Abstract
Background
Disruption of cellular metabolite levels can adversely impact development. Specifically, loss-of-function of the C. elegans NAD+ salvage biosynthesis gene PNC-1 results in an array of developmental phenotypes. Intriguingly, PNC-1 and its functional equivalent in vertebrates are secreted, but the contributions of the extracellular enzymes are poorly understood. We sought to study the tissue-specific requirements for PNC-1 expression and to examine the role of the secreted isoform.
Results
A thorough analysis of PNC-1 expression did not detect expression in tissues that require PNC-1 function. Limited expression of both the secreted and intracellular PNC-1 isoforms provided function at a distance from the tissues with phenotypes. We also find that the secreted isoform contributes to in vivo PNC-1 activity. Furthermore, uv1 cell survival has the most stringent requirements in terms of PNC-1 expression pattern or level.
Conclusion
Using careful promoter analysis and a restricted expression approach we have shown that both the secreted and the intracellular PNC-1 isoforms function cell non-autonomously, and that the PNC-1a isoform is functionally relevant in vivo. Our work suggests a model where PNC-1 function is provided cell non-autonomously by a mix of intra and extracellular activity, most likely requiring NAD+ salvage metabolite transport between tissues.
Keywords: NAD+ salvage biosynthesis, Nampt, Pnc1, nicotinic acid, vitamin B3, nicotinamide
Introduction
Nicotinamide adenine dinucleotide (NAD+) is central to metabolism in all cells, both as a co-factor in redox reactions, including oxidative phosphorylation and glycolysis, and as a substrate of NAD+ consumer enzymes. Differentiation of muscle and immune cells in mammals is disrupted when NAD+ biosynthesis is perturbed (Fulco et al., 2008; Skokowa et al., 2009; Yin et al., 2012), highlighting the importance of NAD+ and its biosynthesis in development. Similarly, we have demonstrated that perturbing NAD+ biosynthesis results in very specific effects during development of the invertebrate C. elegans, establishing C. elegans as an important model for studying NAD+ biosynthesis and metabolism during development (Vrablik et al., 2009; Vrablik et al., 2011).
In its role as a cofactor, NAD+ is interconverted between its oxidized and reduced forms but these reactions do not affect the total concentration of NAD+ available to the cell. However, NAD+ consumers hydrolyze NAD+, creating a need to replenish the key molecule via biosynthesis (Landry et al., 2000; Tanny and Moazed, 2001; Kim et al., 2004). NAD+ may be resynthesized from nicotinamide (NAM), a byproduct of NAD+ consumers, in a process called salvage biosynthesis (Rongvaux et al., 2003). It can also be synthesized from dietary nicotinic acid (NA) via the Preiss-Handler pathway (Preiss and Handler, 1958a; Preiss and Handler, 1958b), from dietary nicotinamide riboside (NR) (Bieganowski and Brenner, 2004), or synthesized de novo from tryptophan (Gazzaniga et al., 2009). Two types of pathways carry out salvage biosynthesis. Vertebrates use nicotinamide phosphoribosyltranferase (Nampt) to convert nicotinamide into nicotinamide mononucleotide (NMN), which is then converted into NAD+ by NMN adenyltransferase (Nmnat). Invertebrates convert nicotinamide (NAM) into nicotinic acid (NA) using a nicotinamidase, which is then fed into the Preiss-Handler pathway (Magni et al., 1999; Rongvaux et al., 2003). Although Nampt and nicotinamidases are enzymatically distinct, both catalyze the conversion of NAM to the next metabolite in their respective NAD+ salvage biosynthesis pathways and both would be expected to modulate NAM levels, making them biologically functionally analogous.
C. elegans uses a nicotinamidase to catalyze the first step of salvage NAD+ biosynthesis (van der Horst et al., 2007; Vrablik et al., 2009; French et al., 2010). The genomic locus of the C. elegans nicotinamidase gene pnc-1 is complex, encoding two isoforms that encode proteins which differ only in regards to the presence of a cleavable signal peptide, thus creating a secreted PNC-1a isoform and an identical intracellular PNC-1b isoform. The PNC-1a isoform is expressed from a distinct promoter (Vrablik et al., 2009), and we show in this study that two promoters appear to direct expression of PNC-1b (Fig. 1). We sought to understand the tissue specific functions of PNC-1 and the contributions of the secreted and intracellular isoforms to development. Given the role of salvage biosynthesis in the maintenance of NAD+ levels and its importance in general metabolism, it came as a surprise that neither expression of Nampt in mice nor nicotinamidases in Drosophila and C. elegans is ubiquitous (Chintapalli et al., 2007; Revollo et al., 2007b; Vrablik et al., 2009). Like C. elegans, which encodes a secreted PNC-1 isoform, vertebrates express an intracellular (iNampt) and extracellular (eNampt) (Yonezawa et al., 2006; Revollo et al., 2007a; Ocón-Grove et al., 2010). NAD+ biosynthetic capacity might be provided to tissues that lack Nampt or nicotinamidase expression by cell-autonomous expression of enzymes in other NAD+ biosynthetic pathways. Alternatively, exchange of metabolites such as NAM, NMN and NA between tissues could contribute to NAM clearance and NAD+ biosynthesis in tissues that don’t express the key salvage enzyme. Alternatively, there is evidence that extracellular conversion of substrate to product and transport of product to tissues in need could play a role (Revollo et al., 2007b; Vrablik et al., 2009; Vrablik et al., 2011). Although Nampt is critical for NAD+ salvage (Rongvaux et al., 2002; van der Veer et al., 2005), the specific role of eNampt is contentious and the need for extracellular activity of eNampt has been controversial (Revollo et al., 2007b; Hara et al., 2011).
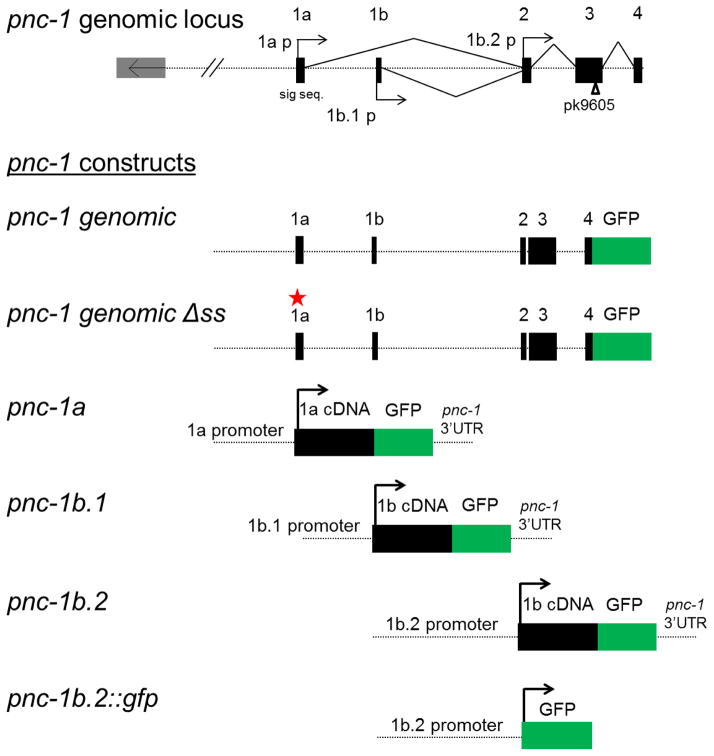
Figure 1.
The genomic organization of the pnc-1 locus showing intron-exon structure and promoter regions. The constructs used in this study are also diagrammed. The location of the point mutation in pnc-1(pk9605) is indicated by an open arrowhead. The red star indicates the mutated signal sequence in pnc-1 ΔSS. Solid boxes and right pointing arrows represent coding regions and promoters, respectively. The 1a exon contains the first 17 signal sequence amino acids and the 1b exon the remaining three signal sequence amino acids and cleavage site. In all other respects, PNC-1 expressed from the three promoters is identical.
PNC-1 has multiple functions in promoting proper development of specific tissues in C. elegans. For example, adequate PNC-1-mediated NAD+ biosynthesis is required to prevent a delay in development of the gonad with respect to the soma (Vrablik et al., 2009). The body wall muscles also require PNC-1 activity during development for optimal function during adulthood. In this tissue, PNC-1 activity is required both to promote biosynthesis of NAD+ and to prevent accumulation of NAM (Vrablik et al., 2011). In addition, PNC-1 promotes egg-laying and survival of uv1 (uterine-vulval) cells located within the gonad by preventing accumulation of NAM that is toxic to the egg-laying muscles and the uv1 cells (Vrablik et al., 2009; Vrablik et al., 2011). In our preliminary expression analysis we were surprised to find no PNC-1 expression in the tissues that manifest pnc-1 phenotypes (Vrablik et al., 2009). Given this disconnect between sites of PNC-1 expression and functional requirement, the presence of a secreted PNC-1a isoform in C. elegans and also the controversy over the role of the secreted eNampt in vertebrate metabolism, we addressed the following questions: Can specific PNC-1 developmental functions be attributed to specific expression sites? Does PNC-1 function cell non-autonomously? And is the secreted PNC-1a isoform necessary and / or sufficient for any specific PNC-1 function?
Results
Is PNC-1 expressed in tissues that exhibit phenotypes?
We previously examined expression of the pnc-1 isoform-specific promoters 1a and 1b.1 and were surprised to find no expression in the body-wall muscle and limited expression in the gonad, two tissues where the global absence of PNC-1 function has developmental consequences (Vrablik et al., 2009). This suggests that pnc-1 may have cell non-autonomous functions. We first aimed to ensure that we are not missing potential regulatory elements that direct pnc-1 expression in the tissues that display phenotypes in the null mutant by constructing several additional GFP reporter genes. We included the native 3′ regulatory region in new translational reporters using the 1a and 1b promoters. We also made transcriptional and translational reporters to test for activity of a predicted promoter upstream of exon 2, in the region between exon 1b and exon 2 (Fig. 1), a promoter region we have dubbed 1b.2. Finally, we made a translational fusion reporter gene in the context of the entire genomic region. This pnc-1 genomic construct includes 1.4 kbp upstream of the first exon and all introns, with the exception of a portion of intron 2 that could not be cloned (Fig. 1).
We examined two to four individual transgenes for each construct and identified each GFP-positive cell (Table 1). We consistently observed expression predominantly in the head, specifically in a large number of neurons, pharyngeal muscle cells and hypodermal cells (Fig. 2A,B,C,D). The promoter for the secreted isoform has the most restricted expression pattern, with expression largely confined to the head, consistent with previous analysis (Vrablik et al., 2009). Transgenes encoding the secreted PNC-1a isoform occasionally had faint diffuse expression in the gonad extracellular space (Fig. 2E, F and Table 1), suggesting provision of the secreted isoform to this tissue by other cells. We find that the intracellular PNC-1b isoform is expressed from two different promoters with overlapping but not identical expression patterns (Table 1). The genomic construct, which has the most regulatory elements presumably in their intact orientation, has the broadest expression pattern, which is broader than three individual promoters together (Table 1). In the animals with the pnc-1 genomic and pnc-1b.2 promoter transgenes, we detected expression in the anterior cells of the intestine, a tissue not previously reported to express PNC-1 (Fig. 2B). Nonetheless, expression of PNC-1 is not ubiquitous. In particular, we failed to detect expression in several tissues that manifest pnc-1 mutant phenotypes, including the gonad and the gonadal uv1 cells, the body wall muscles, and the egg-laying muscles. Therefore we considered whether the PNC-1 activity that promotes development of the gonad, survival of the uv1 cells or function of the muscle could be provided by non-autonomous expression of pnc-1 in other tissues.
Table 1.
pnc-1 global and promoter-specific expression patterns.
| Cell type 1 | Promoter | ||||
|---|---|---|---|---|---|
| Genomic | 1a | 1b.1 | 1b.2 | ||
| Head | |||||
| Neurons | ADF/L, AFD, AIA, ASG/H/I/K/L, AVA, IL1/2, Retrovesicular ganglion | x | (2) | ||
| ASE, ASJ, AWA/B | x | x | x | ||
| AUA | x | x | |||
| CEP | x | x | x | ||
| IL/OLQso (sheath cells) | x | x | |||
| I2/3, M3 | x | ||||
| I4, M1 | x | x | |||
| I5, M2 | x | ||||
| Other | pm3 | x | x | x | |
| pm4 | x | x | |||
| pm6VR, pm7VR/D, pm8, ventral and dorsal gland cell | x | ||||
| pm7 | x | ||||
| vpi | x | ||||
| hyp1 | x | x | |||
| hyp2/3 | x | ||||
| hyp4 | x | ||||
| hyp6 | x | x | |||
| arcade cells | x | x | x | ||
|
| |||||
| Mid-body | |||||
| Neurons | CANL, ventral cord neurons (occasional) | x | |||
| Other | ccPL+AL, ccDR+ccDL | x | |||
| Intestinal cells | x | x | |||
| vulF (occasional, diffuse) | x | ||||
| uv1 cells | - | - | - | - | |
| Gonad | - | -2 | - | - | |
| Body wall or vulva muscle | - | - | - | - | |
|
| |||||
| Tail | |||||
| Neurons | PHA/B | x | x | x | |
| PVC | x | ||||
| PVT | x | ||||
| Phso1/2 (phasmid sheath cell) | x | x | |||
| Other | RecB | x | |||
| RecF, RecU | x | ||||
Every cell observed for individual promoters is recorded; for the pnc-1 genomic construct some expressing cells may have been overlooked due to brighter neighboring expression sites. Expression of the pnc-1 ΔSS signal sequence mutant was identical to that of the pnc-1 genomic with the additional expression sites of ccPR, ccAR, RecB and Phso1/2. These may correspond to sites of expression of PNC-1a because retention of previously exported GFP due to mutation of the signal sequence would cause expression sites to be revealed. The transgenic arrays used to analyze PNC-1 expression are as follows: pnc-1 genomic, psEx216-9; pnc-1 ΔSS, psEx246-7; pnc-1a, psEx213-5 and 238; pnc-1b.1, psEx222-5; pnc-1b.2, psEx207-8.
Expression previously seen in ASK, DTC, uv2 and rectal gland cells using a pnc-1a::gfp reporter with an unc-54 3′UTR (Vrablik et al., 2009).
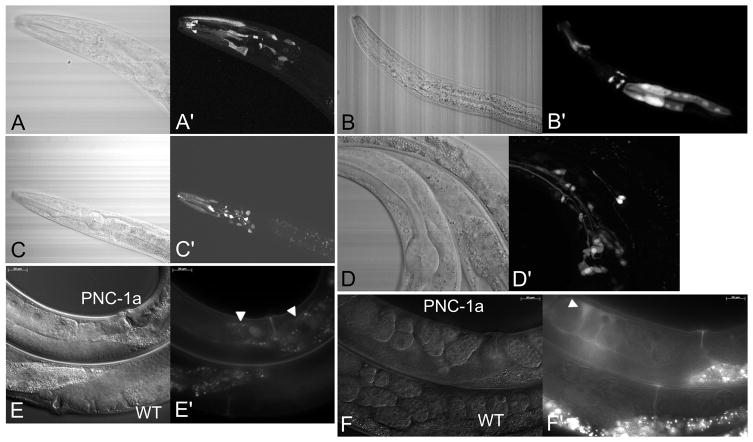
Figure 2.
GFP reporter expression for pnc-1 transgenes. A,A′) pnc-1a transgenic L2 animal, B,B′) pnc-1b.2 transgenic L2 animal, C,C′) pnc-1b.1 transgenic L2 animal, D,D′) pnc-1 genomic transgenic L4 animal. The majority of expression for all constructs was neuronal and in the head and clearly not ubiquitous. pnc-1b.2 expression was also seen in the intestine (B′), a previously unrecognized expression site. E and F) Expression of PNC-1a::GFP results in extracellular fluorescence. Both E and F show one pnc-1a transgenic animal (upper) and one wild-type animal (lower). Transgenic E) young adult hermaphrodites and F) wild-type gravid adult hermaphrodites have faint, diffuse extracellular PNC-1a::GFP in the uterus (white arrowheads in E′ and F′) that is distinct from the autofluorescence visible in both transgenic and non-transgenic animals. Confocal images A to D were captured using an Olympus Fluoview FV1000 microscope and 60x objective, images E and F were captured using a Zeiss Axioplan 2 microscope and x40 oil immersion DIC optic. Confocal GFP images are Z-stacks assembled from scans using ImageJ (Schneider et al., 2012) and corresponding white light images are single pass images from the white light channel of that same scan.
Can PNC-1 function cell non-autonomously?
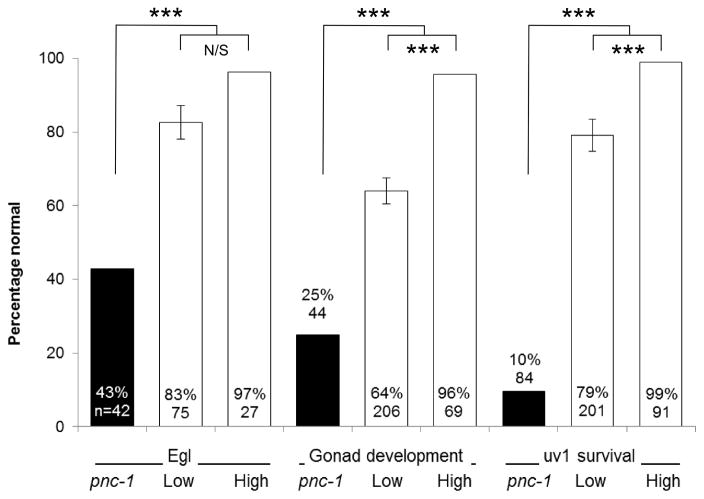
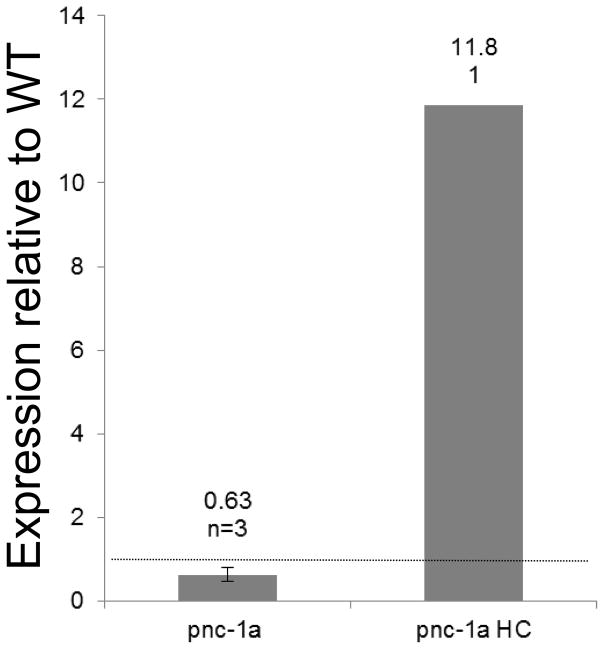
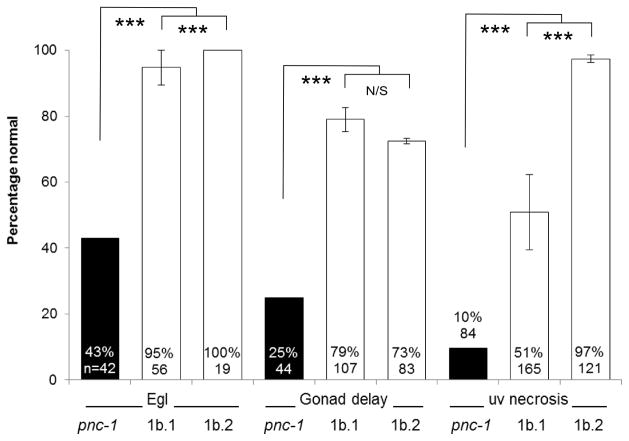
We speculated that the secreted PNC-1a isoform may be critical for providing function to tissues that don’t express PNC-1. To address this hypothesis, we first asked if the secreted PNC-1a isoform was sufficient to provide function to the tissues that have detectable phenotypes in the absence of pnc-1 function. We tested the ability of transgenes that express the secreted PNC-1a isoform from the native pnc-1a promoter to rescue the egg-laying defect, the gonad developmental delay and the uv1 necrosis of the pnc-1 null mutants. These pnc-1a transgenes, which express pnc-1 message at levels comparable to wild type (Fig. 3B), partially rescued all three phenotypes (Fig. 3A). We also engineered animals to express a higher amount of PNC-1a by injecting the vector at a 60-fold higher concentration. This “high copy” array directed higher levels of mRNA transcription (Fig. 3B) and rescued each phenotype almost completely (Fig. 3A). Thus, the expression of the secreted PNC-1a isoform from endogenous expression sites (Fig 2A, E, F and Table 1) is sufficient to provide function to the egg-laying muscles, the gonad and the uv1 cells (Fig. 3A).
Figure 3.
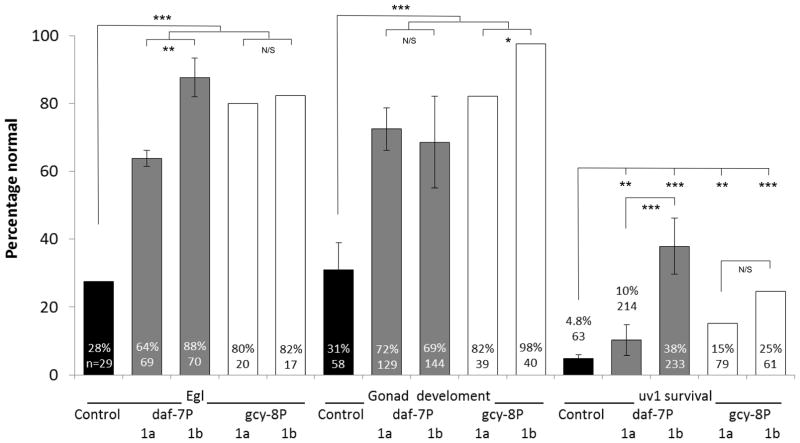
Cell non-autonomous function of PNC-1 isoforms. A) Percentage of pnc-1(pk9605) mutant populations with normal egg-laying (Egl), gonad development and uv1 cells is compared for strains with control transgene or pnc-1a transgenes (Fig. 1) injected at either a low concentration (1a, 1 ng.ml−1, transgenic arrays psEx213-5, 220-1 and 228 (Table 2)) or at a high concentration (1aHC, 60 ng.ml−1, transgenic array psEx238 (Table 2)). Control array psEx236 was used. B) qPCR analysis comparing pnc-1 message levels in pnc-1a arrays injected at two different concentrations. Arithmetic means of ΔΔCt values were calculated for each type of array from the averages of three biological replicates per line. Arrays assayed were pnc-1a: psEx213, 215 and 220; and pnc-1a HC: psEx238. Error bar is S.D. C) The secreted PNC-1a and the intracellular PNC-1b isoforms were expressed individually in the ASI or AFD neurons using the daf-7 promoters (pnc-1a transgenic arrays psEx283-6, pnc-1b transgenic arrays psEx287-290, see Table 2) and gcy-8 promoters (transgenic arrays psEx291 and 292 for pnc-1a and pnc-1b, respectively, see Table 2), respectively, and assayed for rescue of Egl, gonad developmental delay and uv1 necrosis. Control was unc-119(ed3); pnc-1(pk9605). Number of animals assayed and average percentage of the population that were normal for each phenotype for each experiment are shown. gcy-8p::pnc-1a and gcy-8p::pnc-1b experiments were not compared to each other statistically because we had only a single transgenic line for each construct. Error bars are s.e.m. Percentages were analyzed using Fisher’s Exact test. *, ** and *** represent P values of <0.05, <0.01 and <0.001, respectively. n.s. is not significant.
As a more rigorous test of whether the PNC-1 can function cell non-autonomously we expressed the secreted and intracellular isoforms from the highly restricted daf-7 and gcy-8 promoters which are expressed solely in the ASI and AFD neurons, respectively (Ren et al., 1996; Yu et al., 1997; Crook et al., 2010). Both the AFD and ASI neurons are sites of pnc-1 expression from the pnc-1 genomic transgene (Table 1). These transgenes will result in PNC-1 expression as far from the egg-laying muscles, gonad and uv1 cells as possible.
We first confirmed that our constructs were being expressed in the expected neurons and no other cells. Expression was restricted to AFD in all gcy-8 promoter lines; we observed GFP in no other cells. gcy-8P::pnc-1b was expressed diffusely within the cytoplasm of AFD in all transgenic animals. gcy-8P::pnc-1a expression was visible in AFD in only a minority of transgenic animals. In animals where GFP was visible, it was restricted to distinct cytoplasmic puncta. This pattern is consistent with localization of the protein to the secretory apparatus and secretion of the protein into the pseudocoelomic space. The daf-7 promoter-driven constructs directed a lower level of GFP expression but once again expression was completely restricted to only the intended cell, ASI. daf-7P::pnc-1b was detected solely in the ASI neurons in a majority of transgenic animals in all transgenic lines. However, GFP from the daf-7P::pnc-1a transgenes was not detected in any of the four transgenic lines examined, although it clearly functions, again consistent with secretion of this protein into the pseudocoelom.
We then tested these constructs for the ability to rescue the egg-laying (Egl), gonad developmental delay and uv1 necrosis phenotypes. We predicted that the secreted isoform of PNC-1 would show the most robust cell non-autonomous rescue. However, we found that the secreted and intracellular isoforms were both equally able to rescue the Egl and gonad developmental delay phenotypes when expressed in AFD or ASI neurons (Fig. 3C). Thus, both secreted PNC-1a and the intracellular PNC-1b isoform are sufficient for egg-laying muscle function and gonad development when expressed from limited sites. For the uv1 cells, only PNC-1b expressed in the ASI was able to rescue uv1 cell necrosis, and this rescue was not robust relative to the other phenotypes (Fig. 3C). These data show that expression of the secreted PNC-1a from its endogenous promoter or expression of either PNC-1 isoform from two restricted neuronal promoters is sufficient to provide function cell non-autonomously to the egg-laying muscles and the gonad.
Is the secreted PNC-1a isoform necessary in vivo?
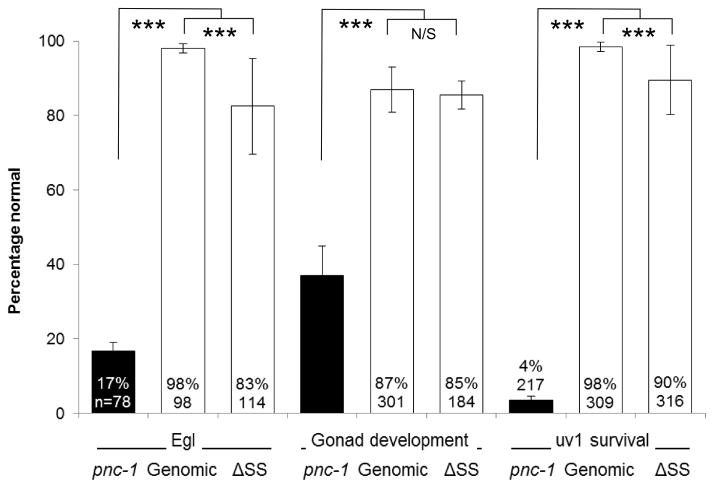
We have shown that expression of the secreted PNC-1a or intracellular PNC-1b isoforms at a distance is sufficient to provide function to other tissues. This still leaves open the question about the importance of the secreted isoform. Is the secretion of PNC-1a necessary for development? To address this question we mutated three codons in the first exon of the pnc-1 genomic transgene to inactivate the pnc-1a signal sequence (see Materials and Methods) and compared the activity of the pnc-1 genomic transgene and the new pnc-1 ΔSS transgene (Fig. 1). We predicted that the pnc-1 genomic arrays would rescue each phenotype and that the pnc-1 ΔSS arrays would fail to fully rescue one or more of the phenotypes tested, due to the lack of secretion of the PNC-1a isoform.
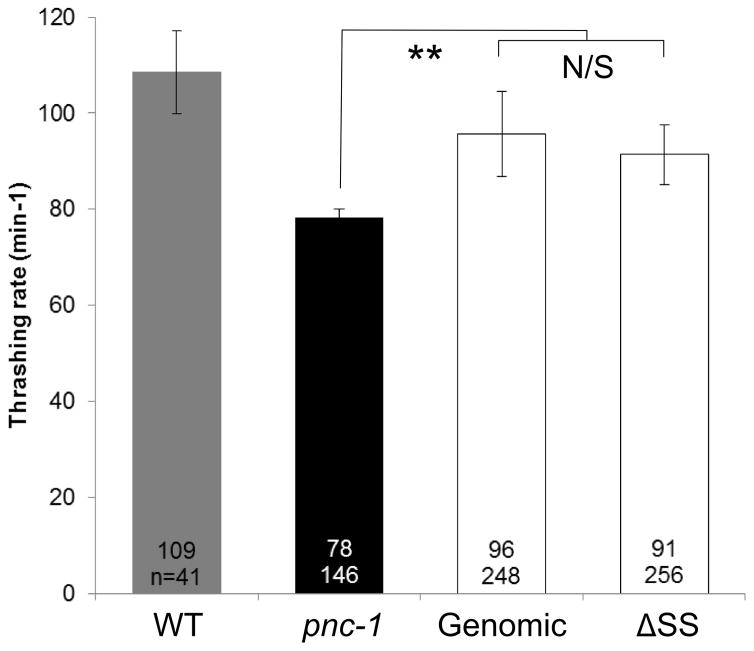
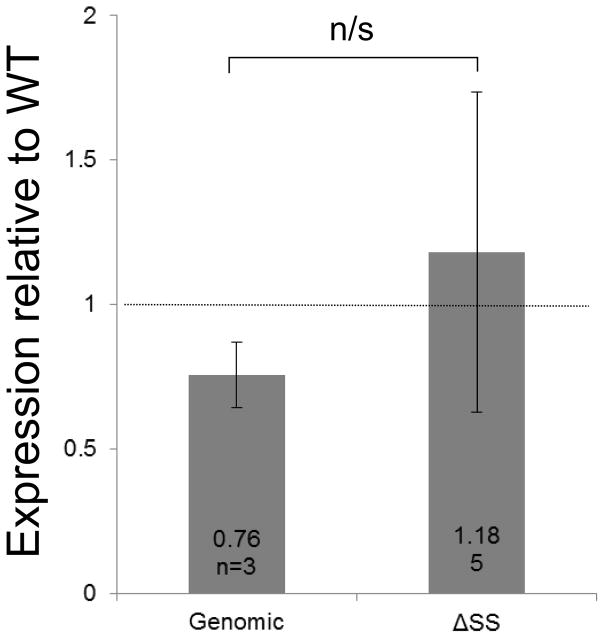
We found that our pnc-1 genomic arrays, which express pnc-1 message at levels comparable to wild type (Fig. 4C), rescued each of the tested phenotypes as expected (Fig. 4A,B). The pnc-1 ΔSS arrays rescued the gonad developmental delay and the body wall muscle thrashing rate as well as the arrays that encoded the secreted version (Fig. 4A,B). Thus, the secreted isoform is not strictly necessary in vivo for optimal PNC-1 activity as it applies to the gonad or the body wall muscle. In the case of the Egl and uv1 necrosis phenotypes, the pnc-1 ΔSS arrays provided robust rescue activity. However, this activity was significantly lower than the arrays with an intact signal sequence (note significant difference between values in Fig. 4A). Moreover, when we compared the standard deviations for the pnc-1 genomic and pnc-1 ΔSS arrays using the F-test we found that they were significantly different for the Egl and uv1 necrosis phenotypes, with values of p= 0.009 and 0.039, respectively, indicating more variability in rescue in the absence of the secreted version. pnc-1 mRNA levels expressed from the genomic and ΔSS arrays were not significantly different (Fig. 4C), suggesting that variation in expression levels between transgene types does not account for the differences in function. The less robust and more variable rescue with the ΔSS arrays suggests that while the secreted PNC-1a isoform is not formally necessary for function it does contribute to optimal function in vivo. In particular, the secreted isoform appears important in phenotypes that are caused by accumulation of NAM indicating the secreted isoform may play an important role in clearance of NAM.
Figure 4.
Is the secreted PNC-1a isoform necessary? Percentage of pnc-1(pk9605) mutant populations with (A) normal egg-laying (Egl), gonad development and uv1 cells and (B) average thrashing rate (expressed as number of body bends per minute) is compared for strains carrying control pnc-1 genomic transgenes (Genomic, transgenic arrays psEx251-2, 265-6, 268, see Table 2) or experimental genomic transgenes that have a mutated signal sequence (ΔSS, transgenic arrays psEx249, 253, 271, 276-8, see Table 2). Number of animals assayed and average result for each experiment are shown. Controls were arrays psEx264 and 273-5, with wild-type non-transgenic animals as positive controls for thrashing rate. Error bars are s.e.m. for a) and S.D. for b). Percentages were analyzed using Fisher’s Exact test and thrashing rate was analyzed using Students T-test. *, ** and *** represent P values of <0.05, <0.01 and <0.001, respectively. n.s. is not significant. (C) qPCR analysis comparing pnc-1 message levels in genomic and ΔSS arrays. Arithmetic means of ΔΔCt values were calculated for each type of array from the averages of three biological replicates per line. Arrays assayed were pnc-1 genomic: psEx251, 252 and 266; and pnc-1 ΔSS: psEx 249, 253, 272, 277 and 278. Error bars are S.D. Means were analyzed using Students T-test. n.s. is not significant.
Can functions be mapped to specific expression sites?
Given that PNC-1b is expressed from two promoters, we sought to test if expression from one or the other was more relevant to PNC-1 activity in vivo. We tested the pnc-1b.1 and pnc-1b.2 promoter translational fusion transgenes (Fig. 1) for their ability to rescue the Egl, gonad developmental delay and uv1 necrosis phenotypes. We observed significantly higher rescue of uv1 necrosis when PNC-1b was expressed from the pnc-1b.2 promoter, compared with the partial rescue from its expression from the pnc-1b.1 promoter (Fig. 5). This differential rescue suggests that PNC-1b expression from sites or at a level specific to the pnc-1b.2 promoter is important for the survival of the uv1 cells. Expression from both promoters was predominantly neuronal with the most obvious difference unique to pnc-1b.2 being expression in the intestine and some pharyngeal cells (Fig. 2B, C and Table 1). We did not see any preference for expression in a particular cell or tissue in terms of the egg-laying muscles or the gonad.
Figure 5.
Functional analysis of two PNC-1b.promoters. The intracellular PNC-1b isoform was expressed at high levels under the control of its two endogenous promoters, 1b.1 (transgenic array psEx222-5, see Table 2) and 1b.2 (transgenic array psEx217-9, see Table 2), and assayed for rescue of Egl, gonad developmental delay and uv1 necrosis. Number of animals assayed and average result for each experiment are shown. Control array psEx236 was used. Error bars are s.e.m. Percentages were analyzed using Fisher’s Exact test. *, ** and *** represent P values of <0.05, <0.01 and <0.001, respectively. n.s. is not significant.
Discussion
PNC-1-mediated NAD+ salvage biosynthesis and clearance of nicotinamide are critical for optimal development and physiology of multiple tissues in C. elegans, including the gonad, the uv1 cells within the gonad, the body wall muscle and the egg-laying muscle (Vrablik et al., 2009; Vrablik et al., 2011). Previous work suggested that PNC-1 was not expressed by body wall muscle cells, and that while the development of the entire gonad was affected in the mutant, the gene was expressed in only a few cells in the gonad (Vrablik et al., 2009). That observation led to the suggestion that PNC-1 might function cell non-autonomously.
Because the expression pattern of PNC-1 was central to our hypothesis that it functions cell non-autonomously, we sought to perform a more rigorous expression analysis. We returned to transgenic reporter gene analysis to search for elements that might direct muscle or gonad expression. We successfully identified another promoter in the genomic locus and fully characterized the expression patterns of all three promoters in isolation as well as expression from a genomic transgene with all three promoters intact, which displayed the broadest expression pattern. Our results are consistent with previous observations that PNC-1 is predominantly expressed in the nervous system (Vrablik et al., 2009), and we revealed expression in the intestine for the first time. It is still formally possible that we have missed a relevant enhancer sequence that drives gonad expression. However, despite extensive effort with multiple transgenes, transgenic lines and different combinations of pnc-1 regulatory sequences, we were unable to detect pnc-1 expression broadly within the gonad or body wall muscle cells. We did detect the secreted PNC-1a::GFP in the uterus, further supporting a potential cell non-autonomous function. In seeking a second method to confirm PNC-1 expression patterns, we have raised antibodies to PNC-1, but these reagents have not been useful in immunohistochemistry experiments (unpublished data).
The presence of multiple promoters suggests the possibility for regulation involving the specific isoforms, i.e. providing PNC-1a versus PNC-1b activity to different tissues in response to demand. While the issue of biologically relevant regulation has yet to be addressed, our analysis does suggest promoter-specific function. Specifically, function to the uv1 cells is provided more efficiently by expression of PNC-1b from the 1b.2 promoter compared with the 1b.1 promoter.
We also sought to experimentally test the hypothesis that PNC-1 can function cell non-autonomously. Our approach was to express the secreted PNC-1a and intracellular PNC-1b isoforms from promoters with a restricted expression pattern. We note that despite having the most restricted expression pattern of the three pnc-1 promoters, 1a-driven expression of secreted PNC-1a was able to robustly rescue all the phenotypes we examined. We suggest that even though this promoter is most restricted in terms of number of tissues we could detect expression from, the pattern of protein localization is likely broader given that the secreted protein would have access to the entire animal via the pseudocoelomic space (see Fig. 2E,F). More surprisingly, expression of the intracellular PNC-1b from either of its endogenous promoters was able to provide function to the egg-laying muscles and gonad, although they differed in their ability to provide function to the uv1 cells. As a more rigorous test of our cell non-autonomous hypothesis we used two promoters to drive expression solely in a single pair of neurons. Expression of either secreted PNC-1a or intracellular PNC-1b in the ASI or AFD neurons was able to robustly rescue the egg-laying and gonad developmental delay phenotypes, although they were either ineffective or much less effective at rescuing the uv1 necrosis phenotype. These data provide strong support for our hypothesis that PNC-1 can function cell non-autonomously and revealed an unexpected role for the intracellular form in providing non-autonomous function. Cell non-autonomous function by the intracellular isoform suggests that the substrate and product of PNC-1 are moving between cells (see below).
Even though intracellular PNC-1 can function at a distance, we were still interested in understanding if secretion of PNC-1 is important to function in the animal, especially given the controversy over the role of secreted eNampt in vertebrates. Although salvage synthesis itself is accomplished via distinct proteins in vertebrates and invertebrates, secretion of the protein which metabolizes NAM is conserved between species. The C. elegans system thus provides a great experimental system to test if secretion is necessary for the organism. To determine if the secretion of PNC-1a is necessary we compared the rescue of two pnc-1 genomic transgenes that were identical except for the presence or absence of a functional pnc-1a signal sequence. We found that pnc-1 genomic transgenes lacking the PNC-1a signal sequence were still able to robustly rescue, which is consistent with our surprising result regarding cell non-autonomous function of intracellular PNC-1b. However, for two NAM-induced phenotypes our evidence hints that the secretion of PNC-1a was biologically and functionally relevant. The transgenic lines that lacked the signal sequence were mildly but statistically significantly reduced in activity and more variable in activity. The simple model we had originally envisioned, where the secreted PNC-1a provides the cell non-autonomous function and the intracellular PNC-1b provides the cell autonomous function is not supported. Instead, we have shown that intracellular PNC-1b is sufficient while the PNC-1a isoform makes a minor contribution, specifically in preventing accumulation of NAM.
Conditions that restore NAD+ biosynthetic capacity to the animal rescue gonad development (Vrablik et al., 2011) whereas NAM conversion to NA is likely responsible for rescue of uv1 cells (Vrablik et al., 2009). PNC-1 provides both of these activities, but how does PNC-1 provide function to a range of tissues when expressed at a distance from these tissues and in a limited number of cells? And why does uv1 cell survival seem to be the phenotype with the most stringent requirement for PNC-1 function, in terms of both sensitivity to the loss of the secreted isoform and requirement for a specific expression site (or level)? We have considered several possibilities for how PNC-1 functions at a distance. First, it may be that distant cells actually direct the development of the gonad or the activity of the muscle. Under this scenario those cells would have to be AFD and ASI acting redundantly. We consider it unlikely that these two pairs of sensory neurons have such distinct functions and previous experiments looking at function of these cells revealed no such activities (Ren et al., 1996; Schackwitz et al., 1996; Kimura et al., 2004; Clark et al., 2006; Beverly et al., 2011). Second, it may be that metabolites are moved between cells and metabolic activity by the intracellular isoform in only two neurons, including their processes which extend to ¼ the length of the animal is adequate to provide NA to the whole animal or at least to key control cells in the vicinity of these neurons. Both the PNC-1 substrate NAM and product NA move between cells both in yeast and in metazoans (Llorente and Dujon, 2000; Houtkooper et al., 2010; Belenky et al., 2011; Nikiforov et al., 2011). A nicotinic acid permease for NA import has been identified in yeast (Llorente and Dujon, 2000), but no known exporter and no NAM transporters have been identified. It is not surprising that widespread expression of intracellular PNC-1 could help prevent NAM, produced from NAD+ hydrolyzing enzymes in many cell types, from accumulating globally. However it was a surprise that uptake and processing of NAM by only two cells would be sufficient to keep NAM levels from accumulating to levels that are toxic to uv1 cells at a distance. Similarly, production of NA or a downstream metabolite in a pair of neurons appears to be sufficient to supply NAD+ biosynthetic precursors more broadly to the animal. In the case of the secreted isoform, it is likely that PNC-1a converts NAM to NA in the pseudocoelomic space, either to supply NA to nearby cells that can further process it to NAD+ or to reduce extracellular NAM to sub-toxic levels.
Why do uv1 cells have the most stringent requirement for PNC-1 activity? It is likely that the uv1 cells are peculiarly sensitive to NAM or that NAM levels in their vicinity are higher. However, the need to metabolize NAM also likely relies disproportionally on PNC-1 relative to the need to synthesize NAD+ because there are other NAD+ biosynthetic pathways that can contribute to the production of NAD+ (Wang et al. submitted), but there is not the same flexibility in terms of mechanisms to clear toxic buildup of NAM. This disparity may explain the importance of secreted PNC-1a or proximal intracellular PNC-1b activity to uv1 survival. Further work on the mechanism by which NAM causes uv1 necrosis may help answer resolve these different possibilities.
In summary, the picture of how NAD+ salvage activity is provided to different tissues in C. elegans is more complex than we first thought. Although we have shown that PNC-1 can function cell non-autonomously, secretion is clearly not vital to cell non-autonomous PNC-1 function. It is likely that PNC-1 functions in a variety of roles depending on the tissue in question, its metabolic needs, including its requirement for NAD+ and its sensitivity to NAM, and this ability to provide function to and from a range of tissues may provide the robustness we would expect for a pathway as vital as NAD+ biosynthesis.
Materials and Methods
C. elegans strains and maintenance
Strains were maintained under standard conditions at 20 °C (Brenner, 1974). We used the following strains; N2 (wild type), MH968 unc-119(ed3) III, HV723 pnc-1(pk9605) IV and HV554 unc-119(ed3) III; pnc-1(pk9605) IV. To determine the expression patterns of pnc-1 and its individual promoters we generated extrachromosomal arrays by injecting hermaphrodites with LiCl purified plasmid DNA at concentrations between 10 and 110 ng.μl−1 (Table 2).
Table 2.
Constructs, primers and transgenes used in this study.
| Construct | Vector | Parent plasmid |
Primers (5′-3′)1 | Transgene psEx# |
Background/ coinjection marker(ng.μl−1)2 |
ng. μl−1 |
|---|---|---|---|---|---|---|
| pnc-1 genomic | pMC1 | pPD95.67 | Restriction linker F GCATGCGCTAGCCTGCAGGCGGCCGCTCTAGA Restriction linker R TCTAGAGCGGCCGCCTGCAGGCTAGCGCATGC pnc-1 F (Pst I) AACTGCAGAATCGCCAACTGGATCTTC pnc-1 R (Not I) AAGCGGCCGCCTTCTTCACGATCCTTTGAACCCATTC pnc-1 Fa (Sph I) GCATGCCAGGCTTTTCTTGACTTTCTGTTACTTTTT pnc-1 Ra (Nhe I) GCTAGCATTTTACCTCTCTAAACTCCGAAAAC pnc-1 F9 (Nhe I) AAGCTAGCGTAAAATGAATGTTTCCGTGTTGTTGTTGTG pnc-1 R18 (Pst I) AACTGCAGCCCAAGCCAAGATTCCATAATCCTCT |
216-9 | unc-119; pnc-1/unc-119(+)(60) | 20 |
| 251-2, 265-6, 268 | pnc-1/sur-5::gfp(60) and pBS KS-(39) | 1 | ||||
|
| ||||||
| pnc-1 genomic ΔSS | pMC11 | pPD95.67 |
pnc-1 F17 (Kas I) GAGGCGCCACATCTCTTCCC pnc-1 R25 (EcoR V) CTGGATATCGTTTTAAACGCTCTAAACGAG pnc-1 F18 (mutagenesis linker) GCATTTATTGaGCaCAagTGGGTGA3 pnc-1 R26 (mutagenesis linker) TCACCCActTGtGCtCAATAAATGC |
246-7 | unc-119/unc-119(+)(60) | 20 |
| 249, 253, 271, 276-8 |
pnc-1/sur-5::gfp(60) and pBS KS-(39) |
1 | ||||
|
| ||||||
| pnc-1a | pMC4 | pPD95.69 | Common to pMC4/5/6 pnc-1 F16 (EcoR I) AAGAATTCAAGTAATAATAAAATATTGTGCATAATTTTGTAAT pnc-1 R24 (Spe I) AAACTAGTGAGGAGTACACGGCGCGT pPD118.20 R1 CCAAGCGAGGACAATTCTCATCG GFP F7 ATGGGTTCAAAGGATCGTGAAGAAGATGAGTAAAGGAGAAGAACTTTTCACTGG pnc-1a/b CM R4 CCAGTGAAAAGTTCTTCTCCTTTACTCATCTTCTTCACGATCCTTTGAACCCAT Specific to pMC4 pnc-1a/b CM F8 ATGTTTCCCTGCCCAAAGCTTATATTCC pnc-1 F3 (Pst I) AACTGCAGTTTTCCAGGCTTTTCTTGACTTTCTGTTAC pnc-1 R4 (Xba I) AATCTAGACAAGAGAATTTGATCTAAGAATGTATTCTCCAG |
213-5, 220-1, 228 | unc-119; pnc-1/unc-119(+)(60) | 1 |
| 238 | “ | 60 | ||||
|
| ||||||
| pnc-1b.1 | pMC5 | pPD95.69 | Common to pMC5 and 6 pnc-1a/b CM F9 CAGCTAATTCCATTTTTGCTCCTAGC Specific to pMC5 pnc-1 F4 (Pst I) AACTGCAGGTGAGTTTTGGTGGATAGGACGGT pnc-1 R5 (Nhe I) AAGCTAGCCTTCGTTTCCGCGGAAACGAGTG |
222-5 | pnc-1/- | 10 |
|
| ||||||
| pnc-1b.2 | pMC3 | pPD95.69 |
pnc-1 F2 (Pst I) CTGCAGGTAAAATGAATGTTTCCGTGTTGTTGTTGTG pnc-1 R3 (Sal I) GTCGACCATCTACAACAATACAAACTATTTGCGAACAAC |
207-8 | unc-119/unc-119(+)(60) | 20 |
| pMC6 | 217-9 | pnc-1/- | 10 | |||
|
| ||||||
| daf-7 pnc-1a/ b | pMC13/14 | TU#61/2 |
daf-7 CF12 (Sph I) GCATGCGTAAACACTTTTCAGATTTTTCAAATTTCAG daf-7 CR14 (Xma I) CCCGGGGAACATGGCTGAACTTCAAGC |
pnc-1a: 283-6 | unc-119; pnc-1/unc-119(+)(60) | 10 |
|
pnc-1b: 287–290 |
||||||
|
| ||||||
| gcy-8 pnc-1a/ b | pMC15/16 | TU#61/2 |
gcy-8 F1 (Sph I) GCATGCCTTCTCCTTGACGAGTCTTCC gcy-8 R1 (Xma I) CCCGGGTTTGATGTGGAAAAGGTAGAATCG |
pnc-1a: 291 | unc-119; pnc-1/unc-119(+)(60) | 10 |
| pnc-1b: 292 | ||||||
Restriction enzymes in parentheses refer to the enzymes used in the initial cloning step with fragments generated using these primers. Subsequent sub-cloning steps are detailed in the methods section.
Coinjection markers used were either pDP#MM016B (unc-119(+)) DNA (Maduro and Pilgrim, 1995) or sur-5::gfp DNA (pTG96_1 (Gu et al., 1998)) and pBluescript KS- to a total of 100ng.ul−1.
Nucleotides in lowercase are those changed to inactivate the pnc-1a signal sequence.
Transgene construction
The pnc-1 genomic locus and the transgenes used in this study are shown in Figure 1. For a full list of primers and construction of transgenic animals see Table 2. Four control arrays (psEx264 and psEx273-5) in a pnc-1(pk9605) background were generated by injecting pnc-1(pk9605) hermaphrodites with 60 ng.μl−1 of sur-5::gfp (Gu et al., 1998) and 40 ng.μl−1 pBluescript KS-DNA; one control array (psEx236) in an unc-119(ed3); pnc-1(pk9605) background was generated by injecting pnc-1(pk9605) hermaphrodites with 60 ng.μl−1 of unc-119(+) DNA (Maduro and Pilgrim, 1995). All constructs were confirmed by sequencing.
pMC1: we first added a multiple cloning site, containing Nhe I, Pst I and Not I, to pPD95.67 (Addgene plasmid 1490) using Sph I/ Xba I. We then sequentially cloned a fragment containing the upstream sequence to the end of exon 1b using pnc-1F1/Sph I and pnc-1 R2/Nhe I, a second fragment containing intron1b to exon2 using pnc-1 F2/ Nhe I and pnc-1 R18/Pst I and a final fragment containing exon 3 to the end of exon 4 using pnc-1 F/Pst I and pnc-1 R/Not I. We were unable to clone the 1 kbp intron 2 using multiple strategies, possibly due to the presence of a predicted hairpin loop structure.
pMC11: we mutated the exon 1a signal sequence (Fig. 1) by extension overlap PCR, changing the hydrophobic glycine-leucine-isoleucine amino acids to polar glutamic acid-histidine-lysine amino acids to inactivate the signal sequence, and then used this to replace the original sequence by Kas I/ EcoR V restriction cloning.
pMC4 and 5: the pnc-1a and -1b cDNAs were each translationally fused by extension overlap PCR to a non-nuclear localized GFP from pPD118.20 (Addgene plasmid 1592). After replacing the unc-54 3′UTR of pPD95.69 (Addgene plasmid 1491) with the 185 bp long pnc-1 3′UTR using EcoR I/ Spe I, these cDNA::gfp fusions were inserted upstream by Xma I/ EcoR I. Finally, the 1a and 1b.1 promoters were cloned upstream of their respective cDNAs by PstI/ XbaI and PstI/ NheI, respectively.
pMC3 and 6: a 3 kbp intron between exon 1b and exon 2 was first cloned as a transcriptional fusion into pPD95.69 using pnc-1 F2/ PstI and pnc-1 R3/ SalI. Once the presence of a third promoter was confirmed, it was subcloned upstream of the pnc-1b::gfp described above by Pst I/ Xma I to create the pnc-1b.2 transgene.
pMC13 to 16: the pnc-1a and -1b cDNAs were each placed under the control of either 1.2kbp of the daf-7 promoter (pMC13 and 14, respectively) or 2.1kbp of the gcy-8 promoter (pMC15 and 16, respectively). daf-7 is expressed solely in the ASI neurons (Ren et al., 1996; Schackwitz et al., 1996; Crook et al., 2010) and gcy-8 is expressed solely in the AFD neurons (Yu et al., 1997). The respective promoters were cloned using SphI/ XmaI upstream of either pnc-1a or pnc-1b cDNAs translationally fused with GFP.
Phenotypic Assays
Egg-laying
For each strain 20 to 30 L4 hermaphrodites were placed onto individual plates and scored for the “bag of worms” Egl phenotype after two, three and four days at 20 °C. Sterile adults and those that died from anything other than bagging were removed from the analysis. The Egl phenotype is expressed as the percentage of the remaining egg-laying adults that had not bagged by day four.
Gonad delay
Gonad development was scored in the mid-L4 stage by examining stage of uterine development relative to the stage of vulval development as described previously (Vrablik et al., 2009). In wild-type animals, the lumen in both arms of the uterus is open and they are joined above the mid-L4 stage vulva, which has a stereotypical and easily recognizable “Christmas tree” morphology. In the pnc-1 mutants, the uterine lumen is not open or is not connected between the two arms by the time the vulva displays the typical mid-L4 stage “Christmas tree” morphology. The phenotype is reported as the percentage of L4 animals with a fully open and connected uterine lumen.
uv1 necrosis
In pnc-1 mutants approximately 95% of the four uv1 cells at the uterine-vulval junction die by necrosis soon after they are specified (Huang and Hanna-Rose, 2006; Vrablik et al., 2009). As it is technically challenging to accurately score the actual number of dying uv1 cells on both sides of the animal by DIC, the late L4 animals were scored for presence or absence of necrotic uv1 cells. The phenotype is reported as percentage of animals without any necrotic uv1 cells.
Muscle function
Muscle function was assessed by measuring thrashing rate, the number of body bends per minute, as described previously (Vrablik et al., 2011). Wild-type non-transgenic animals were used as positive controls.
qPCR analysis of pnc-1 transgene expression
Total RNA was extracted using TRIzol® Reagent (Life Technologies) according to the manufacturer’s instructions. 2 μg total RNA was reverse transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems).
Real-time quantitative PCR amplifications were carried out using SYBR Green (PerfeCTa SYBR Green Super Mix with ROX, Quanta Biosciences) in a StepOnePlus Real-Time PCR System (Life Technologies). Reaction profile was 95°C for 10 min then 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds, followed by a melting curve analysis was carried out (60°C to 95°C) to verify the specificity of amplicons. pnc-1 specific primers were: pnc-1-F tcaacgataataacgggcggtcca and pnc-1-R ccaggcaagtaaaccgaacgcaaa. Three internal reference genes; cdc-42, pmp-3 and tba-1, were used as described previously (Hoogewijs et al., 2008). Three technical replicates were carried out for each of three biological replicates and the ΔCt value for each sample was calculated by subtracting the geometric mean of the reference gene Ct values from that of pnc-1. The ΔΔCt values for each biological replicate were calculated by subtracting the experimental pnc-1 ΔCt value from the wild-type ΔCt value and then transformed using 2ΔΔCt (Livak and Schmittgen, 2001). Finally, transformed ΔΔCt values were normalized using the percentage transgenic animals in each biological replicate.
Bullet points.
PNC-1 expression is not detected in tissues with phenotypes, suggesting non-autonomous function
Either the secreted PNC-1a isoform or the intracellular PNC-1b isoform is sufficient to provide function in vivo
The secreted PNC-1 isoform contributes to in vivo activity
Intracellular and extracellular PNC-1 isoforms function cell non-autonomously in C. elegans development and cell survival.
Acknowledgments
Grant Sponsor and Number: NIH grant R01GM086786 to WHR
This work was funded by NIH grant R01GM086786 to WHR. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). The authors would like to thank Kang Zhou and Sherry Coven for their work in cloning the pnc-1 genomic region and Coral Martinez for her work cloning the pnc-1a and -1b isoforms.
References
- Belenky P, Stebbins R, Bogan KL, Evans CR, Brenner C. Nrt1 and Tna1-independent export of NAD+ precursor vitamins promotes NAD+ homeostasis and allows engineering of vitamin production. PLoS ONE. 2011;6:e19710. doi: 10.1371/journal.pone.0019710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverly M, Anbil S, Sengupta P. Degeneracy and neuromodulation among thermosensory neurons contribute to robust thermosensory behaviors in Caenorhabditis elegans. J Neurosci. 2011;31:11718–11727. doi: 10.1523/JNEUROSCI.1098-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieganowski P, Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell. 2004;117:495–502. doi: 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Clark DA, Biron D, Sengupta P, Samuel AD. The AFD sensory neurons encode multiple functions underlying thermotactic behavior in Caenorhabditis elegans. J Neurosci. 2006;26:7444–7451. doi: 10.1523/JNEUROSCI.1137-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook M, Grant K, Grant WN. Failure of Parastrongyloides trichosuri daf-7 to complement a Caenorhabditis elegans daf-7 (e1372) mutant: Implications for the evolution of parasitism. Int J Parasitol. 2010;40:1675–1683. doi: 10.1016/j.ijpara.2010.07.003. [DOI] [PubMed] [Google Scholar]
- French JB, Cen Y, Vrablik TL, Xu P, Allen E, Hanna-Rose W, Sauve AA. Characterization of nicotinamidases: steady state kinetic parameters, classwide inhibition by nicotinaldehydes, and catalytic mechanism. Biochemistry. 2010;49:10421–10439. doi: 10.1021/bi1012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzaniga F, Stebbins R, Chang SZ, McPeek MA, Brenner C. Microbial NAD metabolism: lessons from comparative genomics. Microbiol Mol Biol Rev. 2009;73:529–541. doi: 10.1128/MMBR.00042-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T, Orita S, Han M. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol Cell Biol. 1998;18:4556–4564. doi: 10.1128/mcb.18.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara N, Yamada K, Shibata T, Osago H, Tsuchiya M. Nicotinamide phosphoribosyltransferase/visfatin does not catalyze nicotinamide mononucleotide formation in blood plasma. PloS ONE. 2011;6:e22781. doi: 10.1371/journal.pone.0022781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren J. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Molecular Biol. 2008;9:1–8. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Cantó C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocrine Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Hanna-Rose W. EGF signaling overcomes a uterine cell death associated with temporal mis-coordination of organogenesis within the C. elegans egg-laying apparatus. Dev Biol. 2006;300:599–611. doi: 10.1016/j.ydbio.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gévry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Miyawaki A, Matsumoto K, Mori I. The C. elegans thermosensory neuron AFD responds to warming. Curr Biol. 2004;14:1291–1295. doi: 10.1016/j.cub.2004.06.060. [DOI] [PubMed] [Google Scholar]
- Landry J, Slama JT, Sternglanz R. Role of NAD+ in the deacetylase activity of the SIR2-like proteins. Biochem Biophys Res Commun. 2000;278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Llorente B, Dujon B. Transcriptional regulation of the Saccharomyces cerevisiae DAL5 gene family and identification of the high affinity nicotinic acid permease TNA1 (YGR260w) FEBS Lett. 2000;475:237–241. doi: 10.1016/s0014-5793(00)01698-7. [DOI] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magni G, Amici A, Emanuelli M, Raffaelli N, Ruggieri S. Enzymology of NAD+ synthesis. Adv Enzymol. 1999;73:135–182. doi: 10.1002/9780470123195.ch5. [DOI] [PubMed] [Google Scholar]
- Nikiforov A, Dölle C, Niere M, Ziegler M. Pathways and subcellular compartmentation of NAD biosynthesis in human cells: FROM ENTRY OF EXTRACELLULAR PRECURSORS TO MITOCHONDRIAL NAD GENERATION. J Biol Chem. 2011;286:21767–21778. doi: 10.1074/jbc.M110.213298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocón-Grove OM, Krzysik-Walker SM, Maddineni SR, Hendricks GL, Ramachandran R. NAMPT (visfatin) in the chicken testis: influence of sexual maturation on cellular localization, plasma levels and gene and protein expression. Reproduction. 2010;139:217–226. doi: 10.1530/REP-08-0377. [DOI] [PubMed] [Google Scholar]
- Preiss J, Handler P. Biosynthesis of Diphosphopyridine Nucleotide: I. Identification of intermediates. J Biol Chem. 1958a;233:488–492. [PubMed] [Google Scholar]
- Preiss J, Handler P. Biosynthesis of Diphosphopyridine Nucleotide: II. Enzymatic aspects. J Biol Chem. 1958b;233:493–500. [PubMed] [Google Scholar]
- Ren P, Lim CS, Johnsen R, Albert PS, Pilgrim D, Riddle DL. Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science. 1996;274:1389–1391. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- Revollo JR, Grimm AA, Imai S. The regulation of nicotinamide adenine dinucleotide biosynthesis by Nampt/PBEF/visfatin in mammals. Curr Opin Gastroenterol. 2007a;23:164–170. doi: 10.1097/MOG.0b013e32801b3c8f. [DOI] [PubMed] [Google Scholar]
- Revollo JR, Körner A, Mills KF, Satoh A, Wang T, Garten A, Dasgupta B, Sasaki Y, Wolberger C, Townsend RR, Milbrandt J, Kiess W, Imai S-i. Nampt/PBEF/Visfatin regulates insulin secretion in β cells as a systemic NAD biosynthetic enzyme. Cell Metabol. 2007b;6:363–375. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A, Andris F, Van Gool F, Leo O. Reconstructing eukaryotic NAD metabolism. Bioessays. 2003;25:683–690. doi: 10.1002/bies.10297. [DOI] [PubMed] [Google Scholar]
- Rongvaux A, Shea RJ, Mulks MH, Gigot D, Urbain J, Leo O, Andris F. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur J Immunol. 2002;32:3225–3234. doi: 10.1002/1521-4141(200211)32:11<3225::AID-IMMU3225>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Schackwitz WS, Inoue T, Thomas JH. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron. 1996;17:719–728. doi: 10.1016/s0896-6273(00)80203-2. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skokowa J, Lan D, Thakur BK, Wang F, Gupta K, Cario G, Brechlin AM, Schambach A, Hinrichsen L, Meyer G, Gaestel M, Stanulla M, Tong Q, Welte K. NAMPT is essential for the G-CSF-induced myeloid differentiation via a NAD+-sirtuin-1-dependent pathway. Nat Med. 2009;15:151–158. doi: 10.1038/nm.1913. [DOI] [PubMed] [Google Scholar]
- Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc Natl Acad Sci U S A. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A, Schavemaker JM, Pellis-van Berkel W, Burgering BMT. The Caenorhabditis elegans nicotinamidase PNC-1 enhances survival. Mech Ageing Dev. 2007;128:346–349. doi: 10.1016/j.mad.2007.01.004. [DOI] [PubMed] [Google Scholar]
- van der Veer E, Nong Z, O’Neil C, Urquhart B, Freeman D, Pickering JG. Pre–B-Cell Colony–Enhancing Factor regulates NAD+-dependent protein deacetylase activity and promotes vascular smooth muscle cell maturation. Circ Res. 2005;97:25–34. doi: 10.1161/01.RES.0000173298.38808.27. [DOI] [PubMed] [Google Scholar]
- Vrablik TL, Huang L, Lange SE, Hanna-Rose W. Nicotinamidase modulation of NAD+ biosynthesis and nicotinamide levels separately affect reproductive development and cell survival in C. elegans. Development. 2009;136:3637–3646. doi: 10.1242/dev.028431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrablik TL, Wang W, Upadhyay A, Hanna-Rose W. Muscle type-specific responses to NAD+ salvage biosynthesis promote muscle function in Caenorhabditis elegans. Dev Biol. 2011;349:387–394. doi: 10.1016/j.ydbio.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Veer Evd, Frontini MJ, Thibert V, O’Neil C, Watson A, Szasz P, Chu MWA, Pickering JG. Intrinsic directionality of migrating vascular smooth muscle cells is regulated by NAD+ biosynthesis. J Cell Sci. 2012;1:5770–5780. doi: 10.1242/jcs.110262. [DOI] [PubMed] [Google Scholar]
- Yonezawa T, Haga S, Kobayashi Y, Takahashi T, Obara Y. Visfatin is present in bovine mammary epithelial cells, lactating mammary gland and milk, and its expression is regulated by cAMP pathway. FEBS Lett. 2006;580:6635–6643. doi: 10.1016/j.febslet.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Yu S, Avery L, Baude E, Garbers DL. Guanylyl cyclase expression in specific sensory neurons: A new family of chemosensory receptors. Proc Natl Acad Sci U S A. 1997;94:3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]