Abstract
Transition path sampling simulations have proposed that human heart lactate dehydrogenase (LDH) employs protein promoting vibrations (PPVs) on the femtosecond (fs) to picosecond (ps) time scale to promote crossing of the chemical barrier. This chemical barrier involves both hydride and proton transfers to pyruvate to form l-lactate, using reduced nicotinamide adenine dinucleotide (NADH) as the cofactor. Here we report experimental evidence from three types of isotope effect experiments that support coupling of the promoting vibrations to barrier crossing and the coincidence of hydride and proton transfer. We prepared the native (light) LDH and a heavy LDH labeled with 13C, 15N, and nonexchangeable 2H (D) to perturb the predicted PPVs. Heavy LDH has slowed chemistry in single turnover experiments, supporting a contribution of PPVs to transition state formation. Both the [4-2H]NADH (NADD) kinetic isotope effect and the D2O solvent isotope effect were increased in dual-label experiments combining both NADD and D2O, a pattern maintained with both light and heavy LDHs. These isotope effects support concerted hydride and proton transfer for both light and heavy LDHs. Although the transition state barrier-crossing probability is reduced in heavy LDH, the concerted mechanism of the hydride–proton transfer reaction is not altered. This study takes advantage of triple isotope effects to resolve the chemical mechanism of LDH and establish the coupling of fs-ps protein dynamics to barrier crossing.
Graphical abstract
INTRODUCTION
Enzyme catalysis involves stochastic and ligand-induced dynamic motions of protein structure on time scales of atomic vibrations in femtoseconds (fs) to conformational changes in milliseconds (ms) or longer. Large enzyme structural motions linked to catalytic turnover (kcat) are usually in the ms range and are associated with substrate binding, product release, and conformational sampling within the catalytic cycle.1–4 Chemical transition states have lifetimes corresponding to local atomic vibrational frequencies on the femtosecond (fs) to picosecond (ps) time scale. Recent studies with transition path sampling (TPS) computational methods and experimental approaches support the coupling of fs-ps time scale protein promoting vibrations (PPVs) to chemical barrier crossing.5–19 Experimental investigation of PPVs has relied on measuring “heavy enzyme” kinetic isotope effects (HE-KIEs) on the reaction rates and the chemical steps.12–19 In this approach, a heavy enzyme is generated by labeling all or some of the amino acids with 13C, 15N, and nonexchangeable 2H (D) to perturb the bond vibrations without affecting the electrostatics of the protein (based on the Born–Oppenheimer approximation). Heavy enzymes typically show a normal HE-KIE, i.e., a slower rate for the chemical step than the corresponding light (native) enzyme, interpreted as mass-dependent contributions of PPVs in crossing the chemical barrier.12–15 As an exception, TPS simulations found that Escherichia coli dihydrofolate reductase (ecDHFR), lacks PPVs in catalyzing the hydride transfer reaction.9 Experimental studies with heavy ecDHFR support the absence of PPVs as predicted by the TPS simulations.16–19
Lactate dehydrogenase (LDH) has been the subject of experimental and computational studies of protein dynamics in enzyme catalysis.6,8,10,20–31 LDH catalyzes the final step of anaerobic glycolysis, the reduction of pyruvate to L-lactate using reduced nicotinamide adenine dinucleotide (NADH) as the hydride donor. This reaction is reversible with an equilibrium strongly favoring the reduction of pyruvate to l-lactate. The Michaelis complex of LDH undergoes a stochastic conformational search to find reactive conformations on the ms-ns time scale.22,29 Faster fs–ps protein dynamics are proposed to contribute to the chemical reaction catalyzed by human heart LDH,6,8,10 but coupling of fast motions to chemical barrier crossing has not previously been experimentally tested. The reduction of pyruvate to l-lactate (referred to as the LDH reaction hereafter) involves a hydride transfer from C4 of the dihydronicotinamide ring of NADH to the carbonyl C2 of pyruvate and a proton transfer from a protonated, conserved His residue (His193 in human heart LDH) to the carbonyl oxygen of pyruvate (Figure 1B). Previous quantum mechanics/molecular mechanics (QM/MM) calculations explored the free energy surfaces of the hydride and proton transfer reactions to investigate the reaction mechanism.25–28 Those studies could not resolve concerted or stepwise hydride and proton transfer mechanism as both showed similar energetics.25 QM/MM calculations with the TPS method reported that human heart LDH employs PPVs to promote crossing of the chemical barrier (Figure 1A).6,8,10 In both the light and heavy human heart LDH (l-LDH and h-LDH, respectively), the hydride transfer preceded the proton transfer in reactive trajectories.10 However, h-LDH displayed greater variation and longer average time lags (ca. 200 fs instead of 25 fs for l-LDH) between the hydride and proton transfer.10 There is limited experimental evidence to support concerted or stepwise hydride–proton transfer mechanism for LDHs.32 Here we use multiple isotope effects to determine if the l- and h-LDH enzymes have concerted or stepwise mechanisms.
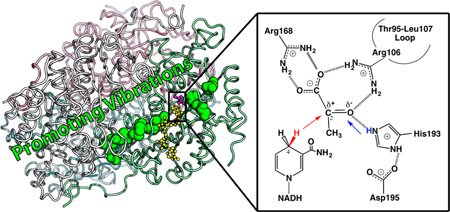
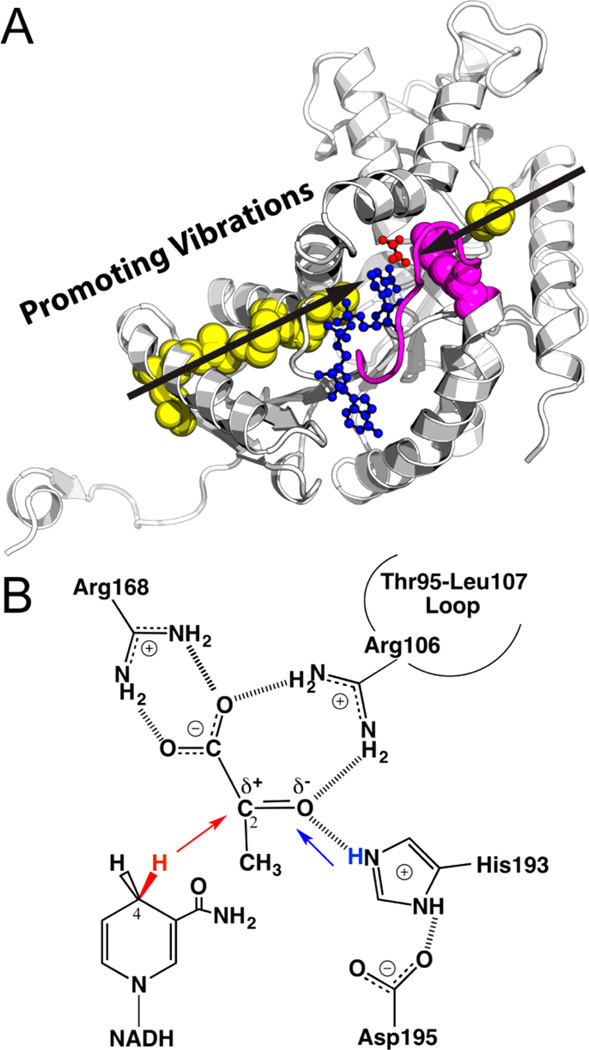
Figure 1.
(A) Active site and promoting vibrations of human heart LDH (one monomer from PDB code 1I0Z). The Thr95-Leu107 loop (magenta) encloses the active site and brings Arg106 (magenta sphere) into hydrogen bond contact with ligands to assist the hydride–proton transfer (illustrated in panel B). Transition path sampling simulations proposed LDH employs promoting vibrations (black arrows) to catalyze the hydride–proton transfer reaction. The distances between the hydride donor (NADH in blue) and acceptor (substrate analogue oxamate in red) are compressed by stochastic promoting vibrations that span the protein architecture (involving residues shown as spheres). (B) The chemical reaction catalyzed by LDH involves a hydride (red) transfer from NADD to the carbonyl carbon of pyruvate and a proton (blue) transfer from a protonated, conserved His residue to the carbonyl oxygen of pyruvate. The present study provides experimental evidence supporting concerted hydride–proton transfer mechanism in human heart LDH, and illustrates mass-dependent barrier crossing, consistent with the presence of promoting vibrations in LDH catalysis.
Concerted and stepwise reaction mechanisms can be experimentally distinguished by dual-label KIE experiments.33 In LDH-catalyzed reaction, the hydride transfer can be studied by measuring the [4R-2H]NADH (NADD) KIE and the proton transfer can be studied by measuring the solvent isotope effect (SIE, with D2O). In a concerted mechanism, if the proton transfer dominates the observed SIE on the kinetic parameters, the D2O SIE will increase the observed NADD KIE (and vice versa) in dual-label experiments. Equal or smaller NADD KIEs in D2O do not necessarily exclude the concerted mechanism due to possible complication of the D2O SIEs by other solvent sensitive steps.32,33 Previous SIEs measured on the steady-state kinetic parameters of a thermophillic LDH from Bacillus stearothermophilus (bsLDH) did not support a concerted mechanism.32 However, TPS simulations predicted that differences in the active sites of bsLDH and human heart LDH can lead to different reaction mechanisms.23
Here we report experiments that support concerted hydride–proton transfer for human heart LDH and demonstrate that mass-sensitive PPVs are coupled to the hydride–proton transfer chemistry. As a prelude to these studies, we optimized expression and purification methods for the l-LDH and h-LDH needed to perform single turnover (STO) experiments to examine the chemical step. We measured triple isotope effects on the STO rates: light vs heavy LDH (HE-KIEs), NADH vs NADD (cofactor KIEs), and H2O vs D2O (SIEs). Synergistic triple isotope effects suggest all three isotope effects arise from the same step (i.e., the hydride–proton transfer chemical step) at low pH. Comparing single KIE/SIE experiments, the dual label experiments showed larger magnitudes for both NADD KIEs and SIEs, suggesting concerted hydride–proton transfer mechanism for both l- and h-LDHs. The normal HE-KIEs support predictions from previous TPS simulations that LDH employs PPVs in catalyzing the hydride–proton transfer reaction.
RESULTS AND DISCUSSION
High-Yield Production of LDH
Despite the interest in studying human LDH for its clinical significance34–41 and enzyme catalysis–dynamics relationship,6,8,10,23,29 the only protocol published for preparing recombinant human LDH required affinity chromatography with oxamate agarose in the presence of NADH.42,43 We designed an LDH expression sequence with a cleavable His6 tag at the N-terminus, which was optimized for expression in Escherichia coli. The His6 tag is cleaved by a tobacco etch virus (TEV) protease,44 which leaves only a single Ser to the N-terminus of LDH sequence (Figure S1 in the Supporting Information). The protocol described in Material and Methods permitted overexpression of LDH in both rich and D2O minimal media, providing up to 130 mg l-LDH or 50 mg h-LDH from 1 L cell culture. The specific activity of l-LDH is 196 units/mg at pH 7, comparable to published data for the native human heart LDH,45,46 suggesting the extra N-terminal Ser does not significantly affect LDH catalysis. The molecular weights of purified l- and h-LDH were determined by protein mass-spectrometry to be 36.7 kDa and 40.8 kDa (11.2% mass increase), respectively, confirming 99.3% heavy isotope enrichment of 13C, 15N, and nonexchangable 2H in h-LDH (Figure S2). This high-yield protocol may be useful for investigating other isozymes of human LDH.
LDH isozyme expression in human tissues provides an example of specialized function to foster metabolic cooperation between organs.36,47–49 Five LDH isozymes exist at different concentrations in the major organ systems, making measurements of extracellular LDH isozymes diagnostic for organ-specific injuries and diseases.34–41,49 The high-yield LDH preparation protocols and mechanistic studies described here add to knowledge of the human heart LDH.
Exposure of Hydride Transfer by Single Turnover Kinetic Experiments
Although computational studies have investigated the hydride and proton transfer chemistry in LDH,6,8,10,21,23,25–28 experimental data is limited. Specific chemical bond changes within a multistep enzymatic reaction can be masked by other steps in steady state kinetic measurements. We measured STO kinetics to reduce steady-state masking of the chemical step, and varied the pH from 4 to 10 at 25 °C. LDH was incubated with 1/5 equiv of NADH, and rapidly mixed with 200 equiv of pyruvate on a stopped-flow instrument. The reaction rates were measured by the decay of the 340 nm UV absorbance following NADH consumption upon the rapid mixing. The time traces of the STO reaction were fit to a single exponential equation (eq 1), where St and S∞ are the 340 nm UV absorbance at time t and after the reaction is complete, respectively; A is the amplitude of the absorbance change due to the reaction; and kSTO is the observed rate constant of the single turnover reaction.
| (1) |
The enzyme was unstable at pH < 4.3 and at pH > 9 (Figure 2A). In the pH range of 4.3–9, kSTO reached a maximum near pH 5 (Figure 2A). The increase of kSTO with decreasing pH is likely due to protonation of His193, the proton donor in the hydride–proton transfer mechanism of LDH (Figure 1B). Although the normal pKa of a His residue is around 6, previous LDH studies suggested the enzyme environment, particularly the negatively charged carboxylate group of Asp168 in close proximity (Figure 1A), stabilizes the protonated state of His193 and thus raises its pKa.50,51 The maximum kSTO (741 ± 18 s−1) was observed at pH 5.5 for NADH, similar to the rate of the chemical step previously estimated for bsLDH (>620 s−1).51
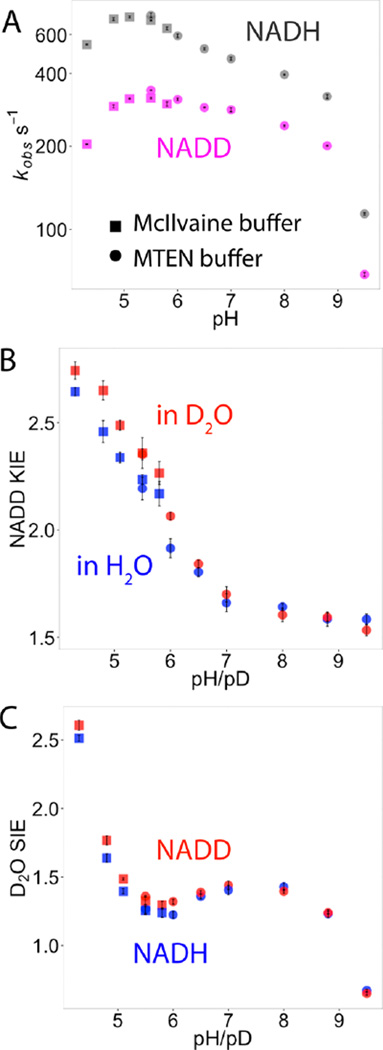
Figure 2.
pH profiles of the single-turnover (STO) rate constants, NADD kinetic isotope effects (KIEs), and D2O solvent isotope effects (SIEs) of human heart LDH at 25 °C. The kinetic experiments were conducted in two buffer systems (square, McIlvaine buffer; round, MTEN buffer; see Materials and Methods) to cover the full pH range where LDH is active. STO rates and KIEs measured in both buffer systems are similar within experimental error at pH/pD = 5.5. (A) The STO rate constants of both NADH (gray) and NADD (magenta) reached maximal values around pH 5. (B, C) Both NADD KIE and D2O SIE increased until precipitation of the enzyme at pH < 4.3. Large NADD KIEs and D2O SIEs at low pH indicate exposure of the hydride–proton transfer chemical step in the STO kinetic experiments. The individual KIEs (blue) with either NADD or D2O labeling are smaller than the NADD and D2O dual-label KIEs (red), supporting concerted hydride–proton transfer mechanism for human heart LDH.
Although the STO rate decreased at pH < 5, NADD KIE increased monotonically at lower pH. The maximum NADD KIE (2.64 ± 0.02, Figure 2B and Table 1) was obtained at pH 4.3. This NADD KIE is similar to the values measured for native and mutant bsLDHs (2.4 to 2.8),52,53 and within the range of intrinsic KIEs calculated for a rabbit muscle LDH (2.44 to 3.38)21 and bsLDH (2.6–5).26,54 The large NADD KIE at low pH suggests the hydride transfer chemistry dominates the STO kinetics under those conditions. The NADD KIE decreased with increasing pH, until it reached a minimum of ca. 1.6 at pH > 8 (Figure 2B).
Table 1.
NADD KIEsa and D2O SIEsb on the STO Rate Constants of l- and h-LDHs at 25 °C in the pH Range 4.3 to 5.8
| pH/pD | |||||||
|---|---|---|---|---|---|---|---|
| isotope effect | enzyme | condition | 4.3 | 4.8 | 5.1 | 5.5 | 5.8 |
| NADD KIEa | l-LDH | in H2O | 2.64 ± 0.02 | 2.46 ± 0.05 | 2.34 ± 0.02 | 2.24 ± 0.02 | 2.17 ± 0.06 |
| in D2O | 2.74 ± 0.04 | 2.65 ± 0.04 | 2.49 ± 0.02 | 2.36 ± 0.07 | 2.26 ± 0.05 | ||
| h-LDH | in H2O | 2.71 ± 0.04 | 2.42 ± 0.06 | 2.26 ± 0.06 | 2.12 ± 0.05 | 2.05 ± 0.01 | |
| in D2O | 2.80 ± 0.03 | 2.63 ± 0.03 | 2.64 ± 0.01 | 2.41 ± 0.06 | 2.18 ± 0.08 | ||
| D2O SIEb | l-LDH | with NADH | 2.51 ± 0.02 | 1.64 ± 0.03 | 1.40 ± 0.02 | 1.26 ± 0.03 | 1.24 ± 0.03 |
| with NADD | 2.61 ± 0.04 | 1.77 ± 0.03 | 1.486 ± 0.008 | 1.33 ± 0.03 | 1.30 ± 0.03 | ||
| h-LDH | with NADH | 2.75 ± 0.04 | 1.96 ± 0.06 | 1.31 ± 0.03 | 1.20 ± 0.04 | 1.23 ± 0.04 | |
| with NADD | 2.84 ± 0.04 | 2.12 ± 0.02 | 1.54 ± 0.02 | 1.368 ± 0.005 | 1.308 ± 0.009 | ||
NADD KIE is the ratio between the STO rate constants of NADH and NADD , for the same enzyme in the same solvent.
D2O SIE is the ratio between the STO rate constants in H2O buffer and in D2O buffer , for the same enzyme with the same cofactor.
Concerted Hydride and Proton Transfer of Human Heart LDH
We probed the hydride–proton transfer mechanism of LDH by dual-label STO experiments to measure the NADD KIEs, D2O SIEs, and their interactions of l-LDH at 25 °C in the pH range of 4.3–9.5. NADD KIEs in both H2O and D2O decreased with increasing pH/pD (Figure 2B). The magnitudes of NADD KIEs in H2O and D2O are within experimental errors at each pH/pD for pH/pD > 6. At lower pH/pD values, NADD KIEs are larger in D2O than in H2O (Figure 2B and Table 1). Similarly, D2O SIEs with NADD as the cofactor are larger than the SIEs with NADH as the cofactor for pH < 6 (Figure 2C and Table 1).
The D2O SIE changed from an inverse value at high pH/pD (0.672 ± 0.009 for NADH at pH/pD = 9.5) to a large normal SIE at low pH/pD (2.51 ± 0.02 for NADH at pH/pD = 4.3). The nonmonotonic pH/pD profile of D2O SIE (Figure 2C) probably involved contributions from other solvent sensitive steps such as protein conformational changes and/or desolvation steps,55 in addition to the proton transfer from His193 to the substrate. However, a large D2O SIE of 2.5 (at pH/pD = 4.3, Figure 2C) indicates the proton transfer makes important contribution to the intrinsic chemical transition state(s).32 Furthermore, if the normal SIEs at pH < 6 were dominated by other solvent sensitive steps, the increased energy barriers of those steps in D2O would make the hydride transfer less rate limiting, thus leading to smaller NADD KIEs in D2O than in H2O. Similarly, the SIEs would also be smaller when NADD is used as a cofactor instead of NADH. Our STO experiments showed larger dual-label isotope effects than the individual NADD KIEs and D2O SIEs for pH/pD < 6 (Figure 2B and C). These results suggest the proton transfer step dominates the observed D2O SIEs and the hydride and proton transfer is concerted under conditions where His193 is predominately protonated.
Promoting Vibrations Affect the Barrier-Crossing Probability but Not the Mechanism of Hydride–Proton Transfer
Previous TPS simulations on LDH were conducted with His193 fully protonated, and the results predicted that heavy isotope labeling of LDH will disrupt the PPVs and thus slow the hydride–proton transfer reaction.10 The simulations also predicted similar reaction paths but different time lags between the hydride and proton transfer reactions for l- and h-LDHs. To test these predictions, we measured the kSTO values and triple isotope effects including the HE-KIE, NADD KIE, and D2O SIE at pH < 6, where the His193 is predominantly protonated and the chemical step is most exposed in the STO kinetics.
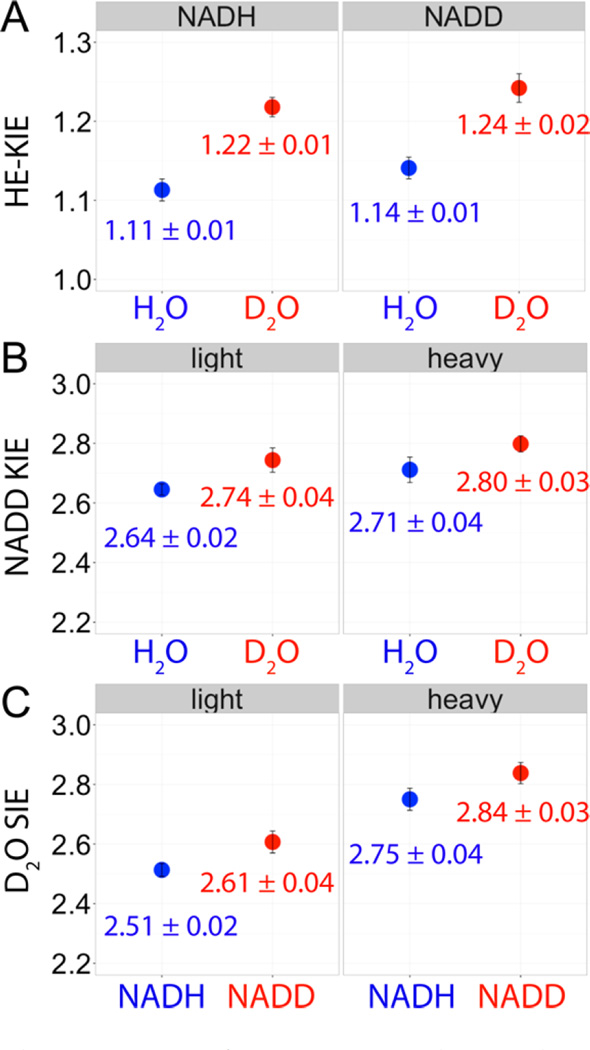
Similar to the NADD KIE and D2O SIE, the HE-KIE also reached a maximum at pH 4.3. Heavy enzyme isotope effects are expected to be small, as they are secondary isotope effects., Experimentally, all the HE-KIEs are small but normal (>1) for pH < 6 (Figure 3). At pH 4.3, the HE-KIE was larger with NADD as the cofactor than NADH, and it was larger in D2O than in H2O (Figure 4A). Due to exchange with solvent deuterium, the relative mass difference (mass h-LDH / l-LDH) decreases slightly in D2O relative to H2O. However, HE-KIE was increased in D2O, where this effect occurs with both NADH and NADD (Figure 4A). The observation that triple isotope effects of heavy LDH, NADD, and D2O are synergistic (Figure 4) suggests all the three isotope effects arise from the same step, i.e., the hydride–proton transfer chemical step, at pH 4.3.
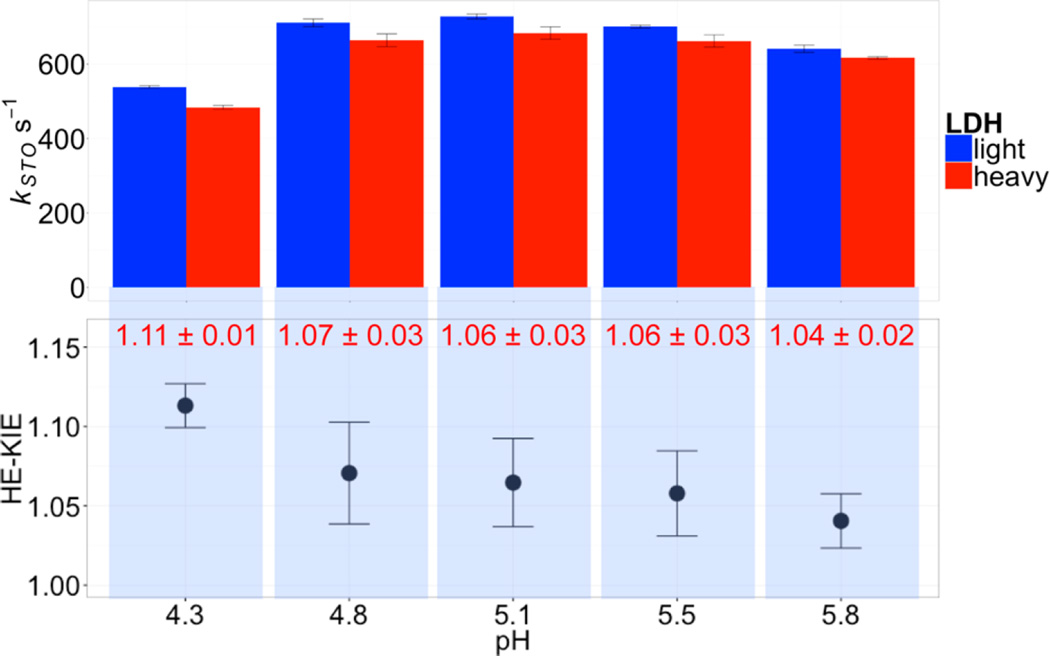
Figure 3.
STO rate constants (upper panel) and heavy enzyme KIEs (HE-KIEs, lower panel) of human heart LDH at 25 °C. HE-KIEs (lower panel) were calculated as the ratio between the STO rate constants of light and heavy LDHs at each pH, with NADH as the cofactor in H2O solvent. The HE-KIEs measured at each pH are shown as red numbers. Heavy LDH shows consistently slower STO rate constants than light LDH in the pH range of 4.3 to 5.8, with the maximal effect observed at the lowest pH (11% slower) where the hydride transfer kinetics is maximally exposed (Figure 2).
Figure 4.
Triple isotope effects measured on the STO rates of human heart LDH at 25 °C, pH/pD 4.3. The heavy enzyme, cofactor (NADD), and solvent (D2O) KIEs are synergistic with each other, suggesting that promoting vibrations, hydride transfer, and proton transfer occur in the same step. The protein mass-modulated promoting vibrations affect the barrier crossing probability but not the concerted mechanism of hydride–proton transfer in human heart LDH.
Compared with the l-LDH, kSTO of h-LDH is ca. 11% slower for the hydride transfer reaction with either NADH or NADD as the cofactor at 25 °C, pH 4.3 (Table 1 and Figure 3). These results support the predictions from previous TPS simulations. The role of promoting vibrations in LDH is similar to the heavy enzyme effects reported for PNP, HIV-1 protease, alanine racemase, and pentaerythritol tetranitrate reductase.10–15 Slower atomic bond vibrations in h-LDH lead to lower barrier-crossing probability for the hydride–proton transfer chemical step in hhLDH-catalyzed reaction.
Similar to l-LDH, h-LDH also showed larger dual-label KIEs than the individual NADD KIEs and D2O SIEs at each pH/pD < 6 (Table 1). Thus, both l- and h-LDHs have similar extents of concerted hydride–proton transfer. The longer time lag between the hydride and proton transfer calculated for h-LDH than for l-LDH, as predicted by TPS simulations,10 does not alter the concerted reaction mechanism, as detected by the dual-label KIE experiments.
Roles of fs–ms Protein Dynamics in LDH Catalysis
Although QM/MM simulations have extensively studied the role of LDH dynamics in the hydride–transfer chemistry,6,8,10,21,23,25–28 there have been limited experimental data characterizing the chemical step or testing the coupling of LDH dynamics to chemistry. Spectroscopic experiments have demonstrated the Michaelis complex of LDH to exist in multiple metastable substates that vary in their propensity toward the chemical step.22 LDH catalysis undergoes ns-µs conformational searches for the rare conformations that are chemically competent, allowing the hydride–proton transfer to occur.56–59 QM/MM simulations suggested slow motions guides the enzyme toward chemically competent conformations by favoring specific fast dynamics.29 For example, closure of the Thr95-Leu107 loop (Figure 1A) on the ms time scale isolates the active site from bulk solvent, which is assisted by protein and water rearrangements on the ns time scale.60 Closure of this loop also brings Arg106 into hydrogen bond contact with the ligands,61 where Arg106 participates in the promoting vibrations on the fs-ps time scale to promote the hydride–proton transfer chemistry (Figure 1).10 The present experimental analysis supports the role of promoting vibrations in the hydride–proton transfer chemistry, which provides preliminary but critical evidence toward understanding the link between slow (ns–ms) and fast (fs–ps) dynamics and their overall contribution to LDH catalysis. Based on the present study and previous data, LDH dynamics on the ns-ms time scale stochastically search for chemically competent states within the Michaelis complexes, followed by fs-ps dynamics that promote passage over the hydride–proton transfer reaction barrier.
CONCLUSIONS
The LDH enzyme demonstrates instructive catalysis–dynamics relationships6,8,10,20–31 and plays an important role in metabolism.36,47–49 We optimized the expression and purification methods to produce human heart LDH at a high yield, which may be adapted to other LDH enzymes to facilitate research with this enzyme family. Heavy enzyme, cofactor, and solvent triple isotope effects on the single turnover rates are synergistic with each other for human heart LDH. Coupling of promoting vibrations of the protein to the chemical step of the catalytic process is consistent with this pattern. The dual-label NADD and D2O isotope effects support concerted mechanisms for both light and heavy LDH. The heavy LDH shows slower single turnover rates than light LDH, demonstrating lower barrier-crossing probability for the hydride–proton transfer chemical step. However, the longer time lag between the hydride and proton transfer in heavy LDH, as predicted by previous TPS simulations, does not cause the system to switch from concerted to stepwise reaction mechanism. The current study adds information to the chemical mechanism of human heart LDH and provides experimental evidence for the role of fs-ps protein dynamics in its catalysis, which has been a missing piece in LDH catalysis–dynamics relationship.
MATERIALS AND METHODS
Materials and Software
[U-13C6-1,2,3,4,5,6,6-2H7]Glucose and [15N]ammonium chloride (NH4Cl) were purchased from Cambridge Isotope Laboratories, Inc. Expression and purification of His-tagged TEV protease followed the published procedure.44 The HisTrap HP column was from GE Healthcare Life Sciences. All other chemicals were purchased from Fisher Scientific or Sigma-Aldrich and used without further purification. All the reagent concentrations refer to the final concentrations in the reaction mixture, unless otherwise specified.
Preparation of l- and h-LDH
A His-tag cleavable by a TEV protease44 was fused to the N-terminal of LDH gene (Figure S1). The His-tag cleavage by TEV protease leaves only a single Ser residue at the beginning of the N-terminus of LDH protein. Since the N-terminus of LDH is unstructured and exposed to solvent (Figure S1), an extra Ser residue is unlikely to affect the enzyme catalysis or dynamics, as evidenced by its similar specific activity (196 units/mg) to published data of native LDH.45,46 The His-tagged LDH gene was optimized for expression in Escherichia coli using GeneGPS Expression Optimization Technology (service provided by DNA2.0), which relied on machine learning algorithms for codon optimization.62 The optimized gene was inserted into a pD451-SR vector, and expressed in BL21 (DE3) cells with 30 mg/L kanamycin. The l-LDH was expressed in Terrific Broth.63,64 The h-LDH was expressed in M63 minimum medium in 2H2O (D2O) supplemented with [U-13C6-1,2,3,4,5,6,6-2H7]glucose and [15N]NH4Cl. Both l- and h-LDH were purified and stored in phosphate buffer in 1H2O to maintain critical hydrogen (1H) bond interactions.
After harvesting and lysing the cells, the cell lysate was loaded onto a HisTrap HP column to purify His-tagged LDH, in 50 mM phosphate buffer with 300 mM sodium chloride (NaCl). His-tagged LDH eluted from the column in the presence of 300 mM imidazole. His-tagged TEV protease was added into purified His-tagged LDH in the ratio of 1:25 (by UV absorbance at 280 nm) to cleave the His-tag of LDH. Upon cleavage, the tag-free LDH was purified again on HisTrap HP column to separate from His-tagged TEV protease and uncleaved LDH. The tag-free LDH eluted from the column without imidazole, and it showed smaller molecular weight than the His-tagged LDH based on SDS-PAGE analysis. The molecular weights of purified tag-free l- and h-LDH were determined to be 36.7 kDa and 40.8 kDa (11.2% increase) by ESI mass-spectrometry, in excellent agreement with theoretical molecular weights based on the amino acid sequence. These results demonstrated the His-tag cleavage was successful, and confirmed 99.3% heavy isotope enrichment of 13C, 15N, and nonexchangable 2H in h-LDH (Figure S2). Both l- and h-LDH were stored as ammonium sulfate suspension at 4 °C until use. The ammonium sulfate suspension of enzymes was dialyzed against 100 mM potassium phosphate buffer at 4 °C prior to kinetic experiments. There was no significant loss of enzyme activity for at least 48 h at 4 °C after the dialysis.
Synthesis of NADD
NADD was prepared by adapting Northrop’s method65 in preparing and storing isotopically labeled NADH. 85 mg of NAD was dissolved in 5 mL of 20 mM ammonium bicarbonate buffer (pH 9.5) followed by the addition of 115 µL of ethanol-d6, 2 mg of alcohol dehydrogenase (360 units/mg), and 25 mg of aldehyde dehydrogenase (1.2 units/mg) under constant stirring at 22 °C. The pH was monitored and adjusted using aqueous sodium hydroxide to maintain pH between pH 9–9.5 and the absorption profile of the reaction mixture was measured every 15 min using a UV–vis spectrophotometer. After 1 h, the temperature of the reaction mixture was raised to 37 °C for an additional 30 min. Afterward, Amicon-4 centrifugal filters (10K MW cutoff) were used to filter out the enzymes from the reaction mixture which was stored briefly at 4 °C prior to purification.
The crude reaction mixture was loaded onto a DE52 anionic exchange column equilibrated with 20 mM ammonium bicarbonate buffer (pH 9.5) using FPLC. The crude NADD product was eluted off the column using a linear gradient of 20 mM ammoninium bicarbonate buffer containing 100 mM sodium chloride (pH 9.5) and fractioned manually into 5–10 mL aliquots. Fractions with an absorption ratio of 260 nm/340 nm <2.3 were collected, pooled together, and lyophilized. The isotope purity of the NADD was assessed using NMR and found to be greater than 99.6% relative to the amount of NADH.
Single-Turnover Experiments
The single turnover experiments were conducted at 25 °C on an Applied Photophysics model SX20 stopped-flow instrument, with a 20 µL cell, which has a dead time of 2 ms. Each data set is an average of at least 10 measurements under the same conditions. The single turnover kinetics were measured by monitoring the decrease in UV absorbance of NADH (or NADD) at 340 nm that follows the hydride transfer. LDH was preincubated with NADH or NADD before rapid mixing with pyruvate. The final reaction mixture contained 25 µM LDH, 5 µM NADH or NADD, and 5 mM pyruvate. Decrease in 340 nm Absorbance upon rapid mixing was fit to the single exponential equation (eq 1) to obtain the observed rate constant (kSTO) under different pH conditions.
The kSTO of light LDH with NADH and NADD were conducted in McIlvaine buffer (citrate-phosphate buffer) for pH ≤ 5.8, and in 50 mM MES, 25 mM Tris, 25 mM ethanolamine, and 100 mM sodium chloride (“MTEN buffer”66) for pH ≥ 5.5. The pH values were measured separately by mixing all the reagents, except the protein, in the same amounts as in the final reaction mixture used for STO experiments.
Triple Kinetic Isotope Effects
The heavy enzyme, substrate, and solvent triple kinetic isotope effects were calculated by comparing the STO rates of light vs heavy LDH, NADH vs NADD, in H2O vs D2O, respectively. When D2O is used as solvent, the substrate stock solutions were prepared in D2O, and the enzymes were dialyzed against 100 mM potassium phosphate buffer in D2O and incubated in D2O buffer overnight prior to the experiments. The glass electrode of the pH meter was immersed in D2O for 5 min prior to pD measurements. The pD value was calculated by adding 0.4 to the pH meter reading to account for the solvent isotope effect on the electrode response.
The pH/pD dependence of STO rates and KIEs of l-LDH suggests the hydride transfer chemical step is better exposed at lower pH values (pH < 6). Thus, the STO kinetics of l-LDH and h-LDH were compared under those conditions (Table 1).
Supplementary Material
Acknowledgments
We thank Prof. Robert Callender for his insightful discussions. This work was supported by NIH research program Project GM068036 and training Grant K121GM102779.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Design of His-tagged LDH; protein mass spectra of l- and h-LDHs; table listing the single turnover rate constants for l- and h-LDHs (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Boehr DD, McElheny D, Dyson HJ, Wright PE. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 2.Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, Hubner CG, Kern D. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 3.Callender R, Dyer RB. Acc. Chem. Res. 2015;48:407–413. doi: 10.1021/ar5002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albesa-Jove D, Guerin ME. Curr. Opin. Struct. Biol. 2016;40:23–32. doi: 10.1016/j.sbi.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Núñez S, Antoniou D, Schramm VL, Schwartz SD. J. Am. Chem. Soc. 2004;126:15720–15729. doi: 10.1021/ja0457563. [DOI] [PubMed] [Google Scholar]
- 6.Basner JE, Schwartz SD. J. Am. Chem. Soc. 2005;127:13822–13831. doi: 10.1021/ja043320h. [DOI] [PubMed] [Google Scholar]
- 7.Saen-Oon S, Quaytman-Machleder S, Schramm VL, Schwartz SD. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16543–16548. doi: 10.1073/pnas.0808413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quaytman SL, Schwartz SD. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12253–12258. doi: 10.1073/pnas.0704304104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dametto M, Antoniou D, Schwartz SD. Mol. Phys. 2012;110:531–536. doi: 10.1080/00268976.2012.655337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masterson JE, Schwartz SD. J. Phys. Chem. A. 2013;117:7107–7113. doi: 10.1021/jp400376h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antoniou D, Ge X, Schramm VL, Schwartz SD. J. Phys. Chem. Lett. 2012;3:3538–3544. doi: 10.1021/jz301670s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva RG, Murkin AS, Schramm VL. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18661–18665. doi: 10.1073/pnas.1114900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kipp DR, Silva RG, Schramm VL. J. Am. Chem. Soc. 2011;133:19358–19361. doi: 10.1021/ja209391n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toney MD, Castro JN, Addington TA. J. Am. Chem. Soc. 2013;135:2509–2511. doi: 10.1021/ja3101243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pudney CR, Guerriero A, Baxter NJ, Johannissen LO, Waltho JP, Hay S, Scrutton NS. J. Am. Chem. Soc. 2013;135:2512–2517. doi: 10.1021/ja311277k. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, Antoniou D, Schwartz SD, Schramm VL. Biochemistry. 2016;55:157–166. doi: 10.1021/acs.biochem.5b01241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luk LY, Ruiz-Pernia JJ, Adesina AS, Loveridge EJ, Tunon I, Moliner V, Allemann RK. Angew. Chem., Int. Ed. 2015;54:9016–9020. doi: 10.1002/anie.201503968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Singh P, Czekster CM, Kohen A, Schramm VL. J. Am. Chem. Soc. 2014;136:8333–8341. doi: 10.1021/ja501936d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luk LY, Javier Ruiz-Pernia J, Dawson WM, Roca M, Loveridge EJ, Glowacki DR, Harvey JN, Mulholland AJ, Tuñón I, Moliner V, Allemann RK. Proc. Natl. Acad. Sci. U. S. A. 2013;110:16344–16349. doi: 10.1073/pnas.1312437110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yadav A, Jackson RM, Holbrook JJ, Warshel A. J. Am. Chem. Soc. 1991;113:4800–4805. [Google Scholar]
- 21.Świderek K, Tuñón I, Martí S, Moliner V. ACS Catal. 2015;5:1172–1185. doi: 10.1021/cs501704f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng H, Vu DV, Clinch K, Desamero R, Dyer RB, Callender R. J. Phys. Chem. B. 2011;115:7670–7678. doi: 10.1021/jp2015929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quaytman SL, Schwartz SD. J. Phys. Chem. A. 2009;113:1892–1897. doi: 10.1021/jp804874p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhadin N, Gulotta M, Callender R. Biophys. J. 2008;95:1974–1984. doi: 10.1529/biophysj.108.132464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moliner V, Williams IH. Chem. Commun. 2000:1843–1844. [Google Scholar]
- 26.Turner AJ, Moliner V, Williams IH. Phys. Chem. Chem. Phys. 1999;1:1323–1331. [Google Scholar]
- 27.Ranganathan S, Gready JE. J. Phys. Chem. B. 1997;101:5614–5618. [Google Scholar]
- 28.Moliner V, Turner AJ, Williams IH. Chem. Commun. 1997;0:1271–1272. [Google Scholar]
- 29.Pineda JRET, Antoniou D, Schwartz SD. J. Phys. Chem. B. 2010;114:15985–15990. doi: 10.1021/jp1071296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan X, Schwartz SD. J. Phys. Chem. B. 2015;119:5430–5436. doi: 10.1021/acs.jpcb.5b01840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan X, Schwartz SD. J. Phys. Chem. B. 2016;120:6612–6620. doi: 10.1021/acs.jpcb.6b05119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie M, Seravalli J, Huskey WP, Schowen KB, Schowen RL. Bioorg. Med. Chem. 1994;2:691–695. doi: 10.1016/0968-0896(94)85019-4. [DOI] [PubMed] [Google Scholar]
- 33.O’Leary MH. Annu. Rev. Biochem. 1989;58:377–401. doi: 10.1146/annurev.bi.58.070189.002113. [DOI] [PubMed] [Google Scholar]
- 34.Huijgen HJ, Sanders GT, Koster RW, Vreeken J, Bossuyt PM. Eur. J. Clin. Chem. Clin. Biochem. 1997;35:569–579. [PubMed] [Google Scholar]
- 35.MedlinePlus Medical Encyclopedia. Bethesda, MD: U.S. National Library of Medicine; 2014. [Google Scholar]
- 36.Drent M, Cobben NA, Henderson RF, Wouters EF, van Dieijen-Visser M. Eur. Respir. J. 1996;9:1736–1742. doi: 10.1183/09031936.96.09081736. [DOI] [PubMed] [Google Scholar]
- 37.Nieder C, Marienhagen K, Dalhaug A, Aandahl G, Haukland E, Pawinski A. Clin. Oncol. (R. Coll. Radiol.) 2014;26:447–452. doi: 10.1016/j.clon.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 38.Kawamoto M. Cancer. 1994;73:1836–1841. doi: 10.1002/1097-0142(19940401)73:7<1836::aid-cncr2820730710>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 39.Rotenberg Z, Weinberger I, Sagie A, Fuchs J, Davidson E, Sperling O, Agmon J. Clin. Chem. 1988;34:668–670. [PubMed] [Google Scholar]
- 40.Fogh-Andersen N, Sorensen P, Moller-Petersen J, Ring TJ. Clin. Chem. Clin. Biochem. 1982;20:291–294. [PubMed] [Google Scholar]
- 41.Carda-Abella P, Perez-Cuadrado S, Lara-Baruque S, Gil-Grande L, Nunez-Puertas A. Cancer. 1982;49:80–83. doi: 10.1002/1097-0142(19820101)49:1<80::aid-cncr2820490118>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 42.O’Carra P, Barry S. FEBS Lett. 1972;21:281–285. doi: 10.1016/0014-5793(72)80183-2. [DOI] [PubMed] [Google Scholar]
- 43.Read JA, Winter VJ, Eszes CM, Sessions RB, Brady RL. Proteins: Struct., Funct., Genet. 2001;43:175–185. doi: 10.1002/1097-0134(20010501)43:2<175::aid-prot1029>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 44.Blommel PG, Fox BG. Protein Expression Purif. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilkinson JH, Withycombe WA. Biochem. J. 1965;97:663–668. doi: 10.1042/bj0970663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nisselbaum JS, Bodansky O. J. Biol. Chem. 1961;236:323–327. [PubMed] [Google Scholar]
- 47.Berg JM, Tymoczko JL, Stryer L. Biochemistry. 5th. New York: W H Freeman; 2002. [Google Scholar]
- 48.Rider CC, Taylor CB. Isoenzymes in Metabolic Regulation. Vol. 5. New York: Springer; 1980. [Google Scholar]
- 49.Eventoff W, Rossmann MG, Taylor SS, Torff HJ, Meyer H, Keil W, Kiltz HH. Proc. Natl. Acad. Sci. U. S. A. 1977;74:2677–2681. doi: 10.1073/pnas.74.7.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holbrook JJ, Ingram VA. Biochem. J. 1973;131:729–738. doi: 10.1042/bj1310729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clarke AR, Wilks HM, Barstow DA, Atkinson T, Chia WN, Holbrook JJ. Biochemistry. 1988;27:1617–1622. doi: 10.1021/bi00405a034. [DOI] [PubMed] [Google Scholar]
- 52.Wilks HM, Halsall DJ, Atkinson T, Chia WN, Clarke AR, Holbrook JJ. Biochemistry. 1990;29:8587–8591. doi: 10.1021/bi00489a013. [DOI] [PubMed] [Google Scholar]
- 53.Clarke AR, Wigley DB, Chia WN, Barstow D, Atkinson T, Holbrook JJ. Nature. 1986;324:699–702. doi: 10.1038/324699a0. [DOI] [PubMed] [Google Scholar]
- 54.Ferrer S, Tunon I, Marti S, Moliner V, Garcia-Viloca M, Gonzalez-Lafont A, Lluch JM. J. Am. Chem. Soc. 2006;128:16851–16863. doi: 10.1021/ja0653977. [DOI] [PubMed] [Google Scholar]
- 55.Quinn DM, Sutton LD. In: Enzyme Mechanism from Isotope Effects. Cook PF, editor. Boca Raton, FL: CRC Press, Inc.; 1991. pp. 73–126. [Google Scholar]
- 56.Reddish MJ, Peng H-L, Deng H, Panwar KS, Callender R, Dyer RB. J. Phys. Chem. B. 2014;118:10854–10862. doi: 10.1021/jp5050546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng H-L, Deng H, Dyer RB, Callender R. Biochemistry. 2014;53:1849–1857. doi: 10.1021/bi500215a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nie B, Deng H, Desamero R, Callender R. Biochemistry. 2013;52:1886–1892. doi: 10.1021/bi3017125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deng H, Vu DV, Clinch K, Desamero R, Dyer RB, Callender R. J. Phys. Chem. B. 2011;115:7670–7678. doi: 10.1021/jp2015929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pineda JR, Callender R, Schwartz SD. Biophys. J. 2007;93:1474–1483. doi: 10.1529/biophysj.107.106146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clarke AR, Wigley DB, Chia WN, Barstow D, Atkinson T, Holbrook JJ. Nature. 1986;324:699–702. doi: 10.1038/324699a0. [DOI] [PubMed] [Google Scholar]
- 62.Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, Gustafsson C. PLoS One. 2009;4:e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Redei GP. Encyclopedia of Genetics, Genomics, Proteomics and Informatics. Springer Netherlands: Dordrecht; 2008. pp. 1950–1950. [Google Scholar]
- 64.Tartoff KD, Hobbs CA. Bethesda Research Labs Focus. 1987;9:12. [Google Scholar]
- 65.Northrop DB, Duggleby RG. Anal. Biochem. 1987;165:362–364. doi: 10.1016/0003-2697(87)90282-x. [DOI] [PubMed] [Google Scholar]
- 66.Ellis KJ, Morrison JF. Methods Enzymol. 1982;87:405–426. doi: 10.1016/s0076-6879(82)87025-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.