Abstract
We investigated the effect of the protein corona on the function of nanoparticle (NP) antibody (Ab) conjugates in dipstick sandwich immunoassays. Ab specific for Zika virus nonstructural protein 1 (NS1) were conjugated to gold NPs, and another anti-NS1 Ab was immobilized onto the nitrocellulose membrane. Sandwich immunoassay formation was influenced by whether the strip was run in corona forming conditions, i.e., in human serum. Strips run in buffer or pure solutions of bovine serum albumin exhibited false positives, but those run in human serum did not. Serum pretreatment of the nitrocellulose also eliminated false positives. Corona formation around the NP-Ab in serum was faster than the immunoassay time scale. Langmuir binding analysis determined how the immobilized Ab affinity for the NP-Ab/NS1 was impacted by corona formation conditions, quantified as an effective dissociation constant, KDeff. Results show that corona formation mediates the specificity and sensitivity of the antibody–antigen interaction of Zika biomarkers in immunoassays, and plays a critical but beneficial role.
Graphical abstract
INTRODUCTION
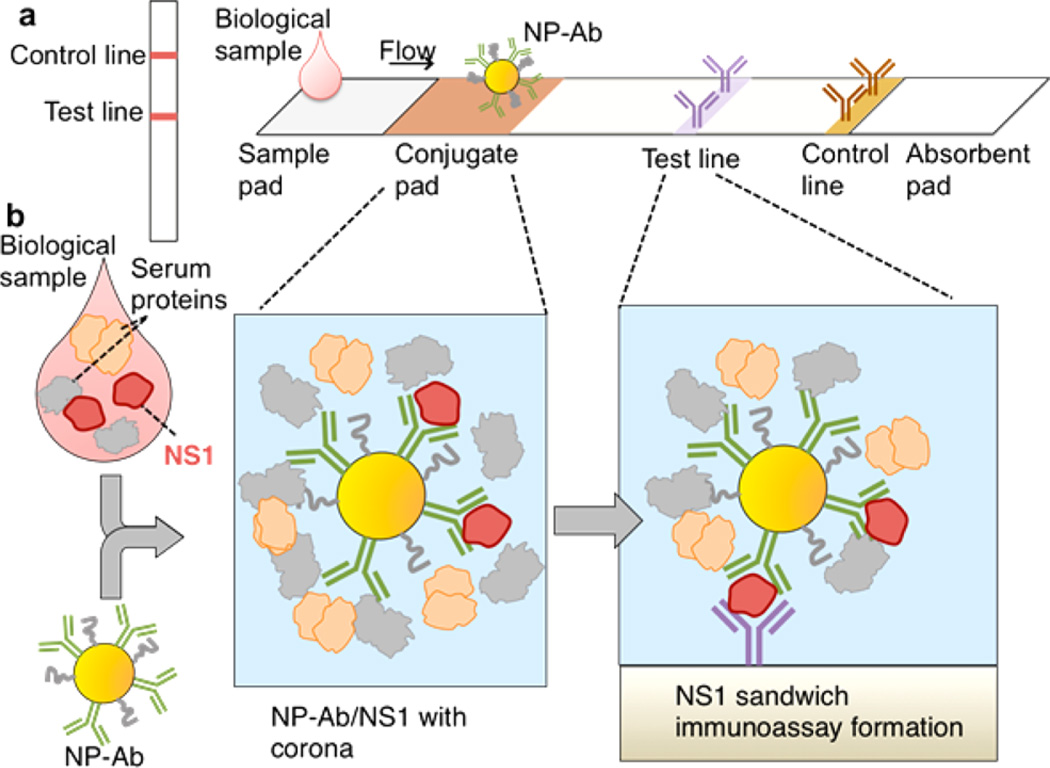
It is widely known that when nanoparticles (NPs) are introduced into biological environments such as blood, serum, or intracellular fluid, the proteins present in these fluids form a protein corona around the NPs.1 Protein coronas mediate the interactions between NPs and their environment, influencing processes such as association with cell surfaces and intracellular uptake.2–6 Often protein coronas have negative impacts on these processes, as the corona around the NP can actually obscure its targeting ligand, reducing the specificity of interaction of the targeting species on the NP surface.7–9 The impact of the protein corona has been studied predominantly for aspects of nanomedicine concerned with the transport, blood circulation, and cell interactions of nanomaterials such as targeting and cell uptake. It is now known that the protein corona mediates interactions of NPs with cell receptors and other binding species, influencing the specificity of cell uptake,8,10–12 and can have both negative and beneficial side effects.13–15 However, another situation where the protein corona has a significant impact is in point of care immunoassays used to detect pathogens and other biomarkers in blood, serum, and other body fluids. In lateral flow immunoassays (Figure 1a),16,17 a biological fluid is added to the sample pad where it wicks to the conjugate pad through capillary action.18,19 In the conjugate pad, the biological fluid is mixed with gold NPs conjugated to antibodies that are selected against the biomarker of interest. Then, the NP-Ab suspended in serum continue wicking through the nitrocellulose membrane where two antibodies have been immobilized, an Ab against the biomarker of interest in the test line, and an Ab against the Ab on the gold NPs in the positive control band. If the biomarker of interest is present in the fluid, it binds simultaneously to antibodies conjugated to the NP (NP-Ab), as well as to the specific antibodies immobilized on the test line. Due to the optical properties of the NPs, a color appears on the test line due to the accumulation of the NPs. In the positive control band, there is NP-Ab binding to an anti-IgG Ab immobilized on the paper.
Figure 1.
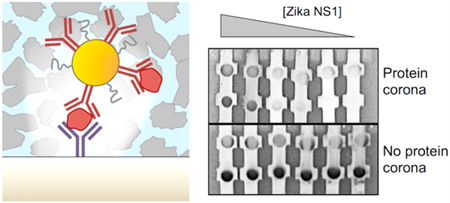
Protein coronas influence sandwich immunoassays. (a) Schematic of a lateral flow immunoassay, which gives rise to two colored lines for a positive test, one line for a negative test. (b) Protein corona formation occurs when the NP-Ab conjugates are mixed with the biomarker NS1 in the biological sample such as serum, and can influence the formation of the sandwich immunoassay at the test line.
For the assay to be successful, the biomarker must be able to bind to the antibodies on the NP and also those on the test line. This binding must be accomplished in the presence of all the proteins, ions, and small molecules that are present in the biological fluid. Here, these proteins are present at high concentrations and thus can form coronas that can potentially obscure the critical antibody–antigen interactions and prevent binding events (Figure 1b). If the corona prevents binding, it can result in false negatives, which can have serious consequences for a diagnostic assay. On the other hand, the corona composition could also lead to false positives, leading to a nonspecific diagnostic. This can appear when other proteins in the sample can bind to the NP-Ab. Several methods, including the addition of sucrose or Tween, or adding capture negative bands in the nitrocellulose, have been used to remove false positives. However, false negatives typically remain a major challenge for dipstick and lateral flow immunoassays. Surface effects20,21 from both the NP and the nitrocellulose substrate and the capillary flow regime of the entire system can further complicate the environment of the test. Lateral flow and dipstick point of care immunoassays have been utilized on a wide range of biological fluids, not only blood and serum, but also urine, saliva, semen, and breast milk, which can result in coronas with different compositions and physical properties. Despite its importance, the protein corona has been largely overlooked in the context of lateral flow and dipstick immunoassays. Thus, there is a need to understand how the protein corona impacts antibody–antigen binding in these sandwich immunoassays.
Here, we studied how the protein corona impacts antibody–antigen binding in sandwich immunoassays for the Zika virus (ZIKV) biomarker nonstructural protein 1 (NS1) in human serum. We probed the properties of coronas that form around the NP-Ab conjugates and quantify the ability of the NP-Ab to bind to Zika NS1 and cause a spot to appear at the test line. We found that the protein corona resulting from the serum influences the outcome of the test. We present an approach to quantify the binding affinity of the Ab to the ligand NS1 using a modified Langmuir isotherm, and show that the binding affinities are modified by the protein coronas. Results show that the corona that forms around the NP-Ab conjugate can have important ramifications, and is a positive side effect. Furthermore, these results underscore the importance of the protein corona in LF/dipstick assays that require the use of serum and other bodily fluids.
RESULTS AND DISCUSSION
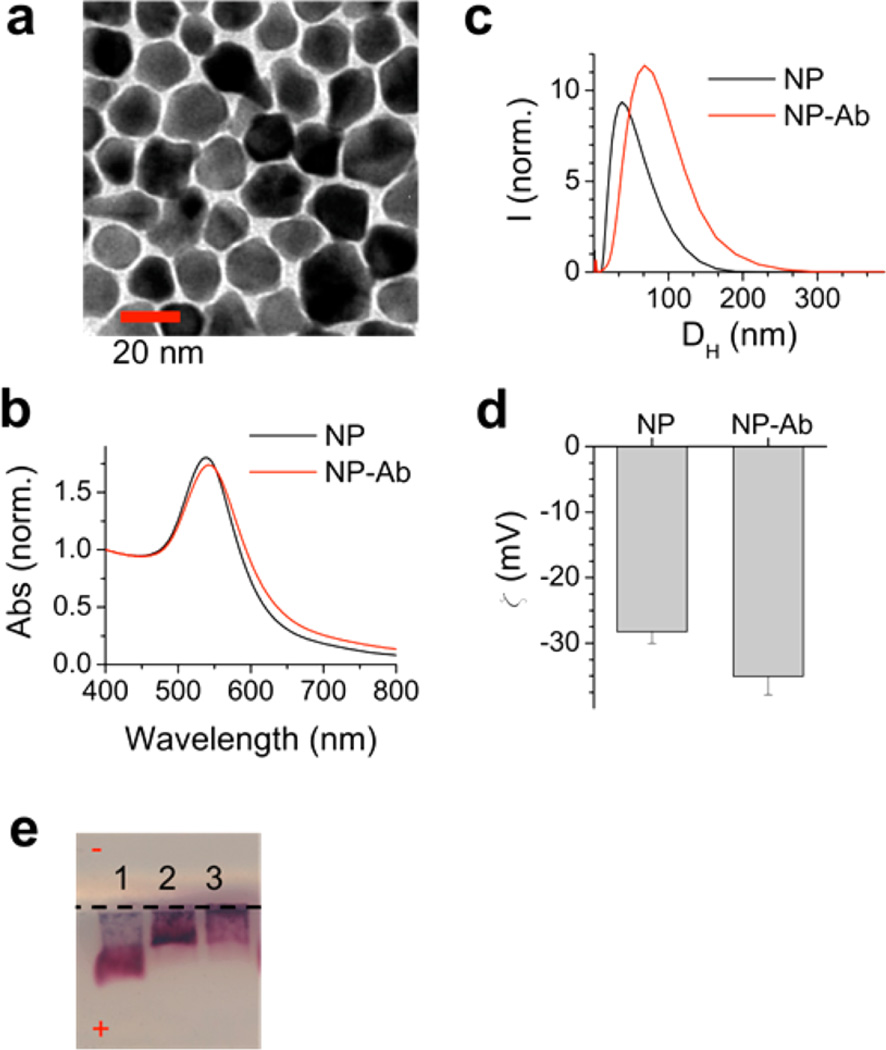
Au star-shaped NPs were synthesized using literature methods, resulting in nanostars with short arms.22,23 TEM imaging showed that the NPs had a mean diameter of 14.5 ± 3.1 nm (Figure 2a). Optical absorption spectra of the NPs showed an SPR peak at 536 ± 5 nm (Figure 2b, black). Measurement of the hydrodynamic diameter (DH) by dynamic light scattering (DLS) showed that the NPs had DH = 32.6 ± 2.6 nm (Figure 2c, black line), and zeta potential measurements determined that the NPs were negatively charged with a zeta potential of −28.3 ± 1.8 mV (Figure 2d). NPs have a known molar extinction coefficient24 (ε536 = 5.7 × 108 M−1 cm−1) which allowed quantification of the NP concentration from the optical absorption spectrum.
Figure 2.
NP-Ab conjugate characterization. (a) TEM image of the NPs synthesized, scale bar = 20 nm. (b) Optical absorption spectra of the NPs (black), NP-Ab conjugates (red). (c) DLS spectra of the NPs (black), NP-Ab conjugates (red). (d) Zeta potential measurements of bare NPs, NP-Abs. (e) Gel electrophoresis of Lane 1, NPs as synthesized; Lane 2, NPs-Abs; and Lane 3, NP-Abs with PEG-thiol backfill.
NPs were conjugated to monoclonal Abs selected for Zika virus nonstructural protein 1 (NS1), a biomarker for the disease (Materials and Methods). Antibodies were selected by injecting mice with Zika NS1 protein, and immortalizing the B cells capable of binding to the Zika NS1, but not to NS1 from other similar viruses (such as Dengue, Yellow Fever, Japanese Encephalitis, West Nile, or tick-borne encephalitis). The anti-NS1 antibodies were conjugated to the NPs by a covalent attachment method (Materials and Methods). Briefly, we used a heterobifunctional linker with a hydrazide on one end and a dithiol on the other, linked by a short polyethylene glycol (PEG) chain (hydrazide dithiolalkanearomatic PEG6-NHNH2). We also explored the use of a noncovalent attachment method where Abs were added to the NPs at an incubation ratio of 100:3, and the Abs were allowed to adsorb onto the surface of the NPs (Supporting Information, Figure S4).25–27
After bioconjugation, the NP surfaces were modified with thiolated PEG of a MW = 5 kDa as a backfill, which is used to reduce nonfavorable Ab–NP and Ab–Ab interactions on the NP surface.28–30 Following this, free Abs were separated by centrifugation. DLS showed that the NPs-Ab conjugates had a DH = 71.6 ± 6.2 nm, and they were larger than the NPs (Figure 2c, red) and that the charge of the NPs, with a zeta potential of −30.1 ± 1.5 mV, became more negative (Figure 2d) relative to the plain NPs, confirming successful Ab conjugation. Gel electrophoresis showed a mobility shift (Figure 2e, lane 3) relative to the NP (lane 1). The NP SPR red-shifted and broadened slightly upon Ab conjugation (Figure 2b, red).
Previous work from our lab has found that PEG backfills improve Ab functionality of the NP-Ab conjugate and reduce nonspecific binding of the conjugate in lateral flow assays, as well as other uses of NP-biomolecule conjugates.31,32 PEG backfill of the NPs did not result in a mobility shift in gel electrophoresis (Figure 2e, lanes 2 and 3) due to the mild backfill conditions, confirming that the Abs were not displaced from the NP. In addition, fluorescence spectroscopy and ELISA of the supernatant after PEG backfill also confirmed that there was negligible Ab in the supernatant, confirming that the Abs were not displaced (Figure S5, Supporting Information). The Ab coverage on the NPs was determined by measuring the change in concentration of free Ab in solution after conjugation to the NPs using fluorescence spectroscopy and ELISA. Ab coverage was determined to be 4.3 ± 2.2 Abs/NP, equivalent to an Ab footprint of 232.6 nm2 per Ab using the calculated surface area of a NP of 1000 nm2.24 The dimensions of a typical IgG antibody of 14.5, 8.5, and 4 nm,33 which has an average footprint of 81.3 nm2, suggest that the coverage is less than a monolayer. However, it is of note that even if the Ab had a high coverage resulting in a multilayer on the NPs, it does not preclude the NP-Ab from being able function in an immunoassay.
We also measured whether or not the Ab would be displaced from the NP-Ab conjugates upon incubation with the various solutions used in the experiments. We measured the amount of Ab in the supernatant by fluorescence spectroscopy and ELISA after exposure to HS, HSA, BSA, and PBS and found the displaced Abs to be negligible for both covalently attached and noncovalently attached Abs (Figure S5, Supporting Information).
Immmunoassays Run in Different Biological Fluids
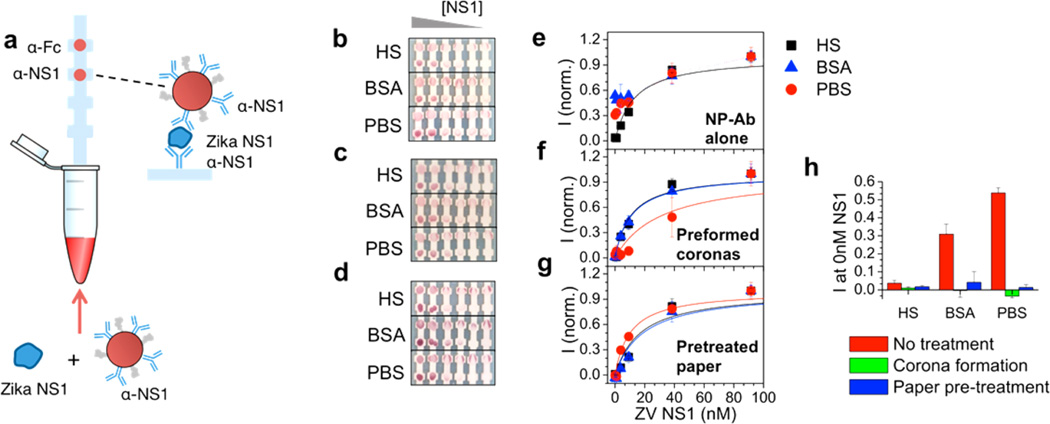
First, the strips were run with the purified NP-Ab in different biological fluids: human serum (HS), pure bovine serum albumin (BSA), and phosphate buffered saline (PBS). The dipstick assay consisted of a nitrocellulose strip onto which anti-Zika NS1 monoclonal antibodies were immobilized on the test area (Figure 3a). Also, a control antibody (anti-Fc) was immobilized on the control area. NP-Ab was mixed with NS1 at varying concentrations in solution, into which the nitrocellulose strip was partially submerged. Upon contact with the nitrocellulose, the fluid migrated up the strip by capillary action to a wick (not depicted for clarity). For the strip run only in PBS (Figure 3b, bottom), the test line intensity was present even when there was no NS1 in the sample (Figure 3h, red). This indicates that the NP-Ab conjugate nonspecifically bound to immobilized Abs on the nitrocellulose. The NP-Ab does not bind to the nitrocellulose outside of the test line, so this was not simply due to NP-Ab nitrocellulose interactions, but evidently the PBS modifies electrostatic screening in a way that affects the specificity of protein–protein or protein–NP interactions. The rightmost strip in Figure 3b in PBS constitutes a false positive. ImageJ34 was used to analyze the test line intensities as a function of NS1 concentration, and the test line intensity showed a concentration dependence that did not reach baseline at 0 nM NS1 (Figure 3e, red circles; Figure 3h).
Figure 3.
Sandwich immunoassay results. (a) Schematic of dipstick immunoassays used. Dipstick immunoassay results as a function of ZIKV NS1 concentration (L–R): 91.6 nM, 38.4 nM, 9.2 nM, 3.8 nM, 0.9 nM, 0 nM for (b) NP-Abs in PBS, BSA, and HS; (c) NP-Abs with preformed coronas in PBS, BSA, and HS; (d) for test strips pretreated with HS and then run in PBS, BSA, and HS. Top dot: control line. Bottom dot: test line. (e) Test line intensity vs NS1 concentration for NP-Abs in PBS (red circles), BSA (blue triangles), and HS (black squares), and fits to KDeff (lines). (f) Test line intensity vs NS1 concentration curves for NP-Abs with preformed protein coronas in PBS (red circles), BSA (blue triangles), and HS (black squares), and fits to KDeff (lines). (g) Test line intensity vs NS1 concentration curves for strips pretreated with HS and then run in in PBS (red circles), BSA (blue triangles), and HS (black squares), and fits to KDeff (lines). (h) Intensity of the test strips at 0 nM NS1 (intensity of false positives) under no treatment (red), preformed coronas (green), and paper HS pretreatment (blue).
The strips were then run in a 10 mg/mL solution of BSA in PBS, which is widely used as a blocking agent for nonspecific adsorption. Again, a band at the test line was present for no NS1 present (Figure 3b, middle), indicating that the NP-Ab was nonspecifically binding to the immobilized antibodies on the test line. Like with PBS, the rightmost strip in Figure 3b in BSA constitutes a false positive. The intensity decreased with NS1 concentration but did not reach baseline at 0 nM NS1 (Figure 3e, blue triangles, and Figure 3h).
When the strips were run with HS (Figure 3b, top), the test line intensity decreased with decreasing NS1 concentration (Figure 3e, black squares). The intensity was zero for no NS1 present (Figure 3h), indicating no false positive and no nonspecific adsorption on the test line. This shows that the proteins present in HS influence the Ab-NS1 interactions, and impact the specificity of the sandwich formation. Because pure BSA does not remove the nonspecific interactions, the removal of false positives is due to the exposure to the complex mixture of HS.
We performed a control experiment to probe whether or not the positive test lines were a result of NS1 adsorbing to the NP-Ab conjugate. We changed the Ab on NP to an anti-Dengue NS1 antibody, which cannot bind to Zika NS1 (Supporting Information, Figure S6a). For strips run with NP-anti-Dengue Abs, the test line intensity remained at the baseline for all Zika NS1 concentrations, indicating that the NP-Ab is not interacting with the NS1 nonspecifically via adsorption to cause the NP-Ab to accumulate at the test line (Supporting Information, Figure S6c, black squares). In addition, we confirmed that the NP-anti-Dengue was indeed functional by using it in an assay with Dengue NS1 and anti-Dengue immobilized Abs (Supporting Information, Figure S6b). Here, the NP-anti-Dengue Ab did bind to the Dengue NS1 and exhibited an expected concentration dependence (Figure S6c, red circles). This shows that the NPs do not accumulate at the test line regardless of the type of Ab on its surface, suggesting that the test lines are not caused by the NS1 nonspecifically adsorbing to the NP. We repeated this for NP-Abs that were noncovalently conjugated, and the results were the same (Supporting Information, Figure S6d). This shows the NS1 causing a positive test line due to nonspecific adsorption, confirming that the specific Ab-NS1 interaction is necessary for formation of a sandwich immunoassay. Furthermore, this confirms that the Abs are not displaced from the NP in the HS, because if the Abs were displaced from the NP, then the behavior in both of these assays would be identical, where the NP would accumulate at the test line regardless of the type of Ab.
Properties of Protein Coronas That Form Around the NP-Ab
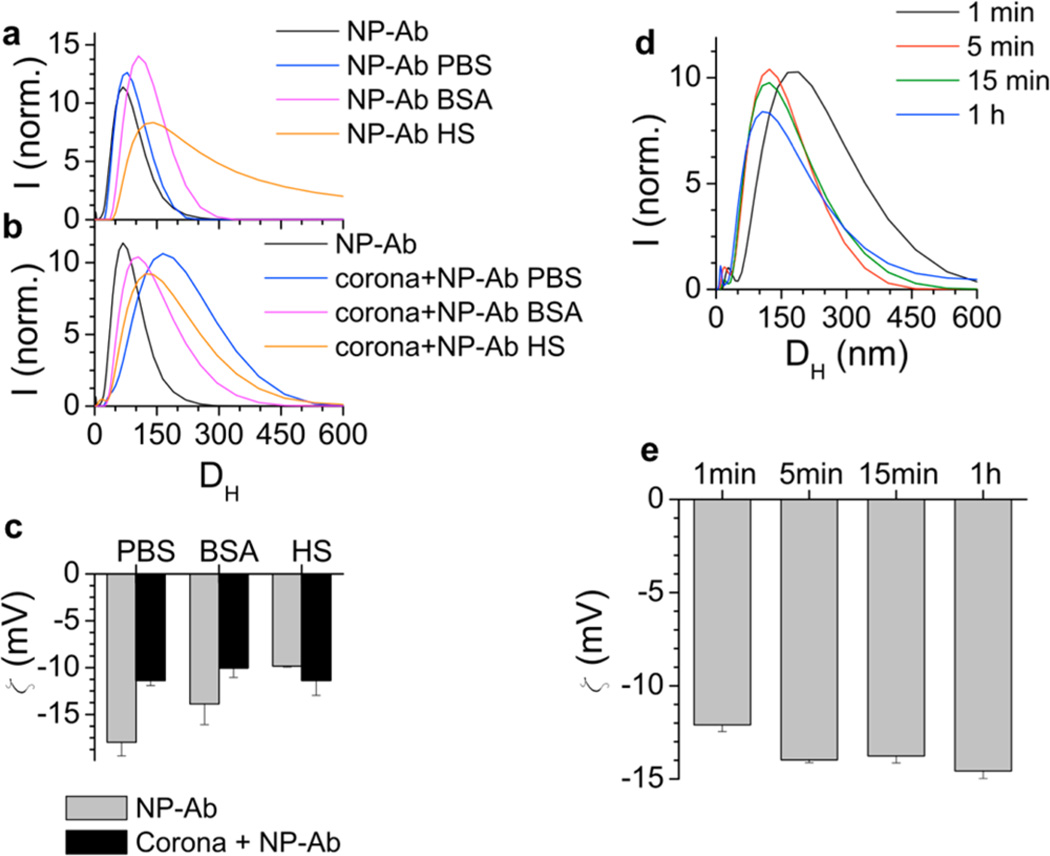
In the strips run with HS, the NP-Ab is in an environment with a high concentration of proteins and also many small molecules, favoring formation of a protein corona. Because corona formation is most likely responsible for modifying the Ab-NS1 interactions in the sandwich immunoassays, we probed the properties of protein coronas that form around the NP-Ab when in HS. NP-Ab were incubated with HS to form protein coronas. The NP-Ab with the coronas were separated from the free serum proteins in HS by spin centrifugation. DLS showed that the DH increased after incubation in HS from 71.6 ± 6.2 nm to 128.9 ± 11.2 nm (Figure 4a, orange), suggesting formation of a large and multilayered protein corona. Zeta potential measurements of the NP-Ab with the coronas (Figure 4c) showed a change to a less negative zeta potential (black compared to gray), suggesting that the charge of the species changed due to corona formation. NP-Ab exposed to BSA showed a shift to a larger DH (Figure 4a, pink), but the increase in size was not as large as for the NP-Ab in HS. The effect on the SPR was negligible, as the absorption spectrum for NP-Ab showed no major change after incubation in HS after 1 h (Supporting Information, Figure S3). The PEG backfill was not able to avoid protein corona formation. Some reports in the literature have shown that PEGylation can prevent coronas, though there is evidence that PEGylated NPs still form coronas.29,35,36 However, given the mild conditions for backfill, corona formation was still expected.
Figure 4.
Properties of coronas that form around NP-Ab in HS. (a) DLS of NP-Ab in water (black), PBS (blue), BSA (pink), and HS (orange). (b) DLS of NP-Ab with preformed coronas in water (black), PBS (blue), BSA (pink), and human serum (orange). (c) Zeta potential measurements of NP-Ab in PBS, BSA, and HS (gray) and NP-Ab with preformed coronas in PBS, BSA, and HS (black). (d) DLS of NP-Ab exposed to HS as a function of time. (e) Zeta potential of NP-Ab exposed to HS as a function of time.
Then, we examined the behavior of the NP-Abs with preformed coronas in the different fluids. NP-Ab were incubated in HS and allowed to form a protein corona using literature methods (Materials and Methods).15 DLS showed that DH was larger compared to NP-Ab (71.6 ± 6.2 nm), and was 172.8 ± 15.0, 106.5 ± 15.6, and 123.3 ± 18.1 in PBS, BSA, and HS (Figure 4b, blue, pink, and orange, respectively). The zeta-potential of the NP-Ab with coronas (Figure 4c, black) showed a change upon corona formation, though the net trend in charge was not systematic. These results show that the preformed corona is larger than the NP-Ab, by as much as 57.3 nm, suggesting a multilayer structure, which is relatively stable in PBS, BSA, and HS. The zeta potential of the NP-Ab with a corona was less negative for both PBS and BSA, and more negative in HS.
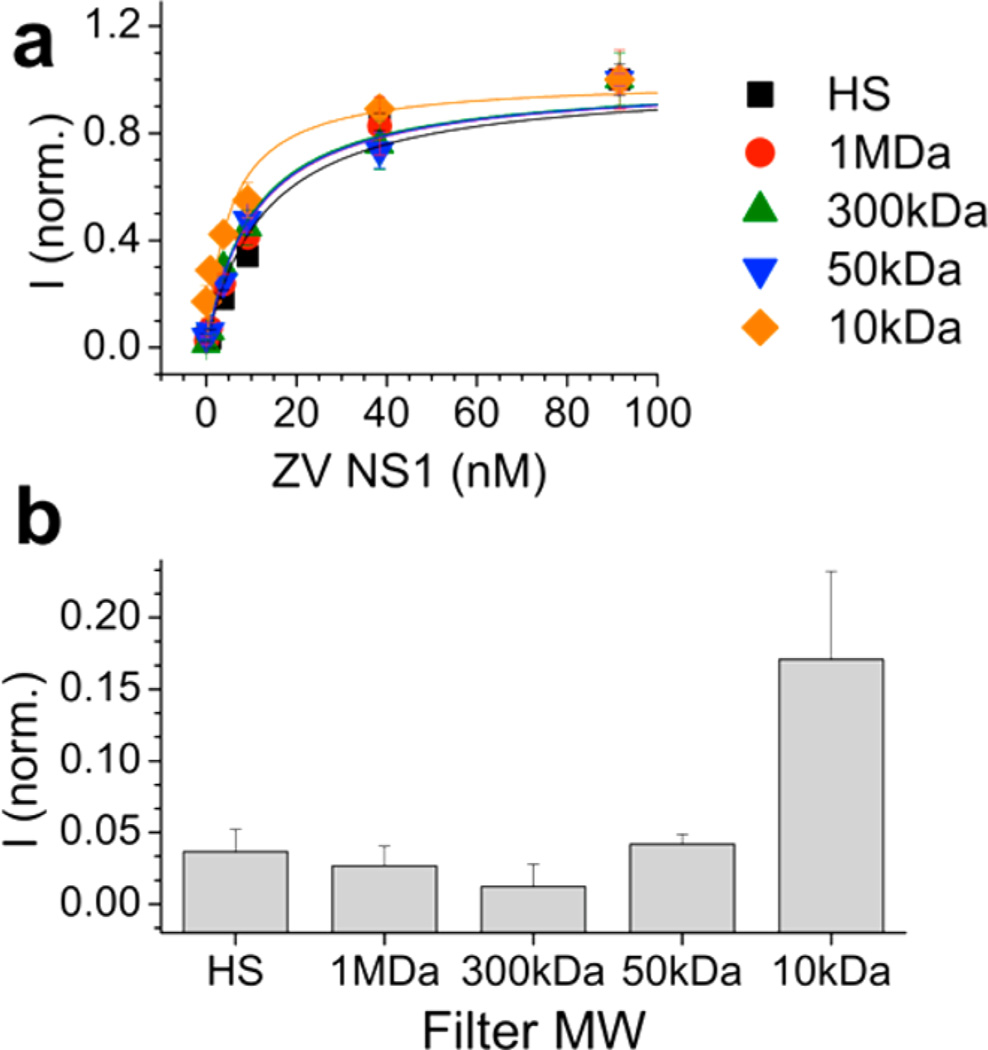
We filtered the HS with different molecular weight cutoff (MWCO) filters to roughly determine which proteins were responsible for removing nonspecific adsorption in the test line. The main components of HS are albumins (MW 67 kDa), globulins, and regulatory proteins of various molecular weights. Strips were run in HS that had been passed through MWCO filters of 1 MDa, 300 kDa, 50 kDa, and 10 kDa, and the test line intensity as a function of NS1 concentration was examined (Figure 5a). The HS that had been passed through the 1 MDa filter (red circles) resembled the strip run in the full HS (black squares), and did not have a false positive at 0 nM NS1 (Figure 5b). However, for the strips run in HS that had been passed through 300 kDa, 50 kDa, and 10 kDa filters, the concentration dependence increasingly resembled the PBS and BSA curves in Figure 3e, and the test line intensity at 0 nM NS1 increased, indicating a false positive signal. Therefore, the high-molecular-weight proteins or multiprotein complexes in the HS are responsible for mediating nonspecific adsorption and contribute to minimizing the false positive binding observed at 0 nM NS1.
Figure 5.
Treatment of HS with molecular weight cutoff (MWCO) filters. (a) Test line intensities of dipstick assays run with HS that has been passed through MWCO filters of 1 MDa (red circles), 300 kDa (green triangles), 50 kDa (blue triangles), 10 kDa (orange diamonds) and no filter, full HS (black squares). Fits to KDeff were performed for the full HS (black dotted line) and 1 MDa HS (red dotted line). (b) Test line intensity at 0 nM ZIKV NS1 for test strips run in HS and HS after passing through MWCO filters. Error bars indicate average of three independent measurements.
Kinetics of Corona Formation
The behavior of protein coronas is dynamic as their formation occurs over time, and their composition and size evolve.37–39 The dipstick sandwich assays used here have running times of ~20 min, which is a typical time scale for POC assays. To investigate the kinetics of corona formation, we probed corona formation as a function of time to see if it would be changing over the time scale of the sandwich immunoassay. NP-Ab were incubated with HS for different times and the size of the resulting corona was measured as a function of time by DLS (Figure 4d). The DH was larger than the original NP-Ab (71.6 ± 6.2 nm), and similar at all times, measuring 181.5 ± 15.0 nm, 116.9 ± 10.6 nm, 116.9 ± 10.6 nm, and 111.3 ± 9.6 nm at 1 min, 5 min, 15 min, and 1 h, respectively), indicating that the corona formed faster than the timescale of a few minutes. Zeta potential measurements of the NP-Ab exposed to HS at different times (Figure 4e) showed that the zeta potential was roughly the same (−12.1 ± 0.4 mV, −14.0 ± 0.2 mV, −13.8 ± 0.4 mV, and −14.6 ± 0.4 mV measured at 1 min, 5 min, 15 min, and 1 h, respectively), also supporting that corona formation was faster than the time scale of the measurement. This corroborates reports from the literature that protein coronas form rapidly around NPs, and apparently here it occurs faster than the time scale of the sandwich immunoassay. Thus, by the time the NP-Ab/NS1 has migrated to the test line, it most likely already has a corona formed around it. Consequently, the protein corona is unavoidable in an immunoassay that requires at least a few minutes to run. The first step in a dipstick assay is NS1 binding to the NP-Ab conjugate, which for the dipstick assay here occurs in the serum solution. However, this time scale is most likely on the order of seconds, and we were not able to conduct DLS and gel assays faster than this time scale to probe how corona formation impacts the NS1 binding to the NP-Ab conjugate.
Preformed Coronas on NP-Ab Affect Test Line Assay
We then investigated the effect of making preformed coronas around the NP-Ab on the sandwich immunoassay. NP-Ab with preformed coronas exhibited markedly different behavior in the immunoassays than NP-Ab without protein coronas. For assays run in PBS and pure BSA with no NS1, the test line intensity above baseline was zero, showing that there was no nonspecific adsorption (Figure 3c, bottom and center). Image analysis showed that the test line intensity increased with NS1 concentration and saturated above 30 nM, similar to the strips run in HS (Figure 3f, red circles and blue triangles, respectively). Assays for NP-Ab coronas run in HS showed behavior similar to the NP-Abs with no preformed coronas (Figure 3c, left, and Figure 3f, black squares). This shows that forming a corona around the NP-Ab can reduce false positives, and the passivation is similar to when the test is run in HS.
Pretreating the Paper with NP-Ab Affects Test Line Assay
In addition, the serum could be impacting the surface effects of the nitrocellulose to mitigate the nonspecific adsorption occurring in the assay. To investigate the effect of protein adsorption on the nitrocellulose, we pretreated the nitrocellulose strips with HS before running the assays. Strips were first put in contact with HS, which was allowed to run through the whole strip. Excess HS was washed by running PBS through the strips. Then, strips were put into a tube with the NP-Ab conjugates with NS1 at different concentrations spiked into HS, PBS, or BSA. The strip was not allowed to dry between the solution transfer, so the kinetics of the migration were slightly different from when dry strips are put in fluid contact with the NP-Ab + antigen solution.40,41 However, this difference in kinetics was assumed to have a minor effect on test line intensity. Test line intensity for all running solutions (PBS, BSA, HS) increased with increasing NS1 concentration (Figure 3d and Figure 3g), though there were slight differences in the dose response curve for the three solutions. Assays run with BSA no longer showed false positives when no NS1 was present, though there was a residual intensity for the strip run in PBS. However, in comparison to no serum pretreatment (Figure 3b), the nonspecific adsorption on the test line was mitigated. This shows that the nitrocellulose also introduces nonspecific adsorption effects and impacts the Ab-NS1 interactions due to exchange between proteins in the nitrocellulose to the NP-Ab complexes, or due to HS corona formation around the nitrocellulose surface. Thus, both the NP and nitrocellulose introduce surface effects that can impact sandwich immunoassays.
Langmuir Model Analysis of Test Line Intensities
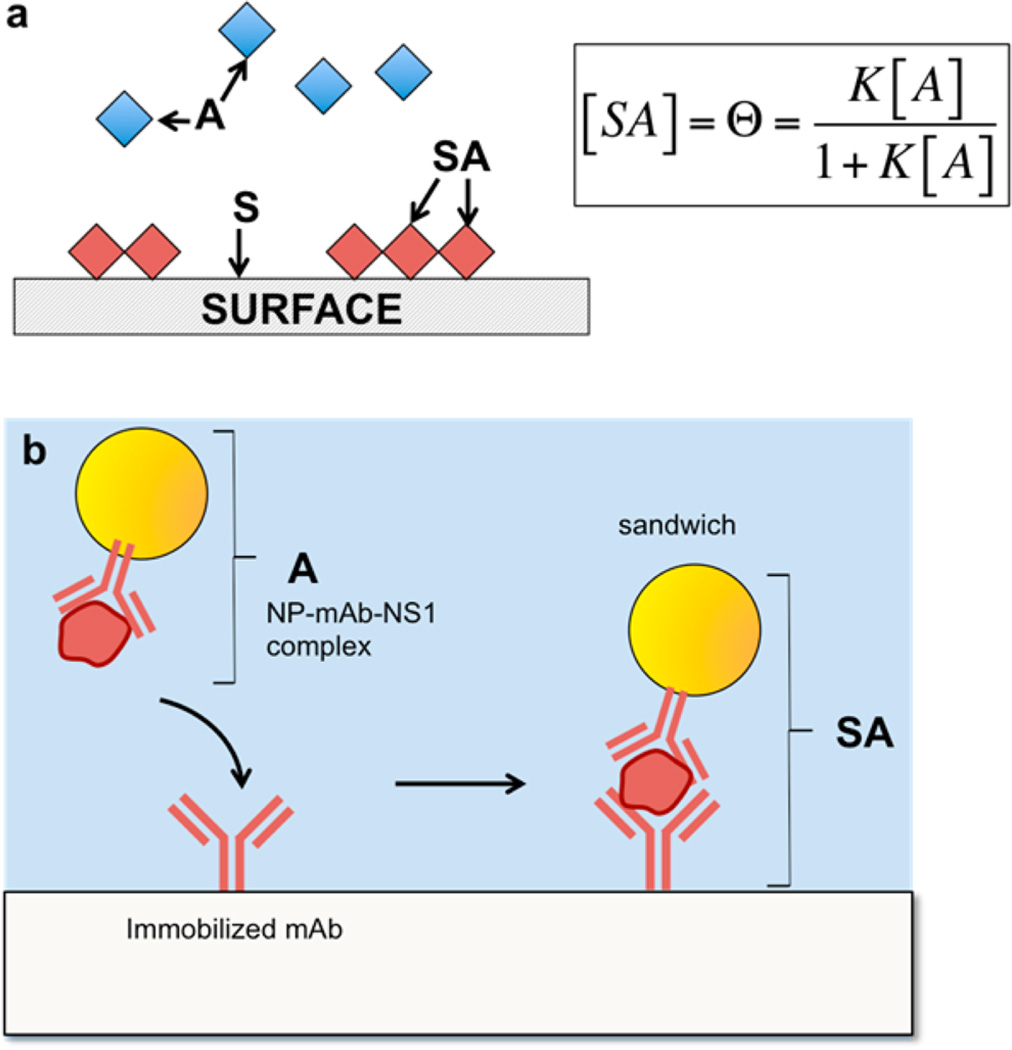
Clearly, both the NP and the nitrocellulose introduce surface effects that influence the Ab-NS1 binding interaction. To quantify the impact of protein corona formation and serum pretreatments on the immunoassays, we utilized a modified Langmuir model to determine a binding affinity of the Ab-NS1 interaction.28,42 The model utilizes the surface adsorption model where the free species is the NP-Ab/antigen complex, and the binding event is described by the binding to the immobilized Ab on the test line, or successful sandwich immunoassay formation. Thus, for a free species A, to bind to a surface, S, to form a surface immobilized species, SA (Figure 6a)
| (1) |
the fraction of occupied sites, Θ, can be measured as a function of the concentration of A to obtain an expression containing KD, the equilibrium dissociation constant
| (2) |
where n is the total number of surface sites, and KD is the equilibrium dissociation constant defined as
| (3) |
which describes the affinity of A for the surface S. In a dipstick or lateral flow assay, there are two binding events. First, the antigen binds to the NP-Ab. Then, the NP-Ab-antigen complex binds to the immobilized antibody on the test line. Here, the Langmuir surface binding model can describe the second binding event, so A is taken to represent the NP-Ab/NS1 complex and S is the immobilized antibody. When the NP-Ab/NS1 binds to immobilized antibody, the sandwich immunoassay represents the species SA (Figure 6b). The concentration of SA is proportional to the test line intensity, which is proportional to the number of NPs at the test line due to Ab-NS1 binding.
Figure 6.
Langmuir binding model. (a) Schematic of the free species A adsorbing onto a surface S, forming a surface bound species, SA. (b) Analogous schematic with the NP-Ab conjugate bound to NS1 binding to surface bound antibody on the test line.
It should be of note that this model makes several assumptions. The Langmuir model assumes that surface bound species SA are independent and do not influence binding of adjacent sites, and that the surface binding can result only in a monolayer. This model also assumes equilibrium binding conditions, which is a large approximation as the conditions are under flow and occur over relatively short time scales.42 The binding affinity constant here is an effective binding constant, KDeff, as it lumps together the binding of the NS1 to the NP-Ab with the binding of the NP-Ab/NS1. With these caveats, we use KDeff only to compare how the binding affinity changes with the different corona conditions, and do not use it as a way to obtain absolute binding affinities. To obtain KDeff values, test line intensities vs NS1 concentration were fit to eq 3 (Table 1). KDeff for NP-Ab run in PBS and BSA (Figure 4e, red circles and blue triangles) were not fit as they would not yield a meaningful value because their test intensity was nonzero at 0 nM NS1, and had no NS1 concentration dependence.
Table 1.
LOD, KDeff, and R2 Values Obtained from Fitting the Test Line Intensities As a Function of ZIKV NS1 Concentration in Figure 3 to the Modified Langmuir Binding Isotherm Model for Bare NPs, Preformed Coronas around the NP-Ab, and HS Pretreatment of the Nitrocellulose
| run media |
LOD (nM) |
KDeff (nM) |
R2 | |
|---|---|---|---|---|
| Bare-NP-Ab | HS | 1.6 | 12.7 | 0.96 |
| BSA | --- | --- | --- | |
| PBS | --- | --- | --- | |
| Preformed coronas on NP-Ab | HS | 0.61 | 10.5 | 0.97 |
| BSA | 15.9 | 31.1 | 0.87 | |
| PBS | 1.1 | 11.0 | 0.98 | |
| Paper pretreatment | HS | 0.8 | 17.2 | 0.93 |
| BSA | 2.3 | 10.2 | 0.93 | |
| PBS | 0.2 | 19.0 | 0.98 |
For the strip run in HS with no pretreatments (Figure 3e, black squares), KDeff = 12.7 nM. Comparison of the values to antibody–antigen interactions for NS1 shows that the affinity of the NP-Ab-NS1 interactions was similar to Ab-NS1.43 For NP-Abs with premade coronas, the KDeff was 11.0 nM when in PBS and 31.1 nM in BSA, which is on the same order of magnitude as the NP-Ab run in HS with no preformed corona. This shows that the preformed corona can restore the specificity of the Ab-NS1 interactions to an affinity similar to the native antibody–antigen interactions.
For the HS pretreated paper, the KDeff for strips run in PBS was 19 nM and BSA 10.2 nM, again showing that the specificity of the Ab-NS1 interaction could be restored by passivating the surface of the nitrocellulose with serum proteins. KDeff differs slightly between PBS vs BSA vs HS, showing that small differences in binding affinities remain even in pretreated paper.
Limits of detection (LODs) were also obtained from the fits, and were found to in the 1–20 nM NS1 order of magnitude range.32,44
Fits of the MWCO filter data (Table 2) could also yield KDeff of 12.7 nM for 1 MDa filtered HS. Data for strips run in HS filtered through 300 kDa, 50 kDa, and 10 kDa filters were 10.7, 10.2, 10.6, and 5.2 nM, respectively.
Table 2.
LODs and KDeff Values Obtained by Langmuir Isotherm Fits of Test Line Intensities (Figure 5a) and Corresponding R2 valuesa
| MWCO filter | LOD (nM) | KDeff (nM) | R2 |
|---|---|---|---|
| Full HS (no filter) | 1.6 | 12.7 | 0.96 |
| 1 MDa | 1.1 | 10.7 | 0.98 |
| 300 kDa | 1.0 | 10.2 | 0.98 |
| 50 kDa | 0.88 | 10.6 | 0.98 |
| 5 kDa | 4.7 | 5.2 | 0.89 |
Full HS and HS filtered with 1 MDa, 300 kDa, 50 kDa, and 10 kDa MWCO filters were fit to obtain KDeff.
CONCLUSIONS
In sandwich immunoassays, the antigen of interest must bind to a matched pair of Abs conjugated a NP and immobilized on the nitrocellulose substrate in a complex environment of full serum. Serum proteins result in protein corona formation around the NP-Ab and also adsorb to the nitrocellulose, and both of these phenomena impact the antibody–antigen interactions at the test line. Here we find that the serum proteins reduce false positives, which suggests that the protein corona plays a critical but beneficial role in the assay. These results call attention to the importance of the protein corona in sandwich immunoassays, which are used in numerous point of care and medical lab assays. Because nonspecific adsorption can influence rapid diagnostic assay results, it can impact treatment, quarantining, and therapy, and thus can have significant medical outcomes.
Furthermore, immunoassays are utilized in a broad range of different biological fluids, not just serum but also blood, urine, saliva, sweat, and others. The protein and small molecule compositions of these fluids vary greatly, and thus can result in formation of different coronas. Because of this, a better understanding of protein corona properties and formation in these different biological fluids is important, which will be the focus of future work. In addition, comparison of affinity constants of the Ab-NS1 interaction, KDeff, is possible via application of a Langmuir binding model, though due to the model’s approximations, most likely only relative changes in KDeff are prudent.
Nonspecific adsorption is problematic even for relatively simple systems of NP–protein or NP–DNA conjugates in solution. In comparison, LF and dipstick assays are much more complex with many inorganic-biological interfaces, resulting in numerous opportunities for nonspecific adsorption. This can have either beneficial or negative side ramifications on immunoassay outcomes. Quantification of these interface effects is important for not only understanding them, but also developing routes for eliminating negative influences of the interface. Despite their prevalent commercial and clinical use, lateral flow immunoassays face many challenges for use in the field. Not only do they suffer from nonspecific adsorption, which can result in faint or smeared test lines, complicating the diagnosis, but also face complications in their stability and robustness. There are numerous efforts to improve their performance by optimizing not just the NP-Ab properties but also the assay materials, housings, and mode of operation. We hope that results from this work will aid in improving lateral flow immunoassays for their widespread deployment.
MATERIALS AND METHODS
Reagents
Au chloride trihydrate was purchased from Sigma-Aldrich (CAS: 16961–25–4). Bis(sulfatophenyl)phenylphosphine dehydrate (BPS) was purchased from Aldrich (CAS:308103–66–4). N-(2-Hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid) (HEPES) was purchased from United States Biochemical Company (CAT: 16926). Sodium citrate was from Mallinckrodt Chemicals and 5kD mPEG was from nanocs. The heterobifunctional linker with a hydrazide on one end and a dithiol on the other, hydrazide dithiolalkanearomatic PEG6-NHNH2, was from Sensopath Technologies. Fluorescent goat anti-mouse IgG (H+L) Secondary Antibody, DyLight 650 conjugate, was purchased from Pierce. Goat anti-mouse IgG, Fc, was purchased from Millipore (AQ127). Zika NS1 native protein was from Native Antigen. Bovine serum albumin (BSA) was from Sigma (CAT: A9418), and diluted in 1× PBS to prepare a 10 mg/mL solution. Phosphate. Phosphate buffer saline (1× PBS, pH 7.4) was from Gibco (CAT: 10010–049). Filtered human serum was obtained by filtering 1 mL of human serum from Sigma-Aldrich (H4522) through a 0.2 µm cellulose acetate syringe filter (Pall, Acrodisc 25 mm Syringe Filter, with 0.2 µm HT Tuffryn Membrane).
In the tests involving filtered human serum at different cutoff molecular weights, the human serum was filtered through Vivaspin 4 mL centrifuge filters: 1 000 000 MWCO, PES Membrane, 300 000 MWCO, PES Membrane, 50 000 MWCO, PES Membrane, and 10 000 MWCO, PES Membrane for the 1 MDa, 300 kDa, 30 kDa, and 10 kDa filtered human serums, respectively. After filtration, the Nanodrop was used to measure the final concentration of protein in the serum. The final protein concentration was 38.6, 7.7, 3.2, 0.7, and 0.65 for the 0.2 µm, 1 MDa, 300 kDa, 50 kDa, and 10 kDa, respectively.
Antibodies
Hybridoma cells producing antibodies against Zika NS1 were obtained by injection of mice with recombinant native NS1 protein of Zika (purchased from Native Antigen). After hybridomas were screened using ELISA and FACS analysis of infected cells, selected cell cultures were harvested and concentrated using Millipore centrifugal units (30 kDa MW). Protein L columns were used to purify the kappa light chain mouse antibodies that were specific to Zika NS1. After purification, the antibodies were buffer-exchanged into PBS, concentrated, and stored at 4 °C. A NanoDrop 2000 UV–vis spectrophotometer at 280 nm was used to calculate the concentration of the purified antibody, and a TapeStation with a P200 ScreenTape from Agilent Technologies was used to confirm the purity of the monoclonal antibodies.
Synthesis and Bioconjugation of Gold Nanostars
To synthesize magenta star-shaped NPs, 200 µL of 140 mM HEPES (pH 7.4) was mixed with 800 µL of 18 MΩ deionized (Milli-Q) water, followed by the addition of 16 µL of 25 mM HAuCl4·3H2O and further vortexing. The solution sat undisturbed for 1 h, during which time the NPs formed. Afterward, ~0.5 mg BPS was added for NP stabilization, the solution was vortexed and left undisturbed for 1 h. Prior to antibody conjugation, the NPs were separated from excess reagents by centrifugation at 12 000 rcf for 12 min 2 times.
For covalent, directional conjugation, a heterobifunctional linker consisting of a hydrazide on one end and a dithiol on the other, linked by a short polyethylene glycol (PEG) chain (hydrazide dithiolalkane aromatic PEG6-NHNH2) was attached to the antibodies. First, 300 µL of a 1 mg/mL solution of antibody in 40 mM HEPES (pH 7.4) was mixed with to 30 µL of 100 mM sodium periodate (NaIO4, Sigma) and allowed to react while agitating for 45 min at room temperature in the dark, thus oxidizing the carbohydrate moieties on the Fc region of the antibody to aldehydes. Then, 1.5 mL of PBS and an excess of linker (2 µL of 33.3 mg/mL in ethanol) were added to the oxidized antibodies and agitated for 30 min, during which time the aldehyde group of the antibodies bound to the hydrazide on the linker. To remove unreacted linker, antibodies were buffer-exchanged into PBS using a 10 kDa centrifugal filter column (Amicon) and resuspended to a 1 mg/mL solution in PBS.
Prior to antibody conjugation, the NPs were separated from excess reagents by centrifugation at 12 000 rcf for 12 min. The resulting NP pellet was resuspended in 100 µL of 40 mM HEPES at pH 7.7 and 300 µL of Milli-Q water, followed by the addition of 10 µL of 1 mg/mL of the modified antibody, vortexed, and further agitated overnight, to enable antibody binding to the nanostars. In order to avoid nonspecific binding on the nanostars, 10 µL of 0.1 mM mPEG was added, and the solution was vortexed and further agitated for 20 min, to enable mPEG to passivate any bare gold surfaces. Finally, NPs were centrifuged for 12 min at 10 000 rcf twice to separate excess reagents, and then used in the immunoassays.
Corona Formation around Nanoparticles
A protein corona was formed around the antibody-conjugated Au NPs by mixing 20 µL of the NP pellet with 500 µL of filtered human serum (HS) vortexing and incubating the mixture during 1 h. Afterward, the NP mixture was centrifuged for 12 min at 10 000 rcf to separate excess serum proteins, the supernatant was discarded and the pellet was further washed three times by resuspending the NPs in 0.5 mL of PBS and centrifuging at 10 000 rcf for 12 min, in order to minimize the presence of noncorona proteins in the NP pellet.
NP Characterization
Optical absorption spectra of the NP were obtained on a Cary 500i UV–vis-NIR Dual-Beam Spectrophotometer from Agilent Technologies. Morphology of the NP was characterized with a FEI Tecnai G2 TEM at 120 kV. ImageJ was used to process the images and measure the dimensions of the NP. In addition, a Zetasizer Nano ZS from Malvern Instruments was used to measure the hydrodynamic diameter (DH) and the ζ of the NP. Agarose gel electrophoresis was used to confirm the antibody and mPEG binding on the NPs; in short, 1% agarose gels were prepared and NPs were loaded by mixing 8 µL of concentrated NPs with 4 µL of 50% glycerol in Milli-Q water. Fluorescence spectroscopy was used to quantify the amount of antibodies bound per nanoparticle, by a supernatant-loss method.24
Running the Assays
Antibodies were immobilized on the nitrocellulose by manually pipetting 0.3 µL of a 2 mg/mL solution of antibodies onto the nitrocellulose paper, where they automatically were immobilized, and further allowed to dry for at least 30 min. In the test band, monoclonal antibodies against Zika-NS1 protein were immobilized. The positive control was spotted with goat antibodies that could bind to the crystallizable fragment (Fc) of the mouse IgG antibodies on the NP. Afterward, the nitrocellulose was attached to the absorbent pad. The nitrocellulose was placed inside a test solution containing 4 µL of 50% w/v sucrose in water and 8 µL of 1% v/v Tween 80 in PBS, 1 µL of the Ab-NP conjugates, and the analyte (total volume of 30 µL, prepared by spiking Zika NS1 protein into filtered human serum), and the tests were run by letting the solution migrate through the strip via capillary action until the samples dried. Strips were run in phosphate buffered saline (PBS), or bovine serum albumin at 10 mg/mL in PBS, or filtered human serum.
In order to form a protein corona around the nitrocellulose, filtered human serum was allowed to migrate through the nitrocellulose by capillary action followed by the migration of PBS through the nitrocellulose, in order to wash the pores from unbound human serum proteins. After that, the tests were placed inside the test solutions containing NPs, sucrose, Tween, and the analyte solution, and allowed to run through capillary action until the samples dried.
Image Analysis of the Nitrocellulose Strips
Once the tests had been dried, images of the finished tests were scanned and quantified with ImageJ.34 For the limit of detection analysis, gray values of the detection areas were obtained by subtracting the grayscale values from the detection areas from the grayscale values of the background, and normalized as follows: , where gray0 is the measured gray value of the blank, graymax is the gray value of the highest concentration point (at saturation), and gray is the gray value at each concentration. After normalization, gray values were plotted and fitted in a Langmuir equation of the form , where [antigen] is the concentration of antigen present in the 30 µL of sample in the solution, and KDeff represents the effective binding constant in a Langmuir-like system. The limit of detection was measured as the concentration of antigen capable of showing a signal at 5× the value of the standard deviation of the blank.
Supplementary Material
Acknowledgments
This work was funded by NIH NIAID (AI100190). H.d.P. was supported by the MIT/SUTD International Design Centre, a Rafael del Pino Fellowship and a Tata Fellowship. Irene Bosch was supported by the Food Drug Administration ORISE Fellowship and by AEDES project, Colombia. We thank Lee Gehrke for support and advice. We thank Justina Tam for experimental support. We thank CMSE at MIT for use of their facilities.
ABBREVIATIONS
- Ab
antibody
- BSA
bovine serum albumin
- DLS
dynamic light scattering
- LOD
limit of detection
- NP
nanoparticle
- NS1
non structural protein 1
- PEG
polyethylene glycol
- TEM
transmission electron microscopy
- ZIKV
Zika virus
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Test strip images, optical absorption spectra, and ELISA and fluorescence data (PDF)
ORCID
Kimberly Hamad-Schifferli: 0000-0002-4839-3179
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
REFERENCES
- 1.Cedervall T, Lynch I, Lindman S, Berggård T, Thulin E, Nilsson H, Dawson KA, Linse S. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patra A, Ding T, Engudar G, Wang Y, Dykas MM, Liedberg B, Kah JCY, Venkatesan T, Drum CL. Component-Specific Analysis of Plasma Protein Corona Formation on Gold Nanoparticles Using Multiplexed Surface Plasmon Resonance. Small. 2016;12(9):1174–1182. doi: 10.1002/smll.201501603. [DOI] [PubMed] [Google Scholar]
- 3.Treuel L, Jiang X, Nienhaus GU. New views on cellular uptake and trafficking of manufactured nanoparticles. J. R. Soc. Interface. 2013;10(82):20120939. doi: 10.1098/rsif.2012.0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zensi A, Begley D, Pontikis C, Legros C, Mihoreanu L, Büchel C, Kreuter J. Human serum albumin nanoparticles modified with apolipoprotein A-I cross the blood-brain barrier and enter the rodent brain. J. Drug Targeting. 2010;18(10):842–848. doi: 10.3109/1061186X.2010.513712. [DOI] [PubMed] [Google Scholar]
- 5.Saha K, Rahimi M, Yazdani M, Kim ST, Moyano DF, Hou S, Das R, Mout R, Rezaee F, Mahmoudi M, et al. Regulation of Macrophage Recognition through the Interplay of Nanoparticle Surface Functionality and Protein Corona. ACS Nano. 2016;10(4):4421–4430. doi: 10.1021/acsnano.6b00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrán-Berdón AL, Pozzi D, Caracciolo G, Capriotti AL, Caruso G, Cavaliere C, Riccioli A, Palchetti S, Laganà A. Time Evolution of Nanoparticle–Protein Corona in Human Plasma: Relevance for Targeted Drug Delivery. Langmuir. 2013;29(21):6485–6494. doi: 10.1021/la401192x. [DOI] [PubMed] [Google Scholar]
- 7.Mirshafiee V, Mahmoudi M, Lou K, Cheng J, Kraft ML. Protein corona significantly reduces active targeting yield. Chem. Commun. 2013;49(25):2557–2559. doi: 10.1039/c3cc37307j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salvati A, Pitek AS, Monopoli MP, Prapainop K, Bombelli FB, Hristov DR, Kelly PM, Aberg C, Mahon E, Dawson KA. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 2013;8(2):137–143. doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- 9.Jiang X, Weise S, Hafner M, Röcker C, Zhang F, Parak WJ, Nienhaus GU. Quantitative analysis of the protein corona on FePt nanoparticles formed by transferrin binding. J. R. Soc., Interface. 2010;7(Suppl 1):S5–S13. doi: 10.1098/rsif.2009.0272.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleischer CC, Payne CK. Nanoparticle Surface Charge Mediates the Cellular Receptors Used by Protein–Nanoparticle Complexes. J. Phys. Chem. B. 2012;116(30):8901–8907. doi: 10.1021/jp304630q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pitek AS, O’Connell D, Mahon E, Monopoli MP, Baldelli Bombelli F, Dawson KA. Transferrin Coated Nanoparticles: Study of the Bionano Interface in Human Plasma. PLoS One. 2012;7(7):e40685. doi: 10.1371/journal.pone.0040685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston H, Brown D, Kermanizadeh A, Gubbins E, Stone V. Investigating the relationship between nanomaterial hazard and physicochemical properties: Informing the exploitation of nanomaterials within therapeutic and diagnostic applications. J. Controlled Release. 2012;164(3):307–313. doi: 10.1016/j.jconrel.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 13.Hamad-Schifferli K. Exploiting the novel properties of protein coronas: emerging applications in nanomedicine. Nanomedicine. 2015;10(10):1663–1674. doi: 10.2217/nnm.15.6. [DOI] [PubMed] [Google Scholar]
- 14.Kah JCY, Chen J, Zubieta A, Hamad-Schifferli K. Exploiting the Protein Corona around Gold Nanorods for Loading and Triggered Release. ACS Nano. 2012;6(8):6730–6740. doi: 10.1021/nn301389c. [DOI] [PubMed] [Google Scholar]
- 15.Cifuentes-Rius A, de Puig H, Kah JCY, Borros S, Hamad-Schifferli K. Optimizing the Properties of the Protein Corona Surrounding Nanoparticles for Tuning Payload Release. ACS Nano. 2013;7(11):10066–10074. doi: 10.1021/nn404166q. [DOI] [PubMed] [Google Scholar]
- 16.Parolo C, Merkoçi A. Paper-based nanobiosensors for diagnostics. Chem. Soc. Rev. 2013;42:450–457. doi: 10.1039/c2cs35255a. [DOI] [PubMed] [Google Scholar]
- 17.Wong R, Tse H. Lateral Flow Immunoassay. New York: Springer; 2009. [Google Scholar]
- 18.Quesada-González D, Merkoci A. Nanoparticle-based lateral flow biosensors. Biosens. Bioelectron. 2015;73:47–63. doi: 10.1016/j.bios.2015.05.050. [DOI] [PubMed] [Google Scholar]
- 19.Polo E, del Pino P, Pelaz B, Grazu V, de la Fuente JM. Plasmonic-driven thermal sensing: ultralow detection of cancer markers. Chem. Commun. (Camb) 2013;49(35):3676–3678. doi: 10.1039/c3cc39112d. [DOI] [PubMed] [Google Scholar]
- 20.Park S, Hamad-Schifferli K. Nanoscale Interfaces to Biology. Curr. Opin. Chem. Biol. 2010;14(5):616–622. doi: 10.1016/j.cbpa.2010.06.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.You C-C, Chompoosor A, Rotello VM. The biomacromolecule-nanoparticle interface. Nano Today. 2007;2(3):34. [Google Scholar]
- 22.Xie J, Lee JY, Wang DIC. Seedless, Surfactantless, High-Yield Synthesis of Branched Gold Nanocrystals in HEPES Buffer Solution. Chem. Mater. 2007;19(11):2823–2830. [Google Scholar]
- 23.Dam DHM, Lee JH, Sisco PN, Co DT, Zhang M, Wasielewski MR, Odom TW. Direct Observation of Nanoparticle-Cancer Cell Nucleus Interactions. ACS Nano. 2012;6(4):3318–3326. doi: 10.1021/nn300296p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Puig H, Tam JO, Yen C-W, Gehrke L, Hamad-Schifferli K. The extinction coefficient of gold nanostars. J. Phys. Chem. C. 2015;119(30):17408–17415. doi: 10.1021/acs.jpcc.5b03624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Sayed IH, Huang X, El-Sayed MA. Surface Plasmon Resonance Scattering and Absorption of anti-EGFR Antibody Conjugated Gold Nanoparticles in Cancer Diagnostics: Applications in Oral Cancer. Nano Lett. 2005;5(5):829–834. doi: 10.1021/nl050074e. [DOI] [PubMed] [Google Scholar]
- 26.Sokolov K, Aaron J, Hsu B, Nida D, Gillenwater A, Follen M, MacAulay C, Adler-Storthz K, Korgel B, Descour, et al. Optical Systems for in Vivo Molecular Imaging of Cancer. Technol. Cancer Res. Treat. 2003;2(6):491–504. doi: 10.1177/153303460300200602. [DOI] [PubMed] [Google Scholar]
- 27.Mandl A, Filbrun SL, Driskell JD. Asymmetrically Functionalized Antibody–Gold Nanoparticle Conjugates to Form Stable Antigen-Assembled Dimers. Bioconjugate Chem. 2016 doi: 10.1021/acs.bioconjchem.6b00459. [DOI] [PubMed] [Google Scholar]
- 28.de Puig H, Federici S, Baxamusa SH, Bergese P, Hamad-Schifferli K. Quantifying the Nanomachinery of the Nanoparticle-Biomolecule Interface. Small. 2011;7(17):2477–2484. doi: 10.1002/smll.201100530. [DOI] [PubMed] [Google Scholar]
- 29.Walkey CD, Olsen JB, Guo H, Emili A, Chan WC. Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J. Am. Chem. Soc. 2012;134(4):2139–2147. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- 30.Sacchetti C, Motamedchaboki K, Magrini A, Palmieri G, Mattei M, Bernardini S, Rosato N, Bottini N, Bottini M. Surface Polyethylene Glycol Conformation Influences the Protein Corona of Polyethylene Glycol-Modified Single-Walled Carbon Nanotubes: Potential Implications on Biological Performance. ACS Nano. 2013;7(3):1974–1989. doi: 10.1021/nn400409h. [DOI] [PubMed] [Google Scholar]
- 31.Park S, Brown KA, Hamad-Schifferli K. Changes in Oligonucleotide Conformation on Nanoparticle Surfaces by Modification with Mercaptohexanol. Nano Lett. 2004;4(10):1925–1929. [Google Scholar]
- 32.Yen C-W, de Puig H, Tam JO, Gomez-Marquez J, Bosch I, Hamad-Schifferli K, Gehrke L. Multicolored silver nanoparticles for multiplexed disease diagnostics: distinguishing dengue, yellow fever, and Ebola viruses. Lab Chip. 2015;15(7):1638–1641. doi: 10.1039/c5lc00055f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan YH, Liu M, Nolting B, Go JG, Gervay-Hague J, Liu GY. A Nanoengineering Approach for Investigation and Regulation of Protein Immobilization. ACS Nano. 2008;2(11):2374–2384. doi: 10.1021/nn800508f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics Int. 2004;11(7):36–42. [Google Scholar]
- 35.Walkey CD, Olsen JB, Song F, Liu R, Guo H, Olsen DWH, Cohen Y, Emili A, Chan WCW. Protein Corona Fingerprinting Predicts the Cellular Interaction of Gold and Silver Nanoparticles. ACS Nano. 2014;8(3):2439–2455. doi: 10.1021/nn406018q. [DOI] [PubMed] [Google Scholar]
- 36.Welsher K, McManus SA, Hsia C-H, Yin S, Yang H. Discovery of Protein- and DNA-Imperceptible Nanoparticle Hard Coating Using Gel-Based Reaction Tuning. J. Am. Chem. Soc. 2015;137(2):580–583. doi: 10.1021/ja511297d. [DOI] [PubMed] [Google Scholar]
- 37.Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. Time Evolution of the Nanoparticle Protein Corona. ACS Nano. 2010;4(7):3623–3632. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- 38.Dell’Orco D, Lundqvist M, Oslakovic C, Cedervall T, Linse S. Modeling the Time Evolution of the Nanoparticle-Protein Corona in a Body Fluid. PLoS One. 2010;5(6):e10949. doi: 10.1371/journal.pone.0010949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lundqvist M, Stigler J, Cedervall T, Berggård T, Flanagan MB, Lynch I, Elia G, Dawson K. The Evolution of the Protein Corona around Nanoparticles: A Test Study. ACS Nano. 2011;5(9):7503–7509. doi: 10.1021/nn202458g. [DOI] [PubMed] [Google Scholar]
- 40.Fu E, Liang T, Houghtaling J, Ramachandran S, Ramsey SA, Lutz B, Yager P. Enhanced Sensitivity of Lateral Flow Tests Using a Two-Dimensional Paper Network Format. Anal. Chem. 2011;83(20):7941–7946. doi: 10.1021/ac201950g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu E, Ramsey SA, Kauffman P, Lutz B, Yager P. Transport in two-dimensional paper networks. Microfluid. Nanofluid. 2011;10(1):29–35. doi: 10.1007/s10404-010-0643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergese P, Oliviero G, Alessandri I, Depero LE. Thermodynamics of mechanical transduction of surface confined receptor/ligand reactions. J. Colloid Interface Sci. 2007;316(2):1017–1022. doi: 10.1016/j.jcis.2007.08.048. [DOI] [PubMed] [Google Scholar]
- 43.Choi DH, Lee SK, Oh YK, Bae BW, Lee SD, Kim S, Shin Y-B, Kim M-G. A dual gold nanoparticle conjugate-based lateral flow assay (LFA) method for the analysis of troponin I. Biosens. Bioelectron. 2010;25(8):1999–2002. doi: 10.1016/j.bios.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 44.Badu-Tawiah AK, Lathwal S, Kaastrup K, Al-Sayah M, Christodouleas DC, Smith BS, Whitesides GM, Sikes HD. Polymerization-based signal amplification for paper-based immunoassays. Lab Chip. 2015;15(3):655–659. doi: 10.1039/c4lc01239a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.