Abstract
Here, we present an affinity membrane chromatography technique for purification of monoclonal and polyclonal antibodies from cell culture media of hybridomas and ascites fluids. The m-NBST method utilizes the nucleotide-binding site (NBS) that is located on the Fab variable domain of immunoglobulins to enable capturing of antibody molecules on a membrane affinity column via a small molecule, tryptamine, which has a moderate binding affinity to the NBS. Regenerated cellulose membrane was selected as a matrix due to multiple advantages over traditionally used resin-based affinity systems. Rituximab was used for proof of concept experiments. Antibody purification was accomplished by first capture of injected samples while running equilibration buffer (50 mM sodium phosphate pH 7.0), followed by elution achieved by running a gradient of mild elution buffer (3 M NaCl in 50 mM phosphate pH 7.0). The results indicate that the m-NBST column efficiency for Rituximab was >98%, with a purity level of >98%. The quality and the capacity of this small molecule membrane affinity purification method is further evaluated for a number of parameters such as: injection concentrations, volumes, wash/bind time, elution gradient, antibody/protein-contaminant combinations, effects of injection buffer, post-purification antigen binding activity of antibodies, and column reusability and stability.
Introduction
Antibodies are employed in a vast array of applications including clinical, academic and industrial, while new implementations are continuously being explored in academic research.1-6 In applications relating to clinical use, antibodies are required to have very high purity.7-9 Typically, such purification processes necessitate four to five independent downstream processing steps including primary recovery steps, adsorption of antibodies, polishing steps and finally buffer exchange and concentration steps.7,10-12 More than 50-80% of the total cost of protein production is reportedly due to these downstream steps.9,13 The leading method for antibody purification, whether for clinical or other applications, is affinity chromatography, where protein A (or protein G) functions as the capture molecule.9,12,14-16 In a single step, protein A (or G) will capture the antibody by binding to its Fc domain, while the contaminants, such as proteins, DNA, and other impurities from the cell culture process, are removed by washing away. Despite this method's high yield and purity (>90%),12,17 several problems exist associated with its use, i) high production cost, ii) leaching of protein A (or G) fragments, iii) requirement of low pH elution buffers that may denature antibodies, etc.18-26 Alternative to protein A (or G) methods, several approaches have been investigated in the search for simple, selective and stable capture molecules.27-29 Generally, the simpler the capture molecules, the more stable the column would be to harsh chemical procedures for elution and cleaning. Therefore, small molecules and peptides are being examined, by us and others, to be used as an alternative to protein A (or G) for antibody purification with varying success.27,28,30-32 Amino acid derivatives33,34, dyes9, and thiophilic compounds35 are some of the small molecules that are mostly proposed for capturing antibodies. Additionally, computational studies were performed to determine small molecules or peptides, binds on different regions of antibodies, to be used for purification purposes.36-39
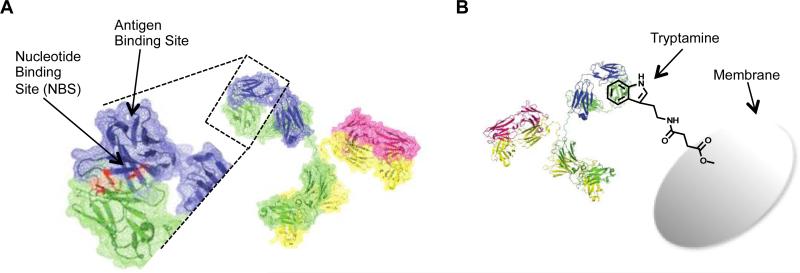
Recently, we reported a new type of affinity chromatography method for antibody purification that was developed in our laboratory by utilizing the not-so-known nucleotide-binding site (NBS) on the antibody.28 The NBS is located between heavy and light chains of the variable region of the Fab arms, and although it has no known function, it is a highly conserved region in almost all antibodies (Figure 1-A).40-42 In previous studies, we extensively characterized the nucleotide-binding site using molecular modeling, and identified the four critical residues, two tyrosine residues on the variable region of light chain (Tyr42 and Tyr103) and one tyrosine (Tyr103) and one tryptophan (Trp118) on the variable region of heavy chain.40,41 Through computational and experimental screening, we identified that the NBS has moderate binding affinities to a set of small hydrophobic, ring structured molecules such as tryptamine and indole-3-butyric acid (IBA) with Kd = 1-8 μM.43 In earlier studies, we demonstrated a wide-range of uses for the NBS from biosensor applications to allergic response inhibition.41-50 Targeting the NBS using the NBS ligands provides nearly limitless applications in site-specific conjugation of various functionalities (drugs, fluorescent tags, biotin tags, reactive sulfur, etc.) to antibodies. Furthermore, we also demonstrated that NBS ligand functionalized resins provide an easy and stable affinity chromatography method for antibody purification.28
Figure 1.
A) Location of the nucleotide-binding site (NBS) is shown in red on the crystal structure of the antibody Fab variable domain (pdb: 2OSL, Rituximab Fab structure). The NBS consists of four residues, namely two tyrosine residues on the variable region of the light chain (Tyr42 and Tyr103) and one tyrosine (Tyr103) and one tryptophan residue on the variable region of heavy chain (Trp118). Positions of the residues determined based on IMGT numbering.58 B) Cartoon representation of antibody capture with tryptamine-conjugated membrane. Membrane conjugated tryptamine will recognize the NBS, and thereby capture the antibody on the column, meanwhile contaminant proteins and impurities will be washed away with the EQ Buffer. When EQ buffer is changed to ELS buffer containing a high salt concentration, antibody would be eluted from the column delivering high recovery and purity.
In this paper, we describe an affinity membrane chromatography technique for antibody purification that uses NBS ligand functionalized membranes for antibody capture. We implemented the NBS targeting approach to the membrane chromatography platform, because it has several advantages over the resin-based affinity chromatography methods. Membrane provides a well-controlled macro-porous polymeric stationary phase, which leads to lower pressure drops, allowing higher flow rates.14,51 Membrane based chromatography generally can be distinguished from resin-based chromatography through its interaction between a solute and a matrix (immobilized ligand) and does not take place in the dead-ended pores of particle, but mainly in the through pores of a membrane. As a result of convective flow of the solution through the pores, the mass transfer resistance is reduced and rapid processing, which improves the adsorption, washing, elution and regeneration steps, can be achieved.14,52 The binding efficiency is typically independent of the feed flow-rate over a wide range; as a result, very high flow-rates may be used.53,54 Consecutively, a larger sample size can be processed in a relatively short time with high recovery of antibody.14 Additionally, easy packing and scale up facilities of membranes make them more preferable in antibody purification systems.51,55,56 Synthesis and production of membranes is relatively easy and not costly, making their replacement reasonable, which eliminates requirements for cleaning and equipment revalidation, making them especially attractive for pharmaceutical applications.53,57 All of these features and advantages over resin-based systems make membranes a better candidate to be used in affinity chromatography systems. Here, we demonstrate a small molecule affinity membrane chromatography method by conjugating a NBS ligand, tryptamine, to regenerated cellulose (RC) membranes to produce an NBS targeting affinity membrane column (m-NBST) for selective capture and purification of antibodies (Figure 1-B).
Materials and methods
Materials
RC 60 (Regenerated Cellulose) Membrane Filters (1.0 μm, Diameter 47 mm) were purchased from Whatman™ (Germany). Tryptamine, N,N-diisopropylethylamine (DIEA), Sodium phosphate monobasic monohydrate, and mouse ascites fluid (clone NS-1) were all purchased from Sigma-Aldrich (St. Louis, MO). Bovine serum albumin, Fraction V was purchased from EMD Chemicals (Gibbstown, NJ). HRP-conjugated goat anti-human IgG Fcγ-specific was purchased from Jackson ImmunoResearch (West Grove, PA). 2-(1H-Benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU), Amicon Ultra centrifugal filters (0.5 mL, 10K), and Coomassie R-250 were purchased from EMD Millipore (Billerica, MA). Tris-Gly running buffer, transfer buffer, and tris buffered saline (TBS) were purchased from Boston Bioproducts (Ashland, MA). Amplex Red assay kit and Quant-iT PicoGreen dsDNA high-sensitivity assay kit were purchased from Invitrogen (Grand Island, NY). The third-generation CHO host cell protein (HCP) enzyme-linked immunosorbent assay (ELISA) kit was purchased from Cygnus Technologies (Southport, NC). RPMI-1640 media was purchased from Cell-Gro (Manassas, VA), and fetal bovine serum (FBS) was from Hyclone (Thermo Scientific, Rockford, IL). Guard column holder and guard cartages (0.4 × 2 cm) were purchased from IDEX Health and Science (Oak Harbor, WA). Rituximab was provided by Dr. Alexander N. Starodub at the Indiana University Health Goshen Center for Cancer Care, IN.
Membrane functionalization
Hydroxyl groups of the RC membrane were reacted with succinic anhydride for 2 h with addition of DIEA to obtain carboxyl groups on the membrane as a functional group. Carboxylated membranes were washed with DMF and DCM, and dried with airflow. Then, carboxyl groups were activated utilizing HBTU with addition of DIEA. Excess amounts of HBTU were removed by washing with DMF and DCM from the membranes to eliminate the cross reaction of HBTU with amine group of the tryptamine molecule. The tryptamine molecule was conjugated to the membrane in DMF solution under basic conditions during an overnight incubation while agitating (Figure S1-A). Functionalized membranes were washed with DMF and DCM at least three times in order to remove excess amounts of Tryptamine molecules, and then membranes were dried with airflow. The dried membranes were stored at RT.
Characterization of functionalized membranes
Functionalized membranes were characterized by FTIR (Figure S1-B). Peaks at 1788 cm−1 and 1662 cm−1 can confirm the carboxylation of membrane. Peaks between 1400-1600 cm−1 representing C-C stretches in an aromatic ring can be observed in tryptamine-functionalized membrane. Among these functional group changes, the FTIR spectrum was the same throughout the modification of RC membrane, indicating that the general membrane structure remained as stable as it initially was.
Packing column
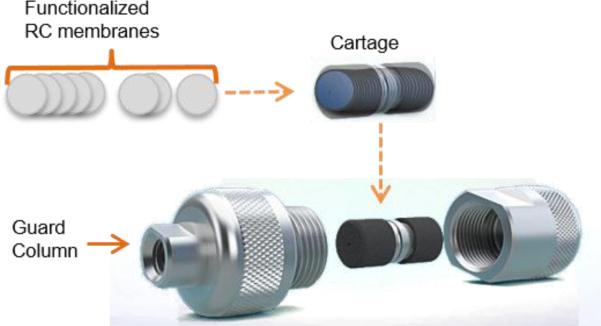
Membranes were cut into 4 mm diameter circles using a Uni-core puncher (4 mm). Post-reaction of membranes with tryptamine, all membranes were dried with airflow. Over 200 membrane circles were packed into 2 cm × 4 mm cartage and then the cartage was placed into a guard column before attaching to HPLC system (Figure 2). Packed column was equilibrated while running EQ buffer through the column for 1 h, then ELS buffer for another 1 h. In order to ensure the column has equilibrated, EQ Buffer was injected to the column and run under the same gradient condition that is used for antibody injections at 0.5 mL/min. This step was repeated until no change was observed on the chromatograms between the following EQ buffer injections. Pressure on the column was 14-17 bar at 0.5 mL/min. The pressure drop across the column ranged between 1 to 3 bar over the course of the NaCl elution gradient at a constant flow rate of 0.5 mL/min.
Figure 2.
Representation of the m-NBST column packing with tryptamine functionalized RC membranes. Several circular pieces of tryptamine functionalized RC membranes with 4 mm diameter were packed in a 2 cm long cartage, then this cartage was placed inside a guard column and was connected to the HPLC system.
To confirm the quality and consistency of the chromatographic operations we measured the integrity of the column bed. For this purpose, an acetone pulse was injected on the tryptamine column to determine the theoretical number of plates, which is determined as N=107.09 (Figure S2). The HETP (height equivalent to the theoretical plates) value was calculated by taking the column length (2 cm) and dividing it by the calculated theoretical number of plates to get HETP=0.019 cm. HETP ≤ 0.02 cm represents good packing of column. Another good packing criteria, peak asymmetry, 1.096, was determined using Agilent Technologies ChemStation LC software, and found within the satisfactory range between 0.8 and 1.4.
Buffers and gradient used for affinity separation
An Agilent Technologies 1200 Series HPLC system was used in all chromatographic injections. Elution from affinity membrane beds predominantly employs gradient, or step changes, in eluent composition to selectively elute products. 50 mM phosphate buffer at pH 7.0 was used as an equilibration buffer (EQ) and 3 M NaCl in 50 mM phosphate buffer at pH 7.0 was used as an Elution Buffer (ELS). pH of both buffers were adjusted using NaOH stock solution. Neutral pH (7.0) was chosen for both EQ and ELS buffers to provide optimal conditions for antibody storage after elution with a salt gradient without any need for buffer exchange. Unless otherwise noted, following a sample injection, while the antibody was still captured on the column, the contaminants were washed away for three minutes with an EQ buffer. The captured antibody was then eluted using ELS buffer with a linear gradient going from 0 to 100% over 10 minutes. At the end of the gradient, the column was cleaned with 100% ELS buffer for two minutes, and then re-equilibrated with the EQ Buffer for five minutes. HPLC method details for the m-NBST column injections are provided in Table 1.
Table 1.
HPLC method for the m-NBST column
| Time (min) | EQ Buffer (%) | ELS Buffer (%) | Flow Rate (mL/min) | Observed Pressure (bar) |
|---|---|---|---|---|
| 0 | 100 | 0 | 0.5 | 14 |
| 3 | 100 | 0 | 0.5 | 14 |
| 13 | 0 | 100 | 0.5 | 17 |
| 15 | 0 | 100 | 0.5 | 17 |
| 15.5 | 100 | 0 | 0.5 | 17 |
| 30 | 100 | 0 | 0.5 | 14 |
Determination of antibody recovery by ELISA
The flow through and elution fractions collected from the m-NBST column were diluted 100-fold in a 0.05 M carbonate-bicarbonate buffer pH 9.6 to a final volume of 100 μL and directly adsorbed on a high-binding Costar 96 well plate for 1.5 h. at room temperature. The surface was subsequently blocked with 2.5 g of BSA in 50 mL of phosphate-buffered saline (PBS) pH 7.4 and 0.05% Tween20 for 45 min. Total antibody in each well was determined using an HRP-conjugated secondary antibody and was quantified using an Amplex Red assay kit (570 nm excitation and 592 nm emission).
Determination of antibody purity by SDS-PAGE
The purity of antibody in the elution fractions was determined by SDS-PAGE under reducing conditions, using 10% polyacrylamide gel with Tris-Glycine running buffer. Sample preparation was done by adding 5 μL of gel loading buffer to 15 μL of concentrated flow through or elution fraction and boiling for 5 min. Gels were Coomassie blue stained using Coomassie R-250. The purity of the product was calculated as the fraction of the total area and intensity equivalent to the IgG bands at 25 kDa and 50 kDa. The antibody purity was determined by densitometric analysis of Coomassie-stained gels using ImageJ software.
Influence of impurities on antibody recovery and purity
The effect of impurities such as BSA, cell culture supernatant, cell lysate, and mouse ascites on m-NBST affinity column was tested by mixing them with antibody sample at various concentrations and analyzing the chromatograms. To study the effect of BSA on the column's performance, samples containing 0.5 mg/mL Rituximab in increasing concentrations of BSA (0, 0.5, 1, 1.5, 2, 3, 5, 10, 15, 20 mg/mL) were prepared in 50 mM sodium phosphate buffer at pH 7.0.
Determination of binding activity of purified antibody
Binding activity of purified antibody using m-NBST affinity column was determined by flow cytometry experiments. For CD-20 expression assays, cells were incubated with Rituximab in binding buffer (1.5% BSA in PBS pH 7.4) on ice for 1 h and washed twice. We verified that IM9 cells expressed CD-20 cell-surface receptor, and that it is available for Rituximab binding (Figure S8). Thus, binding activity of purified Rituximab was tested on IM9 cells. Briefly, 5×105 cells were incubated into each well. After 24 h incubation, cells were washed using PBS, and blocked with 1.5% BSA in PBS for 30 min. Rituximab was incubated with cells on ice for 1 h, then fluorescein conjugated anti-human IgG antibody was used to detect bound Rituximab antibodies on ice. Samples were washed twice and analyzed on a Guava easyCyte 8HT flow cytometer (Millipore).
Specificity of affinity chromatography to active antibodies
Rituximab was denatured using 4 M guanidine hydrochloride (GndCl) and by storing the antibody at room temperature for three hours. Different ratios of denatured and native antibody was mixed and injected into the m-NBST affinity column. Peak areas of flow through (0.5-3 min) and elution fractions from various chromatograms were measured and compared.
Residual host cell DNA content
The double-stranded DNA (dsDNA) content within the flow through and elution fractions was quantified via Quant-iT PicoGreen dsDNA high sensitivity assay kit. Cell culture supernatants and mouse ascites fluid were injected on the column using the standard purification gradient. 20 μL of each collected fraction was added to 200 μL of diluted Quant-iT PicoGreen dye reagent (1:200 dilutions in the provided buffer). The solutions were mixed and allowed to incubate for 5 min at room temperature in a 96 well plate protected from light. The amount of dsDNA present in the samples was determined based on dye fluorescence with a 485 nm excitation and 523 nm emission. This fluorescence was converted to nanograms per microliter of dsDNA based on a standard curve. Data represents the means (±SD) of triplicate experiments.
Residual host cell protein content
A third generation CHO HCP ELISA kit from Cygnus Technologies was used to quantify the HCP (host cell protein) content present in the flow through and elution collected fractions post m-NBST column purification. We followed the recommended high-sensitivity assay protocol as provided by the manufacturer. Briefly, 100 μL of anti-CHO:HRP matrix was added to each well followed by 50 μL of standards, controls, and samples. The plate was covered and incubated on a rotator at room temperature for 2 h. Following incubation the plate was washed with four cycles of 350 μL of wash solution. 100 μL of 3,3’,5,5’ tetramethyl benzidine (TMB) substrate was then added to the wells and incubated for 30 min without rotating. 100 μL of stop solution was added to quench the enzymatic reaction. The amount of residual host cell protein content in the samples quantified by reading the absorbance at 450 nm subtracting off the zero standard as a blank. Data represents the means (±SD) of triplicate experiments.
Results and discussion
Selection of membrane and preparation of stationary phase
The preparation of a membrane for antibody purification purposes requires several steps: i) selection of a suitable membrane, ii) uniform activation of conjugation sites on the membrane, and iii) immobilization of an appropriate NBS ligand as the affinity agent on the membrane.59,60 There are several kinds of commercially available microporous membranes that have been used for protein purification systems with regenerated cellulose (RC), polyethersulfone, and polyvinylidene fluoride.52,60,61 Regenerated cellulose (RC) has been selected as a membrane material for our system due to its specific features such as its strength while wet, extreme chemical resistance, and high mechanical stability. One other advantage of RC membranes is their ability of sterilizing by all methods. Native and derivatized cellulose membranes are soluble only in some strong acids.62 Hydrophilic property of the RC membrane is also an advantage to be used in antibody purification system, eliminating non-specific interactions between the membrane and antibodies or other ingredients (Figure 4-B). We used 1 μm pore sized membranes in order to achieve high flow rates while keeping the pressure low.
Figure 4.
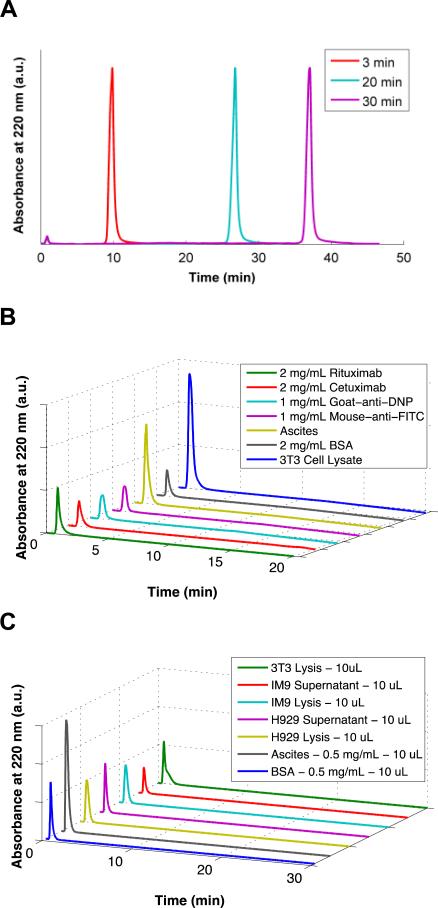
A) Chromatograms demonstrating the effects of changing EQ Buffer wash time on retention of Rituximab by the m-NBST column. Antibody eluted at 10, 27, and 37 min with 3, 20 and 30 min EQ wash times, respectively indicating that retention of the antibody on the column is not due to a size exclusion phenomenon. B) Control column packed with RC membranes without tryptamine modification showed no capture of antibody (Rituximab, Cetuximab, Goat-anti-DNP, and Mouse-anti-FITC) or contaminants (Ascites, BSA, and Cell Lysate). C) The m-NBST column did not display any nonspecific binding for an array of contaminants: 3T3 Lysis, IM9 Supernatant, IM9 Lysis, H929 Supernatant, H929 Lysis, Ascites, and BSA.
The chemical activation of the conjugation sites on RC membranes is similar to activation of chromatographic resins. Since RC membrane has hydroxyl groups, we used succinic anhydride to generate carboxylic acid groups to be further used as functional moieties for conjugation (Figure S1-A). Then, carboxyl groups were activated utilizing HBTU with the addition of DIEA. The tryptamine molecule was conjugated to the membrane in DMF under basic conditions (Figure 1-B). Functionalization of membranes was characterized by FTIR (Figure S1-B). Peaks at 1788 cm−1 and 1662 cm−1 confirm the carboxylation of membrane. Peaks between 1400-1600 cm−1 representing C-C stretches in an aromatic ring can be observed in tryptamine functionalized membrane. Among these functional group changes, the FTIR spectrum was the same throughout the modification of RC membrane, indicating that the general membrane structure conserved as it originally was.
The major limitation of membrane chromatography is that ligand-protein association kinetics limits what the flow-rate can be set to. In order to overcome this limitation and allow an increased interaction time to promote association, we used several stacks of thin membranes. Therefore, we functionalized small pieces (4 mm diameter circles) of RC membranes with tryptamine molecule, and packed the columns (4 mm × 2 cm) with nearly 235 tryptamine-functionalized membranes to construct the m-NBST column (Figure 2). Then, we tested m-NBST column on the HPLC system (Agilent Technologies 1200 Series) for antibody capturing and purification.
Capture and purification of antibody via m-NBST column
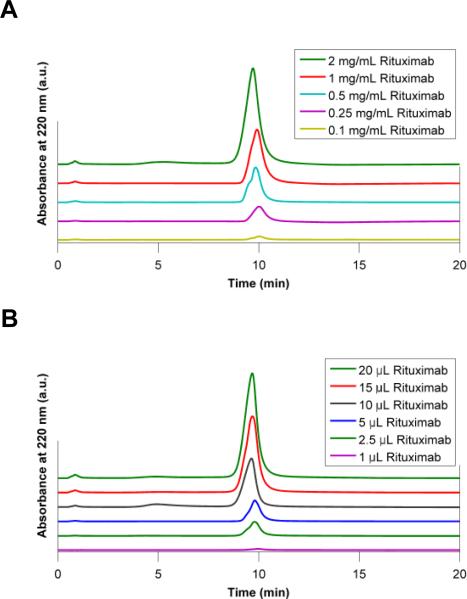
To evaluate antibody capture efficiency of the m-NBST affinity column, we used Rituximab, a chimeric anti-CD20 pharmaceutical antibody. Different amounts of antibody (indicated next to the chromatograms in Figure 3) dissolved in EQ buffer were injected into the column, successfully captured during EQ Buffer run, and eluted using the ELS buffer gradient. Antibody recovery was quantified by peak integration. We tested the column-loading limit using two approaches: i) by increasing the concentration of the antibody in samples while keeping the total volume at 10 μL (Figure 3-A); and ii) by increasing the injection volume while keeping the antibody concentration constant at 0.5 mg/mL (Figure 3-B). Increasing the amount of antibody injected into the column did not have an affect on capture efficiency and consistently yielded >98% antibody recovery. The largest amount of antibody injected on the column was 20 μg of Rituximab (10 μL of 2 mg/mL) into a column volume of 250 μL without any sign of exceeding the column's antibody capture capacity. Thus, 10% breakthrough dynamic loading capacity of the m-NBST column was determined as 0.9 g/L (Figure S3).
Figure 3.
Chromatograms demonstrating the effect of changing injection conditions on the m-NBST column's capture efficiency. Antibody capture profile of m-NBST column was tested with A) increasing concentrations of the antibody at 10 μL and B) increasing volume of antibody at 0.5 mg/mL. Rituximab peak was observed during 65% - 75% of elution buffer gradient at 10 minutes.
To demonstrate that the antibody capture observed with the tryptamine modified membrane column is attributed to the affinity of tryptamine molecules to the antibody, and was not due to size exclusion or another intrinsic property of the RC membrane, we tested antibody capturing and elution properties of the column at various wash times of 3, 20 and 30 minutes. The antibody was retained on the column throughout the EQ wash under all conditions, and was eluted consistently for 7 minutes into the ELS gradient, leading to elution times of 10, 27 and 37 minutes, respectively (Figure 4-A). Since the delay in antibody elution time matches exactly the delay we instilled on the ELS gradient start time, we conclude that the retention of the antibody on the column is not due to a size exclusion effect of the membrane column but strictly a result of affinity capture.
In order to further validate our conclusion, we packed a control column with RC membranes that lack the tryptamine modification. Various contaminants (Ascites, BSA and 3T3 Cell Lysates) and several monoclonal antibodies (Rituximab, Cetuximab, goat-anti-DNP and mouse-anti-FITC) were injected to the non-modified membrane packed column. As expected, neither antibodies nor impurities were captured in the control column, all injected samples eluted in the flow through (0.5 – 3 minutes) further validating our hypothesis (Figure 4-B).
Specificity of m-NBST affinity chromatography column
To assess the specificity of m-NBST column in antibody capture, various contaminants were injected. None of the non-antibody proteins or other biological molecules were retained on the column, and all eluted within the flow through peak (0.5 – 3 min) following injection (Figure 4-C). The results validate that the m-NBST column has a high selectivity only for antibodies without cross-selectivity for other proteins from culture conditions. Flow through fraction of contaminant injections were collected and analyzed on a SDS-PAGE gel for further inspection of potential contaminants (Figure S4). According to results of the control gel, all of the impurities has distinct bands that are not overlapping with heavy and light chain bands of the antibody at 50 kDa and 25 kDa. Additionally, we were able to efficiently capture Rituximab upon mixing with the diverse contaminants indicating that these contaminants do not have any residual negative impact on the affinity of tryptamine column to antibodies. Taken together, these results show that antibody capture on the m-NBST column is a result of specific interactions between the immobilized tryptamine on the membrane and the antibody.
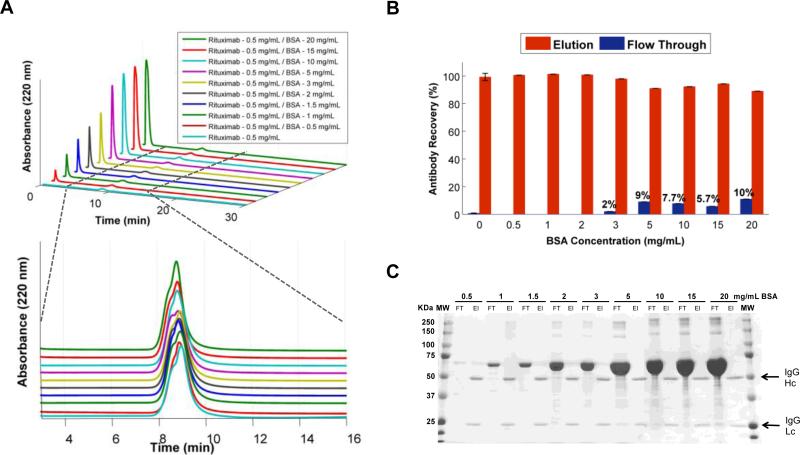
In order to show the capability of tryptamine modified membrane based chromatography to separate antibodies from challenging impurities, we tested antibody samples spiked with various amounts of BSA contamination. Since BSA is the major impurity in the cell culture supernatants and ascites fluid, and is also known to aggressively adhere to surfaces through non-specific interactions; we identified it as a definitive method to test the column. To accurately interpret the experimental challenges we exposed the column, it is important to note that the extreme BSA concentrations were much higher in the samples than the typical biological fluids antibodies are isolated from (normal albumin range in serum is 20 – 50 g/L; thus, in cell media supplemented with 10% FBS is 2-5 g/L).63-65 We collected flow through and elution fractions from the BSA contaminated antibody samples (0.5 mg/mL) for ELISA and SDS-PAGE analysis (Figure 5-A). Our analysis revealed some reduction in the column's ability to capture antibody at elevated BSA concentrations, as some levels of antibody were detected in the flow through (Figure 5-B). According to ELISA, at higher BSA concentrations, the column achieved 90-98% antibody recovery compared to >98% at lower but more relevant BSA concentrations. More importantly, we verified that even at the highest concentrations of BSA contamination, elution peak contained only pure antibody and did not contain any detectable BSA (Figure 5-C). All BSA was found in the flow through fractions of each injection.
Figure 5.
Effect of BSA contamination on antibody recovery and purity of the m-NBST column. A) Chromatograms of Rituximab (0.5 mg/mL) premixed with increasing BSA content; elution times for BSA and Rituximab are indicated as 1-3 min and 8-10 min, respectively. B) ELISA results illustrating percent antibody in the flow through and elution fractions. Data represents the means (±SD) of triplicate experiments. C) SDS-PAGE analysis showed zero indication of BSA contamination in recovered antibody fractions.
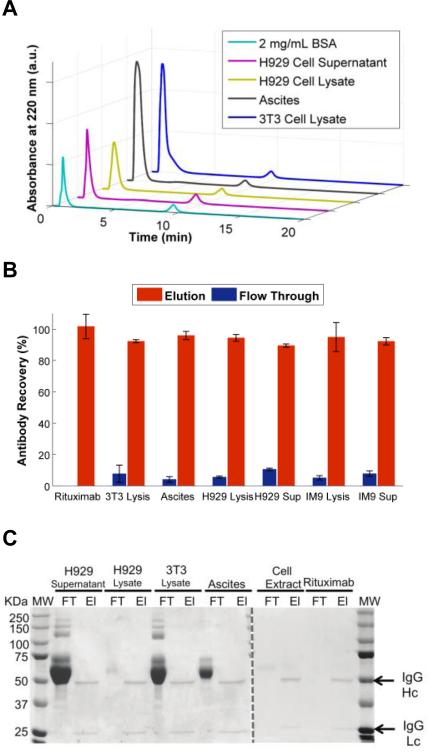
We further investigated the column efficiency for isolating the antibody from more typical contaminants that would arise from common sources along with BSA: conditioned cell culture supernatants, mammalian cell lysates and ascites fluid. Samples of 0.5 mg/mL antibody were mixed with these contaminants and subsequently purified on the tryptamine column (Figure 6-A). The flow through and elution peaks were collected and analyzed for antibody recovery and purity using ELISA and SDS-PAGE. ELISA results showed no significant loss in antibody recovery from any of the tested contaminant sources (Figure 6-B). SDS-PAGE results showed that all contaminants eluted within the flow through fraction, and none of the impurities were detectable within the elution fraction (Figure 6-C). Combined, these results further validated our earlier findings that tryptamine column successfully separates various contaminant proteins and other molecules from the culture media and yield highly pure antibody (>98%).
Figure 6.
Effect of contaminants from various sources on antibody recovery and purity achieved by the m-NBST column. A) Chromatograms of Rituximab (0.5 mg/mL) contaminated with 2 mg/mL BSA, H929 Cell Supernatant, H929 Cell Lysate, Mouse Ascites and 3T3 Cell Lysate. B) ELISA results illustrating percent antibody in flow through and elution fractions. Data represents the means (±SD) of triplicate experiments. C) SDS-PAGE analysis showed absence of no protein contamination in recovered antibody fractions.
Removal of host cell proteins and host cell DNA
Therapeutic antibodies are typically produced in bioreactors using mammalian cells. As a consequence, cellular components from the culture media are the largest source of contaminants, which include host cell proteins (HCPs) and DNA. Therefore, we investigated presence of host cell DNA in the flow through and elution fractions of antibody purified from various sources including cell culture supernatant, lysates and ascites; as determined via binding of fluorescent dye to dsDNA present. This fluorescence was converted to nanograms per microliter of dsDNA by using a standardized curve (Figure S6) and then normalized to antibody content in each fraction. Table 1 shows a summary of DNA content in the collected flow through and elution fractions with log reduction value (LRV). LRV was calculated by taking the logarithm of the ratio of load (sum of flow through and elution) to elution fractions. The results demonstrated that DNA flows through the column leaving a very low level of DNA in the purified antibody elution fraction with around 2-4 of LRV, running congruently with protein A DNA clearance values.18,66
Furthermore, residual HCP content in each collected fraction was analyzed via a broadly reactive HCP ELISA assay (Figure S7). The concentration of HCP present in the initial rituximab/contaminant source solution (sum of flow through and elution fractions) was compared to the purified product. The LRV values were >7 and around 2 when Rituximab was purified from 3T3 cell conditioned media and from other impurities, respectively. These LRV values are comparable to that of protein A chromatography.18,67 A summary of the HCP content in the collected fractions is shown in Table 2. These results further support the high level of purity (>98%) that the tryptamine column technique can attain.
Table 2.
DNA content in flow through and elution fractions
| Sample | Flow Through DNA (ng/mg mAb) | Elution DNA (ng/mg mAb) | LRV |
|---|---|---|---|
| 3T3 | 3×108 | 2×105 | 3.11 |
| Cell Extract | 9×108 | 9×104 | 3.98 |
| Ascites | 1×108 | 1×105 | 2.99 |
| H929 Lysates | 2×107 | 7×104 | 2.38 |
| H929 Supernatant | 1×108 | 5×104 | 3.47 |
| IM9 Lysates | 1×107 | 7×104 | 2.18 |
| IM9 Supernatant | 2×108 | 2×105 | 2.95 |
Effect of NaCl concentration on antibody capture
To evaluate the efficiency of antibody capture by tryptamine column depending on the ionic strength of the sample injection buffer, we tested various concentration of NaCl in the sample injection buffer. For this experiment, we prepared antibody samples in EQ buffer that contained increasing NaCl concentrations and injected them into the column (Figure S5-A). All injections were run using the same gradient and buffers as the earlier injections in order to be able to purely and accurately test the effect of ionic strength of the sample solution on the column. Antibody recovery was quantified by comparing the peak integrations, at 220 nm, of the flow through and elution fractions (Figure S5-B). The integrations from each peak were summed to confirm that not only was the entire injected antibody sample eluted from the column but also that addition of NaCl did not promote irreversible antibody capture on the column. Since high salt concentration of the ELS buffer drives antibody elution, we expected to see a decrease in tryptamine column capture efficiency with increasing NaCl concentration. We observed that capture efficiency dropped to 65% at 2.5 M NaCl.
Retaining binding activity of purified antibody after m-NBST column purification
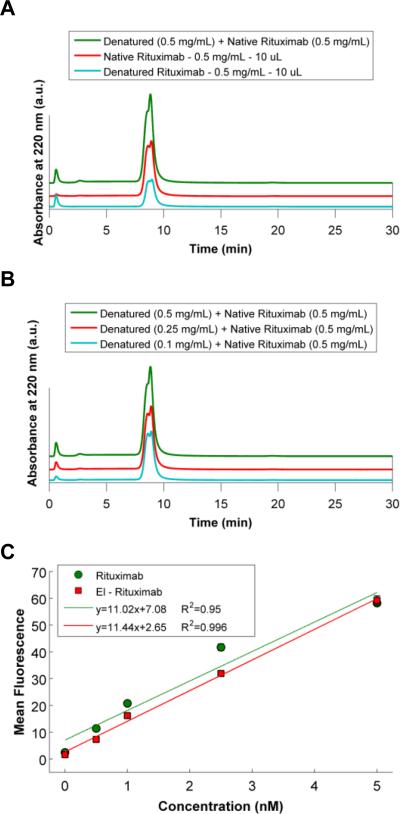
To demonstrate that the NBS affinity-based chromatography method is specific for active antibodies, we designed an experiment where we purified the active antibody from a solution containing both denatured and active antibody. Rituximab was chemically denatured using 4 M GndCl, incubating at room temperature for three hours. Then, GndCl buffer was exchanged to PBS prior to mixing denatured antibody with active antibody. 10 μL samples of denatured antibody, active antibody and a 1:1 mixture of denatured and active antibody were injected into the column, and flow through and elution peak integrations were compared (Figure 7-A). ~49% of Rituximab was determined from the integrated area of the chromatographs to be denatured during the 3 h incubation with GndCl. The sum of flow through fractions of denatured antibody (205.5) and active antibody (31.5) perfectly matched the flow through of mixture sample (234.5). A similar trend was also observed with elution fractions. The elution fraction area of denatured antibody (1589.1) and active antibody (3223) sum (4812.1) gave similar integrated area of mixture sample (4378). Furthermore, to show specificity of the column to active antibody, increasing concentrations of denatured antibody was mixed with same amount of active antibody, and we were able to elute increasing peaks of flow through fractions with increasing amount of the denatured antibody (Figure 7-B). These results demonstrate specificity of the tryptamine column for active antibody from a solution containing a mixture of active and damaged or denatured antibody.
Figure 7.
Chromatograms illustrating specificity of the m-NBST column to active, full-length antibodies. A) Comparison of native, denatured and 1:1 mixture of native and denatured antibodies. Denatured antibodies eluted in the flow through fraction, and non-denatured native antibodies were captured by the m-NBST column, and eluted with NaCl gradient. B) Mixture of native antibodies with increasing concentration of denatured antibodies. C) Flow cytometry results showing the binding activity of native and purified antibodies via the m-NBST column.
Purification of active Rituximab from a solution of active and denatured antibody
Next, we evaluated the binding activity of the purified antibody, which was accomplished through the analysis of antibody binding to cell lines that expressed the antigen protein. Rituximab targets human CD20 and was evaluated using the CD20 expressing multiple myeloma cell line IM-9.40,68 Both native and purified Rituximab was incubated with IM-9 cell lines at increasing concentrations for 3 hours on ice. Binding was detected using a secondary Fc specific fluorescein labeled antibody from goat. The slope of the mean fluorescence curve was used to determine the binding activity. Purified antibody shows similar binding activity (slope: 11.44 R2 = 0.996) with comparison to native antibody (slope: 11.02 R2 = 0.95) (Figure 7-C). These results demonstrated that the antibodies that were purified through the tryptamine column had native levels of binding to the cells. This further validated the tryptamine column purification and confirmed that the antibody activity (including both antigen detection and Fc recognition) was fully retained post purification.
Column stability and reusability
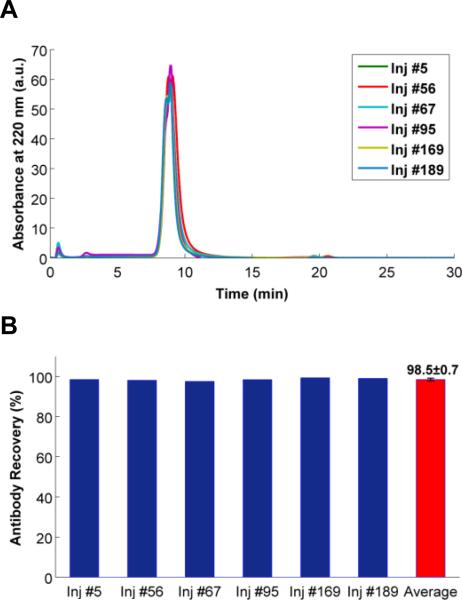
Through the experiments, over 200 injections were performed on m-NBST column. Results of Rituximab injections without mixing with any contaminants were shown in Figure 8-A. On the basis of elution time, peak elution profile, and peak intensity there was no discernable negative impact on the antibody purification efficiency. The m-NBST affinity column yielded reproducible results without a loss in performance in antibody recovery (98.5 ± 0.7) even after 189 injections (Figure 8-B). To test the stability and full elution capability of the column, EQ buffer was injected after a few cycles of antibody injections. The same EQ Buffer chromatograms were obtained before and after sample injections indicating that any antibody or other proteins did not remain in the column, everything eluted during the elution and re-equilibration steps (Figure S9). Furthermore, within these ~200 cycles of injections, the column was taken off from the HPLC system, kept at room temperature in EQ Buffer, and attached again after several weeks, which did not affect the column stability or performance.
Figure 8.
Evaluation of the m-NBST column durability according to conservation of antibody recovery after a high number of injections. A) Overlaid chromatograms of only Rituximab injections (0.5 mg/mL, 10 μL) on the m-NBST column over the course of the 189 injections. B) Percent antibody recovery based on 220 nm peak integration of the Rituximab injections. Average represents the mean (±SD) of the five Rituximab injections.
Conclusion
Here, we demonstrated m-NBST column's competence to purify antibodies from a variety of complex media using NBS selective affinity. The m-NBST affinity column achieved over 98% antibody recovery with over 98% purity from separations performed on antibody samples spiked with a variety of contaminants to reflect potential actual situations. Since the m-NBST column uses mild elution conditions with a neutral pH, it does not require any adjustments to pH or buffer exchange for the purified antibody following the single purification step. The results of the binding experiments to cells that express CD20 receptor with the purified antibody using flow cytometry established that antibody samples purified via the m-NBST affinity column preserve their antigen binding activity and affinity. The results of the binding experiment validate that antibody elution facilitated by high ionic-strength buffer (3 M NaCl) has no plausible negative impact on antibody structure and function.
Even though the m-NBST column has a low dynamic capacity, 0.9 g/L, the dynamic capacity is usually around 1 g/L or higher for most of the small molecule resin-based columns, a high production rate can still be achieved by running multiple cycles in a short amount of time to process a batch of antibody owing to ability to run high flow rates on membrane based columns.34,69-72 Moreover, increased number of injection cycles is not a concern because our results proved that the m-NBST column retained its function without any loss in its performance even after 200 injections. This is a significantly high number compared to any protein based column, especially considering that most of the protein based affinity columns start showing signs of degradation after just 10 cycles of injections, and require cleaning cycles regularly.12,21,73 Additionally, if desired, the m-NBST column material can be easily replaced since membrane production is inexpensive compared to resin production, and this will also eliminate requirement for cleaning cycles. Therefore, utilization of small molecules on membrane instead of biomolecules on resin lowers the production and maintenance costs of the column. Although the maximum binding capacity of the m-NBST column is still far from the binding capacity values of Protein A affinity membrane chromatography74,75, neutral pH buffer conditions for loading and elution of the antibodies, high recovery and purity levels of the m-NBST column are very encouraging to promote more research on small molecule based affinity membrane chromatography. Taken together, the m-NBST affinity column can serve as a model in the design of small molecule affinity membrane column utilizing the NBS for capturing antibodies and provides an alternative to the protein A affinity purification method.
In this demonstration of the proof-of-principle for the m-NBST column for antibody purification, we used Rituximab; nevertheless, we anticipate other pharmaceutical antibodies to also deliver similar results on the m-NBST column since NBS is a universal site conserved on all antibody Fab. Consequently, the m-NBST affinity chromatography column can potentially be used for the purification of antibody fragments such as Fab, Fab2 and ScFv. We observed the column performance to vary due to the minor changes in the binding affinity of the nucleotide analogue to these antibodies and their fragments. Therefore, column optimization studies, such as changing the density of the capture molecule on the membrane surface to regulate the overall avidity or evaluate alternative nucleotide analogues that has a higher affinity for the NBS of all antibody isotypes and species, might be needed. Optimization studies with different nucleotide analogs on various matrixes are currently ongoing in our laboratory to further improve the NBS column to make it a universally effective method for the purification of any and all antibodies. In conclusion, we envision that in the long term, the simplicity and easy scalability of the m-NBST column will provide a benefit to increase the availability of antibody based treatment and diagnostic systems to patients, as well as life-sciences research.
Supplementary Material
Table 3.
HCP content in flow through and elution fractions
| Sample | Flow Through HCP (ng/mg mAb) | Elution HCP (ng/mg mAb) | LRV |
|---|---|---|---|
| 3T3 | 1×107 | 2×10−1 | 7.82 |
| Cell Extract | 8×105 | 6×103 | 2.17 |
| Ascites | 1×106 | 3×103 | 2.59 |
| H929 Lysates | 3×106 | 4×103 | 2.78 |
| H929 Supernatant | 1×106 | 3×103 | 2.77 |
| IM9 Lysates | 2×105 | 8×103 | 1.48 |
| IM9 Supernatant | 6×105 | 1×103 | 2.59 |
Acknowledgements
This work was supported by the National Science Foundation (NSF) (grant award number CBET – 1263713) and the National Institute of Health (NIH) (grant award number R01AI108884). The authors thank the Notre Dame the Center for Environmental Science and Technology for the use of FTIR.
Footnotes
Electronic Supplementary Information (ESI) available: Membrane functionalization and characterization by FTIR; Acetone pulse injection on m-NBST column to assess column-packing characteristics; Determination of Dynamic Binding Capacity of m-NBST column; SDS-PAGE demonstrating flow through collections of impurity injections; Effect of NaCl to antibody capture efficiency by m-NBST affinity column; Quant-iT™ PicoGreen dsDNA High Sensitivity Assay Kit standard curve; Host cell protein content standard curve; Screening of CD20 expression for Rituximab binding on IM9 and H929 cell lines; and Comparison of EQ Buffer Chromatograms.
References
- 1.Lobo E, Hansen R, Balthasar J. J. Pharm. Sci. 2004;93:2645–2668. doi: 10.1002/jps.20178. [DOI] [PubMed] [Google Scholar]
- 2.Lu B, Smyth MR, O'Kennedy R. Analyst. 1996;121:29R–32R. doi: 10.1039/an996210029r. [DOI] [PubMed] [Google Scholar]
- 3.Gray JJ. Curr. Opin. Struct. Biol. 2004;14:110–115. doi: 10.1016/j.sbi.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Makaraviciute A, Ramanaviciene A. Biosensors and Bioelectronics. 2013;50:460–471. doi: 10.1016/j.bios.2013.06.060. [DOI] [PubMed] [Google Scholar]
- 5.Ferreira NS, Sales MGF. Biosensors and Bioelectronics. 2014;53:193–199. doi: 10.1016/j.bios.2013.09.056. [DOI] [PubMed] [Google Scholar]
- 6.Zhang W, Patel K, Schexnider A, Banu S, Radadia AD. ACS nano. 2014;8:1419–1428. doi: 10.1021/nn405240g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Low D, O'Leary R, Pujar NS. Journal of Chromatography B. 2007;848:48–63. doi: 10.1016/j.jchromb.2006.10.033. [DOI] [PubMed] [Google Scholar]
- 8.Pogue GP, Vojdani F, Palmer KE, Hiatt E, Hume S, Phelps J, Long L, Bohorova N, Kim D, Pauly M. Plant biotechnology journal. 2010;8:638–654. doi: 10.1111/j.1467-7652.2009.00495.x. [DOI] [PubMed] [Google Scholar]
- 9.Roque ACA, Lowe CR, Taipa MÂ. Biotechnol. Prog. 2004;20:639–654. doi: 10.1021/bp030070k. [DOI] [PubMed] [Google Scholar]
- 10.Gottschalk U. Biotechnol. Prog. 2008;24:496–503. doi: 10.1021/bp070452g. [DOI] [PubMed] [Google Scholar]
- 11.Thömmes J, Kula M. Biotechnol. Prog. 1995;11:357–367. [Google Scholar]
- 12.Hober S, Nord K, Linhult M. J. Chromatogr. B. 2007;848:40–47. doi: 10.1016/j.jchromb.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 13.Roque ACA, Silva CSO, Taipa MA. J. Chromatogr. A. 2007;1160:44–55. doi: 10.1016/j.chroma.2007.05.109. [DOI] [PubMed] [Google Scholar]
- 14.Zou H, Luo Q, Zhou D. J. Biochem. Biophys. Methods. 2001;49:199–240. doi: 10.1016/s0165-022x(01)00200-7. [DOI] [PubMed] [Google Scholar]
- 15.Shukla AA, Hubbard B, Tressel T, Guhan S, Low D. J. Chromatogr. B. 2007;848:28–39. doi: 10.1016/j.jchromb.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 16.Kelley B. Biotechnol. Prog. 2007;23:995–1008. doi: 10.1021/bp070117s. [DOI] [PubMed] [Google Scholar]
- 17.Lim JAC, Sinclair A, Kim DS, Gottschalk U. BioProcess Int. 2007;5:60–64. [Google Scholar]
- 18.Naik AD, Menegatti S, Gurgel PV, Carbonell RG. J. Chromatogr. A. 2011;1218:1691–1700. doi: 10.1016/j.chroma.2010.11.071. [DOI] [PubMed] [Google Scholar]
- 19.Jiang C, Liu J, Rubacha M, Shukla AA. J. Chromatogr. A. 2009;1216:5849–5855. doi: 10.1016/j.chroma.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Feng H, Jia L, Li H, Wang X. Biomed. Chromatogr. 2006;20:1109–1115. doi: 10.1002/bmc.654. [DOI] [PubMed] [Google Scholar]
- 21.Hahn R, Shimahara K, Steindl F, Jungbauer A. J. Chromatogr. A. 2006;1102:224–231. doi: 10.1016/j.chroma.2005.10.083. [DOI] [PubMed] [Google Scholar]
- 22.Verdoliva A, Marasco D, De Capua A, Saporito A, Bellofiore P, Manfredi V, Fattorusso R, Pedone C, Ruvo M. Chembiochem. 2005;6:1242–1253. doi: 10.1002/cbic.200400368. [DOI] [PubMed] [Google Scholar]
- 23.Calmettes P, Cser L, Rajnavölgyi E. Arch. Biochem. Biophys. 1991;291:277–283. doi: 10.1016/0003-9861(91)90135-6. [DOI] [PubMed] [Google Scholar]
- 24.Arakawa T, Tsumoto K, Ejima D. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 2014;1844:2032–2040. doi: 10.1016/j.bbapap.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Arakawa T, Philo JS, Tsumoto K, Yumioka R, Ejima D. Protein Expr. Purif. 2004;36:244–248. doi: 10.1016/j.pep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 26.Lee JM, Park HK, Jung Y, Kim JK, Jung SO, Chung BH. Anal. Chem. 2007;79:2680–2687. doi: 10.1021/ac0619231. [DOI] [PubMed] [Google Scholar]
- 27.Fassina G, Ruvo M, Palombo G, Verdoliva A, Marino M. J. Biochem. Biophys. Methods. 2001;49:481–490. doi: 10.1016/s0165-022x(01)00215-9. [DOI] [PubMed] [Google Scholar]
- 28.Alves NJ, Stimple SD, Handlogten MW, Ashley JD, Kiziltepe T, Bilgicer B. Anal. Chem. 2012;84(18):7721–8. doi: 10.1021/ac300952r. [DOI] [PubMed] [Google Scholar]
- 29.Follman DK, Farner RL. J. Chromatogr. A. 2004;1024:79–85. doi: 10.1016/j.chroma.2003.10.060. [DOI] [PubMed] [Google Scholar]
- 30.Lin C, Saito K, Boysen RI, Campi EM, Hearn MT. Separation and Purification Technology. 2016;163:199–205. [Google Scholar]
- 31.Yang H, Gurgel PV, Carbonell RG. J. Chromatogr. A. 2009;1216:910–918. doi: 10.1016/j.chroma.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Lund LN, Gustavsson P, Michael R, Lindgren J, Nørskov-Lauritsen L, Lund M, Houen G, Staby A, Hilaire PMS. J. Chromatogr. A. 2012;1225:158–167. doi: 10.1016/j.chroma.2011.12.074. [DOI] [PubMed] [Google Scholar]
- 33.Luo Q, Zou H, Zhang Q, Xiao X, Ni J. Biotechnol. Bioeng. 2002;80:481–489. doi: 10.1002/bit.10391. [DOI] [PubMed] [Google Scholar]
- 34.Platis D, Labrou NE. Journal of separation science. 2008;31:636–645. doi: 10.1002/jssc.200700481. [DOI] [PubMed] [Google Scholar]
- 35.Bøg-Hansen TC. Mol. Biotechnol. 1997;8:279–281. doi: 10.1007/BF02760781. [DOI] [PubMed] [Google Scholar]
- 36.Lin C, Boysen RI, Campi EM, Saito K, Hearn MT. Journal of Molecular Recognition. 2016;29:334–342. doi: 10.1002/jmr.2535. [DOI] [PubMed] [Google Scholar]
- 37.Batalha IL, Zhou H, Lilley K, Lowe CR, Roque AC. J. Chromatogr. A. 2016;1457:76–87. doi: 10.1016/j.chroma.2016.06.032. [DOI] [PubMed] [Google Scholar]
- 38.Camperi S, Iannucci N, Albanesi G, Eberhardt MO, Etcheverrigaray M, Messeguer A, Albericio F, Cascone O. Biotechnol. Lett. 2003;25:1545–1548. doi: 10.1023/a:1025464918453. [DOI] [PubMed] [Google Scholar]
- 39.D'Agostino B, Bellofiore P, De Martino T, Punzo C, Rivieccio V, Verdoliva A. J. Immunol. Methods. 2008;333:126–138. doi: 10.1016/j.jim.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 40.Alves NJ, Champion MM, Stefanick JF, Handlogten MW, Moustakas DT, Shi Y, Shaw BF, Navari RM, Kiziltepe T, Bilgicer B. Biomaterials. 2013;34:5700–5710. doi: 10.1016/j.biomaterials.2013.03.082. [DOI] [PubMed] [Google Scholar]
- 41.Alves NJ, Mustafaoglu N, Bilgicer B. Biosens. Bioelectron. 2013;49:387–393. doi: 10.1016/j.bios.2013.05.052. [DOI] [PubMed] [Google Scholar]
- 42.Handlogten MW, Kiziltepe T, Moustakas DT, Bilgicer B. Chem. Biol. 2011;18:1179–1188. doi: 10.1016/j.chembiol.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 43.Alves NJ, Kiziltepe T, Bilgicer B. Langmuir. 2012;28:9640–9648. doi: 10.1021/la301887s. [DOI] [PubMed] [Google Scholar]
- 44.Alves NJ, Mustafaoglu N, Bilgicer B. Bioconjug. Chem. 2014;25:6534–6541. doi: 10.1021/bc500211d. [DOI] [PubMed] [Google Scholar]
- 45.Mustafaoglu N, Alves NJ, Bilgicer B. Langmuir. 2015;31:9728–9736. doi: 10.1021/acs.langmuir.5b01734. [DOI] [PubMed] [Google Scholar]
- 46.Mustafaoglu N, Alves NJ, Bilgicer B. Biotechnol. Bioeng. 2015;112:1327–1334. doi: 10.1002/bit.25558. [DOI] [PubMed] [Google Scholar]
- 47.Handlogten MW, Kiziltepe T, Serezani AP, Kaplan MH, Bilgicer B. Nat. Chem. Biol. 2013;9:789–795. doi: 10.1038/nchembio.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajagopalan K, Pavlinkova G, Levy S, Pokkuluri PR, Schiffer M, Haley BE, Kohler H. Proc. Natl. Acad. Sci. U. S. A. 1996;93:6019–6024. doi: 10.1073/pnas.93.12.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Russ M, Lou D, Kohler H. J. Immunol. Methods. 2005;304:100–106. doi: 10.1016/j.jim.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 50.Lac D, Feng C, Bhardwaj G, Le H, Tran J, Xing L, Fung G, Liu R, Cheng H, Lam KS. Bioconjug. Chem. 2015;27:159–169. doi: 10.1021/acs.bioconjchem.5b00574. [DOI] [PubMed] [Google Scholar]
- 51.Orr V, Zhong L, Moo-Young M, Chou CP. Biotechnol. Adv. 2013;31:450–465. doi: 10.1016/j.biotechadv.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 52.Govender S, Jacobs E, Bredenkamp M, Swart P. J. Chromatogr. B. 2007;859:1–8. doi: 10.1016/j.jchromb.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 53.Ghosh R. J. Chromatogr. A. 2002;952:13–27. doi: 10.1016/s0021-9673(02)00057-2. [DOI] [PubMed] [Google Scholar]
- 54.Kosior A, Antošová M, Faber R, Villain L, Polakovič M. J. Membr. Sci. 2013;442:216–224. [Google Scholar]
- 55.Warner T, Nochumson S. Pharmaceutical Technology. 2002 Sep;:36–42. [Google Scholar]
- 56.Hou Y, Brower M, Pollard D, Kanani D, Jacquemart R, Kachuik B, Stout J. Biotechnol. Prog. 2015;31:974–982. doi: 10.1002/btpr.2113. [DOI] [PubMed] [Google Scholar]
- 57.Sadavarte R, Spearman M, Okun N, Butler M, Ghosh R. Biotechnol. Bioeng. 2014;111:1139–1149. doi: 10.1002/bit.25193. [DOI] [PubMed] [Google Scholar]
- 58.Lefranc MP, Pommie C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G. Dev. Comp. Immunol. 2003;27:55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 59.Feng Z, Shao Z, Yao J, Huang Y, Chen X. Polymer. 2009;50:1257–1263. [Google Scholar]
- 60.Boi C. J. Chromatogr. B. 2007;848:19–27. doi: 10.1016/j.jchromb.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 61.Boi C, Busini V, Salvalaglio M, Cavallotti C, Sarti GC. J. Chromatogr. A. 2009;1216:8687–8696. doi: 10.1016/j.chroma.2009.05.045. [DOI] [PubMed] [Google Scholar]
- 62.Klein E. J. Membr. Sci. 2000;179:1–27. [Google Scholar]
- 63.Horwich TB, Kalantar-Zadeh K, MacLellan RW, Fonarow GC. Am. Heart J. 2008;155:883–889. doi: 10.1016/j.ahj.2007.11.043. [DOI] [PubMed] [Google Scholar]
- 64.Quinlan GJ, Martin GS, Evans TW. Hepatology. 2005;41:1211–1219. doi: 10.1002/hep.20720. [DOI] [PubMed] [Google Scholar]
- 65.Kim Y, Im Y, Ha N, Im D. Prostaglandins Other Lipid Mediat. 2007;83:130–138. doi: 10.1016/j.prostaglandins.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 66.Butler MD, Kluck B, Bentley T. J. Chromatogr. A. 2009;1216:6938–6945. doi: 10.1016/j.chroma.2009.08.049. [DOI] [PubMed] [Google Scholar]
- 67.Shukla AA, Hinckley P. Biotechnol. Prog. 2008;24:1115–1121. doi: 10.1002/btpr.50. [DOI] [PubMed] [Google Scholar]
- 68.Stefanick JF, Ashley JD, Bilgicer B. ACS Nano. 2013;7(9):8115–8127. doi: 10.1021/nn4033954. [DOI] [PubMed] [Google Scholar]
- 69.Ehrlich G, Bailon P. J. Biochem. Biophys. Methods. 2001;49:443–454. doi: 10.1016/s0165-022x(01)00212-3. [DOI] [PubMed] [Google Scholar]
- 70.Wei Y, Xu J, Zhang L, Fu Y, Xu X. RSC Advances. 2015;5:67093–67101. [Google Scholar]
- 71.Gong Y, Zhang L, Li J, Feng S, Deng H. Bioconjug. Chem. 2016;27:1569–1573. doi: 10.1021/acs.bioconjchem.6b00170. [DOI] [PubMed] [Google Scholar]
- 72.Fahrner RL, Iyer HV, Blank GS. Bioprocess Eng. 1999;21:287–292. [Google Scholar]
- 73.Jagschies G, Sofer GK, Hagel L. Handbook of process chromatography: development, manufacturing, validation and economics. Academic Press; 2007. [Google Scholar]
- 74.Dancette OP, Taboureau J, Tournier E, Charcosset C, Blond P. Journal of Chromatography B: Biomedical Sciences and Applications. 1999;723:61–68. doi: 10.1016/s0378-4347(98)00470-8. [DOI] [PubMed] [Google Scholar]
- 75.Ma Z, Ramakrishna S. J. Membr. Sci. 2008;319:23–28. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.