Abstract
Plant biomass has become an important source of bio-renewable energy in modern society. The molecular structure of plant cell walls is difficult to characterize by most atomic-resolution techniques due to the insoluble and disordered nature of the cell wall. Solid-state NMR (SSNMR) spectroscopy is uniquely suited for studying native hydrated plant cell walls at the molecular level with chemical resolution. Significant progress has been made in the last five years to elucidate the molecular structures and interactions of cellulose and matrix polysaccharides in plant cell walls. These studies have focused on primary cell walls of growing plants in both the dicotyledonous and grass families, as represented by the model plants Arabidopsis thaliana, Brachypodium distachyon, and Zea mays. To date, these SSNMR results have shown that 1) cellulose, hemicellulose, and pectins form a single network in the primary cell wall; 2) in dicot cell walls, the protein expansin targets the hemicellulose-enriched region of the cellulose microfibril for its wall-loosening function; and 3) primary wall cellulose has polymorphic structures that are distinct from the microbial cellulose structures. This article summarizes these key findings, and points out future directions of investigation to advance our fundamental understanding of plant cell wall structure and function.
Keywords: Cellulose, matrix polysaccharide, expansin, lignin, magic-angle spinning, multidimensional correlation, structural polymorphism, cellulose-pectin interactions
Graphical Abstract
1. Plant cell walls: an emerging system in energy and biomaterials
Plants have been a fundamental component of human society for many millennia, providing the source of food, energy, construction and clothing. In recent years, the necessity to reduce our dependency on fossil fuels has made the conversion of plant biomass to an economically viable renewable energy an important area of research. Lignocellulose materials from plant cell walls, which are not a food source, can be converted into solid fuel, liquid fuels, or biogas [1]. A better understanding of the plant cell wall structure on the molecular level is important for this effort, by giving insight into how to increase cell wall mass per hectare, how to reduce cell wall recalcitrance to enzymatic degradation, and how to optimize cell wall composition to increase the yield of fermentation of monosaccharides to fuel [1–4]. Elongating plants cells are surrounded by a primary cell wall whose composition differs from that of secondary cell walls when plants cease growth. Understanding the molecular structural basis for how primary cell walls provide both mechanical strength and extensibility to the cell is thus also a fundamental question of interest in plant sciences.
The molecular structures and intermolecular interactions of plant cell wall polymers have been difficult to study using microscopy and diffraction methods due to the insoluble and mixture nature of the cell wall. Recently, magic-angle-spinning (MAS) multidimensional correlation SSNMR spectroscopy began to be employed to investigate the structure, dynamics, and intermolecular interactions of wall polysaccharides and proteins in important model plants [5]. This SSNMR approach is made possible by the ease of 13C enrichment of plants. By growing plants in liquid culture in the dark with 13C-labeled glucose as the only carbon source, ~100% 13C enrichment of the plants can be achieved [6]. Both the dicot model plant, Arabidopsis thaliana, and two grass model plants, Brachypodium distachyon and Zea mays, have been investigated, and are further complemented by analysis of genetic mutants of Arabidopsis and partially extracted Arabidopsis cell walls with altered physical and chemical properties. The choice of these model plants with fully sequenced genomes and extensive collections of mutants facilitates NMR structural studies, and the focus on primary cell walls makes the structural findings independent of tissue type. This review summarizes the complexity of plant cell walls, key findings from the SSNMR results to date, and future topics of interest in plant cell wall research that are amenable to SSNMR spectroscopy.
2. Challenges: structural and dynamical heterogeneity of plant cell walls
A major challenge in SSNMR studies of plant cell walls is the structural heterogeneity of the cell wall. This heterogeneity is manifested on three levels. First, the constituent polymers and intermolecular networks differ between primary and secondary cell walls, and between dicots and grasses [7]. Cellulose, the main polysaccharide in all plant cell walls, exists as relatively thin microfibrils of 3–5 nm diameter in primary walls but form larger bundles of 10–20 nm diameters in secondary cell walls [8]. Although cellulose is the most crystalline component of the cell wall, the packing and conformation of the β (1–>4) D-glucan chains in the microfibril are still not well understood because of the extensive interaction of cellulose microfibrils with matrix polysaccharides. This feature does not exist in cellulose produced by algae, bacteria and marine animals, which do not interact with other biopolymers and are thus highly crystalline. The composition of matrix polysaccharides also differs significantly between different cell walls. For example, the main hemicelluloses are xyloglucan (XyG) in dicot primary walls but glucuronoarabinoxylan (GAX) and mixed-linkage glucan (MLG) in grass cell walls. Both dicot and grass cell walls also contain polyanionic pectins and structural proteins, but their amount is much lower in grasses. The secondary cell walls are distinguished from primary cell walls by having xylans and glucomannans as the main matrix polysaccharides and by possessing lignin [7], a highly crosslinked phenolic polymer that interacts with cellulose to render secondary cell walls recalcitrant to chemical pretreatment and enzymatic degradation [9, 10]. These chemical compositional differences complicate resonance assignment of solid-state NMR spectra.
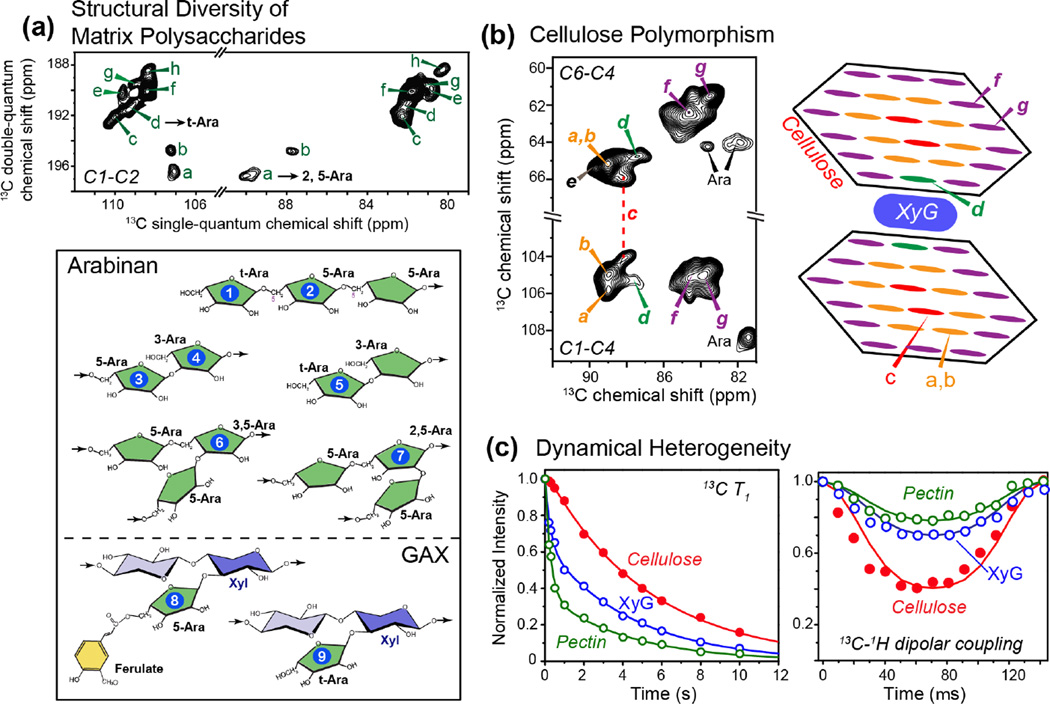
The second level of structural heterogeneity is the fact that each type of sugar can have multiple linkages with other sugars in matrix polysaccharides. Fortunately, under favorable conditions, these linkages can be resolved in multidimensional MAS spectra. For example, arabinose is present in rhamnogalacturonan (RG) I and GAX, which undergo large-amplitude motions to give rise to narrow 13C linewidths of 0.2–0.5 ppm at high magnetic fields [5]. In the Brachypodium cell wall, we resolved nine sets of arabinose signals and five sets of xylose (Xyl) signals [11]. The arabinose signals can be assigned to different covalent linkages at C1, C2, C3, and C5 positions of the sugar (Fig. 1a), depending on whether the arabinose resides in GAX or RG I. Similarly, the multiple sets of Xyl chemical shifts can be assigned to different linkages and substitutions of Xyl in the GAX xylan backbone and in the XyG sidechains. For example, Xyl structures can include t-Xyl in GAX, 3,4-Xyl with Ara substitution at the O3 position, and t-Xyl in XyG, all of which display distinct chemical shifts.
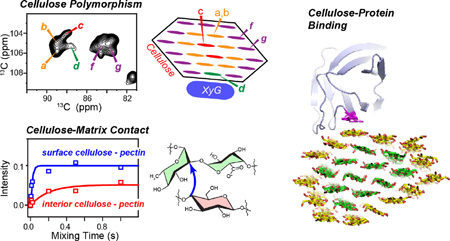
Figure 1.
Structural and dynamical heterogeneity of primary plant cell walls. (a) Structural heterogeneity of arabinose in Brachypodium cell walls [11]. Eight sets of arabinose signals (a–h) are resolved (top), many of which can be assigned to the different linkage types shown at the bottom. (b) Conformational polymorphism of cellulose in Brachypodium cell walls [23]. Seven sets of cellulose signals are partially resolved, with five forms attributed to interior crystalline cellulose (type a–e) and two types to surface cellulose (type f and g). Type-d cellulose can be assigned to XyG-proximal cellulose and is unique to plant cell walls. (c) Representative 13C T1 relaxation data (left) and 13C-1H dipolar coupling data (right) of cellulose (red), hemicellulose XyG (blue) and pectins (green) in Arabidopsis cell walls. Cellulose is mostly rigid while pectins and hemicellulose are partly mobile. The pectin 13C T1 relaxation is double-exponential, with the slow component (~ 5 s) resembling the cellulose 13C T1, indicating that this slow relaxation originates from cellulose-containing pectin segments. The fast-decaying component of ~0.5 s likely results from interfibrillar pectins [15].
Third, each cell-wall polysaccharide can exhibit polymorphic structures. For example, the cellulose microfibrils of primary cell walls have been estimated to contain between 18 and 36 hydrogen-bonded and parallel packed chains, each of which containing tens of thousands of D-glucose rings [12]. Structural heterogeneity perpendicular to the microfibril axis exists due to the distinct environments of interior chains and surface chains, and conformational heterogeneity along the chain axis is also suspected [5]. Thus, it is not surprising that 13C MAS spectra of primary wall cellulose show relatively broad linewidths of 2–3 ppm at low magnetic fields of 9.4–14.1 Tesla. However, at higher fields of 18.8–21.1 Tesla, the linewidths decrease to 0.7–1.0 ppm, allowing the resolution of at least seven sets of chemical shifts in 2D 13C-13C correlation spectra (Fig. 1b). This high-field resolution enhancement indicates that the cellulose linewidths are not entirely inhomogeneous but contain significant homogeneous contributions due to 13C-13C scalar couplings and residual 13C-13C and 13C-1H dipolar couplings. Interestingly, for 11% 13C-enriched wood cellulose, spin-echo-detected 13C T2 values have been reported to be 15–20 times longer than the apparent T2’s, indicating that the linewidths of this wood cellulose are predominantly inhomogeneous and will not decrease at higher magnetic fields [13]. This means that the high-field resolution enhancement of the 13C-labeled primary wall cellulose is due to the 100% 13C enrichment, which increases the contribution of field-independent dipolar and scalar couplings to linewidths. In addition, the linewidth differences also imply that the conformation distribution of primary wall cellulose is narrower than that of secondary wall cellulose. Overall, these results show that high-field multidimensional SSNMR spectroscopy can significantly overcome the challenge of cell-wall heterogeneity and resolve the signals of the multitude of structurally polymorphic polysaccharides in intact primary cell walls.
The structural polymorphism of the wall polysaccharides implies heterogeneous molecular motions. Cellulose is the rigid scaffold of the cell wall, as confirmed by long NMR relaxation times and near-rigid-limit dipolar order parameters. In comparison, matrix polysaccharides are much more mobile. For example, while 13C T1 relaxation times of cellulose are 5–10 s at 14.1–18.8 Tesla, matrix polysaccharides exhibit much shorter 13C T1 relaxation times of ~1 s [6, 14, 15]. Interestingly, the pectin T1 relaxation is heterogeneous with at least two components: a fast component of ~0.4 s and a slow component of ~5 s (Fig. 1c). This strongly suggests that some pectins fill the interfibrillar space and are highly dynamic while other pectin segments are immobilized by contact with cellulose. This in turn implies that pectins may contribute to the crosslinking of cellulose microfibrils in the cell wall. In addition to relaxation times, the dipolar order parameters of wall polysaccharides also differ substantially, from ~0.9 for cellulose to 0.4–0.6 for matrix polysaccharides. In the Brachypodium cell wall, the dipolar order parameters of GAX are ~0.4, which are smaller than those of Arabidopsis matrix polysaccharides. Thus, GAX in grass cell walls is more mobile than matrix polysaccharides in dicot walls [11]. The dynamic heterogeneity of wall polysaccharides allows the use of relaxation and dipolar filters to suppress either the mobile or the rigid component’s signals to simplify the NMR spectra [11, 16]. However, the mobility of interfibrillar polysaccharides makes it difficult to measure the distances of these matrix polysaccharide segments to cellulose. Thus, structural interpretations of intermolecular cross peaks should take into account this bimodal nature of the matrix polysaccharides.
3. Recent progress in investigating plant cell wall structure by SSNMR
3.1 Structural polymorphism of plant cellulose
Cellulose is the most abundant biopolymer on earth: it is the dominant component of plant cell walls, whose estimated annual worldwide production is ~160 billion metric tons [17]. Highly crystalline cellulose of bacteria, algae, and marine animals have been extensively studied using X-ray diffraction, neutron and SSNMR [18–21]. These studies indicate the coexistence of two major allomorphs, Iα and Iβ, in these native cellulose [18]. Crystal structures [19, 22] show that the Iα allomorph is composed of identical chains with two alternating glucose units while the Iβ allomorph contains two types of chains in alternating sheets. In comparison, plant cell wall cellulose has been more difficult to characterize because of its low crystallinity and the difficulty of isolating these microfibrils without perturbing their native structure.
Our recent analysis of the high-field 2D 13C-13C correlation MAS spectra revealed that plant primary-wall cellulose is structurally polymorphic and differs from the Iα and Iβ structures of algal, bacterial and animal cellulose. This polymorphism is manifested as seven sets of 13C chemical shifts [23], all of which differ significantly from the Iα and Iβ chemical shifts. Five of these conformations (types a–e) can be assigned to interior cellulose, while two types (f and g) can be assigned to surface cellulose [23]. An additional minor surface cellulose (h) is identified in the maize cell wall but is absent or unresolved in Arabidopsis and Brachypodium cell walls. Each cellulose form has at least one 13C chemical shift that differs from the chemical shifts of the other forms by more than 1.0 ppm. One of the reasons for these distinct chemical shifts is the C6 hydroxymethyl conformation. DFT calculations of 13C chemical shifts indicate that the C6 exocyclic group mainly adopts the trans-gauche (tg) conformation in four of the five types of interior cellulose, except for type d, which can adopt both tg and gt conformations.
We attribute the structural polymorphism of plant cell wall cellulose to matrix polysaccharides, which are expected to affect chain packing, hydrogen bonding and local conformation. [23, 24]. The chemical shift multiplicity is present in Arabidopsis, Brachypodium and Zea mays cell walls, all of which share the same sets of 13C chemical shifts (Fig. 1b); thus the cellulose polymorphism is conserved among many plants [5]. Interestingly, two genetic mutants of Arabidopsis, one removing XyG and the other mutating cellulose synthase [4, 6], showed a smaller number of cellulose cross peaks but did not alter the chemical shifts of the remaining peaks, suggesting that the cellulose structural distribution is sufficiently robust to resist partial changes in the matrix polysaccharide composition of the cell wall and changes in cellulose synthase activity.
The spatial locations of the different cellulose structures have been probed from intermolecular cross peaks in long-mixing-time spectra and from water-edited 2D correlation spectra [23]. These data indicate that the different types of cellulose mix radially in each microfibril, instead of being separated into different microfibrils or domains along the fibril axis (Fig. 1b). Cellulose f and g interact with pectins on the microfibril surface and are well hydrated, while cellulose a and b do not contact matrix polysaccharides but are in contact with surface cellulose and water. Cellulose c exhibits much weaker cross peaks with both surface cellulose and water, thus is embedded in the core of microfibrils. Finally, cellulose d shows strong cross peaks with surface cellulose but is poorly hydrated. This cellulose d is unique to plants: its 13C chemical shifts differ significantly from those of all other published celluloses; it was previously observed to interact with hemicellulose and is targeted by the wall-loosening protein expansin [25]; and in the XyG-depleted Arabidopsis mutant xxt1xxt2xxt5, cellulose d signals disappear [23]. Thus, cellulose d is important for wall mechanical properties.
These results lead to a three-layer cross-sectional model of the cellulose microfibril in plant primary walls, where the core is separated from the surface by an intermediate layer of glucan chains (Fig. 1b). This structural model is consistent with the relative intensities of surface and interior cellulose in quantitative 13C NMR spectra, which shows that the average cellulose microfibril in primary walls contains at least 24 chains [5]. Since recent biochemical data, electron microscopy and molecular dynamics simulation have converged on the model that the cellulose synthase complex contains 18 enzymes [26–28], the larger cellulose chain number deduced from the SSNMR data suggests that multiple cellulose microfibrils coalesce [29], as also suggested by AFM data of onion cell walls [30, 31].
3.2 Cellulose-matrix polysaccharide interactions: changing the paradigm of wall structure
An important question in plant cell wall research is how cellulose interacts with matrix polysaccharides to form a load-bearing network that not only provides stiffness to plant cells but is also capable of loosening during rapid plant growth [32]. The prevailing structural model, based on extraction and in-vitro binding assays, depicts cellulose microfibrils as hydrogen-bonded to and crosslinked by hemicellulose, and this cellulose-hemicellulose network is embedded in a second phase formed by well-hydrated anionic pectins. Thus, in this “tethered network” model, hemicellulose is an essential structural component of the wall and a target for wall loosening [33]. However, long-mixing-time 2D and 3D SSNMR spectra show few XyG-cellulose cross peaks, suggesting that XyG interacts with cellulose only at limited positions [6, 14]. This finding subsequently found support in biomechanical assays where an XyG-specific endoglucanase (XEG) that hydrolyzes the majority of XyG failed to cause wall creep, while an enzyme (Cel12A) that hydrolyzes both cellulose and a minor component of XyG that is resistant to XEG digestion induced significant creep. This result indicates that only a minor fraction of XyG is involved in wall mechanics [34]. The resulting “biomechanical hotspot” model depicts cellulose as interacting with XyG at limited sites to cause wall loosening [34, 35].
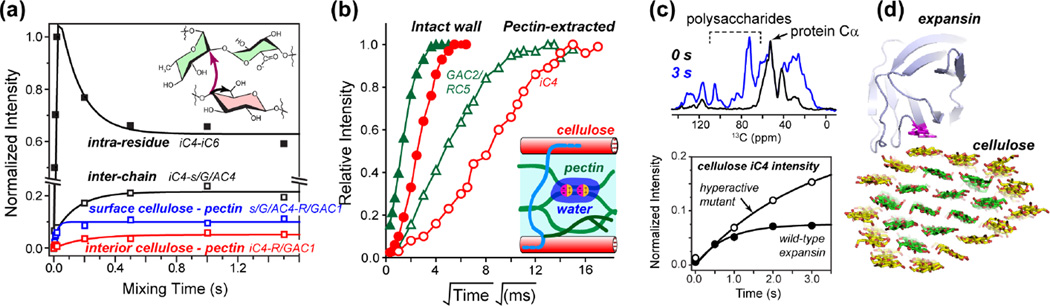
In contrast to the sparseness of cellulose-XyG cross peaks, the 2D and 3D correlation spectra show many cellulose-pectin cross peaks, whose equilibrium intensities represent 25–50% of inter-chain cross peaks within the cellulose microfibrils (Fig. 2a), suggesting that 25–50% of cellulose is in sub-nanometer contact with pectins [36]. This extensive cellulose-pectin spatial contact was unexpected, because it contradicts in vitro binding assays that indicate low affinity between cellulose and pectins [37]. However, four independent pieces of NMR data support this conclusion. First, the cellulose-pectin cross peaks are confirmed in relaxation-compensated difference 2D 13C correlation spectra that removed intramolecular cross peaks while retaining intermolecular cross peaks [38]. Second, water-to-polysaccharide 1H spin diffusion was found to be correlated for pectins and cellulose, suggesting that the two polymers are in close proximity (Fig. 2b). Third, the cellulose-pectin cross peaks are reproduced in never-dried cell walls at both ambient and low temperatures, indicating that the spatial contact is not a hydration artifact and is not caused by temperature-induced changes in molecular packing [15, 39]. Fourth, similar cross peaks are observed between intact Arabidopsis cell wall and a 40% homogalactoronan (HG)-extracted cell wall, indicating that molecular crowding cannot explain the cellulose-pectin spatial contacts [15]. 13C T1 and 1H T1ρ relaxation times suggest that the mobile portion of pectins is easily extracted by trans-1, 2-cyclohexanediaminetetraacetic acid (CDTA) and by sodium bicarbonate, while the more rigid pectins remain in the wall, likely binding to cellulose. Together, these data indicate that cellulose-pectin molecular contacts are an inherent feature of intact hydrated primary cell walls. We attribute the discrepancy between this intermolecular contact and the weak cellulose-pectin affinity in in vitro binding assays to the inability of the latter to fully mimic the wall structure: during cell-wall biosynthesis, pectins may become entrapped between or within cellulose microfibrils, leading to close contacts and reduced mobility of the cellulose-bound pectins that cannot be readily mimicked in vitro.
Figure 2.
Intermolecular interactions of cell wall polysaccharides and proteins from SSNMR. (a) Polysaccharide-polysaccharide through-space contacts from 1H-driven 13C spin diffusion (PDSD) buildup curves [36]. Representative buildup curves are shown for intra-residue iC4–iC6 and inter-chain iC4-s/G/AC4 cross peaks within cellulose (black), intermolecular s/G/AC4-R/GAC1 cross peaks between surface cellulose and pectins (blue), and between interior cellulose iC4 and pectin R/GAC1 (red). (b) Hydration of wall polymers obtained from water-polysaccharide 1H spin diffusion [39]. Water polarization transfer to pectins GAC2/RC5 (green) is faster than to cellulose iC4 in both intact and extracted cell walls. However, partial extraction of pectins significantly slows down water polarization transfer to all polymers, which can be attributed to the removal of Ca2+ ions, which immobilize water in the intact cell wall. (c) Expansin binding to dicot cell walls from 13C-15N filtered 13C spin diffusion experiments [25]. Representative 1D 13C spectra without spin diffusion, which show only the protein 13C signals, and with 3 s spin diffusion, which show both protein and polysaccharides signals, indicate that cellulose signals are enhanced by polarization transfer from wild-type expansin and a hyperactive mutant expansin. Representative protein-cellulose (iC4) magnetization transfer curves are shown for an active expansin mutant versus wild-type expansin. These data indicate that the functional binding target of the expansin is cellulose. (d) Structural model of the expansin-cellulose complex [25].
These SSNMR results, together with biomechanical assays and microscopy evidence, shift the paradigm of the plant primary wall structure. In the new single-network model, cellulose has limited interactions with hemicellulose but extensive contacts with pectins. To understand how this single polysaccharide network is loosened during plant cell growth, we employed 2D correlation experiments under dynamic nuclear polarization (DNP) [40–42] to determine the binding target of expansin [32, 43] in Arabidopsis cell walls. A bacterial analog of the plant expansin [44] was expressed in Escherichia coli and enriched with 13C and 15N, while the cell wall is enriched only in 13C. Using a 13C-15N dipolar filter, we selected the protein 13C polarization, then allowed it to transfer to the adjacent polysaccharides. DNP was necessary because the concentration at which the protein is active is only ~0.1% of the wall polysaccharide mass, and excess protein causes non-specific binding. With polarization transfer from electrons and nuclei, the NMR sensitivity is sufficiently enhanced to enable the detection of the polysaccharides bound to the trace amount of expansin. The spectra show that expansin binds cellulose for its wall-loosening activity (Fig. 2c): a hyper-active expansin mutant shows strong cellulose cross peaks while an inactive mutant shows diminished polarization transfer to cellulose. In contrast, expansin binding to pectins anti-correlates with activity. Furthermore, the expansin-bound cellulose, selectively detected by polarization transfer from the 13C, 15N-labeled protein, is enriched with XyG and has different chemical shifts from bulk cellulose [25], in excellent agreement with biomechanical data [34]. In grass cell walls, the wall-loosening protein is not yet possible to express recombinantly and thus cannot be isotopically enriched. However, by tagging Mn2+ to purified grass expansin and exploiting paramagnetic relaxation enhancement NMR, we found that expansin binds the hemicellulose GAX instead of cellulose in the grass cell wall (unpublished results). This different binding may explain why dicot and grass cell walls have non-interchangeable expansins for wall loosening.
4. Perspectives: underexplored areas
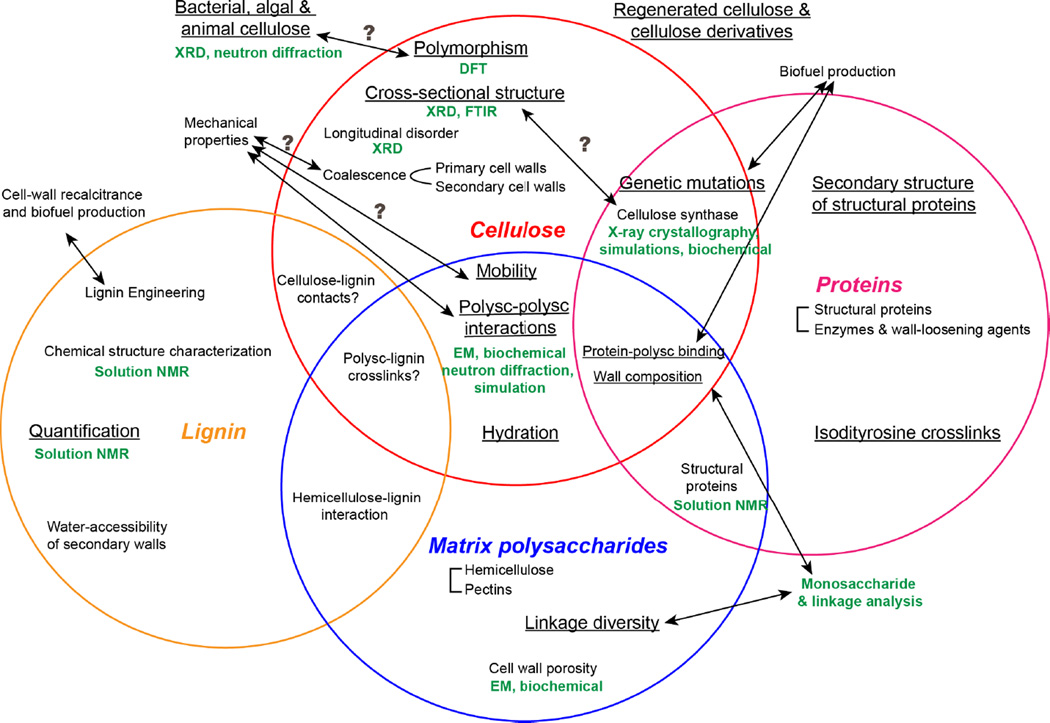
Although major progress has been made in the last five years in SSNMR investigations of plant cell wall structure, many open questions remain, as summarized in Fig. 3. For cellulose, it is still unclear how the NMR-detected cross-sectional structure is related to the cellulose synthase structure. The latest structural results that the cellulose synthase complex likely contains a hexamer of trimers of the enzyme [26, 27, 45, 46] may be reconciled with the NMR results that more than 24 chains exist in each microfibril by the hypothesis that multiple microfibrils coalesce, but this hypothesis needs to be tested experimentally. Second, the location and structure of the XyG- and expansin-interacting cellulose d are still not fully understood, and the nature of cellulose e is elusive. Third, the structures of disordered and crystalline cellulose should be investigated in greater detail through distance and torsion angle experiments to complement the extensive chemical shift data.
Figure 3.
Topics of importance in plant cell wall structure, organized according to the cell wall polymers, including cellulose, matrix polysaccharides (abbreviated as “polysc”), proteins, and lignin. Topics that have been studied using SSNMR are underlined. Complementary techniques are shown in green.
Lignin, the abundant aromatic polymer in secondary cell walls [47], increases the stiffness of plant cell walls and limits biomass conversion to biofuel [48]; thus lignin re-engineering is an important area of research [1, 49]. SSNMR has been used to quantify the amount of lignin in secondary cell walls [50–52], however lignin interactions with polysaccharides and proteins on the molecular level have not been studied (Fig. 3). Dupree and coworkers recently conducted the first SSNMR study of Arabidopsis secondary cell walls in the dry state and identified intermolecular cross peaks between cellulose and non-polysaccharide components. But it is not known whether cross peaks in the 50–60 ppm region result from lignin or structural proteins [24]. Since plant secondary cell walls are the major feedstock for biofuel production, their structural study will be important in the future.
Structural proteins in plant cell walls, including hydroxyproline-rich proteins, glycine-rich proteins, proline-rich proteins, and arabinogalactan proteins, are another under-studied class of macromolecules. These proteins have been proposed to form covalent crosslinks with each other and with polysaccharides, but direct evidence for this interaction is scarce [53]. Using 13C-2H REDOR and [ring-4-13C] Tyr- and [ring-2H4] Tyr-labeled soybean cell walls, Schaefer and coworkers found that a quarter of tyrosine residues are involved in isodityrosine crosslinks between proteins, while lysine is not involved in covalent crosslinks [54]. 2D 13C-13C correlation spectra of 13C-labeled Arabidopsis cell walls indicate that structural proteins mainly adopt random coil and β-sheet conformations, and are well separated from polysaccharides since no intermolecular cross peaks were detected [36]. Further structural investigations of cell-wall proteins will be of interest to understand how structural proteins interact with polysaccharides to participate in wall functions.
5. Concluding remarks
Plant cell walls are complex, insoluble and disordered biomaterials where the constituent macromolecules exhibit heterogeneous structures and dynamics. These properties make plant cell walls challenging systems for most structural techniques. Solid-state NMR is uniquely suited for molecular-level investigations of the cell wall structure. By combining isotopic enrichment with multidimensional MAS NMR, we have made significant advances in understanding the three-dimensional structural network formed by polysaccharides in intact primary cell walls of several model plants of the dicot and grass families, and moreover have related this structural information to the mechanism of wall loosening. Future topics of interest include the structures of secondary cell walls, lignin-cellulose interactions, higher-resolution structures of cellulose in the wall, and the structures of functionally important cell-wall proteins.
Acknowledgments
This work is supported by the Center for Lignocellulose Structure and Formation, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Basic Energy Sciences under Award # DE-SC0001090. The authors thank Dr. Yuning Chen for helpful discussions.
Biographies

Dr. Tuo Wang started his graduate research at Iowa State University in 2010 and moved to Massachusetts Institute of Technology (MIT) in 2014. He received his Ph.D. in Physical Chemistry from MIT in 2016 under the supervision of Professor Mei Hong. He continued at Mei Hong’s group for his postdoctoral training and will join the faculty at Louisiana State University in 2017. His research focuses on the development and application of SSNMR methods to elucidate the structure and dynamics of plant cell walls, membrane peptides, and amyloid fibrils.

Pyae Phyo obtained her B.A. (chemistry) from Berea College in KY, USA. Under the mentorship of Prof. Jay Baltisberger and Prof. Philip Grandinetti, she conducted SSNMR studies of silicate glasses to extract chemical shift anisotropy and J-coupling. In 2015, Pyae started her Ph.D. program in physical chemistry at MIT under the mentorship of Prof. Mei Hong. Her research focuses on the development and application of advanced SSNMR methods to understand plant cell wall structural dynamics.

Dr. Mei Hong received her Ph.D. from the Department of Chemistry, University of California Berkeley in 1996, and conducted postdoctoral research at MIT. She joined the Chemistry faculty at Iowa State University in 1999 and moved back to MIT as a Professor of Chemistry in 2014. Her research focuses on the development and application of solid-state NMR spectroscopy to elucidate the structure and dynamics of biological macromolecules, with special emphasis on membrane-bound ion channels, curvature-inducing membrane proteins, and plant cell walls.
References
- 1.Loque D, Scheller HV, Pauly M. Engineering of plant cell walls for enhanced biofuel production. Curr. Opin. Plant Biol. 2015;25:151–161. doi: 10.1016/j.pbi.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Chuck GS, Tobias C, Sun L, Kraemer F, Li C, Dibble D, Arora R, Bragg JN, Vogel JP, Singh S, Simmons BA, Pauly M, Hake S. Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proc. Natl. Acad. Sci. U. S. A. 2011;108(42):17550–17555. doi: 10.1073/pnas.1113971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Somerville C, Youngs H, Taylor C, Davis SC, Long SP. Feedstocks for Lignocellulosic Biofuels. Science. 2010;329(5993):790–792. doi: 10.1126/science.1189268. [DOI] [PubMed] [Google Scholar]
- 4.Harris DM, Corbin K, Wang T, Gutierrez R, Bertolo AL, Petti C, Smilgies DM, Estevez JM, Bonetta D, Urbanowicz BR, Ehrhardt DW, Somerville CR, Rose JKC, Hong M, DeBolt S. Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 2012;109(11):4098–4103. doi: 10.1073/pnas.1200352109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang T, Hong M. Solid-state NMR investigations of cellulose structure and interactions with matrix polysaccharides in plant primary cell walls. J. Exp. Bot. 2016;67(2):503–514. doi: 10.1093/jxb/erv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dick-Perez M, Zhang YA, Hayes J, Salazar A, Zabotina OA, Hong M. Structure and interactions of plant cell wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry-Us. 2011;50(6):989–1000. doi: 10.1021/bi101795q. (*The authors investigated the structure, dynamics and intermolecular interactions of polysaccharides in Arabidopsis primary cell walls using 2D and 3D correlation SSNMR and 13C-labeled cell walls. The spectra show extensive cellulose-pectin cross peaks but only a few cellulose-xyloglucan cross peaks, leading to the proposal of a single-network model of the primary cell wall. The authors also examined a xyloglucan-deficient mutant cell wall and found significantly faster polysaccharide motions compared to wild-type cell walls. This is the first multidimensional SSNMR study of intact plant cell walls.)
- 7. Cosgrove DJ, Jarvis MC. Comparative structure and biomechanics of plant primary and secondary cell walls. Front. Plant. Sci. 2012;3 doi: 10.3389/fpls.2012.00204. (*The authors reviewed the compositional and mechanical differences between primary and secondary plant cell walls).
- 8.Fernandes AN, Thomas LH, Altaner CM, Callow P, Forsyth VT, Apperley DC, Kennedy CJ, Jarvis MC. Nanostructure of cellulose microfibrils in spruce wood. Proc. Natl. Acad. Sci. U.S.A. 2011;108(47):E1195–E1203. doi: 10.1073/pnas.1108942108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mansfield SD, Kim H, Lu FC, Ralph J. Whole plant cell wall characterization using solution-state 2D NMR. Nat. Protoc. 2012;7(9):1579–1589. doi: 10.1038/nprot.2012.064. [DOI] [PubMed] [Google Scholar]
- 10.Cheng K, Sorek H, Zimmermann H, Wemmer DE, Pauly M. Solution-State 2D NMR Spectroscopy of Plant Cell Walls Enabled by a Dimethylsulfoxide-d(6)/1-Ethyl-3-methylimidazolium Acetate Solvent. Anal. Chem. 2013;85(6):3213–3221. doi: 10.1021/ac303529v. [DOI] [PubMed] [Google Scholar]
- 11. Wang T, Salazar A, Zabotina OA, Hong M. Structure and dynamics of Brachypodium primary cell wall polysaccharides from two-dimensional 13C solid-state nuclear magnetic resonance spectroscopy. Biochemistry-Us. 2014;53(17):2840–2854. doi: 10.1021/bi500231b. (*This paper reported the first study of the primary cell wall of Brachypodium a model plant of grasses, showing that the cell wall mainly contains cellulose and GAX, which interact with each other to form a single structural network.)
- 12.Klemm D, Heublein B, Fink HP, Bohn A. Cellulose: Fascinating biopolymer and sustainable raw material. Angew. Chem. Int. Edit. 2005;44(22):3358–3393. doi: 10.1002/anie.200460587. [DOI] [PubMed] [Google Scholar]
- 13.Lesage A, Bardet M, Emsley L. Through-bond carbon-carbon connectivities in disordered solids by NMR. J. Am. Chem. Soc. 1999;121:10987–10993. [Google Scholar]
- 14.Dick-Perez M, Wang T, Salazar A, Zabotina OA, Hong M. Multidimensional solid-state NMR studies of the structure and dynamics of pectic polysaccharides in uniformly 13C-labeled Arabidopsis primary cell walls. Magn. Reson. Chem. 2012;50(8):539–550. doi: 10.1002/mrc.3836. [DOI] [PubMed] [Google Scholar]
- 15.Wang T, Park YB, Cosgrove DJ, Hong M. Cellulose-Pectin Spatial Contacts Are Inherent to Never-Dried Arabidopsis thaliana Primary Cell Walls: Evidence from Solid-State NMR. Plant Physiol. 2015;168(3):871. doi: 10.1104/pp.15.00665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Komatsu T, Kikuchi J. Selective Signal Detection in Solid-State NMR Using Rotor-Synchronized Dipolar Dephasing for the Analysis of Hemicellulose in Lignocellulosic Biomass. J Phys. Chem. Lett. 2013;4(14):2279–2283. (*The authors used dipolar filters to remove the cellulose signals for selective detection of mobile matrix polysaccharides.)
- 17.Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 2008;54:559–568. doi: 10.1111/j.1365-313X.2008.03463.x. [DOI] [PubMed] [Google Scholar]
- 18.Atalla RH, Vanderhart DL. Native cellulose: a composite of two distinct crystalline forms. Science. 1984;223(4633):283–285. doi: 10.1126/science.223.4633.283. [DOI] [PubMed] [Google Scholar]
- 19.Nishiyama Y, Langan P, Chanzy H. Crystal structure and hydrogen-bonding system in cellulose Iβ from synchrotron X-ray and neutron fiber diffraction. J. Am. Chem. Soc. 2002;124(31):9074–9082. doi: 10.1021/ja0257319. [DOI] [PubMed] [Google Scholar]
- 20.Nishiyama Y, Kim UJ, Kim DY, Katsumata KS, May RP, Langan P. Periodic disorder along ramie cellulose microfibrils. Biomacromolecules. 2003;4(4):1013–1017. doi: 10.1021/bm025772x. [DOI] [PubMed] [Google Scholar]
- 21.Kono H, Numata Y. Structural investigation of cellulose Iα and Iβ by 2D RFDR NMR spectroscopy: determination of sequence of magnetically inequivalent D-glucose units along cellulose chain. Cellulose. 2006;13:317–326. [Google Scholar]
- 22.Nishiyama Y, Sugiyama J, Chanzy H, Langan P. Crystal structure and hydrogen bonding system in cellulose Iα, from synchrotron X-ray and neutron fiber diffraction. J. Am. Chem. Soc. 2003;125(47):14300–14306. doi: 10.1021/ja037055w. [DOI] [PubMed] [Google Scholar]
- 23. Wang T, Yang H, Kubicki JD, Hong M. Cellulose Structural Polymorphism in Plant Primary Cell Walls Investigated by High-Field 2D Solid-State NMR Spectroscopy and Density Functional Theory Calculations. Biomacromolecules. 2016;17(6):2210–2222. doi: 10.1021/acs.biomac.6b00441. (*Using 2D SSNMR experiments at high magnetic fields and DFT calculations, the authors investigated the cellulose structure in several plant primary cell walls. Cellulose is found to be highly polymorphic, with at least seven distinct structures, most of which differ from bacterial and tunicate cellulose. The seven cellulose polymorphs coexist in the same microfibril and their lateral locations are determined using long-mixing 2D 13C-13C correlation experiments and water-edited experiments.)
- 24. Dupree R, Simmons TJ, Mortimer JC, Patel D, Iuga D, Brown SP, Dupree P. Probing the Molecular Architecture of Arabidopsis thaliana Secondary Cell Walls Using Two- and Three-Dimensional 13C Solid State Nuclear Magnetic Resonance Spectroscopy. Biochemistry-Us. 2015;54(14):2335–2345. doi: 10.1021/bi501552k. (*The authors investigated polysaccharide structures in 13C-labeled dry secondary cell walls of Arabidopsis and found that xylan adopts a distribution of conformations. Long-range cross peaks are reported between cellulose and non-polysaccharide polymers.)
- 25. Wang T, Park YB, Caporini MA, Rosay M, Zhong LH, Cosgrove DJ, Hong M. Sensitivity-enhanced solid-state NMR detection of expansin's target in plant cell walls. Proc. Natl. Acad. Sci. U.S.A. 2013;110(41):16444–16449. doi: 10.1073/pnas.1316290110. (*Using dynamic nuclear polarization NMR, the authors determined the binding site of expansin in Arabidopsis cell walls. Expansins bind xyloglucan-enriched regions of the cellulose microfibril to cause wall loosening. This is the first proof of the molecular binding target of expansin in plant cell walls.)
- 26.Hill JL, Hammudi MB, Tien M. The Arabidopsis Cellulose Synthase Complex: A Proposed Hexamer of CESA Trimers in an Equimolar Stoichiometry. Plant Cell. 2014;26(12):4834–4842. doi: 10.1105/tpc.114.131193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandavasi VG, Putnam DK, Zhang Q, Petridis L, Heller WT, Nixon BT, Haigler CH, Kalluri U, Coates L, Langan P, Smith JC, Meiler J, O'Neill H. A Structural Study of CESA1 Catalytic Domain of Arabidopsis Cellulose Synthesis Complex: Evidence for CESA Trimers. Plant Physiol. 2016;170(1):123–135. doi: 10.1104/pp.15.01356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarvis MC. Cellulose Biosynthesis: Counting the Chains. Plant Physiol. 2013;163(4):1485–1486. doi: 10.1104/pp.113.231092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman RH, Hill SJ, Harris PJ. Wide-angle x-ray scattering and solid-state nuclear magnetic resonance data combined to test models for cellulose microfibrils in mung bean cell walls. Plant Physiol. 2013;163(4):1558–1567. doi: 10.1104/pp.113.228262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang T, Mahgsoudy-Louyeh S, Tittmann B, Cosgrove DJ. Visualization of the nanoscale pattern of recently-deposited cellulose microfibrils and matrix materials in never-dried primary walls of the onion epidermis. Cellulose. 2014;21(2):853–862. [Google Scholar]
- 31.Zhang T, Zheng YZ, Cosgrove DJ. Spatial organization of cellulose microfibrils and matrix polysaccharides in primary plant cell walls as imaged by multichannel atomic force microscopy. The Plant journal : for cell and molecular biology. 2016;85(2):179–192. doi: 10.1111/tpj.13102. [DOI] [PubMed] [Google Scholar]
- 32.Cosgrove DJ. Catalysts of plant cell wall loosening. F1000Res. 2016;5 doi: 10.12688/f1000research.7180.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cosgrove DJ. Re-constructing our models of cellulose and primary cell wall assembly. Curr. Opin. Plant Biol. 2014;22C:122–131. doi: 10.1016/j.pbi.2014.11.001. (*This review presents the most updated model of plant primary cell walls based on SSNMR, biochemical, AFM, and MD simulation results. The author also summarized evidence for the emerging hexamer-of-trimer structural model of the cellulose synthase complex.)
- 34.Park YB, Cosgrove DJ. A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol. 2012;158(4):1933–1943. doi: 10.1104/pp.111.192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park YB, Cosgrove DJ. Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol. 2015;56(2):180–194. doi: 10.1093/pcp/pcu204. [DOI] [PubMed] [Google Scholar]
- 36.Wang T, Zabotina O, Hong M. Pectin-cellulose interactions in the Arabidopsis primary cell wall from two-dimensional magic-angle-spinning solid-state nuclear magnetic resonance. Biochemistry-Us. 2012;51(49):9846–9856. doi: 10.1021/bi3015532. [DOI] [PubMed] [Google Scholar]
- 37.Zykwinska A, Thibault JF, Ralet MC. Competitive binding of pectin and xyloglucan with primary cell wall cellulose. Carbohydr. Polymers. 2008;74(4):957–961. [Google Scholar]
- 38. Wang T, Williams JK, Schmidt-Rohr K, Hong M. Relaxation-compensated difference spin diffusion NMR for detecting 13C-13C long-range correlations in proteins and polysaccharides. J. Biomol. NMR. 2015;61(2):97–107. doi: 10.1007/s10858-014-9889-0. (*The authors use a relaxation-compensated modification of 2D proton-driven 13C spin diffusion technique for clean detection of long-range cellulose-pectin cross peaks).
- 39. White PB, Wang T, Park YB, Cosgrove DJ, Hong M. Water-polysaccharide interactions in the primary cell wall of Arabidopsis thaliana from polarization transfer solid-state NMR. J. Am. Chem. Soc. 2014;136(29):10399–10409. doi: 10.1021/ja504108h. (*Using 1H polarization transfer NMR, the authors investigated the hydration of intact and sequentially digested Arabidopsis cell walls. Water polarization transfers to pectins before cellulose. Partial removal of pectins slowed down the polarization transfer, but this is restored upon digestion of XyG. These results indicate that pectins are essential for wall hydration and support the single-network model of the cell wall.)
- 40.Ni QZ, Daviso E, Can TV, Markhasin E, Jawla SK, Swager TM, Temkin RJ, Herzfeld J, Griffin RG. High Frequency Dynamic Nuclear Polarization. Acc Chem Res. 2013 doi: 10.1021/ar300348n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Can TV, Ni QZ, Griffin RG. Mechanisms of dynamic nuclear polarization in insulating solids. J Magn Reson. 2015;253:23–35. doi: 10.1016/j.jmr.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao SY, Lee M, Wang T, Sergeyev IV, Hong M. Efficient DNP NMR of membrane proteins: sample preparation protocols, sensitivity, and radical location. J. Biomol. NMR. 2016;64(3):223–237. doi: 10.1007/s10858-016-0023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407(6802):321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- 44.Georgelis N, Tabuchi A, Nikolaidis N, Cosgrove DJ. Structure-function analysis of the bacterial expansin EXLX1. J Biol Chem. 2011;286:16814–16823. doi: 10.1074/jbc.M111.225037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sethaphong L, Haigler CH, Kubicki JD, Zimmer J, Bonetta D, DeBolt S, Yingling YG. Tertiary model of a plant cellulose synthase. Proc Natl Acad Sci USA. 2013;110(18):7512–7517. doi: 10.1073/pnas.1301027110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nixon BT, Mansouri K, Singh A, Du J, Davis JK, Lee JG, Slabaugh E, Vandavasi VG, O'Neill H, Roberts EM, Roberts AW, Yingling YG, Haigler CH. Comparative Structural and Computational Analysis Supports Eighteen Cellulose Synthases in the Plant Cellulose Synthesis Complex. Sci. Rep. 2016;6:28696. doi: 10.1038/srep28696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 48.Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. Lignin Biosynthesis and Structure. Plant Physiol. 2010;153(3):895–905. doi: 10.1104/pp.110.155119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vanholme R, Morreel K, Ralph J, Boerjan W. Lignin engineering. Curr. Opin. Plant Biol. 2008;11(3):278–285. doi: 10.1016/j.pbi.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 50.Fu L, McCallum SA, Miao J, Hart C, Tudryn GJ, Zhang F, Linhardt RJ. Rapid and accurate determination of the lignin content of lignocellulosic biomass by solid-state NMR. Fuel. 2015;141:39–45. doi: 10.1016/j.fuel.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bernardinelli OD, Lima MA, Rezende CA, Polikarpov I, deAzevedo ER. Quantitative 13C MultiCP solid-state NMR as a tool for evaluation of cellulose crystallinity index measured directly inside sugarcane biomass. Biotechnol. Biofuels. 2015;8:110. doi: 10.1186/s13068-015-0292-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson RL, Schmidt-Rohr K. Quantitative solid-state 13C NMR with signal enhancement by multiple cross polarization. J Magn Reson. 2014;239:44–49. doi: 10.1016/j.jmr.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 53.Showalter AM. Structure and function of plant cell wall proteins. Plant Cell. 1993;5(1):9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cegelski L, O'Connor RD, Stueber D, Singh M, Poliks B, Schaefer J. Plant Cell-Wall Cross-Links by REDOR NMR Spectroscopy. J. Am. Chem. Soc. 2010;132(45):16052–16057. doi: 10.1021/ja104827k. (*The authors conducted 13C-2H REDOR experiments on l-[ring-d4] and l-[ring-4-13C] tyrosine labeled soybean cell walls. The data show that 25% of the tyrosines are part of isodityrosine cross-links between protein chains.)