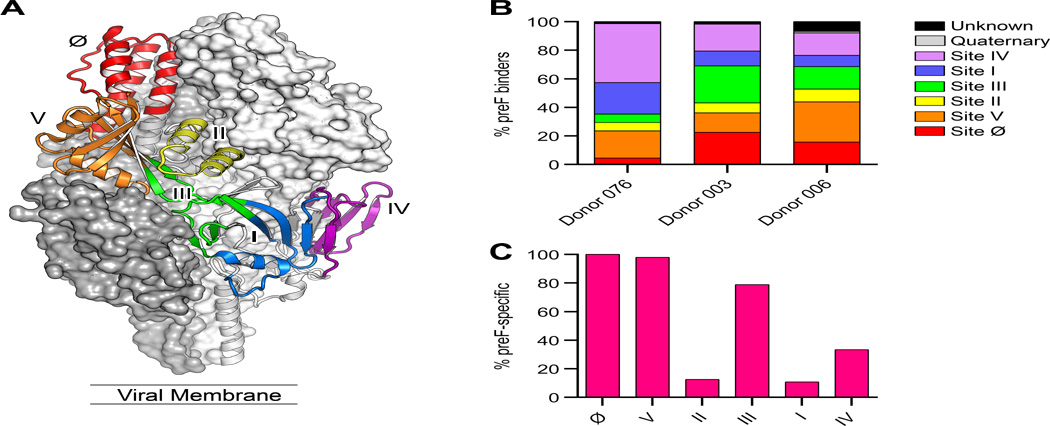
Fig. 3.
Antibodies isolated from adult donors recognize six antigenic sites spanning the surface of preF. (A) Structure of RSV preF with one protomer shown as a ribbon colored by antigenic site. (B) Percentage of antibodies targeting each antigenic site within each donor repertoire. The antigenic sites recognized by 215 antibodies with higher than 2 nM affinity for preF were mapped using a combination of antibody binning and patch variant mutational analysis. (C) Percentage of preF-specific antibodies targeting each antigenic site.
Analysis of the preF- and postF-binding activities of the antibodies targeting each antigenic site (Fig. 3C and fig. S5) revealed that three sites are almost exclusively found on preF (Ø, III, and V). Antibodies targeting site Ø and site III have been previously described (10, 17), and these sites are located on the top and side of the preF protein, respectively. Between 4 and 22% of antibodies from each donor recognized site Ø and between 6 and 26% recognized site III. A relatively large proportion of antibodies from each donor (14–28%) recognized the previously undescribed site V (Fig. 3B). The majority of site V antibodies competed with D25, MPE8 and motavizumab, which was unexpected given the distance between the epitopes recognized by these three antibodies. The patch mutant analysis revealed that site V antibodies interact with the α3 helix and β3/β4 hairpin of preF. This region is located between the epitopes recognized by D25, MPE8, and motavizumab, explaining the unusual competition profile observed for this group of antibodies (fig. S6). Interestingly, two-thirds of the site V-directed antibodies utilized the same VH–VL germline pair (VH1–18 and VK2–30) and had CDRH3 lengths of 14 or 15 amino acids (Data file S2). In addition, many of these antibodies shared certain somatic mutations in both the VH and VL genes (Data file S2), suggesting a common binding mode. In addition to the three primarily preF-specific sites, approximately one-third of the antibodies that recognized antigenic site IV were preF-specific, likely due to contacts with β22, which dramatically rearranges during the transition from pre- to postF. In summary, the epitope mapping data show that the large majority of isolated antibodies target six dominant antigenic sites, approximately half of which are exclusively expressed on preF.
Highly potent neutralizing antibodies target preF-specific epitopes
The 364 IgGs were tested for neutralizing activity against RSV subtypes A and B using a previously described high-throughput neutralization assay (15). For all three donor repertoires, 70–80% of the isolated antibodies showed neutralizing activity, and 19–38% neutralized with high potency (IC50 ≤ 0.05 µg/ml) (Fig. 4A, B). Notably, several clonally unrelated antibodies were ≥ 5.0-fold more potent than D25 and ≥ 100-fold more potent than palivizumab (Fig. 4A). Interestingly, there was no correlation between neutralization potency and level of SHM (P=0.89, r=0.0082) (fig. S7, Data file S1), suggesting that extensive SHM is not required for potent neutralization of RSV. Consistent with the binding cross-reactivity data, the majority of neutralizing antibodies showed activity against both subtype A and B (fig. S8).
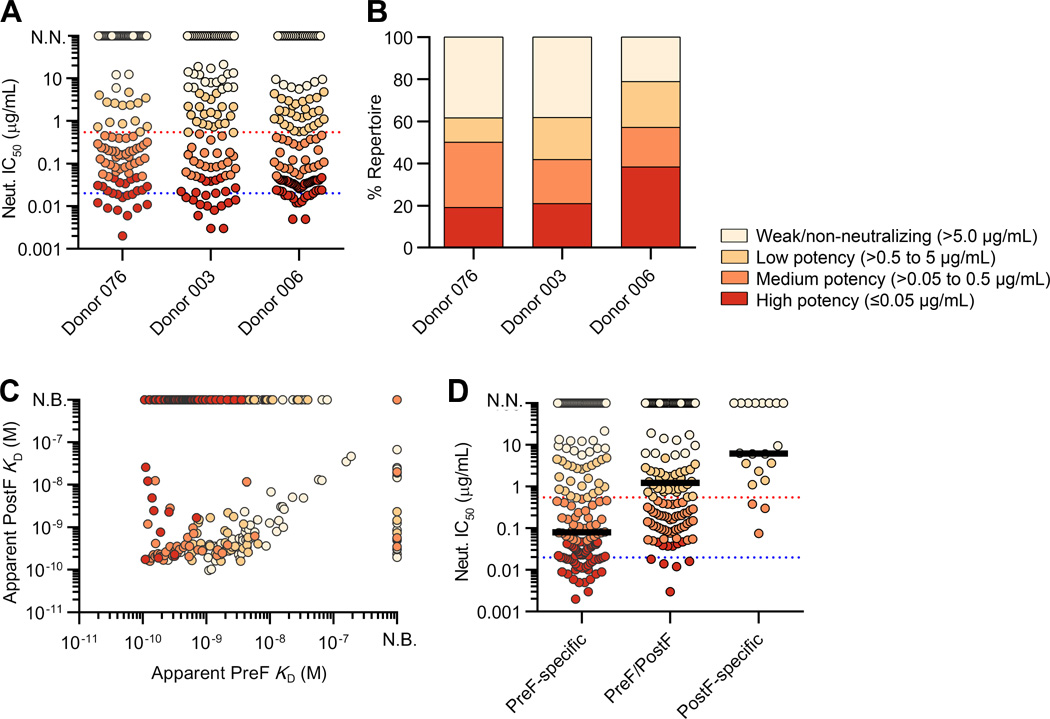
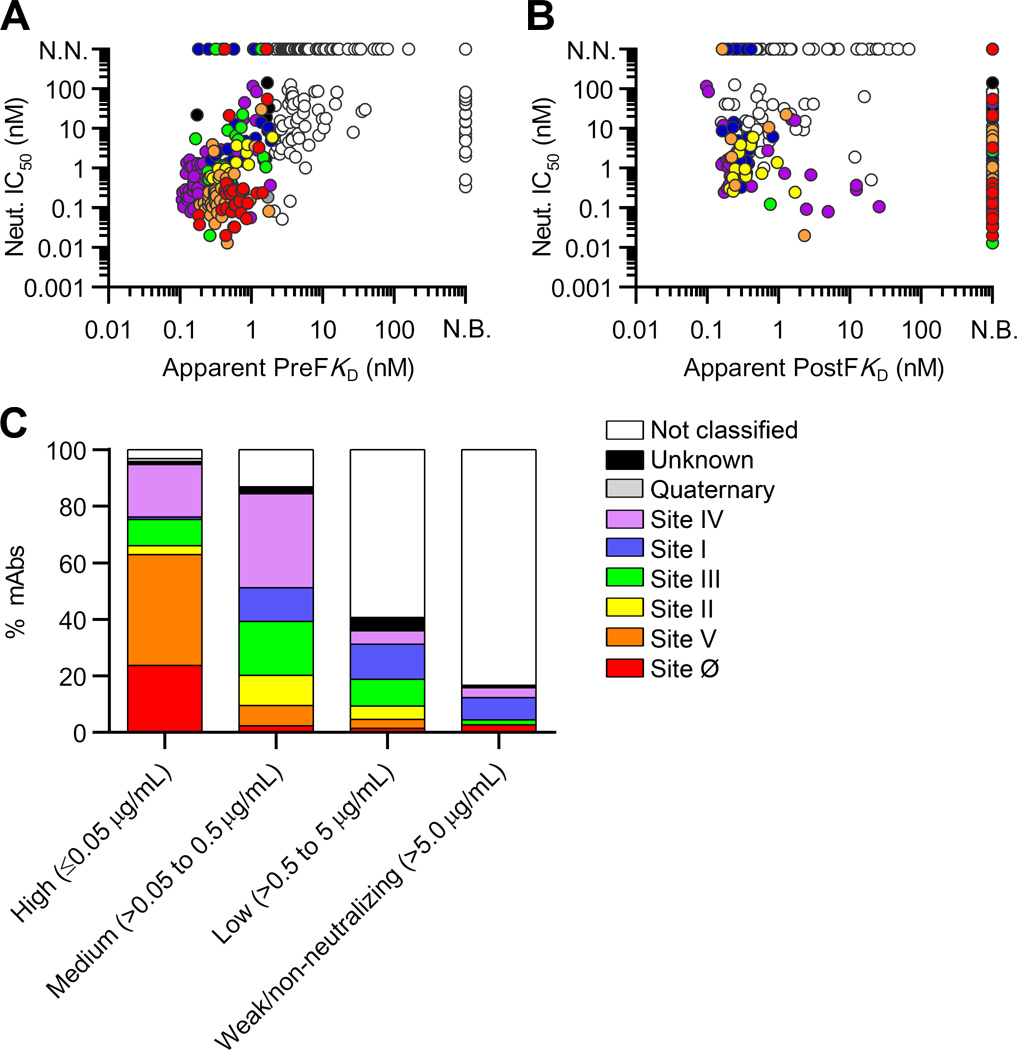
Fig. 4.
The majority of potent neutralizing antibodies recognize preF-specific sites. (A) Neutralization IC50s for the antibodies isolated from each donor repertoire. Data points are colored based on neutralization potency. Red and blue dotted lines depict motavizumab and D25 IC50s, respectively. N.N., non-neutralizing. (B) Percentage of neutralizing antibodies in each donor repertoire, grouped by potency. (C) Apparent binding affinities for preF and postF are plotted for each antibody and colored according to neutralization potency. (D) Neutralization IC50s are plotted for preF-specific, postF-specific, and cross-reactive antibodies. Red and blue dotted lines depict motavizumab and D25 IC50s, respectively. Black bar depicts median IC50.
We next analyzed the relationship between preF- and postF-binding affinity and neutralization potency (Fig. 4C). This analysis revealed that greater than 85% of highly potent antibodies (IC50 < 0.05 µg/ml) were specific for preF (fig. S9). Furthermore, preF-specific antibodies were more than 8-fold more potent than pre- and postF cross-reactive antibodies and 80-fold more potent than antibodies that specifically recognized postF (Fig. 4D). Importantly, there was a positive correlation between preF binding and neutralization (P<0.001, r=0.24), and the apparent preF KDs generally corresponded well with the neutralization IC50s (Fig. 5A). In contrast, there was no correlation between postF binding and neutralization (P=0.44, r=−0.07) (Fig. 5B). In addition, relatively few antibodies neutralized with IC50s lower than 100 pM, which is consistent with the previously proposed ceiling to affinity maturation (47, 48).
Fig. 5.
The most potent neutralizing antibodies bind with high affinity to preF and recognize antigenic sites Ø and V. (A) Neutralization IC50 is plotted against apparent preF KD and colored according to antigenic site. N.N., non-neutralizing; N.B., non-binder. (B) Neutralization IC50 is plotted against apparent postF KD and colored by antigenic site. (C) Antibodies are grouped according to neutralization potency and colored by antigenic site.
We next analyzed the relationship between neutralization potency and antigenic site (Fig. 5C). Over 60% of the highly potent neutralizing antibodies targeted antigenic sites Ø and V, which represent two of the three prefusion-F specific sites. In contrast, antibodies targeting sites III and IV showed a wide range of neutralization potencies, and antibodies targeting sites I and II were generally moderate to non-neutralizing. Similar results were obtained for subtype B viruses (fig. S10). Interestingly, a subset of site IV-directed antibodies neutralized with substantially lower potency than would be expected based on preF binding affinity (Fig. 5A). There are several possible explanations for this observation, including differences in (i) the accessibility of different sites on the crowded surface of the virion (ii) the sensitivity of different sites to the antibody angle of approach, and (iii) the mechanisms of neutralization for preF-specific antibodies compared with antibodies that are reactive with both preF and postF.
Several antibodies cross-neutralize RSV and HMPV
Given that the RSV and human metapneumovirus (HMPV) F proteins share 33% amino acid identity, and certain RSV F-specific antibodies cross-neutralize HMPV (17, 49), we next tested the antibodies in our panel for neutralizing activity against HMPV. Of the 364 antibodies tested, nine neutralized HMPV and two showed highly potent activity against both HMPV and RSV (Table 1). Sequence analysis revealed that the nine antibodies comprise five clonal families, which do not show convergent VH germline gene usage, CDRH3 lengths, or somatic mutations (Data file S3). Nearly all of the cross-neutralizing antibodies bound exclusively to preF and competed with MPE8 (antigenic site III) (Table 1). This result was not unexpected, as MPE8 has been previously shown to cross-neutralize four pneumoviruses, including RSV and HMPV (17). Although HMPV F was not used for B cell sorting, all three donor repertoires contained antibodies that cross-neutralized HMPV, suggesting that highly conserved epitopes are relatively immunogenic in the context of natural RSV and/or HMPV infection.
Table 1.
A subset of anti-RSV F antibodies cross-neutralize human metapneumovirus.
| Name | HMPV- A1 IC50 (µg/ml) |
RSV-A2 IC50 (µg/ml) |
Prefusion RSV F KD (M) |
Postfusion RSV F KD (M) |
RSV F Binding Site |
|---|---|---|---|---|---|
| ADI-14448 | 0.05 | 0.04 | 3.8 × 10−10 | N.B. | III |
| ADI-15614 | 0.22 | 0.03 | 4.1 × 10−10 | N.B. | III |
| ADI-14441 | 37.8 | >25 | 8.6 × 10−9 | N.B. | III* |
| ADI-14501 | 31.4 | 12.4 | 1.6 × 10−8 | N.B. | III* |
| ADI-15657 | 11.9 | >25 | 5.7 × 10−9 | N.B. | III* |
| ADI-15665 | 13.5 | 12.7 | 8.3 × 10−9 | N.B. | III* |
| ADI-15647 | 20.3 | >25 | 7.8 × 10−9 | N.B. | III* |
| ADI-15623 | 0.37 | 0.05 | 2.1 × 10−9 | N.B. | III* |
| ADI-18992 | 6.1 | 2.5 | 7.6 × 10−10 | 1.5 × 10−9 | IV* |
| MPE8 Control | 0.07 | 0.04 | – | – | – |
N.B., non-binder.
Binding site assignment based on competition only.
Discussion
An in-depth understanding of the human antibody response to RSV infection will aid the development and evaluation of RSV vaccine candidates. Although previous studies have coarsely mapped the epitopes targeted by RSV-specific neutralizing antibodies in human sera (4, 8), the specificities and functional properties of antibodies induced by natural RSV infection have remained largely undefined. Here, we have used preF- and postF-stabilized proteins (11, 15), a high-throughput antibody isolation platform, and a structure-guided collection of prefusion F mutants to clonally dissect the human memory B cell response to RSV F in three naturally infected adult donors.
In the repertoires analyzed, the ratio of preF-specific antibodies to those that recognize both pre- and postF varied slightly among the three donors, with an average ratio of approximately 1:1. These values are somewhat lower than those reported for human sera, which showed approximately 70% of anti-F serum binding is specific for preF (8). This discrepancy may be the result of differences between the levels of individual antibodies in serum, differences in the B cell phenotypes achieved for a particular specificity, or variation between donors. Despite these minor differences, the results of both studies suggest that preF-specific epitopes and epitopes shared by pre- and postF are immunogenic during natural RSV infection, whereas the unique surfaces on postF are substantially less immunogenic.
Our repertoire analysis reveals that the large majority of RSV F-specific antibodies target six dominant antigenic sites on prefusion RSV F: Ø, I, II, III, IV, and V. We defined these sites based on previously determined structures, resistance mutations, and secondary structure of the preF protein. It is important to note that the nomenclature for describing RSV F antigenic sites has evolved over time (6, 50–56), and previous mapping efforts were based on the postfusion conformation of F and did not include surfaces present exclusively on preF. The crystal structure of preF has provided critical information about F structure and function as well as new reagents to map antibody binding sites on the unique surfaces of preF and surfaces shared with postF. We therefore propose an update to the nomenclature system for antigenic sites on RSV F, building on previous information but now including structurally defined regions of preF. To a first approximation, each antibody can be assigned primarily to one of these sites. However, it is likely that antibody epitopes cover the entire surface of F and that there are antibodies that bind two or more adjacent antigenic sites within a protomer and quaternary antibodies that bind across protomers. Thus, the antigenic site nomenclature will never be precise enough to describe the entire spectrum of RSV F-specific antibodies, but should be considered as a rationally defined three-dimensional guide to major antigenic sites of preF.
Importantly, the results show that the most potently neutralizing antibodies target antigenic sites Ø and V, both of which are located near the apex of the preF trimer. These findings are consistent with results obtained from human sera mapping, which determined that the majority of neutralizing activity can be removed by pre-incubation with preF (4, 8) and that preF-specific sites other than site Ø make up a considerable fraction of preF-specific neutralizing antibodies (8). Although antigenic site Ø has been shown to be a target of potently neutralizing antibodies (8, 10), the interaction of antibodies with site V is less well understood. Interestingly, we found that the majority of site V-directed antibodies share several convergent sequence features, suggesting that it may be possible to rapidly detect these types of antibodies in human samples using high-throughput sequencing technology (57). This may prove to be particularly useful for profiling antibody responses to RSV vaccine candidates that aim to preserve the apex of the preF trimer.
A limitation of our repertoire analysis is the relatively small number of donors studied. We chose to isolate a large number of monoclonal antibodies from each donor, rather than to include a large number of donors with limited numbers of antibodies isolated from each. As a result, intra-donor comparisons can be made with high certainty, but inter-donor comparisons are less robust. However, the high degree of similarity among the three donor repertoires (Fig. 3B), combined with the overall agreement of our findings with those of the sera analysis published previously (8), suggest that the antibody repertoires analyzed here are likely representative of naturally infected adult donors.
This extensive panel of antibodies provides new opportunities for passive prophylaxis. More than 30 of these antibodies neutralize RSV more potently than D25, which served as the basis for MEDI8897—a monoclonal antibody that is currently in clinical trials for the prevention of RSV in young, at risk children (58). Additionally, we have isolated several antibodies that cross-neutralize HMPV, including one that neutralizes RSV with a potency comparable to D25. The identification of a cross-neutralizing antibody with potency equivalent to D25 suggests that cross-neutralization of HMPV does not necessarily occur at the expense of potent RSV neutralization.
Although passive prophylaxis with highly potent antibodies may substantially reduce the RSV disease burden in select populations, an effective vaccine would produce the greatest benefit at the lowest cost. The development of an RSV vaccine has presented a number of unique challenges, and selection of the optimal vaccination strategy will be of the utmost importance. The in-depth analysis of the human antibody response to natural RSV infection presented here provides insights for the development of such a vaccine. Importantly, our results suggest that immunization of pre-immune donors with preF immunogens will likely boost neutralizing responses, whereas the use of postF immunogens would be expected to expand B cell clones with moderate or weak neutralizing activity. Similarly, immunization of RSV naïve infants with preF immunogens would be expected to activate naïve B cells targeting epitopes associated with substantially more potent neutralizing activity compared to postF immunogens. In addition, the ideal RSV vaccine should preserve antigenic sites Ø and V, since these sites are targeted by the most highly potent antibodies elicited in response to natural RSV infection. Finally, the reagents described here provide a useful set of tools for the evaluation of clinical trials, which will be critical for selecting the optimal RSV vaccination strategy from the many currently under investigation (59).
Materials and Methods
For the detailed Materials and Methods, please see the Supplementary Materials.
Supplementary Material
Acknowledgments
We thank Tushar Jain for guidance on statistical analyses, Todd Boland for assistance with antibody sequence analysis, Emilie Shipman for assistance with protein expression, Wen Li for technical assistance, Cody Williams and S.M. Eagol for assistance with figure preparation, and Margaret Ackerman for use of the magnetic microplate washer (BioTek) and the FLEXMAP 3D flow cytometer (Luminex). PBMC processing was carried out in DartLab, the Immune Monitoring and Flow Cytometry Shared Resource, supported by a National Cancer Institute Cancer Center Support Grant to the Norris Cotton Cancer Center (P30CA023108-37) and an Immunology COBRE Grant (P30GM103415-15) from the National Institute of General Medical Sciences. All the IgGs were sequenced by Adimab's Molecular Core and produced by the High Throughput Expression group. Biolayer interferometry binding experiments were performed by Adimab's protein analytics group. Funding: Support for this work was provided by the National Institute of General Medical Sciences of the National Institutes of Health award T32GM008704 (M.S.A.G.) and P20GM113132 (J.S.M.) and intramural funding from the National Institute of Allergy and Infectious Diseases to support work at the Vaccine Research Center (B.S.G.). This work was partially supported by grant SAF2015-67033-R to J.A.M. from Plan Nacional I+D+i.
Competing interests: L.M.W. is an inventor on pending patent applications describing the RSV antibodies (Anti-respiratory syncytial virus antibodies, and methods of their generation and use, USSN 62/411,500, USSN 62/411,508, and USSN 62/411,510). M.C., J.S.M and B.S.G. are inventors on patent applications regarding prefusion-stabilized F proteins and their use (Prefusion RSV F proteins and their use, PCT/US2014/026714). L.M.W., C.C., and E.G. have an equity position in Adimab, LLC.
Footnotes
Author contributions: L.M.W., M.S.A.G., and J.S.M. wrote the manuscript. L.M.W., J.S.M., P.F.W., and B.S.G. analyzed results, edited the manuscript, and provided intellectual oversight. C.C., M.S.A.G., M.C., J.O.N., L.M.W., S.M., and E.G. planned and performed experiments. L.W. performed the statistical analyses.
Data and materials availability: GenBank accession numbers for the antibody variable region gene sequences reported in this study can be found in Data file S1.
Materials and methods
Fig. S1. Purification of pre- and postF sorting probes.
Fig. S2. Representative gating strategy for RSV F-specific B cell sorting.
Fig. S3. Anti-RSV F antibodies lack polyreactivity.
Fig. S4. Generation and validation of preF patch panel mutants.
Fig. S5. Antibody binding affinities for pre- and postF.
Fig. S6. Antigenic site V resides between the epitopes recognized by D25, MPE8 and motavizumab.
Fig. S7. Degree of somatic hypermutation does not correlate with neutralization potency.
Fig. S8. Neutralization of RSV subtype B.
Fig. S9. Neutralizing activities of preF-specific, cross-reactive, and postF-specific antibodies.
Fig. S10. Relationship between subtype B neutralization and epitope.
Table S1. Antigenic sites targeted by prototypic RSV antibodies.
Table S2. Efficiency of binder rescue from B cell sorting.
Table S3. Competition profile of antibodies used in binning experiments.
Data file S1. Binding, neutralization, epitope assignment, limited sequence features and GenBank accession codes for the isolated antibodies.
Data file S2. Site V-directed antibodies show convergent sequence features.
Data file S3. Antibody variable gene sequences of HMPV cross-neutralizing antibodies. References (60–62)
References and Notes
- 1.Rogovik AL, Carleton B, Solimano A, Goldman RD. Palivizumab for the prevention of respiratory syncytial virus infection. Can Fam Physician. 2010;56:769–772. [PMC free article] [PubMed] [Google Scholar]
- 2.Graham BS. Biological challenges and technological opportunities for respiratory syncytial virus vaccine development. Immunol Rev. 2011;239:149–166. doi: 10.1111/j.1600-065X.2010.00972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groothuis JR, Simoes EA, Hemming VG. Respiratory syncytial virus (RSV) infection in preterm infants and the protective effects of RSV immune globulin (RSVIG). Respiratory Syncytial Virus Immune Globulin Study Group. Pediatrics. 1995;95:463–467. [PubMed] [Google Scholar]
- 4.Magro M, Mas V, Chappell K, Vazquez M, Cano O, Luque D, Terron MC, Melero JA, Palomo C. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc Natl Acad Sci U S A. 2012;109:3089–3094. doi: 10.1073/pnas.1115941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson S, Oliver C, Prince GA, Hemming VG, Pfarr DS, Wang SC, Dormitzer M, O'Grady J, Koenig S, Tamura JK, Woods R, Bansal G, Couchenour D, Tsao E, Hall WC, Young JF. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J Infect Dis. 1997;176:1215–1224. doi: 10.1086/514115. [DOI] [PubMed] [Google Scholar]
- 6.Beeler JA, van Wyke Coelingh K. Neutralization epitopes of the F glycoprotein of respiratory syncytial virus: effect of mutation upon fusion function. J Virol. 1989;63:2941–2950. doi: 10.1128/jvi.63.7.2941-2950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karron RA, Buonagurio DA, Georgiu AF, Whitehead SS, Adamus JE, Clements-Mann ML, Harris DO, Randolph VB, Udem SA, Murphy BR, Sidhu MS. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci U S A. 1997;94:13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ngwuta JO, Chen M, Modjarrad K, Joyce MG, Kanekiyo M, Kumar A, Yassine HM, Moin SM, Killikelly AM, Chuang GY, Druz A, Georgiev IS, Rundlet EJ, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Nason MC, Capella C, Peeples ME, Ledgerwood JE, McLellan JS, Kwong PD, Graham BS. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med. 2015;7:309ra162. doi: 10.1126/scitranslmed.aac4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.T. I.-R. S. Group. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics. 1998;102:531–537. [PubMed] [Google Scholar]
- 10.McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLellan JS, Yang Y, Graham BS, Kwong PD. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol. 2011;85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swanson KA, Settembre EC, Shaw CA, Dey AK, Rappuoli R, Mandl CW, Dormitzer PR, Carfi A. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci U S A. 2011;108:9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liljeroos L, Krzyzaniak MA, Helenius A, Butcher SJ. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc Natl Acad Sci U S A. 2013;110:11133–11138. doi: 10.1073/pnas.1309070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krarup A, Truan D, Furmanova-Hollenstein P, Bogaert L, Bouchier P, Bisschop IJ, Widjojoatmodjo MN, Zahn R, Schuitemaker H, McLellan JS, Langedijk JP. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun. 2015;6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, Zhou T, Graepel KW, Kumar A, Moin S, Boyington JC, Chuang GY, Soto C, Baxa U, Bakker AQ, Spits H, Beaumont T, Zheng Z, Xia N, Ko SY, Todd JP, Rao S, Graham BS, Kwong PD. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwakkenbos MJ, Diehl SA, Yasuda E, Bakker AQ, van Geelen CM, Lukens MV, van Bleek GM, Widjojoatmodjo MN, Bogers WM, Mei H, Radbruch A, Scheeren FA, Spits H, Beaumont T. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med. 2010;16:123–128. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corti D, Bianchi S, Vanzetta F, Minola A, Perez L, Agatic G, Guarino B, Silacci C, Marcandalli J, Marsland BJ, Piralla A, Percivalle E, Sallusto F, Baldanti F, Lanzavecchia A. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature. 2013;501:439–443. doi: 10.1038/nature12442. [DOI] [PubMed] [Google Scholar]
- 18.Magro M, Andreu D, Gomez-Puertas P, Melero JA, Palomo C. Neutralization of human respiratory syncytial virus infectivity by antibodies and low-molecular-weight compounds targeted against the fusion glycoprotein. J Virol. 2010;84:7970–7982. doi: 10.1128/JVI.00447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor G, Stott EJ, Furze J, Ford J, Sopp P. Protective epitopes on the fusion protein of respiratory syncytial virus recognized by murine and bovine monoclonal antibodies. J Gen Virol. 1992;73(Pt 9):2217–2223. doi: 10.1099/0022-1317-73-9-2217. [DOI] [PubMed] [Google Scholar]
- 20.Calder LJ, Gonzalez-Reyes L, Garcia-Barreno B, Wharton SA, Skehel JJ, Wiley DC, Melero JA. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology. 2000;271:122–131. doi: 10.1006/viro.2000.0279. [DOI] [PubMed] [Google Scholar]
- 21.Gilman MS, Moin SM, Mas V, Chen M, Patel NK, Kramer K, Zhu Q, Kabeche SC, Kumar A, Palomo C, Beaumont T, Baxa U, Ulbrandt ND, Melero JA, Graham BS, McLellan JS. Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein. PLoS Pathog. 2015;11:e1005035. doi: 10.1371/journal.ppat.1005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joyce MG, Wheatley AK, Thomas PV, Chuang GY, Soto C, Bailer RT, Druz A, Georgiev IS, Gillespie RA, Kanekiyo M, Kong WP, Leung K, Narpala SN, Prabhakaran MS, Yang ES, Zhang B, Zhang Y, Asokan M, Boyington JC, Bylund T, Darko S, Lees CR, Ransier A, Shen CH, Wang L, Whittle JR, Wu X, Yassine HM, Santos C, Matsuoka Y, Tsybovsky Y, Baxa U, Mullikin JC, Subbarao K, Douek DC, Graham BS, Koup RA, Ledgerwood JE, Roederer M, Shapiro L, Kwong PD, Mascola JR, McDermott AB. Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses. Cell. 2016;166:609–623. doi: 10.1016/j.cell.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Truck J, Ramasamy MN, Galson JD, Rance R, Parkhill J, Lunter G, Pollard AJ, Kelly DF. Identification of antigen-specific B cell receptor sequences using public repertoire analysis. J Immunol. 2015;194:252–261. doi: 10.4049/jimmunol.1401405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parameswaran P, Liu Y, Roskin KM, Jackson KK, Dixit VP, Lee JY, Artiles KL, Zompi S, Vargas MJ, Simen BB, Hanczaruk B, McGowan KR, Tariq MA, Pourmand N, Koller D, Balmaseda A, Boyd SD, Harris E, Fire AZ. Convergent antibody signatures in human dengue. Cell Host Microbe. 2013;13:691–700. doi: 10.1016/j.chom.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson KJ, Liu Y, Roskin KM, Glanville J, Hoh RA, Seo K, Marshall EL, Gurley TC, Moody MA, Haynes BF, Walter EB, Liao HX, Albrecht RA, Garcia-Sastre A, Chaparro-Riggers J, Rajpal A, Pons J, Simen BB, Hanczaruk B, Dekker CL, Laserson J, Koller D, Davis MM, Fire AZ, Boyd SD. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe. 2014;16:105–114. doi: 10.1016/j.chom.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson FW, Collier AM, Clyde WA, Jr, Denny FW. Respiratory-syncytial-virus infections, reinfections and immunity. A prospective, longitudinal study in young children. N Engl J Med. 1979;300:530–534. doi: 10.1056/NEJM197903083001004. [DOI] [PubMed] [Google Scholar]
- 27.Moody MA, Haynes BF. Antigen-specific B cell detection reagents: use and quality control. Cytometry A. 2008;73:1086–1092. doi: 10.1002/cyto.a.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Habibi MS, Jozwik A, Makris S, Dunning J, Paras A, DeVincenzo JP, de Haan CA, Wrammert J, Openshaw PJ, Chiu C. I. Mechanisms of Severe Acute Influenza Consortium, Impaired Antibody-mediated Protection and Defective IgA B-Cell Memory in Experimental Infection of Adults with Respiratory Syncytial Virus. Am J Respir Crit Care Med. 2015;191:1040–1049. doi: 10.1164/rccm.201412-2256OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bornholdt ZA, Turner HL, Murin CD, Li W, Sok D, Souders CA, Piper AE, Goff A, Shamblin JD, Wollen SE, Sprague TR, Fusco ML, Pommert KB, Cavacini LA, Smith HL, Klempner M, Reimann KA, Krauland E, Gerngross TU, Wittrup KD, Saphire EO, Burton DR, Glass PJ, Ward AB, Walker LM. Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science. 2016;351:1078–1083. doi: 10.1126/science.aad5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 32.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, James J, Air GM, Capra JD, Ahmed R, Wilson PC. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyd SD, Gaeta BA, Jackson KJ, Fire AZ, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, Nadeau KC, Egholm M, Miklos DB, Zehnder JL, Collins AM. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J Immunol. 2010;184:6986–6992. doi: 10.4049/jimmunol.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, Wyatt R, Choe H, Farzan M, Kwong PD. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A. 2004;101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan CH, Hadlock KG, Foung SK, Levy S. V(H)1–69 gene is preferentially used by hepatitis C virus-associated B cell lymphomas and by normal B cells responding to the E2 viral antigen. Blood. 2001;97:1023–1026. doi: 10.1182/blood.v97.4.1023. [DOI] [PubMed] [Google Scholar]
- 37.Godoy-Lozano EE, Tellez-Sosa J, Sanchez-Gonzalez G, Samano-Sanchez H, Aguilar-Salgado A, Salinas-Rodriguez A, Cortina-Ceballos B, Vivanco-Cid H, Hernandez-Flores K, Pfaff JM, Kahle KM, Doranz BJ, Gomez-Barreto RE, Valdovinos-Torres H, Lopez-Martinez I, Rodriguez MH, Martinez-Barnetche J. Lower IgG somatic hypermutation rates during acute dengue virus infection is compatible with a germinal center-independent B cell response. Genome Med. 2016;8:23. doi: 10.1186/s13073-016-0276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, Mehta A, Razavi B, Del Rio C, Zheng NY, Lee JH, Huang M, Ali Z, Kaur K, Andrews S, Amara RR, Wang Y, Das SR, O'Donnell CD, Yewdell JW, Subbarao K, Marasco WA, Mulligan MJ, Compans R, Ahmed R, Wilson PC. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand CJ, Taylor WM, Lim S, Huang M, Qu X, Lee JH, Salgado-Ferrer M, Krammer F, Palese P, Wrammert J, Ahmed R, Wilson PC. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med. 2015;7:316ra192. doi: 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu M, Yang G, Wiehe K, Nicely NI, Vandergrift NA, Rountree W, Bonsignori M, Alam SM, Gao J, Haynes BF, Kelsoe G. Polyreactivity and autoreactivity among HIV-1 antibodies. J Virol. 2015;89:784–798. doi: 10.1128/JVI.02378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, Artyomov MN, Pietzsch J, Connors M, Pereyra F, Walker BD, Ho DD, Wilson PC, Seaman MS, Eisen HN, Chakraborty AK, Hope TJ, Ravetch JV, Wardemann H, Nussenzweig MC. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelly RL, Sun T, Jain T, Caffry I, Yu Y, Cao Y, Lynaugh H, Brown M, Vasquez M, Wittrup KD, Xu Y. High throughput cross-interaction measures for human IgG1 antibodies correlate with clearance rates in mice. MAbs. 2015:0. doi: 10.1080/19420862.2015.1043503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y, Roach W, Sun T, Jain T, Prinz B, Yu TY, Torrey J, Thomas J, Bobrowicz P, Vasquez M, Wittrup KD, Krauland E. Addressing polyspecificity of antibodies selected from an in vitro yeast presentation system: a FACS-based, high-throughput selection and analytical tool. Protein Eng Des Sel. 2013;26:663–670. doi: 10.1093/protein/gzt047. [DOI] [PubMed] [Google Scholar]
- 44.Bowley DR, Labrijn AF, Zwick MB, Burton DR. Antigen selection from an HIV-1 immune antibody library displayed on yeast yields many novel antibodies compared to selection from the same library displayed on phage. Protein Eng Des Sel. 2007;20:81–90. doi: 10.1093/protein/gzl057. [DOI] [PubMed] [Google Scholar]
- 45.Wu H, Pfarr DS, Johnson S, Brewah YA, Woods RM, Patel NK, White WI, Young JF, Kiener PA. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J Mol Biol. 2007;368:652–665. doi: 10.1016/j.jmb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 46.McLellan JS, Chen M, Chang JS, Yang Y, Kim A, Graham BS, Kwong PD. Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J Virol. 2010;84:12236–12244. doi: 10.1128/JVI.01579-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foote J, Eisen HN. Kinetic and affinity limits on antibodies produced during immune responses. Proc Natl Acad Sci U S A. 1995;92:1254–1256. doi: 10.1073/pnas.92.5.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batista FD, Neuberger MS. Affinity dependence of the B cell response to antigen: a threshold, a ceiling, and the importance of off-rate. Immunity. 1998;8:751–759. doi: 10.1016/s1074-7613(00)80580-4. [DOI] [PubMed] [Google Scholar]
- 49.Schuster JE, Cox RG, Hastings AK, Boyd KL, Wadia J, Chen Z, Burton DR, Williamson RA, Williams JV. A broadly neutralizing human monoclonal antibody exhibits in vivo efficacy against both human metapneumovirus and respiratory syncytial virus. J Infect Dis. 2015;211:216–225. doi: 10.1093/infdis/jiu307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernie BF, Cote PJ, Jr, Gerin JL. Classification of hybridomas to respiratory syncytial virus glycoproteins. Proc Soc Exp Biol Med. 1982;171:266–271. doi: 10.3181/00379727-171-41509. [DOI] [PubMed] [Google Scholar]
- 51.Cote PJ, Jr, Fernie BF, Ford EC, Shih JW, Gerin JL. Monoclonal antibodies to respiratory syncytial virus: detection of virus neutralization and other antigen-antibody systems using infected human and murine cells. J Virol Methods. 1981;3:137–147. doi: 10.1016/0166-0934(81)90048-3. [DOI] [PubMed] [Google Scholar]
- 52.Walsh EE, Hruska J. Monoclonal antibodies to respiratory syncytial virus proteins: identification of the fusion protein. J Virol. 1983;47:171–177. doi: 10.1128/jvi.47.1.171-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson LJ, Bingham P, Hierholzer JC. Neutralization of respiratory syncytial virus by individual and mixtures of F and G protein monoclonal antibodies. J Virol. 1988;62:4232–4238. doi: 10.1128/jvi.62.11.4232-4238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scopes GE, Watt PJ, Lambden PR. Identification of a linear epitope on the fusion glycoprotein of respiratory syncytial virus. J Gen Virol. 1990;71(Pt 1):53–59. doi: 10.1099/0022-1317-71-1-53. [DOI] [PubMed] [Google Scholar]
- 55.Arbiza J, Taylor G, Lopez JA, Furze J, Wyld S, Whyte P, Stott EJ, Wertz G, Sullender W, Trudel M, et al. Characterization of two antigenic sites recognized by neutralizing monoclonal antibodies directed against the fusion glycoprotein of human respiratory syncytial virus. J Gen Virol. 1992;73(Pt 9):2225–2234. doi: 10.1099/0022-1317-73-9-2225. [DOI] [PubMed] [Google Scholar]
- 56.Lopez JA, Bustos R, Orvell C, Berois M, Arbiza J, Garcia-Barreno B, Melero JA. Antigenic structure of human respiratory syncytial virus fusion glycoprotein. J Virol. 1998;72:6922–6928. doi: 10.1128/jvi.72.8.6922-6928.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med. 2015;21:86–91. doi: 10.1038/nm.3743. [DOI] [PubMed] [Google Scholar]
- 58.National Library of Medicine. doi: 10.1080/15360280801989377. ( NCT02290340, https://clinicaltrials.gov/) [DOI] [PubMed]
- 59.PATH, RSV Vaccine Snapshot. 2016 http://sites.path.org/vaccinedevelopment/files/2016/07/RSV-snapshot-July_13_2016.pdf. [Google Scholar]
- 60.Graham BS, Perkins MD, Wright PF, Karzon DT. Primary respiratory syncytial virus infection in mice. J Med Virol. 1988;26:153–162. doi: 10.1002/jmv.1890260207. [DOI] [PubMed] [Google Scholar]
- 61.Hotard AL, Shaikh FY, Lee S, Yan D, Teng MN, Plemper RK, Crowe JE, Jr, Moore ML. A stabilized respiratory syncytial virus reverse genetics system amenable to recombination-mediated mutagenesis. Virology. 2012;434:129–136. doi: 10.1016/j.virol.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anderson LJ, Hierholzer JC, Stone YO, Tsou C, Fernie BF. Identification of epitopes on respiratory syncytial virus proteins by competitive binding immunoassay. J Clin Microbiol. 1986;23:475–480. doi: 10.1128/jcm.23.3.475-480.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.