Abstract
Metabolomics, or the comprehensive profiling of small molecule metabolites in cells, tissues, or whole organisms, has undergone a rapid technological evolution in the past two decades. These advances have led to application of metabolomics for defining predictive biomarkers for incident cardiometabolic diseases, and increasingly, as a blueprint for understanding their pathophysiologic mechanisms. Progress in this area and challenges for the future are reviewed here.
Introduction
The term “metabolomics” emerged at the dawn of the third millennium to describe attempts to measure all of the small molecule metabolites in a biological system, or at least a large number of metabolites at one time. In reality, metabolomics is nothing more than analytical chemistry, which of course has a much longer history. The advent of metabolomics has been fueled by major improvements in instrument technology, most notably in the sensitivity and mass range of mass spectrometers, and associated gas and liquid chromatography techniques. Today, the most advanced systems deployed in a non-targeted mode (sometimes referred to as “shotgun” metabolomics), are able to detect up to 10,000 independent spectral features in a single biological specimen (Zamboni et al., 2015; Patti et al., 2012; Jin et al., 2016). However, in the best of such studies, only about one-third of the detected peaks can be linked to a specific chemical structure in an unambigious fashion, and only after many weeks of work. Non-targeted metabolomics is typically used to compare two biological conditions, e.g. drug-treated or genetically engineered cells compared to control cells. As such, non-targeted metabolomics is best suited as a discovery tool for identifying metabolites that change in response to manipulation of a biological system (relative concentration) rather than providing the exact concentration of a known metabolite (real concentration). Targeted metabolomics, which focuses on measurement of known metabolites in clusters with similar chemical structures (e.g. amino acids, acylcarnitines, organic acids, etc) is a more quantitative tool, as it often involves the use of stable isotope-labeled metabolites (usually 2H- or 13C-labeled) as internal standards to allow the quantity of a targeted analyte to be deduced by the ratio of its peak area to that of the labeled standard added at a known concentration (referred to as isotope dilution mass spectrometry) (Newgard et al., 2009; Ferrara et al., 2008; Bain et al., 2009). The evolution of targeted and non-targeted methods for static profiling of metabolites (referring to measurement of metabolite levels at one specific time point) has been complemented by advances in metabolic flux analysis, in which heavy atoms from stable isotope-labeled substrates are detected as they label downstream metabolic products, in experiments involving multiple time points (Zamboni et al., 2015; Buescher et al., 2015; Fan et al., 2014; Alves et al., 2015). When used together, these assembled tools provide a deep and dynamic view of metabolic functions of cells, tissues/organs, and even whole animals and humans.
But the purpose of this article is not to review metabolomics technologies. Several reviews describing key technological advances in this domain have appeared recently and should be considered by interested readers (Zamboni et al., 2015; Patti et al., 2012; Bain et al., 2009; Buescher et al., 2015; Alves et al, 2015). Instead, this piece attempts to review the application of metabolomics to prominent metabolic diseases and conditions, particularly obesity, diabetes, and cardiovascular diseases, from the perspective of the physiologist/physician/biologist, rather than from that of the analytical biochemist/technologist. In that context, the piece seeks to answer a specific question—In the past 15 years, what has metabolomics contributed to our understanding of complex cardiometabolic diseases, either in terms of our ability to detect and diagnose these conditions, or via new insights into disease mechanisms?
Understanding Metabolic Diseases--Why Metabolomics?
Modern human society is encumbered with a pandemic of chronic diseases and conditions in which metabolic dysregulation plays a key role in pathogenesis and progression, including obesity, diabetes, and cardiovascular disease. Increasingly, dysregulated metabolism is also being recognized as a major contributor to diseases not traditionally considered as “metabolic” in origin, such as cancer, cognitive disorders, and respiratory pathologies. The successful sequencing of the human genome seemed to herald a new age of personalized medicine, in which genomic variation would be used to predict the impact of specific therapeutic interventions, leading to optimal management of disease in a given individual. However, many of our most prevalent chronic diseases are polygenic, including diabetes and cardiovascular diseases, and approaches such as genome-wide association studies (GWAS) have thus far explained only a small fraction of these diseases and made modest contributions to mechanism-based intervention strategies (O’Rahilly, 2009; Newgard and Attie, 2010). Moreover, better methods must be developed to probe the interaction of genetics with environmental factors such as diet, the gut microbiome, and physical activity.
Comprehensive metabolite profiling, or “metabolomics” defines the chemical phenotype of human subjects and animal models, and as such has unique potential for defining biomarkers that predict disease incidence, severity, and progression, and for casting new light on underlying mechanistic abnormalities. Advantages of metabolomics relative to other “omics” technologies include: 1) Humans have been estimated to contain about 6500 discrete small molecule metabolites (Wishart et al., 2013), although new and more sensitive measurement technologies are gradually revealing a larger number of chemical species over time (Zamboni et al., 2015). Nevertheless the number of metabolites is likely to remain smaller than the estimated 25,000 genes, 100,000 transcripts, and 1,000,000 proteins found in humans; 2) Metabolomics measures chemical phenotypes that are downstream from genomic, transcriptomic, and proteomic variability, thus providing a highly integrated profile of biological status; 3) Metabolomics also serves as a precise and non-invasive tool to discern mechanisms of action and possible toxicological effects of drug therapies, and to separate contributions of genetics, microbiome activity, and nutrition on overall metabolic phenotypes.
Metabolomics reveals associations of metabolites with cardiometabolic diseases and predicts disease and intervention outcomes
Metabolomics applied to type 2 diabetes
The association between cardiometabolic diseases and certain lipid metabolites commonly measured in clinical chemistry laboratories, including triglycerides, cholesterol, and total non-esterified fatty acids (NEFA), has long been recognized, leading some to conclude that cardiometabolic diseases are driven by perturbed lipid homeostasis. Application of metabolomics has led to a broader appreciation of metabolites that associate with these maladies, and in some cases that predict intervention outcomes and/or future disease development.
As one example, targeted metabolomics has identified a signature of dysregulated metabolism of branched-chain amino acids (BCAA) in subjects with various forms of cardiometabolic disease. The finding first emerged in a study of obese, insulin resistant compared to lean, insulin sensitive subjects (Newgard et al., 2009). Principal components analysis (PCA) performed on data sets comprised of plasma and urine amino acids, acylcarnitines, organic acids, and fatty acids identified a cluster of metabolites comprised of BCAA, aromatic amino acids (Phe, Tyr), Glu/Gln, Met, and C3 and C5 acylcarnitines that strongly associated with insulin resistance as measured by the HOMA-IR score. In one sense, this finding was simply a re-discovery, given that association of BCAA and aromatic amino acids with obesity and insulin resistance was reported 40 years earlier by Felig, Marliss and Cahill (Felig et al., 1969). However, the broader spectrum of analytes measured with metabolomics afforded two new insights: 1) The association of the BCAA-related metabolite cluster with insulin resistance was stronger than observed for several lipid-related clusters, including one principal component comprised of fatty acids and ketones, and another comprised of medium-chain acylcarnitines derived from fatty acid oxidation (Newgard et al., 2009); 2) The clustering of glutamate/glutamine, C3 and C5 acylcarnitines with BCAA defined a signature comprising metabolites generated during BCAA catabolism, suggesting fundamental alteration of BCAA metabolism in insulin resistant states. Glutamate is produced in the first step of BCAA catabolism, the transamination reaction catalyzed by branched chain aminotransferase (BCAT), whereas C3 and C5 acylcarnitines are derived from three carbon and five carbon acyl CoA intermediates produced by mitochondrial metabolism of the carbon skeletons of BCAA (Figure 1). In addition to the positive association between BCAA, aromatic amino acids, and insulin resistance, glycine was found to have a strong negative association with insulin resistance when measured as HOMA score (Newgard et al., 2009), or by hyperinsulinemic/euglycemic clamp (Thalacker-Mercer et al., 2014).
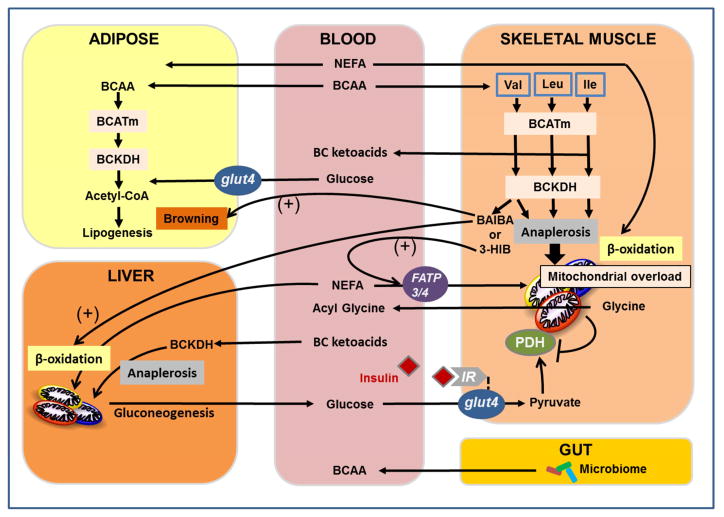
Figure 1. Emergent mechanisms of branched-chain amino acid (BCAA) metabolism in cardiometabolic disease pathogenesis unveiled with metabolomics.
Several mechanisms contribute to accumulation of BCAA in plasma of obese, insulin resistant humans, including increased de novo production of BCAA by the gut microbiome and reduced utilization of BCAA in liver and adipose tissue. BCAA utilization does not appear to be suppressed in skeletal muscle, and under obese conditions, elevated BCAA induce a decrease in skeletal muscle glycine levels, removing a potential escape valve for excess acyl CoAs, Combined substrate pressure from elevated BCAA and lipids in obesity contribute to accumulation of incompletely oxidized fatty acids in mitochondria (“mitochondrial overload”) and reduced efficiency of glucose disposal. In addition, valine catabolism yields two new BCAA-derived factors that contribute to energy balance and metabolic homeostasis--β-aminoisobutyric acid (BAIBA), which stimulates thermogenesis and browning of white fat, and 3-hydroxyisobutryate (3-HIB), which stimulates trans-endothelial and muscle uptake of fatty acids. See text for details and discussion.
The strong association of the BCAA-related metabolite cluster with insulin resistance was confirmed in multiple studies, including a cross-sectional study of subjects with metabolic syndrome and varying BMI (Huffman et al., 2009), a study in Chinese and Asian-Indian subjects residing in Singapore in which BMI was controlled (average BMI of 24) (Tai et al., 2010), a study on subjects at extremes of insulin sensitivity in the Insulin Resistance Atherosclerosis Study (IRAS) (Palmer et al., 2015), and in studies of the effects of combined aerobic and resistance training in insulin resistant subjects (Glynn et al., 2015). In all of these studies, the BCAA-related factor was more correlated with insulin resistance than any lipid-related factor. Also, IRAS cohort subjects that converted from prediabetes to diabetes during follow-up experienced an increase in BCAA and a drop in glycine across this transition (Palmer et al., 2015). Importantly, these findings have been confirmed via application of metabolomics to large cross-sectional cohorts. Thus, in 7098 young Finns (mean age 31 years), nuclear magnetic resonance (NMR) spectroscopy was used to profile 39 circulating metabolites and lipids, with the findings of highly significant correlations between BCAA, aromatic amino acids, ketones, and fatty acid composition and saturation with insulin resistance measured by HOMA-IR (p < 0.0005 for 20 metabolites) (Wurtz et al., 2012). Similarly, metabolomics profiling of 447 metabolites in 2204 female subjects in the TwinsUK cohort found a strong association of BCAA and their metabolites with type 2 diabetes and impaired fasting glucose levels (Menni et al., 2013). Several groups have also shown that BCAA and related metabolites are associated with coronary artery disease (CAD), even when controlled for diabetes (Shah, et al., 2010; Magnusson et al., 2013; Bhattacharya et al., 2014).
Targeted and non-targeted metabolomics studies have also revealed that BCAA and their metabolites are prognostic for incident type 2 diabetes and obesity interventions outcomes. Thus, elevated plasma levels of Leu, Ile, Val, Phe and Tyr were associated with up to a 5-fold risk for future development of type 2 diabetes in the Framingham Heart and Malmo Diet and Cancer study cohorts (Wang et al., 2011). Analysis in the Framingham cohort compared 189 subjects that developed type 2 diabetes over the course of follow-up versus 189 matched controls that were diabetes free. Furthermore, targeted metabolomics was performed on baseline blood samples taken from 500 subjects in the weight loss maintenance (WLM) trial prior to a six-month behavioral/dietary (DASH diet) intervention. At baseline, the factor score for a BCAA-related principal component very similar to that associated with insulin resistance strongly predicted improved insulin sensitivity, whereas lipid-related factors or the amount of weight lost had little or no predictive association (Shah et al., 2012). Thereafter, NMR profiling of 1680 subjects from the Cardiovascular Risk in Young Finns study identified a strong association of the three BCAA, Tyr and Phe with development of insulin resistance over a 6-year follow up period (Wurtz et al., 2013). This association was most pronounced in men. Finally, a recent study confirms a strong association between BCAA, Tyr and Phe with insulin resistance in 429 Chinese subjects, and a close association with these metabolites with future development of diabetes (Chen et al., 2016).
Another means of aligning metabolomics markers with disease is to study their pattern of response to efficacious disease interventions. As an example, obese subjects with type 2 diabetes undergoing gastric bypass (GBP) surgery have a much more dramatic decline in circulating BCAA, C3 and C5 acylcarnitines, Phe, and Tyr than found in response to dietary intervention, despite equal weight loss (Laferrère et al., 2011). In another study, a similar large drop in BCAA and related metabolites was observed in gastric bypass and gastric sleeve forms of surgical intervention (Magkos et al., 2013). These findings are significant because the surgical methods induce more dramatic improvements in glucose homeostasis than life style interventions (Laferrère et al., 2011; Clifton, 2010). In aggregate, work from recent years has established that BCAA and related metabolites are associated with insulin resistance, diabetes, and CAD, that they are predictive of diabetes development, that they are predictive of intervention outcomes, and that they are highly and uniquely responsive to therapeutic interventions.
Interestingly, these findings may not extend to all medical therapies. Thus, treatment of 30 insulin sensitive and 30 insulin resistant subjects with a single dose of the sulfonylurea drug, glibizide, which lowers blood glucose by stimulating insulin secretion, effectively lowered BCAA and aromatic amino acid levels in insulin sensitive, but not insulin resistant subjects (Walford et al., 2013). In contrast, upon treatment of the same subjects with the insulin sensitizing drug metformin (two days of twice-daily injections), lowering of glucose and insulin and an increase in the levels of BCAA and aromatic amino acids occurred only in insulin resistant subjects. Further studies of longer duration will be required to fully understand the impact of various diabetes therapies on metabolomics profiles.
More recently, application of non-targeted metabolomics has expanded the range of metabolites that predict risk of future diabetes to include other amino acids, their metabolites, and lipids. For example, comprehensive lipidomic profiling of the same 189 diabetic and 189 non-diabetic subjects from the Framingham cohort previously subjected to LC-MS/MS-based metabolomics profiling revealed that higher diabetes risk is associated with triacylglyerols (TG) that contain fatty acids of lower carbon number and double bond content, whereas subjects with TG containing fatty acids with higher carbon number and double bonds had lower diabetes risk (Rhee et al., 2011). In addition, application of more comprehensive LC-MS/MS profiling to these subjects identified 2-aminoadipic acid (2-AAA) as a metabolite strongly correlated with incident T2D, with subjects in the top quartile for this metabolite having a >4-fold increase in risk of disease (Wang et al., 2013). 2-AAA is thought to be derived from lysine catabolism, but surprisingly, 2-AAA levels were not associated with BCAA or aromatic amino acids in this study, suggesting that this marker reports on an independent disease risk mechanism. Finally, application of a combined GC/MS and LC-MS/MS nontargeted metabolomics approach to plasma samples from 399 subjects from the Relationship of Insulin Sensitivity to Cardiovascular Risk (RISC) study confirmed that BCAA were among the positively associated and glycine among the negatively associated metabolites with insulin resistance measured by hyperinsulinemic clamp (Gall et al., 2010). However, other metabolites, most notably α– hydroxybutyrate, linoleoyl-glycerophosphocholine, and oleic acid, were more strongly associated with the insulin resistant state compared to BCAA or glycine, and furthermore were selectively correlated with impaired glucose tolerance across multiple cohorts (Gall et al., 2010; Cobb et al., 2016).
Metabolomics applied to Gestational Diabetes and Offspring Outcomes
All of the studies discussed above were performed in adult humans, but metabolomics has also recently been applied to pediatric populations to search for markers of early stage insulin resistance and prediction of future diabetes, and also to pregnant women in search of biomarkers that assess risk of conversion from gestational diabetes to type 2 diabetes, and that may predict outcomes in offspring. Findings in pediatric cohorts have been inconsistent (Frohnert and Rewers, 2016). Thus, two studies showed a positive correlation between BCAA, BMI and HOMA-IR (Newbern et al., 2014; Perng et al., 2014), and another an association with obesity (McCormack et al., 2013), but still others found no significant differences in BCAA levels in normal weight compared to obese adolescents (Wahl et al., 2012), and even a positive correlation between BCAA/BCAA metabolites and insulin sensitivity (Si) (Michaliszyn, et al., 2012). However, in a group of 17 pre- or early-pubertal children (age 8–13 years), baseline BCAA levels were found to be associated with HOMA-IR measured 18 months later, suggesting that, as in adults, BCAA levels may be predictive of future disease risk in adolescent subjects (McCormack et al., 2013). More work with larger cohorts of adolescent subjects is needed, and it remains possible that biomarkers other than BCAA and related metabolites will have stronger associations with pediatric insulin resistance and type 2 diabetes.
Studies linking maternal metabolic status to offspring outcomes are just beginning to emerge. One study found positive associations of maternal triglycerides, leucine/isoleucine, ketone and lactate levels and negative association of glycine levels with various measures of offspring body weight, although these were attenuated when corrected for glycemic status of the mothers (Scholtens et al., 2013). In another study, children of obese mothers were shown to have higher BCAA levels than those of lean mothers (Perng et al., 2014). Importantly, maternal metabolomics profiling provided an improved ability to predict newborn size outcomes beyond traditional risk factors, including maternal glucose (Scholtens et al., 2016). Future studies conducted over longer periods of follow up should shed further light on the relationship of maternal metabolic status to that of their offspring.
The use of metabolomics to predict the transition from gestational diabetes (GDM) to full-blown type 2 diabetes (which occurs in approximately 30% of GDM cases) is more immediately promising. A recent study used a plate assay technology to measure 163 metabolites, and complemented this with direct amino acid analysis by LC-MS/MS and fatty acid analysis by GC/MS. These methods were used to compare 122 incident cases of type 2 diabetes with 122 non-cases of incident T2D matched by age, BMI and race/ethnicity from a cohort of 1035 pregnant women with GDM. Remarkably, metabolites significantly elevated in women with incident T2D included all three BCAA, Tyr, and 2-AAA, whereas glycine was negatively associated with diabetes risk (Allalou et al., 2016). Interestingly, multiple fatty acids were decreased in the subjects destined for incident diabetes as well. In contrast, metabolomic profiling of 96 women with GDM versus 96 women matched by age, BMI and gravidity with normal glucose tolerance found 6 metabolites to be different between the groups, anthranilic acid, alanine, glutamate, creatinine, allantoin and serine, but no differences in BCAA, aromatic amino acids, or glycine (Bentley-Lewis et al., 2015). Similarly, few metabolites associated with fasting plasma glucose levels in 400 pregnant women of European descent from the Hyperglycemia and Adverse Pregnancy Outcome (HAPO) cohort (Scholtens et al., 2013). However, many more metabolites were associated with glucose levels at the one-hour time point of an oral glucose tolerance test in these subjects, including positive associations with leucine/isoleucine, glutamate/glutamine, phenylalanine, α-hydroxybutryate, and multiple acylcarnitines and fatty acids. Finally, a recent study comparing a cohort of GDM and normoglycemic mothers using a combined LC-MS and GC-MS approach revealed a striking (7-fold) increase in a furan fatty acid metabolite 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid (CMPF) in the GDM subjects (Prentice et al., 2014; Retnakaran et al., 2016). Furthermore, CMPF levels were even higher in a subset of GDM subjects that progressed to type 2 diabetes, and were elevated in separate analysis of subjects with type 2 diabetes compared to non-diabetic subjects (Prentice et al., 2014). Evidence for the involvement of CMPF in diabetes pathogenesis is discussed later in this article.
Metabolomics Applied to Prediction of Cardiovascular Events
Metabolomics has also been applied to predict incident cardiovascular events (Shah et al., 2012a). A principal component factor comprised of short to medium chain dicarboxylated acylcarnitines (DC-AC, including C4-DC, C5-DC, and C6-DC) predicted incident death/MI within 2 years in patients referred for diagnostic catheterization in the CATHGEN study, a finding confirmed in a validation cohort (Shah et al., 2010). These findings were expanded via a cross-sectional survey of 2023 subjects from the CATHGEN biorepository, demonstrating association of death/MI with a DC-AC factor (p = 0.005) essentially identical to the predictive signature from the case-control study (Shah et al., 2012b). In addition, studies of 478 CATHGEN subjects who underwent coronary artery bypass grafting (CABG) revealed that the same short-chain DC-AC principal component was associated with adverse outcome in univariate analysis (P=0.002), and remained independently predictive of adverse outcomes in multivariable time-to-event analysis (P<0.001) (Shah et al., 2012c). In a follow-up study, integrated omics analysis including metabolomics, GWAS, transcriptomics and epigenetics in the CATHGEN cohort found that the DC-AC diagnostic signature maps with high significance to several SNPs associated with genes involved in ER stress and the protein unfolding response (Kraus et al., 2015). Those SNPs in turn are independently associated with CVD events in time-to-event analysis. Transcriptomic and epigenetic profiling in the same subjects also identified associations between ER stress genes and the DC-AC cluster (Kraus et al., 2015).
Metabolomics has also been used to reveal a link between the diet, the gut microbiome, host metabolism, and biomarkers of incident CVD events (Wang et al., 2011). Using a non-targeted LC-MS approach, 18 analytes were found to be associated with cardiovascular events, including 3 involved in choline metabolism--choline, betaine, and trimethylamine N-oxide (TMAO). Nutrients such as phosphatidylcholine, carnitine, and choline are metabolized by gut bacteria to generate trimethylamine (TMA). TMA is absorbed and carried by the portal circulation to the liver, where it is metabolized to TMAO by hepatic flavin monooxygenases. Several large studies have confirmed a link between TMAO and cardiovascular events, and cause and effect relationships between TMAO and cardiovascular diseases is emerging, as discussed below.
Metabolomics applied to fatty liver and NASH
Overstorage of fat in the liver leads to the clinical syndrome of non-alcoholic fatty liver disease (NAFLD), which if left untreated develops an inflammatory component and evolves to non-alcoholic steatohepatitis (NASH). This in turn can progress to more advanced forms of liver disease, including cirrhosis and hepatocellular carcinoma (Lallukka and Yki-Jarvinen, 2016). In the United States, approximately 30% of adults have NAFLD, and approximately 4% have NASH, but this varies across ethnic groups. The incidence of NAFLD is highest among Mexican-Americans (estimated at 45%), intermediate in European-Americans, and lowest in African-Americans (24%) (Sherif et al., 2016). Important insight into this imbalance in incidence across ethnic groups emerged with the identification of a variant allele (I148M) of the gene encoding patatin-like phospholipase A3 (PNPLA3) strongly associated with liver fat and highly prevalent in Hispanic individuals within a multi-ethnic cohort of subjects in the Dallas Heart Study (Romeo et al., 2008). The exact biological function of PNPLA3 remains unknown, but the recombinant I148M PNPLA3 variant protein has hydrolytic activity against various esterified lipids (Huang et al., 2011), and exhibits enhanced propensity for association with lipid droplets (Smagris et al., 2015). Remarkably, recent evidence suggests that subjects with NAFLD induced by the PNPLA3 mutation are relatively protected against insulin resistance and type 2 diabetes whereas NAFLD that develops independent of PNPLA3 mutations (“metabolic NAFLD”) is strongly predictive of future diabetes risk (Lallukka and Yki-Jarvinen, 2016; Luukkonen et al., 2016). To investigate this further, comprehensive lipidomics analysis was performed on subjects with NAFLD with and without the PNPLA3 mutation (Luukkonen et al., 2016). The severity of steatosis was associated with insulin resistance as measured by HOMA-IR in subjects with the “normal” PNPLA3 allele, but no such relationship was found in subjects with the I148M PNPLA3 mutation. Remarkably, metabolic NAFLD with insulin resistance was associated with preferential storage of saturated fats and ceramides in liver, whereas the PNPLA3 NAFLD group had lower levels of these lipids and increased levels of polyunsaturated fats (Luukkonen et al., 2016). Obviously, there is huge opportunity imbedded in understanding how lipid composition of excess liver fat influences systemic glucose homeostasis and insulin action (and vice-versa), and this is likely to be an intensive area of investigation in the coming years.
New metabolic regulatory and pathophysiologic mechanisms emerging from metabolomics studies
New insights into mechanisms of insulin resistance
Whereas the potential utility of metabolomics in the realm of disease prediction should be apparent from the foregoing summary, perhaps the most exciting windfall that has emerged from application of this technology is the blueprint that it provides for defining new physiologic and pathophysiologic mechanisms. As an early example, targeted metabolomics was used to show increases in a broad array of fatty acid-derived acylcarnitines, and a concurrent decrease in TCA cycle intermediates in skeletal muscle of obese rodents (An et al., 2004; Koves et al., 2005; Koves et al., 2008). Genetic obesity or high fat feeding results in upregulation of enzymes of fatty acid beta-oxidation, but in sedentary animals, this occurs absent a concurrent increase in enzymes of the TCA cycle and electron transport chain. This “disconnect” between the β-oxidation pathway and the terminal oxidative machinery results in incomplete fatty acid oxidation, leading to accumulation of mitochondrial-derived acyl CoA and acylcarnitine metabolites. The potential pathophysiologic relevance of this finding was demonstrated in two studies in mice lacking expression of malonyl CoA decarboxylase or carnitine acetyltransferase (CrAT). In the former case, increases in levels of malonyl CoA, an allosteric inhibitor of carnitine palmitoyltransferase-1 (CPT-1), resulted in reduced flux of fatty acids through the β-oxidative pathway and a decrease in muscle acylcarnitine levels in diet-induced obesity, accompanied by improved glucose tolerance (Koves et al., 2008). In the latter case, knockout of CrAT, which catalyzes conversion of short chain acyl CoAs to membrane permeant acylcarnitines, resulted in accumulation of incompletely oxidized acyl CoA metabolites, exacerbating glucose intolerance and insulin resistance (Muoio et al., 2012).
Significant effort has also been applied to investigation of the strong association of BCAA and related metabolites with metabolic diseases, and evidence for a role of BCAA in disease pathogenesis has started to emerge. Thus, rats fed a high-fat (HF) diet supplemented with BCAA develop insulin resistance despite eating less food and gaining less weight than rats fed HF diet alone (Newgard et al., 2009). In contrast, rats pair-fed on HF diet to match the caloric intake of the HF/BCAA group do not become insulin resistant. Targeted metabolomics revealed that the HF/BCAA group exhibited accumulation of a broad spectrum of acylcarnitines, to the same extent as observed in heavier HF-fed animals (Newgard et al., 2009). These findings are consistent with a model in which excess BCAA contribute to impaired efficiency of fatty acid oxidation, resulting in accumulation of incompletely oxidized lipid species (Newgard, 2012).
Conversely, feeding obese rodents with a BCAA restricted diet improves insulin sensitivity (White et al., 2016; Fontana et al., 2016), and possible mechanisms to explain this finding are emerging. In studies comparing Zucker obese rats fed a standard chow diet or an isonitrogenous diet in which BCAA content was lowered by 45%, feeding of the BCAA restricted diet resulted in improved whole-animal insulin sensitivity as measured by hyperinsulinemic/euglycemic clamp, as well as enhanced muscle glucose uptake and glycogen synthesis (White et al., 2016). This improvement in glucose metabolism was accompanied by normalization of multiple short and medium-chain acyl CoA species in skeletal muscle. In addition, muscle glycine levels are dramatically reduced in Zucker-obese compared to Zucker-lean rats, but are completely normalized in Zucker-obese rats by feeding of the BCAA-restricted diet, providing the first evidence of a direct connection between BCAA supply and glycine levels, helping to explain the consistent observation of high BCAA levels and low glycine levels in epidemiological studies. The restoration of the muscle glycine pool is accompanied by increased excretion of acetylglycine in the urine, suggesting that formation of acylglycine adducts may constitute a means by which BCAA restriction lowers muscle acyl CoA levels and restores muscle insulin sensitivity. Consistent with this, BCAA restriction lowers the respiratory exchange ratio in Zucker-obese rats, indicative of enhanced efficiency of fatty acid oxidation. These findings are aligned with a model wherein elevated circulating BCAA contribute to development of obesity-related insulin resistance by interfering with lipid oxidation in skeletal muscle (Figure 1). BCAA-dependent depletion of the skeletal muscle glycine pool may contribute to this effect by slowing acyl-glycine export to the urine (White et al., 2016). Interestingly, protein restricted diets have recently been shown to increase circulating levels of FGF21, a hormone that increases lipid oxidation and improves glucose homeostasis (Laeger et al., 2014). However, induction of FGF21 does not seem to mediate the salutory effects of BCAA restriction on metabolic state, as FGF21 levels increase in response to generalized protein restriction, but not in response to specific restriction of BCAA (Fontana et al., 2016).
Two new amino acid-related mechanisms for regulation of metabolic homeostasis have recently emerged via application of metabolomics to rodent models of transgenic overexpression of the transcriptional co-regulator PGC-1α. PGC-1α overexpression in skeletal muscle mimics the effects of exercise by inducing browning of white adipose tissue. Profiling of media from cultured muscle cells by LC-MS identified 4 metabolites that were significantly increased by forced overexpression of PGC-1α, including β-aminoisobutyric acid (BAIBA), which is derived from valine metabolism (Roberts et al., 2014). BAIBA increases expression of PPARα in white adipose and liver, induces thermogenic genes in adipose tissue, and activates fatty acid oxidation in hepatocytes. Administration of BAIBA to mice limits weight gain, enhances glucose tolerance, and induces browning of white fat. Exercise increases circulating BAIBA concentrations in rodents and humans, and it is proposed that BAIBA represents an endocrine factor that modifies liver and adipose metabolism in response to physical activity (Roberts et al., 2014). More recently, a different valine metabolite, 3-hydroxyisobutryate (3-HIB), was reported to be induced by PGC-1α overexpression and shown to function as an activator of trans-endothelial fatty acid transport (Jang et al., 2016). Like the BCAA, 3-HIB levels were found to be elevated in db/db mice and in humans with type 2 diabetes, and administration of 3-HIB to rodents resulted in lipid accumulation in muscle and development of insulin resistance. Adding complexity, sedentary mice with muscle specific overexpression of PGC-1α fed on a high fat diet actually have impaired glucose tolerance relative to non-transgenic littermates (Choi et al., 2008), and mice of the two genotypes exhibit similar improvements in body weights and glucose control in response to a combined caloric restriction/exercise intervention (Wong et al., 2015). Targeted metabolomic profiling of muscles and plasma from these latter animals identified a principal component comprised of increased levels of ceramides, medium-chain acylcarnitines, and Val, Met, His, and ornithine, and decreased levels of the TCA cycle intermediates malate, fumarate and succinate that correlated with changes in energy balance, muscle insulin sensitivity and glucose tolerance. BAIBA or 3-HIB were not measured in these studies.
How to reconcile these seemingly disparate findings? One unifying model requires that the regulatory significance of BAIBA or 3-HIB is different dependent on nutritional status (see Figure 1). Thus, in lean animals or humans, 3-HIB may act as a paracrine factor to increase fatty acid uptake by muscle during exercise, and BAIBA may function to mobilize and oxidize lipids as needed to support the increase in physical activity. By inducing valine catabolism, exercise increases the concentration of both mediators to engage these healthful effects. In contrast, whereas BAIBA levels are negatively correlated with fasting glucose, HOMA-IR, and circulating triglycerides and cholesterol in the Framingham cohort (Roberts et al., 2014), 3-HIB levels are positively correlated with blood glucose in a cohort of diabetic and normal subjects (Jang et al., 2016). Does this suggest that elevated BCAA in sedentary and metabolically unhealthy subjects are preferentially metabolized to yield 3-HIB rather than BAIBA, and that the consequence of this in a mileu of elevated lipids is to promote lipid overstorage and impair insulin action in muscle? Further studies will be required to investigate this issue and others related to these fascinating new mechanisms.
Serving as an important framework for these findings are recent discoveries relating to the mechanisms that cause BCAA and related metabolites to increase in the obese condition. BCAA catabolism is regulated at several early steps, including transport of BCAA into cells through the large neutral amino acid transporter LAT-1, transamination catalyzed by branched-chain aminotransferase (BCAT), and conversion of branched chain keto acids to acyl CoAs by the branched chain ketoacid dehydrogenase (BCKDH) complex. BCKDH is a multi-subunit enzyme complex of a design and subunit composition similar to that of the pyruvate dehydrogenase (PDH) complex. Like PDH, BCKDH activity is regulated by phosphorylation and dephosphorylation catalyzed by specific kinase (branched-chain ketoacid dehydrogenase kinase, BDK) and phosphatase (PPM1K) enzymes. Obesity has complex effects on regulation of BCAA catabolism that vary across key metabolic tissues and organs. In adipose tissue, obesity causes concerted suppression of all of the enzymes in the catabolic pathway at the transcriptional level (She et al., 2007; Herman et al., 2010; Hsaio et al., 2011). This mechanism does not seem to be active in liver, where instead hepatic BCAA catabolism is suppressed by increased inhibitory phosphorylation of BCKDH, secondary to increased expression of BDK and decreased expression of PPM1K (She et al., 2007). Remarkably, neither of these mechanisms appear to be operative in skeletal muscle (She et al., 2007) and BCKDH activity is actually increased in muscle of Zucker-obese compared to Zucker-lean rats (White et al., 2016). This differential regulation may contribute to mitochondrial substrate overload in skeletal muscle in obese animals by several mechanisms, including elevations in circulating BCAA due to reduced utilization by liver and adipose tissue, generation of anaplerotic substrates from BCAA catabolism in muscle that “fill up” the TCA cycle and make it more difficult for fatty acids to be completely oxidized (Newgard, 2012; White et al., 2016) and 3-HIB-stimulated trans-endothelial lipid transport (Jang et al., 2016). Interestingly, 2-AAA, a lysine-derived metabolite strongly associated with risk of type 2 diabetes discussed earlier (Wang et al., 2013), is also metabolized by an enzyme complex that resembles PDH and BCKDH known as α-ketoadipate dehydrogenase. Little is known about the regulation of this enzyme complex, but its similarity to PDH and BCKDH suggests that it may be downregulated in obesity, thereby contributing to the accumulation of its substrate, 2-AAA.
Another factor that may contribute to accumulation of BCAA in obesity is the gut microbiome. BCAA are “essential” amino acids, in the sense that humans are unable to synthesize them de novo, and it had been assumed that they are primarily obtained from the diet, particularly meat and dairy products in which they are enriched. However, some gut bacteria are able to synthesize BCAA from the simple organic acid precursors pyruvate and ketobutyrate. Transcriptomic analysis of microbiota from human twins discordant for obesity revealed induction of the entire de novo pathway for BCAA synthesis in the microbiota of obese compared to lean twins (Ridaura et al., 2013). Induction of this pathway correlated with increased production of BCAA by the microbiota of obese subjects, and their transplantation into germ free mice caused a significant increase in circulating levels of BCAA and related metabolites in host animals. Remarkably, just two weeks after transplant, mice colonized with microbiota from obese subjects were glucose intolerant and exhibited a marked increase in levels of a broad spectrum of short chain, medium chain, and long chain acylcarnitines in skeletal muscle compared to animals that received microbiota from lean twins (Ridaura et al., 2013). More recently, another group has extended these observations by performing metabolomics and microbiome profiling in 277 non-diabetic subjects, and inclusion of 75 subjects with type 2 diabetes for comparative analysis (Pedersen et al., 2016). A strong correlation between BCAA levels and insulin resistance was confirmed in this study, and the specific bacterial species P. copri and B. vulgatus were identified as those primarily responsible for driving BCAA biosynthesis in insulin resistant subjects. Indeed, transplantation of P. copri into germ free mice raised circulating levels of BCAA and caused insulin resistance and glucose intolerance. Taken together, these studies suggest that in addition to regulation of energy balance and adiposity, the gut microbiome can have a substantive impact on metabolic homeostasis that contributes to disease pathogenesis. Further studies will be required to delineate the relative contributions of genetics, diet, gut microbiome, and selective suppression of BCAA catabolism in host tissues to the observed increases in BCAA and related metabolites in obese and insulin resistant subjects.
Metabolomics Applied to Pancreatic Islet Biology
Metabolomics has also been used to study pancreatic islet biology, including mechanisms leading to development of β-cell dysfunction in diabetes. In one set of studies involving rat insulinoma (INS-1) derived cell lines with different capacities for glucose-stimulated insulin secretion (GSIS), application of NMR-based mass isotopomer analysis was used to calculate relative flux rates of pyruvate through the oxidative enzyme PDH and the anaplerotic enzyme pyruvate carboxylase (PC). The studies showed that variable GSIS across the panel of INS-1-derived cell lines and in islets is tightly correlated to pyruvate anaplerosis and cycling activity rather than pyruvate oxidation (Lu et al., 2002; Alves et al., 2015). The significance of this finding is that anaplerotic metabolism of pyruvate through PC can produce excess TCA cycle intermediates that exit the mitochondria to engage with cytosolic enzymes. Indeed, surprisingly, flux through PC-catalyzed anaplerosis is more active in glucose-stimulated primary islet cells than flux through the oxidative enzyme PDH (Schuit et al., 1997). Systematic study of several possible “pyruvate cycling” pathways implicated the cytosolic NADP-dependent isocitrate dehydrogenase (ICDc or IDH1) as a key extra-mitochondrial step, based on studies in which knockdown of the citrate/isocitrate carrier (responsible for export of citrate and isocitrate from the mitochondria) or IDH1 resulted in significant impairment of GSIS (Jensen et al., 2008; Joseph et al., 2006; Ronnebaum et al., 2006). The IDH1 reaction appears to play two key roles in mediating insulin release by producing NADPH needed for the glutathione reductase reaction (Ferdaoussi et al., 2015; Ivarsson, et al., 2005), and generating α-ketoglutarate that can be transaminated to glutamate, a building block of glutathione (Figure 2). These pathways help maintain a robust pool of reduced glutathione (GSH), that in turn drives reduction of the desumoylation enzyme SENP1, thereby triggering insulin granule exocytosis via desumoylation of secretory granule proteins (Ferdaoussi et al., 2015). Intermediates in this pathway, including isocitrate, NADPH and GSH (but not NADH or α-ketoglutarate) induce exocytosis in patch clamped rodent and human β-cells (Ferdaoussi et al., 2015; Ivarsson et al., 2005), an effect that is lost in mouse islets lacking expression of SENP1 or in human islets with shRNA-mediated suppression of SENP1 expression (Ferdaoussi et al., 2015). Importantly, isocitrate and NAPDH rescue exocytosis in glucose unresponsive β-cells from humans with type 2 diabetes (Ferdaoussi et al., 2015).
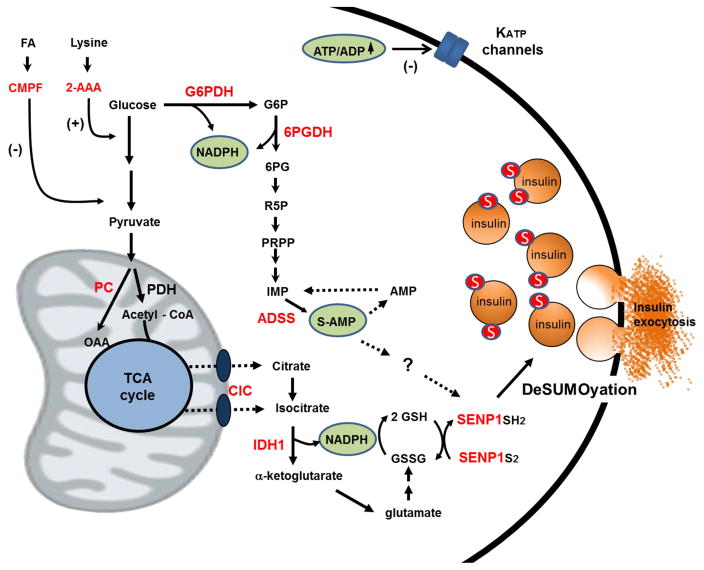
Figure 2. Emergent mechanisms of glucose-stimulated insulin secretion (GSIS) from metabolomics studies.
Application of metabolomics methods to the pancreatic islet has resulted in identification of mechanisms that may complement the classical KATP-channel-dependent pathway for GSIS. This includes a pathway initiated by anaplerotic metabolism of glucose-derived pyruvate through pyruvate carboxylase (PC), egress of citrate, isocitrate, and a-ketoglutarate from the mitochondria to the cytosol via the citrate/isocitrate carrier (CIC), engagement of isocitrate with the cytosolic, NADP-dependent isoform of isocitrate dehydrogenase (IDH1) and reduction of glutathione to GSH by glutathione reductase. A second pathway involving metabolism of glucose through the pentose monophosphate shunt, including the first two NAPDH producing steps glucose-6-phosphate dehydrogenase (G6PDH) and 6-phosphogluconate dehydrogenase (6PGDH), results in a sharp increase in adenylosuccinate (S-AMP) produced from IMP via the adenylosuccinate synthase (ADSS) reaction. Importantly, intermediates generated by either the isocitrate/GSH (isocitrate, NADPH, GSH) or S-AMP (S-AMP) pathways stimulate exocytosis in permeabilized human β-cells and rescue loss of glucose regulation in β-cells from humans with type 2 diabetes. Also for both pathways, the effects on exocytosis require expression of the insulin granule desumoylating enzyme SENP1. Also shown, metabolomics has identified two metabolites associated with risk of type 2 diabetes that modulate β-cell function, 2-aminoadipic acid (2-AAA), which enhances insulin secretion at basal glucose levels, and the fatty acid furan metabolite, 3-carboxy-4-methyl-5-propyl-2- furanpropanoic acid (CMPF), which impairs glucose metabolism and insulin secretion. See text for details and discussion.
Metabolomics was also used to demonstrate glucose-induced changes in an array of purine and nucleotide pathway intermediates in islet cells, although the functional relevance of these findings was not initially defined (Huang and Joseph, 2014; Lorenz et al., 2013; Spegel et al., 2013; El-Azzouny et al., 2014). To investigate this further, a targeted, quantitative LC-MS/MS method was developed and used to demonstrate a robust decrease in inosine monophosphate (IMP), and an equally striking increase in adenylosuccinate (SAMP) in glucose-stimulated 832/13 cells (Gooding et al., 2015). IMP is converted to S-AMP by adenylosuccinate synthase (ADSS), suggesting that this enzyme could play a regulatory role in β-cell glucose sensing (Figure 2). Indeed, pharmacologic or molecular suppression of ADSS caused impairment of GSIS with an attendant decrease in S-AMP, effects that could be overcome by addition of adenine. Moreover, siRNA-mediated suppression of ADSL, a more proximal enzyme in the S-AMP biosynthetic pathway, impaired GSIS and lowered S-AMP levels, independent of decreases in other adenine nucleotides or GTP. Infusion of S-AMP into patch-clamped β-cells from non-diabetic humans revealed that it is equipotent to glucose for stimulation of exocytosis. Moreover, similar to intermediates in the isocitrate/GSH pathway, S-AMP rescued exocytosis in glucose-unresponsive β-cells from humans with type 2 diabetes. Finally, the stimulatory effect of S-AMP on exocytosis in patch-clamped human β-cells is impaired by shRNA-mediated suppression of SENP1 (Gooding et al., 2015).
The new pyruvate/isocitrate/GHS and S-AMP pathways of GSIS are viewed as complementary to the classical initiating pathway, involving inhibition of ATP-sensitive K channels (KATP channels), membrane depolarization and activation of voltage-gated Ca2+ channels (Henquin et al., 2003) (Figure 2). It remains to be determined if the newly described pathways are additive or synergistic with each other and/or with the KATP channel-dependent pathway in control of GSIS. The source of NADPH for reducing GSH also remains to be defined. Suggesting a role for the pentose monophosphate shunt, it has been reported that inhibition of the first enzyme in the pathway, glucose-6-phosphate dehydrogenase (G6PDH) by dehydroepiandrosterone (DHEA) (Spegel et al., 2013), or the second enzyme in the pathway, 6-phosphogluconate dehydrogenase (6PGDH) by siRNA or the chemical inhibitor 6-AN (Goehring et al., 2011), results in impaired GSIS. The effects of 6PGDH inhibition were ascribed to accumulation of early PPP intermediates and activation of p-ERK (Goehring et al., 2011), whereas DHEA treatment caused a decrease in GSH levels, although effects on NADPH were not reported (Spegel et al., 2013). These findings are not consistent with other work showing that shRNA-mediated suppression of IDH1 is sufficient to ablate the glucose-induced increment in GSH and GSH:GSSG ratio in islet cells (Ferdaoussi et al., 2015). In addition, the finding that S-AMP has a direct effect to activate exocytosis in normal islets, and is able to rescue dysfunctional exocytosis in human T2D islets seems more aligned with the concept that S-AMP is the key regulator of GSIS produced in the pentose shunt (Gooding et al., 2015). Elegant metabolic flux methods have recently been reported for calculating the relative contributions of various pathways to total NADPH production (Fan et al., 2014). For example, absolute pentose pathway flux is measured by the classical method of the difference between 14CO2 production from [1-14C] and [6-14C] glucose. The contribution of the pentose shunt to NADPH synthesis is then measured by fractional enrichment (FE) of the NADPH and G6P pools in cells incubated with [3-D]glucose (Fan et al., 2014). Using these methods, a surprisingly large contribution of folate metabolism to total NAPDH production was recently reported in HEK293T cells (Fan et al., 2014). Application of similar methods to pancreatic islet cells should allow the fractional contributions of various NADPH producing pathways to be quantified. Performance of such flux experiments in the presence and absence of strategic suppression of contributory enzymes such as G6PDH and IDH1 will help to define key operative pathways for control of GSIS.
Two metabolites that were discussed earlier in the context of their utility for prediction of diabetes, 2-AAA and CMPF, may contribute to diabetes pathogenesis by altering β-cell function. Paradoxically, administration of 2-AAA to mice actually lowers rather than raises circulating glucose levels, which seems to be mediated by an effect of 2-AAA to stimulate insulin secretion, even at low glucose levels (Wang et al., 2013). This raises the possibility that the link between elevated levels of 2-AAA and diabetes risk in humans could be development of insulin resistance secondary to chronic hyperinsulinemia. This hypothesis requires further investigation. In contrast, CMPF seems to have a direct effect to induce β-cell dysfunction (Liu et al., 2016). Thus, treatment of diet-induced obese (DIO) or ob/ob mice with CMPF caused a decrease in glucose-stimulated insulin secretion and a worsening of glucose intolerance in these models. The decrease in β-cell function in CMPF-treated mice was associated with a decrease in glucose metabolism and an increase in fatty acid oxidation, accompanied by an increase in generation of reactive oxygen species. CMPF also caused an impairment in insulin granule maturation that led to a higher insulin content and an increase in proinsulin:insulin ratio. The precise molecular target(s) of CMPF in mediating these effects remain to be defined. Interestingly, a clear increment in CMPF levels was observed in pre-diabetic subjects that progressed to overt diabetes, whereas those from this cohort that remained in the prediabetic state had stable CMPF levels (Liu et al., 2016). This finding implicates CMPF as a “tipping factor” that may help to explain why some individuals can remain in the prediabetic state for many years before suddenly progressing to type 2 diabetes.
Metabolomics Applied to Transgenic Mouse Models
As described earlier, new mechanisms linking BCAA metabolism and metabolic homeostasis were unveiled in studies of mice with muscle-specific overexpression of the transcriptional co-regulator PGC-1α (Roberts et al., 2014; Jang et al., 2016; Choi et al., 2008; Wong et al., 2015). Metabolomics has also helped to identify metabolic pathways regulated by other transcription factors and co-regulators in transgenic mouse models. Such studies can delineate the specific pathways affected, the tissues and organs in which regulation occurs, and in some cases, novel metabolites that mediate biological effects. For example, a series of studies have been conducted on transgenic mice with global knockout of the transcriptional co-activators SRC-1, SRC-2, and SRC-3. Metabolic profiling contributed to our understanding that SRC2 regulates glucose-6-phosphatase expression in liver (Chopra et al., 2008), SRC1 integrates hepatic fatty acid and amino acid metabolism in liver and other tissues (Louet et al., 2010), whereas SRC3 knockout has focused metabolic effects in muscle, and a particular signature to cause accumulation of very-long chain acylcarnitines (York et al., 2012). This latter finding led to the discovery that SRC3 controls expression of the mitochondrial carnitine-acylcarnitine translocase (CACT), which mediates entry of long-chain acylcarnitines into the mitochondrial matrix during fatty acid oxidation. Mutations in the gene encoding this protein underlie human CACT metabolic myopathy.
Another novel mechanism has emerged from studies in mice with genetic manipulation of GLUT-4 expression in adipose tissue. Thus, mice with adipose-specific GLUT-4 knockout develop insulin resistance in liver and muscle (Abel, et al., 2001), whereas transgenic overexpression of GLUT-4 causes improvement in glucose tolerance, despite an increase in lipogenesis and circulating fatty acids (Shepherd et al., 1993). Surprisingly, the effect of GLUT-4 overexpression to improve glucose disposal depends on lipogenic activity in adipose tissue, as the effect on glucose homeostasis is eliminated in mice with dual knockout of the key lipogenic transcription factor ChREBP and GLUT-4 (Herman et al., 2012). This led to the hypothesis that specific lipids may mediate the glucoregulatory effects of GLUT4 manipulation in adipose tissue. To test this idea, comprehensive, nontargeted lipidomic analysis was performed on adipose tissue and plasma from GLUT-4 overexpressing and control mice (Yore et al., 2014). Among 5 lipids found to be enriched in the samples from GLUT-4 transgenic mice, four were identified as fatty acid-hydroxy fatty acid esters, comprised of a typical long chain fatty acid (palmitate (PA), oleate (OA), stearate (SA), or palmitoleate (PA)) esterified to hydroxylated versions of one of the same four fatty acids. Among the 16 possible fatty acid-hydroxy fatty acid (FAHFA) dimeric species, 6 were increased in serum of GLUT-4 transgenic compared to wild-type mice, and the most clearly upregulated form, PA-hydroxy SA (PAHSA), was studied in more detail. Serum and adipose tissue PAHSA levels were positively correlated with insulin sensitivity in humans. A single oral bolus of PAHSA given to mice 30 minutes prior to a glucose tolerance test enhanced glucose clearance, and an effect of PAHSA to enhance insulin-stimulated GLUT4 translocation was demonstrated in cultured adipocytes. Remarkably, PAHSA also caused increases in circulating insulin and GLP-1, and suppressed inflammation, suggesting multiple sites of action (Yore et al., 2014).
Fatty acid-hydroxy fatty acid esters are not the first lipids to be implicated as therapeutic agents in metabolic disease. The monomeric fatty acid, palmitoleate (cis-16:1n7) was reported to decrease hepatic fat and enhance skeletal muscle insulin sensitivity in mice (Cao et al., 2008). However, palmitoleate does not have the same strong associations with metabolic control in humans as suggested for PAHSA. Omega-3-fatty acids (polyunsaturated fats found in fish oils) are also reported to be protective against metabolic diseases (Oh et al., 2010). Interestingly, both PAHSA and omega-3-fatty acids are ligands for the G-protein coupled receptor, GPR120, possibly explaining their common effects to suppress inflammation and promote GLUT-4 translocation (Yore et al., 2014; Oh et al., 2010). This work has highlighted the power of untargeted metabolomics for discovery of new mechanisms, but several important issues remain to be addressed, including the enzymatic pathways responsible for synthesis of FAHFA and their modes of regulation, the levels of FAHFA that might be present in different foods and supplements, and the effect of administration of PAHSA in human subjects with metabolic diseases.
Metabolomics Applied to Cardiovascular Disease Mechanisms
Recent studies have demonstrated surprising efficacy of “diabetes drugs” such as empagliflozin, an inhibitor of renal glucose reuptake by the sodium-glucose co-transporter SGLT2, and liraglutide, a GLP-1 analogue, for reducing risk of cardiovascular events in diabetic subjects (Zinman et al., 2015; Marso et al., 2016). These findings highlight the strong metabolic interconnections between obesity, diabetes, and cardiovascular diseases, and suggest that metabolic signatures that predict incident cardiovascular events, including the short-chain dicarboxylacylcarnitine (Shah et al., 2010; Shah et al., 2012b; Shah et al., 2012c) and TMAO (Wang et al., 2011) signatures discussed earlier, may serve as biomarkers for engagement of diabetes drugs for mitigation of cardiovascular risk.
As discussed earlier, the gut microbiome may play an important role in production of metabolites such as BCAA that contribute to dysregulation of metabolic pathways and eventual development of type 2 diabetes. More recently, strong evidence for a key role of the microbiome in regulating biological functions relevant to cardiovascular diseases has also been accumulating. TMAO, produced via sequential metabolism of choline and carnitine by gut microbes and host enzymes, is a strong predictive biomarker of atherosclerotic burden and cardiovascular events. Consistent with a cause/effect relationship, supplementation of mouse diets with TMAO or choline results in an increase in atherosclerotic plaque size in apo E−/− mice that is proportional to the increase in circulating TMAO levels. Moreover, the effect of dietary choline to enhance plaque formation in apo E−/− mice is abrogated by generalized ablation of the microbiome with broad-spectrum antibiotics (Wang et al., 2011). More recently, TMAO has also been associated with risk of thrombotic events (MI or stroke) in a cohort of 4007 clinically referred subjects (Zhu et al., 2016). Extending these findings, TMAO enhances platelet function in response to several agonists, including ADP, thrombin, and collagen, and also increases the rate of thrombus formation in the FeCl3 mouse model of experimental thrombosis. Choline supplementation also enhances thrombosis in this model in a microbiome-dependent manner. Progress is occurring in identifying particular bacterial species that are strong producers of trimethylamines (TMA), the precursors of TMAO formation, suggesting potential therapeutic strategies. Indeed, inhibition of bacterial TMA lyases with a choline analogue, 3,3-dimethyl-1-butanol (DMB), was recently shown to reduce circulating TMAO and inhibit plaque formation in the apo e−/− mouse model (Wang et al., 2015). The effects of DMB on mitigating the thrombotic effects of TMAO remain to be demonstrated, and potential long-term side effects of the agent may emerge, but the approach has the potential to be one of the first broadly impactful examples of “drugging the microbiome”. If this occurs, it should be remembered that the approach is based on an original discovery made with non-targeted metabolomics.
Integration of Metabolomics and Genetics for Understanding Cardiometabolic Diseases
Human genome-wide association studies have identified loci that map with cardiovascular risk and diabetes, but these explain only a small fraction of these diseases. This is likely due to the fact that both type 2 diabetes and cardiovascular diseases are driven by a mixture of genetic susceptibility, environmental factors, and as has been illustrated herein, the gut microbiome, conspiring to perturb metabolic homeostasis and health. With the emergence of metabolite clusters that predict incident cardiometabolic disease and intervention outcomes, we are provided with intermediate phenotypes that present fresh opportunity to define underlying genetic architecture. This is particularly true given evidence that metabolic profiles are heritable. Thus, metabolomic analysis of eight multiplex families with familial early-onset CVD identified specific amino acids, fatty acids, and acylcarnitines with high heritability scores, even after adjustment for variables such as diabetes, hypertension, and BMI (Shah et al., 2009). Other studies have combined metabolomics profiling and genotyping in general population cohorts such as KORA and TwinsUK (Illig et al., 2010; Suhre et al., 2011; Shin et al., 2014). The most recent of these studies performed GWAS analysis and measured 333 known metabolites by a combined GC/MS and LC-MS/MS approach across 7824 subjects in the two cohorts (Shin et al., 2014). Overall, 145 independent SNP/metabolite associations with genome wide significance have emerged from these studies. New biological insights and disease detection strategies are implied by this work. For example, changes in carnitine levels were associated with SNP rs7094971 in the SLC16A9 gene, also known as MCT9, which has homology to monocarboxylic acid transporters (Suhre et al., 2011). Expression of this gene in Xenopus oocytes demonstrated that it encodes a carnitine transporter. In addition, SNPs in several genes involved in aromatic amino acid metabolism, including TDO2 (tryptophan 2,3-dioxygenase), IDO1 (indoleamine 2,3-dioxygenase), TAT1 (T-type amino acid transporter1), and SLC7A5 (LAT1 amino acid transporter) were associated with multiple tryptophan metabolites, suggesting the potential for new blood-borne markers of altered tryptophan metabolism linked to mood disorders (Shin et al., 2014). Similarly, other recent studies have integrated GWAS or exome array analysis and metabolomics profiling in 2076 Framingham Heart subjects and 1528 validation cohort subjects from the Atherosclerosis Risk in Communities study to define novel coding variants that influence levels of metabolites such as histidine, phenylalanine, and ureidopropionate, and cholesterol ester and triglyceride metabolites (Rhee et al., 2013). Finally, as discussed earlier, a metabolite cluster (prinicipal component) consisting of several short-chain dicarboxylated acylcarnitines that predict future cardiovascular events maps to genes associated with the protein unfolding response and ER stress pathways (Rhee et al., 2016). Further efforts to integrate metabolomics with genomics and other “omics” data sets could yield new information about genetic pathways that control intermediate phenotypes contributing to polygenic metabolic diseases.
Summary and Conclusions
As the newest member of the stable of “omics” methodologies for comprehensive molecular profiling, metabolomics has undergone a rapid technological evolution, and is now increasingly applied to human epidemiology and fundamental mechanistic research in almost equal measure. Consistent with its theoretical advantage of measuring the chemistry of biological systems, and therefore being distal to genomic, transcriptomic and proteomic variation, metabolomics has proven its utility for identifying new biomarkers of cardiometabolic diseases, and even more impressively, for detecting future disease events and intervention outcomes. As new and unanticipated associations emerge between cardiometabolic diseases and specific metabolites, this has led in multiple cases to demonstration of their role in disease pathogenesis, and to new insights into biological pathways that contribute to complex human diseases and disorders. Because a significant portion of metabolomics-based research to date has been descriptive, the full potential of metabolomics for defining new detection strategies and pathogenic mechanisms of cardiometabolic diseases remains to be realized. It is hoped that this perspective will encourage increased application of metabolomics to metabolic disease research, and more specifically, the use of disease-associated metabolomics signatures as a blueprint for defining novel disease mechanisms and targets.
Acknowledgments
Work described in this article from the author’s laboratory is currently supported by grants from the National Institutes of Health (grants DK046492; PO1-DK058398; R24-DK085610, and DK078669) and was previously supported by a sponsored research agreement with the CVMED unit, Pfizer. The author is grateful to all members of his laboratory and the metabolomics core laboratory at the Stedman Center/Duke Molecular Physiology Institute for their contributions to the cited works. Special thanks to Dr. Mette Jensen and Dr. Jie An for their help in preparation of schematic figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Abel ED, Peroni O, Kim JK, Kim YB, Boss O, Hadro E, Minnemann T, Shulman GI, Kahn BB. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729–733. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- Allalou A, Nalla A, Prentice KJ, Liu Y, Zhang M, Dai FF, Ning JX, Osborne LR, Cox BJ, Gunderson EP, Wheeler MB. A predictive metabolic signature for the transition from gestational diabetes to type 2 diabetes. Diabetes. 2016;65:2529–2539. doi: 10.2337/db15-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves TC, Pongratz RL, Zhao X, Yarborough O, Sereda S, Shirihai O, Cline GW, Mason G, Kibbey RG. Integrated, step-wise, mass-isotopomeric flux analysis of the TCA cycle. Cell Metabolism. 2015;22:936–947. doi: 10.1016/j.cmet.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J, Muoio DM, Shiota M, Fujimoto Y, Cline GW, Shulman GI, Stevens R, Millington D, Newgard CB. Hepatic expression of malonyl CoA decarboxylase reverses muscle, liver, and whole-animal insulin resistance. Nature Medicine. 2004;10:268–274. doi: 10.1038/nm995. [DOI] [PubMed] [Google Scholar]
- Bain JR, Stevens RD, Wenner BR, Ilkayeva O, Muoio DM, Newgard CB. Metabolomics applied to diabetes research: Moving from information to knowledge. Diabetes. 2009;58:2429–2443. doi: 10.2337/db09-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley-Lewis R, Huynh J, Xiong G, Lee H, Wenger J, Clish C, Nathan D, Thadhani R, Gerszten R. Metabolomic profiling in the prediction of gestational diabetes mellitus. Diabetologia. 2015;58:1329–1332. doi: 10.1007/s00125-015-3553-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Granger CB, Craig D, Haynes C, Bain J, Stevens RD, Hauser ER, Newgard CB, Kraus WE, Newby LK, Shah SH. Validation of the association between a branched chain amino acid metabolite profile and extremes of coronary artery disease in patients referred for cardiac catheterization. Atherosclerosis. 2014;232:191–196. doi: 10.1016/j.atherosclerosis.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher JM, Antoniewicz MR, Boros LG, Burgess SC, Brunengraber H, Clish CB, DeBerardinis RJ, Feron O, Frezza C, Ghesquiere B. A roadmap for interpreting (13)C metabolite labeling patterns from cells. Curr Opin Biotechnol. 2015;34:189–201. doi: 10.1016/j.copbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Gerhold K, Mayers JR, Wiest MM, Watkins SM, Hotamisligil GS. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 2008;134:933–944. doi: 10.1016/j.cell.2008.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ni Y, Ma X, Bao Y, Liu J, Huang F, Hu C, Xie G, Zhao A, Jia W, Jia W. Branched-chain and aromatic amino acid profiles and diabetes risk in Chinese populations. Scientific Reports. 2016;6:20594. doi: 10.1038/srep20594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CS, Befroy DE, Codella R, Kim S, Reznick RM, Hwang YJ, Liu ZX, Lee HY, Distefano A, Samuel VT, et al. Paradoxical effects of increased effects of increased expression of PGC-1alpha on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proc Natl Acad Sci USA. 2008;105:19926–19931. doi: 10.1073/pnas.0810339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra AR, Louet JF, Saha P, An J, Demayo F, Xu J, York B, Karpen S, Finegold M, Moore D, et al. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke’s disease. Science. 2008;322:1395–1399. doi: 10.1126/science.1164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton P. Diabetes: treatment of type 2 diabetes mellitus with bariatric surgery. Nature Rev Endocrinol. 2010;6:191–193. doi: 10.1038/nrendo.2010.23. [DOI] [PubMed] [Google Scholar]
- Cobb J, Eckhart A, Motsinger-Reif A, Carr B, Groop L, Ferrannini E. A-hydroxybutryic acid is a selective metabolite biomarker of imparied glucose tolerance. Diabetes Care. 2016;39:988–995. doi: 10.2337/dc15-2752. [DOI] [PubMed] [Google Scholar]
- El-Azzouny M, Evans CR, Treutelaar MK, Kennedy RT, Burant CF. Increased Glucose Metabolism and Glycerolipid Formation by Fatty Acids and GPR40 Receptor Signaling Underlies the Fatty Acid Potentiation of Insulin Secretion. Journal of Biological Chemistry. 2014;289:13575–13588. doi: 10.1074/jbc.M113.531970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felig P, Marliss E, Cahill GF. Plasma amino acid levels and insulin secretion in obesity. N Engl J Med. 1969;281:811–816. doi: 10.1056/NEJM196910092811503. [DOI] [PubMed] [Google Scholar]
- Ferdaoussi M, Dai X, Jensen MV, Wang R, Peterson BS, Huang C, Ilkayeva O, Smith N, Miller N, Hajmrle C, et al. Isocitrate-to-SENP1 signaling amplifies insulin secretion and rescues dysfunctional beta-cells. J Clin Invest. 2015;125:3847–3860. doi: 10.1172/JCI82498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara CT, Wang P, Neto EC, Stevens RD, Bain JR, Wenner BR, Ilkayeva OR, Keller MP, Blasiole DA, Kendziorski C, et al. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptomic profiling. PLoS Genetics. 2008;4:e1000034. doi: 10.1371/journal.pgen.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Cummings NE, Arriola Apelo SI, Neuman JC, Kasza I, Schmidt BA, Cava E, Spelta F, Tosti V, Syed FA, et al. Decreased consumption of branched-chain amino acids improves metabolic health. Cell Reports. 2016;16:1–11. doi: 10.1016/j.celrep.2016.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohnert BI, Rewers MJ. Metabolomics in childhood diabetes. Pediatric Diabetes. 2016;17:3–14. doi: 10.1111/pedi.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall WE, Beebe K, Lawton KA, Adam KP, Mitchell MW, Nakhle PJ, Ryals JA, Milburn MV, Nannipieri M, Camastra S, et al. Alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS One. 2010;28:e10883. doi: 10.1371/journal.pone.0010883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring I, Sauter NS, Catchpole G, Assmann A, Shu L, Zien KS, Moehlig M, Pfeiffer AFH, Oberholzer J, Willmitzer L, et al. Identification of an intracellular metabolic signature impairing beta-cell function in the rat beta cell line INS-1E and human islets. Diabetologia. 2011;54:2584–2594. doi: 10.1007/s00125-011-2249-7. [DOI] [PubMed] [Google Scholar]
- Gooding JR, Jensen MV, Dai XQ, Wenner BR, Lu D, Arumugam R, Ferdaoussi M, MacDonald PE, Newgard CB. Adenylosuccinate (S-AMP) is an insulin secretagogue derived from glucose-induced purine metabolism. Cell Reports. 2015;13:157–167. doi: 10.1016/j.celrep.2015.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn EL, Piner LW, Huffman KM, Slentz CA, Elliot-Penry L, AbouAssi HN, White PJ, Bain JR, Muehlbauer MJ, Ilkayeva OR, et al. Impact of combined resistance and aerobic exercise training on branched-chain amino acid turnover, glycine metabolism, and insulin sensitivity in overweight humans. Diabetologia. 2015;58:2324–2335. doi: 10.1007/s00125-015-3705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henquin JC, Ravier MA, Nenquin M, Jonas JC, Gilon P. Hierarchy of the beta-cell signals controlling insulin secretion. Eur J Clin Invest. 2003;33:742–750. doi: 10.1046/j.1365-2362.2003.01207.x. [DOI] [PubMed] [Google Scholar]
- Herman MA, She P, Peroni OD, Lynch CJ, Kahn BB. Adipose tissue branched chain amino acid (BCAA) metabolism modulates circulating BCAA. J Biol Chem. 2010;285:11348–11356. doi: 10.1074/jbc.M109.075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman MA, Peroni OD, Villoria J, Schon MR, Abumrad NA, Bluher M, Klein S, Kahn BB. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 2012;484:333–338. doi: 10.1038/nature10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao G, Chapman J, Ofrecio JM, Wilkes J, Resnik JL, Thapar D, Subramaniam S, Sears DD. Multi-tissue, selective PPARg modulation of insulin sensitivity and metabolic pathways in obese rats. Am J Physiol Endocrinol Metab. 2011;300:E164–E174. doi: 10.1152/ajpendo.00219.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Cohen JC, Hobbs HH. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J Biol Chem. 2011;286:37085–37093. doi: 10.1074/jbc.M111.290114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Joseph JW. Assessment of the metabolic pathways associated with glucose-stimulated biphasic insulin secretion. Endocrinology. 2014;155:1653–1666. doi: 10.1210/en.2013-1805. [DOI] [PubMed] [Google Scholar]
- Huffman KM, Shah SH, Stevens RD, Bain JR, Muehlbauer M, Slentz CA, Tanner CJ, Kuchibhatla M, Houmard JA, Newgard CB, et al. Relationships between circulating metabolic intermediates and insulin action in overweight to obese, inactive men and women. Diabetes Care. 2009;32:1678–1683. doi: 10.2337/dc08-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illig T, Gieger C, Zhai G, Römisch-Margl W, Wang-Sattler R, Prehn C, Altmaier E, Kastenmüller G, Kato BS, Mewes HW, et al. A genome-wide perspective of genetic variation in human metabolism. Nature Genetics. 2010;42:137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivarsson R, Quintens R, Dejonghe S, Tsukamoto K, In’t Veld P, Renstrom E, Schuit FC. Redox control of exocytosis: regulatory role of NAPDH, thioredoxin, and glutaredoxin. Diabetes. 2005;54:2132–2142. doi: 10.2337/diabetes.54.7.2132. [DOI] [PubMed] [Google Scholar]
- Jang C, Oh SF, Rowe GC, Liu L, Chan MC, Rhee J, Hoshino A, Kim B, Ibrahim A, Baca LG, et al. A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nature Med. 2016;22:421–426. doi: 10.1038/nm.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MV, Joseph JW, Ronnebaum SM, Burgess SC, Sherry AD, Newgard CB. Metabolic cycling in control of glucose-stimulated insulin secretion. Am J Physiol Endocrinol Metab. 2008;295:E1287–1297. doi: 10.1152/ajpendo.90604.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Banton S, Tran VT, Konomi JV, Li S, Vos MB. Amino acid metabolism is altered in adolescents with nonalcoholic fatty liver disease—an untargeted, high resolution metabolomics study. J Pediatrics. 2016;172:14–19. doi: 10.1016/j.jpeds.2016.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph JW, Jensen MV, Ilkayeva O, Palmieri F, Alarcon C, Rhodes CJ, Newgard CB. The mitochondrial citrate carrier plays a regulatory role in glucose-stimulated insulin secretion. J Biol Chem. 2006;281:35624–35632. doi: 10.1074/jbc.M602606200. [DOI] [PubMed] [Google Scholar]
- Koves TR, Li P, An J, Akimoto T, Slentz D, Ilkayeva O, Dohm GL, Yan Z, Newgard CB, Muoio DM. Peroxisome proliferator-activated receptor-gamma co-activator 1alpha-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem. 2005;280:33588–98. doi: 10.1074/jbc.M507621200. [DOI] [PubMed] [Google Scholar]
- Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JR, Newgard CB, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metabolism. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Kraus WE, Stevens R, Craig D, Bain JR, Burns E, Haynes C, Kwee L, Krupp D, Muehlbauer M, Hauser ER, et al. Metabolomic quantitative trait loci (mQTL) mapping implicates ubiquitin proteasome system in cardiovascular disease pathogenesis. PLoS Genetics. 2015;11:e1005553. doi: 10.1371/journal.pgen.1005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeger T, Henagan TM, Albarado DC, Redman LM, Bray GA, Noland RC, Munzberg H, Hutson SM, Gettys TW, Schwartz MW, Morrison CD. FGF21 is an endocrine signal of protein restriction. J Clin Invest. 2014;124:3913–3922. doi: 10.1172/JCI74915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laferrère B, Arias S, Swerdlow N, Gorroochurn P, Bose M, Bawa B, Teixeira J, Stevens RD, Wenner BR, Bain JR, Muehlbauer MJ, et al. Differential metabolic impact of gastric bypass surgery versus dietary intervention in obese diabetic subjects despite identical weight loss. Science Transl Med. 2011;3:80re2. doi: 10.1126/scitranslmed.3002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallukka S, Yki-Jarvinen H. Non-alcoholic fatty liver disease and risk of type 2 diabetes. Best Pract Res Clin Endocrinol Metab. 2016;30:385–395. doi: 10.1016/j.beem.2016.06.006. [DOI] [PubMed] [Google Scholar]
- Liu Y, Prentice KJ, Eversley JA, Hu C, Batchuluun B, Leavey K, Hansen JB, Wei DW, Cox B, Dai FF, et al. Rapid elevation in CMPF may act as a tipping point in diabetes development. Cell Reports. 2016;14:2889–2990. doi: 10.1016/j.celrep.2016.02.079. [DOI] [PubMed] [Google Scholar]
- Lorenz MA, El Azzouny MA, Kennedy RT, Burant CF. Metabolome response to glucose in the beta-cell line INS-1 832/13. J Biol Chem. 2013;288:10923–10935. doi: 10.1074/jbc.M112.414961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louet JF, Chopra AR, Sagen JV, An J, York B, Tannour-Louet M, Saha PK, Stevens RD, Wenner BR, Ilkayeva OR, et al. The coactivator SRC-1 is an essential coordinator of hepatic glucose production. Cell Metabolism. 2010;12:606–618. doi: 10.1016/j.cmet.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Mulder H, Zhao P, Burgess SC, Jensen MV, Kamzolova S, Newgard CB, Sherry AD. 13C NMR isotopomer analysis reveals a connection between pyruvate cycling and glucose-stimulated insulin secretion. Proc. Natl. Acad. Sci USA. 2002;99:2708–2713. doi: 10.1073/pnas.052005699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luukkonen PK, Zhou Y, Sadevirta S, Leivonen M, Arola J, Oresic M, Hyotylainen T, Yki-Jarvinen H. Hepatic ceramides dissociate steatosis and insulin resistance in patients with non-alcoholic fatty liver disease. J Hepatology. 2016;64:1167–1175. doi: 10.1016/j.jhep.2016.01.002. [DOI] [PubMed] [Google Scholar]
- Magkos F, Bradley D, Schweitzer GG, Finck BN, Eagon JC, Ilkayeva O, Newgard CB, Klein S. Effect of roux-en-Y bypass and laparoscopic adjustable gastric banding on branched-chain amino acid metabolism. Diabetes. 2013;62:2757–2761. doi: 10.2337/db13-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson M, Lewis GD, Ericson U, Orho-Melander M, Hedblad B, Engstrom G, Ostling G, Clish C, Wang TJ, Gerszten RE, et al. A diabetes predictive amino acid score and future cardiovascular disease. Eur Heart J. 2013;34:1982–1989. doi: 10.1093/eurheartj/ehs424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marso SP, Daniels GH, Brown-Frandsen K, Kristensen P, Mann JF, Nauck MA, Nissen SE, Pocock S, Poulter NR, Ravn LS, et al. Liraglutide and cardiovascular outcomes in type 2 diabetes. New Engl J Med. 2016;375:311–322. doi: 10.1056/NEJMoa1603827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack SE, Shaham O, McCarthy MA, Deik AA, Wang TJ, Gerszten RE, Clish CB, Mootha VK, Grinspoon SK, Fleischman A. Circulating branched-chain amino acid concentrations are associated with obesity and future insulin resistance in children and adolescents. Pediatric Obesity. 2013;8:52–61. doi: 10.1111/j.2047-6310.2012.00087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menni C, Fauman E, Erte I, Perry JRB, Kastenmuller G, Shin SY, Petersen AK, Hyde C, Psatha M, Ward KJ, et al. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes. 2013;62:4270–4276. doi: 10.2337/db13-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaliszyn SF, Sjaarda LA, Mihalik SJ, Lee S, Bacha F, Chace DH, De Jesus VR, Vockley J, Arsianian SA. Metabolomic profiling of amino acids and beta-cell function relative to insulin sensitivity in youth. J Clin Endocrinol Metab. 2012;97:2119–2124. doi: 10.1210/jc.2012-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muoio DM, Noland RC, Kovalik JP, Seiler SE, Davies MN, DeBalsi KL, Ilkayeva OR, Stevens RD, Kheterpal I, Zhang J, et al. Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell Metabolism. 2012;15:764–77. doi: 10.1016/j.cmet.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbern D, Gumus Balikcioglu P, Balikcioglu M, Bain J, Muehlbauer M, Stevens R, Ilkayeva O, Dolinsky D, Armstrong S, Irizarry K, Freemark Sex differences in biomarkers associated with insulin resistance in obese adolescents: metabolomic profiling and principal components analysis. J Clin Endocrinol Metab. 2014;99:4730–4739. doi: 10.1210/jc.2014-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq A, Shah SH, Arlotto M, Slentz CA, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metabolism. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB, Attie AD. Getting biological about the genetics of diabetes. Nature Medicine. 2010;16:388–391. doi: 10.1038/nm0410-388. [DOI] [PubMed] [Google Scholar]
- Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metabolism. 2012;15:606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DY, Talukdar S, Bae EJ, Imamura T, Morinaga H, Fan W, Li P, Lu WJ, Watkins SM, Olefsky JM. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell. 2010;142:687–698. doi: 10.1016/j.cell.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rahilly S. Human genetics illuminates the paths to metabolic disease. Nature. 2009;19:307–314. doi: 10.1038/nature08532. [DOI] [PubMed] [Google Scholar]
- Palmer ND, Stevens RD, Antinozzi PA, Anderson A, Bergman RN, Wagenknecht LE, Newgard CB, Bowden DW. A metabolomics profile associated with insulin resistance and conversion to diabetes in the insulin resistance atherosclerosis study. J Clin Endocrin Metab. 2015;100:E463–468. doi: 10.1210/jc.2014-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti GJ, Yanes O, Siuzdak G. Metabolomics: the apogee of the omics trilogy. Nature Rev. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BAH, Forslund K, Hildebrand F, Prifti E, Falony G, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- Perng W, Gillman MW, Fleisch AF, Michalek RD, Watkins SM, Isganaitis E, Patti ME, Oken E. Metabolomic profiles and childhood obesity. Obesity. 2014;22:2570–2578. doi: 10.1002/oby.20901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice KJ, Luu L, Allister EM, Liu Y, Jun LS, Sloop KW, Hardy AB, Wei L, Jia W, Fantus IG, Sweet DH, Sweeney G, Retnakaran R, Dai FF, Wheeler MB. The furan fatty acid metabolite CMPF is elevated in diabetes and induces β-cell dysfunction. Cell Metabolism. 2014;19:653–666. doi: 10.1016/j.cmet.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Retnakaran R, Ye C, Kramer CK, Connelly PW, Hanley AJ, Sermer M, Zinman B. Evaluation of circulating determinants of beta-cell function in women with and without gestational diabetes. J Clin Endocrinol Metab. 2016;101:2683–2691. doi: 10.1210/jc.2016-1402. [DOI] [PubMed] [Google Scholar]
- Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, Yang E, Farrell L, Fox CS, O’Donnell CJ, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest. 2011;121:1402–1411. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee EP, Ho JE, Chen M-H, Shen D, Chang S, Larson MG, Ghorbani A, Shi X, Helenius IT, O’Donnell CJ, et al. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metabolism. 2013;18:130–143. doi: 10.1016/j.cmet.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee EP, Yang Q, Yu B, Liu X, Cheng S, Deik A, Pierce KA, Bullock K, Ho JE, Levy D, et al. An exome array study of the plasma metabolome. Nature Comm. 2016;7:12360. doi: 10.1038/ncomms12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Cultured gut bacterial consortia from twins discordant for obesity modulate adiposity and metabolic phenotypes in gnotobiotic mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts LD, Bostrom P, O’Sullivan JF, Schinzel RT, Lewis GD, Dejam A, Lee YK, Palma MJ, Calhoun S, Georgiadi A, et al. beta-Aminoisobutyric Acid Induces Browning of White Fat and Hepatic beta-Oxidation and Is Inversely Correlated with Cardiometabolic Risk Factors. Cell Metabolism. 2014;19:96–108. doi: 10.1016/j.cmet.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnebaum S, Joseph JW, Burgess SC, Lu D, Ilkayeva O, Stevens R, Becker TC, Muehlbauer J, Sherry AD, Newgard CB, et al. A pyruvate cycling pathway involving cytosolic NAPD-dependent isocitrate dehydrogenase regulates glucose-stimulated insulin secretion. J Biol Chem. 2006;281:31041–31049. doi: 10.1074/jbc.M511908200. [DOI] [PubMed] [Google Scholar]
- Scholtens DM, Muehlbauer MJ, Stevens RD, Daya NR, Dyer AR, Lowe LP, Metzger BE, Newgard CB, Bain JR, Lowe WL. Metabolomics reveals broad-scale metabolic perturbations in hyperglycemic mothers during pregnancy. Diabetes Care. 2013;37:158–166. doi: 10.2337/dc13-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtens DM, Bain JR, Reisetter AC, Muehlbauer MJ, Nodzenski M, Stevens RD, Ilkayeva O, Lowe LP, Metzger BE, Newgard CB, Lowe WL., Jr Metabolic networks and metabolites underlie associations between maternal glucose during pregnancy and newborn size at birth. Diabetes. 2016;65:2039–2050. doi: 10.2337/db15-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuit F, De Vos A, Farfari S, Moens K, Pipeleers D, Brun T, Prentki M. Metabolic fate of glucose in purified islet cells. Glucose-regulated anaplerosis in beta cells. J Biol Chem. 1997;272:18572–18579. doi: 10.1074/jbc.272.30.18572. [DOI] [PubMed] [Google Scholar]
- Shah SH, Hauser ER, Bain JR, Muehlbauer MJ, Haynes C, Stevens RD, Wenner BR, Dowdy ZE, Granger CB, Ginsburg GS, et al. High heritability of metabolomic profiles in families burdened with premature cardiovascular disease. Mol Syst Biol. 2009;5:258–266. doi: 10.1038/msb.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SH, Bain JR, Crosslin DR, Muehlbauer MJ, Stevens RD, Haynes C, Dungan J, Newby LK, Hauser ER, et al. Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events. Circulation Cardiovascular Genetics. 2010;3:207–214. doi: 10.1161/CIRCGENETICS.109.852814. [DOI] [PubMed] [Google Scholar]
- Shah SH, Crosslin DR, Haynes CS, Nelson S, Turer CB, Stevens RD, Muehlbauer MJ, Wenner BR, Bain JR, Laferrère B, et al. Branched-chain amino acid levels are associated with improvement in insulin resistance with weight loss. Diabetologia. 2012;55:321–330. doi: 10.1007/s00125-011-2356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation. 2012;126:1110–1120. doi: 10.1161/CIRCULATIONAHA.111.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SH, Sun JL, Stevens RD, Bain JR, Muehlbauer MJ, Pieper KS, Haynes C, Hauser ER, Kraus WE, Granger CB, et al. Baseline metabolomics profiles predict cardiovascular events in patients at risk for coronary artery disease. American Heart Journal. 2012;163:844–850. doi: 10.1016/j.ahj.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Shah AA, Craig DM, Sebek JK, Haynes C, Stevens RD, Muehlbauer MJ, Granger CB, Hauser ER, Newby LK, Newgard CB, et al. Metabolite profiles predict adverse events following coronary artery bypass grafting. J of Thoracic and Cardio Surgery. 2012;143:873–878. doi: 10.1016/j.jtcvs.2011.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She P, Van Horn C, Reid T, Hutson SM, Cooney RN, Lynch CJ. Obesity-related elevations in plasma leucine are associated with alterations in enzymes involved in branched-chain amino acid metabolism. Am J Physiol Endocrinol Metab. 2007;293:E1552–E1563. doi: 10.1152/ajpendo.00134.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd PR, Gnudi L, Tozzo E, Yang H, Leach F, Kahn BB. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J Biol Chem. 1993;268:22243–22246. [PubMed] [Google Scholar]
- Sherif ZA, Saeed A, Ghavimi S, Nouraie SM, Laiyemo AO, Brim H, Ashktorab H. Global epidemiology of nonalcoholic fatty liver disease and perspectives on US minorities. Dig Dis Sci. 2016;61:1214–1225. doi: 10.1007/s10620-016-4143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SY, Fauman EB, Petersen A-K, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang T-P, et al. An atlas of genetic influences on human blood metabolites. Nature Genetics. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagris E, BasuRay S, Li J, Huang Y, Lai KM, Gromada J, Cohen JC, Hobbs HH. PNPLA3 I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology. 2015;61:108–118. doi: 10.1002/hep.27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spegel P, Sharoyko VV, Goehring I, Danielsson AP, Malmgren S, Nagorny CL, Andersson LE, Koeck T, Sharp GW, Straub SG, et al. Time-resolved metabolomics analysis of beta-cells implicates the pentose phosphate pathway in the control of insulin release. Biochem J. 2013;450:595–605. doi: 10.1042/BJ20121349. [DOI] [PubMed] [Google Scholar]
- Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wägele B, Altmaier E, Deloukas P, Erdmann J, Grundberg E, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai ES, Tan ML, Stevens RD, Low YL, Muehlbauer MJ, Goh DL, Ilkayeva O, Wenner B, Bain JR, Lee JJ, et al. Insulin resistance is associated with a metabolic profile of altered protein metabolism in Chinese and Asian-Indian men. Diabetologia. 2010;53:757–767. doi: 10.1007/s00125-009-1637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalacker-Mercer AE, Ingram KH, Guo F, Ilkayeva O, Newgard CB, Garvey WT. BMI, RQ, diabetes, and sex affect the relationships between amino acids and clamp measures of insulin action in humans. Diabetes. 2014;63:791–800. doi: 10.2337/db13-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl S, Yu Z, Kleber M, Singmann P, Holzapfel C, He Y, Mittelstrass K, Polonikov A, Prehn C, Romisch-Margl W, Adamski J, Shure K, Grallert H, Illig T, Wang-Sattler R, Reinehr T. Childhood obesity is assocciated with changes in the serum metabolite profile. Obes Facts. 2012;5:660–670. doi: 10.1159/000343204. [DOI] [PubMed] [Google Scholar]
- Walford GA, Davis J, Warner AS, Ackerman RJ, Billings LK, Chamarthi B, Fanelli RR, Hernandez AM, Huang C, Khan SQ, et al. Branched chain and aromatic amino acids change acutely following two medical therapies for type 2 diabetes mellitus. Metabolism. 2013;62:1772–1778. doi: 10.1016/j.metabol.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, et al. Metabolic profiles and the risk of developing diabetes. Nature Medicine. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Ngo D, Psychogios N, Dejam A, Larson MG, Rhee EP, Sinha S, McCabe E, Fox CS, Magnusson M, et al. 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest. 2013;123:4309–4317. doi: 10.1172/JCI64801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, et al. Gut flora metabolism of phosphtidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Roberts AB, Buffa JA, Levison BS, Zhu W, Org E, Gu X, Huang Y, Zamanian-Daryoush M, Culley MK, et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. 2015;163:1585–1595. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PJ, Lapworth AL, An J, Wang L, McGarrah RW, Stevens RD, Ilkayeva O, George T, Muehlbauer MJ, Bain JR, et al. Branched-chain amino acid restriction in Zucker-obese rats improves muscle insulin sensitivity by enhancing efficiency of fatty acid oxidation and acyl-glycine export. Molecular Metabolism. 2016;22:538–551. doi: 10.1016/j.molmet.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KE, Mikus CR, Slentz DH, Seiler SE, DeBalsi KL, Ilkayeva OR, Crain KR, Kinter MT, Kien CL, Stevens RD, Muoio DM. Muscle-Specific Overexpression of PGC-1alpha Does Not Augment Metabolic Improvements in Response to Exercise and Caloric Restriction. Diabetes. 2015;64:1532–1543. doi: 10.2337/db14-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz P, Makinen VP, Soininen P, Kangas AJ, Tukiainen T, Kettunen J, Savolainen MJ, Tammelin T, Viikari JS, Ronnemaa T, et al. Metabolic signatures of insulin resistance in 7098 young adults. Diabetes. 2012;61:1372–1380. doi: 10.2337/db11-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz P, Soininen P, Kangas AJ, Ronnemaa T, Lehtimaki T, Kahonen M, Viikari JS, Raitakari OT, Ala-Korpela M. Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care. 2013;36:648–655. doi: 10.2337/dc12-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yore MM, Syed I, Moraes-Vieira PM, Zhang T, Herman MA, Homan EA, Patel RT, Lee J, Chen S, et al. Discovery of a Class of Endogenous Mammalian Lipids with Anti-Diabetic and Anti-inflammatory Effects. Cell. 2014;159:318–332. doi: 10.1016/j.cell.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York B, Reineke EL, Sagen JV, Nikolai BC, Zhou S, Louet JF, Chopra AR, Chen X, Reed G, Noebels J, et al. Ablation of steroid receptor coactivator-3 resembles the human CACT metabolic myopathy. Cell Metabolism. 2012;15:752–763. doi: 10.1016/j.cmet.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni N, Saghatelian A, Patti GJ. Defining the metabolome: size, flux, and regulation. Molecular Cell. 2015;58:699–706. doi: 10.1016/j.molcel.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M, et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 2016;165:111–124. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinman B, Wanner C, Lachin JM, Fitchett D, Bluhmki E, Hantel S, Mattheus M, Devins T, Johansen OE, Woerle HJ, et al. Empagliflozin, cardiovascular outcomes, and mortality in type 2 diabetes. New Engl J Med. 2015;373:2117–2128. doi: 10.1056/NEJMoa1504720. [DOI] [PubMed] [Google Scholar]