Abstract
In this study, lentivirus-mediated RNA interference (RNAi) was applied to inhibit latent membrane protein 2A (LMP2A) gene expression, in order to explore the effects of LMP2A silencing on the growth of an Epstein-Barr virus-associated gastric carcinoma (EBVaGC) cell line in vitro. Lentivirus-mediated RNAi technology was employed to specifically knock down the LMP2A gene in the EBV-positive gastric carcinoma cell line GT38. After infection, reverse transcription-quantitative polymerase chain reaction, western blotting, flow cytometry and colony formation assays were conducted to evaluate the expression of LMP2A and the biological behavior of the GT38 cell line in vitro. The results showed that the expression of the LMP2A gene was clearly downregulated in the infected cells, which indicated that a highly efficient and stable lentivirus vector was successfully constructed. In the GT38 cells in which the expression of LMP2A was downregulated, the proliferation and colony formation of the cells was significantly inhibited. In addition, it was found that the cell cycle of the GT38 cells was arrested in the G0/G1 phase and the apoptosis rate was increased. These results indicate that lentivirus-mediated RNAi knockdown of LMP2A inhibits the growth of the EBVaGC cell line GT38 in vitro, and suggests that LMP2A is a potential target for gene therapy in the treatment of EBVaGC.
Keywords: latent membrane protein 2A, gastric carcinoma, RNA interference, lentivirus, proliferation
Introduction
Epstein-Barr virus (EBV) is a human gamma herpes virus that can exist in humans for a long time without producing any symptoms (1). A variety of human malignancies such as Burkitt's lymphoma, nasopharyngeal carcinoma and gastric cancer (GC) are reported to be associated with EBV (2). EBV-associated gastric carcinoma (EBVaGC) accounts for 10–18% of gastric carcinoma cases, and there are estimated to be >75,000 new cases of EBVaGC each year worldwide (3,4). As a type of GC, which is one of the most common types of malignant tumors, EBVaGC is very difficult to remove and the complete elimination of tumor cells by surgical, radiotherapeutic and chemotherapeutic methods is challenging. Therefore, the development of new therapeutic approaches for the inhibition of tumor cell growth or survival to treat EBVaGC is of critical importance.
EBVaGC is closely associated with EBV infection, and EBVaGC cells have been found to express a well-defined set of latent viral genes, including latent membrane protein 2A (LMP2A) (5). It has been revealed that ~40% EBVaGC expresses LMP2A, whose expression closely correlates with a poor survival outcome (6). LMP2A consists of a 27-amino-acid carboxy-terminal cytoplasmic domain, a 119-amino-acid tyrosine-rich amino-terminal cytoplasmic domain and 12 hydrophobic transmembrane domains, with the cytoplasmic domain playing a role as a signaling effector (7). LMP2A has various functions, one of which is to activate the phosphoinositide 3-kinase (PI3K)/Akt, nuclear factor (NF)-κB, β-catenin, signal transducers and activators of transcription (STAT) and Syk tyrosine kinase pathways in epithelial cells (8–10). LMP2A also plays an important role in cell transformation activities, such as the induction of cell growth, enhancement of cell adhesion and cell motility, as well as the inhibition of epithelial cell differentiation (11,12). Thus, the aforementioned findings indicate that LMP2A may be a potential target for gene therapy for EBVaGC treatment.
RNA interference (RNAi) is an efficient tool that can cause post-transcriptional silencing of gene expression and induce loss-of-function phenotypes (13). Lentivirus vectors have been developed to be a powerful technology for the achievement of a significant level of gene transfer in vitro (14). In the present study, lentivirus-mediated RNAi was used to inhibit LMP2A gene expression, and the effects of LMP2A-silencing on cell growth, cell cycle and cell apoptosis of EBVaGC cell line GT38 were studied.
Materials and methods
Cell line and culture
The EBV-positive human gastric carcinoma cell line GT38 was bought from American Type Culture Collection (Manassas, VA, USA). Cells were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; ExCell Bio, Shanghai, China), 50 U/ml penicillin G and 50 U/ml streptomycin (Gibco) in a humidified atmosphere of 5% CO2 at 37°C. The medium was changed every 2 days and the cell line was passaged every 4–5 days.
Construction of lentivirus vectors
In order to produce lentivirus expressing RNAi specific for the LMP2A gene (GU979791), RNAi Designer software (rnaidesigner.thermofisher.com/rnaiexpress/; BLOCK-iTTM RNAi; Thermo Fisher Scientific, Inc.) was used to identify the RNAi sequence for human LMP2A (CTCCCAATATCCATCTGCT), and then a scrambled sequence (TTCTCCGAACGTGTCACGT) was created as a negative control construct (control RNAi) that should have no homology with the human genome. DNA oligos with the target sequence were chemically synthesized, annealed, double digested with AgeI and EcoRI, and inserted into the pLenR-GPH expression vector (GeneChem Co., Ltd., Shanghai, China) using T4 DNA ligase (Invitrogen; Thermo Fisher Scientific, Inc.), following the manufacturer's guidelines. The ligated vector was transformed into competent Escherichia coli DH5a cells (Invitrogen; Thermo Fisher Scientific, Inc.). Restriction enzyme analysis and DNA sequencing were performed to identify the correct transformant. The sequences were cloned into the pGCSIL-Green Fluorescent Protein (GFP) vector (GeneChem Co., Ltd.) to generate lentivirus vectors. 293T cells (Shanghai Research Institute of Chinese Academy of Sciences, Beijing, China) were used to generate lentiviruses after their transfection into the expression vectors and package vectors with the use of Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). After 48 h, supernatants containing the lentiviruses pGCSIL-LMP2A-shRNA-LV and pGCSIL-neg-shRNA-LV were harvested and the remaining cells were removed by filtering with 0.45 µm filters. Ultracentrifugation (4,000 × g at 4°C for 10 min) was then performed to concentrate the lentiviruses and the titer was finally determined by 293T cell infection assay.
Infection of lentivirus
In this assay, 5×103 GT38 cells in the logarithmic growth phase were seeded in each well of a 96-well microplate and cultured overnight. The lentiviruses were then diluted with 0.2 ml RPMI complete medium containing Polybrene (10 µg/ml) and added to infect the seeded cells for 12 h at 37°C. The virus-containing medium was then changed with fresh culture medium. Fluorescence microscopy (IX-53; Olympus Corporation, Tokyo, Japan) was used to detect GFP in the successfully infected cells, and the percentage of GFP-positive cells was used to measure the infection efficiency of the cells. At 5 days after the infection, analysis of LMP2A expression, cell proliferation and cell apoptosis was performed. GT38 cells treated differently were divided into three groups in subsequent assays: Blank control group (CON group; cells without infection), negative control (NC group; cells were infected with pGCSIL-neg-shRNA-LV) and the LMP2A knockdown group (KD group; cells were infected with pGCSIL-LMP2A-shRNA-LV).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Shanghai, China) and reverse transcribed using the SuperScript III First-strand Synthesis System kit (cat. no. 18080051; Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. PCR amplification was carried out using Platinum SYBR Green qPCR SuperMix UDG (Invitrogen). The reaction volume contained DNA Polymerase 10X buffer (2 µl), MgCl2 (1.5 mM), dNTP mix (2.5 mM), 10 pmol each primer, DNA template (1 µg), Taq DNA Polymerase (2–3 µl) and nuclease-free water to a final volume of 20 µl. The integrity of RNA was detected by the parallel amplification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. The specific primer pairs were as follows: LMP2A (280 bp), sense: ATGACTCATCTCAACACATA and antisense: CATGTTAGGCAAATTGCAA; GAPDH (450 bp), forward: 5′-CTCAGACACCATGGGGAAGGTGA-3′ and reverse: 5′-ATGATCTTGAGGCTGTTGTCATA-3′. The following thermal cycling conditions were used: 95°C for 5 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 15 sec and 72°C for 1 min, and the final extension was 72°C for 5 min. The 2−ΔΔCq method was applied to analyze the data (15).
Western blot analysis
Protein was extracted from the cells using lysis buffer (150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40, and 50 mM Tris, pH 8.0), with the addition of 2 mM phenylmethylsulfonyl fluoride. The six-well plates of lentivirus-infected cells were put on ice, freshly prepared lysis buffer (100 µl/well) was added and the cells were incubated for 10 min. A protein assay kit was used to determine the protein concentration. Equal amounts of protein samples (10 µg per condition) were prepared in loading buffer and boiled for 10 min, then the boiled samples were loaded to 15% SDS-PAGE gels and separated. Separated proteins were transferred from the gel to polyvinylidene difluoride (PVDF) membranes at 100 V for 1 h. The PVDF membrane with protein samples was blocked using blocking buffer which was freshly prepared with PBS containing 5% skimmed milk powder. After blocking, anti-LMP2A (cat. no. ab59026; Abcam, Cambridge, UK) and anti-GAPDH (cat. no. 5174; Cell Signaling Technology, Inc., Danvers, MA, USA) primary antibodies and anti-rabbit horseradish-peroxidase (HRP)-conjugated IgG (cat. no. 7071; Cell Signaling Technology, Inc.) and anti-mouse HRP-conjugated IgG (cat. no. 7072; Cell Signaling Technology, Inc.) secondary antibodies were diluted to 1:1,000 concentration with PBS and Tween 20 (PBST) buffer before they were successively incubated with the PVDF membrane for 60 min at room temperature. Prior to each step, the PVDF membrane was washed 3 times with PBST. Protein bands were then detected using enhanced chemiluminescence (Pierce ECL Substrate; Thermo Fisher Scientific, Inc.), and gel analysis was performed using Image J software (version 1.8.0; National Institutes of Health, Bethesda, MD, USA).
Proliferation assay
Cell proliferation was measured using a Cell Counting kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). After lentivirus-infected cells had been seeded in 96-well plates for 20, 44, 68, 92 or 116 h, 10 µl CCK-8 solution was added to each well and incubated for 1 h at 37°C. Absorbance was measured at 490 nm using an enzyme immunoassay analyzer (model 680; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Colony formation assay
A colony formation assay was performed to analyze the effect of LMP2A silencing on the colony formation ability of GT38 cells. In this assay, 8×102 cells were seeded in 6-cm dishes and cultured with RPMI-1640 supplemented with 10% FBS in an atmosphere of 5% CO2 and 95% humidity at 37°C. After 2 weeks, the cell colonies were washed twice with PBS, and were fixed with 4% paraformaldehyde for 15 min. The fixed colonies were then stained with Giemsa for 20 min, and washed twice with ddH2O. The colonies that consisted of ≥50 cells were manually counted.
Cell cycle analysis
Cells were harvested with 0.25% trypsin, fixed with 70% ethanol, and then resuspended in 20 mg/ml propidium iodide (PI). Flow cytometry (FCM) using a FACSCalibur™ flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) was used to detect DNA content, and the FCM data were used to determine the relative proportions of cells in the individual cell-cycle phases.
Apoptosis assay
In order to differentiate intact cells from apoptotic cells, the cells were stained with Annexin V-fluorescein isothiocyanate (FITC) and PI. Briefly, a total of 1.0×106 cells were washed twice with ice-cold PBS and put into binding buffer (5 µl Annexin V-FITC and 5 µl PI) for a 30 min incubation. Finally, FCM was used to analyze the Annexin V-FITC/PI staining.
Statistical analysis
All experiments were performed in triplicate, and data are shown as the mean ± standard deviation where applicable. Statistically significant differences between the control and treatment groups were determined by one-way analysis of variance followed by Tukey's test using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA), and P<0.05 was considered to indicate a statistically significant result.
Results
Successfully construction of LMP2A-RNAi-LV
A lentivirus vector system derived from HIV-1 was used to knock down the LMP2A gene by expressing short hairpin RNAs (shRNAs) directed against LMP2A. pGCSIL-LMP2A-shRNA-LV and pGCSIL-neg-shRNA-LV were constructed successfully, carrying GFP as a reporter gene. When GT38 cells were infected once with the constructed lentiviruses, >95% of the infected GT38 cells expressed GFP 72 h after the infection (Fig. 1), indicating that highly efficient and stable lentivirus vectors had been successfully constructed.
Figure 1.
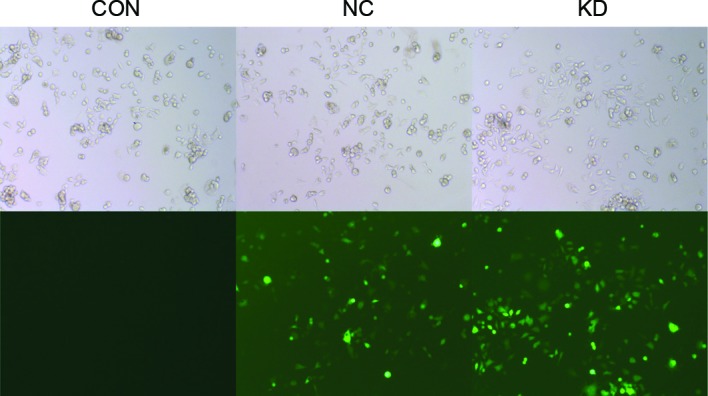
Lentiviral vectors encoding LMP2A RNAi or control RNAi were constructed and transfected into GT38 cells, with untreated GT38 cells serving as a blank control. The cells were observed using bright-field and fluorescence microscopy (magnification, ×100). LMP2A, latent membrane protein 2A; RNAi, RNA interference; CON, blank control; NC, negative control; KD, LMP2A knockdown.
LMP2A-RNAi-LV downregulates LMP2A expression in GT38 cells
The expression of LMP2A in GT38 cells was examined by RT-qPCR and western blot analysis following infection with LMP2A-specific RNAi-expressing lentivirus. Compared with the NC group, the mRNA and protein levels of LMP2A in the KD group were significantly reduced (P<0.05, n=6; Fig. 2). The mRNA level was decreased by 65.4%, while the protein level was reduced by 50.8%. The CON group exhibited no significant difference compared with the NC group. These data show that the expression of the LMP2A gene in GT38 cells was efficiently downregulated following the lentivirus infection.
Figure 2.

mRNA and protein levels of LMP2A in GT38 cells infected with LMP2A-RNAi-LV. (A) The mRNA level of of LMP2A was detected by RT-qPCR [the 2−ΔΔCt method (14) was applied to indicate the relative mRNA level of the NC and KD groups, compared with the CON group]. The mRNA levels of LMP2A in the KD group were significantly reduced compared with that of the NC group (*P<0.05). Data are presented as the mean plus standard deviation (n=6). (B) The protein level of LMP2A was assessed by western blotting. LMP2A, latent membrane protein 2A; RNAi, RNA interference; CON, blank control; NC, negative control; KD, LMP2A knockdown.
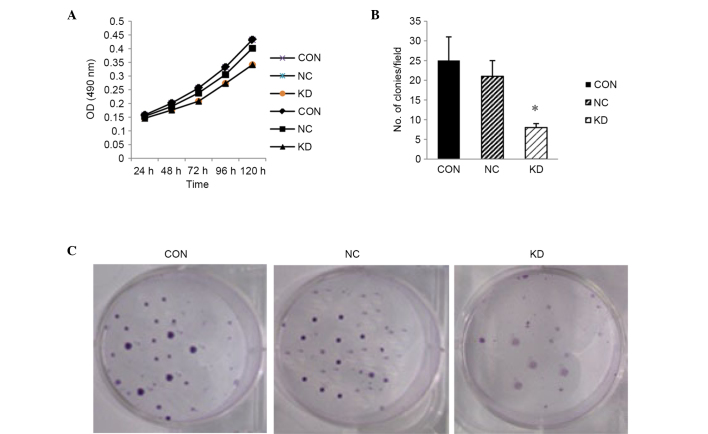
Knockdown of LMP2A inhibits the proliferation and clonogenicity of GT38 cells
To determine the effect of downregulation of LMP2A on cell growth in vitro, a cell proliferation assay was performed after LMP2A RNAi using the CCK-8 method. The results show that cell proliferation in the KD group was inhibited (Fig. 3A), compared with that in the CON and NC groups. A colony formation assay was then performed to confirm the inhibitory effect of LMP2A RNAi. The results show that when compared with the CON and NC groups, the colony formation ability of LMP2A RNAi cells was significantly decreased (P<0.05, n=6; Fig. 3B and C), suggesting that knockdown of the LMP2A gene results in the inhibition of cell growth.
Figure 3.
Suppression of cell growth in LMP2A RNAi-treated GT38 cells. (A) Cell proliferation was determined by CCK-8 assay. Cell proliferation in the KD group was inhibited at each time point (24, 48, 72, 96 and 120 h) compared with the CON and NC groups. (B) Colony formation assay was performed, and the cell proliferation inhibitory effect of LMP2A RNAi was confirmed (*P<0.05 vs. CON and NC), indicating that the clone number in the KD group was greatly reduced after LMP2A RNAi. Data are presented as the mean plus standard deviation (n=6). (C) Colonies of GT38 cells that were cultured in 6-cm dishes for 2 weeks and stained with Giemsa. LMP2A, latent membrane protein 2A; RNAi, RNA interference; CCK-8, Cell Counting kit-8; CON, blank control; NC, negative control; KD, LMP2A knockdown; OD, optical density.
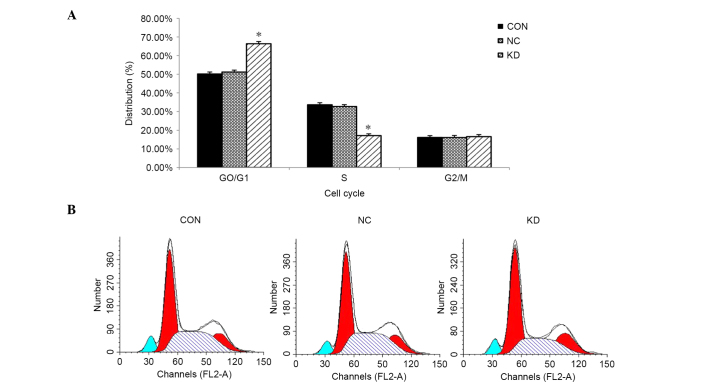
GT38 cells are arrested in the G0/G1 phase after knockdown of LMP2A
FCM analysis was conducted to evaluate whether downregulation of LMP2A affects the cell cycle of GT38 cells. The results show an increase in the proportion of cells in the G0/G1 phase in the KD group compared with the NC group (66.35±1.18 vs. 51.19±1.07%, respectively; P<0.05, n=6; Fig. 4). Compared with the control group, a greater proportion of LMP2A RNAi cells remained in the G0/G1 phase while fewer cells were in the S phase. The results indicate that downregulation of LMP2A expression in GT38 cells arrests their cell cycle in the G0/G1 phase.
Figure 4.
LMP2A RNAi treatment changed the cell cycle distribution of GT38 cells. (A) Proportion of cells in the G0/G1 phase significantly increased while the proportion in the S phase decreased in the KD group after treatment with LMP2A RNAi, compared with the CON and NC groups (*P<0.05). Data are presented as the mean plus standard deviation (n=6). (B) FCM analysis showed that LMP2A RNAi arrests GT38 cells in the G0/G1 phase of the cell cycle. LMP2A, latent membrane protein 2A; RNAi, RNA interference; FCM, flow cytometry; CON, blank control; NC, negative control; KD, LMP2A knockdown.
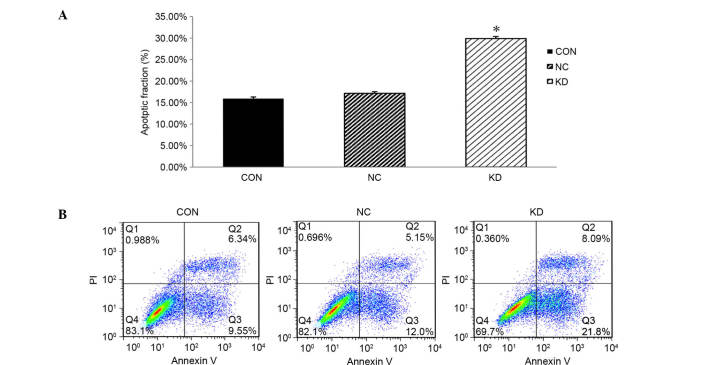
Knockdown of LMP2A induces apoptosis of GT38 cells
FCM analysis was applied to evaluate the rate of apoptosis, aiming to further investigate the effects of downregulation of LMP2A expression on the cell death of GT38 cells. Apoptosis rates in the CON and NC groups were 15.89±0.41 and 17.15±0.39%, respectively, while the apoptosis rate of the KD group was 29.89±0.48%; the percentage of apoptosis in the KD group was significantly increased compared with that in the CON and NC groups (P<0.05, n=6; Fig. 5). These data indicate that downregulation of LMP2A significantly increases the apoptosis rate of GT38 cells compared with that in the control group, demonstrating that suppression of cell proliferation may be caused by cell death resulting from LMP2A RNAi.
Figure 5.
LMP2A RNAi treatment induced GT38 cells into apoptosis which was detected by FCM using Annexin V-FITC/PI double staining. (A) Compared with the CON and NC groups, LMP2A RNAi significantly promoted cell apoptosis in the KD group (*P<0.05). Data are presented as the mean plus standard deviation (n=6). (B) FCM data show that the apoptotic rate of cells with downregulation of LMP2A is significantly increased. LMP2A, latent membrane protein 2A; RNAi, RNA interference; FCM, flow cytometry; FITC, fluorescein isothiocyanate; PI, propidium iodide; CON, blank control; NC, negative control; KD, LMP2A knockdown.
Discussion
LMP2A has been detected in the majority of EBVaGC samples (16). EBVaGC has been reported constitute 10–18% of gastric carcinoma cases and has a very poor survival outcome (17). LMP2A has been found to play important roles in epithelial cells, such as the enhancement of cell adhesion and cell motility, inhibition of epithelial cell differentiation and induction of anchorage-independent growth; these functions are reported to be correlated with the activation of the PI3K/Akt, NF-κB, β-catenin, STAT and Syk tyrosine kinase pathways (18–20). In addition, LMP2A is also reported to be involved in the regulation of the proliferation, apoptosis and self-renewal ability of cancer stem cells (21). Through the findings outlined above, LMP2A can be considered as a potential target of gene therapy for EBVaGC treatment. However, the detailed mechanism by which LMP2A expression downregulation inhibits EBVaGC cell growth has not been clarified.
In the present study, an LMP2A-specific RNAi-expressing lentivirus was constructed and used to infect the human gastric carcinoma cell line GT38. This was conducted to downregulate LMP2A expression, in order to investigate the effects of LMP2A silencing on cell growth, the cell cycle and cell apoptosis of EBVaGC cells. The results indicate that when LMP2A is downregulated, the proliferation of GT38 cells is significantly decreased, while the cell apoptosis is markedly increased, and a greater proportion of GT38 cells are arrested in the G0/G1 phase of the cell cycle. The inhibition of cell growth may be caused by cell apoptosis resulting from LMP2A RNAi. Although, the mechanisms by which downregulation of the LMP2A gene results in growth inhibition and apoptosis promotion in the GT38 cell line are not clear, there is evidence that when LMP2A is activated, gastric carcinoma cells would be protected from apoptosis and their proliferating ability promoted, all of which are regulated by a variety of signaling pathways (22–25) Thus, it is hypothesized that LMP2A gene silencing suppresses cell proliferation of the EBVaGC cell line GT38 following such mechanisms, and LMP2A can be considered as a potential therapeutic target in the treatment of EBVaGC. For further investigation, future studies may be conducted to attempt to clarify the mechanisms by which LMP2A modulate those pathways to affect cell biological functions, which may indicate new methods for creating novel therapies against EBVaGC.
RNAi has become a powerful tool for molecular biological research with efficient ability for gene functional analysis, and also can be considered as a potential therapeutic strategy for various diseases, including cancers (26). RNAi is considered to be a novel method for gene therapy against cancer, particularly when combined with lentivirus. The exploration of target genes that when downregulated could efficiently inhibit EBVaGC cell growth may increase the possibility of clinical application of gene-silencing therapy. During the present study, a highly efficient and stable lentivirus vector system was successfully constructed, which efficiently knocked down the LMP2A gene in the infected cell line GT38. The results also revealed that after LMP2A was knocked down by lentivirus-mediated RNAi, the cell proliferation and growth potency of EBVaGC cells were decreased significantly. Thus, a new strategy involving reduction of the expression of LMP2A by lentivirus-mediated RNAi is provided with the potential for treating human EBVaGC.
In conclusion, the results of the present study show that the knockdown of LMP2A by lentivirus-mediated RNAi significantly suppresses cell growth and induces apoptosis in the EBVaGC cell line GT38. This provides an attractive anticancer therapeutic strategy for the treatment of human EBVaGC.
Acknowledgements
This study was supported by the Key Scientific Research Program of Wuxi Municipal Health Bureau (grant no. Z201509), the Natural Science Foundation of Jiangsu Province (cat. no. BK20161152), the National Natural Science Foundation of China (grant nos. 31670857, 81672975, 81522039 and 31400720) and the Key Research and Development Program of China (grant no. 2016YFC0904702).
References
- 1.Kieff E. Epstein-Barr virus and its replication. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. 3rd. Lippincott-Raven; Philadelphia, PA: 1996. pp. 2343–2396. [Google Scholar]
- 2.Shah KM, Young LS. Epstein-Barr virus and carcinogenesis: Beyond Burkitt's lymphoma. Clin Microbiol Infect. 2009;15:982–988. doi: 10.1111/j.1469-0691.2009.03033.x. [DOI] [PubMed] [Google Scholar]
- 3.Shin JY, Kim JO, Lee SK, Chae HS, Kang JH. LY294002 may overcome 5-FU resistance via down-regulation of activated p-AKT in Epstein-Barr virus-positive gastric cancer cells. Bmc Cancer. 2010;10:425. doi: 10.1186/1471-2407-10-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takada K. Epstein-Barr virus and gastric carcinoma. Mol Pathol. 2000;53:255–261. doi: 10.1136/mp.53.5.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seo JS, Jun SM, Kwon SW, Oh IH, Kim TG, Lee SK. Establishment and characterization of gastric carcinoma cell clones expressing LMP2A of Epstein-Barr virus. Int N Mol Med. 2010;25:11–16. [PubMed] [Google Scholar]
- 6.Iizasa H, Nanbo A, Nishikawa J, Jinushi M, Yoshiyama H. Epstein-Barr virus (EBV)-associated gastric carcinoma. Viruses. 2012;4:3420–3439. doi: 10.3390/v4123420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukuda M, Longnecker R. Epstein-Barr virus latent membrane protein 2A mediates transformation through constitutive activation of the Ras/PI3-K/Akt Pathway. J Virol. 2007;81:9299–9306. doi: 10.1128/JVI.00537-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J, Lin WH, Chen SY, Longnecker R, Tsai SC, Chen CL, Tsai CH. Syk tyrosine kinase mediates Epstein-Barr virus latent membrane protein 2A-induced cell migration in epithelial cells. J Biol Chem. 2006;281:8806–8814. doi: 10.1074/jbc.M507305200. [DOI] [PubMed] [Google Scholar]
- 9.Morrison JA, Raab-Traub N. Roles of the ITAM and PY motifs of Epstein-Barr virus latent membrane protein 2A in the inhibition of epithelial cell differentiation and activation of {beta}-catenin signaling. J Virol. 2005;79:2375–2382. doi: 10.1128/JVI.79.4.2375-2382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan YR, Vatsyayan J, Chang YS, Chang HY. Epstein-Barr virus latent membrane protein 2A upregulates UDP-glucose dehydrogenase gene expression via ERK and PI3K/Akt pathway. Cell Microbiol. 2008;10:2447–2460. doi: 10.1111/j.1462-5822.2008.01221.x. [DOI] [PubMed] [Google Scholar]
- 11.Allen MD, Young LS, Dawson CW. The Epstein-Barr virus-encoded LMP2A and LMP2B proteins promote epithelial cell spreading and motility. J Virol. 2005;79:1789–1802. doi: 10.1128/JVI.79.3.1789-1802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fotheringham JA, Coalson NE, Raab-Traub N. Epstein-Barr virus latent membrane protein-2A induces ITAM/Syk- and Akt-dependent epithelial migration through αV-integrin membrane translocation. J Virol. 2012;86:10308–10320. doi: 10.1128/JVI.00853-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 14.O'Rourke JP, Newbound GC, Kohn DB, Olsen JC, Bunnell BA. Comparison of gene transfer efficiencies and gene expression levels achieved with equine infectious anemia virus- and human immunodeficiency virus type 1-derived lentivirus vectors. J Virol. 2002;76:1510–1515. doi: 10.1128/JVI.76.3.1510-1515.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Han J, Chen JN, Zhang ZG, Li HG, Ding YG, Du H, Shao CK. Sequence variations of latent membrane protein 2A in Epstein-Barr virus-associated gastric carcinomas from Guangzhou, southern China. PloS One. 2012;7:e34276. doi: 10.1371/journal.pone.0034276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukayama M, Ushiku T. Epstein-Barr virus-associated gastric carcinoma. Pathol Res Pract. 2011;207:529–537. doi: 10.1016/j.prp.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda M, Longnecker R. Epstein-Barr virus latent membrane protein 2A mediates transformation through constitutive activation of the Ras/P13-K/Akt pathway. J Virol. 2007;81:9299–9306. doi: 10.1128/JVI.00537-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scholle F, Bendt KM, Raab-Traub N. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation and activates Akt. J Virol. 2000;74:10681–10689. doi: 10.1128/JVI.74.22.10681-10689.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart S, Dawson CW, Takada K, Curnow J, Moody CA, Sixbey JW, Young LS. Epstein-Barr virus-encoded LMP2A regulates viral and cellular gene expression by modulation of the NF-kappaB transcription factor pathway. Proc Natl Acad Sci USA. 2004;101:15730–15735. doi: 10.1073/pnas.0402135101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong QL, Hu LJ, Cao JY, Huang YJ, Xu LH, Liang Y, Xiong D, Guan S, Guo BH, Mai HQ, et al. Epstein-Barr virus-encoded LMP2A induces an epithelial-mesenchymal transition and increases the number of side population stem-like cancer cells in nasopharyngeal carcinoma. PloS Pathog. 2010;6:e1000940. doi: 10.1371/journal.ppat.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan YY, Hsiao JR, Chang KC, Chang JS, Chen CW, Lai HC, Wu SY, Yeh TH, Chang FH, Lin WH, et al. Epstein-Barr virus latent membrane protein 2A promotes invasion of nasopharyngeal carcinoma cells through ERK/Fra-1-mediated induction of matrix metalloproteinase 9. J Virol. 2012;86:6656–6667. doi: 10.1128/JVI.00174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Portis T, Longnecker R. Epstein-Barr virus (EBV) LMP2A mediates B-lymphocyte survival through constitutive activation of the Ras/PI3K/Akt pathway. Oncogene. 2004;23:8619–8628. doi: 10.1038/sj.onc.1207905. [DOI] [PubMed] [Google Scholar]
- 24.Pal AD, Basak NP, Banerjee AS, Banerjee S. Epstein-Barr virus latent membrane protein-2A alters mitochondrial dynamics promoting cellular migration mediated by Notch signaling pathway. Carcinogenesis. 2014;35:1592–1601. doi: 10.1093/carcin/bgu069. [DOI] [PubMed] [Google Scholar]
- 25.Pang MF, Lin KW, Peh SC. The signaling pathways of Epstein-Barr virus-encoded latent membrane protein 2A (LMP2A) in latency and cancer. Cell Mol Biol Lett. 2009;14:222–247. doi: 10.2478/s11658-008-0045-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izquierdo M. Short interfering RNAs as a tool for cancer gene therapy. Cancer Gene Ther. 2005;12:217–227. doi: 10.1038/sj.cgt.7700791. [DOI] [PubMed] [Google Scholar]