Abstract
Background
In animal pathogenic bacteria, horizontal gene transfer events (HGT) have been frequently observed in genomic regions that encode functions involved in biosynthesis of the outer membrane located lipopolysaccharide (LPS). As a result, different strains of the same pathogen can have substantially different lps biosynthetic gene clusters. Since LPS is highly antigenic, the variation at lps loci is attributed to be of advantage in evading the host immune system. Although LPS has been suggested as a potentiator of plant defense responses, interstrain variation at lps biosynthetic gene clusters has not been reported for any plant pathogenic bacterium.
Results
We report here the complete sequence of a 12.2 kb virulence locus of Xanthomonas oryzae pv. oryzae (Xoo) encoding six genes whose products are homologous to functions involved in LPS biosynthesis and transport. All six open reading frames (ORFs) have atypical G+C content and altered codon usage, which are the hallmarks of genomic islands that are acquired by horizontal gene transfer. The lps locus is flanked by highly conserved genes, metB and etfA, respectively encoding cystathionine gamma lyase and electron transport flavoprotein. Interestingly, two different sets of lps genes are present at this locus in the plant pathogens, Xanthomonas campestris pv. campestris (Xcc) and Xanthomonas axonopodis pv. citri (Xac). The genomic island is present in a number of Xoo strains from India and other Asian countries but is not present in two strains, one from India (BXO8) and another from Nepal (Nepal624) as well as the closely related rice pathogen, Xanthomonas oryzae pv. oryzicola (Xoor). TAIL-PCR analysis indicates that sequences related to Xac are present at the lps locus in both BXO8 and Nepal624. The Xoor strain has a hybrid lps gene cluster, with sequences at the metB and etfA ends, being most closely related to sequences from Xac and the tomato pathogen, Pseudomonas syringae pv. tomato respectively.
Conclusion
This is the first report of hypervariation at an lps locus between different strains of a plant pathogenic bacterium. Our results indicate that multiple HGT events have occurred at this locus in the xanthomonad group of plant pathogens.
Background
LPS is an important constituent of the outer membrane of gram-negative bacteria. Variation in LPS composition can have profound consequences for these cells by potentially providing resistance against bacteriophages and antimicrobial compounds as well as facilitating evasion of the host immune system in animal pathogens. Extreme variation at LPS gene clusters has been reported in animal pathogenic bacteria. Recently, eleven highly divergent gene clusters were reported to occupy an LPSspecific locus in Pseudomonas aeruginosa, an opportunistic human pathogen [1]. The acquisition by horizontal gene transfer of a new LPS biosynthetic gene cluster in Vibrio cholerae is considered as a major cause for the cholera epidemic that originated in India in 1992 [2]. In plant pathogenic bacteria, LPS is an important virulence factor and mutations in the genes involved in LPS production result in severe virulence deficiency [3-8]. LPS has been shown to induce resistance in plants against pathogens [9,10] and in some recent studies, LPS is found to induce expression of plant defense genes [11,12] as well as an oxidative burst reaction in cell cultures [13]. Since LPS recognition appears to be an important aspect of plant defense responses, variation in lps gene repertoire is to be expected within different strains of plant pathogenic bacteria.
The genus Xathomonas includes a number of plant pathogenic bacteria. Two related members of this genus, Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoor) cause diseases of rice [14]. They exhibit different tissue specificities with Xoo growing in the xylem vessels while Xoor grows within the intercellular spaces of the parenchymatous tissue. Xoo causes bacterial leaf blight, the most serious bacterial disease of rice. This disease is prevalent in many rice growing countries in Asia, extending from the Indian subcontinent to Japan and Korea. DNA fingerprinting studies using multi-locus RFLP and PCR probes have indicated that there is extensive genetic diversity within Xoo strains isolated from various countries [15-19]. In India, multi-locus RFLP profiling has indicated that one lineage of Xoo (called the BXO1 lineage, based on the type strain for this group) is widely distributed within the country. Strains within the BXO1 lineage cluster together at about the 90 % similarity level in a dendrogram. A second group of strains is quite diverse, both at the haplotypic and pathotypic level, and clusters with the BXO1 group at about the 55% similarity level [19].
In previous research, we have reported a 5.5 kb region in the genome of Xoo strain BXO1 and demonstrated that it encodes three genes that are involved in biosynthesis of LPS and extracellular polysaccharide (EPS) as well as in virulence [8]. All the three genes have atypical G+C content, as compared to the rest of the Xoo genome. In this study, we have completed the entire sequence of this 12.2 kb genomic locus and indicate that it encodes three additional genes, wxoD, wzt and wzm, that are postulated to be involved in LPS biosynthesis and transport. These newly described genes also have atypical G+C content and all the six genes at this locus exhibit altered codon usage pattern, as compared to other Xoo genes. We present evidence that this locus is present in many, but not all, Xoo strains and that it is absent in Xoor. Our results indicate that there is substantial variation at this locus among various xanthomonads. The possible significance of these results is discussed.
Results
Genetic organization of a Xoo lps locus
In an earlier study, a novel Xoo locus was reported to be required for LPS and extracellular polysaccharide (EPS) production as well as virulence. A 35 kb cosmid, pSD5, that complements mutations in this region was isolated [8]. Partial sequence (5.5 kb) of this locus indicated that the region has atypical G+C content and contains three genes which encode a predicted sugar nucleotide epimerase and two predicted glycosyl transferases. We report here the complete 12.2 kb sequence and genomic organization of this locus in Xoo strain BXO1 (Fig. 1). The insert in the pSD5 cosmid includes 7 EcoRI fragments (0.6, 2.2, 3.5, 4.0, 6.0, 9.0 and 10 kb). We subcloned all the fragments into pBlueScript. Based on the end sequences of the inserts in the subclones and pSD5, the lps locus was mapped to four of these EcoRI fragments (0.6, 4, 3.5 and 9 kb). The previously obtained sequence was found to include all of the 3.5 kb and part of the 4 kb fragment and the remaining sequence of this region was obtained by sequencing the 0.6 kb and the 9 kb fragment (Please refer Methods). A total sequence of 13.18 kb was constituted by joining 6.14 kb of previously obtained sequence [8] and 7.04 kb of new sequence. The 13.18 kb sequence includes 12.2 kb of the lps locus and some flanking regions. The additional sequence of the lps locus encodes three putative genes which encode a predicted O-antigen acetylase, a predicted ABC transporter permease and a predicted ATP-binding protein and three insertion sequence (IS) elements.
Figure 1.
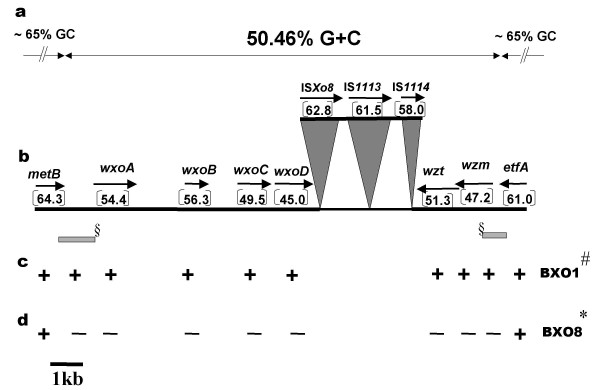
Genetic organization of a locus encoding LPS biosynthetic genes in Xoo strain BXO1. a. Overall G+C content of the locus and the flanking regions. The G+C content of the genomic island was calculated without including the sequences of IS elements. The overall G+C content of the genome is ~65%. b. Organization and G+C content of individual genes and transposases of IS elements. IS1114 encodes a truncated ORF. Arrows indicate transcriptional orientation. c. and d. Presence (+) and absence (-) of genes/PCR products in particular strains. § Indicates PCR products obtained using primer pairs directed against either metB and wxoA or etfA and wzm. # similar results were obtained with all Xoo strains tested excepting BXO8 and Nepal 624. * similar results were obtained with the Nepal 624 strain.
All of the putative genes have been named as per Bacterial Polysaccharide Genes Nomenclature (BPGN) [20]. The first three genes, wxoA (encodes a predicted epimerase), wxoB and wxoC (both encode predicted glycosyl transferases) have been described earlier. The fourth gene is wxoD and encodes a predicted 327 amino acids long protein. A BLAST [21] search reveals strong homology to acetyltransferases that are involved in LPS modification and the best match is with an acetyltransferase from Mesorhizobium loti (MAFF303099; 34% identity and 46% similarity at amino acid level). Interestingly, no homologs of this gene have been reported in any other xanthomonad. The fifth gene, wzt, encodes a predicted 436 amino acid long protein. A BLAST search reveals homology to functions involved in LPS transport. The best match is with the ATPase component of an ABC-type polysaccharide transport system from Burkholderia fungorum (ZP_00033174.1; 47% identity and 65% similarity at amino acid level). The sixth gene, wzm, encodes a predicted 437 amino acid long protein which is homologous to integral membrane protein components of ABC transporter systems that are involved in LPS transport. The best match is with a permease component of the ABC-type polysaccharide export system from Pseudomonas fluorescens PfO-1 (ZP_00085342.1; 50% identity and 65% similarity at amino acid level). The start codon of wzt overlaps with the stop codon of wzm. Homologs of wzt and wzm are typically present in many lps gene clusters. Interestingly, two complete Insertion Sequence (IS) elements (ISXo8 and IS1113) and one truncated IS element (IS1114) interrupt this cluster between the genes, wxoD and wzt. ISXo8 is a novel 1320 bp long insertion sequence and a BLAST search shows homology to transposase of ISRSO17 encoded by Ralstonia solanacearum (CAD17626; 51% identity and 63% similarity at amino acid level). A complete copy of the IS1113 element (AF482989) and a truncated copy of the IS1114 element (AF232058) are also present as indicated in Fig. 1. The presence of IS elements is a marked feature of many lps loci [22]. Transcriptional orientation suggests the possibility that ORFs wxoA, wxoB, wxoC and wxoD might constitute one operon and that ORFs wzm and wzt might be transcribed together. The overlap between the start codon of wzt and the stop codon of wzm also suggests that these two genes are co-transcribed.
The lps locus is flanked by metB, which encodes a predicted cystathionine gamma lyase, and etfA which encodes a predicted electron transport flavoprotein. The genome sequences of Xanthomonas campestris pv. campestris (Xcc; infects crucifer plants like cabbage, cauliflower, mustard, etc.) and Xanthomonas axonopodis pv. citri (Xac; infects citrus plants) have been obtained [23]. The Xoo metB gene (a partial sequence of 642 bp is available) exhibits within the sequenced region, 91% and 88% nucleotide identity to metB genes of Xac (AE012010.1) and Xcc (AE012157.1), respectively. The Xoo etfA gene (a partial sequence of 328 bp is available) exhibits within the sequenced region, 93% and 91% nucleotide sequence identity, respectively, with etfA genes in Xac (AE012009.1) and Xcc (AE012159.1). Interestingly, the lps biosynthetic gene cluster of Xcc, which comprises fifteen genes, is also located between the metB and etfA genes [24]. In Xac, this gene cluster is missing at this locus and is replaced by a set of fourteen genes, several of which are homologous to functions involved in LPS synthesis and transport. The gene clusters present at this locus in Xcc, Xac and Xoo have distinct nucleotide sequences, gene numbers (15 genes in Xcc, 14 genes in Xac, 6 genes in Xoo) and gene organization.
The Xoo lps cluster is a genomic island
a) Atypical G+C content
The average G+C content of Xoo and other Xanthomonads is estimated to be around 65% [25], while the average G+C content of the lps locus is 50.46% (excluding the IS elements) [Fig. 1]. The variation is much more marked among the genes, from as low as 45.0% (wxoD) to 56.3% (wxoC). Atypical G+C content is a characteristic feature of "genomic islands" that are believed to be acquired by horizontal gene transfer. The transposase genes encoded by ISXo8 and IS1113 have a G+C content that is >61%, a value which is typical for the genomes of Xoo and other xanthomonads. The G+C content of metB and etfA genes that flank the genomic island have G+C content of 64.3% and 61% respectively (within the partial sequences that have been obtained) which is typical of the Xoo genome.
b) Altered codon usage
An additional hallmark of a genomic island is the altered codon usage. Here we present a simple and graphical way of calculating and representing the codon usage differences and refer to it as Codon Usage Pattern or CUP (Please refer Methods). Eight aminoacids, i.e., Glycine, Valine, Threonine, Leucine, Arginine, Serine, Proline and Alanine, were selected to study CUP because they have atleast four synonymous codons. The percentage of synonymous codons that end with G or C was calculated for each aminoacid and gene. This analysis was conducted for six genes of the lps island and six genes from elsewhere in the Xoo genome (please refer Methods). We show that CUP of the genes present in the genomic island is dramatically different from the typical Xoo genes (Fig. 2). The %G+C at third codon position of synonymous codons for amino acid Glycine is only 52.5 % for genes present in the lps locus, while it is 78 % in case of Xoo genes that are located elsewhere in the genome. Similarly, for amino acids Valine, Alanine, Threonine, Serine, Arginine, Leucine and Proline the values are 46.6, 47, 59, 52, 53, 57 and 34.6 % respectively for genes at the lps locus, while the values are 84, 77.5, 89.5, 79.5, 75.6, 90.3 and 86.16 % for the respective aminoacids in case of the typical Xoo genes. Altered codon usage is a characteristic feature of horizontally acquired genes and CUP clearly indicates that the Xoo lps cluster is a genomic island (Fig. 2).
Figure 2.
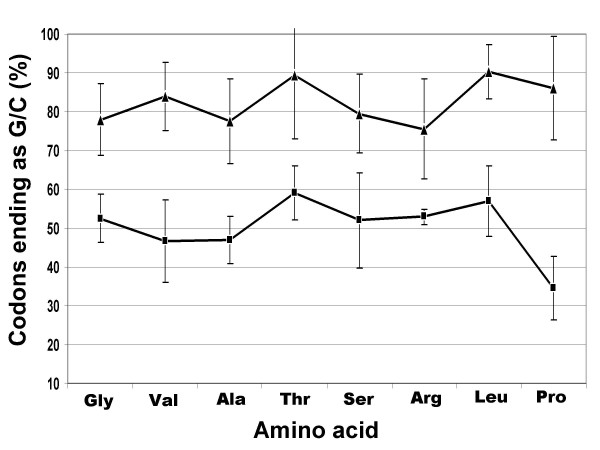
Genes encoded in the lps locus exhibit altered Codon Usage Pattern (CUP). Eight amino acids, each of which has atleast four synonymous codons, are represented on the X-axis. The % of codons ending with G/C for each of these amino acids is represented on the Y-axis as mean ± SD. The lower line represents CUP for eight aminoacids of the six genes (excluding transposase ORFs) encoded in the lps locus. The upper line represents CUP of six Xoo genes from elsewhere in the genome (Please refer Methods).
The lps locus is present in the genomes of many, but not all, Xoo strains
The presence of the genomic island in different Xoo strains was assessed by PCR using gene specific primers, for all the six lps genes, as described in the Methods. The list of strains used in the study is given in the Table 1 and the list of gene specific primers is given in Table 2. In order to confirm that the genomic island is present at the same genomic location in all strains, PCR was also performed using two primer pairs that are designed to amplify fragments from metB to wxoA and wzm to etfA, respectively. The analysis included nine Indian Xoo strains representing different geographic locations and the BXO1 and non BXO1 groups. The list also includes twelve Xoo strains from different Asian countries and a Xoor strain, BXOR1, from India. Our study revealed that the genomic island is present in the majority (7/8) of Xoo strains that we have examined from India (Fig. 1). Four BXO1 group strains (BXO4, BXO7, BXO13 and BXO479) and three of the non-BXO1 strains (BXO5, BXO6 and BXO20) have the genomic island. The genomic island is also present in two strains each from China, Malaysia, Indonesia, Philippines, Korea and one strain from Nepal (Fig. 1, Table 1). The lps locus is present, in all these strains, between the metB and etfA genes. Interestingly, we find that the genomic island is not present (as judged by PCR [Fig. 3A] and Southern hybridisation [Fig. 3B]; see Methods) in the genomes of Xoo strains BXO8 and Nepal624, as well as the Xoor strain, BXORI. The results obtained with the probes directed against the wxoA gene are presented but similar results were obtained using probes that are specific for the other five genes. The blots used above were reprobed as a positive control with a metB specific probe and the results gave an expected size band in BXO1 (lane 1), and different sized bands in BXO8 (lane 3), Nepal624 (lane 4) and BXOR1 (lane 2) indicating that the metB gene is present but located in different EcoRI fragments (Fig. 3C).
Table 1.
List of strains used in the study
| Xanthomonas oryzae pv. oryzae (Xoo) strains used in the present study | ||
| From India | ||
| Strain Name | Location | Source |
| 1) BXO1 | Chinsuria, West Bengal | Lab collection |
| 2) BXO4 | Kapurthala, Punjab | " |
| 3) BXO5 | Ferozpur, Punjab | " |
| 4) BXO6 | Patiala, Punjab | " |
| 5) BXO7 | Titabar, Assam | " |
| 6) BXO8 | Nellore, Andhra Pradesh | " |
| 7) BXO13 | Marutheru, Andhra Pradesh | " |
| 8) BXO20 | Pantnagar, Uttar Pradesh | " |
| 9) BXO479 | Nawgam, Gujarat | " |
| From other countries – (gift from Dr. Jan Leach) | ||
| 1) China Xoo NX2 | ||
| 2) China Xoo #B21 | ||
| 3) Korea Xoo #197 | ||
| 4) Korea Xoo #220 | ||
| 5) Nepal Xoo #537 | ||
| 6) Nepal Xoo #624 | ||
| 7) Malaysia Xoo #90 | ||
| 8) Malaysia Xoo #101 | ||
| 9) Indonesia Xoo #16 | ||
| 10)Indonesia Xoo #40 | ||
| 11)Philippines PXO86 | ||
| 12)Philippines PXO99A9 | ||
| Xanthomonas oryzae pv. oryzicola (Xoor) strain used in the present study | ||
| 1) BXOR1 | Rajendranagar, Andhra Pradesh | lab collection |
Table 2.
Plasmids and primers used in this study
| Plasmids | Relevant characteristics | Reference or source |
| pUFR034 | IncW Nmr Mob+ mob (P) lacZ alpha Par+ cos (8.7 kb) | [44] |
| Pbluescript | Apr | Stratagene |
| pSD5 | pUFR034 + a 35 kb insert from the BXO1 genome | [8] |
| pBP1 | pBluescript + a 2.2 kb EcoRI fragment from pSR1 | This study |
| pBP2 | pBluescript + a 3.5 kb EcoRI fragment from pSR1 | This study |
| pBP3 | pBluescript + a 4.0 kb EcoRI fragment from pSR1 | This study |
| pBP4 | pBluescript + a 9.0 kb EcoRI fragment from pSR1 | This study |
| pBP5 | pBluescript + a 10 kb EcoRI fragment from pSR1 | This study |
| Primers specific to ORFs present in the genomic island of BXO1 | ||
| WxoA | Forward primer CCAAGCGACCAGAGGTGCTTGACG | |
| Reverse primer GAGGAGCACCATCCGCTACCGCCC | ||
| WxoB | Forward primer GTTTTTGTTGGTACTGGGTGCGAG | |
| Reverse primer GTACGCCACGGTCAGATCGGCTGC | ||
| WxoC | Forward primer CTACTGATGTTGTTCGCAAGGTGG | |
| Reverse primer GGCGACTCACCTGCATATCGAGCC | ||
| WxoD | Forward primer GTGCTGGTGAGCCATCATTTTG | |
| Reverse primer TTACTCACCGGCCATAATCCTTTT | ||
| Wzt | Forward primer GACATCGCTATCGAAGTAAAAGGT | |
| Reverse primer TCAGGTGCTGTTTGAAGTAGCGGAC | ||
| Wzm | Forward primer CATCGGCAAACCCCTTTCGGGT | |
| Reverse primer CGCAGCTTACTGATGGAACCCT | ||
| Primers used for TAIL PCR | ||
| designed against metB gene sequence of BXO1 | ||
| CglL1 | CTTCGACGCAGCCAAGCGTTTC | |
| CglL2 | CTGCGAGAAGACCGAGCTGTTCAC | |
| CglL3 | CGAATCGCTCGGTGGTGTTGAA | |
| designed against etfA gene sequence of BXO1 | ||
| EtfL1 | TCGGCCAGACCGGCAAGATCAT | |
| EtfL2 | AGCTGTACATGGCCATCGGCAT | |
| EtfL3 | AGCATCTGACCGGCATCAAGGA | |
| Other primers described in the paper | ||
| Pbp1 | AGCGTGCTGGTGAGCCATCA | |
| Pbp2 | GCAGCAAAAATGCTGTCATAACCA |
Figure 3.
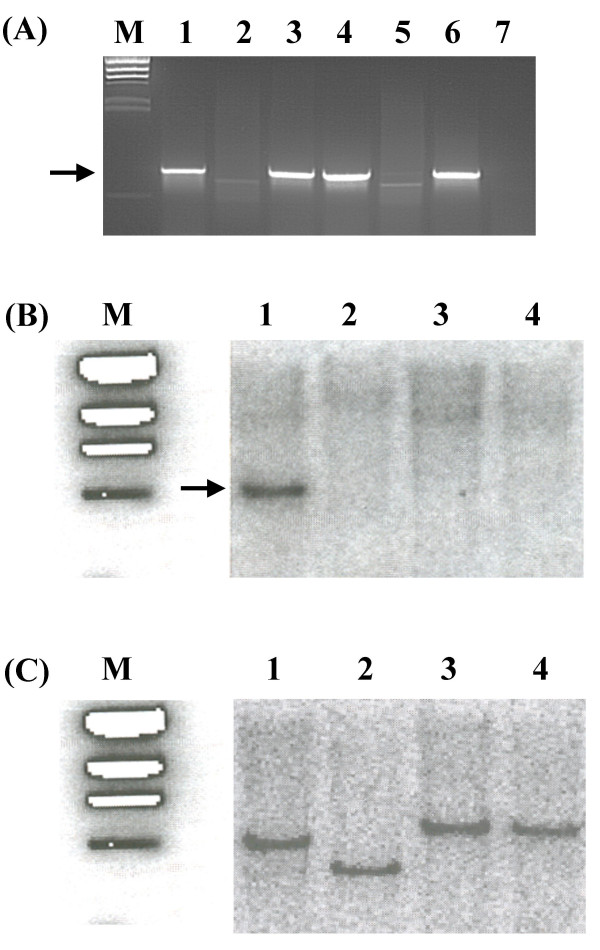
The lps locus is absent from the genomes of Xoo strains BXO8, Nepal624 and Xoor strain BXORI. (A) PCR analysis using primers that are specific to wxoA gene. M is the λ HindIII Marker lane. An expected band of 1 kb (indicated by arrow) is present in the Xoo strains, BXO1 (lane 1), BXO5 (lane 3), BXO6 (lane 4) and BXO20 (lane 6) but absent in BXO8 (lane 5), Nepal624 (lane 7) and BXORI (lane 2). (B) Southern hybridization analysis of EcoRI digested genomic DNA using α-32-P labeled wxoA specific probe (see Methods). A 4 kb band can be seen (indicated by arrow) in BXO1 (lane 1) but not in BXORI (lane 2), BXO8 (lane 3) and Nepal624 (lane 4). Similar results were obtained for wxoB, wxoC, wxoD, wzm and wzt genes. (C) The blot from (B) was deprobed and was hybridized with α-32P labeled probe specific to the metB gene. A specific band can be seen in all the lanes. Note the sizes of the bands indicating that metB is present in different EcoRI fragments in BXO1, BXORI and BXO8/Nepal624.
BXO8 and Nepal 624 have sequences related to Xac at the lps locus
What are the sequences present at this genomic location in the Xoo strains that lack the lps locus? Thermal Asymmetric Interlaced (TAIL) PCR is an efficient technique for isolation of target DNA segments adjacent to known sequences [26]. TAIL-PCR and sequencing using primers directed against the conserved flanking metB and etfA genes suggests that sequences which are significantly similar to the Xac lps gene cluster are present at this genomic location in both of these strains. Next to metB, a wzm homolog is present in BXO8 (a partial sequence of 398 bp is available) and Xac with 69.2% identity at nucleotide level within the sequenced region. Next to etfA, a putative integral membrane protein encoding gene is present in both BXO8 (a partial sequence of 405 bp is available) and Xac with 91.3% identity at nucleotide level within the sequenced region. The BXO8 and Nepal624 strains exhibit 100% nucleotide sequence identity within the sequenced region. TAIL- PCR analysis of the Xoor strain indicates that it has a hybrid lps gene cluster. Next to metB, a unique wzm gene is located (a partial sequence of 548 bp is available) which exhibits 62.8% nucleotide identity to wzm gene of Pseudomonas syringae pv. tomato strain DC3000 (AE016859.1). Next to etfA, a putative inner membrane protein encoding gene is located (a partial sequence of 402 bp is available) which exhibits 97% and 92% nucleotide sequence identity, respectively, with similarly located genes in BXO8 and Xac. Because the BXO8 and Nepal624 strains have different sequences at the lps locus, as compared to other Xoo strains, we inoculated these strains along with appropriate controls onto leaves of the susceptible rice cultivar Taichung Native-1. We find that BXO8 and Nepal624 strains are able to cause typical bacterial leaf blight disease symptoms that are indistinguishable from those elicited by other Xoo strains (data are not shown).
Presence of inverse repeats at the 3' ends of metB and etfA genes that flank the lps locus
We have performed an alignment using BLAST2 [27] of the nucleotide sequences derived from the metB and etfA genes in BXO1 and BXO8. The homology breakpoints appear to localise to the 3' regions of metB and etfA genes, exactly 18 bp upstream of their respective stop codons. Upto the break points, within the sequenced region at either end of the lps locus, the nucleotide sequence is identical in BXO1, BXO8 and Nepal624. The DNA sequence immediately preceding the break points was examined manually for presence of direct or inverse repeats. Interestingly, we could find three inverted repeats (I, II and III) within the 3' regions of metB and etfA near the homology breakpoints between BXO1 and BXO8 (Fig. 4). The first repeat is the smallest one (5 bp) and the third repeat is the largest (11 bp). The second repeat is 6 bp long and is 7 bp from the first repeat on the metB side and 9 bp from the first repeat on the etfA side. The distance between the second and third repeats is 4 bp in metB and etfA. We also found similarly located inverse repeats in the metB and etfA genes of Xac, Xcc and Xoor. A consensus sequence of the repeats was derived (Fig. 4) by scoring a nucleotide if it is present in a majority of repeats.
Figure 4.
Invert repeat sequences flanking the lps gene cluster in the BXO1 strain of Xoo. The horizontal arrows represent the ORFs of metB and etfA. I, II, III represent three different invert repeats in the 3' regions of metB and etfA genes. The vertical arrows represent the breakpoints of homology between BXO1 and BXO8. The distances of the break points from the stop codons of the metB and etfA genes are indicated. Dashed lines indicate the remainder of the metB and etfA genes. The sequence of the corresponding inverse repeats in Xac, Xcc and Xoor are also indicated along with the derived consensus sequence for the repeats. The numbers in brackets indicate distances in bp between individual repeats.
Relationship between BXO8 and Nepal 624 strains
The TAIL PCR results indicate that the BXO8 and Nepal624 strains have identical sequences in place of the BXO1 lps locus. As both the strains are from the Indian subcontinent, there is the possibility that these are identical/nearly identical to each other. We therefore performed DNA fingerprinting analysis of the BXO8 and Nepal624 strains using the IS1112 insertion element as a probe. This probe is highly informative and can clearly differentiate the BXO1 and non BXO1 group of strains in India [19]. The following strains were also included in the analysis: BXO1, three non BXO1 group strains (BXO5, BXO6, BXO20) and BXORI. The hybridisation pattern revealed that BXO8 and Nepal624 are quite distinct from each other (Fig. 5). We could score 42 unique bands and the data generated were used to calculate pairwise similarity coefficients and cluster analysis was performed to generate a dendrogram using UPGMA (please refer Methods). The similarity coefficient between BXO8 and Nepal624 is only 56%. The dendrogram (Fig. 6) indicates that BXO8 clusters with BXO#s 5, 6 and 20 at about the 58% similarity level while Nepal624 clusters with all these four strains at about the 53% similarity level. All of the Xoo strains cluster with each other at about the 51% similarity level. Although the bootstrap values for these clusters are low, it is clear that the BXO8 and Nepal624 strains are not closely related to each other. As expected for an outgroup strain, BXOR1 clusters with Xoo strains at the 29% similarity level and the bootstrap value for this cluster is a high 96.8%.
Figure 5.
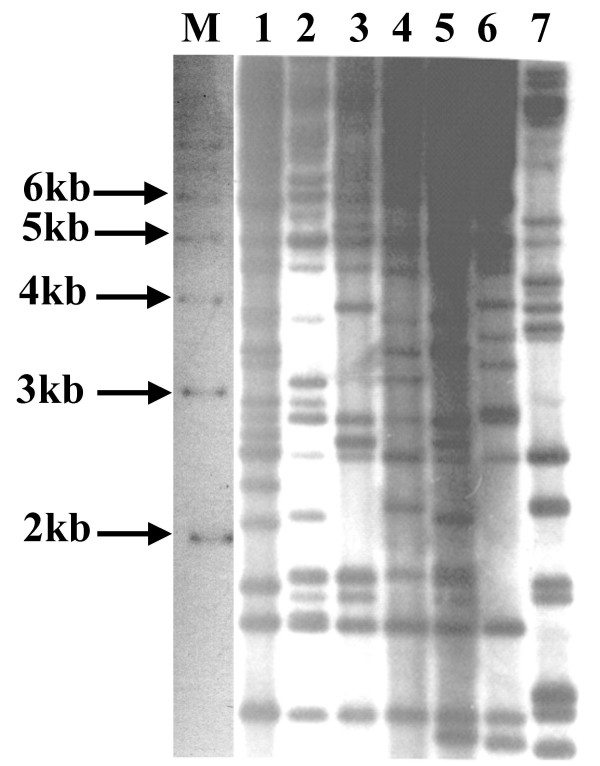
Restriction fragment length polymorphism analysis of Xoo strains. Southern analysis of EcoRI-digested genomic DNA was performed using α-32P labeled IS1112 as a probe (see Methods). Lanes: 1; BXO1, 2; BXO5, 3; BXO6, 4; BXO8, 5; BXO20, 6; Nepal624, 7; Xoor strain BXORI. M; indicates the size of molecular weight markers in kb.
Figure 6.
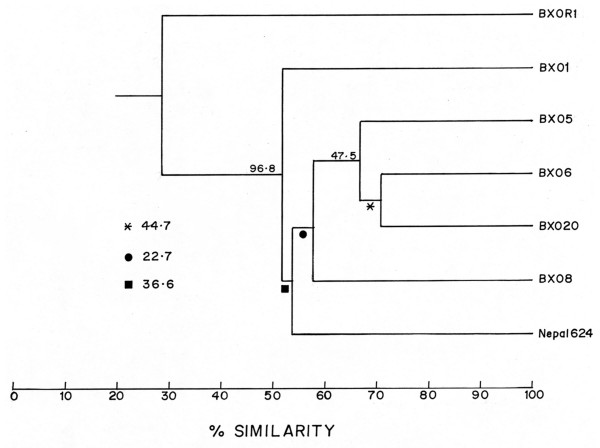
Cluster analysis of Xoo strains. The dendrogram was constructed using the UPGMA option of PHYLIP on the basis of restriction fragment length polymorphism data obtained with IS1112 probe. Numbers or symbols at the internal branches indicate bootstrap values for clusters. The BXORI (Xoor) strain constitutes the outgroup.
Discussion
We report here the complete sequence and genomic organization of the lps locus in the BXO1 strain of Xoo. Three of the genes in this locus i.e., wxoA, wxoB and wxoC were shown in an earlier study to be required for lipopolysaccharide production and virulence [8]. The predicted proteins encoded by the three new genes i.e., wxoD, wzt and wzm described in the present study are homologous to functions involved in lipopolysaccharide modification and transport. The wxoD gene encodes a predicted O-antigen acetylase which is homologous to similar functions encoded in phage genomes and other bacteria. O-antigen is the most variable part of LPS. Acetylation of O-antigen is shown to confer resistance to anitimicrobial peptides in Proteus mirabilis [28] and determines serotype in many bacterial pathogens [29-31]. The other two genes, wzm and wzt, are typically present in most lps gene clusters [including those of Xac and Xcc][23] as tandem genes and encode functions involved in LPS transport. The wzm and wzt genes of BXO1 have overlapping ORFs, an arrangement that is also seen in wzm and wzt genes of the lps loci in other bacteria including Xac. IS elements are frequently found interrupting many lps loci [22] and in BXO1, three IS elements interrupt the gene cluster between wxoD and wzt genes.
The complete genome sequences of more than 150 bacteria are now available [32] and studies have revealed the presence of DNA segments with G+C content and codon usage different from the rest of the genome. These regions are referred to as genomic islands and are believed to be acquired by horizontal gene transfer [33,34]. Another feature of genomic islands is their absence from the genomes of closely related strains. Our study clearly indicates that the lps locus of Xoo strain BXO1 fulfils all of the above criterion and constitutes a genomic island. The G+C content of this lps locus, excluding the IS elements, is 50%. The transposases encoded by ISXo8 and IS1113 have a G+C content that is >61%. This value, which is typical for the genomes of Xoo and other xanthomonads [25], suggests the possibility that these elements have transposed into the lps locus after it's transfer into the Xoo genome. The presence of this genomic island in Xoo strains that are distributed across a vast segment of the Asian continent suggests that it was introduced into the Xoo genome early in the evolution of this pathogen.
The BXO8 and Nepal624 strains do not have the lps locus that is present in the other Xoo strains. The related xanthomonad, Xoor, also has an lps locus that is different from the BXO1 lps locus. Also, different gene clusters are present at this locus in Xac and Xcc (Fig. 7). This indicates that multiple HGT events have occurred at this locus among xanthomonads. One HGT event occurred early in (or possibly at the time of) the evolution of the Xoo pathogen. This led to the introduction of the genomic island described in Figure 1. Two separate HGT events are likely to have occurred in the lineages that gave rise to BXO8 and Nepal624 Xoo strains. This is inferred from the observation that BXO8 and Nepal624 are quite unrelated in their genomic background. Another HGT can be inferred to have occurred in the Xoor strain wherein sequences that are most closely related to Pseudomonas syringae pv. tomato have been introduced at one end of the lps cluster. At least one more HGT has occurred to differentiate the lps gene clusters in Xcc and Xac.
Figure 7.
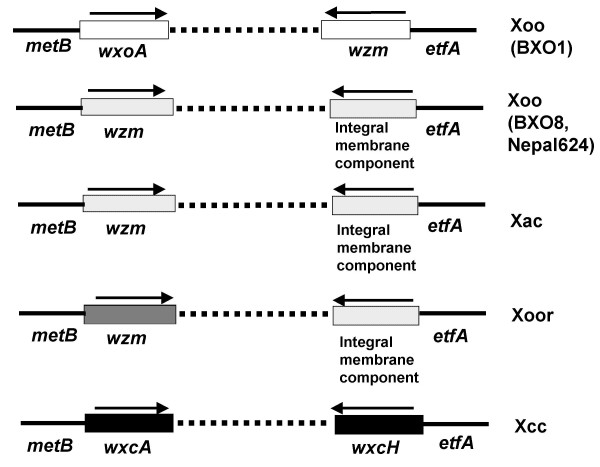
Variation in lps gene clusters within the xanthomonads. The genes that are adjacent to metB and etfA in different xanthomonads are indicated. Dashed lines represent the remainder of the lps cluster. Empty and filled boxes represent sequences specific to Xoo and Xcc respectively. Boxes with dots indicate that the sequences are either from or related to Xac genes. Box with stripes represents sequences that are related to Pseudomonas syringae pv. tomato. Arrows indicate transcriptional orientation. The wzm gene encodes a predicted ABC transporter permease protein, wxoA encodes a predicted epimerase, wxcA encodes a glycosyl transferase and wxcH encodes a hypothetical protein.
The presence of invert repeats in the regions that flank the lps locus is likely to be significant. The presence of these repeats in the metB and etfA genes is especially striking as both genes encode completely different functions. The location of the repeats flanking the Xoo lps locus suggests that they might be involved in promoting recombination during HGT and/or gene regulation. A short inverted repeat sequence (GGCCAATCGA) flanking the lipopolysaccharide gene cluster has been reported in Mycobacterium avium subsp. paratuberculosis [35]. Another conserved sequence, called JUMPstart has been found located in intergenic regions upstream of polysaccharide biosynthetic gene clusters in several animal pathogenic bacteria like Escherichia coli strain K5, Vibrio cholera, etc. This sequence was implicated to be involved in gene regulation and has also been suggested to have a role in recombination [22,36].
As LPS is highly immunogenic, lps loci of animal pathogenic bacteria are under intense host selection and extreme variation is reported in lps specific gene clusters [22]. The observation that the two Xoo strains have different lps gene clusters suggests that the plant pathogenic bacteria are also under selection to vary their LPS. Alterations in LPS composition might result in resistance against predators like bacteriophages [4,10] or reduced susceptibility to certain anti-microbial compounds [7] in the host/environment. Most importantly, it might help in evasion of the host defense response.
Conclusions
These results provide, for the first time, evidence for substantial variation in lps biosynthetic gene clusters within different strains of a plant pathogenic bacterium. The results also indicate that multiple HGT events have occurred at this locus in various xanthomonads and provide a new parallel in the mechanisms that plant and animal pathogenic bacteria can employ to generate variability in cell surface molecules.
Methods
Complete sequencing of the lps locus in the BXO1 strain of Xoo
The lps locus was cloned as part of a 35 kb cosmid clone, pSD5. The insert includes 0.6, 2.2, 3.5, 4.0, 6.0, 9.0 and 10 kb fragments upon EcoRI (New England Biolabs [NEB], Beverly, MA) digestion and all the fragments were subcloned in to pBlueScript (Stratagene, La Jolla, CA). Most of the sequence obtained in this study was generated by sequencing the 9 kb subclone, pBP4, using a modified shotgun sequencing procedure. Here, pBP4 was digested with EcoRI and the 9 kb fragment was gel eluted. Then the fragment was partially digested (1.5–2.5 kb) using a blunt-end cutter, HaeIII (NEB) and cloned into pMOS (Amersham Pharmacia Biotech, Buckinghamshire, England). The inserts were amplified from random clones by colony PCR using vector primers and were sequenced using an ABI Prism 3700 automated DNA sequencer (Applied Biosystems, Foster City, CA). After editing, the assembly of the sequence data was done using GeneTools (BioTools, Alberta, Canada) and Blast2 [27]. Multiple single strand sequences (3–8 X coverage) were generated for each region in the sequence. Contig assembly was confirmed by restriction fragment analysis of a 12.5 kb PCR amplified product containing the lps locus that was obtained using long range PCR (Triple Master™, Eppendorf, Hamburg, Germany) with BXO1 genomic DNA as template. The sizes of the fragments corresponded to the sizes that are predicted by in silico analysis of the sequence (data are not shown). The ORF's were assigned using ORF finder [37] and genes were named as per Bacterial Polysaccharide Genes Nomenclature [20]. Two primers, Pbp1 and Pbp2 (Table 2), were used to derive the sequence of the 0.6 kb EcoRI fragment which is also a part of the lps locus. The Pbp1 primer binds just after the wxoC ORF (which forms part of the 3.5 kb EcoRI fragment) and Pbp2 binds within the wxoD ORF (which forms part of the 9.0 kb EcoRI fragment). A 0.67 kb PCR amplified fragment is obtained from BXO1 genomic DNA using Pbp1 and Pbp2. The band was gel eluted and was sequenced using Pbp1 and Pbp2. The sequence was found to include the 0.6 kb EcoRI fragment. In addition, the sequences of all six ORFs were confirmed by sequencing of PCR amplified fragments from genomic DNA using specific sets of gene specific primers (see the list of primers in Table 2).
Codon Usage Pattern
For each gene the frequency of codon usage for different aminoacids was calculated using a web based program [38]. Further, eight aminoacids i.e., Glycine, Valine, Threonine, Leucine, Arginine, Serine, Proline and Alanine that have atleast four synonymous codons were selected and the percentage of synonymous codons that end with G or C was calculated for each aminoacid and gene. The pattern was calculated for a group of genes by plotting mean values ± SD corresponding to a particular aminoacid. The first group was chosen to include genes that encode proteins which participate in diverse functions and are present at different locations in the Xoo genome outside the lps locus. These genes encode: a putative siderophore receptor (AF325732), Xanthomonas adhesin like protein (AF288222), a putative phytase (AY151260), rpfF (AF411962), shikimate dehydrogenase (AF258797) and secreted xylanase (AF331922). The second group comprised the six genes (excluding transposases) encoded in the Xoo lps gene cluster (AF337647).
Screening of Xoo strains and Xoor for the presence of the genomic island
Specific oligonucleotide primer pairs were designed and used to amplify gene specific fragments for each of the ORFs encoded in the BXO1 genomic island (see the list of primers given in Table 2). DNA sequencing was used to confirm the authenticity of the PCR product obtained with each primer pair using BXO1 genomic DNA as template. Southern hybridizations were performed using these gene specific PCR products as probes. Genomic DNA was isolated from Xoo and Xoor strains according to the procedure described by Leach et. al. [16]. The DNA was then digested with EcoRI (NEB) according to supplier's instructions. Digested genomic DNA was separated on a 0.8% agarose gel and vacuum transferred to a Hybond N+ filter (Amersham) using 0.4% NaOH as described by Sambrook et al. [39]. Probes were labelled with α-32P dATP using random primer labelling kit (Board of Radiation Technology, Mumbai, India). Prehybridization, hybridisation and washings were done at 68°C as described by Yashitola et al [19]. Membranes were then exposed to phoshoimager plates and images captured using a Fuji FLA-3000 phosphoimager system (Fuji, Japan).
To screen for the presence of the genomic island in different strains, a procedure for colony PCR was standardized. A portion of a single colony (or 10λ of a saturated culture that has approximately 1 × 109 colony forming units/ml) was lysed in 100λ of 0.01 N NaOH by boiling for 10 minutes. After spinning at 13 K for 1 min., 2λ of supernatant was used as template for PCR using the gene specific primers described above. The products were separated by electrophoresis on 1.5% agarose gels and visualized by ethidium bromide staining.
TAIL-PCR and sequence analysis
Specific primers were designed against the conserved metB and etfA gene sequences (Table 2) and the protocol for TAIL-PCR was as originally described by Liu and Whittier [26]. Sequencing of TAIL-PCR products was done using either the cglL3 or etfL3 primer. Homology searches were done using BLAST [21] through NCBI [40] and FASTA [41] through EMBL-EBI [42]. BLAST2 [27] was used to identify the homology break points in the genomic regions that flank the lps locus of BXO1 and BXO8. The sequences that were present upstream of the break points were manually examined and three repeat sequences were identified in the 3' coding regions of metB and etfA genes. Similar repeat sequences were identified in the corresponding regions of BXO8, Nepal624, BXORI, Xac and Xcc. A consensus was derived by aligning these repeat sequences and a particular nucleotide was scored if it is present in a majority of repeats.
DNA fingerprinting and data analysis
The Xoo IS element, IS1112 [16], was used as the hybridisation probe. This probe has been previously used to detect genetic variability in Xoo strains from different countries [15-19]. DNA isolation and Southern hybridisation was done as described in the section on screening of Xoo and Xoor strains for the presence of genomic island. The presence or absence of particular bands was scored as 1 or 0, respectively. The data were analysed using the Dice coefficient option in the program WINDIST [43] to generate distance matrices. The data were used to construct a dendrogram using the NEIGHBOR program in PHYLIP (phylogeny inference software package; University of Washington, Seattle) using the UPGMA (unweighted pair group method of averages) option. To test the robustness of the dendrogram, bootstrap analysis was carried out using the WinBoot program [43] with 2,000 iterations.
GenBank submissions
The nucleotide sequences obtained in this study have been deposited in GenBank with the following Accession numbers: Sequence of lps locus from BXO1 (AF337647); Sequences of TAIL-PCR products from metB end of BXO8 (AY319936), Nepal624 (AY319938) and BXORI (AY319940); Sequences of TAIL-PCR products from etfA end of BXO8 (AY319937), Nepal624 (AY319939) and BXORI (AY319941).
Authors' contributions
PBP carried out all the aspects of the work and drafted the manuscript. RVS conceived the study, and participated in its design and coordination. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Jan Leach and Marietta Ryba-White for providing Xoo strains. PBP was supported by a Senior Research Fellowship from the University Grants Commission (UGC), Government of India and currently has a Senior Research Fellowship from the Council of Scientific and Industrial Research. Meher Sultana and N. Nagesh are thanked for their help in oligosynthesis and sequencing.
Contributor Information
Prabhu B Patil, Email: prabhu@ccmb.res.in.
Ramesh V Sonti, Email: sonti@ccmb.res.in.
References
- Raymond CK, Sims EH, Kas A, Spencer DH, Kutyavin TV, Ivey RG, Zhou Y, Kaul R, Clendenning JB, Olson MV. Genetic variation at the O-antigen biosynthetic locus in Pseudomonas aeruginosa. J Bacteriol. 2002;184:3614–3622. doi: 10.1128/JB.184.13.3614-3622.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooi FR, Bik EM. The evolution of epidemic Vibrio cholerae strains. Trends Microbiol. 1997;5:161–165. doi: 10.1016/S0966-842X(96)10086-X. [DOI] [PubMed] [Google Scholar]
- Drigues P, Demery-Lafforgue D, Trigalet A, Dupin P, Samain D, Asselineau J. Comparative studies of lipopolysaccharide and exopolysaccharide from a virulent strain of Pseudomonas solanacearum and from three avirulent mutants. J Bacteriol. 1985;162:504–509. doi: 10.1128/jb.162.2.504-509.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonejans E, Expert D, Toussaint A. Characterization and virulence properties of Erwinia chrysanthemi lipopolysaccharide-defective, phi EC2-resistant mutants. J Bacteriol. 1987;169:4011–4017. doi: 10.1128/jb.169.9.4011-4017.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titarenko E, Lopez-Solanilla E, Garcia-Olmedo F, Rodriguez-Palenzuela P. Mutants of Ralstonia (Pseudomonas) solanacearum sensitive to antimicrobial peptides are altered in their lipopolysaccharide structure and are avirulent in tobacco. J Bacteriol. 1997;179:6699–6704. doi: 10.1128/jb.179.21.6699-6704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley MT, Gabriel DW, Marlow GC, Roberts PD. The opsX locus of Xanthomonas campestris affects host range and biosynthesis of lipopolysaccharide and extracellular polysaccharide. J Bacteriol. 1993;175:5839–5850. doi: 10.1128/jb.175.18.5839-5850.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow JM, Osbourn AE, Wilson TJ, Daniels MJ. A locus determining pathogenicity of Xanthomonas campestris is involved in lipopolysaccharide biosynthesis. Mol Plant-Microbe Interact. 1995;8:768–777. doi: 10.1094/mpmi-8-0768. [DOI] [PubMed] [Google Scholar]
- Dharmapuri S, Yashitola J, Vishnupriya MR, Sonti RV. Novel genomic locus with atypical G+C content that is required for extracellular polysaccharide production and virulence in Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact. 2001;14:1335–339. doi: 10.1094/MPMI.2001.14.11.1335. [DOI] [PubMed] [Google Scholar]
- Graham TL, Sequeira L, Huang TS. Bacterial lipopolysaccharides as inducers of disease resistance in tobacco. Appl Environ Microbiol. 1977;34:424–432. doi: 10.1128/aem.34.4.424-432.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeman M, Van Pelt JA, Den Ouden FM, Heinsbroek M, Bakker PAHM, Schippers B. Induction of systemic resistance against Fusarium wilt of radish by lipopolysaccharides of Pseudomonas fluorescens. Phytopathology. 1995;85:1021–1027. [Google Scholar]
- Newman MA, Daniels MJ, Dow JM. Lipopolysaccharide from Xanthomonas campestris induces defense-related gene expression in Brassica campestris. Mol Plant-Microbe Interact. 1995;8:778–780. doi: 10.1094/mpmi-8-0778. [DOI] [PubMed] [Google Scholar]
- Coventry HS, Dubery IA. Lipopolysaccharides from Burkholderia cepacia contribute to an enhanced defensive capacity and the induction of pathogenesis-related proteins in Nicotiana tabacum. Physiological and Molecular Plant Pathology. 2001;58:149–158. doi: 10.1006/pmpp.2001.0323. [DOI] [Google Scholar]
- Meyer A, Puhler A, Niehaus K. The lipopolysaccharides of the phytopathogen Xanthomonas campestris pv. campestris induce an oxidative burst reaction in cell cultures of Nicotiana tabacum. Planta. 2001;213:214–222. doi: 10.1007/s004250000493. [DOI] [PubMed] [Google Scholar]
- Swings J, Van den Mooter M, Vauterin L, Hoste B, Gillis M, Mew TW, Kersters K. Reclassification of the causal agents of bacterial leaf streak (Xanthomonas campestris pv. oryzicola) of rice as pathovars of Xanthomonas oryzae (ex Ishiyama 1922) Sp. Nov., nom.rev. Int J Syst Bacteriol. 1990;40:309–311. [Google Scholar]
- Adhikari TB, Cruz CMV, Zhang Q, Nelson RJ, Skinner DZ, Mew TW, Leach JE. Genetic diversity of Xanthomonas oryzae pv. oryzae in Asia. Appl Environ Microbiol. 1995;61:966–971. doi: 10.1128/aem.61.3.966-971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach JE, White FF, Rhoads ML, Leung H. A repetitive DNA sequence differentiates Xanthomonas campestris pv. oryzae from other pathovars of X. campestris. Mol Plant-Microbe Interact. 1990;3:238–246. [Google Scholar]
- Leach JE, Rhoads ML, Vera Cruz CM, White FF, Mew TW, Leung H. Assessment of genetic diversity and population structure of Xanthomonas oryzae pv. oryzae with a repetitive DNA element. Appl Environ Microbiol. 1992;58:2188–2195. doi: 10.1128/aem.58.7.2188-2195.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai O, Horino K, Miyajima K, Kaku H. Genetic diversity of Xanthomonas oryzae pv. oryzae strains from Sri Lanka. Phytopathology. 2000;90:415–421. doi: 10.1094/PHYTO.2000.90.4.415. [DOI] [PubMed] [Google Scholar]
- Yashitola J, Krishnaveni D, Reddy APK, Sonti RV. Genetic diversity within the population of Xanthomonas oryzae pv. oryzae in India. Phytopathology. 1997;87:760–765. doi: 10.1094/PHYTO.1997.87.7.760. [DOI] [PubMed] [Google Scholar]
- Reeves PR, Hobbs M, Valvano MA, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz CR, Rick PD. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/S0966-842X(97)82912-5. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PP, Wang L. Genomic organization of LPS-specific loci. Curr Top Microbiol Immunol. 2002;264:109–135. [PubMed] [Google Scholar]
- da Silva AC, et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature. 2002;417:459–463. doi: 10.1038/417459a. [DOI] [PubMed] [Google Scholar]
- Vorholter FJ, Niehaus K, Puhler A. Lipopolysaccharide biosynthesis in Xanthomonas campestris pv. campestris : a cluster of 15 genes is involved in the biosynthesis of the LPS O-antigen and the LPS core. Mol Genet Genomics. 2001;266:79–95. doi: 10.1007/s004380100521. [DOI] [PubMed] [Google Scholar]
- Starr MP. The genus Xanthomonas. In: Starr MP, Stolp H, Truper GH, Balows A, Schelegel GH, editor. In The Prokaryotes. Vol. 2. Berlin: Springer-Verlag; 1981. pp. 742–763. [Google Scholar]
- Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-J. [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. Blast 2 sequences – a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1016/S0378-1097(99)00149-4. [DOI] [PubMed] [Google Scholar]
- Gun JS. Bacterial modification of LPS and resistance to antimicrobial peptides. J Endotoxin Res. 2001;7:57–62. doi: 10.1179/096805101101532558. [DOI] [PubMed] [Google Scholar]
- Newton GJ, Daniels C, Burrows LL, Kropinski AM, Clarke AJ, Lam JS. Three-component-mediated serotype conversion in Pseudomonas aeruginosa by bacteriophage D3. Mol Microbiol. 2001;39:1237–1247. doi: 10.1046/j.1365-2958.2001.02311.x. [DOI] [PubMed] [Google Scholar]
- Verma NK, Brandt JM, Verma DJ, Lindberg AA. Molecular characterization of the O-acetyl transferase gene of converting bacteriophage SF6 that adds group antigen 6 to Shigella flexneri. Mol Microbiol. 1991;5:71–75. doi: 10.1111/j.1365-2958.1991.tb01827.x. [DOI] [PubMed] [Google Scholar]
- Hellerqvist CG, Linberg B, Svensson S, Holme T, Lindberg AA. Structural studies on the O-specific side-chains of the cell-wall lipopolysaccharide from Salmonella typhimurium 395 MS. Carbohydr Res. 1968;8:43–55. doi: 10.1016/S0008-6215(00)81689-4. [DOI] [Google Scholar]
- Entrez Genome http://ncbi.nlm.nih.gov/genomes/MICROBES/Complete.html
- Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Ann Rev Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Bull TJ, Sheridan JM, Martin H, Sumar N, Tizard M, Taylor JH. A novel mycobacterial insertion sequence (IS 1612), inserted into an acetylase gene (mpa) in Mycobacterium avium subsp. silvaticum but not in Mycobacterium avium subsp. paratuberculosis. Veterinary Microbiol. 2000;77:453–463. doi: 10.1016/S0378-1135(00)00330-8. [DOI] [PubMed] [Google Scholar]
- Hobbs M, Reeves PR. The JUMPstart sequence: a 39 bp element common to several polysaccharide gene clusters. Mol Microbiol. 1994;12:855–856. doi: 10.1111/j.1365-2958.1994.tb01071.x. [DOI] [PubMed] [Google Scholar]
- ORF Finder htpp://ncbi.nlm.nih.gov/gorf/orfig/cgi
- The Sequence Manipulation Suite htpp://bioinformatics.org/sms/codon_usage.html
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. 2. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY; 1989. [Google Scholar]
- National Centre for Biotechnology Information (NCBI) BLAST htpp://ncbi.nlh.nih.gov
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Bioinformatics Institute htpp://ebi.ac.uk/services
- Yap I, Nelson RJ. WinBoot: A program for performing bootstrap analysis of binary data to determine the confidence limits of UPGMA-based dendrograms. IRRI Discussion Papers Series No 14 International Rice Research Institute, Manila, the Philippines. 1996.
- DeFeyter R, Kado CI, Gabriel DW. Small stable shuttle vectors for use in Xanthomonas. Gene. 1990;88:65–72. doi: 10.1016/0378-1119(90)90060-5. [DOI] [PubMed] [Google Scholar]


