Abstract
Lasso peptides are a class of bioactive ribosomally synthesized and post-translationally modified peptides (RiPPs), with a threaded knot structure that is formed by an isopeptide bond attaching the N-terminus of the peptide to a side chain carboxylate. Some lasso peptide biosynthetic clusters harbor an enzyme that specifically hydrolyzes the isopeptide bond to yield the linear peptide. We describe here the 2.4 Å resolution structure of a lasso peptide isopeptidase revealing a topologically novel didomain architecture consisting of an open β-propeller appended to an α/β hydrolase domain. The 2.2 Å resolution cocrystal structure of an inactive variant in complex with a lasso peptide reveals deformation of the substrate, and reorganization of the enzyme active site, which exposes and orients the isopeptide bond for hydrolysis. Structure-based mutational analysis reveals how this enzyme recognizes the lasso peptide substrate by shape complementarity rather than through sequence specificity. The isopeptidase gene can be used to facilitate genome mining, as a network-based mining strategy queried with this sequence identified 87 putative lasso peptide biosynthetic clusters, 65 of which have not been previously described. Lastly, we validate this mining approach by heterologous expression of two clusters encoded within the genome of Asticcaucalis benevestitus, and demonstrate that both clusters produce lasso peptides.
Graphical Abstract
Introduction
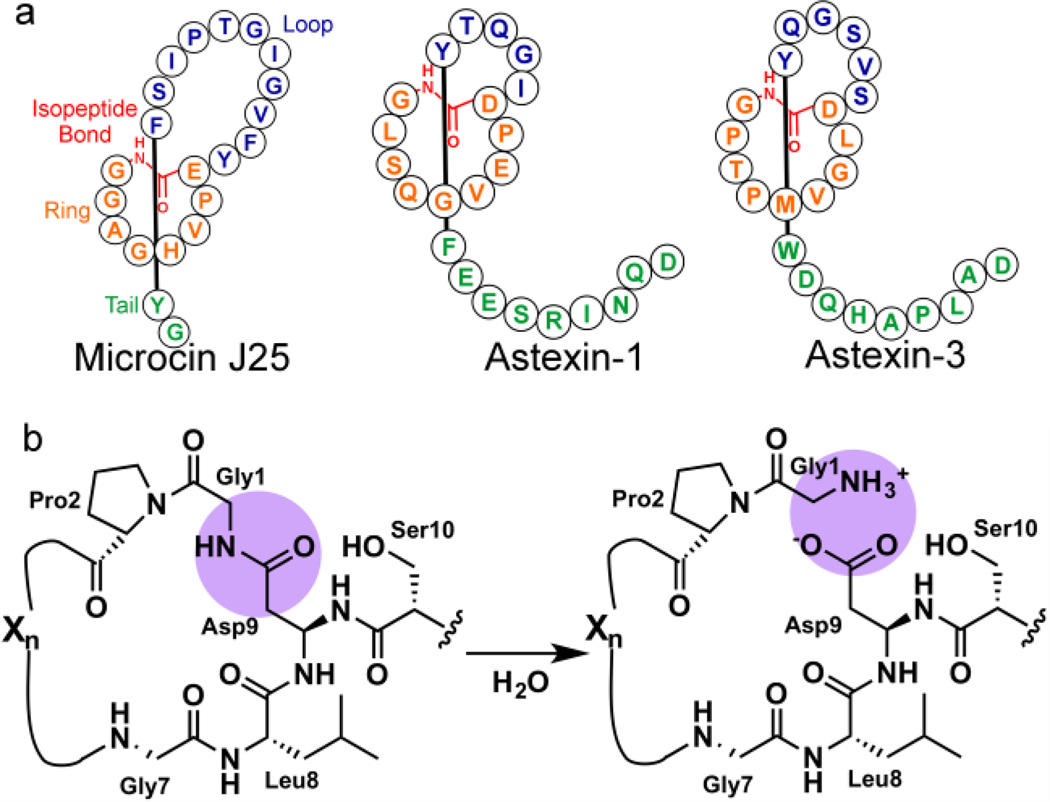
The knowledge base on the biosynthesis of ribosomally synthesized and post-translationally modified peptide (RiPP) natural products is quickly growing.1 However, little is known about the breakdown or degradation of RiPPs. A new mechanism for RiPP catabolism was recently identified in a subset of biosynthetic clusters encoding for lasso pep-tides,2 a class of bioactive peptides characterized by a threaded structure resembling a slipknot.3 The characteristic structure of lasso peptides is defined by an isopeptide bond between the N-terminal α-amine and the side chain carboxylate of an Asp or Glu residue, with the linear C-terminal tail threaded through the resultant macrolactam ring (Figure 1A). The biosynthesis of lasso peptides requires a gene encoding the peptide precursor (A-gene) and two enzymes (encoded for by the B-gene and C-gene) necessary for installing the post-translational modifications. In particular, the B-protein excises the leader sequence from the precursor peptide, and the C-protein installs the isopeptide linkage. Many lasso peptide gene clusters also include an ABC transporter for export of the mature peptide, serving as an immunity factor.4–6 Previous genome mining efforts in our laboratory identified additional genes often located adjacent to lasso peptide gene clusters, including genes annotated as proteases located downstream of the two lasso biosynthetic clusters in the freshwater bacterium Asticcacaulis excentricus.2 The atxE1 protease gene is located adjacent to the biosynthetic cluster for astexin-1, while the atxE2 gene is found adjacent to the cluster for astexin-2 and astextin-3. No-tably, these clusters lack the ABC transporter found in many lasso peptide clusters.
Figure 1.
Lasso peptide structure and cleavage. a) Schematic of representative lasso peptides microcin J25, astexin-1, and astexin-3. b) AtxE2 hydrolyzes the isopeptide bond of astexin-3
The organization of a putative protease adjacent to the lasso peptide biosynthetic machinery suggested that the enzyme might play a role in the catabolism of lasso peptides. Incubation of the cognate lasso peptides astexin-2 and astexin-3 with purified AtxE2 resulted in an increase in mass of the peptide by 18 Da and mass spectral analysis of the product confirmed that the lasso peptide had been linearized. These, and additional biochemical data, demonstrated that AtxE2 is an isopeptidase that can hydrolyze the lasso peptides astexin-2 and astexin-3, encoded within the same biosynthetic cluster, into linear products (Figure 1B).2 Hydrolysis of a lasso peptide into a linear species by isopeptidases may be a self-immunity mechanism to limit the build-up of intracellular concentration of the bioactive lasso product. Alternatively, we have previously proposed that lasso peptides with associated isopeptidases may be functioning analogously to siderophores.2,7 In this scenario, the isopeptidase may serve as a factor that releases cargo bound to the lasso peptide. Presumptive homologs of the AtxE2 isopeptidase are found in other proteobacterial lasso peptide clusters, and likely represent a canonical catabolism gene for these natural products.2
The isopeptidases seemingly have strict specificity for their substrates, as AtxE1 and AtxE2, encoded within two different biosynthetic clusters in Asticcacaulis excentricus, do not function on the lasso substrates produced by the other cluster.2 However, analysis of the substrate tolerance of AtxE2 against a panel of Ala variants of astexin-2 and astexin-3 shows the isopeptidase to be broadly tolerant of amino acid substitutions within the substrate.8 AtxE2 cleaved all alanine variants of astexin-2 and -3 with the exception of the Val11Ala variant of astexin-3. Lastly, the isopeptidases are only functional on threaded substrates, i.e. lasso peptides in which the C-terminal tail is inserted through the isopeptide-bonded ring.2
Sequence analysis using fold-recognition software identifies AtxE2 as a member of the S9 peptidase family, whose members include prolyl oligopeptidases (POPs), dipeptidyl peptidase IVs, and acylamino-acid-releasing enzyme.9 However, S9 peptidases generally hydrolyze peptide bonds within linear, not macrocyclic, substrates, and there are no known family members that can cleave isopeptide bonds. Moreover, the active site cavity of many S9 peptidases would be far too small to accommodate a threaded lasso peptide substrate. In order to reconcile these seemingly contradictory perspectives on substrate recognition and tolerance, we determined the crystal structure of AtxE2 both in isolation, and in complex with an astexin-3 substrate, revealing an open α-propeller domain that is distinct from canonical S9 peptidases. Binding of the lasso substrate occurs through topological, rather than sequence, complementarity. Substrate binding by AtxE2 results in a deformation near the isopeptide bond, and a reorganization of the active site into a conformation that is productive for hydrolysis. Kinetic characterization and mutational analysis provide a rationale for the range of substrate tolerance for this new class of isopeptidases. The presence of the isopeptidase gene may be used to demarcate new lasso peptide biosynthetic clusters, and a network-based genome mining approach identified 87 potential biosynthetic clusters, greatly expanding the list of this subfamily of lasso peptides.
Results
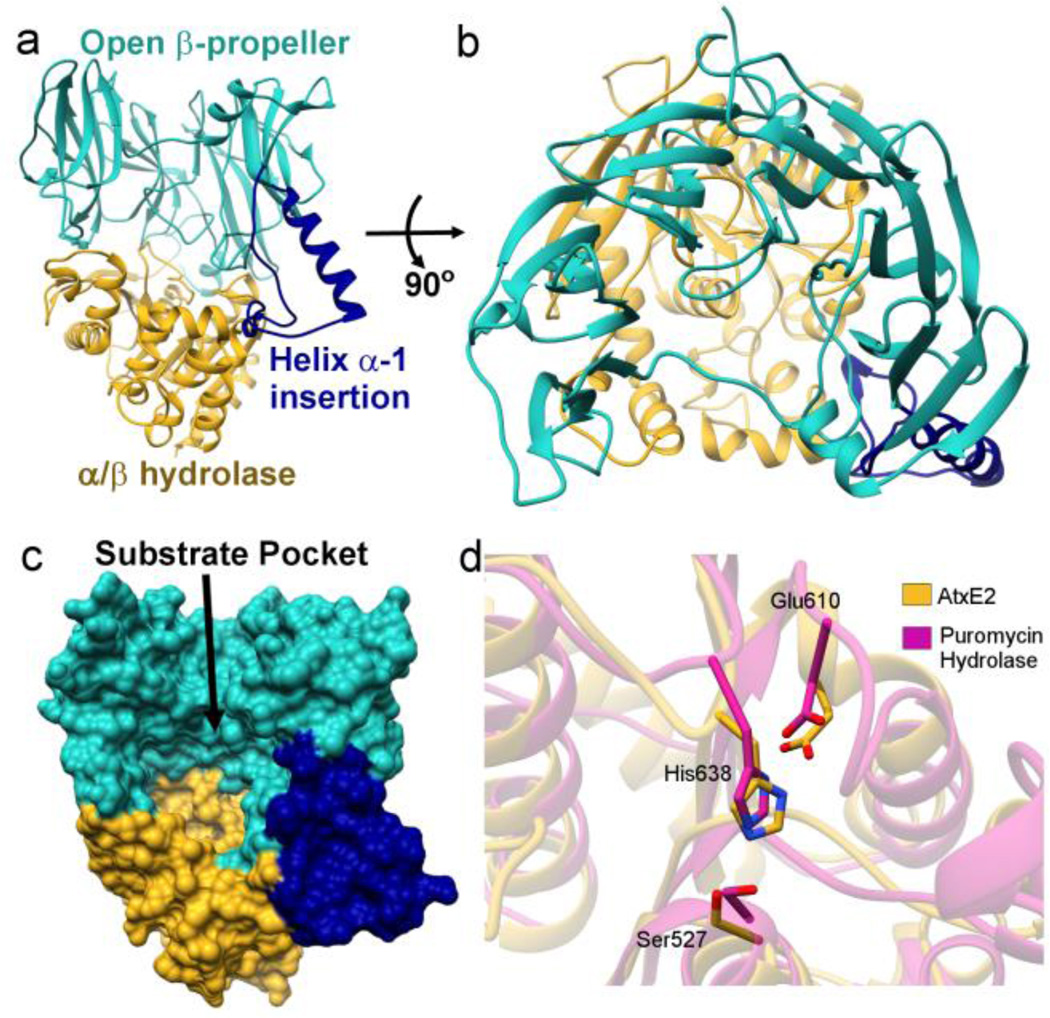
The crystal structure of AtxE2 was determined to a highest resolution cutoff of 2.4 Å, using crystallographic phases determined from data collected on selenomethionine labeled protein (Figure 2A, Table S2). Despite the prior software prediction of homology to S9 peptidases, the AtxE2 structure reveals a unique topology. The overall structure consists of two domains with residues Ala25 through Tyr397 forming an N-terminal open β-propeller domain, and residues Arg398 through Gln686 forming a C-terminal α/β hydrolase domain. The N-terminal open β-propeller domain of AtxE2 (Figure 2A and 2B) consists of six repeats of stacked β-strands that form an enclosure that resembles an inverted cup. A helical insert (helix α-1, numbered according to Figure S1), consisting of Arg189 through Ile229, forms one side of the cup and defines an open cavity adjacent to the active site triad. A short loop containing helix α-2 connects the next blade of the propeller to complete the circular shape.
Figure 2.
Structure of AtxE2. a,b) Ribbondiagram of ligand-free AtxE2. c) The surface model of AtxE2 reveals the largelasso peptide-binding pocket. d) The catalytic triad of AtxE2 (gold) is well conserved when compared to puromycin hydrolase (purple). Residues were numbered according to AtxE2. However, nearby α-helices and loops are moved away from the triad in AtxE2.
The arrangement of the secondary structural elements in the AtxE2 open β-propeller domain contrasts with the typical seven-bladed propeller observed in canonical S9 peptidase family members. A topology diagram of AtxE2 (Figure S2) indicates the location of both the α-1 and α-2 helices and their absence in the structurally similar S9-peptidase family member puromycin hydrolase from Streptomyces morookaense (PDB Code:3AZQ).10 The additional helices increase the amino acid length of the third blade of the propeller. Furthermore, the fourth blade of propeller is completely absent in AtxE2 and is replaced by a short linker to connect the fifth blade. These modifications in the secondary structure produce the substantial structural changes that alter the canonical β-propeller to generate the newly observed open β-propeller fold (Figure S3).
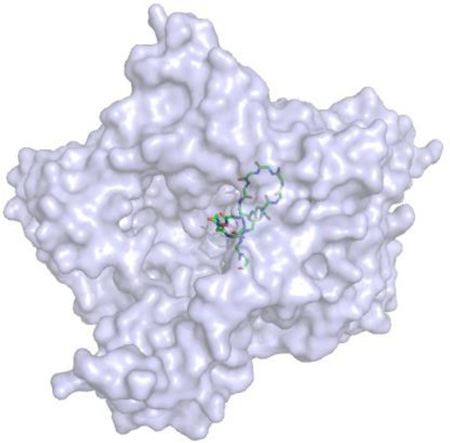
The expectation that AtxE2 would form an S9-like protein fold is likely due to the presence of the C-terminal hydrolase domain, as structure-based analysis of this domain using the DALI server11 identifies similarities with the hydrolase domains of puromycin hydrolase from Streptomyces mo-rookaense10 (RMSD 2.9 Å over 249 Cα atoms) and a prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis12 (PDB Code: 2DCM, RMSD 2.4 Å over 240 Cα atoms) (Figure 2D). The hydrolase domain houses the Ser527-His638-Glu610 catalytic triad that forms the active site. In spite of the overall conservation of the C-terminal hydrolase domain, there are significant differences in the orientation of secondary structural elements (Figure S4). Specifically when AtxE2 is superimposed upon puromycin hydrolase, loop-1, and helices α-3, α-4, and α-5 are pushed back from the catalytic Ser. These movements presumably are necessary to accommodate the bulky threaded lasso peptide substrate. The extended helix α-1 establishes immediacy between the N- and C-terminal domains, and the interaction interface between the three establishes the large substrate-binding pocket (Figure 2C). This large pocket has a volume of 8988 Å3 as measured by CASTp using a probe size of 2.5 Å.13 This would easily accommodate the astexin-3 lasso peptide substrate8, which as a volume of 2457 Å3 as calculated by Chimera.14
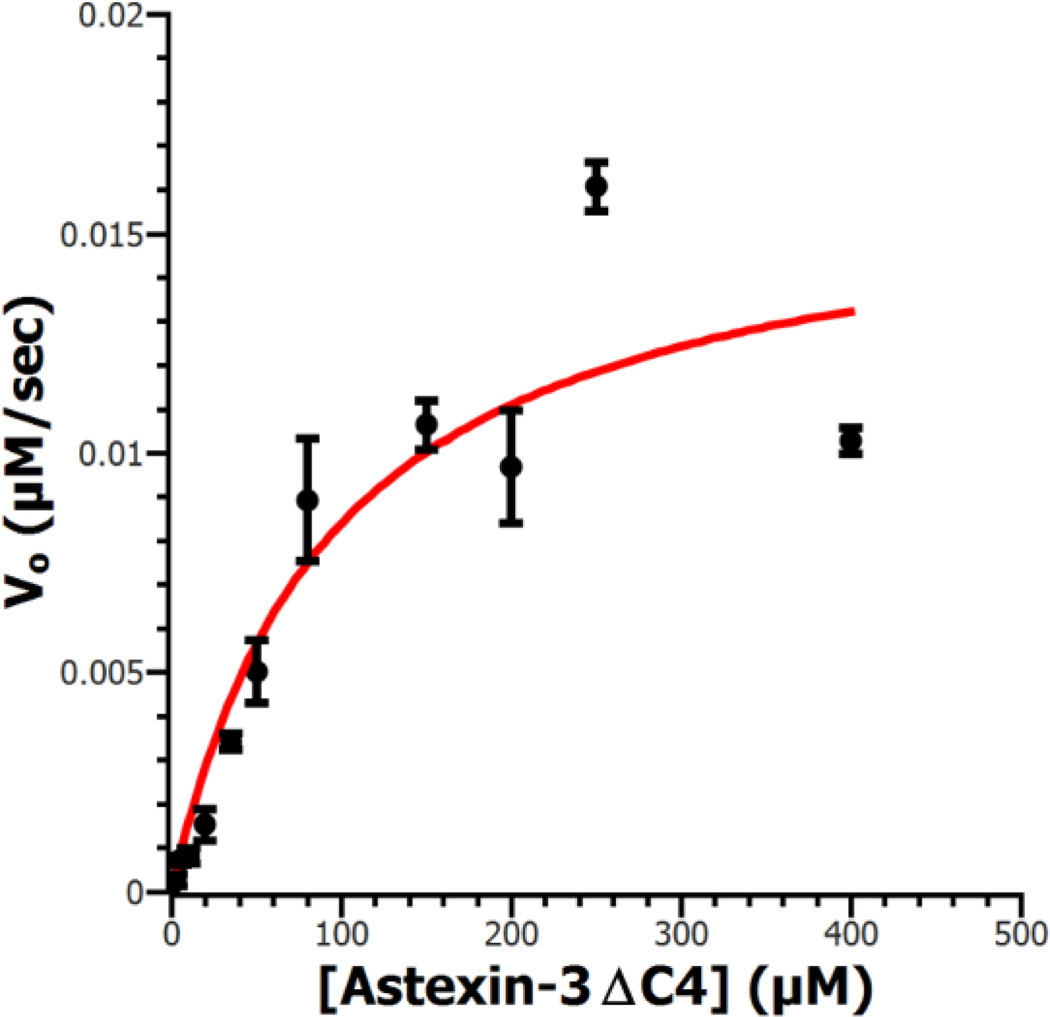
In order to understand the basis of specificity for isopeptide hydrolysis, we sought to determine the structure of an inactive Ser527Ala AtxE2 variant2 in complex with the lasso peptide astexin-3. Attempts to obtain cocrystals of AtxE2 with full-length astexin-3 were unsuccessful. Prior experiments demon-strated that AtxE2 could process astexin-3 substrates that were truncated at the C-terminal tail, which is the most flexible region of the lasso peptide.2,8 Treatment of astexin-3 with car-boxypeptidase Y yielded a truncated lasso peptide that lacked the C-terminal 4 amino acids (astexin-3ΔC4) (Figure S5). We obtained kinetic parameters for AtxE2 using astexin-3ΔC4 as a substrate by monitoring a decrease in fluorescence that occurs upon lasso peptide hydrolysis (Figure 3). The KM of 75 µM ± 23 µM for astexin-3ΔC4 was similar to the previously deter-mined KM of 131 µM ± 34 µM for the full length astexin-3. The kcat of 1.36 ± 0.16 s−1 was approximately three-fold larger compared to full-length astexin-3. These kinetic parameters suggest that the catalytic efficiency of AtxE2 is not compromised upon removal of the 4 residues at the C-terminus of astexin-3.
Figure 3.
Kinetic constants for astexin-3ΔC4 hydrolysis were similar to value of full length astexin-3, indicating the tail of the lasso peptide does not enhance activity.
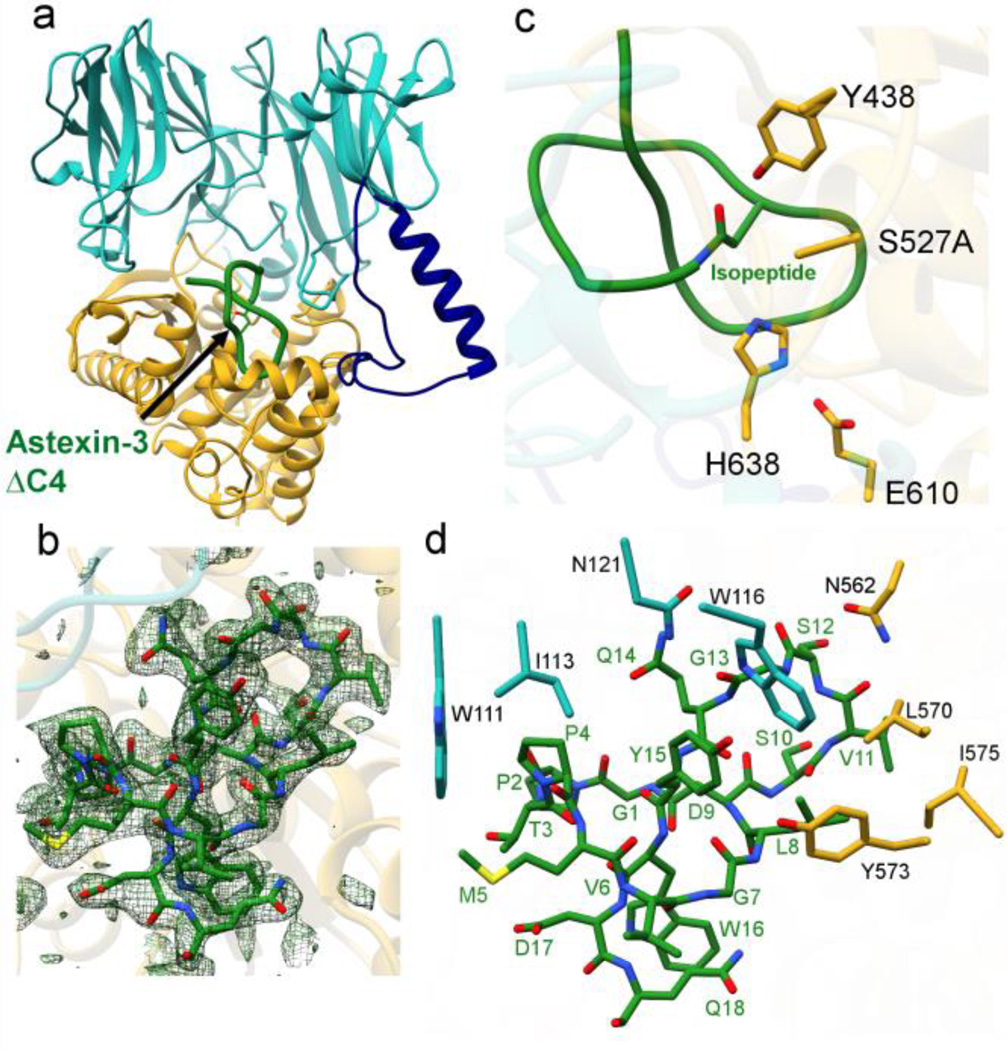
Crystallization efforts of inactive AtxE2 in complex with astexin-3ΔC4 yielded the 2.2 Å structure of the binary com-plex (Figure 4A and 4B). A comparison of the unliganded AtxE2 structure with that of the astexin-3ΔC4 complex reveals that a small active site loop, encompassing Gln437 through Gly442, shifts towards the substrate, resulting in a movement of Tyr438 to within 3.3 Å of the carbonyl oxygen of the isopeptide bond (Figure S6). The orientation of Tyr438 upon binding substrate suggests that this residue may stabilize the oxyanion in the tetrahedral intermediate that forms upon attack of the catalytic Ser onto the isopeptide carbonyl carbon. To probe the role of this residue, we generated the Tyr438Phe variant, which lacked any detectable activity (Figure 5). The movement of this loop to orient Tyr438 appears be induced by binding of the substrate, specifically by the threaded C-terminal tail. In the AtxE2 cocrystal structure, the first few residues that are threaded through the lasso (Trp16 and Asp17) form a solvent-excluded pocket that directs Tyr438 towards the isopeptide linkage.
Figure 4.
Structure of AtxE2-astexin-3 complex a) Overall struc-ture of AtxE2 with astexin-3ΔC4 bound (green) indicates that the lasso peptide binds into the large pocket of AtxE2. b) The structure of the binary complex was refined without astexin-3ΔC4 and the Fo-Fcdifference maps scaled to 2 σ (green mesh) was overlaid into astexin-3ΔC4. c) The catalytic triad of AtxE2 is well positioned for hydrolysis of the isopeptide bond (green). d) Interactions between astexin-3ΔC4 (green labels) and AtxE2 (black labels).
Figure 5.
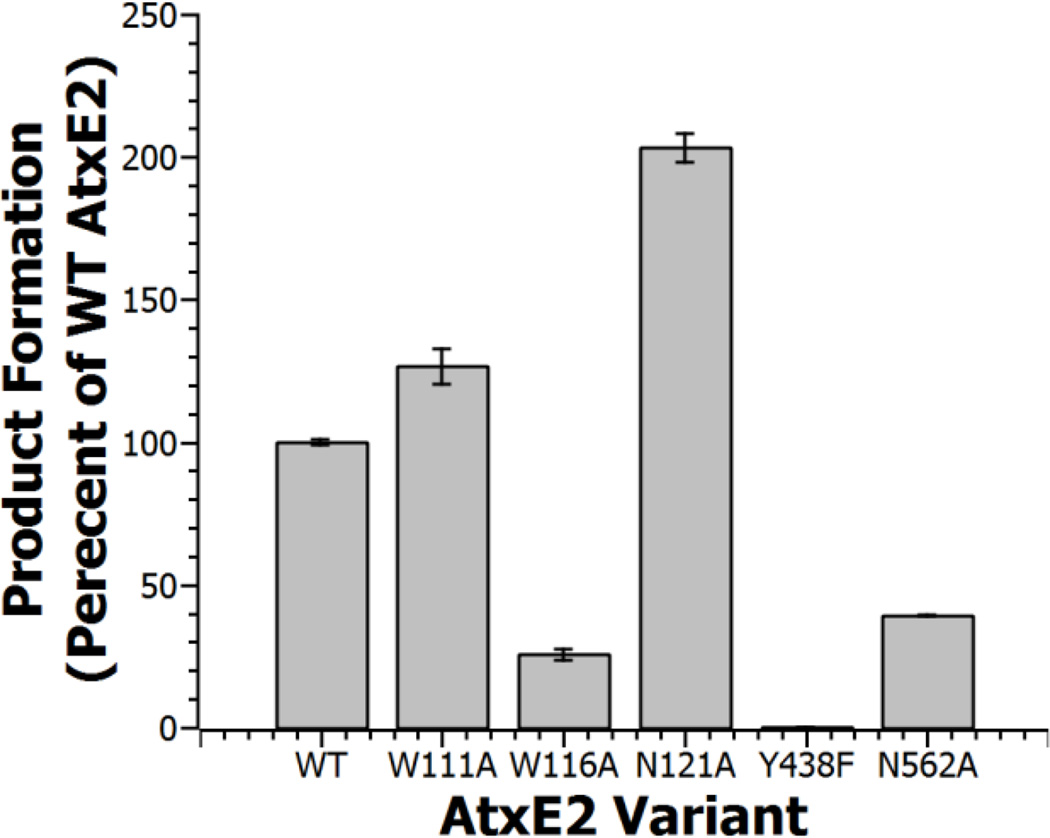
Analysis of production formation of AtxE2 variants after 1 hour revealed that Trp116 and Asn562 are important for activity in AtxE2. Substitution of Tyr438, important for stabilization of the alkoxide intermediate, leads to loss of detectable activity. Amounts of product were monitored by HPLC, integrated, and normalized to WT production.
In the cocrystal structure, much of the lasso peptide is contained within the large binding pocket adjacent to the catalytic triad in AtxE2 (Figure 4A). The isopeptide bond between Gly1-Asp9 of astexin-3ΔC4 is located within the active site of the 3α/β hydrolase domain, where the catalytic Ser527 alkoxide would be poised for nucleophilic attack and the aforementioned Tyr438 would act to stabilize the negative charge on the oxyanion that develops in the transition state (Figure 4C). The Ser10-Gln14 loop of the substrate fits into a small and slightly acidic pocket (Figure S7) lined with residues Asn121, His214, Thr531, Thr553, Ala558, Asn562, Ile566, Leu570, and Ala572 (Figure 4A and 4B). This further explains why AtxE2 cannot process unthreaded substrates.2 The loop region of an unthreaded lasso would lack shape complementarity while the extended tail would clash with the binding pocket of the macrolactam ring.
Inspection of the isopeptidase-peptide interface reveals only a few specific interactions between astexin-3ΔC4 and AtxE2 (Figure 4D). Trp111 and Ile113 of AtxE2 create a hydropho-bic pocket for Pro4 of astexin-3ΔC4 while Leu570, Tyr573, and Ile575 of AtxE2 form a pocket for Leu8 of the peptide; Asn121 and Asn562 form hydrogen-bonding interactions with the backbone of Gly13 and Val11 of astexin-3ΔC4, respectively. Lastly, Trp116 can hydrogen bond with the backbone oxygen of Gly13 and may form stacking interactions with Tyr15 of the substrate. Several residues within astexin-3ΔC4, including Met5 and Val6 are entirely solvent exposed. The absence of extensive side-chain specific interactions explains the tolerance AtxE2 for the panel of astexin-3 Ala variants, almost all of which are substrates for the enzyme.8
To confirm the importance of the observed interactions, we generated the Trp111Ala, Trp116Ala, Asn121Ala, and Asn562Ala variants of AtxE2, and tested both the wild-type and enzyme variants for product formation in an endpoint assay (Figure 5, Figure S8). The hydrolytic product for each variant was observed by HPLC, the corresponding peaks were integrated, and normalized with respect to product formation by wild-type AtxE2. Surprisingly, the Trp111Ala variant has activity comparable to wild type AtxE2 while the Asn121Ala variant appeared to even enhance activity. These data indicate that mutations within the AtxE2 hydrophobic pocket that engage Pro4 of the substrate, and at residues that hydrogen bond with Gly13 of the substrate do not inactivate the enzyme. However, both Trp116Ala and Asn562Ala AtxE2 variants had greatly reduced activity indicating the importance of the hydrogen-bonding interactions provided by these residues.
The lack of extensive hydrogen-bonding interactions between AtxE2 and the lasso peptide substrate suggests that entropy, rather an enthalpy, should be the major binding driving force for binding. To test this hypothesis, isothermal titration calorimetry was carried out to measure the thermodynamics of binding between the catalytically inactive AtxE2 Ser527Ala variant and asetxin-3 (Figure S9). Minimal heat was released upon ligand addition, consistent with our hypothesis that binding is entropically driven and that enthalpy does not significantly contribute to binding.
A superposition of the previously determined solution NMR structure of astexin-32 with that of astexin-3ΔC4 bound in the AtxE2 cocrystal structure reveals major differences in the structure of the lasso peptide (Figure S10). In the solution structure, Val11, Ser12, and Gly13 of the loop protect the isopeptide bond. As a result of these interactions, the isopeptide bond of astexin-3ΔC4 in the solution conformation would be inaccessible to AtxE2. The cocrystal structure illustrates how AtxE2 opens the lasso peptide to expose the isopeptide bond for hydrolysis. Specifically, the Ser10-Gln14 loop is moved away from the isopeptide bond and positioned into a pocket in the α/β hydrolase domain, while the threaded tail is similarly directed away from the isopeptide bond. Similarly, the structure of the loop of microcin J25 is altered upon binding to the ferric citrate transporter FhuA, which is responsible for its cellular import.15
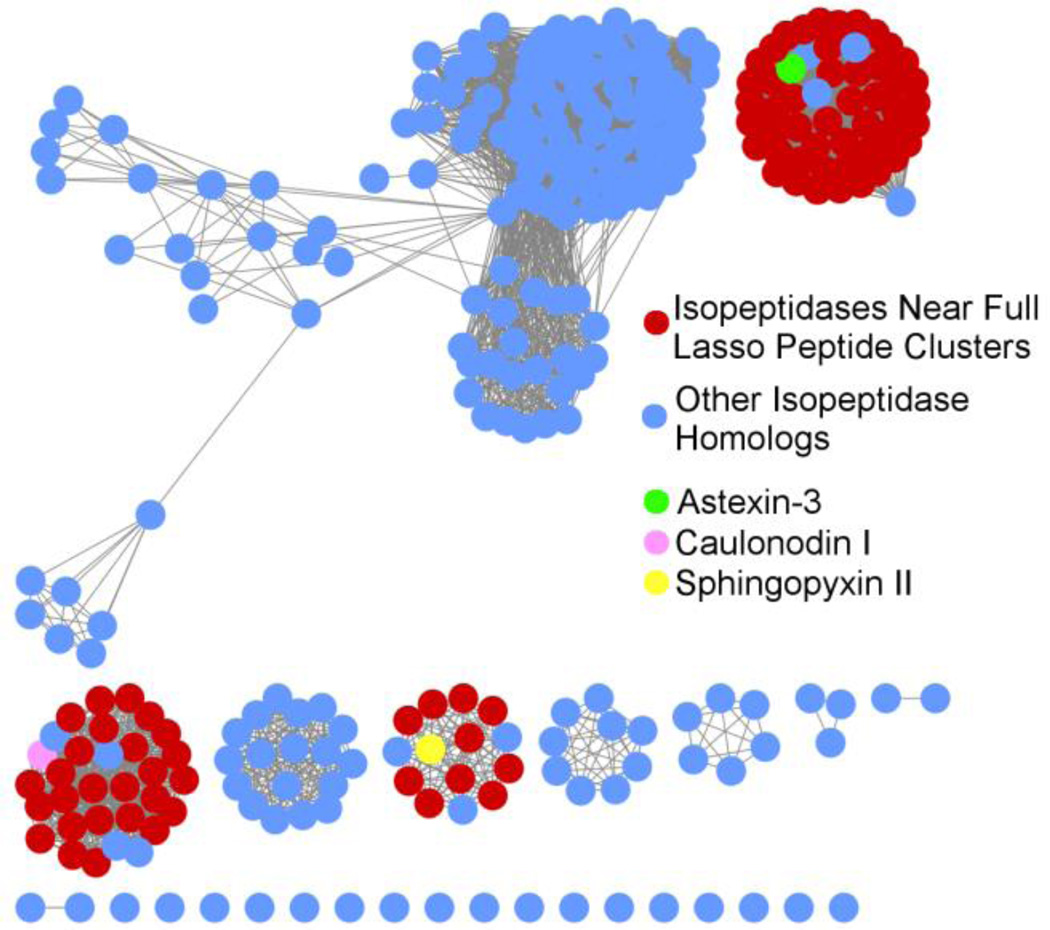
In order to determine the incidence of the isopeptidase in lasso peptide biosynthetic clusters, we created a sequence similarity network of homologs16 using the AtxE2 sequence as a query (Figure 6). A network was derived using an alignment score of e−85, corresponding to at least 30% sequence identity between connected nodes. This result was then used for a genome neighborhood network analysis, in order to identify those sequence similar clusters that contained an isopeptidase homolog adjacent to lasso peptide biosynthetic machinery. A total of 98 unique putative lasso peptide isopeptidase homologs with <95% sequence identity were identified using this criterion and they clustered into three groups within the sequence similarity network (Figure 6). The persistence of the isopeptidase as a regulatory or catabolic mechanism appears to be most prevalent amongst the Alphaproteobacteria with some examples in Gammaproteobacteria as well.17
Figure 6.
A sequence similarity network was derived from AtxE2 using a similarity score of e−85, where connected nodes represent genes that are related by at least 30% at the primary sequence level. Red nodes indicate proteins located near complete putative lasso peptide clusters, while blue nodes indicate proteins not located near complete putative lasso peptide clusters. Examples of three isopeptidases located near known lasso peptide clusters are indicated: astexin-3 (green), caulonodin I (pink), and sphingopyx-in II (yellow).
In order to analyze the conservation of important sequence elements across these homologs, we mapped the alignment of sequences onto the structure of AtxE2. Notably, these 98 sequences showed very few regions with high sequence conservation (Figure S11). As expected, the catalytic Ser-His-Asp/Glu is highly conserved, as is the catalytically important Tyr438. Otherwise, there is little conservation in the sequence of residues that form the binding pocket for the lasso peptide. This lack of sequence conservation is consistent with the broad range of sequences, as well as macrolactam ring sizes, found in lasso peptides that may be catabolized by isopeptidase homologs.
Previous genome mining efforts to discover lasso peptides in our laboratory have relied on pattern matching to the lasso peptide precursor followed by searches for conserved motifs in the B- and C-enzymes in the vicinity of the precursor.18,19 B-enzymes are comprised of an N-terminal domain that resembles the PqqD protein fused to a cysteine protease domain, and are responsible for the cleavage of the leader peptide from the core peptide. C-enzymes are homologous to asparagine synthetase and form the isopeptide bond in the lasso peptide. Marahiel and co-workers have used an approach that relies on the relatively unique B-protein as the seed for genome mining searches.17 We sought to determine if the presence of the catabolic isopeptidase gene could be used as an alternative seed in genome mining. To this end, we carried out a comparative search effort to explore whether the isopeptidase-centric mining approach could identify clusters not previously detected. Manual inspection of the genomic context of all 98 the isopeptidases in these three clusters revealed that 87 contained a complete cluster with an A-peptide precursor, B-protein, C-protein, and a TonB-dependent transporter (Table S3). However, 19 of the 87 clusters contain a mis-annotation of either the B- or C-protein (Table S3). Three examples are shown and re-annotated in Figure S12. The original improper annotation would have precluded identification of these putative lasso peptide clusters using the biosynthetic enzymes as a search sequence. Of the 87 clusters identified, 65 have not been previously described.
Nearly all of the 87 gene clusters share the architecture found in the astexin-2 and -3 gene cluster with the A-, B-, and C-genes found in one operon and the isopeptidase and TonB-dependent transporter in a separate divergently transcribed operon. We have previously demonstrated2 that these proteobacterial gene clusters form a phylogenetic clade distinct from antimicrobial lasso peptide gene clusters. The finding of dozens of new proteobacterial clusters, nearly all with the same architecture, underscores how well-conserved these clusters are in Proteobacteria. Despite the homogeneity in cluster architecture, some of the newly identified clusters exhibit features that have not been observed before in lasso peptides. In Sphingopyxis sp. 113P3, the cluster has the canonical architecture, but also includes a second isopeptidase and TonB-dependent transporter. In Sphingopyxis sp. H071, the precursor gene is not located upstream of the B- and C-genes, but is located instead between the C-gene and the isopeptidase E-gene. Lastly, the cluster from Sphingomonas hengshuiensis remarkably includes a HipA/HipB toxin/antitoxin pair sandwiched between the two operons in the lasso peptide gene cluster.
Our data suggest that the genome mining efforts using the protein sequences of catabolic genes may serve as another useful tool for the identification of RiPP biosynthetic clusters, especially in cases such as lasso peptides where the biosynthetic enzymes are distantly related to metabolic enzymes. To validate this approach, we carried out the heterologous expression of the two lasso peptides predicted to be expressed by the organism Asticcaucalis benevestitus (Figure S13); we have named these compounds benenodin-1 and benenodin-2. Given that robust heterologous expression systems exist for proteobacterial lasso peptide gene clusters,17,18 many of the additional clusters found here can likely be expressed in a similar fashion.
Discussion
The lasso peptide isopeptidase is a unique member of the serine protease family as it carries out the hydrolysis of an isopeptide (rather than peptide) bond, and only on threaded lasso peptide substrates. The structure represents a topology that is constructed to accommodate the complex substrate. The comparison of the substrate astexin-3ΔC4 from the cocrystal structure with the structure of the lasso peptide in solution illustrates how the enzyme remodels the substrate to access the isopeptide bond for hydrolysis. The Ser10-Gln14 isopeptide loop, which is converted from a linear to a loop topology during lasso maturation, is proposed to play a role in the biosynthetic process.8 Our studies indicate this loop also plays an important function in the catabolism of lasso peptides, as binding to the isopeptidase repositions this loop away from the isopeptide bond and into a binding pocket. Hence, it is likely that both the RiPP biosynthetic and catabolic enzymes utilize the same epitope in the substrate for recognition.
The tolerance of AtxE2 to substitutions at nearly all residues within the substrate is accommodated by the lack of extensive specific contacts between the enzyme and substrate. The interactions observed in the crystal structure show that the shape of the lasso peptide rather than the composition of individual residues may be the primary recognition motif. Thus, AtxE2 may only engage substrates based on the number of amino acids within the lasso macrolactam ring and the size of the loop. Enzymes involved in the biosynthesis of RiPPs are shown to be tolerant of substitutions within the core peptide,20–22 and this tolerance is thought to provoke diversity in the resultant scaffolds. While the biosynthetic enzymes can process substrates with mutations along the core, they generally are not permissive to changes in the size of the ring. One exception are the caulosegnins I–III, for which the identical biosynthetic enzymes generate rings of 8 and 9 residues.23 The broad tolerance of the isopeptidase likely is necessary to allow for hydrolysis of point mutants that may occur in the substrate, which is also gene encoded.8
Hydrolysis of linear peptide substrates by canonical serine proteases occurs through an acyl-enzyme intermediate and substrate specificity is partially mediated by the complementarity between the enzyme specificity pocket and the amino acid located at the P1 position,24 i.e. the residue preceding the labile peptide bond.25 Although AtxE2 does not hydrolyze a peptide bond, the enzyme may use a similar means for substrate specificity. Specifically, formation of an acyl-enzyme adduct at Ser527 via the side chain of Asp9 of astexin-3 would result in the deposition of the Ser10-Gln14 isopeptide loop in a large pocket analogous to the specificity pocket of serine proteases, suggesting complementarity between the isopeptide loop of the substrate determines the specificity of the lasso isopeptidase. Lastly, reorganization of the region harboring Tyr438 to position this residue for transition state stabilization is partially facilitated by the threaded tail of the lasso substrate. Similar active site reorganizations are observed in the structurally and chemically unrelated ubiquitin cysteine isopeptidase USP7, where the active site His-Cys dyad is oriented only upon substrate binding.26
Our bioinformatic studies showed that isopeptidase homologs are quite common across Alphaproteobacteria, facilitating the identification of 87 lasso biosynthetic clusters using the AtxE2 isopeptidase sequence as a search query, and suggesting a further strategy to quickly identify lasso peptide gene clusters in Proteobacteria. Mining efforts using motifs from biosynthetic enzymes have been fruitful for RiPP genome mining of lanthipeptides,27,28 cyanobactins,29 and thioazole/oxazole modified microcins,30,31 largely because of the unique nature of some of their constituent modification enzymes. In contrast, the lasso biosynthetic enzymes are not unique in sequence and show moderate sequence similarity to transglutaminase (B-protein) and asparagine synthetase (C-protein), which complicates straightforward identification of putative clusters. Another complication is that the B- and C-proteins are commonly misannotated, whereas the larger isopeptidase gene with its conserved α/β hydrolase domain is more commonly correctly annotated. The identification of multiple new biosynthetic clusters by this method suggests that an approach using sequences of precursor peptides, biosynthetic enzymes, as well as those of catabolic genes, should produce more candidates and eliminate false positives in genome mining. The validation and further characterization of the 87 biosynthetic clusters identified through this work is currently underway.
Experimental Section
Peptide and protein production, crystallization, and kinetic and end-point assays
Details may be found in the SI Materials and Methods.
Structure determination, and refinement
Diffraction data was indexed, scaled, and integrated using a combination of HKL-2000 and autoPROC32 packages. Crystallographic phases were obtained from selenomethionine labeled AtxE2 using the phenix.autosol.33 Initial model building was completed with phenix.autobuild.33 This model was used to obtain crystallographic phases for both the ligand-free AtxE2 and the astexin-3ΔC4 complex. Manual refinement was completed with iterations of COOT34 and REFMAC5.35 Refinement parameters are listed in Table S2.
Supplementary Material
Acknowledgments
We thank Keith Brister and the staff at Life Sciences Collaborative Access Team (LS-CAT) at Argonne National Labs, IL., for facilitating X-ray data collection. J.R.C. is supported in part by the Hager Fellowship from the Department of Biochemistry. This work was supported by a grant from the NIH to AJL (GM107036) and JDK was supported in part by a training grant from the NIH (GM7388).
Footnotes
ASSOCIATED CONTENT
Supplementary methods, tables, and figures are available free of charge via the Internet at http://pubs.acs.org. Ligand-free AtxE2 and AtxE2 S527A with astexin3ΔC4 bound were deposited to the Protein Data Bank as accession codes 5TXC and 5TXE respectively.
REFERENCES
- 1.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RD, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maksimov MO, Link AJ. J. Am. Chem. Soc. 2013;135:12038–12047. doi: 10.1021/ja4054256. [DOI] [PubMed] [Google Scholar]
- 3.Wilson KA, Kalkum M, Ottesen J, Yuzenkova J, Chait BT, Landick R, Muir T, Severinov K, Darst SA. J. Am. Chem. Soc. 2003;125:12475–12483. doi: 10.1021/ja036756q. [DOI] [PubMed] [Google Scholar]
- 4.Yan K-P, Li Y, Zirah S, Goulard C, Knappe TA, Marahiel MA, Rebuffat S. ChemBioChem. 2012;13:1046–1052. doi: 10.1002/cbic.201200016. [DOI] [PubMed] [Google Scholar]
- 5.Duquesne S, Destoumieux-Garzón D, Zirah S, Goulard C, Peduzzi J, Rebuffat S. Chem. Biol. 2007;14:793–803. doi: 10.1016/j.chembiol.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Solbiati JO, Ciaccio M, Farías RN, González-Pastor JE, Moreno F, Salomón Ra. J. Bacteriol. 1999;181:2659–2662. doi: 10.1128/jb.181.8.2659-2662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegemann JD, Zimmermann M, Xie X, Marahiel MA. Acc. Chem. Res. 2015;48:1909–1919. doi: 10.1021/acs.accounts.5b00156. [DOI] [PubMed] [Google Scholar]
- 8.Maksimov MO, Koos JD, Zong C, Lisko B, Link AJ. J. Biol. Chem. 2015;290:30806–30812. doi: 10.1074/jbc.M115.694083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polgár L. Cell. Mol. Life Sci. 2002;59:349–362. doi: 10.1007/s00018-002-8427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matoba Y, Nakayama A, Oda K, Noda M, Kumagai T, Nishimura M, Sugiyama M. Proteins Struct. Funct. Bioinforma. 2011;79:2999–3005. doi: 10.1002/prot.23139. [DOI] [PubMed] [Google Scholar]
- 11.Holm L, Rosenström P. Nucleic Acids Res. 2010;38:545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito K, Nakajima Y, Xu Y, Yamada N, Onohara Y, Ito T, Matsubara F, Kabashima T, Nakayama K, Yoshimoto T. J. Mol. Biol. 2006;362:228–240. doi: 10.1016/j.jmb.2006.06.083. [DOI] [PubMed] [Google Scholar]
- 13.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. Nucleic Acids Res. 2006;34:116–118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 15.Mathavan I, Zirah S, Mehmood S, Choudhury HG, Goulard C, Li Y, Robinson CV, Rebuffat S, Beis K. Nat. Chem. Biol. 2014;10:340–342. doi: 10.1038/nchembio.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, Whalen KL. Biochim. Biophys. Acta - Proteins Proteomics. 2015;1854:1019–1037. doi: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hegemann JD, Zimmermann M, Zhu S, Klug D, Marahiel MA. Biopolymers. 2013;100:527–542. doi: 10.1002/bip.22326. [DOI] [PubMed] [Google Scholar]
- 18.Maksimov MO, Pelczer I, Link AJ. Proc. Natl. Acad. Sci. U. S. A. 2012;109:15223–15228. doi: 10.1073/pnas.1208978109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maksimov MO, Link AJ. J. Ind. Microbiol. Biotechnol. 2014;41:333–344. doi: 10.1007/s10295-013-1357-4. [DOI] [PubMed] [Google Scholar]
- 20.Donia MS, Ravel J, Schmidt EW. Nat. Chem. Biol. 2008;4:341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Sher D, Kelly L, Shi Y, Huang K, Knerr PJ, Joewono I, Rusch D, Chisholm SW, van der Donk WA. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10430–10435. doi: 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sardar D, Pierce E, McIntosh JA, Schmidt EW. ACS Synth. Biol. 2015;4:167–176. doi: 10.1021/sb500019b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegemann JD, Zimmermann M, Xie X, Marahiel MA. J. Am. Chem. Soc. 2013;135:210–222. doi: 10.1021/ja308173b. [DOI] [PubMed] [Google Scholar]
- 24.Schechter I, Berger A. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 25.Di Cera E. IUBMB Life. 2009;61:510–515. doi: 10.1002/iub.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, Shi Y. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 27.Yu Y, Zhang Q, van der Donk WA. Protein Sci. 2013;22:1478–1489. doi: 10.1002/pro.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, Doroghazi JR, Zhao X, Walker MC, van der Donk WA. Appl. Environ. Microbiol. 2015;81:4339–4350. doi: 10.1128/AEM.00635-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leikoski N, Liu L, Jokela J, Wahlsten M, Gugger M, Calteau A, Permi P, Kerfeld CA, Sivonen K, Fewer DP. Chem. Biol. 2013;20:1033–1043. doi: 10.1016/j.chembiol.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 30.Dunbar KL, Chekan JR, Cox CL, Burkhart BJ, Nair SK, Mitchell DA. Nat. Chem. Biol. 2014;10:823–829. doi: 10.1038/nchembio.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox CL, Doroghazi JR, Mitchell DA. BMC Genomics. 2015;16:778. doi: 10.1186/s12864-015-2008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Lohkamp B, Scott WG, Cowtan K. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.