Abstract
It is well established that transcriptional silencing of critical tumor suppressor genes by DNA methylation is a fundamental process in the initiation of lung cancer. However, the involvement of microRNAs (miRNAs) in restoring abnormal DNA methylation patterns in lung cancer is not well understood. Therefore, and since miRNA-101 is complementary to the 3′-untranslated region of DNA methyltransferase 3A (DNMT3A), we investigated whether miRNA-101 could restore normal DNA methylation patterns in lung cancer cell lines. Bioinformatics has indicated that DNMT3A is a major target of miR-101. In addition, the overexpression of miR-101 downregulates DNMT3A. Using a methylation-specific polymerase chain reaction assay, we demonstrated that methylation of the phosphatase and tensin homolog (PTEN) promoter was reduced in A549 cells transfected with miR-101, but not in the transfected control. Furthermore, overexpression of miR-101 and silencing of DNMT3A suppressed lung cell proliferation and S/G2 transition, and increased apoptosis through the PTEN/AKT pathway in vitro. Furthermore, we observed the opposite phenomenon in A549 cells transfected with a miR-101 inhibitor. Subsequent investigation revealed that overexpression of miR-101 significantly inhibited the tumorigenicity of A549 cells in a nude mouse xenograft model. These results demonstrate that miR-101 affects lung cancer progression through the PTEN/AKT signaling pathway by targeting DNMT3A in lung cells, suggesting that miR-101 may be a novel potential therapeutic strategy in lung cancer treatment.
Keywords: miR-101, DNA methyltransferase 3A, methylation, proliferation, apoptosis, PTEN-AKT signaling pathway, lung cancer
Introduction
DNA methylation in mammalian cells is a conserved epigenetic silencing mechanism, which is associated with numerous significant biological processes (1–3). Aberrant patterns of DNA methylation affect the expression of oncogenes or tumor suppressor genes (TSGs), encoding proteins involved in genomic instability, malignant cell growth, cell differentiation and metastasis (4–6). Therefore, DNA methylation plays a crucial role in tumorigenesis. Similar to other members of the DNA methyl transferase (DNMT) family, abnormal levels of DNMT3A have been identified in numerous types of malignancies (7,8). High expression of DNMT3A has been observed in non-small cell lung carcinomas (NSCLCs) (9). However, the mechanism underlying DNMT3A's role in lung cancer requires further investigation.
MicroRNAs (miRNAs) are a class of non-protein-coding, endogenous, small RNAs that cause mRNA degradation or inhibition of mRNA translation by interacting with the 3′-untranslated region (3′-UTR) of the target gene mRNA (10). miRNAs of 22 nucleotides (nt) play significant regulatory roles in various fundamental biological processes, including development, differentiation and apoptosis, sharing common pathways with cancer (11–14). Loss and gain of miRNA functions contribute to the development of cancer through the upregulation of oncogenes and silencing of TSGs. miR-101 acts as a tumor suppressor, which regulates growth, apoptosis, migration and invasion of various tumor cells (15–17). However, miR-101 has not been clearly identified as a selective regulator of DNMT3A in NSCLC.
In the present study, we analyzed targets of miR-101 using bioinformatics, and observed that miR-101 targeted DNMT3A. Then, using dual-luciferase reporter assays, we further demonstrated that DNMT3A is a novel target of miR-101. We also identified that methylation of the phosphatase and tensin homolog (PTEN) promoter was reduced in A549 cells transfected with miR-101. Furthermore, in vitro experiments proved that re-expression of miR-101 inhibited NSCLC proliferation significantly, arrested the NSCLC cell cycle at the S/G2 phase, and induced cell apoptosis. Silencing of DNMT3A by miR-101 or siDNMT3A not only downregulated the cell cycle-related proteins phospho-AKT (p-AKT), cyclin A2 (CCNA2), cyclin-dependent kinase 2 (CDK2) and the cell apoptosis-related protein B-cell lymphoma 2 (Bcl-2), but also significantly upregulated bax. The same results were observed in vivo. These findings demonstrate that miR-101 suppresses the progression of A549 cells by targeting DNMT3A via the PTEN/AKT pathway.
Materials and methods
Cell line culture
A549 cells were cultured in RPMI-1640 medium (PAA Laboratories GmbH, Pasching, Austria) supplemented with 10% fetal bovine serum (FBS; PAA Laboratories GmbH). Cells were maintained at 37°C in a humidified chamber with 95% air and 5% CO2.
Plasmid constructions
The vector pcDNA™ 6.2-GW/EmGFP-miR (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to generate vectors of re-expression of miR-101. First, EcoRI and HindIII sites were inserted into the multiple cloning site of the vector. Then, the gene coding for miR-101 was chemically synthesized and cloned into pcDNA™6.2-GW/EmGFP-miR, between the EcoRI and HindIII sites (Pri-miR-101-S5′-TGCCCTGGCTCAGTTATCACAGTGCTGATGCTGTCTATTCTAAAGGTACAGTACTGTGATAACTGAAGGATGGCA-3′, Pri-miR-101-A5′-TGCCATCCTTCAGTTAGTTATCACAGTACTGTACCTTTAGAATAGACAGCATCAGCACTGTGATAACTGAGCCAGGGCA-3′). The software programs RegRNA (regulatory RNA motifs and elements finder; http://regrna.mbc.nctu.edu.tw/), TargetScan (http://www.targetscan.org/) and DIANA (http://diana.cslab.ece.ntua.gr/microT/) were used to predict gene-related specified microRNA. Using bioinformatics analyses, we identified the fragments of DNMT3A by miR-101 targeting. Specific fragments of DNMT3A were chemically synthesized (DNMT3A 3′UTR-S 5′-CTATATATATAAAAGGTACTGTTC-3′, DNM T3A 3′UTR-A 5′-TCGAGAACAGTACCTTTTATATATATAGAGCT-3′, DNM T3A 3′UTR-MS 5′-CTATATATATAAAAGACACTGTTC-3′, DNM T3A 3′UTR-MA 5′-TCGAGAACAGTGTCTTTTATATATATAGAGCT-3′). The luciferase-UTR reporter constructions were generated by introducing the wild-type (wt)/mutant (mut) EGFR 3′-UTR, carrying a putative miR-101 binding site, into the pmirGLO Dual-Luciferase miRNA Target Expression vector (Promega Corporation, Madison, WI, USA), between the XhoI and SacI sites.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol solution (Invitrogen Life Technologies) according to the manufacturer's instructions, and RNAse-free DNase was used to remove DNA contamination. Total RNA concentration and quantity were assessed using a DNA/protein analyzer (GeneQuant pro RNA/DNA; GE Healthcare Bio-Sciences, Pittsburgh, PA, USA). cDNA was synthesized from RNA using a PrimeScript™ RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). Specific primers were used to synthesize miR-101 cDNA (miR-101 RT 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTTCAGTT-3′). cDNA was amplified using SYBR Premix Ex Taq™ II (Takara Biotechnology Co., Ltd.). The PCR primers used in this study were miR-101-F 5′-ATCCAGTGCGTGTCGTG-3′ and miR-101-R 5′-TGCTTACAGTACTGTGAT-3′). PCR amplification was performed with an IQ5 Optical System real-time PCR machine. β-actin and U6 were used to normalize mRNA and miRNA, respectively (β-actin-F 5′-CGGGAAGCTTGTCATCAATGG-3′, β-actin-R 5′-GGCAGTGATGGCATGGACTG-3′; U6 RT 5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAAAATATGG-3′, U6-F 5′-TGCGGGTGCTCGCTTCGGCAGC-3′, U6-R 5′-CCAGTGCAGGGTCCGAGGT-3′). Relative quantification of mRNA expression levels was determined using the relative standard curve method according to the manufacturer's instructions (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
MTT assay was performed as described previously (18). Following transfection with miR-101, miR-101-inhibitor, si-DNMT3A or their respective controls, the cells were further cultivated for an additional 1–3 days. Cell viability was assessed using an MTT assay and a FLUOstar OPTIMA microplate reader (BMG Labtech, Aylesbury, UK). Each experiment contained three replicates and was repeated at least twice. The data were summarized as the means ± standard deviation.
Colony formation assay
Post-transfected A549 cells were re-suspended and seeded onto 12-well plates at a density of 2,000 cells/well, incubated for two weeks, and then stained with 0.5% crystal violet for 30 min. Excess dye was rinsed off twice with phosphate-buffered saline (PBS). Images were obtained using the computer software Quantity One® from Bio-Rad Laboratories, Inc.
Cell cycle analysis
Cell cycle analysis was performed as described previously (18). A549 cells were transfected with miR-101 re-expression vector, miR-101 inhibitor, si-DNMT3A or their controls. Cells were harvested by trypsinization, and 1×106 cells were used for analysis after 24, 48 and 72 h. The cells were washed with PBS and fixed in ice-cold ethanol overnight at 4°C. The cells were then washed with PBS and incubated in 1 ml staining solution (20 µg/ml propidium iodide and 10 U/ml RNaseA) for 30 min at room temperature. Cell cycle distributions were assayed by fluorescence-activated cell sorting using a flow cytometer (FACSort; BD Biosciences, Franklin Lakes, NJ, USA).
Cell apoptosis analysis
Cell apoptosis analysis was performed with an Annexin-V/fluorescein isothiocyanate apoptosis detection kit (Invitrogen Life Technologies), according to the manufacturer's instructions. Cells were seeded onto 12-well plates at a density of 1×106 cells per well in triplicate, transfected with DNA vectors or siRNAs for 48 h, and then analyzed using the flow cytometer (BD Biosciences). Apoptotic populations were determined using ModFit software (Verity Software House, Inc., Topsham, ME, USA).
Western blot analysis
Western blot analysis was performed as described previously (18). A549 cells were lysed using RIPA buffer supplemented with protease inhibitor (Invitrogen Life Technologies). Protein concentration was estimated using a quantitative analyzer (GeneQuant pro RNA/DNA). Proteins were then separated with 8 to 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (Invitrogen Life Technologies), transferred to a nitrocellulose membrane, and incubated with DNMT3A, PTEN, p-AKT, AKT, CCNA2, CDK2, Bcl-2, Bax and β-actin antibodies (diluted 1/500; Bioworld Technology, Inc., St. Louis Park, MN, USA). The membrane was washed three times with Tris-buffered saline and Tween-20 and incubated with a goat anti-rabbit antibody (Bioworld, diluted 1/5000). Relative protein expression was then normalized to β-actin levels in each sample.
Dual-luciferase assay
Dual-luciferase assay was performed as described previously. Reporter gene assays were performed 24 h post-transfection using the Dual Luciferase® reporter assay system (Promega Corporation) according to the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla luciferase activity. All experiments were performed at least three times.
DNA extraction and methylation-specific PCR
DNA was extracted using a Qiagen DNeasy tissue kit (Qiagen Inc., Valencia, CA, USA). DNA (1 µg) was placed in 100 µl water and denatured by adding 7 µl 3 M NaOH for 10 min at 37°C. To each denatured DNA solution was added 550 µl freshly prepared sodium bisulfite mixture (Qiagen, Inc.). The resulting mixtures were then incubated at 50°C for 16 h. During bisulfite modification, unmethylated cytosines are deaminated and converted to uracils, whereas 5-methyl-cytosines remain unaltered. DNA samples were then purified by ethanol precipitation and re-suspended in 25–50 µl TE buffer (10 mM Tris/0.1 mM EDTA, pH 7.5). The bisulfite-treated DNA was amplified with methylation-specific primers (using an annealing temperature of 60°C for 40 cycles) or unmethylated-specific primers (using an annealing temperature of 58°C for 40 cycles). The primer sequences were PTENM-F 5′-TTTTTTTTCGGTTTTTCGAGGC-3′, PTE NM-R 5′-CAATCGCGTCCCAACGCCG-3′; PTE NUM-F 5′-TTTTGAGGTGTTTGGGTTTTTGGT-3′, PTENUM-R 5′-ACACAATCACATCCCAACACCA-3′).
Tumorigenicity assay in nude mice
Five-week-old female nude mice were used to analyze tumorigenicity. A549 cells were transfected with lentiviral vector (LV)-miR-101 and control (LV-CN) and re-suspended in PBS, then 1×106 cells were injected subcutaneously into both posterior flanks of nude mice. Tumor size was measured using a vernier caliper every 3 days for 30 days and monitored by bioluminescent imaging using IVIS Spectrum (Xenogen Corp., Alameda, CA, USA). The mice were anesthetized by intra-peritoneal injection with 1% pentobarbital sodium (50 mg/kg). The tumor was removed following induction of deep anesthesia and the incision was closed with surgical staples. Mice were euthanized 3 weeks after the injection. Tumor volumes (V) were calculated by measuring the length (L) and width (W) of tumors, using the formula: V=(LxW2)/2. All animal experiments were approved by the Institutional Animal Care and Use Committee of Xi'an Jiaotong University, China.
Statistical analysis
Each experiment was repeated at least three times. Numerical data are presented as the means ± standard deviation. Unless indicated, the differences between the two groups were analyzed using Student's t-test (two-tailed). All statistical analyses were performed using SPSS 13.0 software (SPSS Inc., Chicago, IL, USA).
Results
miR-101 targets DNMT3A
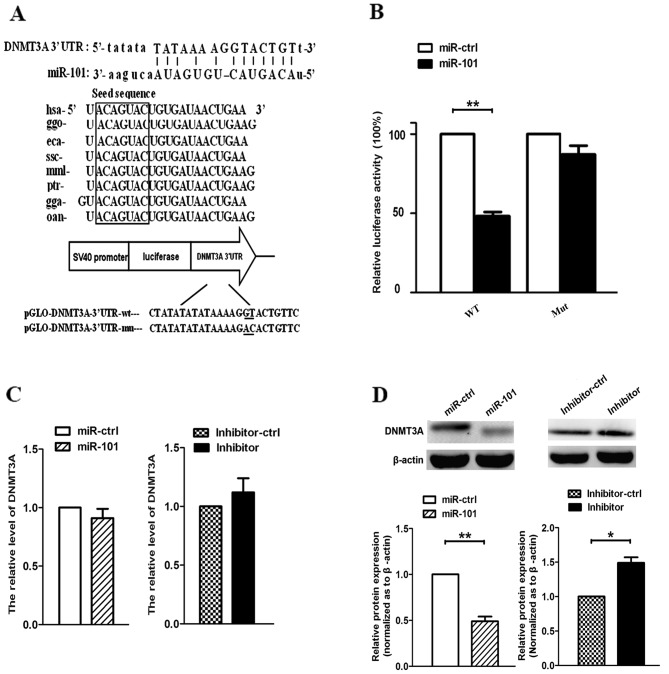
We searched for miR-101 target genes using three computer-aided miRNA target prediction programs: RegRNA, DIANA and TargetScan. As shown in Fig. 1A, we identified an miR-101 binding site at 3891–3912 nt of the DNMT3A 3′-UTR. By comparing the human sequence with those of other species, we observed that the sequence of miR-101 was highly conserved among different species. To determine whether DNMT3A was a target gene of miR-101, we constructed pmirGLO-DNMT3A-3′-UTR-wt and pmirGLO-DNMT3A-3′-UTR-mut. Furthermore, we co-transfected A549 cells with miR-101 or miR-ctrl, and pmirGLO-DNMT3A-3′-UTR-wt or pmirGLO-DNMT3A-3′-UTR-mut. The results revealed that miR-101 suppressed the firefly luciferase activity of pmirGLO-DNMT3A-3′-UTR-wt after 24 h, whereas miR-ctrl did not (Fig. 1B). Next, we demonstrated that re-expression of miR-101 or expression of miR-101 inhibitor did not affect the mRNA expression of DNMT3A (Fig. 1C). However, when cells were transfected with miR-101 and miR-101 inhibitor, the protein levels of DNMT3A were decreased and increased, respectively (Fig. 1D). These data suggest that miR-101 inhibits DNMT3A expression at the translational but not the transcriptional level in A549 cells.
Figure 1.
miR-101 targets DNA methyltransferase 3A (DNMT3A). (A) miR-101 is highly conserved across different species. (B) Luciferase assay in A549 cells. 24-bp region (wt) miR-101 binding sites in the DNMT3A 3′UTR were cloned into pmirGLO Dual-Luciferase miRNA Target Expression vector. Identical mutated constructs were generated. Either miR-ctrl or miR-101 was co-transfected with pmirGLO-DNMT3A-3′ UTR-wt or pmirGLO-DNMT3A-3′UTR-mut into A549 cells and luciferase activity was assayed after 24 h. The normalized firefly luciferase activity was obtained by firefly luciferase activity/Renilla luciferase activity, and the results are shown as the means ± standard deviation (**P<0.01, Student's t-test). (C) DNMT3A mRNA protein expression levels measured by reverse transcription-quantitative polymerase chain reaction and western blot analysis 48 h after transfection with miR-ctrl or miR-101. (D) DNMT3A protein expression levels measured by western blot analysis 48 h after transfection with miR-ctrl or miR-101. β-actin was used as a housekeeping control. wt, wild type; mut, mutant.
DNMT3A affects the expression of a downstream gene
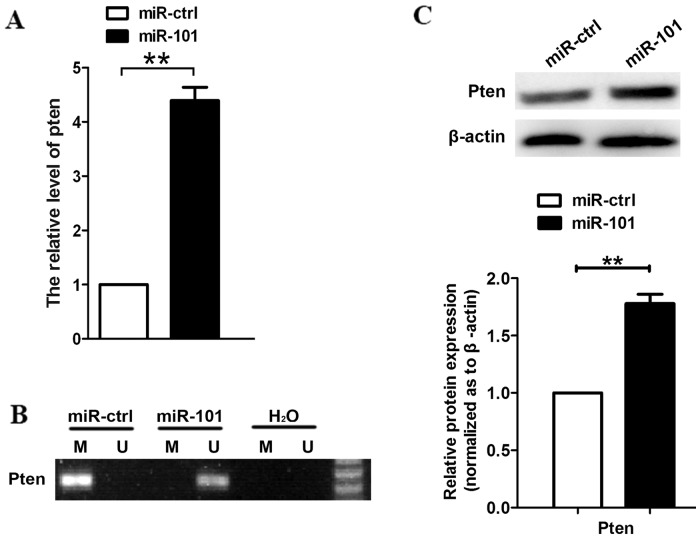
Using RT-qPCR, we measured the transcript levels of PTEN following transfection with miR-101, and noted that PTEN expression was increased (Fig. 2A). Since DNMT3A affects the expression of oncogenes or TSG-encoding proteins, we examined DNA methylation at the upstream region of the PTEN coding sequence using methylation-specific PCR. The results revealed that the CpG sites of PTEN were highly unmethylated in A549 cells transfected with miR-101, but not in miR-ctrl-transfected cells (Fig. 2B). We also observed that protein levels of PTEN were increased upon transfection with miR-101 (Fig. 2C).
Figure 2.
DNA methylation is responsible for the upregulation of phosphatase and tensin homolog (PTEN). (A) Reverse transcription-quantitative polymerase chain reaction (qPCR) analysis was performed to determine the expression level of PTEN following transfection of miR-101 or miR-ctrl. (B) Methylation-specific PCR analysis of the PTEN CpG sites in A549 cells following transfection of miR-101 or miR-ctrl. H2O was used as a negative control. Bands in the ‘M’ and ‘U’ lanes are PCR products obtained with methylation-specific and non-methylation-specific primers, respectively. (C) Western blot analysis was used to assay the expression of PTEN in A549 cells following transfection of miR-101 or its negative control. β-actin was used as a housekeeping control (**P<0.01, Student's t-test).
miR-101 inhibits the growth of A549 cells by suppressing the PTEN/AKT signaling pathway
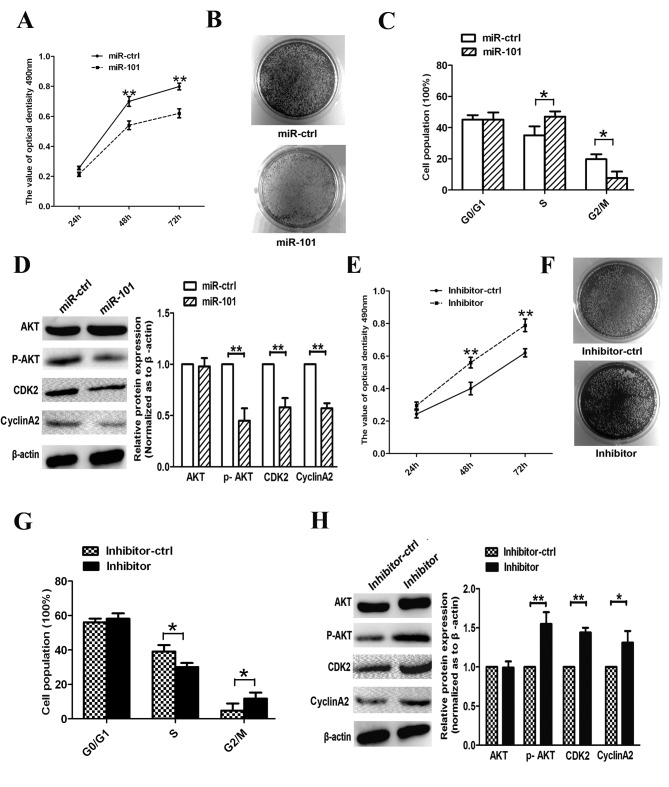
To study the role of miR-101 in A549 cell proliferation, we performed MTT, clone formation and cell cycle assays. The results demonstrated that overexpression of miR-101 inhibits the proliferation of A549 after 48 and 72 h (Fig. 3A); miR-101-transfected cells exhibited fewer and smaller colonies compared with miR-ctrl-transfected cells (Fig. 3B). Moreover, re-expression of miR-101 resulted in a marked repression from the S phase to the G2 phase in A549 cells (Fig. 3C). In addition, we measured the expression of cell cycle regulators associated with the PTEN/AKT pathway. We observed that the expression of p-AKT, CCNA2 and CDK2 was suppressed upon transfection with miR-101 (Fig. 3D), suggesting that miR-101 arrested the cell cycle at the S/G2 transition phase and suppressed cell proliferation in vitro.
Figure 3.
miR-101 decreases A549 cell proliferation and induces S-phase arrest in vitro. (A) The effects of miR-101 on A549 cell proliferation were determined by MTT assay at 24, 48 and 72 h. (B) Representative results of colony formation of A549 cells following miR-101 overexpression. (C) The cell cycle was determined in A549 cells 48 h after transfection. (D) Analysis of the expression of S-phase regulatory protein in A549 cells following transfection with miR-ctrl and miR-101 overexpression construct. (E) MTT was used to assay the effects of miR-101 inhibitor on the proliferation of A549 cells. (F) The growth of A549 cells was detected by colony formation following transfection of inhibitor. (G) The cell cycle was determined by transfecting with inhibitor or its control. (H) Expression of cell cycle regulators 48 h after transfection with inhibitor and control (*P<0.05, **P<0.01, Student's t-test).
To examine the anti-proliferative role of miR-101 in human lung cancer cells, we eliminated endogenous miR-101 in A549 cells using a miR-101 inhibitor. The inhibitor reduced endogenous expression of miR-101 in A549 cells and increased cell viability and colony forming numbers (Fig. 3E and F). We further investigated the effects of miR-101 inhibitor on cell cycle progression in A549 cells and revealed that transfection with this inhibitor decreased the amount of cells in the S phase and increased the percentage of cells in the G2 phase (Fig. 3G). Furthermore, expression levels of p-AKT, CCNA2 and CDK2 were increased in inhibitor-transfected cells (Fig. 3H). These results suggest that endogenous miR-101 plays an essential anti-carcinogenic role in A549 cells during lung cancer progression.
miR-101 induces apoptosis in A549 cells
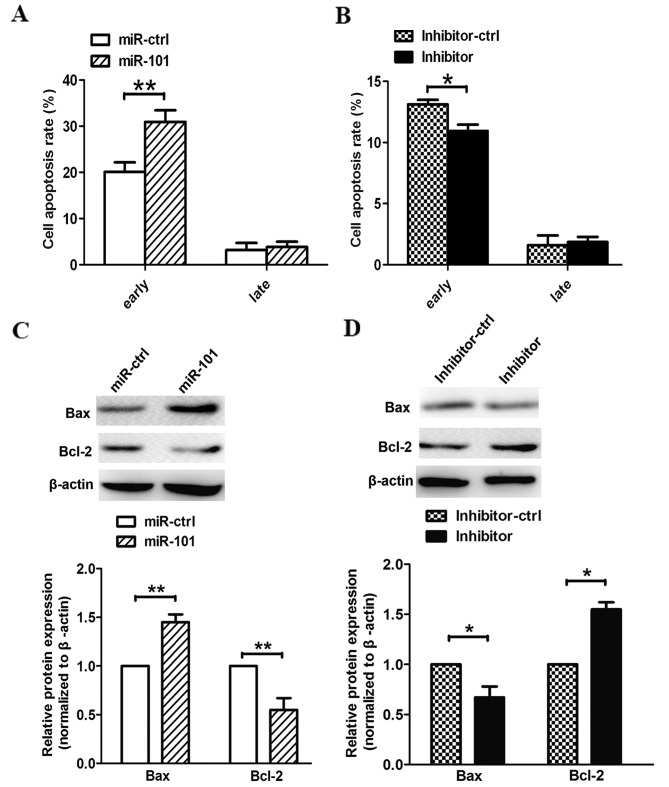
In our in vitro experiments, the overexpression of miR-101 also induced cell apoptosis. Compared with cells transfected with a control vector, pre-miR-101-transfected cells exhibited higher apoptotic rates at an early phase. In addition, cells transfected with the miR-101 inhibitor exhibited lower apoptotic rates than cells transfected with a control vector (Fig. 4A and B). Our data demonstrate that miR-101 induces apoptosis in human lung cancer cells in vitro. Furthermore, we observed that miR-101 modified the expression of apoptosis-associated genes in the PTEN/AKT signaling pathway. As a result, suppression of p-AKT promoted apoptosis by accelerating Bax and inactivating Bcl-2 (Fig. 4C). Notably, we observed the opposite phenomenon when A549 cells were transfected with the miR-101 inhibitor (Fig. 4D).
Figure 4.
miR-101 induces apoptosis in A549 cells. (A and B) Cell apoptosis was detected by Annexin-V/propidium iodide combined labeling flow cytometry in A549 cells 48 h after transfection with miR-101, miR-101 inhibitor, or their respective controls. Apoptotic evaluation was carried out by the percentage of the apoptotic cell number of the total cell number. (C and D) Expression of cell apoptosis-associated proteins downstream of the DNA methyltransferase 3A/AKT signaling pathway were determined in A549 cells 48 h after transfection of miR-ctrl, miR-101, inhibitor-ctrl or inhibitor by western blot analysis. β-actin was used as a housekeeping control. *P<0.05, **P<0.01, Student's t-test.
DNMT3A silencing suppresses lung cancer cell growth and induces G1/S-phase arrest and cell apoptosis
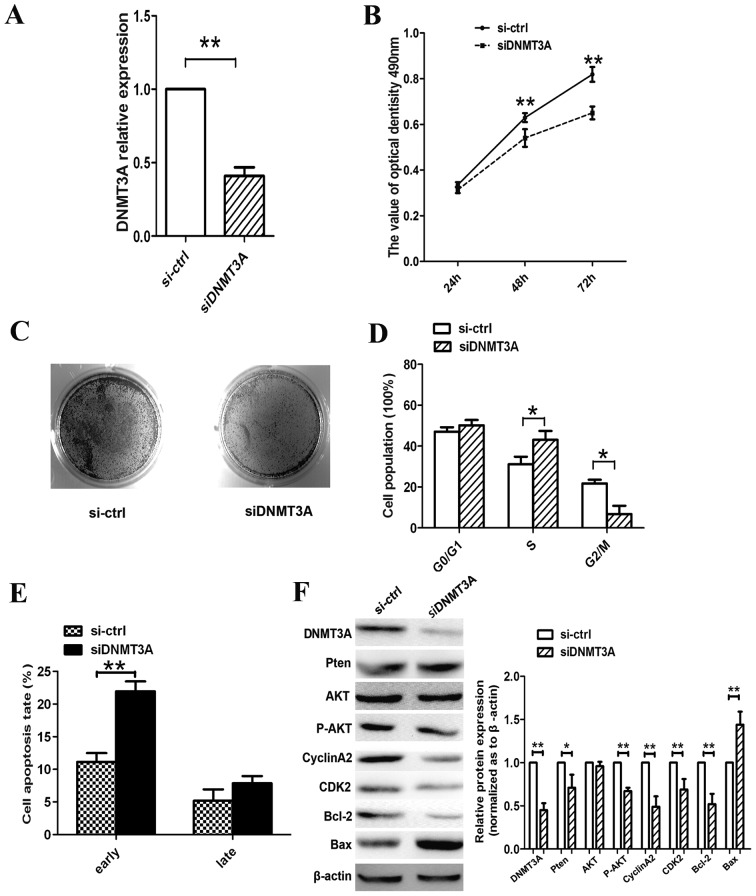
As demonstrated previously, overexpression of miR-101 affects cell growth, cell proliferation, cell cycle and cell apoptosis in A549 lung cancer cells. We also confirmed that DNMT3A was a direct target of miR-101. Therefore, we silenced DNMT3A expression using RNA interference to confirm that DNMT3A is involved in the antitumor effects of miR-101. On the mRNA expression level, siDNMT3A knocked down DNMT3A (Fig. 5A). Moreover, DNMT3A silencing resulted in cell growth suppression, arrest of the S/G2 phase and promotion of cell apoptosis (Fig. 5B-E). These results follow the same trend as those obtained with miR-101-transfected A549 cells.
Figure 5.
Silencing of DNA methyltransferase 3A (DNMT3A) suppresses lung cancer cell growth and induces S-phase arrest and cell apoptosis in A549 cells. (A) Reverse transcription-quantitative polymerase chain reaction analysis was performed to determine the expression of DNMT3A following transfection of siDNMT3A. (B-E) MTT assay was performed and clone formation, cell cycle and cell apoptosis were assessed to determine the effects of siDNMT3A on the growth and apoptosis of A549 cells. (F) Analysis of expression of DNMT3A/AKT signaling pathway regulatory proteins in A549 cells at 48 h after knockdown of DNMT3A. β-actin was used as a housekeeping control (*P<0.05, **P<0.01, Student's t-test).
Furthermore, we examined the expression of genes associated with the PTEN/AKT pathway. As shown in Fig. 5F, the expression levels of DNMT3A and p-AKT and of the cell cycle regulators CCNA2 and CDK2 were reduced, whereas PTEN expression was increased. Moreover, siRNA promoted apoptosis by activating the pro-apoptotic protein Bax and inactivating the anti-apoptotic protein Bcl-2. Therefore, we concluded that miR-101 regulates lung cancer cell progression by directly targeting DNMT3A through the PTEN/AKT signaling pathway.
miR-101 induces growth inhibition of A549 cells in vivo
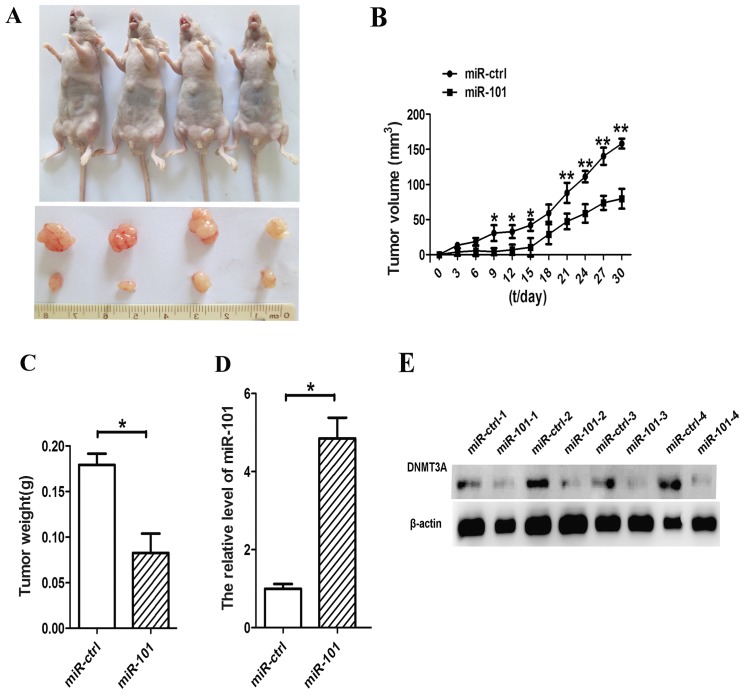
To further confirm the growth inhibitory function of miR-101 in lung cancer, we used lentiviral vectors to stably restore the expression of miR-101 in A549 cells. The LV-miR-101-infected and LV-CN-infected cells were injected subcutaneously into the left and right posterior flank of the same nude mice, respectively. We measured xenograft tumor growth for four weeks. As shown in Fig. 6A, tumor growth was significantly suppressed by LV-miR-101 compared with the control during the experiment. On day 30, the average volume of miR-101-treated tumors was much smaller than that of control tumors (Fig. 6B). The average tumor weights for the control and miR-101 groups on day 30 were 0.18 and 0.07 g, respectively (Fig 6C). Furthermore, expression levels of miR-101 and DNMT3A in tumor tissues were examined by RT-qPCR and western blot analysis. Consistent with the in vitro data, the in vivo data revealed that the expression of miR-101 was increased and the expression of the DNMT3A protein was decreased in miR-101-treated tumors (Fig. 6D and E).
Figure 6.
miR-101 inhibits lung cancer progression in vivo. (A) Small animal imaging analysis was used to assess the tumor volume in situ during the fourth week of tumor development. The flanks of four nude mice were injected with lentiviral vector (LV)-miR-101-transfected cells (left flank) and LV control (right flank), respectively. The lower portion reveals the morphology of mice injected with miR-101 and miR-ctrl. (B) Tumor growth curves. (C) Tumor weight. (D) Expression levels of miR-101 were measured by reverse transcription-quantitative polymerase chain reaction analysis in the tumor tissues from the animals. (E) Expression levels of DNA methyltransferase 3A (DNMT3A) were assessed by western blot analysis in tissues from the animals. β-actin was used as a housekeeping control (*P<0.05, **P<0.01, Student's t-test).
Discussion
Lung cancer is one of the most lethal malignant diseases in the world. NSCLC, constituting ~80% of lung cancer cases, is a primary type of lung cancer. Current treatments including chemotherapy have limited efficacy, leading to poor prognosis and metastasis of lung cancer. Therefore, it is essential to investigate the underlying molecular mechanisms to improve the situation. Previous studies have revealed that hypermethylation of promoters, mediated by DNMTs, is the main reason for epigenetic inactivation of TSGs. Hypermethylation is responsible for the silencing of TSGs involved in tumorigenesis (19,20). DNMT3A, like other DNMT family members, is involved in tumorigenesis, differentiation and metastasis (21,22), but the correlation between DNMT3A and NSCLCs remains largely unknown. In the last ten years, miRNAs have been noted to play a significant role in the initiation and progression of NSCLCs (23,24). miR-101 is a miRNA that regulates a variety of tumor-related biological processes by modulating the expression of several target genes at the transcript or protein level (25,26). Normally, miR-101 inhibits the expression of lung cancer promoting genes (17,27). In this study, we observed a miR-101 binding site at 3891–3912 nt of the DNMT3A 3′-UTR. In addition, dual-luciferase reporter assays demonstrated that miR-101 targeted directly DNMT3A by recognizing the 3′-UTR of the DNMT3A miRNA, and inhibited DNMT3A translation (Fig. 1).
Furthermore, P14, P16, RASSF1A and PTEN are all TSGs; their functions have been investigated in a number of malignant tumors (28–30). These genes are frequently inactivated in numerous human malignancies including lung, breast and esophageal cancers (31–33). DNMT3A regulated the expression of TSGs by methylating their upstream region. Fig. 2 demonstrates that the expression of PTEN was increased and the CpG sites of PTEN were less methylated in A549 cells transfected with miR-101 than in the control cells. The same results were observed in hepatocellular carcinoma cells (7).
PTEN is a phosphatase, and mutations of PTEN are observed in a number of cancers (34). Normally, degradation and inactivation of phosphatidylinositol (3,4,5)-trisphosphate (PIP3) is due to PTEN dephosphorylation (35). The phosphoinositide 3-kinase (PI3K) pathway is one of the most potent pro-survival pathways in the development of cancer (36). Inactivation of PTEN not only leads to accumulation of PIP3, but also increases activity of the kinase AKT, which contributes to oncogenesis in numerous cancers, including glioblastoma, prostate and liver cancers (37,38). AKT, a key downstream effector of the P13K signaling pathway, modulates the function of numerous substrates associated with cell cycle progression and cell apoptosis, either by direct phosphorylation of the target proteins themselves or, indirectly, by regulating protein expression levels (39). In our study, overexpression of miR-101 or siDNMT3A inhibited DNMT3A expression, which resulted in PTEN activation and a decline in AKT phosphorylation. Next, we verified the effects of AKT on the downstream target genes CCNA2 and CDK2, which are key transcriptional factors in the S/G2 phase. From these results, we noted a reduced expression of CDK2 and CCNA2 in A549 cells transfected with miR-101. Furthermore, in order to investigate the role of miR-101 and DNMT3A in the apoptosis of A549 cells, we also measured the expression levels of Bcl-2 and Bax, and demonstrated that the miR-101-induced PI3K-AKT pathway plays a significant role in the regulation of cell apoptosis.
Further animal studies indicated that miR-101 suppressed the growth of lung cancer cells in vivo and decreased DNMT3A expression in treated tumors (Fig. 6). The in vivo studies support our in vitro observations that miR-101 targets DNMT3A and suppresses lung cancer cell growth.
In summary, we investigated the roles of miR-101 and its targeted gene, DNMT3A, in the cell cycle and apoptosis. Our findings suggest that miR-101 may be a novel tumor suppressor that blocks the growth of NSCLC cells through the PTEN/AKT signaling pathway by targeting DNMT3A. Our findings highlight the functional association of miR-101 and its host genes, provide new insight into the regulatory network of the cell cycle and apoptosis, and open possibilities for future therapeutic interventions.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (81402008, 31100921 and 5143827), the Fundamental Research Funds for the Central Universities (08142006), and the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT: 1171).
References
- 1.Ohgane J, Aikawa J, Ogura A, Hattori N, Ogawa T, Shiota K. Analysis of CpG islands of trophoblast giant cells by restriction landmark genomic scanning. Dev Genet. 1998;22:132–140. doi: 10.1002/(SICI)1520-6408(1998)22:2<132::AID-DVG3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 2.Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H, Held WA. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci USA. 2005;102:3336–3341. doi: 10.1073/pnas.0408436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Turek-Plewa J, Jagodziński PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631–647. [PubMed] [Google Scholar]
- 6.Li Y, Tollefsbol TO. Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr Med Chem. 2010;17:2141–2151. doi: 10.2174/092986710791299966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Z, Wu Q, Cheng J, Qiu X, Zhang J, Fan H. Depletion of DNMT3A suppressed cell proliferation and restored PTEN in hepatocellular carcinoma cell. J Biomed Biotechnol. 2010;2010:737535. doi: 10.1155/2010/737535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starlard-Davenport A, Kutanzi K, Tryndyak V, Word B, Lyn-Cook B. Restoration of the methylation status of hypermethylated gene promoters by microRNA-29b in human breast cancer: a novel epigenetic therapeutic approach. J Carcinog. 2013;12:15. doi: 10.4103/1477-3163.115720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Wiemer EA. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43:1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 13.Liu W, Mao SY, Zhu WY. Impact of tiny miRNAs on cancers. World J Gastroenterol. 2007;13:497–502. doi: 10.3748/wjg.v13.i4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oakley EJ, Van Zant G. Unraveling the complex regulation of stem cells: implications for aging and cancer. Leukemia. 2007;21:612–621. doi: 10.1038/sj.leu.2404530. [DOI] [PubMed] [Google Scholar]
- 15.Shen Q, Bae HJ, Eun JW, Kim HS, Park SJ, Shin WC, Lee EK, Park S, Park WS, Lee JY, et al. MiR-101 functions as a tumor suppressor by directly targeting nemo-like kinase in liver cancer. Cancer Lett. 2014;344:204–211. doi: 10.1016/j.canlet.2013.10.030. [DOI] [PubMed] [Google Scholar]
- 16.Xiaoping L, Zhibin Y, Wenjuan L, Zeyou W, Gang X, Zhaohui L, Ying Z, Minghua W, Guiyuan L. CPEB1, a histone-modified hypomethylated gene, is regulated by miR-101 and involved in cell senescence in glioma. Cell Death Dis. 2013;4:e675. doi: 10.1038/cddis.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin J, Wang M, Jin C, Qi Q. miR-101 sensitizes A549 NSCLC cell line to CDDP by activating caspase 3-dependent apoptosis. Oncol Lett. 2014;7:461–465. doi: 10.3892/ol.2013.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Yao J, Zhang X, Guo B, Le X, Cubberly M, Li Z, Nan K, Song T, Huang C. miRNA-302b suppresses human hepatocellular carcinoma by targeting AKT2. Mol Cancer Res. 2014;12:190–202. doi: 10.1158/1541-7786.MCR-13-0411. [DOI] [PubMed] [Google Scholar]
- 19.Yu YY, Chen C, Kong FF, Zhang W. Clinicopathological significance and potential drug target of RUNX3 in breast cancer. Drug Des Devel Ther. 2014;8:2423–2430. doi: 10.2147/DDDT.S71815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D, Cui W, Wu X, Qu Y, Wang N, Shi B, Hou P. RUNX3 site-specific hypermethylation predicts papillary thyroid cancer recurrence. Am J Cancer Res. 2014;4:725–737. [PMC free article] [PubMed] [Google Scholar]
- 21.Ma QL, Wang JH, Wang YG, Hu C, Mu QT, Yu MX, Wang L, Wang DM, Yang M, Yin XF, et al. High IDH1 expression is associated with a poor prognosis in cytogenetically normal acute myeloid leukemia. Int J Cancer. 2015;137:1058–1065. doi: 10.7150/jca.12519. [DOI] [PubMed] [Google Scholar]
- 22.Cao XY, Ma HX, Shang YH, Jin MS, Kong F, Jia ZF, Cao DH, Wang YP, Suo J, Jiang J. DNA methyltransferase3a expression is an independent poor prognostic indicator in gastric cancer. World J Gastroenterol. 2014;20:8201–8208. doi: 10.3748/wjg.v20.i25.8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang P, Ye B, Yang Y, Shi J, Zhao H. MicroRNA-181 functions as a tumor suppressor in non-small cell lung cancer (NSCLC) by targeting Bcl-2. Tumour Biol. 2015;36:3381–3387. doi: 10.1007/s13277-014-2972-z. [DOI] [PubMed] [Google Scholar]
- 24.Mataki H, Seki N, Chiyomaru T, Enokida H, Goto Y, Kumamoto T, Machida K, Mizuno K, Nakagawa M, Inoue H. Tumor-suppressive microRNA-206 as a dual inhibitor of MET and EGFR oncogenic signaling in lung squamous cell carcinoma. Int J Oncol. 2015;46:1039–1050. doi: 10.3892/ijo.2014.2802. [DOI] [PubMed] [Google Scholar]
- 25.Lin C, Huang F, Li QZ, Zhang YJ. miR-101 suppresses tumor proliferation and migration and induces apoptosis by targeting EZH2 in esophageal cancer cells. Int J Clin Exp Pathol. 2014;7:6543–6550. [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, Guo J, Yu L, Cai J, Gui T, Tang H, Song L, Wang J, Han F, Yang C, et al. miR-101 regulates expression of EZH2 and contributes to progression of and cisplatin resistance in epithelial ovarian cancer. Tumour Biol. 2014;35:12619–12626. doi: 10.1007/s13277-014-2585-6. [DOI] [PubMed] [Google Scholar]
- 27.Lei YM, Zu YF, Wang J, et al. Interleukin-1β-mediated suppression of microRNA-101 and upregulation of enhancer of zeste homolog 2 is involved in particle-induced lung cancer. Med Oncol. 2015;32:387. doi: 10.1007/s12032-014-0387-8. [DOI] [PubMed] [Google Scholar]
- 28.Chaar I, Amara S, Elamine OE, Khiari M, Ounissi D, Khalfallah T, Ben Hmida A, Mzabi S, Bouraoui S. Biological significance of promoter hypermethylation of p14/ARF gene: relationships to p53 mutational status in Tunisian population with colorectal carcinoma. Tumour Biol. 2014;35:1439–1449. doi: 10.1007/s13277-013-1198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camacho CV, Mukherjee B, McEllin B, Ding LH, Hu B, Habib AA, Xie XJ, Nirodi CS, Saha D, Story MD, et al. Loss of p15/Ink4b accompanies tumorigenesis triggered by complex DNA double-strand breaks. Carcinogenesis. 2010;31:1889–1896. doi: 10.1093/carcin/bgq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korah R, Healy JM, Kunstman JW, Fonseca AL, Ameri AH, Prasad ML, Carling T. Epigenetic silencing of RASSF1A deregulates cytoskeleton and promotes malignant behavior of adrenocortical carcinoma. Mol Cancer. 2013;12:87. doi: 10.1186/1476-4598-12-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamada K, Kohno T, Takahashi M, Yamazaki M, Yamazaki M, Tashiro H, Sugawara C, Ohwada S, Sekido Y, Minna JD, et al. Two regions of homozygous deletion clusters at chromosome band 9p21 in human lung cancer. Genes Chromosomes Cancer. 2000;27:308–318. doi: 10.1002/(SICI)1098-2264(200003)27:3<308::AID-GCC12>3.3.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi N, Sugimoto Y, Tsuchiya E, Ogawa M, Nakamura Y. Somatic mutations of the MTS (multiple tumor suppressor) 1/CDK4l (cyclin-dependent kinase-4 inhibitor) gene in human primary non-small cell lung carcinomas. Biochem Biophys Res Commun. 1994;202:1426–1430. doi: 10.1006/bbrc.1994.2090. [DOI] [PubMed] [Google Scholar]
- 33.Burbee DG, Forgacs E, Zöchbauer-Muller S, Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 35.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 36.Zhou BP, Liao Y, Xia W, Spohn B, Lee MH, Hung MC. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat Cell Biol. 2001;3:245–252. doi: 10.1038/ncb1101-e245. [DOI] [PubMed] [Google Scholar]
- 37.Sano T, Lin H, Chen X, Langford LA, Koul D, Bondy ML, Hess KR, Myers JN, Hong YK, Yung WK, Steck PA. Differential expression of MMAC/PTEN in glioblastoma multiforme: relationship to localization and prognosis. Cancer Res. 1999;59:1820–1824. [PubMed] [Google Scholar]
- 38.Buontempo F, Ersahin T, Missiroli S, Senturk S, Etro D, Ozturk M, Capitani S, Cetin-Atalay R, Neri ML. Inhibition of Akt signaling in hepatoma cells induces apoptotic cell death independent of Akt activation status. Invest New Drugs. 2011;29:1303–1313. doi: 10.1007/s10637-010-9486-3. [DOI] [PubMed] [Google Scholar]
- 39.Xu N, Lao Y, Zhang Y, Gillespie DA. Akt: A double-edged sword in cell proliferation and genome stability. J Oncol. 2012;2012:951724. doi: 10.1155/2012/951724. [DOI] [PMC free article] [PubMed] [Google Scholar]