Abstract
The centromere–kinetochore complex is a specialized chromatin structure that mediates bipolar attachment of replicated chromosomes to the mitotic spindle, thereby ensuring proper sister chromatid separation during anaphase. The manner in which this important multimeric structure is specified and assembled within chromatin is unknown. Using in vivo cross-linking followed by immunoprecipitation, we show that the Mif2 protein of the budding yeast Saccharomyces cerevisiae, previously implicated in centromere function by genetic criteria, resides specifically at centromeric loci in vivo. This provides definitive evidence for structural conservation between yeast and mammalian centromeres, as Mif2p shares homology with CENP-C, a mammalian centromere protein. Ndc10p and Cbf1p, previously implicated in centromere function by genetic and in vitro biochemical assays, were also found to interact with centromeric DNA in vivo. By examining Mif2p, Ndc10p, and Cbf1p association with centromeric DNA derivatives, we demonstrate the existence of centromeric subcomplexes that may correspond to assembly intermediates. Based on these observations, we provide a simple model for centromere assembly. Finally, given the sensitivity of this technique, its application to other sequence-specific protein–DNA complexes within the cell, such as origins of replication and enhancer–promoter regions, could be of significant value.
Keywords: MIF2, CENP-C, centromere, kinetochore, chromatin immunoprecipitation, CSE4
The centromere–kinetochore complex (hereafter centromere or CKC) is a highly specialized chromatin-based organelle that mediates chromosome attachment to and movement upon the mitotic spindle (Brinkley et al. 1992; Pluta et al. 1995). As mitosis proceeds, the centromeres of sister chromatids ultimately interact with microtubules (MTs) from opposite spindle poles, thereby ensuring that each daughter cell will receive a precise complement of chromosomes. The fidelity of chromosome transmission is enhanced not only by the geometry of stable centromere–MT interactions but also by the action of at least one centromere-based mitotic checkpoint control system that monitors those interactions and delays the onset of anaphase until stable bipolar attachment is achieved (Gorbsky 1995; Rudner and Murray 1996). Thus, centromeres play both important structural and regulatory roles in ensuring faithful chromosome segregation during mitosis. To enact these roles, centromeres must contain many proteins including MT-binding proteins and motors, cohesion factors, checkpoint proteins, and possibly specific architectural and heterochromatin proteins. These proteins must be assembled together at a specific site on the chromosome and function in an integrated fashion with respect to one another and to the cell cycle.
The CKC of mammalian cells has been defined cytologically as a trilaminar disc-shaped structure situated at the primary constriction of metaphase chromosomes (Brinkley et al. 1992; Pluta et al. 1995). These structures typically engage 15–25 MTs and encompass up to several megabase pairs of DNA. Their size, combined with the fact that centromeric regions consist largely of heterochromatic blocks of repetitive satellite DNAs, has made it difficult to define functional centromeric DNA sequences in higher eukaryotes. On the other hand, several conserved protein constituents of mammalian centromeric heterochromatin have been identified. CENP-A (17 kD) contains a carboxy-terminal histone-fold domain similar to that of histone H3 and may define a class of centromere-specific nucleosomes (Sullivan et al. 1994). CENP-B (80 kD) is a sequence-specific DNA-binding protein that localizes to the heterochromatic interior of the centromere (Cooke et al. 1990). CENP-B binds to a subset of the alphoid satellite sequences that comprise human centromeric DNA (Masumoto et al. 1989). CENP-C (140 kD) is a novel, basic protein that localizes to the inner plate of the CKC, a site corresponding to the outermost region of the CKC in which DNA can be detected (Saitoh et al. 1992).
Although CENP-A and CENP-B may be involved in packaging the centromeric DNA, their contribution to centromere function is unclear. However, several observations suggest that CENP-C is an essential component of a functional human centromere. For example, nuclear microinjection of CENP-C-specific antibodies profoundly delays mitosis and causes abnormalities in CKC ultrastructure (Tomkiel et al. 1994). Importantly, these effects correlate with a specific decrease in centromeric CENP-C staining. Conversely, the presence of CENP-C strictly correlates with the ability of a given centromere to support chromosome segregation. Based on cytogenetic analyses of mitotically stable, dicentric or neocentric human chromosomes, only functionally active centromeres contain CENP-C (Earnshaw et al. 1989; Sullivan and Schwartz 1995; du Sart et al. 1997). In contrast, CENP-B and/or the alphoid satellite DNA it recognizes may or may not be found at active centromeres and can also be found at inactive centromeres. Thus, it has been suggested that CENP-C is an important architectural component of the active mammalian centromere.
The CKC of the budding yeast Saccharomyces cerevisiae is functionally similar to those of higher eukaryotes, harboring activities for MT binding (Kingsbury and Koshland 1991), potential anaphase A movement (Guacci et al. 1997), and activation of the mitotic checkpoint (Pangilinan and Spencer 1996). However, the centromere of budding yeast appears to be a much simpler organelle than its mammalian counterpart, engaging only a single MT (Winey et al. 1995). Yeast centromeric ultrastructure has been impossible to visualize by electron microscopy, perhaps because of its small size (<200 bp).
The centromeric DNA (CEN DNA) of S. cerevisiae is one of the best characterized DNA segregation elements, largely owing to the development of sensitive genetic assays for centromere function in this organism (Hegemann and Fleig 1993; Hyman and Sorger 1995). The minimal functional centromere (∼120 bp) is comprised of three conserved centromere DNA elements (CDEs). The central element, CDEII, consists of alternating stretches of A and T residues and is flanked by two highly conserved palindromic motifs, CDEI (8 bp) and CDEIII (25 bp). Of these, CDEIII and at least part of CDEII are essential for centromere function. In vivo, a unique nuclease-resistant chromatin structure encompassing ∼200 bp is associated with CEN DNA and presumably corresponds to the yeast CKC (Bloom and Carbon 1982). Five genes that encode yeast centromere proteins have been identified by a combination of genetic and biochemical approaches. Four of these—NDC10/CBF2 (Goh and Kilmartin 1993; Jiang et al. 1993), CBF3B/CEP3 (Lechner 1994; Strunnikov et al. 1995), CTF13 (Doheny et al. 1993), and SKP1 (Connelly and Hieter 1996; Stemmann and Lechner 1996)—are essential for viability and encode components of a multisubunit complex (CBF3) that binds to CDEIII DNA in vitro (Lechner and Carbon 1991). In addition, CBF1/CEP1/CPF1 encodes a nonessential basic helix–loop–helix protein that binds to CDEI DNA in vitro (Baker and Masison 1990; Cai and Davis 1990; Mellor et al. 1990). Mutations in these five genes impair chromosome segregation, thereby further implicating them as centromere protein genes. However, occupation of CEN DNA in vivo by any of the corresponding proteins has not been demonstrated. Moreover, that none of these proteins shares homology with CENP-A, CENP-B, or CENP-C suggested that yeast and mammalian centromeres have different architectures and distinct mechanisms of assembly.
An indication that this hypothesis is incorrect came with the identification and characterization of the yeast MIF2 gene and protein (Mif2p). By several criteria, MIF2 may also encode a centromere component. MIF2 is essential, and mutations in the gene confer phenotypes consistent with a centromere defect. These include chromosome mis-segregation, disruption of mitotic spindle integrity, stabilization of dicentric minichromosomes, and synergistic increases in chromosome loss or synthetic lethality when combined with mutations in various cis- and trans-acting components of the centromere (Brown et al. 1993; Meluh and Koshland 1995). Interestingly, two regions of Mif2p essential for its function share homology with the two most highly conserved regions of the mammalian centromere protein CENP-C (Brown 1995; Meluh and Koshland 1995). With the completion of the Yeast Genome Project, it is now clear that Mif2p represents the best candidate for a yeast CENP-C homolog. In addition, Mif2p has two features that suggest it interacts with DNA: an “A-T hook” motif common to several chromatin proteins that bind AT-rich DNA [e.g., mammalian HMG-I(Y) proteins], and an acidic domain (Brown et al. 1993). Taken together, these genetic and structural features suggest that Mif2p interacts with both Cbf1p and components of CBF3, possibly while bound to the central AT-rich CDEII element. Thus, like CENP-C, Mif2p may serve as an architectural component of a higher order centromere complex.
Although these observations strongly implicated Mif2p as a yeast centromere protein, they did not exclude other interpretations, especially because Mif2p was not among the handful of CEN DNA-binding proteins previously identified and because the similarity between Mif2p and CENP-C is quite limited. Conceivably, the effects of mif2 mutations on centromere function could reflect an indirect effect on the expression of CBF3 components. In light of the potential implications of structural conservation within the centromeres of diverse organisms, it was imperative to demonstrate that Mif2p directly interacts with the centromere. Here we analyze Mif2p function by indirect immunofluorescence and by an in vivo cross-linking and immunoprecipitation strategy. We show that Mif2p specifically interacts with CEN DNA in vivo in a pattern that correlates with centromere function.
Results
Mif2p is a nuclear protein
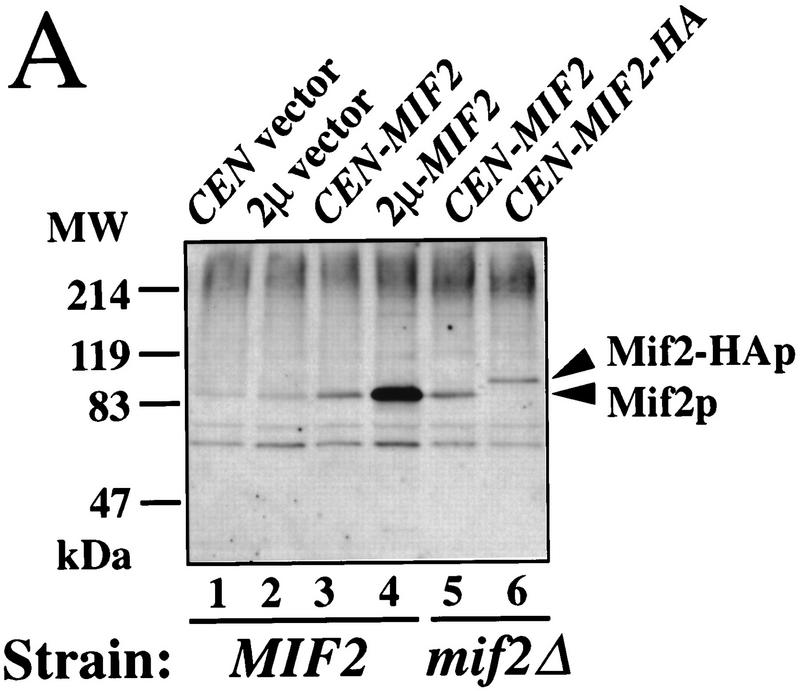
To determine the distribution of Mif2p in cells, antisera specific for the carboxyl terminus of Mif2p (amino acids 364–549), as well as a functional epitope-tagged allele of MIF2, MIF2-HA (hemagglutinin), were generated. Both crude sera recognize a ∼94-kD protein in yeast whole cell extracts (Fig. 1A). This apparent molecular mass is significantly greater than that predicted from the MIF2 coding sequence (maximally 62.5 kD; Brown et al. 1993). However, the detected protein is Mif2p, as its steady-state level increases in strains that overexpress Mif2p and because its relative mobility decreases in extracts prepared from MIF2–HA strains. The aberrant migration may be due to the presence of a highly acidic region in Mif2p (amino acids 178–263), as has been observed for other proteins that contain acidic domains, notably the yeast CDEI-binding protein Cbf1p (Baker and Masison 1990).
Figure 1.
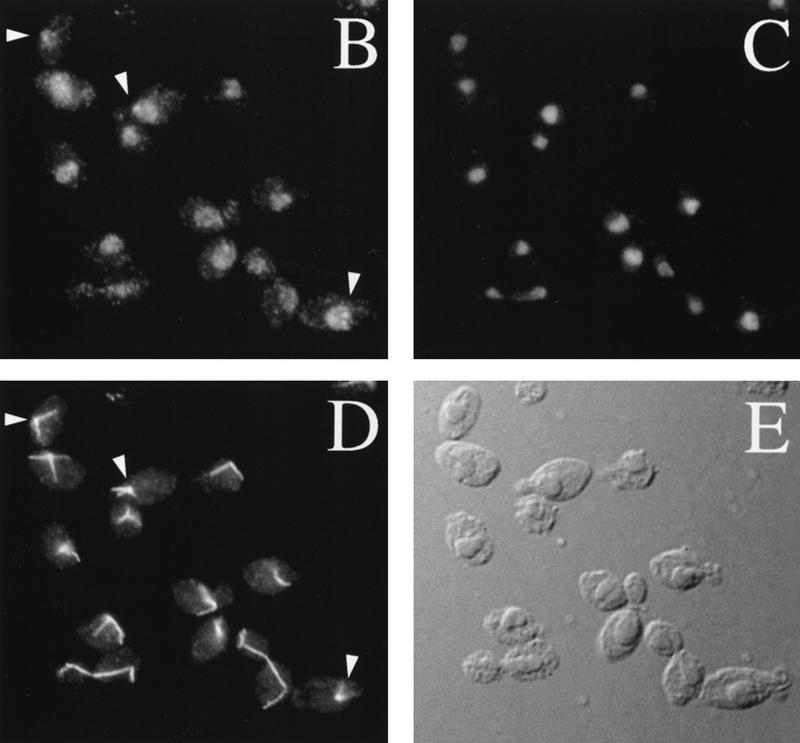
Characterization of Mif2p by Western analysis and indirect immunofluorescence. (A) Western analysis of Mif2p in whole cell extracts from logarithmically growing yeast strains (30°C). Mif2p was detected using anti-Mif2p antiserum and chemiluminescence. Mif2p migrates as at ∼94 kD; Mif2–HAp migrates at ∼102 kD. (Lane 1) Wild-type strain 5371-10-2 with pRS316, CEN6 URA3 vector (Sikorski and Hieter 1989); (lane 2) 5371-10-2 [pRS202], 2μ URA3 vector; (lane 3) 5371-10-2 [pPM4], pRS316–MIF2; (lane 4) 5371-10-2 [pPM44], pRS202–MIF2; (lane 5) PM1202-7B, mif2::HIS3 [pPM4]; (lane 6) PM1203–1C, mif2::HIS3 [pPM102], pRS316–MIF2–HA. Molecular masses of protein standards are indicated (MW). (B–E) Localization of Mif2p in wild-type diploid cells (BP5050; 30°C) by indirect immunofluorescence. (B) Mif2p staining as visualized by affinity-purified anti-Mif2p; (C) Total DNA as detected by DAPI staining; (D) MTs as visualized by anti-α-tubulin; (E) DIC image of field. Brighter foci of Mif2p staining are indicated by arrowheads.
One of the sera was affinity purified and used for indirect immunofluorescence of asynchronously grown wild-type yeast diploid cells. Consistent with the hypothesis that Mif2p is a centromere protein, a dim nuclear staining pattern was observed in cells at all stages of the cell cycle (Fig. 1B). In addition, in most unbudded and small budded cells a brighter focus of staining in the vicinity of the spindle pole body could be seen (arrowheads, Fig. 1B). Brighter staining foci were not evident in cells at later stages of the cell cycle. In cells that overexpress Mif2p or Mif2–HAp, general and bright nuclear staining was observed (data not shown). Thus, Mif2p is a nuclear protein. In addition, the pattern of spindle pole body proximal staining early in the cell cycle is reminiscent of the position of the centromeric DNA at this time as revealed by fluorescence in situ hybridization (Guacci et al. 1997). Lack of focal staining at later times may reflect masking of Mif2p upon assembly of other centromere proteins (see below).
Mif2p interacts with CEN DNA in vivo
Although punctate nuclear staining for Mif2p is consistent with centromere association, such localization does not rigorously distinguish whether Mif2p acts at the centromere, at the spindle pole body, or within the spindle. To further implicate Mif2p as a centromere-associated protein, we first tried a gel mobility shift assay similar to that used to demonstrate in vitro CEN DNA binding by Cbf1p and the CBF3 complex (Doheny et al. 1993; Sorger et al. 1994, 1995). However, our attempts were unsuccessful (data not shown). Conceivably, formation or resolution of higher order structures containing Mif2p is precluded under conditions where Cbf1p– and CBF3–CEN DNA subcomplexes are detected. CBF3 reconstituted from highly purified components and CBF3 affinity-purified from cell extracts exhibit identical CEN-binding properties (Stemmann and Lechner 1996). Therefore, as an alternative approach to study potential Mif2p–CEN DNA interactions, we used the technique of formaldehyde fixation followed by chromatin immunoprecipitation (Dedon et al. 1991; Braunstein et al. 1993; Orlando and Paro 1993; Hecht et al. 1996; Orlando et al. 1997). The choice of formaldehyde, which creates heat-reversible cross-links between proteins, as well as between proteins and nucleic acid, rendered assumptions about the primary nature of any Mif2p–CEN DNA interaction unnecessary.
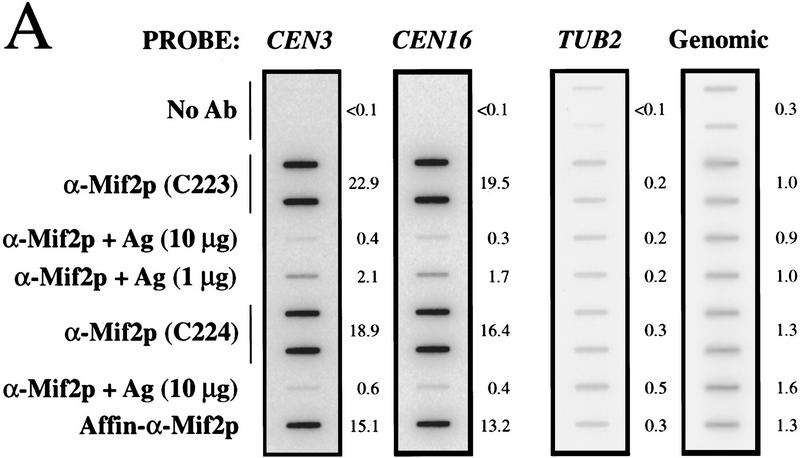
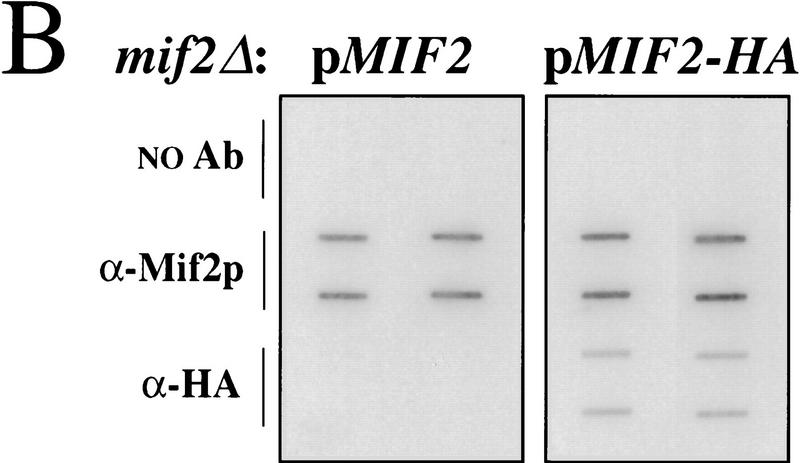
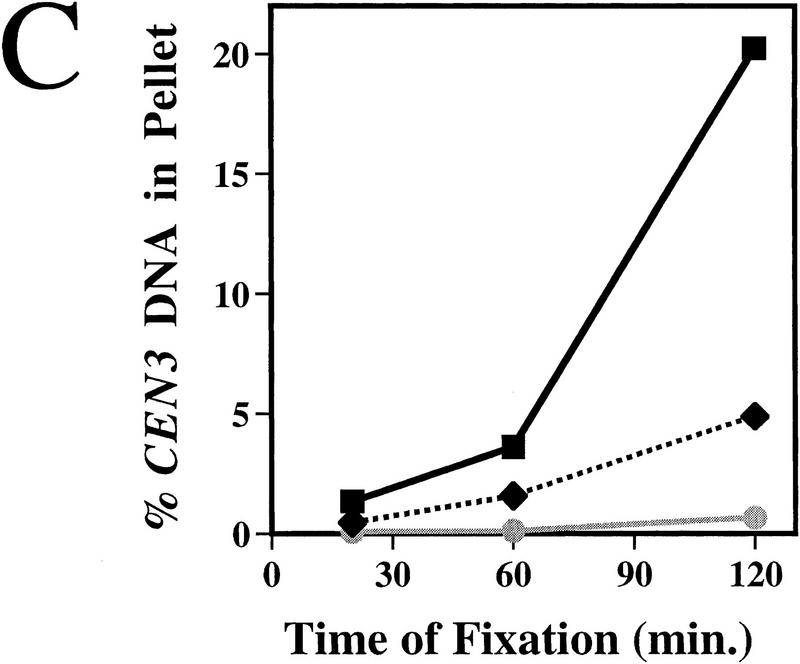
To test whether Mif2p interacts with CEN DNA in vivo, wild-type yeast cells in mid-logarithmic phase were first fixed in situ with 1% formaldehyde. A sonicated chromatin solution prepared from the fixed cells was then subjected to immunoprecipitation with anti-Mif2p antisera. The presence of various DNA sequences in the immunoprecipitates was assessed by slot blot hybridization. Several CEN DNA sequences tested (i.e., CEN3, CEN16, and CEN1) were enriched in Mif2p immunoprecipitates as compared to a mock-treated control (Fig. 2A; data not shown). The enrichment was specific for Mif2p, as it was abrogated by the addition of recombinant Mif2 antigen to the immunoprecipitation reaction (Figs. 2A and 3B). CEN DNA sequences were similarly enriched when affinity-purified Mif2p antibody was used (Fig. 2A) or when a well-characterized anti-HA antibody was used with chromatin prepared from the MIF2–HA strain (Fig. 2B). Finally, for a given CEN DNA locus, the fraction of total material present in Mif2p immunoprecipitates increased with fixation time (Fig. 2C). These data indicate that Mif2p interacts, either directly or indirectly, with CEN DNA in vivo. Importantly, several other genomic loci, including regions with high AT content (Figs. 2A and 4F) were not enriched in Mif2p immunoprecipitates, suggesting that Mif2p interacts exclusively with centromeric regions. Thus, as predicted by its numerous genetic interactions with centromere components, Mif2p is a centromere-associated protein in vivo.
Figure 2.
Coimmunoprecipitation of CEN DNA with Mif2p. (A) Formaldehyde cross-linked chromatin (2 hr fixation) prepared from wild-type strain 5371-10-2 was immunoprecipitated with anti-Mif2p antibodies in the absence or presence of recombinant 6-His–Mif2 fusion protein. Some reactions were prepared in duplicate. Total input material and coimmunoprecipitated DNA were analyzed by slot blot hybridization with the indicated probes. The percent of total input material for a given locus present in an immunoprecipitate is indicated to the right of each panel. The variable low levels of material detected by the TUB2 and total genomic DNA probes probably reflect contaminating RNA, although noncentromeric chromatin may exhibit some insolubility as compared to centromeric chromatin. (B) Cross-linked chromatin (2 hr fixation) prepared from mif2::HIS3 strains expressing either wild-type MIF2 or epitope-tagged MIF2–HA from a centromere-based plasmid was immunoprecipitated with anti-Mif2p antiserum or anti-HA antibody. Coimmunoprecipitated DNA was analyzed by slot blot hybridization using a CEN3-specific probe. (C) Time course of fixation. Cross-linked chromatin prepared from the MIF2–HA strain after various times of fixation was mock-treated (shaded circle) or immunoprecipitated with anti-Mif2p antiserum (black square) or anti-HA (black diamond) and analyzed as in B. The percent of total input CEN3 DNA that coimmunoprecipitated with Mif2–HAp was plotted. Identical results were obtained in B and C using a CEN16 probe (data not shown). That CEN DNA recovery was consistently greater in immunoprecipitates prepared with authentic Mif2p antibodies as compared to anti-HA presumably reflects the polyclonal nature of these sera.
Figure 3.
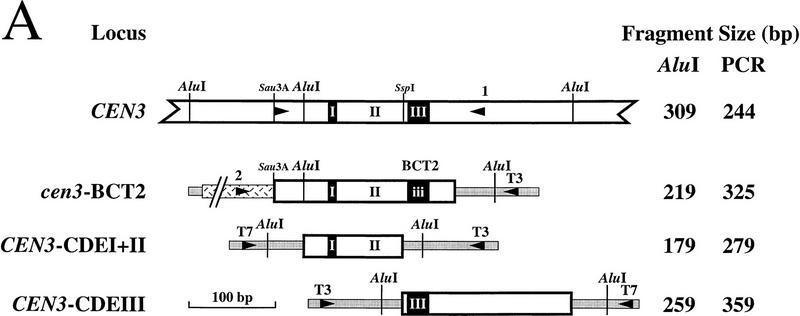
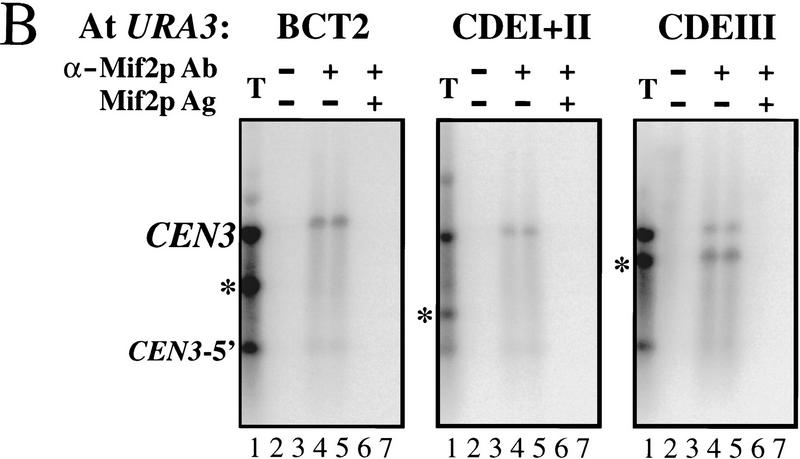
Interaction of Mif2p with segregation-incompetent derivatives of CEN3. (A) Schematic of the CEN3 genomic locus and three segregation-incompetent CEN3 derivatives that were individually integrated at the URA3 locus of wild-type yeast strain MS10. The predicted sizes of CEN3-containing AluI restriction fragments or PCR products are listed. Shading indicates the pBluescript polylinker present in the integrating vector pRS306 (Sikorski and Hieter 1989). Hatching indicates pBR322 sequences present in the cen3–BCT2 construct. Arrowheads designate positions of PCR primers (see Materials and Methods). (B) Crosslinked chromatin (1 hr fixation) prepared from the URA3:: cen3–BCT2, URA3::CEN3–CDEI+II, and URA3:: CEN3–CDEIII strains was mock treated (lanes 2,3) or immunoprecipitated with anti-Mif2p antiserum (C223) in the absence (lanes 4,5) or presence of recombinant Mif2 protein (lanes 6,7). Reactions were prepared in duplicate. Total input material (T) and coimmunoprecipitated DNA were digested with AluI and analyzed by Southern blot hybridization with a CEN3-specific probe. Coimmunoprecipitated DNAs were loaded in sixfold excess relative to the total. The asterisk in each panel indicates the position of the AluI fragment diagnostic for the ectopic locus. Background hybridization reflects prior sonication of the chromatin.
Figure 4.
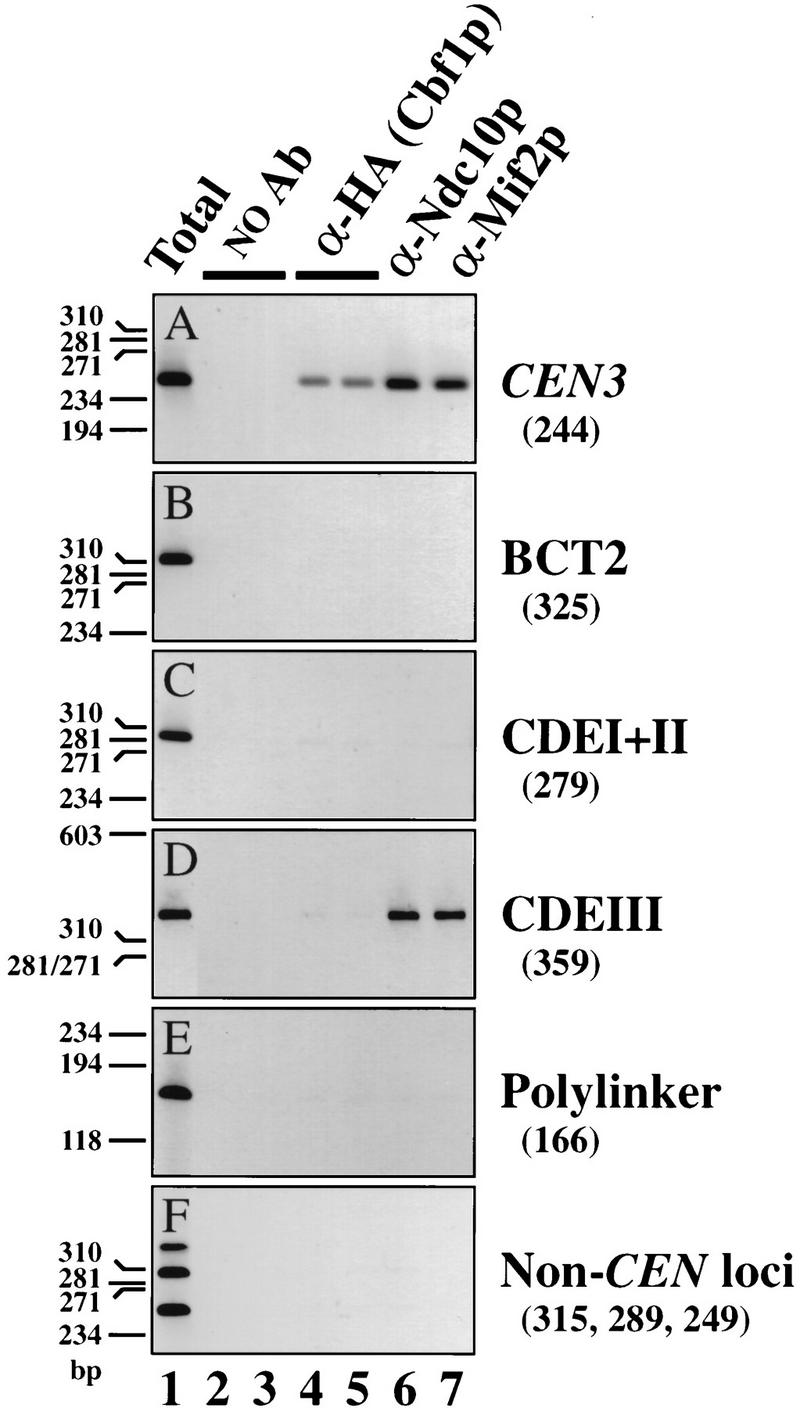
Interaction of centromere proteins Cbf1p and Ndc10p with CEN3 DNA. Cross-linked chromatin (2 hr fixation) was prepared from a set of epitope-tagged CBF1–HA strains in which either pRS306 [vector (polylinker)] or segregation-incompetent CEN3 derivatives (Fig. 3A) were integrated at the URA3 locus. Chromatin was mock-treated (lanes 2,3) or immunoprecipitated with anti-HA [specific for Cbf1-HAp; (lanes 4,5)], an affinity-purified anti-Ndc10p antibody (lane 6), or anti-Mif2p antiserum (lane 7). Aliquots of total input material (≈3 μl chromatin solution) and coimmunoprecipitated DNA (≈30 μl chromatin solution) were analyzed by PCR with primers specific for the authentic CEN3 locus (A), the integrated constructs (B–E), or three noncentromeric loci on yeast chromosome III (F) (see Materials and Methods for details). Shown are negative images of the ethidium bromide-stained products resolved by PAGE. Size standards are indicated to the left of each panel. Expected sizes of the PCR products are in parentheses. A and F are from the URA3::cen3–BCT2 strain; identical results were obtained for the other three strains (data not shown).
CDEIII is both necessary and sufficient for the Mif2p–CEN DNA interaction
To further characterize the Mif2p–CEN DNA interaction, we tested whether Mif2p interacts with segregation-incompetent derivatives of CEN3 that were integrated at the URA3 locus (Fig. 3A). Southern blot (Fig. 3B) or PCR (Fig. 4) analysis of DNA that coimmunoprecipitated with Mif2p allowed the authentic CEN3 locus to be distinguished from each ectopic locus (Fig. 3B, asterisks). A single-base-pair change in the central conserved cytidine residue of CDEIII (cen3–BCT2; McGrew et al. 1986) disrupted the Mif2p–CEN DNA interaction (Figs. 3B, left, and 4B), indicating that intact CDEIII is necessary. Consistent with this, Mif2p failed to interact with a wild-type CEN3–CDEI+II subfragment (Figs. 3B, middle, and 4C). Mif2p is, nonetheless, capable of interacting with CEN DNA within the context of the URA3 locus, as a wild-type CEN3–CDEIII subfragment was enriched in Mif2p immunoprecipitates (Figs. 3B, right, and 4D). Thus, CDEIII is both necessary and sufficient for the Mif2p–CEN DNA interaction in vivo. Moreover, as the cen3–BCT2 (or related) mutation abolishes all known in vivo and in vitro aspects of centromere function (McGrew et al. 1986; Ng and Carbon 1987; Saunders et al. 1988; Kingsbury and Koshland 1991; Lechner and Carbon 1991; Schulman and Bloom 1993; Sorger et al. 1994), we conclude that the interaction of Mif2p with wild-type CEN DNA is biologically relevant.
Ndc10p and Cbf1p also interact with CEN DNA in vivo
To validate our conclusions regarding Mif2p and to initiate a dissection of yeast centromere architecture and assembly, we extended in vivo cross-linking analysis to Ndc10p and Cbf1p, two proteins established previously as centromere components by genetic criteria and various in vitro biochemical assays. Ndc10p could be cross-linked to wild-type CEN DNA (Figs. 4A and 5) but not to several noncentromeric loci (Fig. 4E,F). The Ndc10p–CEN DNA interaction was abolished by the cen3–BCT2 mutation (Fig. 4B). In addition, the wild-type CEN3–CDEIII subfragment (Fig. 4D), but not the complementary CEN3–CDEI+II (Fig. 4C) subfragment, was enriched in Ndc10p immunoprecipitates. Therefore, as with Mif2p, the Ndc10p–CEN DNA interaction occurs through CDEIII and is dependent on the integrity of this element. Thus, Ndc10p is a centromere-associated protein in vivo and its association follows a pattern consistent with the criteria originally used to isolate the CBF3 complex (Ng and Carbon 1987; Lechner and Carbon 1991). By extension, we presume the other CBF3 components—namely, Cbf3Bp, Ctf13p, and Skp1p—are also centromere associated in vivo. Nonetheless, the observation that both Mif2p and Ndc10p can interact with isolated CDEIII motifs in vivo demonstrates the existence of centromeric subcomplexes. The occurrence of such subcomplexes is consistent with previous reports that CDEIII, although incapable of mediating accurate chromosome segregation on its own, is not genetically neutral and can act in cis to reduce the copy number of an otherwise high-copy-number episome (Schulman and Bloom 1993).
Figure 5.
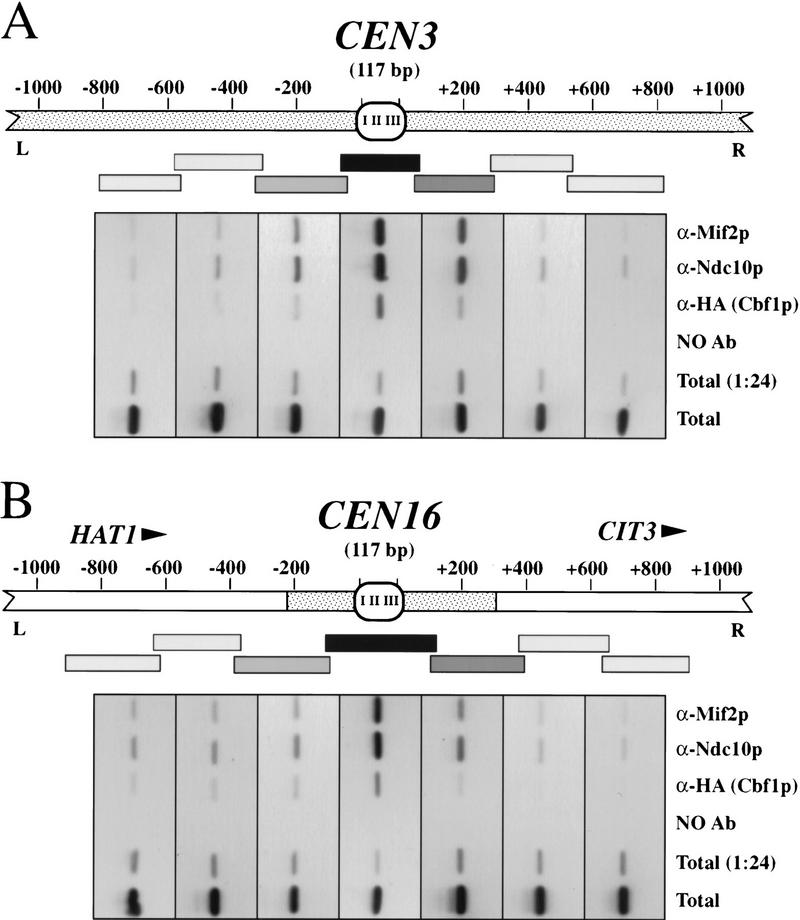
Interactions of Mif2p, Ndc10p, and Cbf1p are limited to the CEN DNA. The CEN3 (A) and CEN16 (B) chromosomal regions are represented. Stippling corresponds to noncoding DNA; orientation and position of open reading frames near CEN16 are indicated. Cross-linked chromatin (2 hr fixation) prepared from a CBF1–HA strain was immunoprecipitated with anti-Mif2p, anti-Ndc10p, anti–HA (specific for Cbf1–HAp), or mock treated. Total input material, a 1:24 dilution, thereof, and coimmunoprecipitated DNAs were analyzed by PCR as in Fig. 4, using primers specific for the indicated series of short, overlapping, CEN-flanking fragments (rectangles). Shading indicates the relative enrichment in the immunoprecipitates of DNA corresponding to those intervals and is based on the negative images of ethidium bromide-stained PCR products shown below each schematic. Data shown are for chromatin prepared from a URA3::cen3–BCT2 strain (A) and a URA3::CEN3–CDEI+II strain (B).
An epitope-tagged version of Cbf1p (Cbf1–HAp) could also be cross-linked to wild-type CEN DNA (Figs. 4A and 5). Thus, this protein is centromere associated in vivo, as predicted from previous studies. However, no interaction was detected between Cbf1–HAp and any segregation-incompetent CEN3 derivative, even though cen3–BCT2 and CEN3–CDEI+II each contain an intact CDEI octamer. In contrast, purified Cbf1p or Cbf1p in crude lysates can readily bind to CDEI-containing fragments in vitro (Baker et al. 1989; Baker and Masison 1990; Cai and Davis 1990; Mellor et al. 1990; Sorger et al. 1995; Kuras et al. 1997). Thus, the Cbf1p–CEN DNA interaction is independent of CDEIII in vitro but dependent on CDEIII in vivo.
Interactions of Mif2p, Ndc10p, and Cbf1p are limited to the centromeric region
It was of interest to determine the extent to which centromere proteins associate with DNA surrounding the CEN DNA proper. Although centromere function is specified by a 120-bp sequence, it is possible that the functional centromeric domain extends beyond the CEN DNA in a sequence-independent fashion. For example, nuclease protection studies indicate that CEN3 is flanked by phased nucleosomes (Bloom and Carbon 1982). Conceivably, one or more centromere proteins also interact with these nucleosomes. To explore this possibility, we analyzed DNA that coimmunoprecipitated with Mif2p, Ndc10p, or Cbf1p for the presence of sequences adjacent to the CEN DNA (Fig. 5A). PCR analysis of ∼2 kb encompassing the CEN3 genomic locus revealed that only the CEN DNA-containing region was strongly enriched in the immunoprecipitates. If a protein participated in interactions beyond the CEN DNA, more than one PCR fragment should have been strongly amplified. Therefore, we conclude that interactions detected at CEN3 are limited to the immediate vicinity of the CEN DNA. Moderate enrichment of the more CEN–proximal regions presumably reflects the heterogeneous nature of the sonicated chromatin. Similar results were obtained for the CEN16 locus (Fig. 5B). The simplest interpretation of these data is that Mif2p, Ndc10p, and Cbf1p interact solely with CEN DNA regions and that any hypothetical centromeric domain that extends beyond the CEN DNA is not defined by these proteins.
Discussion
Mif2p as a centromere protein
Here we show that the S. cerevisiae Mif2 protein, which shares homology with the mammalian centromere protein CENP-C, is a nuclear protein that associates with yeast centromeric DNA in vivo. We demonstrate this association using in vivo formaldehyde cross-linking followed by chromatin immunoprecipitation of CEN DNA fragments with Mif2p-specific antibodies. Importantly, Mif2p had not been implicated in centromere function by in vitro tests. Thus, in vivo cross-linking followed by immunoprecipitation should prove complementary to current in vitro approaches to the analysis of budding yeast centromeres.
The Mif2p–CEN DNA interaction occurs primarily through CDEIII. This result seems inconsistent with previous speculation that Mif2p interacts with CDEII via its potential HMG-I(Y)-related AT-rich DNA-binding motif (Brown et al. 1993; Meluh and Koshland 1995). Our data do not exclude this possibility but simply imply that any such interaction must be dependent on CDEIII. For example, Mif2p might associate with CDEII by a cooperative assembly process that initiates at CDEIII. Assembly would be limited to the CEN DNA, however, as we did not detect interactions outside the immediate centromeric region. How Mif2p interacts with CDEIII in vivo is unclear. The initial interaction could be direct, perhaps through the AT-rich flanks of CDEIII. All 16 CDEIIIs conform to the consensus 5′-tGttTtTG-tTTCCGAAa---aaAa-3′. However, Mif2p has eluded detection as a CDEIII-binding protein in vitro. Alternatively, Mif2p could interact with CDEIII indirectly through components of the CBF3 complex. In this regard, MIF2 and NDC10 show allele-specific genetic interactions (Meluh and Koshland 1995). Also, we have found that mutations in NDC10 substantially diminish the Mif2p–CEN DNA interaction at the nonpermissive temperature (data not shown). Thus, the presence of Mif2p at the centromere might be stabilized by contacts with Ndc10p, as well as with the AT-rich portions of CDEIII and/or CDEII. This model is consistent with our view of the budding yeast centromere as a stereospecific complex (Meluh and Koshland 1995). Finally, a protein capable of binding isolated CDEIII, but that may bind more efficiently to CEN DNA containing both CDEII and CDEIII, has been inferred from in vitro studies of partially reassembled yeast CKCs (Sorger et al. 1994). In light of the preceding discussion, Mif2p would be a good candidate for this hypothetical centromere factor.
Subcomplexes and centromere assembly in budding yeast
We also used in vivo cross-linking to confirm the presence of Ndc10p and Cbf1p on budding yeast centromeres. These two proteins had been regarded almost certainly as centromere proteins by virtue of a large body of genetic and in vitro biochemical data. Analyses of the interactions of Mif2p, Ndc10p, and Cbf1p with segregation-incompetent centromere derivatives produced an unexpected result that may provide insight into centromere assembly in budding yeast. Namely, we found that in vivo the Cbf1p–CEN DNA interaction was dependent on CDEIII, whereas in vitro Cbf1p can bind to CDEI independently of other factors (Baker et al. 1989; Kuras et al. 1997). One explanation for the in vivo result might be that the interaction we detect reflects an indirect cross-link through the CBF3 complex rather than a direct cross-link of Cbf1p to CDEI. This interpretation would agree with genetic arguments for physical association of Cbf1p and CBF3 in vivo (summarized in Meluh and Koshland 1995).
A more interesting explanation for the requirement of CDEIII in Cbf1p–CEN DNA interaction stems from our identification of Mif2p- and Ndc10p-containing subcomplexes on isolated CDEIII DNA. The potential for such subcomplexes was anticipated by the ability of isolated CDEIII motifs to act in cis to reduce plasmid copy number (Schulman and Bloom 1993). Conceivably, CDEIII subcomplexes correspond to authentic centromere assembly intermediates. In such a scenario, centromere assembly would initiate with recruitment of the CBF3 complex and possibly Mif2p to CDEIII (Fig. 6). The resultant subcomplex would then serve as the nucleation site around which a fully functional centromere is assembled if CDEI and CDEII are adjacent. The CDEIII subcomplex could facilitate further assembly directly through protein–protein interactions or indirectly by remodeling nearby chromatin to render the CEN DNA accessible to other centromere factors, such as Cbf1p. We note that a related model based on in vitro analysis of centromere reassembly has been proposed by Sorger et al. (1994).
Figure 6.
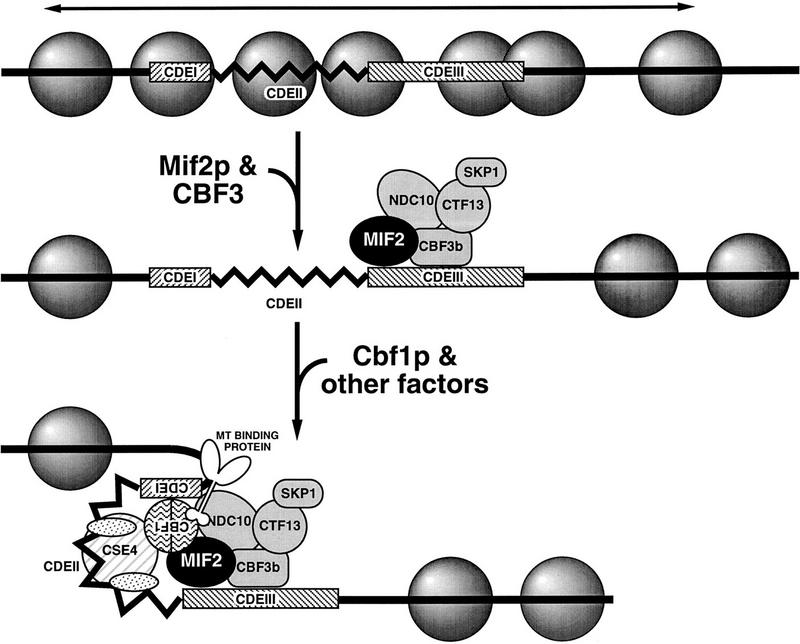
Model for assembly of the budding yeast CKC in vivo. Conserved centromeric DNA elements, CDEI (8 bp) and CDEIII (25 bp), indicated as hatched boxes, flank AT-rich CDEII (78–87 bp). CDEIII is both necessary and sufficient for the interaction of Mif2p and Ndc10p with CEN DNA in vivo. Ndc10p is a component of the multiprotein CDEIII-binding complex, CBF3, as are Cbf3Bp, Ctf13p, and Skp1p. CDEIII is also necessary, though not sufficient, for interaction of the CDEI-binding protein, Cbf1p, with CEN DNA in vivo. The observed in vivo dependence of the Cbf1p–CEN DNA interaction on CDEIII suggests that centromere assembly initiates with binding of factors at CDEIII (e.g., CBF3 components, indicated in gray, and Mif2p), which in turn nucleate further centromere assembly. Nucleosomes are represented by shaded spheres. Potential repressive chromatin that would preclude independent association of some factors (e.g., Cbf1p) is indicated by the horizontal arrow. Other factors comprising the mature CKC might include one or more MT-binding proteins (Kingsbury and Koshland 1991; Sorger et al. 1994), CENP-A-related Cse4p (Stoler et al. 1995), and mitotic checkpoint proteins (Rudner and Murray 1996). Ordered nucleosomal arrays surround the mature CKC in budding yeast (Bloom and Carbon 1982). Genetic evidence for possible looping in the mature centromere is discussed in Meluh and Koshland (1995). For further explanation, see text.
This CDEIII nucleation model for yeast centromere assembly is consistent with the genetic preeminence of CDEIII in centromere specification and function, as well as with additional observations from this and other studies. First, mutations in CDEIII that abolish measurable centromere function (e.g., cen3–BCT2) completely disrupt centromeric chromatin structure as visualized by in vivo footprinting techniques (Saunders et al. 1988). Second, in this study we observed that strains bearing the isolated cen3–CDEIII and associated protein complex do not exhibit a cell cycle delay as judged by FACS and cytological analyses (data not shown). Thus, as would be expected for a normal intermediate in centromere assembly, the CDEIII subcomplex is not itself perceived as aberrant and does not elicit a mitotic checkpoint response. Third, there is some precedent for facilitated binding of Cbf1p to DNA. Specifically, studies addressing the role of Cbf1p in transcriptional activation of sulfur metabolism genes indicate that although Cbf1p can bind cognate recognition sites independently, its in vitro DNA-binding activity is enhanced by association with two other proteins (Kuras et al. 1997). A similar phenomenon might occur at centromeres, as in vitro DNA binding of Cbf1p to various mutant CDEI DNAs does not strictly correlate with the in vivo centromere function of those derivatives (Wilmen et al. 1994). Finally, the requirement for chromatin remodeling in the assembly of fully functional centromeres as invoked by the model is supported by the dependence of centromere function and/or structure on histone levels (Saunders et al. 1990), the histone H3-related Cse4 protein (Stoler et al. 1995), and the chromatin factors Spt4p and Spt6p (Basrai et al. 1996).
As presented, our nucleation model for centromere assembly implies that Mif2p would normally associate with CEN DNA prior to Cbf1p. However, to explain the requirement for CBF1 in a mif2 mutant background, we suggested previously that Cbf1p bound at CDEI enhances centromere association or activity of Mif2p (Meluh and Koshland 1995). As shown here, Mif2p can interact with a CEN3–CDEIII construct in which the nearest CDEI-like element is >1 kb away. Although we have not yet tested whether mutant Mif2p interacts with CEN3–CDEIII, preliminary cross-linking experiments indicate that mutant Mif2p can interact with a CDEI-defective centromere. Therefore, Cbf1p most likely influences the activity, but not the presence of Mif2p at the centromere.
Conservation of structure and function within the centromere
Although centromeres of diverse organisms clearly possess common functional properties, they vary in size and appearance. It has been an open question whether these intricate machines are built from the same or different parts. Prevailing indications are that centromeres differ substantially among organisms at both the DNA and protein level. In contrast, our finding that Mif2p is a centromere protein suggests that centromeres of budding yeast and mammalian cells are structurally as well as functionally conserved.
Although the precise centromeric functions of CENP-C and Mif2p have not been determined, certain functional parallels between the two proteins exist. CENP-C is required for normal centromere assembly and may be an architectural component that mediates interactions at the interface between the centromeric heterochromatin and the outer kinetochore plate to which microtubules attach (Saitoh et al. 1992; Tomkiel et al. 1994). Based on genetic data (Meluh and Koshland 1995), as well as the model for centromere assembly developed here, we speculate that Mif2p also serves an architectural role within the centromere complex, perhaps promoting interactions between factors bound at CDEI and CDEIII. The conserved regions may correspond to surfaces that bind other evolutionarily conserved centromere proteins. Formally, these hypothetical factors could be centromeric chromatin factors that recruit CENP-C or Mif2p, or distal factors (e.g., MT motor or checkpoint proteins) that are themselves recruited to the centromere via CENP-C or Mif2p. Identification of proteins that interact with CENP-C and/or Mif2p will help distinguish between these possibilities.
We believe that additional examples of structural conservation within the centromere will emerge. In budding yeast, aside from Mif2p, the best candidate to date for a conserved centromere protein is Cse4p. Cse4p was originally identified in a genetic screen for chromosome loss mutants and contains a carboxy-terminal histone H3-fold domain similar to that of CENP-A (Stoler et al. 1995). These investigators speculated that like CENP-A, Cse4p is part of a centromere-specific nucleosome. This notion was strengthened by the isolation of CSE4 as a high-copy suppressor of a histone H4 allele that affects chromosome stability and progression through mitosis (Smith et al. 1996). Using our cross-linking assay, we have recently found that Cse4p associates with CEN DNA in vivo (P.B. Meluh, P. Yang, L. Glowczeski, D. Koshland, and M.M. Smith, in prep.), strongly supporting the idea that aspects of centromere structure are conserved even at the most intimate level of chromatin organization.
Thus, the first definitive examples of conserved centromere proteins in yeast—Mif2p and Cse4p—correspond to presumed constituents of mammalian centromeric heterochromatin—CENP-C and CENP-A, respectively. This raises the possibility that structural properties inherent to the chromatin itself can contribute to centromere function, perhaps by providing a unique scaffold for centromere protein assembly or by providing insulation against the negative effects of transcription and/or recombination. An important implication of the observed structural conservation is that paradigms developed in budding yeast will also pertain to mammalian centromeres. Certainly, our findings substantiate the view that the mammalian CKC consists of tandemly repeated MT-binding units similar to the single MT-binding unit represented by the budding yeast centromere (Fitzgerald-Hayes et al. 1982; Zinkowski et al. 1991). Also, the concept of a centromere nucleation complex derived from our observation of CDEIII subcomplexes in vivo may be insightful for thinking about mammalian centromere assembly. Studies suggest that mammalian centromeres assemble on blocks of repetitive DNA at the centromere proper but not on homologous blocks at centromere–distal loci (Grady et al. 1992; for review, see Brinkley et al. 1992; Tyler-Smith and Willard 1993; Pluta et al. 1995). One explanation for these observations is that a CDEIII-like subcomplex at the centromere directs assembly of a mature CKC on the surrounding repetitive DNA and is missing at centromere–distal loci. Thus, although repetitive sequences tend to occur at the centromeres of higher eukaryotes, one cannot rule out the existence of relatively short DNA motifs that would specify a functional centromere.
Conclusion
In summary, we have used cross-linking followed by immunoprecipitation to initiate an in vivo analysis of centromere structure and assembly in budding yeast. Prior to this work, in vivo analysis of yeast centromeric chromatin relied upon nuclease and chemical sensitivity assays, which cannot distinguish direct and indirect effects. Our method does allow such a distinction and thus imparts a kind of molecular cytology to the budding yeast centromere. We note that Saitoh et al. (1997) have recently used a similar technique to demonstrate association of the Mis6 protein with DNA sequences found in the central region of Schizosaccharomyces pombe centromeres. However, in this case, as in many previous applications of in vivo cross-linking (Braunstein et al. 1993; Orlando and Paro 1993; Hecht et al. 1996), the interactions being mapped pertain to repetitive chromatin proteins that bind over many kilobases of a chromosome. Here we describe its successful application to a highly specialized multiprotein complex that assembles on a 120-bp CEN DNA sequence representing <0.002% of the genome. Although the stoichiometry of Mif2p or Ndc10p in the centromere is currently unknown, Cbf1p is thought to bind as a homodimer at CDEI (Mellor et al. 1990; Hegemann and Fleig 1993; Hyman and Sorger 1995). Thus, our ability to detect the presence of Cbf1p underscores the potential sensitivity of this assay in the analysis of specific protein–DNA complexes. The continued application of this methodology in combination with various cis- and trans-acting centromere mutations should reveal many aspects of budding yeast centromere structure and function. Furthermore, a similar strategy could provide equally great insight into the in vivo assembly, structure, and function of other protein–DNA complexes such as promoters, enhancers, and origins of replication.
Materials and methods
Strains
A list of strains used in this study is given in Table 1.
Table 1.
Strains used in these experiments
| Strain
|
Genotype
|
Source
|
|---|---|---|
| 5371-10-2 | MATa ura3 | Brown et al. (1993) |
| 6730 | MATa/MATα ura3/ura3 leu2/+ his3/his3 met2/+sap3/+ mif2::HIS3/+ | Brown et al. (1993) |
| PM1202-7B | MATα ura3 his3 mif2::HIS3 [pPM4; CEN–MIF2] | this study |
| PM1203-1C | MATα ura3 his3 mif2::HIS3 [pPM102; CEN–MIF2–HA] | this study |
| BP5050 | MATa/MATα leu2/+ ade2/+ ade3/+his7/his7 HOM3/+ can1/can1 sap3/sap3 CYC2/+ gal1/gal1 | Guacci et al. (1997) |
| MS10 | MATa ura3-52 leu2-3,112 ade2–101 | Mark Rose (Princeton University, NJ) |
| PM5371-101 | MATa ura3 cbf1::URA3 | Meluh and Koshland (1995) |
| PM1114 | MATa ura3 CBF1–HA | this study |
High copy and HA-tagged alleles of MIF2 and CBF1
All DNA and bacterial manipulations were by standard protocols (Sambrook et al. 1989). Yeast growth media and protocols are described in Rose et al. (1990). Plasmid pPM44 contains a 2.6-kb MIF2 genomic PstI fragment (cloned as a HindIII–SacI fragment from plasmid pPM3; Meluh and Koshland 1995) on the 2μ-based URA3-containing vector pRS202. A closely related vector, pRS426, is described in Christianson et al. (1992). A centromere-based plasmid bearing MIF2–HA (pPM102) was derived from plasmid pPM4 (Meluh and Koshland 1995) by insertion of a 132-bp fragment encoding three copies of the HA epitope after codon 365 of the predicted MIF2 open reading frame. Haploid mif2::HIS3 strains bearing either plasmid pPM4 or pPM102 were obtained by transformation and subsequent dissection of heterozygous mif2::HIS3/+ diploid strain 6730. A 2μ-based plasmid bearing CBF1–HA (pPM81) was derived from pPM35 (Meluh and Koshland 1995) by insertion of a 117-bp fragment encoding three copies of the HA epitope after codon 9 of the predicted CBF1 open reading frame. Haploid CBF1–HA strain PM1114 was derived from cbf1::URA3 (Met−) strain PM5371-101 by one-step gene replacement with the 2.2-kb CBF1–HA fragment from pPM81 and selecting for Met+ transformants.
Generation of Mif2 antisera and immunological techniques
Two anti-Mif2p rabbit antisera (C223 and C224) were raised against a bacterially expressed and Ni2+ affinity-purified (ProBond Resin; Invitrogen, Carlsbad, CA) 6-His-tagged fusion protein containing the carboxy-terminal 186 residues of Mif2p. Anti-Mif2p antiserum C224 was affinity purified against the 6-His–Mif2 fusion protein bound to Immobilon-P PVDF membrane (Millipore Corp., Bedford, MA; Harlow and Lane 1988). For Western blot analysis, yeast whole cell extracts were prepared by mechanically breaking cells in buffer containing 1% SDS, 100 mm NaCl, 5 mm EDTA, 50 mm Tris-HCl at pH 7.4, and 1 mm PMSF. Lysates were mixed with sample buffer, heated briefly at 100°C, and clarified. Proteins were resolved by SDS-PAGE (7.5%), transferred to nitrocellulose, and probed with Mif2p antiserum (C223; 1:2000 dilution). Proteins bound by anti-Mif2p were detected using a goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (Bio-Rad Laboratories, Hercules, CA) and Renaissance chemiluminescence reagent (DuPont NEN, Boston, MA). Indirect immunofluorescence was performed as described (Rose et al. 1990) except that cells were fixed for ≤1 hr, as detection of Mif2p expressed at wild-type levels was dependent on shorter fixation. Mif2p was visualized using affinity-purified anti-Mif2p and a Cy3-conjugated goat anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Microtubules were visualized using the rat monoclonal antibody YOL1/34 directed against yeast α-tubulin (Serotec, Indianapolis, IN) and a FITC-conjugated goat anti-rat secondary antibody. Total DNA was visualized using DAPI. Images were taken on a Zeiss Axioskop microscope using a Micromax cooled CCD camera (Princeton Instruments, Inc., Princeton, NJ) and IPLab Spectrum software (Scanalytics Corp., Vienna, VA).
Segregation-incompetent derivatives of CEN3
The cen3–BCT2-integrating plasmid pPM147 was created by inserting a 0.58-kb SalI DNA fragment containing the cen3–BCT2 allele (211 bp) into the yeast-integrating plasmid pRS306 (URA3; Sikorski and Hieter 1989). The cen3–BCT2 fragment, originally derived from pBCT2 (McGrew et al. 1986), also contains the HindIII–BamHI region of pBR322 (346 bp) 5′ to CDEI. To create plasmids pPM148 and pPM149, which contain wild-type CEN3–CDEI+II and CEN3–CDEIII DNA, respectively, a 0.32-kb BamHI DNA fragment, corresponding to a genomic CEN3-containing AluI fragment to which BamHI linkers had been added, was digested with SspI. The resultant 0.12-kb BamHI–SspI fragment containing CEN3–CDEI+II and the 0.2-kb SspI–BamHI fragment containing CEN3–CDEIII plus 3′-flanking sequence were then individually subcloned into pRS306. Plasmids pRS306, pPM147, pPM148, and pPM149 were each stably integrated at the URA3 locus of wild-type yeast strain MS10 and CBF1–HA strain PM1114 by a standard transformation protocol.
In vivo cross-linking and chromatin immunoprecipitation
Yeast strains were grown in YPD to an OD600 of 1.2–1.5, then treated in situ with 1% formaldehyde. Fixation time varied. Cross-linked chromatin solutions suitable for immunoprecipation were prepared as described by Braunstein et al. (1993). Fixed cells were harvested, washed, and converted to spheroplasts using oxalyticase. Spheroplasts were washed, resuspended in lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris at pH 8.1), supplemented with protease inhibitors (1 mm PMSF, 0.6 μg/ml of leupeptin, 0.8 μg/ml of pepstatin A), and sonicated to fragment chromosomal DNA to an average size of 300–1000 bp. Sonicated material was diluted into immunoprecipitation buffer (final concentration, 0.1% SDS; 1% Triton X-100; 150 mm NaCl; 2 mm EDTA; 20 mm Tris at pH 8.1) supplemented with protease inhibitors. Aliquots of the resultant chromatin solution corresponding to ∼18 OD600 cell equivalents were incubated with antibody overnight at 4°C. Crude anti-Mif2p rabbit antisera were used at a 1:250 or 1:500 dilution. Affinity-purified anti-Mif2p antibody was used at a 1:50 dilution. In some cases, recombinant 6-His–Mif2 antigen was included in the immunoprecipitation at a concentration of ≤6.7 μg/ml. Purified anti-HA monoclonal antibody 12CA5 (BAbCO, Richmond, CA) was used at 4 μg/ml final concentration. The anti-Ndc10p antibody, provided by Anthony Hyman (European Molecular Biology Laboratory, Heidelberg, Germany), was raised and affinity purified against the amino-terminal 444 amino acids of Ndc10p. Following addition of sonicated λ DNA (2 μg), immune complexes were harvested by incubation with protein A–Sepharose CL-4B beads (Pharmacia Biotech, Uppsala, Sweden) for 1–2 hr. Beads were sequentially washed as described by Braunstein et al. (1993), being transferred to a fresh tube prior to the final TE wash (10 mm Tris at pH 8.0, 1 mm EDTA). Immunoprecipitated material was eluted from the beads with 1% SDS, 0.1 m Na2CO3, heated at 65°C for 4–5 hr to reverse formaldehyde cross-links, and ethanol precipitated. Aliquots of total chromatin solution were similarly heat treated and precipitated. Recovered material was treated with proteinase K (Boehringer Mannheim, Germany), extracted with organic solvents, and ethanol precipitated.
Analysis of coimmunoprecipitated DNA
Resultant total input DNA and coimmunoprecipitated DNA samples were analyzed by slot blot or Southern hybridization, or by PCR. For slot blot analysis, DNA samples were diluted in 6× SSC, denatured by heating at 100°C for 10 min, and applied to Nytran membrane filters (Schleicher & Schuell; Keene, NH). For Southern analysis, DNA samples were cut with AluI, treated with RNase, resolved on 2.5% agarose gels, and transferred to GeneScreen Plus (NEN Research Products; Boston, MA). [α-32P]dATP-Labeled hybridization probes were prepared by random hexamer priming using the following templates: a 244-bp CEN3-containing PCR fragment (Sau3A/Primer 1 product; see Fig. 3A); a 345-bp CEN16-containing PCR fragment (see below); a 660-bp BglII fragment from the coding region of TUB2; and sonicated total yeast DNA. Blots were hybridized and washed as described by Church and Gilbert (1984). Hybridized blots were digitally imaged and quantitated using the storage Phospor technology of the Storm 860 imaging system and ImageQuant software (Molecular Dynamics, Inc.; Sunnyvale, CA). PCR reactions (24 cycles) used 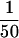 of the immunoprecipitates and
of the immunoprecipitates and 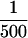 of the total material input and serial dilutions thereof. PCR products (one-third reaction) were separated on 8% polyacrylamide gels and visualized with ethidium bromide. The region encompassing CEN3 (bp 113925–114168 of chromosome III) was amplified with the Sau3A primer (5′-GATCAGCGCCAAACAATATGG-3′) and Primer 1 (5′-AACTTCCACCAGTAAACGTTTC-3′), as indicated in Figure 3A. The cen3–BCT2 mutation was amplified with Primer 2 (5′-CACTATCGACTACGCGATCA-3′; 5′-to pBR322 BamHI site) and the T3 primer (5′-AATTAACCCTCACTAAAGGG-3′). Other segregation incompetent CEN3 derivatives and the pRS306 polylinker were amplified with the T3 and T7 (5′-TAATACGACTCACTATAGGG-3′) primers, which flank the pRS306 polylinker. To test noncentromeric loci on chromosome III, primer pairs were designed that correspond to part of the LEU2 coding region (bps 91019–91267; ∼23 kb to the left of CEN3); an AT-rich intergenic region near PGK1 (bp 138557–138845; ∼24.5-kb to the right of CEN3); and an AT-rich intergenic region near HMRa (bp 291213–291527; ∼177 kb to the right of CEN3). Primers specific for CEN16 (bp 555845–556189 on chromosome XVI) and CEN1 (bp 151379–151681 on chromosome I) were also used. Details regarding primers for amplification of fragments adjacent to CEN3 and CEN16 are available on request.
of the total material input and serial dilutions thereof. PCR products (one-third reaction) were separated on 8% polyacrylamide gels and visualized with ethidium bromide. The region encompassing CEN3 (bp 113925–114168 of chromosome III) was amplified with the Sau3A primer (5′-GATCAGCGCCAAACAATATGG-3′) and Primer 1 (5′-AACTTCCACCAGTAAACGTTTC-3′), as indicated in Figure 3A. The cen3–BCT2 mutation was amplified with Primer 2 (5′-CACTATCGACTACGCGATCA-3′; 5′-to pBR322 BamHI site) and the T3 primer (5′-AATTAACCCTCACTAAAGGG-3′). Other segregation incompetent CEN3 derivatives and the pRS306 polylinker were amplified with the T3 and T7 (5′-TAATACGACTCACTATAGGG-3′) primers, which flank the pRS306 polylinker. To test noncentromeric loci on chromosome III, primer pairs were designed that correspond to part of the LEU2 coding region (bps 91019–91267; ∼23 kb to the left of CEN3); an AT-rich intergenic region near PGK1 (bp 138557–138845; ∼24.5-kb to the right of CEN3); and an AT-rich intergenic region near HMRa (bp 291213–291527; ∼177 kb to the right of CEN3). Primers specific for CEN16 (bp 555845–556189 on chromosome XVI) and CEN1 (bp 151379–151681 on chromosome I) were also used. Details regarding primers for amplification of fragments adjacent to CEN3 and CEN16 are available on request.
Acknowledgments
We gratefully thank Anthony Hyman for generously providing anti-Ndc10p antibody, Alejandro Roth for assistance in the construction of the epitope-tagged MIF2–HA strain, and members of the Koshland laboratory for critical reading of the manuscript. This work was supported by grant GM41718-07 from the National Institutes of Health to D.K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
We note that Tomoyuki et al. (Cell 90: 649–660) and Aparicio et al. (Cell 91: 59–69) have recently applied in vivo cross-linking to the analysis of yeast origins of replication. Also, Starr et al. (J. Cell Biol. 138: 1289–1301) have recently identifed homologs of the Drosophila ZW10 centromere protein in diverse plant and animal species, further supporting the idea of structural conseration within centromeres.
Footnotes
E-MAIL meluh@mail1.ciwemb.edu; FAX (410) 243-6311.
References
- Baker RE, Masison DC. Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol Cell Biol. 1990;16:2458–2467. doi: 10.1128/mcb.10.6.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RE, Fitzgerald-Hayes M, O’Brien TC. Purification of the yeast centromere binding protein CP1 and a mutational analysis of its binding site. J Biol Chem. 1989;264:10843–10850. [PubMed] [Google Scholar]
- Basrai MA, Kingsbury J, Koshland D, Spencer F, Hieter P. Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2838–2847. doi: 10.1128/mcb.16.6.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom KS, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes & Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- Brinkley BR, Ouspenski I, Zinkowski RP. Structure and molecular organization of the centromere-kinetochore complex. Trends Cell Biol. 1992;2:15–21. doi: 10.1016/0962-8924(92)90139-e. [DOI] [PubMed] [Google Scholar]
- Brown MT. Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene. 1995;160:111–116. doi: 10.1016/0378-1119(95)00163-z. [DOI] [PubMed] [Google Scholar]
- Brown MT, Goetsch L, Hartwell LH. MIF2 is required for mitotic spindle integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J Cell Biol. 1993;123:387–403. doi: 10.1083/jcb.123.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Davis RW. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990;61:437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke CA, Bernat RL, Earnshaw WC. CENP-B: A major human centromere protein located beneath the kinetochore. J Cell Biol. 1990;110:1475–1488. doi: 10.1083/jcb.110.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedon PC, Soults JA, Allis CD, Gorovsky MA. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal Biochem. 1991;197:83–90. doi: 10.1016/0003-2697(91)90359-2. [DOI] [PubMed] [Google Scholar]
- Doheny K, Sorger P, Hyman A, Tugendreich S, Spencer F, Hieter P. Identification of essential componenets of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Sart D, Cancilla MR, Earle E, Mao JI, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KH. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nature Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Ratrie H, Stetten G. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Goh PY, Kilmartin J. NDC10: A gene involved in chromosome segregation in S. cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky GJ. Kinetochores, microtubules and the metaphase checkpoint. Trends Cell Biol. 1995;5:143–148. doi: 10.1016/s0962-8924(00)88968-0. [DOI] [PubMed] [Google Scholar]
- Grady DL, Ratliff RL, Robinson DL, McCanlies EC, Meyne J, Moyzis RK. Highly conserved repetitive DNA sequences are present at human centromeres. Proc Natl Acad Sci. 1992;89:1695–1699. doi: 10.1073/pnas.89.5.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Centromere position in budding yeast: Evidence for anaphase A. Mol Biol Cell. 1997;8:957–972. doi: 10.1091/mbc.8.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- Hegemann JH, Fleig UN. The centromere of budding yeast. BioEssays. 1993;15:451–460. doi: 10.1002/bies.950150704. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Sorger PK. Structure and function of kinetochores in budding yeast. Annu Rev Cell Dev Biol. 1995;11:471–495. doi: 10.1146/annurev.cb.11.110195.002351. [DOI] [PubMed] [Google Scholar]
- Jiang W, Lechner J, Carbon J. Isolation and characterization of a gene (CBF2) specifying a protein component of the budding yeast kinetochore. J Cell Biol. 1993;121:513–519. doi: 10.1083/jcb.121.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury J, Koshland D. Centromere-dependent binding of yeast minichromosomes to microtubules in vitro. Cell. 1991;66:483–495. doi: 10.1016/0092-8674(81)90012-x. [DOI] [PubMed] [Google Scholar]
- Kuras L, Barbey R, Thomas D. Assembly of a bZIP-bHLH transcription activation complex: Formation of the yeast Cbf1-Met4-Met28 complex is regulated through Met28 stimulation of Cbf1 DNA binding. EMBO J. 1997;16:2441–2451. doi: 10.1093/emboj/16.9.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J. A zinc finger protein, essential for chromosome segregation, constitutes a putative DNA binding subunit of the Saccharomyces cerevisiae kinetochore complex, Cbf3. EMBO J. 1994;13:5203–5211. doi: 10.1002/j.1460-2075.1994.tb06851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J, Carbon J. A 240 kD multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew J, Diehl B, Fitzgerald-Hayes M. Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:530–538. doi: 10.1128/mcb.6.2.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J, Jiang W, Funk M, Rathjan J, Barnes CA, Hinz T, Hegemann JH, Philippsen P. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 1990;8:4017–4026. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R, Carbon J. Mutational and in vitro protein-binding studies on centromere DNA from Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:4522–4534. doi: 10.1128/mcb.7.12.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- Pangilinan F, Spencer F. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol Biol Cell. 1996;7:1195–1208. doi: 10.1091/mbc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta A, Mackay A, Ainsztein A, Goldberg I, Earnshaw W. The centromere: Hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics: A laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Rudner AD, Murray AW. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Tomkiel J, Cooke CA, Ratrie H, Maurer M, Rothfield NF, Earnshaw WC. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- Saitoh S, Takahashi K, Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Saunders M, Fitzgerald-Hayes M, Bloom K. Chromatin structure of altered yeast centromeres. Proc Natl Acad Sci. 1988;85:175–179. doi: 10.1073/pnas.85.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders MJ, Yeh E, Grunstein M, Bloom K. Nucleosome depletion alters the chromatin structure of Saccharomyces cerevisiae centromeres. Mol Cell Biol. 1990;10:5721–5727. doi: 10.1128/mcb.10.11.5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman IG, Bloom K. Genetic dissection of centromere function. Mol Cell Biol. 1993;13:3156–3166. doi: 10.1128/mcb.13.6.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MM, Yang P, Soledad SM, Boone PW, Goldstein AT, Megee PC. A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol Cell Biol. 1996;16:1017–1026. doi: 10.1128/mcb.16.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Severin FF, Hyman AA. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J Cell Biol. 1994;127:995–1008. doi: 10.1083/jcb.127.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Doheny KF, Hieter P, Kopski KM, Huffaker TC, Hyman AA. Two genes required for the binding of an essential Saccharomyces cerevisiae kinetochore complex to DNA. Proc Natl Acad Sci. 1995;92:12026–12030. doi: 10.1073/pnas.92.26.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmann O, Lechner J. The Saccharomyces cerevisiae kinetochore contains a cyclin-CDK complexing homologue, as identified by in vitro reconstitution. EMBO J. 1996;15:3611–3620. [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes & Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Kingsbury J, Koshland D. CEP3 encodes a centromere protein of Saccharomyces cerevisiae. J Cell Biol. 1995;128:749–760. doi: 10.1083/jcb.128.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Schwartz S. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet. 1995;4:2189–2197. doi: 10.1093/hmg/4.12.2189. [DOI] [PubMed] [Google Scholar]
- Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkiel J, Cooke CA, Saitoh H, Bernat RL, Earnshaw WC. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J Cell Biol. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith C, Willard HF. Mammalian chromosome structure. Curr Opin Genet Dev. 1993;3:390–397. doi: 10.1016/0959-437x(93)90110-b. [DOI] [PubMed] [Google Scholar]
- Wilmen A, Pick H, Niedenthal RK, Sen-Gupta M, Hegemann JH. The yeast centromere CDEI/Cpf1 complex: Differences between in vitro binding and in vivo function. Nucleic Acids Res. 1994;22:2791–2800. doi: 10.1093/nar/22.14.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: a repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]