Abstract
microRNAs (miRNAs) are a class of small RNAs that regulate gene expression. It has been demonstrated that aberrant miRNA expression is associated with cancer development and carcinogenesis. Altered miRNA expression has been suggested to occur in bladder cancer. In other cancer systems, studies have indicated that miR-143, as a tumor suppressor gene, plays essential roles in cancer progression. However, its role in bladder cancer has yet to be elucidated. In the present study, we observed that miR-143 expression was downregulated in human bladder cancer tissues and cells, and that its levels were negatively correlated with bladder cancer clinical stages. We further demonstrated that insulin-like growth factor-1 receptor (IGF-1R) is a functional target of miR-143. Their expression levels were inversely correlated in bladder cancer samples. Overexpression of miR-143 inhibited cell proliferation and promoted chemosensitivity of bladder cancer 5637 cells to gemcitabine. Consistently, small interfering RNA-mediated knockdown of IGF-1R phenocopied miR-143 overexpression. Notably, the expression of IGF-1R is a predictor of patient prognosis. Collectively, our findings indicate that miR-143 is a valuable biomarker for bladder cancer. The miR-143/IGF-1R axis is associated with bladder cancer drug resistance and patient survival.
Keywords: miR-143, bladder cancer, gemcitabine, IGF-1R, tumor growth
Introduction
Bladder cancer is the ninth most common malignancy in the world. In the US, ~15,000 people succumb to this disease every year (1). As a common malignancy, bladder cancer often exhibits the characteristics of frequent recurrence and an undesirable clinical outcome when tumors progress to invasive disease (2). Approximately 90% of bladder cancer cases are urothelial carcinomas, previously named transitional cell carcinomas. The majority of bladder cancers are low-grade, non-muscle-invasive cancers, which account for up to 70–80% of cases, and which seldom progress to the muscle-invasive stage (3,4). However, 10–20% of patients are diagnosed with high-grade and muscle-invasive tumors, exhibiting aggressive clinical behavior (3). Although limited treatment options are available, 31–78% of patients still suffer from disease recurrence within 5 years. In addition, ~20% of the recurring tumors have an increased degree of malignancy (5). Accumulating evidence suggests that genetic and epigenetic factors contribute to uncontrolled cancer cell proliferation and invasion, therefore mediating tumor cell survival, treatment resistance and metastatic spread. Immunotherapy and chemotherapy are commonly used to temporary restrain the recurrence; however, eventually the tumor progresses (6). Methods of targeting these chemotherapy-resistant cancer cells are now emerging as a new and critical focus in the field.
miRNAs are a type of 21- to 25-nucleotide non-coding RNAs which regulate gene expression at the level of mRNA stability and translation (7). miRNAs are post-transcriptional regulators that bind to the 3′-untranslated regions (3′-UTRs) of target mRNAs, usually leading to translational repression and gene silencing (8). It is well established that a number of miRNAs are highly tissue-specific and play significant roles in multiple physiological processes, including cell differentiation and tissue development. Therefore, aberrant expression of miRNA is involved in cellular dedifferentiation, oncogenesis, tumor invasion and metastasis (9). Identification of the targets of miRNAs would help us to understand the function of miRNAs in tumor development and progression. Among them, miR-143 is of particular interest due to its constant downregulation in a wide range of cancer cell lines and tumors, including bladder cancer (10). However, the mechanism of miR-143 action remains largely unknown.
Insulin-like growth factor-1 receptor (IGF-1R) and its ligand play an essential role in regulating cellular proliferation and apoptosis (11). Upregulated expression of IGF-1R has been documented in numerous malignancies, including lung, breast, liver and thyroid cancer (12). Ligand binding to IGF-1R triggers various downstream signaling pathways, including the PI3K/Akt pathway, which is essential for cell survival (13,14). In colorectal cancer, overexpression of miR-143 inhibits cell proliferation, migration and tumor growth and increases chemosensitivity to oxaliplatin treatment in an IGF-1R-dependent manner, suggesting that IGF-1R is a functional target of miR-143 (15). Moreover, miR-143 levels in human blood and tumor tissues are associated with cancer occurrence, metastasis and drug resistance (15), indicating the potential of miR-143 as a biomarker. The role of IGF-1R in cancerous transformation of urothelial cells is not well established, but previous studies have demonstrated that IGF-1R is overexpressed in bladder cancer (16). Given the interrelationship between IGF-1R and miR-143, we speculate that downregulation of miR-143 in bladder cancer may be involved in tumor development via the activation of IGF-1R and other downstream pathways (e.g. PI3K/Akt and MAPK). To confirm this hypothesis, we examined the levels of miR-143 and IGF-1R levels in benign bladder and cancer tissues, as well as cancer cell lines. We also performed a functional study to reveal the role of miR-143 and IGF-1R in bladder cancer cell growth and chemoresistance. Finally, a large bladder cancer data set from The Cancer Genome Atlas (TCGA) was utilized to correlate the expression of IGF-1R to clinical traits.
Materials and methods
Reagents and cell culture
Two human bladder cancer cell lines (T24 and 5637) and an immortalized cell line (SV-HUC-1) from human urinary bladder urothelium were obtained from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cell lines were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 mg/l streptomycin in a humidified incubator with 5% CO2 at 37°C.
Human specimens
Human bladder cancer tissues (n=23) and their matched normal adjacent tissues (n=23) were obtained from patients at the Huai'an First People's Hospital, Jiangsu, China. The study was approved by the Academic and Ethics Advisory Board of Nanjing Medical University with documented patient consent.
Lentivirus infection and transient transfections
High-titer stocks of lentivirus carrying miR-143 or scramble control (miR-SCR) were purchased from GenePharma (Shanghai, China). The lentiviruses were used to infect the T24 and 5637 cells. Infected cells were selected by puromycin (Sigma-Aldrich, St. Louis, MO, USA), and then stable T24 and 5637 cell lines expressing miR-143 and miR-SCR were established. Small interfering RNA (siRNA) duplex oligonucleotides targeting IGF-1R or scramble control were synthesized by GenePharma. T24 cells were transfected with either siIGF-IR or siSCR using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNAs were extracted from cultured cells or human tissues using TRIzol reagent (Invitrogen Life Technologies) according to the manufacturer's instructions. To quantify the mRNA levels of IGF-1R and GAPDH, RNAs were transcribed using a PrimeScript RT reagent kit and oligo dT primer (Takara Biotechnology Co., Ltd., Dalian, China). To measure miR-143 and U6 expression levels, RNAs were transcribed using stem-loop RT primer and the PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.) as previously described (17,18). RT-qPCR was performed using SYBR Premix DimerEraser (Takara Biotechnology Co., Ltd.). Primers were purchased from RiboBio (Shenzhen, China).
Antibodies and western blot analysis
Tissues or cells were harvested and lysed on ice for 30 min in RIPA buffer (Beyotime Institute of Biotechnology, Haimen, China) supplemented with 1 mM phenylmethylsulfonyl fluoride. Lysates were subjected to western blot assay as described previously (19). Antibodies against IGF-1R, p-Akt (Ser-473), Akt, p-ERK (Thr202/Tyr204), ERK (Cell Signaling Technology, Inc., Danvers, MA, USA) and GAPDH (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) were used to check the protein expression.
Cell proliferation and chemosensitivity assays
A total of 4,000 cells per well were plated in 96-well plates, and cultured at 37°C in an incubator with 5% CO2. Cell proliferation was measured at 24, 48, 72 and 96 h using a Cell counting kit 8 (CCK8; Dojindo Molecular Technologies, Inc., Tokyo, Japan) according to the manufacturer's instructions. Results were obtained from three separate experiments with six replications per experiment, and presented as the means ± standard deviation (SD). For chemosensitivity assay, freshly prepared gemcitabine (Sigma-Aldrich) was added at varying concentrations (ranging from 0.64 to 10,000 nM). Cell viability was determined 72 h later by CCK8 kit.
Statistical analysis
Results were mainly presented as mean ± SD, and Graphpad Prism software (GraphPad Software, Inc., La Jolla, CA, USA) was used to calculate the mean and standard deviation. The statistical differences within results among the groups or treatments were analyzed using an unpaired Student's t-test or Pearson correlation. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-143 is downregulated in bladder cancer tissues and cell lines
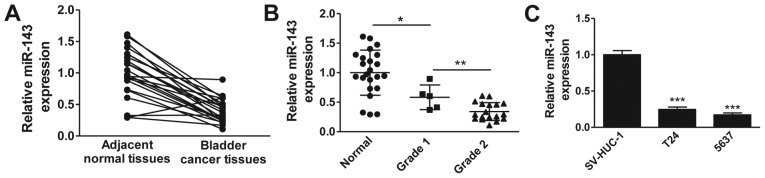
Accumulating evidence suggests a downregulation of miR-143 in multiple cancers. To determine whether miR-143 is downregulated in clinical bladder cancer tissues, we examined the miR-143 expression levels in 20 pairs of bladder cancer tissues and matched adjacent normal tissues by RT-qPCR. As shown in Fig. 1A, the expression levels of miR-143 in bladder cancer tissues were significantly lower than those in adjacent normal tissues. We further analyzed the miR-143 expression levels according to the pathological features of bladder cancer patients. Consistently, miR-143 expression was notably decreased in bladder cancer tissues compared with normal tissues, and the expression of miR-143 was negatively correlated with the level of malignancy, with grade 2 demonstrating the lowest expression (Fig. 1B). We next evaluated the expression levels of miR-143 in widely used bladder cancer cell lines. The result clearly demonstrated that miR-143 expression was markedly downregulated in bladder cancer cells (T24 and 5637) compared with normal uroepithelial cells (SV-HUC-1) (Fig. 1C). Collectively, these data confirm the downregulation of miR-143 in clinical bladder cancer tissues and cancer cell lines.
Figure 1.
Expression of miR-143 in human benign tissues, bladder cancer tissues and cancer cell lines. (A) Reverse transcription-quantitative polymerase chain reaction analysis of miR-143 levels in bladder cancer tissues and adjacent normal tissues from cystectomy patients. (B) Expression of miR-143 in normal bladder tissues, and grade 1 and 2 cancer tissues (*P<0.05 and **P<0.01). (C) miR-143 levels in normal bladder and cancer cell lines. ***P<0.001 vs. SV-HUC-1.
miR-143 targets IGF-1R
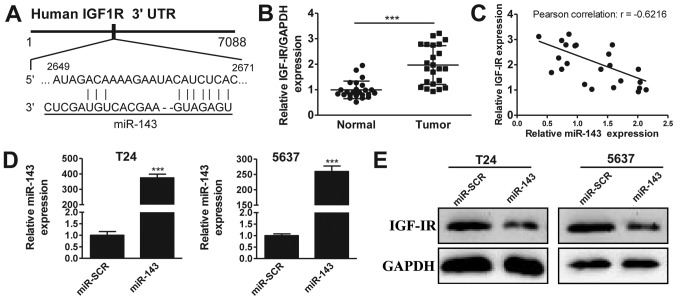
It has previously been suggested that IGF-1R may be a target of miR-143 (20). To investigate this further in bladder cancer, we first performed in silico analysis using TargetScan, observing that miR-143 potentially targets a region of the 3′-UTR sequence of the IGF-1R gene (Fig. 2A). Next, we analyzed the expression of IGF-1R in 20 pairs of bladder cancer and matched adjacent normal tissues. The RT-qPCR results revealed that IGF-1R was overexpressed in bladder cancer tissues compared with normal tissues (Fig. 2B), a pattern opposite to that of miR-143. This suggested that IGF-1R may be a target of miR-143. To strengthen this hypothesis, we plotted the expression data from Figs. 1A and 2B, and observed that the expression levels of miR-143 and IGF-1R were reversely correlated (Fig. 2C, Pearson's correlation scatter plots: R=−0.6216, P<0.05). To confirm that miR-143 targets IGF-1R, we performed overexpression experiments. As shown in Fig. 2C, transfection of miRNA mimics successfully induced the upregulation of miR-143 in T24 (395-fold higher) and 5637 (250-fold higher) cancer cell lines, compared with the scrambled negative control (miR-SCR) group. Significantly, IGF-1R protein levels were markedly decreased by miR-143 overexpression in the two cell lines (Fig. 2D). Together, our data suggest that miR-143 inhibits IGF-IR expression in bladder cancer.
Figure 2.
Insulin-like growth factor-1 receptor (IGF-1R) is a target of miR-143. (A) Schematic of the putative miR-143 binding site in the IGF-1R 3′-untranslated region (3′ UTR). (B) The mRNA levels of IGF-IR in 20 matched pairs of bladder cancer and adjacent normal tissues. (C) Correlation of miR-143 and IGF-IR expression in bladder cancer tissues. (D) Lentiviral-mediated miR-143 overexpression in T24 and 5637 cells. (E) Effect of miR-143 overexpression on IGF-1R levels in T24 and 5637 cells using western blot analysis. ***P<0.001.
Overexpression of miR-143 and knockdown of IGF-1R enhance chemosensitivity to gemcitabine
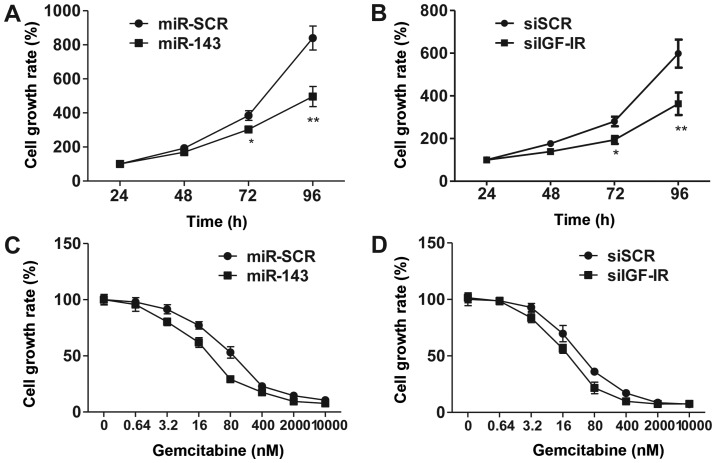
Several studies have suggested that miR-143 functions as a tumor suppressor (21–23). To investigate the effects of overexpression of miR-143, we utilized CCK-8 assay to demonstrate that forced miR-143 expression inhibited 5673 cell proliferation (Fig. 3A). Consistently, knockdown of IGF-1R by siRNA phenocopied miR-143 overexpression in regulating cell proliferation. These results suggest that miR-143 exhibits its function via the inhibition of IGF-1R signaling.
Figure 3.
Improvement of chemosensitivity to gemcitabine by upregulation of miR-143 and knockdown of insulin-like growth factor-1 receptor (IGF-1R). (A and B) Effect of lentiviral-mediated miR-143 overexpression and siRNA-mediated downregulation of IGF-1R on bladder cancer cell proliferation. (C and D) Effect of miR-143 overexpression and knockdown of IGF-1R on cancer cell sensitivity to gemcitabine. *P<0.05, **P<0.01.
IGF-1R is overexpressed in bladder cancer cells, and IGF signaling plays a significant role in promoting cell survival and proliferation. To explore the contribution of IGF-1R and miR-143 in chemotherapy, we treated 5637 cells with various concentrations of gemcitabine, a leading chemotherapy drug used in the treatment of bladder cancer. The results revealed that overexpression of miR-143 and siRNA-mediated knockdown of IGF-1R significantly increased cell sensitivity to gemcitabine (Fig. 3C and D). These data suggested that miR-143 enhances chemosensitivity to gemcitabine through, at least partially, downregulation of IGF-1R.
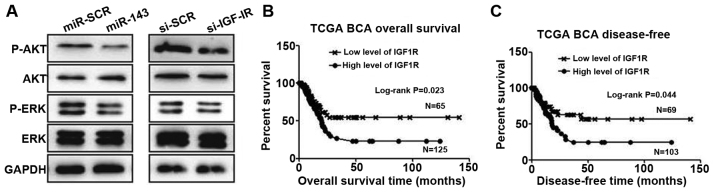
miR-143/IGF-1R axis regulates Akt and extracellular signal-regulated kinase (ERK) pathway activation in 5637 cells
To assess the involvement of downstream signaling molecules, we transfected 5637 cells with miR-143 or negative control and examined Akt and ERK pathway activation. Overexpression of miR-143 significantly inhibited p-Akt and p-ERK levels in 5637 cells (Fig. 4A). Similarly, knockdown of IGF-1R also reduced the expression of these molecules (Fig. 4A). To put these findings into clinical relevance, we speculate that downregulation of miR-134 would benefit cancer cell growth through activation of the IGF-1R, Akt and ERK pathways.
Figure 4.
miR-143/insulin-like growth factor-1 receptor (IGF-1R) axis regulates Akt and extracellular signal-regulated kinase (ERK) signaling pathways in cancer cells and patient survival rate. (A) The levels of p-Akt, Akt, p-ERK and ERK in 5637 cells transfected with miR-143 and siRNA targeting IGF-1R. (B-C) Meta-analysis revealed that higher expression of IGF-1R negatively correlates with bladder cancer (BCA) patient overall survival (B) and disease-free survival. RNA-seq bladder cancer data from The Cancer Genome Atlas (TCGA) was used, and the log-rank test was used for statistical analysis.
Expression of IGF-1R negatively correlates with patient survival
A previous study (24) and our data suggest that IGF-1R is upregulated in bladder cancer cells. To investigate the possibility of IGF-1R as a biomarker for prognosis, we employed the largest TCGA bladder cancer database. After extracting and plotting IGF-1R expression against patient clinical data, we observed that IGF-1R is a suitable marker for predicting patient overall survival, with higher expression demonstrating a short survival time (Fig. 4B). Significantly, IGF-1R levels also predict tumor recurrence (Fig. 4C).
Discussion
It is well known that miRNAs are involved in diverse processes, including apoptosis, proliferation, differentiation and chemosensitivity. Thus, a number of miRNAs have been linked to distinct development defects or cancers (25,26). Previous studies have revealed low miR-143 expression in various types of tumors (27,28). In the present study, we observed decreased expression of miR-143 in bladder cancer tissues and cell lines compared with normal tissues, consistent with the findings of Noguchi et al (28). We observed a negative correlation between miR-143 and IGF-1R mRNA expression in bladder cancer tissues and cell lines, suggesting that the tumor-suppressive role of miR-143 may be attributed to its suppression of IGF-1R. Subsequently, we identified IGF-1R as a functional target for miR-143. Furthermore, chemosensitivity to gemcitabine was improved by upregulating miR-143 and knocking down IGF-1R in 5637 cells.
Notably, miR-143 has been suggested as a biomarker for various types of cancer (29,30). Qian et al (15) indicated that low levels of miR-143 in colorectal cancer tissues were closely associated with cancer stage and metastasis. Consistently, the results of the present study revealed a decreasing expression of miR-143 with the increasing bladder cancer grade, suggesting that miR-143 has a tumor suppressive role in bladder cancer progression.
IGF-1R has been identified as a key regulator of tumor development by regulating cell proliferation, differentiation and survival (31,32). Upregulated expression of IGF-1R has been observed in numerous human malignancies (12). Previous studies have revealed that IGF-1R is also upregulated in human invasive bladder cancer and promotes cell migration and invasion (33). Based on the signaling pathway analysis results, we speculated that miR-143 exerts a tumor suppressor function through the inhibition of IGF-1R, consistent with previous studies demonstrating that miR-143 directly targets the 3′-UTR region to suppress IGF-1R expression (20). Aberrant expression of IGF-1R affects a number of downstream signaling cascades, including the PI3K/Akt and MAPK/ERK pathways (34,35), which are frequently activated in cancer cells. These pathways play critical roles in tumor growth and progression (36,37). The present study indicated that overexpression of miR-143 significantly inhibits Akt and ERK activity, as evidenced by the decreased levels of p-Akt and p-ERK. Consequently, forced expression of miR-143 enhanced the chemosensitivity of 5637 cells to gemcitabine in vitro. To further strengthen the clinical relevance of our study, we analyzed TCGA bladder cancer data, observing that expression of IGF-1R stratifies patients with higher levels of IGF-1R, predicting a poorer patient survival time.
In conclusion, our study demonstrated that miR-143 inhibits bladder cancer cell proliferation and improves chemosensitivity to gemcitabine, at least in part through the inactivation of IGF-1R and downstream pathways in vitro. The expression of IGF-1R negatively correlates with a beneficial clinical outcome of patients. The findings of the present study provide new information to support the possible usage of miR-143/IGF-1R-based therapeutic strategies to treat bladder cancer patients in the future.
Glossary
Abbreviations
- miRNA
microRNA
- miR-143
microRNA-143
- IGF-1R
insulin-like growth factor-1 receptor
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- UTR
untranslated region
- mRNA
messenger RNA
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Dinney CP, McConkey DJ, Millikan RE, Wu X, Bar-Eli M, Adam L, Kamat AM, Siefker-Radtke AO, Tuziak T, Sabichi AL, et al. Focus on bladder cancer. Cancer Cell. 2004;6:111–116. doi: 10.1016/j.ccr.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Jebar AH, Hurst CD, Tomlinson DC, Johnston C, Taylor CF, Knowles MA. FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene. 2005;24:5218–5225. doi: 10.1038/sj.onc.1208705. [DOI] [PubMed] [Google Scholar]
- 4.McConkey DJ, Lee S, Choi W, Tran M, Majewski T, Lee S, Siefker-Radtke A, Dinney C, Czerniak B. Molecular genetics of bladder cancer: emerging mechanisms of tumor initiation and progression. Urol Oncol. 2010;28:429–440. doi: 10.1016/j.urolonc.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babjuk M, Oosterlinck W, Sylvester R, Kaasinen E, Böhle A, Palou-Redorta J. European Association of Urology (EAU): EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder. Eur Urol. 2008;54:303–314. doi: 10.1016/j.eururo.2008.04.051. [DOI] [PubMed] [Google Scholar]
- 6.Lamm DL, Riggs DR, Traynelis CL, Nseyo UO. Apparent failure of current intravesical chemotherapy prophylaxis to influence the long-term course of superficial transitional cell carcinoma of the bladder. J Urol. 1995;153:1444–1450. doi: 10.1016/S0022-5347(01)67427-5. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 10.Dyrskjøt L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL, Andersen CL, Zieger K, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 11.Rubin R, Baserga R. Insulin-like growth factor-I receptor. Its role in cell proliferation, apoptosis, and tumorigenicity. Lab Invest. 1995;73:311–331. [PubMed] [Google Scholar]
- 12.Macaulay VM. Insulin-like growth factors and cancer. Br J Cancer. 1992;65:311–320. doi: 10.1038/bjc.1992.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 14.Baserga R. Controlling IGF-receptor function: a possible strategy for tumor therapy. Trends Biotechnol. 1996;14:150–152. doi: 10.1016/0167-7799(96)10023-8. [DOI] [PubMed] [Google Scholar]
- 15.Qian X, Yu J, Yin Y, He J, Wang L, Li Q, Zhang LQ, Li CY, Shi ZM, Xu Q, et al. MicroRNA-143 inhibits tumor growth and angiogenesis and sensitizes chemosensitivity to oxaliplatin in colorectal cancers. Cell Cycle. 2013;12:1385–1394. doi: 10.4161/cc.24477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rochester MA, Patel N, Turney BW, Davies DR, Roberts IS, Crew J, Protheroe A, Macaulay VM. The type 1 insulin-like growth factor receptor is over-expressed in bladder cancer. BJU Int. 2007;100:1396–1401. doi: 10.1111/j.1464-410X.2007.06931.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X. A PCR-based platform for microRNA expression profiling studies. RNA. 2009;15:716–723. doi: 10.1261/rna.1460509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jing Y, Liu LZ, Jiang Y, Zhu Y, Guo NL, Barnett J, Rojanasakul Y, Agani F, Jiang BH. Cadmium increases HIF-1 and VEGF expression through ROS, ERK, and AKT signaling pathways and induces malignant transformation of human bronchial epithelial cells. Toxicol Sci. 2012;125:10–19. doi: 10.1093/toxsci/kfr256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He J, Qian X, Carpenter R, Xu Q, Wang L, Qi Y, Wang ZX, Liu LZ, Jiang BH. Repression of miR-143 mediates Cr (VI)-induced tumor angiogenesis via IGF-IR/IRS1/ERK/IL-8 pathway. Toxicol Sci. 2013;134:26–38. doi: 10.1093/toxsci/kft101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang R, Xiao T, Fang Z, Sun Y, Li F, Gao Y, Feng Y, Li L, Wang Y, Liu X, et al. MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. J Biol Chem. 2012;287:23227–23235. doi: 10.1074/jbc.M112.373084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregersen LH, Jacobsen A, Frankel LB, Wen J, Krogh A, Lund AH. MicroRNA-143 down-regulates Hexokinase 2 in colon cancer cells. BMC Cancer. 2012;12:232. doi: 10.1186/1471-2407-12-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent OA, Fox-Talbot K, Halushka MK. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene. 2013;32:2576–2585. doi: 10.1038/onc.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metalli D, Lovat F, Tripodi F, Genua M, Xu SQ, Spinelli M, Alberghina L, Vanoni M, Baffa R, Gomella LG, et al. The insulin-like growth factor receptor I promotes motility and invasion of bladder cancer cells through Akt- and mitogen-activated protein kinase-dependent activation of paxillin. Am J Pathol. 2010;176:2997–3006. doi: 10.2353/ajpath.2010.090904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekar D, Islam VI Hairul, Thirugnanasambantham K, Saravanan S. Relevance of miR-21 in HIV and non-HIV-related lymphomas. Tumour Biol. 2014;35:8387–8393. doi: 10.1007/s13277-014-2068-9. [DOI] [PubMed] [Google Scholar]
- 26.Iida K, Fukushi J, Matsumoto Y, Oda Y, Takahashi Y, Fujiwara T, Fujiwara-Okada Y, Hatano M, Nabashima A, Kamura S, Iwamoto Y. miR-125b develops chemoresistance in Ewing sarcoma/primitive neuroectodermal tumor. Cancer Cell Int. 2013;13:21. doi: 10.1186/1475-2867-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han Y, Chen J, Zhao X, Liang C, Wang Y, Sun L, Jiang Z, Zhang Z, Yang R, Chen J, et al. MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS One. 2011;6:e18286. doi: 10.1371/journal.pone.0018286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noguchi S, Mori T, Hoshino Y, Maruo K, Yamada N, Kitade Y, Naoe T, Akao Y. MicroRNA-143 functions as a tumor suppressor in human bladder cancer T24 cells. Cancer Lett. 2011;307:211–220. doi: 10.1016/j.canlet.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Ak S, Tunca B, Tezcan G, Cecener G, Egeli U, Yilmazlar T, Ozturk E, Yerci O. MicroRNA expression patterns of tumors in early-onset colorectal cancer patients. J Surg Res. 2014;191:113–122. doi: 10.1016/j.jss.2014.03.057. [DOI] [PubMed] [Google Scholar]
- 30.Ng EK, Li R, Shin VY, Siu JM, Ma ES, Kwong A. MicroRNA-143 is downregulated in breast cancer and regulates DNA methyltransferases 3A in breast cancer cells. Tumour Biol. 2014;35:2591–2598. doi: 10.1007/s13277-013-1341-7. [DOI] [PubMed] [Google Scholar]
- 31.Hart LS, Dolloff NG, Dicker DT, Koumenis C, Christensen JG, Grimberg A, El-Deiry WS. Human colon cancer stem cells are enriched by insulin-like growth factor-1 and are sensitive to figitumumab. Cell Cycle. 2011;10:2331–2338. doi: 10.4161/cc.10.14.16418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao D, Bakirtzi K, Zhan Y, Zeng H, Koon HW, Pothoulakis C. Insulin-like growth factor-1 receptor transactivation modulates the inflammatory and proliferative responses of neurotensin in human colonic epithelial cells. J Biol Chem. 2011;286:6092–6099. doi: 10.1074/jbc.M110.192534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Z, Xu T, Wang L, Wang X, Zhong S, Xu C, Shen Z. MicroRNA-145 directly targets the insulin-like growth factor receptor I in human bladder cancer cells. FEBS Lett. 2014;588:3180–3185. doi: 10.1016/j.febslet.2014.06.059. [DOI] [PubMed] [Google Scholar]
- 34.Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 35.Park BH, Davidson NE. PI3 kinase activation and response to trastuzumab therapy: what's neu with herceptin resistance? Cancer Cell. 2007;12:297–299. doi: 10.1016/j.ccr.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Vivanco I, Sawyers CL. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 37.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]