Abstract
Neural circuitry has evolved to form distributed networks that act dynamically across large volumes. Collecting data from individual planes, conventional microscopy cannot sample circuitry across large volumes at the temporal resolution relevant to neural circuit function and behaviors. Here, we review emerging technologies for rapid volume imaging of neural circuitry. We focus on two critical challenges: the inertia of optical systems, which limits image speed, and aberrations, which restrict the image volume. Optical sampling time must be long enough to ensure high-fidelity measurements, but optimized sampling strategies and point spread function engineering can facilitate rapid volume imaging of neural activity within this constraint. We also discuss new computational strategies for the processing and analysis of volume imaging data of increasing size and complexity. Together, optical and computational advances are providing a broader view of neural circuit dynamics, and help elucidate how brain regions work in concert to support behavior.
Introduction
In neural circuitry, action potentials are the coin of the realm. To understand how ensemble neural circuitry encodes stimuli, processes information, and guides adaptive behavior, it is essential to observe the spatiotemporal dynamics of action potential activity in populations of cells with single-neuron resolution.
Techniques for recording neural activity have evolved since Emil du Bois-Reymond’s (1818 – 1896) discovery of the action potential using metal electrodes1. Since then, metal electrodes have shrunk in size2,3, and are banded together as tetrodes4 or arrays of electrodes2,3,5, in an attempt to record from more neurons while reducing damage to surrounding tissue. Electrodes offer unparalleled temporal resolution, but they suffer a fundamental limitation: they typically can only reliably isolate spikes from a subset of nearby neurons6. Thus, electrodes alone may not be sufficient for recording densely and comprehensively from large populations of neurons in parallel.
Optical imaging can be more comprehensive and less invasive, as light can be focused and collected by instrumentation located away from the brain. To detect action potential activity, fluorescent indicators can be imaged. Originally, calcium dyes provided the strongest signals7, but the best genetically encoded indicators (e.g., GCaMP68) now rival the best dyes on some metrics9. These and other indicators can be used to infer neural activity with high sensitivity, and in some cases detect individual action potentials, though accuracy is limited by nonlinearities and signal-to-noise ratio. Indicator engineering is still undergoing rapid development, providing an array of new tools for measuring neural activity9. Imaging with fluorescent proteins also enables targeted recording of genetically-defined cell types, and chronic monitoring of the same neurons over months10.
Some neurobiological model systems are relatively transparent, making them particularly amenable to imaging. For example, neural activity in transparent larval zebrafish can be captured with light sheet imaging11. Many other preparations, including the mammalian brain, scatter light so strongly that most imaging approaches are useless beyond shallow depths. To image scattering tissue, two-photon laser scanning microscopy (2PLSM) is the method of choice12, capable of resolving individual neurons and their subcompartments hundreds of microns deep13–19, and is the focus of this review.
Advances in two-photon calcium imaging in vivo have increased image speed and depth, and provided greater flexibility of scan patterns. However, the field-of-view (FOV) of 2PLSM in most implementations has remained limited to <1 mm2, and typically only a single z-plane (one thin optical section) is acquired. This small acquisition region is a critical barrier to progress in systems neuroscience because it limits imaging of neuronal activity across extended circuitry. During behavior, such circuitry processes information20–24 and generates internal dynamics to guide adaptive behavior. Observing activity in one slice at a time is akin to eavesdropping on one side of a telephone conversation: the content is incomplete and often uninterpretable. Observing all sides of the conversation—across a large volume—can reveal correlations, multi-area dynamics, signal transformations, and statistical properties crucial to understanding integrative brain function25,26. In this review, we discuss recent and emerging technical advances for imaging cellular-resolution neural activity from large brain volumes.
Instrumentation challenges
Technology to image large brain volumes must address two problems: optical access and sampling speed. Optical access is the problem of maximizing the brain volume over which individual neuron resolution can be maintained. It is limited by optical aberrations, light scattering, and the physical dimensions of imaging systems. Sampling speed is the problem of measuring activity in as many neurons as possible while maintaining the temporal resolution required by the experiment. It is limited by inertia of the instrumentation and the number of photons that can be collected from a given sample (i.e., pixel or voxel), which limits the signal-to-noise ratio (SNR) and is a function of several parameters including fluorescent probe brightness and the laser dwell time for each sample.
Optical access
An imaging system has optical access to neurons within its field-of-view (FOV) over which individual neurons can be resolved, from the surface of the brain down to the maximum imaging depth (Fig. 1a). In many organisms, this requires surgical preparations such as thinned skull27,28, cranial windows29, cannulas30,31, embedded prisms32,33, or GRIN lenses34,35 (through-skull imaging has been demonstrated in mouse using three-photon excitation at 1.7 µm36). The maximal imaging depth depends on the tissue staining sparseness and brightness37 (sparse and bright staining can aid deep imaging), and thus cannot be precisely defined for a given instrument. Ultimately, imaging depth is limited by aberrations (brain-induced optical distortions that degrade image quality) as well as the attenuation by the opaque brain (loss of both excitation light and detectable fluorescence signal by tissue scattering and absorption) at depth38, and is limited to 5 – 6 attenuation lengths39. However, even at superficial depths, the optics within the microscope itself, due to their physical dimensions and designs, confines us to viewing only a limited FOV.
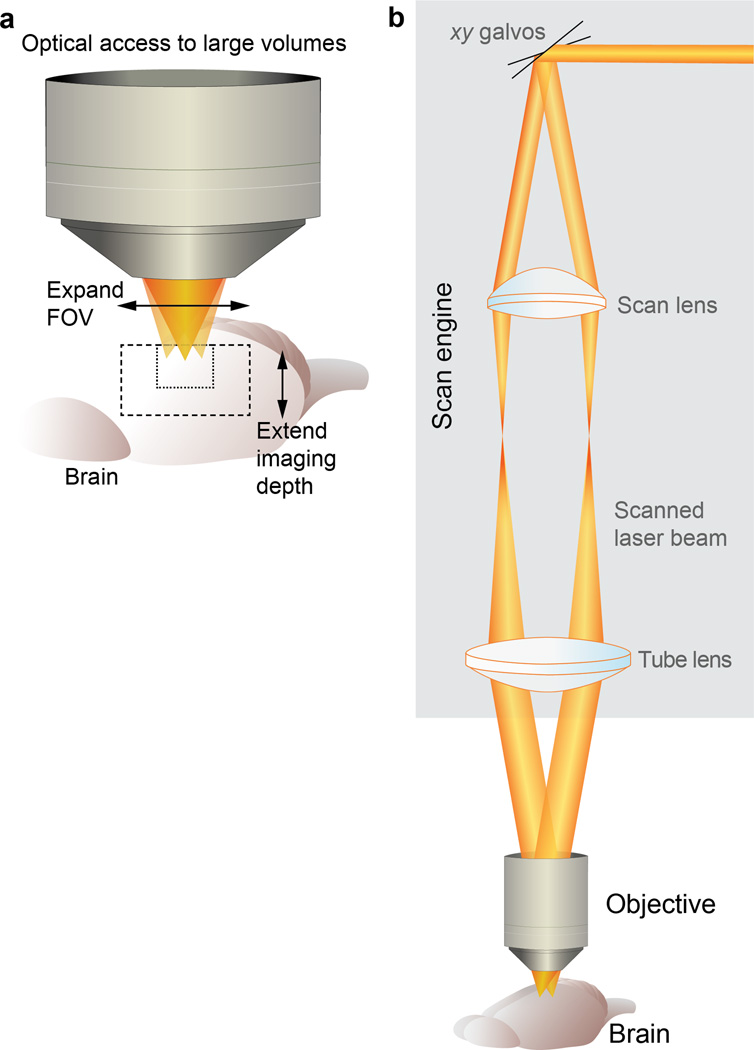
Fig. 1.
Optical access and basic optical layout of a 2PLSM. (a) Obtaining optical access to large brain volumes entails expanding the field of view (FOV) and extending the imaging depth. (b) A 2PLSM is comprised of a scan engine and an objective. In the scan engine, rapidly movable mirrors (galvanometer beam scanners, xy Galvos) reflect the excitation laser beam across a range of angles that are relayed using a scan lens and tube lens to the objective.
The basic optical layout of a laser scanning microscope involves a scan engine and an objective (Fig. 1b) that are constrained by simple geometric optics (Fig. 2). The scan engine generates a beam that pivots around a central point at the back focal plane of the objective (Figs. 1a and 2a). The objective has a numeric aperture (NA) which determines its resolving power, and a focal length (FL), which is commonly expressed as a magnification factor (FL is the focal length of the tube lens divided by magnification). The FOV is defined by the FL and the maximal scan angle (θ) of the laser entering the objective back focal plane (FOV = 2×FL×tan(θ)) (Fig. 2b,c). In practice, θ is typically limited to ~1 – 5 degrees (measured from the central axis) because at higher angles the beam will be clipped by lenses in the objective, resulting in a distorted point spread function (PSF) and poor imaging quality.
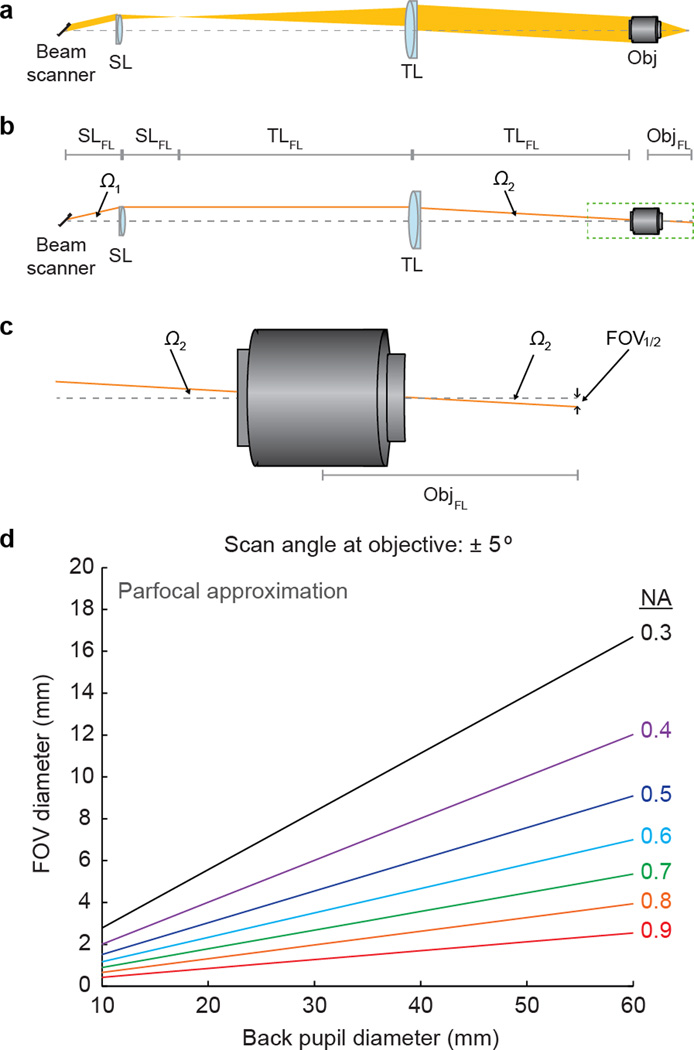
Fig. 2.
Scan engine and objective determine imaging FOV. (a) Scan engines expand the excitation beam and rapidly vary the incidence angle on the back aperture of the objective (Obj) to create a scan pattern. A conventional approach involves beam scanners followed by a scan lens (SL) and a tube lens (TL). (b) The SL is placed at a distance equal to its focal length (FL) from the beam scanner (SLFL). The TL is placed at a distance equal to its FL from the Obj (TLFL). The beam diameter is expanded by a factor of TLFL/SLFL and the beam scanner scan angle (Ω1) is reduced by the reciprocal factor (SLFL/TLFL) to a smaller angle (Ω2). To use the full resolution of the Obj, the beam must be expanded to overfill the back aperture, and this expansion reduces the scan angle at the objective and can reduce the FOV. (c) Half of the width of the FOV (FOV1/2) is equal to Obj focal length (ObjFL, which is commonly expressed as a magnification factor, rather than a FL, by commercial vendors) multiplied by tan(Ω2). (d) FOV increases more rapidly with Obj back aperture diameter for lower NA optics. This relationship is illustrated using the parfocal approximation, and performance of real world systems can vary from the traces illustrated here, but the general relationship still applies.
For a given NA and θ, increases in FOV require increases in FL, which in turn require larger-diameter lenses (Fig. 2d). For example, the diameter of the objective back pupil increases linearly with FL for a particular NA (back pupil diameter, BPD = 2×FL×NA). To use the full NA of the system, the beam diameter must match the BPD, which requires a scan engine that scans a large-diameter beam over a range of θ. Therefore, a straightforward approach to create a large FOV imaging system is to simply scale up microscope objectives and scan engines to larger lenses, and scan at as high angles as possible.
However, there are engineering challenges in this approach. In high-NA imaging systems, even small increases in FOV are accompanied by large increases in BPD (Fig. 2d). Moreover, high θ requires careful correction of optical aberrations. Aberrations are inherent in real-world optics because the path difference for light passing through the center of lenses and light passing through the edges of lenses results in aberrated (distorted) wavefronts. In two-photon imaging, the problem is particularly serious because aberrations not only decrease resolution, but also decrease excitation efficiency. So images become both blurry and dimmer, and these effects can cause rapid degradation of image quality outside of a central FOV region. To correct for these aberrations, as the FOV increases, more lenses are required at a cost of system complexity and light loss. Moreover, the fastest beam scanning elements are often small (to minimize mass and inertia and maximize speed), and can only scan small-diameter beams. Thus, the beam has to be subsequently magnified to use the full NA of the objective. However, the same optics that expand the beam diameter also decrease the scan angle. So aggressive beam expansion reduces θ and thus the FOV. In summary, engineering tradeoffs need to be made to optimize two-photon imaging systems for imaging large FOVs and volumes.
This engineering is facilitated by the modern optical design software and manufacturing, which has allowed individual neurobiology labs to design and build custom optical systems for two-photon imaging40–42. For example, a completely custom two-photon imaging system, using no commercial microscope parts, expanded the area of the FOV >50 fold compared to commercial two-photon imaging systems by optimizing the designs for both the microscope objective and scan engine40.
Microscope objectives critically govern the performance of an imaging system. In traditional biological microscopy, broad achromatic performance over the entire visible light range is a high priority to ensure accurate colocalization of multiple fluorescence labels. Maximizing the FOV, by contrast, is a lower priority. Thus, in commercial biological microscope systems, optical aberrations grow rapidly outside of the central highly corrected region43. By contrast, in large FOV two-photon imaging, achromatic performance may only be needed over a smaller wavelength range (typically in the infrared), since the visible fluorescence light is not imaged but simply sent to a single-channel detector. Therefore, maximizing the FOV involves a rebalancing of the priorities in optical design. For example, engineers can relax requirements for field flatness (since individual optical sections in a brain volume do not need to be perfectly flat) and broad achromatic correction (assuming a relatively narrow band of wavelengths are used for excitation) for the sake of low aberrations for the excitation wavelength across a large FOV (larger range of scan angles, up to 5 degrees).
Scan angles are typically rapidly varied using two low-inertia scan mirrors that are positioned close together, followed by a scan lens and tube lens to magnify the beam diameter to overfill the objective (which also results in a reduction of the scan angle by the inverse of the magnification factor) (Fig. 2b). Although this simple design is widely used, there is a variety of possible scan engines, each with its own selection of engineering tradeoffs. Ideally, scan mirrors should be optically relayed to one another to prevent beam walk in later elements, which can lead to beam clipping, aberrations, and degraded image quality. With close positioning and small scan angles, these negative consequences can be minimized, but such measures can also limit the FOV. Scan engines can include some or all of following: lenses, mirrors, mechanics, piezo actuators, voice-coil actuators, and acousto-optic deflectors. Scan engines optimized for a specific purpose can offer substantial advantages over general-purpose scan engines44. Tsai and colleagues developed a scan engine for use with a 0.28 NA commercial objective (FL = 45 mm) that supports relatively low aberrations over a FOV 10-mm wide41. Voigt and colleagues presented a scan engine with two modular, temporally multiplexed focal plane units and low aberrations to support simultaneous imaging in two non-overlapped regions within a 1.7 mm wide FOV with a 0.8 NA commercial objective (FL = 12.5 mm)45,46. Stirman and colleagues developed a scan engine with two temporally multiplexed beams for simultaneously imaging two regions that can be positioned anywhere within a 3.5-mm wide FOV of a custom objective (NA = 0.43, FL = 27.5 mm)40. Sofroniew et al. presented a scan engine with a fast small-angle resonant scanner followed by large-angle scanners to support high-frame-rate scanning of 0.5-mm wide subfields that can be positioned anywhere within a 5-mm wide FOV of a custom objective (0.6 NA, FL = 21 mm)42. These FOV sizes are the extent over which the PSF remains sufficient for the intended experiment. In some cases the PSF is unchanged over the stated FOV40, while in others it can vary by 2 – 4 fold over the stated FOV41,42,47.
Imaging speed
Imaging speed is particularly important for understanding neural computation, where information-encoding electrical activity occurs on millisecond time scales. Even though commonly used fluorescent indicators of neuronal activity (e.g., the Ca2+ concentration indicators) function as low-pass-filters of these electrical signals, sub-second time resolution is still required. Both individual neurons and neural circuits often extend over hundreds of microns in each dimension. To image the entire volume at sufficient rates so as to capture all the calcium transients, a straightforward approach is to rapidly vary the 3D position of laser focus inside the brain. We discuss these and other volumetric imaging technologies below.
I. Planar imaging
Galvanometer-based optical scanners (or “galvos”), composed of lightweight mirrors mounted on galvanometers controlled by servo drivers, are the most commonly used devices to move the excitation laser focus in the plane perpendicular to optical axis (i.e., lateral or xy plane, although xz plane imaging is also possible using microprisms32,33,48,49). Varying the electric current passing through the galvanometer causes rapid rotation of the mounted mirror, and leads to changes in the direction of the reflected laser beam (Figs. 1b and 2a). These direction changes are converted to positional shifts of the laser focus on the focal plane of the objective (Fig. 3a). A pair of galvos that rotate along orthogonal axes direct the beam in 2D xy plane, usually with a raster (i.e., line-by-line) scanning pattern and sub-millisecond step response time (a few kilohertz linescan rates). Sometimes, one of the galvos is designed to oscillate at a fixed frequency (e.g., 8 kHz or 12 kHz)50, to achieve higher frame rates (e.g., 512×512 pixels at 30 Hz or “video rate”, even higher rate such as 200 Hz can be achieved by simply reducing the number of lines51).
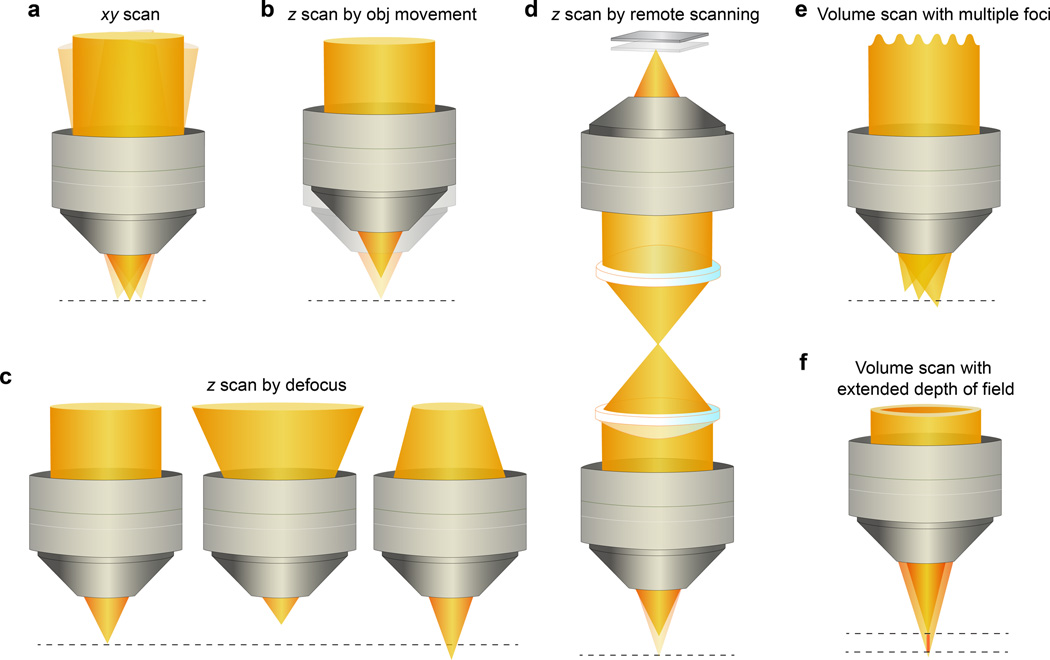
Fig. 3.
2D and 3D scanning strategies. (a) Focus is scanned in the xy plane by varying the direction of the excitation laser at the back pupil of the objective. (b) Focus is moved along z axis by moving the objective relative to the sample. (c) With a stationary imaging objective, focal shift in z can be achieved by changing the divergence of the laser beam. (d) With two objectives and a movable mirror, z position of the focus can be varied without incurring optical aberrations. (e) Multiple foci can be generated by manipulating the laser wavefront, which allows speed increase via multiplexing. (f) Volume can be imaged with an elongated focus with extended depth of field.
One alternative approach to mechanical laser scanning is to use acousto-optic deflectors (AODs) for beam steering. Piezoelectric transducers bonded to one surface of a transparent crystal, when driven at radio frequency (RF), can generate traveling acoustic waves inside the crystal and cause compression and rarefaction of the material, which lead to alterations in its refractive index. Because the resulting refractive index modulations are periodic, the crystal now acts as a diffractive grating and deflects a laser beam passing through, with the beam diffraction direction determined by the period of the grating. Because RF frequency determines grating period, rapid adjustments of the drive frequency allow the crystal to act like a fast tunable deflector. For example, ramping the frequency of the RF signal produces a line scan. 2D steering of the laser focus within the objective focal plane can be accomplished by two AODs arranged with their acoustic gratings at orthogonal directions52–54. Alternatively, an AOD can also be combined with a galvanometer-driven scanning mirror for hybrid 2D scanning55.
Because AOD-based beam deflection involves no movable parts and is not constrained by inertia, 2D scanning at very high rates can be achieved. Raster scanning with an AOD for fast x-scan and a galvo for y-scan, Chen et al. recorded, at >1,000 frames · s−1 (80 scan lines per frame) over 28×9 µm2 area, sound-evoked calcium transients in single spines of mouse primary auditory cortical neurons in vivo56. In addition to raster scanning mode by ramping the RF drive signals, random-accessing operation can be achieved by generating RF drive signal of discretely changing frequencies, which allows selected positions (e.g., discrete compartments along a dendrite) to be accessed at arbitrary sequences with microsecond reposition time, a big advantage when compared with galvo-based raster scanning methods where field positions are sampled sequentially, irrespective whether all the positions are of experimental interest or not. With a pair of AODs programmed for random access, Iyer et al. visualized at 500 Hz over >10 sites (spanning 60×90 µm2) calcium transients that were triggered by back-propagating action potentials along CA1 pyramidal neuron dendrites in brain slices57. Using similar systems for 2D random accessing scans, Otsu et al. measured calcium transients from 30 – 80 spine heads and attached dendritic shafts of Purkinje cells in acute slices at frame rates of 0.5 – 1.5 kHz58, and Grewe et al. obtained fluorescence measurements from 34 – 91 neurons at a 180 – 490 Hz sampling rate from L2/3 of mouse cortices in vivo59.
II. Volume imaging
a. By objective motion
For volume imaging, in addition to scanning the focus in the lateral xy plane, the laser focus also needs to move relative to the sample in the axial z direction. For most neurobiological applications (e.g., in vivo imaging of mouse brains), the simplest way to obtain a 3D image stack is by translating the objective along its optical axis using a mechanical actuator, and taking a 2D raster-scanning image at each axial position (Fig. 3b). With 2D images routinely obtained at video rate and higher, the rate-limiting step in volume imaging is often the time delay between successive image planes that is required for mechanical oscillations to subside and the objective to settle into its appropriate axial position. This dead time between consecutive 2D scans can be minimized by continuously oscillating the objective throughout image acquisition (causing the image planes to tilt relative to the sample60). With fast piezoelectric objective positioners61, reasonable volume rates can be achieved over small axial ranges and have been used to measure calcium transients in volumes of brain tissues in vivo (e.g., 14-plane volume of 60 µm×60 µm×48 µm at 4 Hz62 and 150 µm×150 µm×45 µm volume at 1 Hz63, 3-plane volume at 7.8 Hz rate for 600 µm×600 µm×20 µm64,65, 5-plane volume at 8.5 Hz rate for 80 µm×80 µm×25 µm66, 4-plane volume of 260 µm×260 µm×24 µm at 7 Hz67). A further improvement in speed was made by Gobel et al., who demonstrated a 3D vector-scanning method where the laser focus was steered along a 3D trajectory that maximally sampled cell bodies within the volume. Driving the objective to oscillate axially in a sinusoidal pattern and providing synchronized control signals to the x and y galvos, they generated 3D scanning paths that sampled, at 10 Hz, 375 cell somata within a 250 µm×250 µm×200 µm volume, which allowed them to monitor network activity patterns of neurons and astrocytes in vivo68. Katona et al. increased the volume image rate even further by driving their objective to oscillate at higher frequency (150 – 700 Hz, Roller Coaster scanning69). Even though overdriving reduces the amplitude of z scanning range to tens of microns, this method was successful in detecting spontaneous dendritic events in long dendritic segments in vitro at very high speed (650 µm×650 µm×25 µm at 150 Hz).
b. By divergence control
Moving the laser focus along optical axis without moving objective or sample avoids the problem posed by inertia, but requires controlling the wavefront of the excitation light, because a shift of focus axially relative to the objective corresponds to a change of the beam divergence (Fig. 3c). Devices such as deformable mirrors, spatial light modulators, variable-focus lenses, and AODs have been employed for this purpose. Reflecting the excitation light off a mirror with negative or positive curvature (or a spatial light modulator with proper phase patterns70) produces converging or diverging beam to achieve upward or downward focal offset71. Adjusting the mirror curvature rapidly (e.g., at 5 kHz72) therefore scans the focus axially. Variable-focus lenses have also been used to vary beam divergence for axial scanning. A variety of such lenses are commercially available and operate on different mechanisms, but are typically tuned by either changing the shape73 or the refractive index74 of the lens. Grewe et al. characterized a tunable lens assembly that, by having the curvature of one surface controlled electrically, can generate 700 µm of axial shift, and used it to measure the calcium transients of 40 neurons across two image planes separated by 40 µm at 30 Hz75. More recently, Sheffield et al. used the same approach to image dendrites, somata, and axons of CA1 place cells in behaving mice (15.6 Hz for 2-plane volume, 10.4 Hz for 3-plane volumes extending axially 58 – 284 µm)76. Another class of variable-focus lens uses ultrasound waves in a confined liquid to create a tunable gradient refractive index lens74. Because the ultrasound lens generates continuously varying beam divergence at ~MHz rates, leading to axial displacements that are faster than lateral displacements, axial scan can be used as the fast axis61. Using this approach, Kong et al. demonstrated volume imaging over 40 µm or 130 µm axial ranges with volume rates up to 56 Hz77.
Two orthogonal pairs of AODs can control beam divergence and allow 3D random access78. Each AOD pair consists of two AODs with chirped (i.e., continuously frequency-varied) and counter-propagating acoustic waves79, which deflect the beam laterally as well as alter its divergence, leading to independent lateral and axial focus repositioning. Using such a system to generate axial offsets of up to 50 µm, Reddy et al. monitored dendritic calcium dynamics at both laterally and axially distinct locations on apical dendrites in acute brain slices at up to 10 kHz80. Further refinements increased the axial range (to >137 µm) and improved excitation efficiency by reducing AOD-associated pulse-broadening81. Using AODs with large apertures, Katona et al. built a system that maintained dendrite-resolving resolution in a 290 µm×290 µm×200 µm core volume and single-cell resolution over a 700 µm×700 µm×1,400 µm volume47. The system was used to image the backpropagation of action potential (BAP) at 3D locations spanning 700 µm×700 µm×140 µm in mouse acute hippocampal slices. With sub-millisecond temporal resolution, the latency of BAP-evoked calcium transients was measured along lengths of dendrites. Calcium transients from 532 neurons within a 400 µm×400 µm×500 µm volume in vivo were also monitored at 56 Hz47. Such 3D AOD-based systems have been used to study network activities of neurons in vivo82.
Even though these divergence-control-based axial-scanning methods can generate large axial shifts at high speed, because microscope objectives are designed to produce optimal performance for light of specific divergence (e.g., most commonly used infinity-corrected objectives require collimated incident beam)83, changing beam divergence leads to degraded focus at large z displacements. Even at tens of microns outside the natural z focus depth, defocus methods already suffer from degraded image quality and substantially reduced resolution and signal-to-noise ratio70,75,80,81 (although in principle, AODs can be driven to shape the excitation wavefront in order to correct for aberrations84,85). This can be problematic for experiments where diffraction-limited resolution is required (e.g., imaging synaptic terminals). To address this issue, Botcherby et al. introduced a remote focusing method, where the objective and the sample remain stationary, and axial scanning is carried out by moving a lightweight mirror upstream of the sample objective86,87 (Fig. 3d). Because this mirror is imaged onto the sample by an additional objective (top objective in Fig. 3d) and reflects the excitation light before it enters the imaging objective (bottom objective in Fig. 3d), axial movements of the mirror cause focal shifts in the sample. The aberrations experienced by the converging or diverging beam at the sample objective are canceled by those through the mirror objective, leading to aberration-free axial scanning over 200 µm88. Because of the low inertia of the remote mirror, z scanning can be so fast that z direction is no longer the slow axis, and 3D scanning of planes at arbitrary orientation89, curved surfaces of complex morphology (e.g., to adapt to sample topology90), and complex paths (sampled at >300 Hz88) has been demonstrated. Using galvos to move the remote mirror rapidly, Botcherby et al. measured calcium transients triggered by extracellular electrodes from two dendritic segments >30 µm apart axially and neurons extending 60 µm in z were monitored at 500 Hz and 1 kHz, respectively88. Positioning the remote mirror with a voice coil motor, Rupprecht et al. realized focal shifts >500 µm and demonstrated 6 Hz volume imaging of calcium dynamics (9 planes of 512×256 pixels over 240 µm depth) in adult zebrafish brain91.
c. By multiplexing
All the above methods reach high volume-imaging speed by moving a single focus rapidly in 3D. Another way to increase speed is by utilizing multiple foci to parallelize the imaging process. In principle, N spatially distinct foci can increase imaging speed N fold, if the fluorescent signal from each focus can be unambiguously assigned (Fig. 3e). The conventional multifocal multiphoton microscopy scans a 2D array of foci across the xy plane92,93. Using spatial light modulators for wavefront shaping, the spatial arrangement of multiple foci can be flexibly controlled94. However, because images of the focal plane are captured by an array detector (rather than a single-element detector such as a photomultiplier tube), image depth is limited by sample scattering even with optimized detectors95. But the multifocal approaches can be made resistant to scattering if the fluorescence excited at each focus is temporally separated. Using two axially displaced and temporally delayed foci, Amir et al. demonstrated simultaneous imaging of two focal planes71. With a typical laser for two-photon excitation operating at ~80 MHz repetition rate, the maximal number of temporally distinguishable foci is determined by fluorescence decay lifetime. Using Fluo-4 as calcium sensor (~1 ns lifetime), Cheng et al. simultaneously recorded activities of neurons in mouse neocortex on four imaging planes 90 µm apart at 60 Hz96. Reducing the repetition rate of the laser can increase the number of temporally multiplexed foci97. Combining temporal with spectral multiplexing98 in brain volumes with different fluorescence labels (e.g., GCaMP and RCaMP), the volume imaging rate may be increased further. Temporal multiplexing have also been implemented together with defocus-based z control in multi-area two-photon microscopes to scan laterally and axially displaced volumes simultaneously40,46.
A fundamental difference between imaging experiments in the brain and those in other biological systems (e.g., imaging live cells or developing embryos) is that, in the brain, the structures of interest (e.g., neurons and their subcellular compartments) typically do not move during the few hours of experiment. As a result, it is often unnecessary to constantly monitor and resolve their positions in 3D. Using a spatial light modulator, Yang et al. generated seven axially displaced foci to simultaneously image seven planes in a brine shrimp. In the more scattering mouse brain, they generated three axially shifted foci to characterize the functional property of L2/3, L4, and L5 neurons (170 µm, 350 µm, and 500 µm below pia, respectively) simultaneously at 10 Hz (three planes extending over 500 µm in z were also demonstrated)99. Here no temporal multiplexing was used to separate signals from different foci. But because activity in cortex is sparse, even though neurons at different z depths may overlap in the xy plane and cannot be resolved spatially, they can be segmented through their distinct activity patterns by independent component analysis100 or nonnegative matrix factorization methods101,102.
Taking this approach to the extreme, the speed of volumetric imaging is maximized when no attempt is made to resolve structures in the axial direction. This is equivalent to having axially shifted foci that are so close together as to form a continuous axial profile. One way to generate such an axially extended focus is by using annular illumination at the excitation objective103 (Fig. 3f), which form a focus known in optics as a Bessel beam. Using a phase mask to generate a high-NA Bessel beam, Botcherby et al. demonstrated extended depth-of-field imaging for two-photon fluorescence microscopy104. Using an axicon (a refractive element shaped like a cone)105, Theriault et al. applied the extended depth-of-field imaging to brain slices106 and showed that displacement of the annular illumination on the objective back pupil can be used to recover depth information via stereoscopic imaging104,106. Because a single elongated focus now probes all the structures along the z axis within the extended depth of field, 2D frame rate becomes 3D volume rate. Using an easy-to-adapt SLM-based module to generate Bessel foci, Lu et al. demonstrated in vivo volume imaging of neurons in ferret, mouse, zebrafish, and fly brains over 160 µm in depth at up to 30 Hz volume rate107. Preserving synaptic resolution over large axial and lateral displacements, Bessel-focus-based extended depth-of-field imaging allowed functional characterization of dendritic spines over multiple dendrites extending over 60 µm in depth.
Sampling strategies
Now that we have discussed the instrumentation for imaging neural activity in large volumes, we turn our attention to the sampling strategies. To maximize the number of neurons that can be sampled within a brain volume, optimized scan strategies are required. Scan strategies can be compared by considering the neuron-samples per second, assuming sufficient signal-to-noise ratio (SNR) per sample. The SNR in imaging systems that are shot-noise-limited, increases with higher photon fluxes (shot noise scales as the square root of the number of photons detected). The SNR requirement sets the range of photon counts that must be obtained for each neuron at each time step108, and can vary from experiment to experiment. In some cases, single action potential resolution is needed, which requires high SNR measurements. In other experiments, more qualitative measurements of changes in activity are desired, and SNR can be traded off to image more neurons. Ultimately the SNR sets the minimal dwell time per pixel, which is generally on the order of 0.1 – 1.0 µs for raster scanning, and 10 – 50 µs for sampling each neuron (a single neuron may be sampled by a single pixel or over multiple pixels that are averaged together during data analysis)59,109.
In practice, the achievable SNR is determined by the total number of photons that can be collected from each region of interest. It depends on the concentration, brightness, and photostability of the indicator, as well as the excitation and collection efficiencies of the imaging system. Simply increasing the excitation power may produce initial gain in SNR, but photobleaching of the indicator and the photodamage of the brain tissue may make such increase in SNR unsustainable. For large volume imaging, the excitation and collection efficiencies are typically in the same range as they are for small volume imaging, though the latter can be improved using supplementary collection strategies110,111. Sampling strategy should be optimized for each experiment with the required and achievable SNR in mind.
To compare approaches, we can define a neuronal sample rate U in units of neuron-samples·s−1. That is, either U neurons can be sampled at 1 Hz, or U/2 neurons can be sampled at 2 Hz, or U/500 neurons can be sampled at 500 Hz, and so forth. This quantity depends on the dwell time required to obtain sufficient signals, which is a function of the indicator properties, staining intensity, and the efficiencies of excitation and collection. Therefore, we will refer to actual realized performance in experiments, rather than theoretical values for specific approaches.
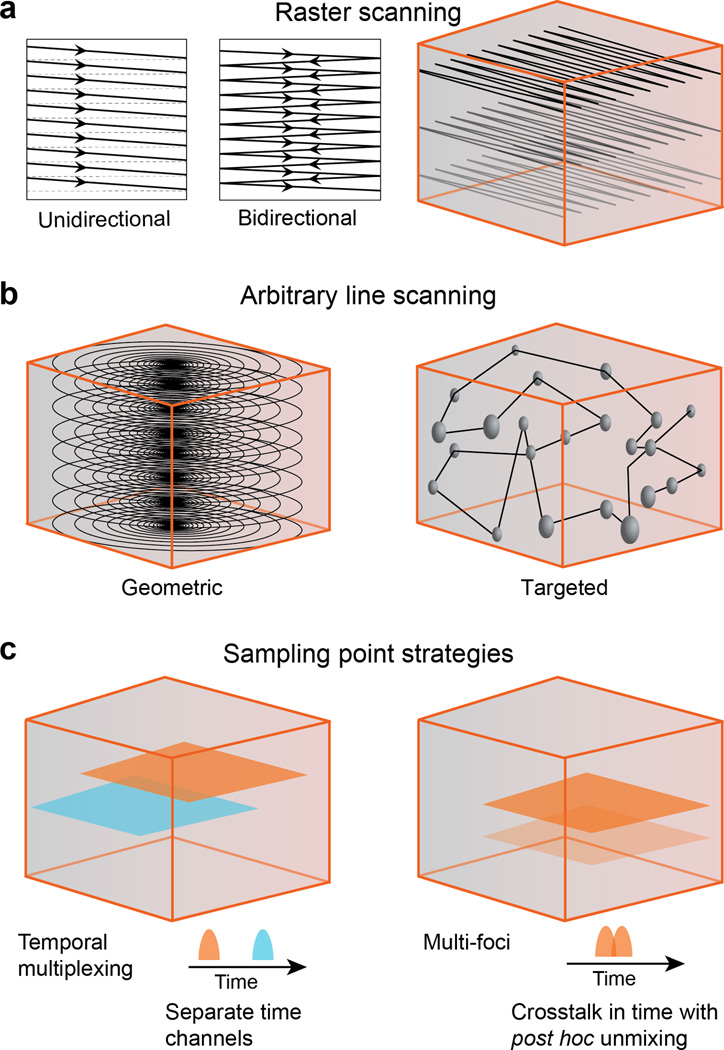
Raster scanning is the most commonly used strategy, but it is also among the least optimal because so many of the samples (i.e., pixels) are of structures that are not of interest, including blood vessels and neuropil. Still, unidirectional and bidirectional raster scanning (Fig. 4a) remains the preferred scan mode in practice for several reasons. First, raster scanning data can be registered post hoc to compensate for movement, which is often necessary in data from awake, behaving animals112. Second, resonant scan mirrors can provide frame rates that are sufficient to reveal neuronal activity dynamics on the time scale of behavior (10s to 100s of ms). Raster scanning with a resonant scanning axis yields about 100 cells at 30 Hz (U = 3,000 neuron-samples·s−1)113,114. Third, raster scanning provides several samples per neuron that can be averaged together to increase SNR.
Fig. 4.
Sampling strategies. (a) Raster scanning evenly samples a single plane of a volume. The scanning can be unidirectional or bidirectional, the latter offers higher speed, but may require additional image processing to reduce artifacts. Volumes can be imaged using multiple raster scanned planes. (b) Arbitrary line scanning can more optimally sample a volume. A geometric approach can be used with no prior knowledge of the anatomy, to sparsely sample a volume. Targeted scanning can use a previously acquired, raster-scanned, multiplane volume, as in a to target an arbitrary line scan to sample specific neurons. (c) Temporal multiplexing involves multiple beams scanning the volume with slight temporal delays between their laser pulses. Fluorescence events are attributed to specific beam if they occur in a small time window after excitation by that beam’s laser pulse, to ensure minimal crosstalk between multiplexed beams. Multi-foci scanning involves splitting laser power of each pulse between multiple foci. There is complete crosstalk between signals from the two pathways, but given sufficient sparsity, it is possible to demix signals from different neurons with high fidelity.
A potentially more optimal strategy is arbitrary line scanning (Fig. 4b). This involves a non-raster scan path that can be either optimized to sample a large volume sparsely or targeted to specific neurons whose locations are identified with a prior raster volume scan. High 2D imaging speed were achieved by using 2D vector scan and constraining the laser scan path to user-defined structures of interest and minimizing the “dead time” in between (e.g., vector-mode scanning115, targeted path scanning116,117, multiple line scanning method118, and heuristically optimal path scanning119). In a brain slice preparation, Sadovsky and colleagues achieved a U of 8,500 (1,000 neurons at 8.5 Hz)119. Gobel et al. used an untargeted arbitrary 3D scan path to sample in vivo 458 neurons at 10 Hz (U = 4,580)68. AODs are well-suited for random access sampling. Using a 3D AOD setup, Katona et al. obtained a random-access sample rate U of 23,800 to 54,30047. Cotton et al. used AODs to realize a U of 50,000 neuron-samples·s−1 (e.g., 500 cells at 100 Hz, or 250 cells at 50 Hz)109. One problem with arbitrary line scans for in vivo preparations is that movement artifacts cannot be corrected for post hoc. Online adjustments of arbitrary line scan paths has yet to be demonstrated (though raster scans have been corrected online120), but fast plane scans of high contrast objects have been used to detect movement and discard data that is corrupted by large movements109.
As described in the previous section, temporal multiplexing40,45,71,96 can multiply U by a factor equal to the number of foci (e.g., a factor of 4 in Cheng et al.96) (Fig. 4c). Finally, in the case of relatively sparse labeling or activity, analytical unmixing of signal generated by multiple foci simultaneously is a possibility99,102. This approach can multiply U by a factor equal to the number of foci.
Other considerations
Via the combination of hardware development and shrewd sampling strategy, we now have a variety of two-photon fluorescence microscopy methods at our disposal for rapid imaging of brain volumes. However, several factors, both practical and fundamental, restrict the usability of these methods in mammalian brains.
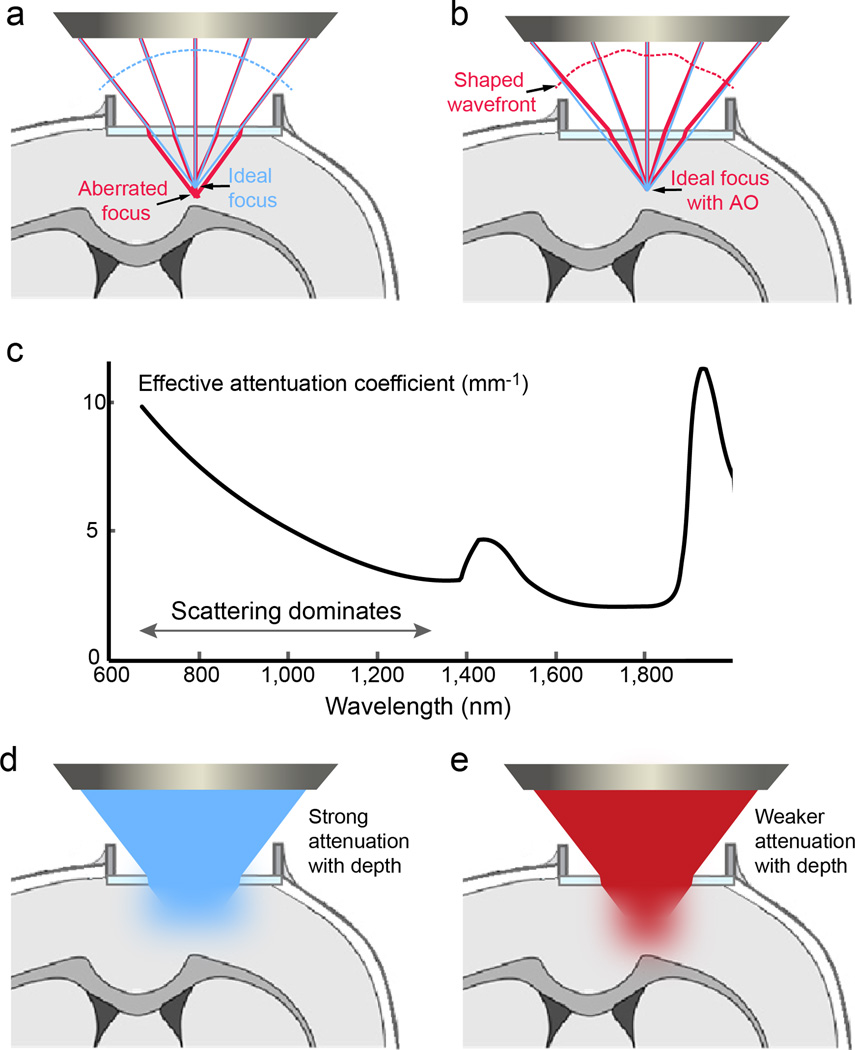
Brain-induced deterioration of image quality due to aberration and scattering places a constraint on volume imaging, especially along its z extent. Even if the imaging module itself operates in an aberration-free manner, the optical inhomogeneities of the mammalian brain distort and scatter the excitation light, which reduces its focal intensity and degrades image resolution at depth38 (Fig. 5). The brain’s distortion effects on the excitation wavefront can be counteracted by adaptive optics, where active shaping of the wavefront is used to cancel out brain-induced aberrations121–127 (Fig. 5a,b). With adaptive optics, diffraction-limited imaging with two-photon fluorescence microscopy can now be achieved deep inside mouse cortex and has been proven to be essential for the accurate characterization of tuning properties of thalamic boutons in the mouse primary visual cortex in vivo128. Combining adaptive optics with the volume imaging methods described above becomes increasingly important as imaging volumes grow in size.
Fig. 5.
Aberration and scattering limit imaging depth. (a) Brain distorts the wavefront of the excitation light and leads to an aberrated focus (formed by orange rays), lowering image resolution and brightness. (b) Shaping the wavefront with adaptive optics cancels out brain-induced aberrations and recovers an ideal, diffraction-limited focus (formed by red rays). Green rays illustrate ideal imaging condition where the brain does not change ray directions. (c) The wavelength dependence of the effective attenuation coefficient (1/mm) (modified from reference 129) indicates that optimal excitation wavelength windows are near 1,300 and 1,700 nm. (d) and (e) Longer wavelength excitation light penetrates scattering brains more effectively than shorter wavelength ones.
The reduction of focal intensity and detection efficiency by tissue scattering and absorption is more difficult to combat, and limits the maximal imaging depth to 5 – 6 attenuation lengths39. Currently, the most practical solutions for reducing scattering are through using longer-wavelength excitation light or red-shifted fluorescent dyes, due to the substantially lower attenuation by scattering at longer wavelength (Fig. 5c–e)129. For example, for neocortex labeled with red dyes in vivo, the characteristic attenuation lengths were found to be 131 µm at 775 nm, 285 µm at 1,280 nm, and 365 µm at 1,675 nm, respectively39,130. Using a red-emitting calcium indicator Cal-590 and 1,050 nm excitation, Tischbirek et al. recorded action potential-dependent calcium transients in all six layers of mouse cortex up to 900 µm below the pia surface in vivo131. Recently developed red genetically encoded calcium indicators also allowed orientation tuning properties of L6 neurons at 850 µm under pia in mouse visual cortex to be characterized in vivo98. Using Alexa680-Dextran and 1,280-nm excitation, Kobat et al. demonstrated in vivo two-photon imaging of brain vasculature down to 1.6-mm depth in mouse cortex132. Using 1,700 nm for three-photon excitation of RFP, subcortical neurons at over 1mm depth have been imaged through intact cortex in vivo39. Conveniently, most of the volumetric imaging methods (with the exception of AOD-based methods, which are usually optimized to work in a narrower wavelength range, due to their strong dispersion) can be used at longer wavelength without any modification.
A final factor that constrains the speed of volume imaging is the heating and damage of the brain by the excitation light. Because of the limited brightness of the probe, with the increase of image speed, the amount of power deposited into the brain also increases, which would eventually affect the physiological events under investigation. Although scattering decreases with wavelengths, increasing water absorption above 1.1 µm may lead to tissue heating129. Ongoing work has been carried out to study how the brain is affected at different power regimes133. Care needs to be always taken to ensure that the physiology of the system is not sacrificed in the pursuit of speed.
Data analysis
Large volumetric imaging presents enormous opportunities for characterizing the neural code, but also several computational challenges.
The most immediate challenge might appear to be the size of the data. A typical dataset of two-photon laser scanning microscopy in the mouse cortex generates around 50GB an hour (512×512 pixels of 4 planes at 8 Hz), and these numbers are likely to grow with new techniques. To give some perspective, however, this is far smaller than even the high-speed video used to capture mouse behavior (500 GB an hour). Such video data are processed immediately with well-vetted algorithms, and the raw data are discarded or highly compressed134.
The unique challenge of functional imaging data is not the sheer size, but the complexity, diversity, and continual evolution of analysis approaches. There is a semi-standard sequence of processing steps that most labs perform. (1) Image registration, which can include global cross-correlation, non-local alignment, or model-based approaches, (2) neuron identification or segmentation, and correction for neuropil contamination, which can be performed using morphological methods or matrix factorization, and (3) spike detection, which can use deconvolution or inverse modeling (See review on “Data quality” in this issue for more details on how to perform these steps). The fluorescence time course or spike train for every neuron can then be used to fit models that relate neuronal responses to stimuli, behavioral parameters, or the responses of other neurons10,25,64,135. There are myriad ways to perform each of these steps, and most methods must be tailored to experiment-specific acquisition properties, indicators, cell types, and spatial or temporal resolutions. The output of each step informs the others – an artifact due to neuropil contamination may only become clear when computing cross-neuron correlations – so most steps must be performed repeatedly, with different parameters, and with substantial manual inspection and intervention.
When repeating multiple steps in a complex pipeline, the time required for each step becomes a severe bottleneck, and for datasets of even 10 s or 100 s of GBs total processing time can grow to hours or days. One strategy for speeding up computation is parallelization, which can be achieved through multi-core processing on a single CPU, distributing computation across multiple machines in a compute cluster, or GPU computing. Several open source libraries well-suited to distributed array computation are available across a variety of languages, though only a subset are currently actively used for processing volumetric imaging data (e.g., Spark136, cluster computing in Python and Scala). In most cases, cluster computing is better suited to flexible workflows that require extensive customization and modification, or require repeated passes over an entire dataset136, whereas GPUs are well-suited to a subset of well-defined operations like image filtering or convolution, or operations that require minimal reading or writing of data137.
Regardless of the platform, different operations demand different strategies for parallelization. For raw data processing, many operations must be performed locally in space but can be distributed across time (e.g. image filtering or registration), whereas others must be performed locally in time but can be distributed across space (e.g. baseline removal or pixel-wise regression), and others require a combination (e.g. space-time matrix factorization, which is a common step in cell segmentation algorithms100,102). This kind of re-representation might be trivial on a single machine, but it can incur substantial costs when data are distributed across a cluster, and efficient processing requires structuring data on disk and in memory to support particular workflows.
After segmentation and time series extraction, data are often much smaller in size, typically less than a couple GBs, but parallelization remains important, in new ways. Fitting neural encoding models25,64, which capture the statistical relationship between stimulus or behavioral parameters and neural responses, often requires nonlinear optimization and calculating appropriate non-parametric statistics (e.g. permutation tests, monte carlo simulations). Fitting these models to hundreds or thousands of neurons at once, or fitting highly complex models, can benefit enormously from parallelization, as has been demonstrated both on CPUs52 and GPUs138. Faster computation can enable new forms of experimentation – such as fitting models online during an experiment and using the fits to present stimuli that test targeted, model-driven hypotheses139,140. When fitting models that examine pair-wise interactions, the number of parameters grows quadratically with the number of neurons, which requires regularization to avoid overfitting141, as well as strategies to avoid explicitly representing neuron-by-neuron matrices, which become unwieldy even on a large compute cluster.
Future prospects
There are fundamental limits to consider when evaluating the future prospects of large volume imaging142. Two key limiting factors are the fluorescent reporters that influence the SNR, and the difficulty of imaging with multiply scattered light. Firstly, brighter and larger signal (ΔF/F) calcium dyes and genetically encoded calcium indicators could aid large volume brain imaging. These improvements would increase the SNR, and thus permit smaller dwell times and faster sampling. Red-shifted indicators can also provide for deeper imaging by causing less scattering for both excitation and emission light on their way to and from the focal point, and less signal loss due to absorption by hemoglobin129. Calcium imaging is an indirect reporter of neuronal activity, and thus there are opportunities for more direct reporters of activity, e.g., reporters of voltage changes or neurotransmitter release143–145. Secondly, new methods to image with multiply scattered light would aid large volume brain imaging. This is particularly true when imaging mammalian cortex. Recent research has revealed that it is possible to focus light into highly scattering tissue146, although it is challenging to account for the fast spatial and temporal variations in the scattering profile of live tissue in real time. Technical progress in indicator and hardware performance can provide potential for these approaches to enhance the imaging of neuronal activity within large volumes.
Thus, ultimately there is ample technological headroom to explore for imaging neural activity dynamics over larger tissue volumes with individual neuron resolution. This headroom can be explored using iterative engineering (e.g., larger imaging optics, faster scanning) and novel approaches (e.g., constrained non-negative matrix factorization for unmixing). It can be helpful to design new technology with a specific experimental question in mind. Such guidance can ensure that the technology can be used right away to advance neuroscience, and moreover, the experimental demands of a specific experiment may actually relax some requirements and allow for a different set of engineering compromises. For example, high axial resolution may be key for some experiments, but low axial resolution can also enable high throughput in sparsely labeled samples. New technology for imaging in larger volumes should be guided by pressing neurobiological questions, and evaluated on the basis of its fitness for its intended use.
Acknowledgments
We thank J. Kuhl and A. Chen for help with illustrations and C. Xu for providing the high-resolution version of Fig. 5c, and N. Sofroniew for providing helpful comments. N.J. and J. F. are supported by Howard Hughes Medical Institute. S.L.S. is supported by grants from the Human Frontier Science Program (CDA00063/2012, RGP0027/2016;), the National Science Foundation (1450824), the Whitehall Foundation, the Klingenstein Foundation, the McKnight Foundation, the Simons Foundation (SCGB 325407SS), and the National Institutes of Health (R01NS091335,R01EY024294).
References
- 1.Meltzer SJ. Emil Du Bois-Reymond. Science. 1897;5:217–219. doi: 10.1126/science.5.110.217. [DOI] [PubMed] [Google Scholar]
- 2.Rothschild RM. Neuroengineering tools/applications for bidirectional interfaces, brain computer interfaces, and neuroprosthetic implants - a review of recent progress. Frontiers in Neuroengineering. 2010;3 doi: 10.3389/fneng.2010.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith SL, Judy JW, Otis TS. An ultra small array of electrodes for stimulating multiple inputs into a single neuron. Journal of Neuroscience Methods. 2004;133:109–114. doi: 10.1016/j.jneumeth.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Harris KD, Henze DA, Csicsvari J, Hirase H, Buzsáki G. Accuracy of Tetrode Spike Separation as Determined by Simultaneous Intracellular and Extracellular Measurements. Journal of Neurophysiology. 2000;84:401–414. doi: 10.1152/jn.2000.84.1.401. [DOI] [PubMed] [Google Scholar]
- 5.Nicolelis MAL, Ribeiro S. Multielectrode recordings: the next steps. Current Opinion in Neurobiology. 2002;12:602–606. doi: 10.1016/s0959-4388(02)00374-4. [DOI] [PubMed] [Google Scholar]
- 6.Henze DA, et al. Intracellular Features Predicted by Extracellular Recordings in the Hippocampus In Vivo. Journal of Neurophysiology. 2000;84:390–400. doi: 10.1152/jn.2000.84.1.390. [DOI] [PubMed] [Google Scholar]
- 7.Hendel T, et al. Fluorescence Changes of Genetic Calcium Indicators and OGB-1 Correlated with Neural Activity and Calcium In Vivo and In Vitro. The Journal of Neuroscience. 2008;28:7399–7411. doi: 10.1523/JNEUROSCI.1038-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen T-W, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grienberger C, Konnerth A. Imaging Calcium in Neurons. Neuron. 2012;73:862–885. doi: 10.1016/j.neuron.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Huber D, et al. Multiple dynamic representations in the motor cortex during sensorimotor learning. Nature. 2012;484:473–478. doi: 10.1038/nature11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Meth. 2013;10:413–420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- 12.Denk W, Strickler J, Webb W. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 13.Denk W, et al. Anatomical and functional imaging of neurons using 2-photon laser scanning microscopy. Journal of Neuroscience Methods. 1994;54:151–162. doi: 10.1016/0165-0270(94)90189-9. [DOI] [PubMed] [Google Scholar]
- 14.Svoboda K, Denk W, Kleinfeld D, Tank DW. In vivo dendritic calcium dynamics in neocortical pyramidal neurons. Nature. 1997;385:161–165. doi: 10.1038/385161a0. [DOI] [PubMed] [Google Scholar]
- 15.Denk W, Svoboda K. Photon Upmanship: Why Multiphoton Imaging Is More than a Gimmick. Neuron. 1997;18:351–357. doi: 10.1016/s0896-6273(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 16.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat Biotech. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 17.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Meth. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 18.Stosiek C, Garaschuk O, Holthoff K, Konnerth A. In vivo two-photon calcium imaging of neuronal networks. Proceedings of the National Academy of Sciences. 2003;100:7319–7324. doi: 10.1073/pnas.1232232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill DN, Varga Z, Jia H, Sakmann B, Konnerth A. Multibranch activity in basal and tuft dendrites during firing of layer 5 cortical neurons in vivo. Proceedings of the National Academy of Sciences. 2013;110:13618–13623. doi: 10.1073/pnas.1312599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Gao E, Burkhalter A. Gateways of Ventral and Dorsal Streams in Mouse Visual Cortex. The Journal of Neuroscience. 2011;31:1905–1918. doi: 10.1523/JNEUROSCI.3488-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marshel JH, Garrett ME, Nauhaus I, Callaway EM. Functional specialization of seven mouse visual cortical areas. Neuron. 2011;72:1040–1054. doi: 10.1016/j.neuron.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andermann ML, Kerlin AM, Roumis DK, Glickfeld LL, Reid RC. Functional specialization of mouse higher visual cortical areas. Neuron. 2011;72:1025–1039. doi: 10.1016/j.neuron.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q, Sporns O, Burkhalter A. Network Analysis of Corticocortical Connections Reveals Ventral and Dorsal Processing Streams in Mouse Visual Cortex. The Journal of Neuroscience. 2012;32:4386–4399. doi: 10.1523/JNEUROSCI.6063-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glickfeld LL, Andermann ML, Bonin V, Reid RC. Cortico-cortical projections in mouse visual cortex are functionally target specific. Nature Neurosci. 2013;16:219–226. doi: 10.1038/nn.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pillow JW, et al. Spatio-temporal correlations and visual signalling in a complete neuronal population. Nature. 2008;454:995–999. doi: 10.1038/nature07140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Averbeck BB, Latham PE, Pouget A. Neural correlations, population coding and computation. Nat Rev Neurosci. 2006;7:358–366. doi: 10.1038/nrn1888. [DOI] [PubMed] [Google Scholar]
- 27.Yang G, Pan F, Parkhurst CN, Grutzendler J, Gan W-B. Thinned-skull cranial window technique for long-term imaging of the cortex in live mice. Nat. Protocols. 2010;5:201–208. doi: 10.1038/nprot.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drew PJ, et al. Chronic optical access through a polished and reinforced thinned skull. Nat Meth. 2010;7:981–984. doi: 10.1038/nmeth.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holtmaat A, et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat. Protocols. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barretto RPJ, et al. Time-lapse imaging of disease progression in deep brain areas using fluorescence microendoscopy. Nat Med. 2011;17:223–228. doi: 10.1038/nm.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bocarsly ME, et al. Minimally invasive microendoscopy system for in vivo functional imaging of deep nuclei in the mouse brain. Biomed Opt Express. 2015;6:4546–4556. doi: 10.1364/BOE.6.004546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chia TH, Levene MJ. Microprisms for In Vivo Multilayer Cortical Imaging. Journal of Neurophysiology. 2009;102:1310–1314. doi: 10.1152/jn.91208.2008. [DOI] [PubMed] [Google Scholar]
- 33.Andermann Mark L, et al. Chronic Cellular Imaging of Entire Cortical Columns in Awake Mice Using Microprisms. Neuron. 2013;80:900–913. doi: 10.1016/j.neuron.2013.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung JC, Mehta AD, Aksay E, Stepnoski R, Schnitzer MJ. In Vivo Mammalian Brain Imaging Using One- and Two-Photon Fluorescence Microendoscopy. J Neurophysiol. 2004;92:3121–3133. doi: 10.1152/jn.00234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levene MJ, Dombeck DA, Kasischke KA, Molloy RP, Webb WW. In vivo multiphoton microscopy of deep brain tissue. J Neurophysiol. 2004;91:1908–1912. doi: 10.1152/jn.01007.2003. [DOI] [PubMed] [Google Scholar]
- 36.Horton N, Wang K, Kobat D, Wise FW, Xu C. Optics in the Life Sciences NT3B.3. Hawaii: Optical Society of America, Waikoloa Beach; 2013. In Vivo Deep Penetration Three-Photon Imaging of Mouse Brain through an Unthinned, Intact Skull. [Google Scholar]
- 37.Theer P, Hasan MT, Denk W. Two-photon imaging to a depth of 1000 µm in living brains by use of a Ti:Al2O3 regenerative amplifier. Opt. Lett. 2003;28:1022–1024. doi: 10.1364/ol.28.001022. [DOI] [PubMed] [Google Scholar]
- 38.Ji N. The practical and fundamental limits of optical imaging in mammalian brains. Neuron. 2014;83:1242–1245. doi: 10.1016/j.neuron.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Horton NG, et al. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat Photon. 2013;7:205–209. doi: 10.1038/nphoton.2012.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stirman JN, Smith IT, Kudenov MW, Smith SL. Wide field-of-view, twin-region two-photon imaging across extended cortical networks. bioRxiv. 2014 [Google Scholar]
- 41.Tsai PS, et al. Ultra-large field-of-view two-photon microscopy. Optics express. 2015;23:13833–13847. doi: 10.1364/OE.23.013833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sofroniew NJ, Flickinger D, King J, Svoboda K. A large field-of-view two-photon microscope with subcellular resolution for in vivo imaging. bioRxiv. 2016 doi: 10.7554/eLife.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng G, Ou X, Horstmeyer R, Yang C. Characterization of spatially varying aberrations for wide field-of-view microscopy. Optics express. 2013;21:15131–15143. doi: 10.1364/OE.21.015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Negrean A, Mansvelder HD. Optimal lens design and use in laser-scanning microscopy. Biomed Opt Express. 2014;5:1588–1609. doi: 10.1364/BOE.5.001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voigt FF, Chen JL, Krueppel R, Helmchen F. A modular two-photon microscope for simultaneous imaging of distant cortical areas in vivo. 2015 93292C-93292C-93297. [Google Scholar]
- 46.Chen JL, Voigt FF, Javadzadeh M, Krueppel R, Helmchen F. Long-Range Population Dynamics of Anatomically Defined Neocortical Networks. eLife. 2016:e14679. doi: 10.7554/eLife.14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katona G, et al. Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat Meth. 2012;9:201–208. doi: 10.1038/nmeth.1851. [DOI] [PubMed] [Google Scholar]
- 48.Heys James G, Rangarajan Krsna V, Dombeck Daniel A. The Functional Micro-organization of Grid Cells Revealed by Cellular-Resolution Imaging. Neuron. 2014;84:1079–1090. doi: 10.1016/j.neuron.2014.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Low RJ, Gu Y, Tank DW. Cellular resolution optical access to brain regions in fissures: Imaging medial prefrontal cortex and grid cells in entorhinal cortex. Proceedings of the National Academy of Sciences. 2014;111:18739–18744. doi: 10.1073/pnas.1421753111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan GY, et al. Video-Rate Scanning Two-Photon Excitation Fluorescence Microscopy and Ratio Imaging with Cameleons. Biophysical Journal. 1999;76:2412–2420. doi: 10.1016/S0006-3495(99)77396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Varga Z, Jia H, Sakmann B, Konnerth A. Dendritic coding of multiple sensory inputs in single cortical neurons in vivo. Proceedings of the National Academy of Sciences. 2011;108:15420–15425. doi: 10.1073/pnas.1112355108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bullen A, Patel SS, Saggau P. High-speed, random-access fluorescence microscopy: I. High-resolution optical recording with voltage-sensitive dyes and ion indicators. Biophysical Journal. 1997;73:477–491. doi: 10.1016/S0006-3495(97)78086-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iyer V, Losavio BE, Saggau P. Compensation of spatial and temporal dispersion for acousto-optic multiphoton laser-scanning microscopy. Journal of Biomedical Optics. 2003;8:460–471. doi: 10.1117/1.1580827. [DOI] [PubMed] [Google Scholar]
- 54.Roorda RD, Hohl TM, Toledo-Crow R, Miesenböck G. Video-Rate Nonlinear Microscopy of Neuronal Membrane Dynamics With Genetically Encoded Probes. Journal of Neurophysiology. 2004;92:609–621. doi: 10.1152/jn.00087.2004. [DOI] [PubMed] [Google Scholar]
- 55.Lechleiter JD, Lin D-T, Sieneart I. Multi-Photon Laser Scanning Microscopy Using an Acoustic Optical Deflector. Biophysical Journal. 2002;83:2292–2299. doi: 10.1016/S0006-3495(02)73989-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X, Leischner U, Rochefort NL, Nelken I, Konnerth A. Functional mapping of single spines in cortical neurons in vivo. Nature. 2011;475:501–505. doi: 10.1038/nature10193. [DOI] [PubMed] [Google Scholar]
- 57.Iyer V, Hoogland TM, Saggau P. Fast Functional Imaging of Single Neurons Using Random-Access Multiphoton (RAMP) Microscopy. Journal of Neurophysiology. 2006;95:535–545. doi: 10.1152/jn.00865.2005. [DOI] [PubMed] [Google Scholar]
- 58.Otsu Y, et al. Optical monitoring of neuronal activity at high frame rate with a digital random-access multiphoton (RAMP) microscope. Journal of Neuroscience Methods. 2008;173:259–270. doi: 10.1016/j.jneumeth.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 59.Grewe BF, Langer D, Kasper H, Kampa BM, Helmchen F. High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nat Meth. 2010;7:399–405. doi: 10.1038/nmeth.1453. [DOI] [PubMed] [Google Scholar]
- 60.Hammond AT, Glick BS. Raising the Speed Limits for 4D Fluorescence Microscopy. Traffic. 2000;1:935–940. [PubMed] [Google Scholar]
- 61.Callamaras N, Parker I. Construction of a confocal microscope for real-time x-y and x-z imaging. Cell Calcium. 1999;26:271–279. doi: 10.1054/ceca.1999.0085. [DOI] [PubMed] [Google Scholar]
- 62.Andermann ML, Kerlin AM, Reid C. Chronic cellular imaging of mouse visual cortex during operant behavior and passive viewing. Frontiers in Cellular Neuroscience. 2010;4 doi: 10.3389/fncel.2010.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kerlin AM, Andermann ML, Berezovskii VK, Reid RC. Broadly Tuned Response Properties of Diverse Inhibitory Neuron Subtypes in Mouse Visual Cortex. Neuron. 2010;67:858–871. doi: 10.1016/j.neuron.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peron Simon P, Freeman J, Iyer V, Guo C, Svoboda K. A Cellular Resolution Map of Barrel Cortex Activity during Tactile Behavior. Neuron. 2015;86:783–799. doi: 10.1016/j.neuron.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 65.Sofroniew NJ, Vlasov YA, Andrew Hires S, Freeman J, Svoboda K. Neural coding in barrel cortex during whisker-guided locomotion. eLife. 2015;4:e12559. doi: 10.7554/eLife.12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seelig JD, Jayaraman V. Neural dynamics for landmark orientation and angular path integration. Nature. 2015;521:186–191. doi: 10.1038/nature14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cossell L, et al. Functional organization of excitatory synaptic strength in primary visual cortex. Nature. 2015;518:399–403. doi: 10.1038/nature14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gobel W, Kampa BM, Helmchen F. Imaging cellular network dynamics in three dimensions using fast 3D laser scanning. Nat Meth. 2007;4:73–79. doi: 10.1038/nmeth989. [DOI] [PubMed] [Google Scholar]
- 69.Katona G, et al. Roller Coaster Scanning reveals spontaneous triggering of dendritic spikes in CA1 interneurons. Proceedings of the National Academy of Sciences. 2011;108:2148–2153. doi: 10.1073/pnas.1009270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dal Maschio M, De Stasi AM, Benfenati F, Fellin T. Three-dimensional in vivo scanning microscopy with inertia-free focus control. Optics letters. 2011;36:3503–3505. doi: 10.1364/OL.36.003503. [DOI] [PubMed] [Google Scholar]
- 71.Amir W, et al. Simultaneous imaging of multiple focal planes using a two-photon scanning microscope. Optics letters. 2007;32:1731–1733. doi: 10.1364/ol.32.001731. [DOI] [PubMed] [Google Scholar]
- 72.Moghimi MJ, Chattergoon KN, Wilson CR, Dickensheets DL. High Speed Focus Control MEMS Mirror With Controlled Air Damping for Vital Microscopy. Journal of Microelectromechanical Systems. 2013;22:938–948. [Google Scholar]
- 73.Blum M, Büeler M, Grätzel C, Aschwanden M. Compact optical design solutions using focus tunable lenses. 2011 81670W-81670W-81679. [Google Scholar]
- 74.Mermillod-Blondin A, McLeod E, Arnold CB. High-speed varifocal imaging with a tunable acoustic gradient index of refraction lens. Optics letters. 2008;33:2146–2148. doi: 10.1364/ol.33.002146. [DOI] [PubMed] [Google Scholar]
- 75.Grewe BF, Voigt FF, van ’t Hoff M, Helmchen F. Fast two-layer two-photon imaging of neuronal cell populations using an electrically tunable lens. Biomed Opt Express. 2011;2:2035–2046. doi: 10.1364/BOE.2.002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sheffield MEJ, Dombeck DA. Calcium transient prevalence across the dendritic arbour predicts place field properties. Nature. 2015:200–204. doi: 10.1038/nature13871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kong L, et al. Continuous volumetric imaging via an optical phase-locked ultrasound lens. Nat Meth. 2015;12:759–762. doi: 10.1038/nmeth.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reddy GD, Saggau P. Fast three-dimensional laser scanning scheme using acousto-optic deflectors. Journal of Biomedical Optics. 2005;10 doi: 10.1117/1.2141504. 064038-064038-064010. [DOI] [PubMed] [Google Scholar]
- 79.Kaplan A, Friedman N, Davidson N. Acousto-optic lens with very fast focus scanning. Optics letters. 2001;26:1078–1080. doi: 10.1364/ol.26.001078. [DOI] [PubMed] [Google Scholar]
- 80.Reddy GD, Kelleher K, Fink R, Saggau P. Three-dimensional random access multiphoton microscopy for functional imaging of neuronal activity. Nat Neurosci. 2008;11:713–720. doi: 10.1038/nn.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kirkby PA, Srinivas Nadella KMN, Silver RA. A compact acousto-optic lens for 2D and 3D femtosecond based 2-photon microscopy. Optics express. 2010;18:13720–13744. doi: 10.1364/OE.18.013720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Froudarakis E, et al. Population code in mouse V1 facilitates readout of natural scenes through increased sparseness. Nat Neurosci. 2014;17:851–857. doi: 10.1038/nn.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sheppard CJR, Gu M. Aberration compensation in confocal microscopy. Appl. Opt. 1991;30:3563–3568. doi: 10.1364/AO.30.003563. [DOI] [PubMed] [Google Scholar]
- 84.Konstantinou G, et al. Dynamic wavefront shaping with an acousto-optic lens for laser scanning microscopy. Optics express. 2016;24:6283–6299. doi: 10.1364/OE.24.006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Akemann W, et al. Fast spatial beam shaping by acousto-optic diffraction for 3D non-linear microscopy. Optics express. 2015;23:28191–28205. doi: 10.1364/OE.23.028191. [DOI] [PubMed] [Google Scholar]
- 86.Botcherby EJ, Juskaitis R, Booth MJ, Wilson T. Aberration-free optical refocusing in high numerical aperture microscopy. Optics letters. 2007;32:2007–2009. doi: 10.1364/ol.32.002007. [DOI] [PubMed] [Google Scholar]
- 87.Botcherby EJ, Juškaitis R, Booth MJ, Wilson T. An optical technique for remote focusing in microscopy. Optics Communications. 2008;281:880–887. [Google Scholar]
- 88.Botcherby EJ, et al. Aberration-free three-dimensional multiphoton imaging of neuronal activity at kHz rates. Proceedings of the National Academy of Sciences. 2012;109:2919–2924. doi: 10.1073/pnas.1111662109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith CW, Botcherby EJ, Booth MJ, Juškaitis R, Wilson T. Agitation-free multiphoton microscopy of oblique planes. Optics letters. 2011;36:663–665. doi: 10.1364/OL.36.000663. [DOI] [PubMed] [Google Scholar]
- 90.Colon J, Lim H. Shaping field for 3D laser scanning microscopy. Optics letters. 2015;40:3300–3303. doi: 10.1364/OL.40.003300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rupprecht P, Prendergast A, Wyart C, Friedrich RW. Remote z-scanning with a macroscopic voice coil motor for fast 3D multiphoton laser scanning microscopy. Biomed Opt Express. 2016;7:1656–1671. doi: 10.1364/BOE.7.001656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bewersdorf J, Pick R, Hell SW. Multifocal multiphoton microscopy. Optics letters. 1998;23:655–657. doi: 10.1364/ol.23.000655. [DOI] [PubMed] [Google Scholar]
- 93.Buist, Müller, Squier, Brakenhoff Real time two-photon absorption microscopy using multi point excitation. Journal of Microscopy. 1998;192:217–226. [Google Scholar]
- 94.Watson BO, Nikolenko V, Yuste R. Two-photon imaging with diffractive optical elements. Frontiers in Neural Circuits. 2009;3 doi: 10.3389/neuro.04.006.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim KH, et al. Multifocal multiphoton microscopy based on multianode photomultiplier tubes. Optics express. 2007;15:11658–11678. doi: 10.1364/oe.15.011658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cheng A, Goncalves JT, Golshani P, Arisaka K, Portera-Cailliau C. Simultaneous two-photon calcium imaging at different depths with spatiotemporal multiplexing. Nat Meth. 2011;8:139–142. doi: 10.1038/nmeth.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sheetz KE, Hoover EE, Carriles R, Kleinfeld D, Squier JA. Advancing multifocal nonlinear microscopy: development and application of a novel multibeam Yb:KGd(WO4)2 oscillator. Optics express. 2008;16:17574–17584. doi: 10.1364/oe.16.017574. [DOI] [PubMed] [Google Scholar]
- 98.Dana H, et al. Sensitive red protein calcium indicators for imaging neural activity. eLife. 2016;5:e12727. doi: 10.7554/eLife.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang W, et al. Simultaneous Multi-plane Imaging of Neural Circuits. Neuron. 2016;89:269–284. doi: 10.1016/j.neuron.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mukamel EA, Nimmerjahn A, Schnitzer MJ. Automated Analysis of Cellular Signals from Large-Scale Calcium Imaging Data. Neuron. 2009;63:747–760. doi: 10.1016/j.neuron.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Maruyama R, et al. Detecting cells using non-negative matrix factorization on calcium imaging data. Neural Networks. 2014;55:11–19. doi: 10.1016/j.neunet.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 102.Pnevmatikakis Eftychios A, et al. Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron. 2016;89:285–299. doi: 10.1016/j.neuron.2015.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Welford WT. Use of Annular Apertures to Increase Focal Depth. Journal of the Optical Society of America. 1960;50:749–753. [Google Scholar]
- 104.Botcherby EJ, Juškaitis R, Wilson T. Scanning two photon fluorescence microscopy with extended depth of field. Optics Communications. 2006;268:253–260. [Google Scholar]
- 105.Thériault G, De Koninck Y, McCarthy N. Extended depth of field microscopy for rapid volumetric two-photon imaging. Optics express. 2013;21:10095–10104. doi: 10.1364/OE.21.010095. [DOI] [PubMed] [Google Scholar]
- 106.Thériault G, Cottet M, Castonguay A, McCarthy N, De Koninck Y. Extended two-photon microscopy in live samples with Bessel beams: steadier focus, faster volume scans, and simpler stereoscopic imaging. Frontiers in Cellular Neuroscience. 2014;8 doi: 10.3389/fncel.2014.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lu R, et al. Video-rate volumetric imaging of brains with synaptic resolution. BioRxiv. 2016 [Google Scholar]
- 108.Wilt Brian A, Fitzgerald James E, Schnitzer Mark J. Photon Shot Noise Limits on Optical Detection of Neuronal Spikes and Estimation of Spike Timing. Biophysical Journal. 104:51–62. doi: 10.1016/j.bpj.2012.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cotton RJ, Froudarakis E, Storer P, Saggau P, Tolias AS. Three-dimensional mapping of microcircuit correlation structure. Frontiers in Neural Circuits. 2013;7 doi: 10.3389/fncir.2013.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Engelbrecht CJ, Göbel W, Helmchen F. Enhanced fluorescence signal in nonlinear microscopy through supplementary fiber-optic light collection. Optics express. 2009;17:6421–6435. doi: 10.1364/oe.17.006421. [DOI] [PubMed] [Google Scholar]
- 111.McMullen JD, Kwan AC, Williams RM, Zipfel WR. Enhancing collection efficiency in large field of view multiphoton microscopy. Journal of Microscopy. 2011;241:119–124. doi: 10.1111/j.1365-2818.2010.03419.x. [DOI] [PubMed] [Google Scholar]
- 112.Dombeck DA, Khabbaz AN, Collman F, Adelman TL, Tank DW. Imaging Large-Scale Neural Activity with Cellular Resolution in Awake, Mobile Mice. Neuron. 2007;56:43–57. doi: 10.1016/j.neuron.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nguyen QT, Callamaras N, Hsieh C, Parker I. Construction of a two-photon microscope for video-rate Ca2+ imaging. Cell Calcium. 2001;30:383–393. doi: 10.1054/ceca.2001.0246. [DOI] [PubMed] [Google Scholar]
- 114.Bonin V, Histed MH, Yurgenson S, Reid RC. Local diversity and fine-scale organization of receptive fields in mouse visual cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:18506–18521. doi: 10.1523/JNEUROSCI.2974-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nikolenko V, Poskanzer KE, Yuste R. Two-photon photostimulation and imaging of neural circuits. Nat Meth. 2007;4:943–950. doi: 10.1038/nmeth1105. [DOI] [PubMed] [Google Scholar]
- 116.Lillis KP, Eng A, White JA, Mertz J. Two-photon imaging of spatially extended neuronal network dynamics with high temporal resolution. Journal of Neuroscience Methods. 2008;172:178–184. doi: 10.1016/j.jneumeth.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Valmianski I, et al. Automatic Identification of Fluorescently Labeled Brain Cells for Rapid Functional Imaging. Journal of Neurophysiology. 2010;104:1803–1811. doi: 10.1152/jn.00484.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lőrincz A, Rózsa B, Katona G, Vizi ES, Tamás G. Differential distribution of NCX1 contributes to spine–dendrite compartmentalization in CA1 pyramidal cells. Proceedings of the National Academy of Sciences. 2007;104:1033–1038. doi: 10.1073/pnas.0605412104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sadovsky AJ, et al. Heuristically optimal path scanning for high-speed multiphoton circuit imaging. Journal of Neurophysiology. 2011;106:1591–1598. doi: 10.1152/jn.00334.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen JL, Pfäffli OA, Voigt FF, Margolis DJ, Helmchen F. Online correction of licking-induced brain motion during two-photon imaging with a tunable lens. The Journal of Physiology. 2013;591:4689–4698. doi: 10.1113/jphysiol.2013.259804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ji N, Milkie DE, Betzig E. Adaptive optics via pupil segmentation for high-resolution imaging in biological tissues. Nature Meth. 2010;7:141–147. doi: 10.1038/nmeth.1411. [DOI] [PubMed] [Google Scholar]
- 122.Tao X, F B, Azucena O, Fu M, Garcia D, Zuo Y, Chen D, Kubby J. Adaptive optics confocal microscopy using direct wavefront sensing. Opt. Lett. 2011;36 doi: 10.1364/OL.36.001062. [DOI] [PubMed] [Google Scholar]
- 123.Tao X, A O, Fu M, Zuo Y, Chen D, Kubby J. Adaptive optics microscopy with direct wavefront sensing using fluorescent protein guide stars. Opt. Lett. 2011;36 doi: 10.1364/OL.36.003389. [DOI] [PubMed] [Google Scholar]
- 124.Ji N, Sato TR, Betzig E. Characterization and adaptive optical correction of aberrations during in vivo imaging in the mouse cortex. Proc. Natl. Acad. Sci. U S A. 2012;109:22–27. doi: 10.1073/pnas.1109202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang C, et al. Multiplexed aberration measurement for deep tissue imaging in vivo. Nature Meth. 2014;11:1037–1040. doi: 10.1038/nmeth.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang K, et al. Rapid adaptive optical recovery of optimal resolution over large volumes. Nat Meth. 2014;11:625–628. doi: 10.1038/nmeth.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang K, et al. Direct wavefront sensing for high-resolution in vivo imaging in scattering tissue. Nat Commun. 2015;6 doi: 10.1038/ncomms8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sun W, Tan Z, Mensh BD, Ji N. Thalamus provides layer 4 of primary visual cortex with orientation- and direction-tuned inputs. Nat Neurosci. 2016;19:308–315. doi: 10.1038/nn.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang K, Horton NG, Charan K, Xu C. Advanced Fiber Soliton Sources for Nonlinear Deep Tissue Imaging in Biophotonics. IEEE Journal of Selected Topics in Quantum Electronics. 2014;20:50–60. [Google Scholar]
- 130.Kobat D, et al. Deep tissue multiphoton microscopy using longer wavelength excitation. Optics express. 2009;17:13354–13364. doi: 10.1364/oe.17.013354. [DOI] [PubMed] [Google Scholar]
- 131.Tischbirek C, Birkner A, Jia H, Sakmann B, Konnerth A. Deep two-photon brain imaging with a red-shifted fluorometric Ca2+ indicator. Proceedings of the National Academy of Sciences. 2015;112:11377–11382. doi: 10.1073/pnas.1514209112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kobat D, et al. Deep tissue multiphoton microscopy using longer wavelength excitation. Opt. Express. 2009;17:13354–13364. doi: 10.1364/oe.17.013354. [DOI] [PubMed] [Google Scholar]
- 133.Podgorski K, Ranganathan G. Brain heating induced by near infrared lasers during multi-photon microscopy. J Neurophysiol. 2016 doi: 10.1152/jn.00275.2016. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Clack NG, et al. Automated Tracking of Whiskers in Videos of Head Fixed Rodents. PLoS Comput Biol. 2012;8:e1002591. doi: 10.1371/journal.pcbi.1002591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ahrens M, Paninski L, Sahani M. Inferring input nonlinearities in neural encoding models. Network: computation in neural systems. 2008;19:35–67. doi: 10.1080/09548980701813936. [DOI] [PubMed] [Google Scholar]
- 136.Freeman J, et al. Mapping brain activity at scale with cluster computing. Nat Meth. 2014;11:941–950. doi: 10.1038/nmeth.3041. [DOI] [PubMed] [Google Scholar]
- 137.Amat F, et al. Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat Meth. 2014;11:951–958. doi: 10.1038/nmeth.3036. [DOI] [PubMed] [Google Scholar]
- 138.Latimer KW, Yates JL, Meister MLR, Huk AC, Pillow JW. Single-trial spike trains in parietal cortex reveal discrete steps during decision-making. Science. 2015;349:184–187. doi: 10.1126/science.aaa4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Grosenick L, Marshel James H, Deisseroth K. Closed-Loop and Activity-Guided Optogenetic Control. Neuron. 2015;86:106–139. doi: 10.1016/j.neuron.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Freeman J, et al. Mapping nonlinear receptive field structure in primate retina at single cone resolution. eLife. 2015;4:e05241. doi: 10.7554/eLife.05241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yatsenko D, et al. Improved Estimation and Interpretation of Correlations in Neural Circuits. PLoS Comput Biol. 2015;11:e1004083. doi: 10.1371/journal.pcbi.1004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Marblestone AH, et al. Physical Principles for Scalable Neural Recording. Frontiers in Computational Neuroscience. 2013;7 doi: 10.3389/fncom.2013.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Marvin JS, et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat Meth. 2013;10:162–170. doi: 10.1038/nmeth.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gong Y, et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science. 2015;350:1361–1366. doi: 10.1126/science.aab0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.St-Pierre F, et al. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat Neurosci. 2014;17:884–889. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Judkewitz B, Wang YM, Horstmeyer R, Mathy A, Yang C. Speckle-scale focusing in the diffusive regime with time reversal of variance-encoded light (TROVE) Nat Photon. 2013;7:300–305. doi: 10.1038/nphoton.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]