Introduction
Mycobacterium tuberculosis is the causative agent of human tuberculosis. The bacterium has the capacity to persist in its human host for decades prior to progressing to active disease. In fact, on balance, humans deal with M. tuberculosis infection quite effectively with only an estimated 5–10% of those infected actually ending up with clinical disease. However, because of the extraordinary penetrance of this infectious agent across the global population, this accounts for in excess of one million deaths due to tuberculosis every year. The combination with HIV in Sub-Saharan Africa is catastrophic and M. tuberculosis is now the leading cause of mortality amongst individuals living with HIV.
The success of M. tuberculosis as an infectious agent is due in large part to its ability to persist in its host, and this is due to the extraordinarily intimate association that is formed between the bacterium and its host cell. We believe firmly that in considering tuberculosis one should consider the infected macrophage as the “minimal unit of infection”. Much of our research interests over the past two decades has centered on understanding how the physiology and metabolism of both bacterium and host cell are influenced and shaped by this enduring association.
Our understanding of the early events following infection is restricted predominantly to data from murine infections and the phagocyte populations in the lung during early M. tuberculosis infection of mice are extremely plastic. Ernst and colleagues conducted detailed analysis of the different phagocytes infected with GFP-expressing M. tuberculosis at different times post infection (1). They showed that M. tuberculosis was differentially distributed in alveolar macrophages, recruited interstitial macrophages, monocytes, dendritic cells, and neutrophils. The potential significance of this phagocyte heterogeneity was illustrated by Leeman and colleagues who demonstrated that bulk depletion of macrophages prior to infection with a lethal challenge dose of M. tuberculosis improved survival of the mice, however, in contrast, specific depletion of activated macrophages was detrimental to the mice (2, 3). One interpretation of these data is that certain macrophages are required to provide a permissive niche for bacterial growth, but that depletion of classically-activated macrophages reduces control of the infection. The idea that disease progression can be influenced both positively and negatively by the relative expansion of distinct subsets of phagocytes was shown elegantly by the work of Antonelli and colleagues (4). They treated mice intra-nasally with the Type 1 IFN inducer Poly (I:C) prior to infection with M. tuberculosis and found that this induced a marked increase in bacterial load in the lungs. More recently, Dorhoi and colleagues showed that Type 1 IFN receptor-deficient mice were partially protected from M. tuberculosis-challenge and exhibited depressed recruitment of inflammatory monocytes to the lung (5). The data all reinforce the perception that the identity and characteristics of the host phagocyte populations present and recruited to the site of infection have a fundamental impact on bacterial survival and growth.
The above studies all focused on initial stages of infection prior to and during development of the acquired immune response. However, phagocyte heterogeneity is also observed in established granulomas in non-human primates (NHP). Flynn and colleagues demonstrated that TB granulomas in macaques contain many diverse types of phagocytes that express different surface markers and activation proteins such as Arg1, Arg2, iNOS and eNOS (6). These data were the basis for a model for granuloma progression driven by macrophage polarization developed by Kirschner and Flynn (7). They argue that macrophage polarization ratios are predictive of granuloma outcome and that stable, necrotic granulomas with low bacterial burden and limited inflammation are characterized by transient intervals of NF-κB activation. The heterogeneity in phagocyte phenotype is also reflected in the heterogeneous balance of lymphocyte subsets in different granulomas in M. tuberculosis-infected macaques (8).
Clearly the dynamics of the M. tuberculosis-phagocyte interaction in vivo are extremely complex. How does one start to unravel the potential significance of this in vivo heterogeneity in phagocyte populations on bacteria fitness and growth, and the progression of disease? In this chapter we explore how the understanding of experimental M. tuberculosis infections of murine bone marrow-derived macrophage in tissue culture, as a defined, manipulable model system can be used to develop and validate tools to probe the extraordinary heterogeneity of the in vivo infection. Finally, and more significantly, how the incorporation of the host phagocyte into drug discovery programs allows identification of metabolic targets unique to the intracellular environment exploited by M. tuberculosis.
Phagocytosis and beyond
M. tuberculosis is internalized by classic phagocytosis. Normally, particles phagocytosed by macrophages are delivered to the acidic, hydrolytic environment of the lysosome, but M. tuberculosis has evolved strategies to arrest phagosome maturation (9). The compartment in which M. tuberculosis resides has a slightly acidified pH (pH 6.4), remains accessible to the endosomal network, and exhibits minimal acquisition of lysosomal hydrolases. Classic activation of the macrophage with interferon gamma (IFN-γ) prior to infection enables the macrophage to overcome this blockade and deliver the bacterium to a more acidic compartment (10, 11). The ultimate killing of M. tuberculosis by activated macrophages is dependent on multiple factors most significantly the production of nitric oxide (NO), the low pH of the lysosome, and the delivery of antimicrobial peptides through the process of autophagy (12–14).
Several publications document M. tuberculosis’s ability to escape the phagosome and access the cytosol of its host cell (15–18). However, this event appears to precipitate the necrotic death of the infected macrophage therefore we feel it is a transient event that may have significance with respect to the pathology observed in late stage disease, but is of less importance to the long-term survival of the pathogen across the phagocyte populations of its host. We believe that temporally and spatially, the intravacuolar population of M. tuberculosis represents a more significant target for therapeutics (19).
M. tuberculosis’s response to the intracellular environment
Schnappinger and colleagues published the first transcriptional profiling analysis of M. tuberculosis in macrophages under differing levels of immune activation (20). These data indicate that M. tuberculosis perceived the environment within the macrophage as hostile environment and exhibited upregulation of expression of genes associated with DNA-damage, cell wall attack, and showed up-regulation of genes linked to fatty acid metabolism as opposed to glucose metabolism.
More recently we have probed M. tuberculosis’s response to the phagosomal environment by transcriptional profiling with two goals in mind (21, 22). The first was to link transcriptional responses to specific environmental cues encountered within the phagosome of the macrophage, and the second was to exploit that information to build fluorescent reporter bacterial strains that could be used to inform us of the functionally-significant heterogeneity of the bacterial populations in different infection models (23). Transcriptional profiling is an extremely powerful approach, but the data generated are always going to represent an average across the bacterial population, and because heterogeneity exists in both the host phagocytes and the bacterial population it is critical that we develop effective means of assessing bacterial fitness and growth across the spectrum of phagocyte populations that the bacterium infects in vivo.
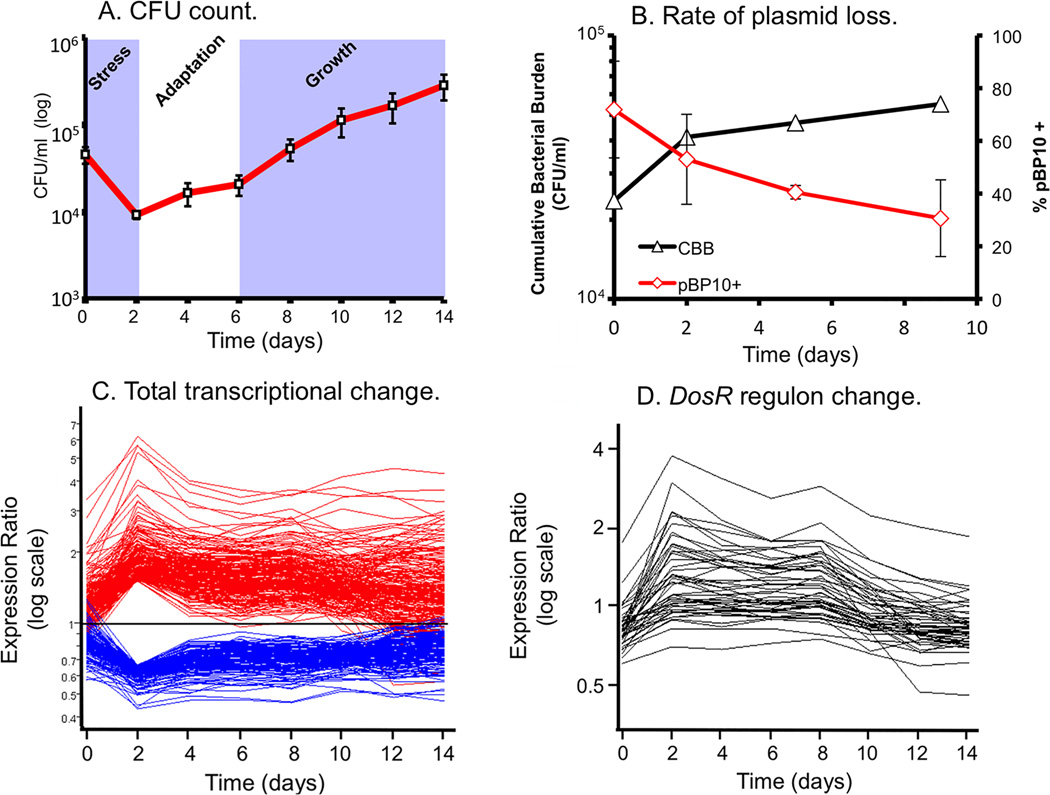
Rohde and colleagues developed a long-term culture infection model that allowed us to maintain M. tuberculosis in macrophage culture for 14 days, Figure 1 (24). Interestingly, the most marked transcriptional response was at two days post-infection, with the profile trending towards the mean over the rest of the infection period. Determination of the CFU counts indicated that there was an initial drop in bacterial viability over the first 2 days confirming that the transition was stressful for M. tuberculosis. Interestingly, analysis of bacterial replication rates utilizing a clock plasmid developed by Gill and coworkers (25) demonstrated that bacterial replication was rapid during early infection, which slowed considerably from days 2–4, and then recommenced from day 4–6 onwards. These data indicate that rapid bacterial growth appears to be inconsistent with survival within the phagosomal environment of the macrophage. If one mapped the temporal changes in transcriptional profile of genes of known association, such as those in the DosR stress regulon, one observes increased expression from day 0–2, at a relatively sustained level of expression from day 2–6, and then decreasing abundance of transcripts from day 6 onwards, as the bacterial population re-enters growth phase.
Figure 1. Life and death dynamics during long-term intracellular survival of M. tuberculosis.
(A) Survival Assays. Resting murine bone-marrow derived macrophages were infected at low MOI (~1:1) with M. tuberculosis CDC1551. Viable CFU were quantified at day 0 and at 2 day intervals post-infection (p.i.) over a 14-day time-course by lysis of monolayers, serial dilution, and plating on 7H10 medium. Error bars indicate standard error of the mean. (B) Replication Clock Plasmid. The percentage of bacteria containing the pBP10 plasmid during growth in resting macrophages was determined by comparing CFU (mean ± s.d.) recovered on kanamycin vs. nonselective media (red). The cumulative bacterial burden (CBB) (black) was determined by mathematical modeling based on total viable CFU and plasmid frequency data. Data shown represents two independent experiments. (C) The “bottleneck” response. Temporal expression profiles of genes differentially regulated at Day 2 p.i., including genes from (A) that were up- (red) or down-regulated (blue) >1.5-fold (shown as ratio of signal intensity relative to control). Note the maximal change in transcript levels at day 2 p.i. followed by majority trending back towards control levels. (D) “Guilt by association” analysis. Genes regulated in synch with known virulence regulons – i.e. the DosR regulon - were identified by using a highly regulated member of this regulon, hspX, in place of synthetic profiles. This figure is reproduced from Rohde et al. (24).
Homolka and colleagues extended these analyses in a comparison between 2 lab strains (CDC1551 and H37Rv) and 15 clinical isolates that represented the genetic diversity of the M. tuberculosis complex that currently circulate within humans across the globe (26). The goal of this study was to identify the “core” or a common transcriptome expressed by M. tuberculosis inside macrophages. Once again the induced expression of genes associated with hypoxia, oxidative and nitrosative stress, cell wall and DNA repair, and fatty acid metabolism across the panel of clinical isolates confirmed the previous, single strain data (20, 21).
Linking environmental cues and responses
The next step in our analysis was to link the transcriptional responses to specific environmental cues. Phagosome maturation is a continuum and it would be logical for M. tuberculosis to have evolved the capacity to sense and respond to gradients linked to the maturation status of its vacuole. In initial studies we blocked phagosomal acidification with the V-ATPase inhibitor concanamycin A and compared the transcriptional profile to bacteria in unperturbed phagosomes (23, 27). In control macrophages M. tuberculosis upregulated the expression of 68 genes 2 hrs post internalization, whereas in the non-acidified phagosomal environment only 38 of these genes were up-regulated. Much of this pH-dependent response was regulated by the two-component regulator PhoPR, which included a locus, AprABC, that regulates the expression of genes linked to the synthesis of triacylglycerol (TAG) and pthiocerol-dimycocerosate (PDIM). In fact, M. tuberculosis exposed to reduced pH exhibits enhanced production of PDIMs such as pthiodiolone and pthiocerol A, which are virulence-associated lipids that are also linked to the utilization of 3-carbon intermediates like propionyl-CoA generated from the breakdown of cholesterol (28). This is discussed in greater depth later in this review.
Chloride is one of the counter-ions that balance the activity of the proton-ATPase responsible for phagosome acidification, and analysis of phagosome maturation using fluorogenic chemical readouts show that the concentration of Cl− increases in proportion to the reduction of pH, reaching a maximal concentration of around 100 mM. We have found that M. tuberculosis senses and responds to Cl− over this physiological concentration range, and that the expression of several genes are up-regulated synergistically by exposure to a combination of lowered pH and increased Cl− concentration (29).
These types of complex multi-genic responses to relatively simple environmental cues are excellent examples of how bacterial regulons are molded by evolutionary pressure to facilitate pathogens, such as M. tuberculosis, to rapidly re-align their physiology to exploit the different environmental niches to which they are exposed within their host.
Construction of reporter strains
We utilized our transcriptional profiling data to select genes that were specifically, and markedly up-regulated in response to environmental cues of interest. We cloned putative promoter regions from these ORFs into a plasmid upstream of M. tuberculosis codon-optimized gfp, and tested for enhanced expression of GFP under appropriate environmental conditions (23, 29). For reasons we did not pursue, many constructs did not work! Nonetheless, we currently have a panel of reporter M. tuberculosis strains that respond to environmental stimuli. In addition to the inducible GFP expression, these reporter plasmids also contain the mCherry gene downstream of the constitutive promoter from smyc (30).
We next developed a murine model of vaccination to probe the phenotype(s) of our reporter strains in the context of both naïve and vaccinated hosts (31). Mice were vaccinated with heat-killed Erdman M. tuberculosis and challenged with three different reporter strains of Erdman M. tuberculosis. The bacterial strains we used responded to pH and chloride ions (rv2390c’::GFP, smyc’::mCherry), NO radicals (hspX’::GFP, smyc’::mCherry), and a replication reporter strain that contained a translational fusion between the single strand binding protein (SSB) with the GFP protein (SSB-GFP, smyc’::mCherry). This latter construct had been used previously in E. coli where replicating bacteria form a green fluorescent puncta which is associated with chromosomal replication (32). In M. tuberculosis, we found that this SSB-GFP positive complex persists for 70–80% of the replication cycle of the bacterium. We sacrificed mice at 14 and 28 days post challenge to quantify and determine the bacterial responses through imaging of thick sections from formaldehyde-fixed tissue.
The response of the rv2390c’::GFP, smyc’::mCherry reporter construct was theorized to correspond to vacuolar maturation. Interestingly, we found that the levels of induction of GFP in this reporter strain were lower in vaccinated versus naïve mice at early time points, which appears counter-intuitive. The data suggest that in mock-treated mice, M. tuberculosis initially encounters a high [Cl−]/low pH environment consistent with increased phagosomal maturation, which trends towards a lower [Cl−]/higher pH environment at later time points. This indication of early stress during the establishment of infection is consistent with published studies in both macrophage and murine infections, which implies that bacteria coming from broth culture have to re-align their physiology to be compatible with intracellular survival (22, 27). During this adjustment period, rapidly growing organisms survive poorly. The reduced expression of rv2390c’::GFP in the presence of an acquired immune response, observed in the current study, could be explained by either of two mechanistically-divergent scenarios. Firstly, while the adjustment in bacterial physiology required to support intracellular survival may come at a cost with respect to bacterial numbers, the re-alignment of bacterial physiology in the surviving bacteria may be accelerated by the presence of a pre-existing immune response to M. tuberculosis. Or, secondly, that bacilli expressing higher levels of rv2390c’::GFP may be the very bacteria that are killed most effectively by the pre-existing immune response. Although the underlying mechanisms may differ the outcome is the same; the accelerated generation of a bacterial population better equipped to survive in its host.
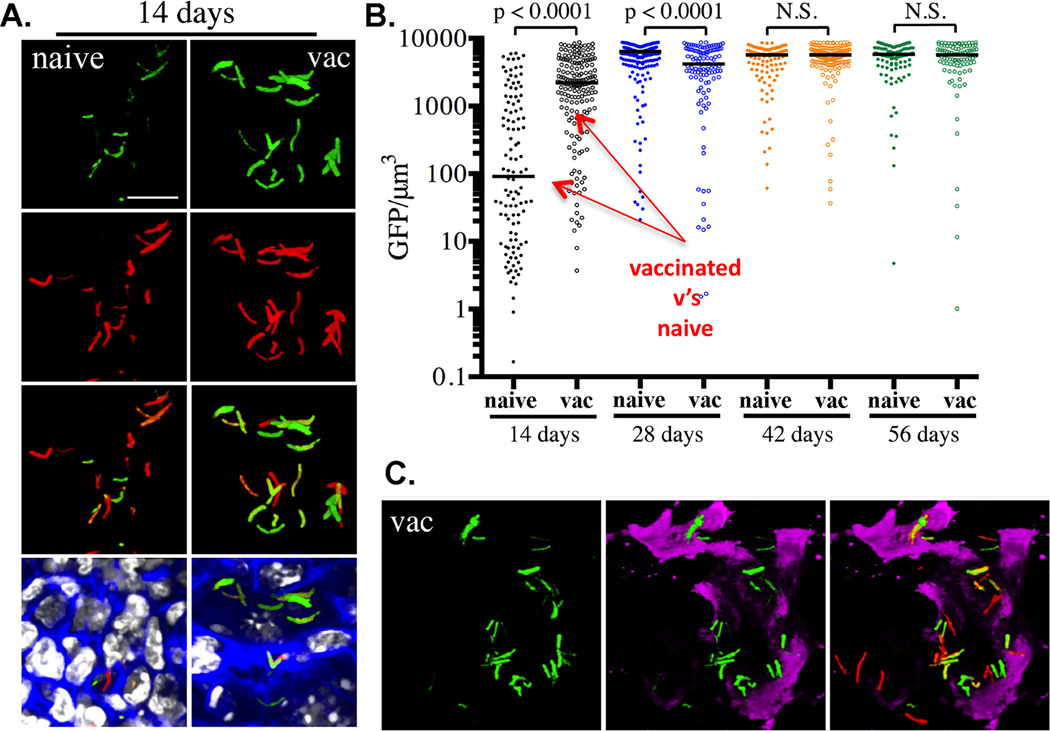
The presence of a pre-existing immune response against M. tuberculosis should lead to immune activation of infected macrophages and the induction of expression of the inducible nitric oxide synthase (NOS2). Our second reporter strain hspX’::GFP, smyc’::mCherry, was selected because of the robustness of the response of hspX expression to NO in culture. In this bacterium the level of expression of GFP was markedly enhanced in the vaccinated mice at day 14 post-infection but equivalent at day 28 post-infection suggesting that expression closely mirrored development of an acquired immune response, Figure 2. Furthermore, the levels of expression of GFP in IFN-γ-deficient and NOS2-deficient mice were considerably lower than those of wild-type mice. In addition, co-labeling of the tissue with antibodies against the NOS2 demonstrated that the regions of tissue positive for NOS2 enzyme corresponded to those regions containing GFP-positive bacilli.
Figure 2. Demonstrating the usefulness of the HspX’::GFP reporter strain in assessing and reporting on the localized induction of iNOS at the site of infection.
PBS-immunized (naïve) and mice vaccinated with heat-killed M. tuberculosis (vac) were infected with hspX’::GFP, smyc’:: mCherry Erdman M. tuberculosis reporter strain. Fluorescence induction of the hspX promoter-dependent GFP is higher at 14 days in the vaccinated animals assessed by confocal microscopy of thick tissue sections (A), that were scored subsequently by Volocity (B). (C) The thick tissue sections were probed with antibodies against murine NOS2 (magenta) demonstrating the co-localization between GFP induction and NOS2 expression at the site(s) of infection. Data shown are detailed in Sukumar et al, (31).
Our final reporter strain used in these studies contained the plasmid SSB-GFP, smyc’::mCherry, which reports on bacterial replication. At 14 days post-challenge, there was considerable diversity in the percentage of actively replicating M. tuberculosis within the various treatment groups, and no statistically significant differences between M. tuberculosis in vaccinated and naive mice were observed. In contrast there were distinct differences in the relative replication status of M. tuberculosis between experimental groups at 28 days post-challenge. Confocal microscopy of infected tissue showed that fewer M. tuberculosis exhibiting SSB-GFP foci were observed in vaccinated mice as opposed to naïve mice. Furthermore, consistent with the increased growth of M. tuberculosis in IFNγ-deficient mice at 28 days post-challenge, we observed a greater number of bacteria positive for SSB-GFP foci in these mice. These data are consistent with the conventional view that vaccination will lead to a reduction in the number of actively-replicating M. tuberculosis in the host.
Moving to the single cell suspension
In these early studies, performed primarily to validate the reporter strains, we relied on histological approaches to visualize and quantify the bacterium’s response to the host immune environment. Moving forward we wanted to exploit these reporters to probe M. tuberculosis’s phenotypes within the context of the different host phagocyte populations utilizing methods similar to those developed by Ernst and colleagues (1, 33).
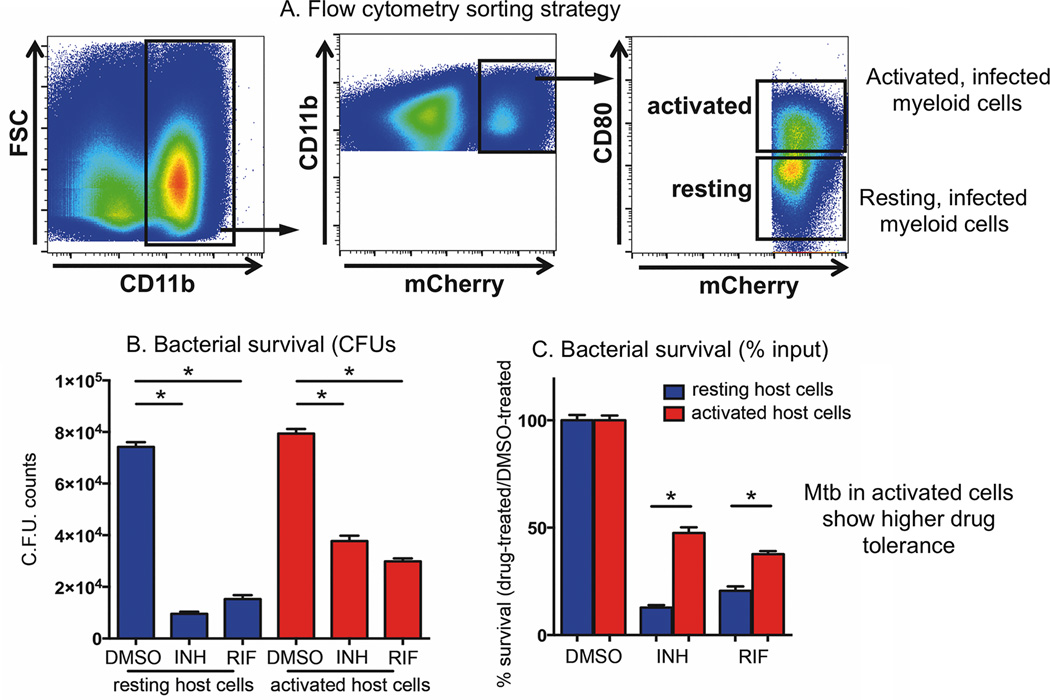
In our first study probing bacterial phenotypes in the context of phagocyte subsets during the course of infection we investigated the impact of the immune status of the host cell on the susceptibility of the intracellular M. tuberculosis in response to different frontline drugs, Figure 3 (34). Mice were infected with M. tuberculosis expressing mCherry for 21 days prior to euthanasia and we generated single cell suspensions from the infected lung tissue. We identified infected phagocytes by gating on cells that were CD11b-positive which contained mCherry expressing M. tuberculosis. We then sorted infected myeloid cells into CD80 high and CD80 low populations to segregate activated versus resting M. tuberculosis-infected cells. Further flow cytometric analysis of these sorted populations showed that the CD80 high cells also expressed high levels of MHC class II and NOS2 confirming their immune activated state. These cells were then established in culture and incubated with isoniazid (INH) or rifampin (RIF) for 24 hr. Bacterial survival was then scored by counting CFUs from these isolated cells. Normalizing the bacterial burden from the drug-treated cell populations to the appropriate control populations revealed that M. tuberculosis in activated myeloid cells exhibited markedly decreased sensitivity to both INH and RIF. We subsequently determined that immune activation of macrophages in tissue culture induced drug tolerance to INH, RIF, pyrazinamide (PZA), and ethambutol (EMB) in the intracellular M. tuberculosis. We went on to demonstrate, with NOS2-deficient mice, that the expression of NO was a dominant driver of this drug tolerant phenotype. However, the stress response induced in drug tolerant M. tuberculosis is multigenic and involved up-regulation of 4 major regulons, including dosR, mprA, phoP, and sigK. The expression of the genes in these regulons is up-regulated in response to exposure to NO, low pH, oxidative stress, hypoxia, membrane damage and nutrient starvation, implying that the acquisition of a drug tolerant phenotype could result from exposure to different host-derived stresses. These data are consistent with the previous report by Manina and colleagues documenting the phenotypic heterogeneity that the immune response to infection induces in M. tuberculosis, and how this links for metabolic activity and drug tolerance (35).
Figure 3. Flow sorting of activated and resting M. tuberculosis-infected host cells demonstrates that the drug sensitivity of M. tuberculosis recovered from in vivo infection correlates inversely with the immune activation status of the host phagocyte.
(A–C) M. tuberculosis recovered from activated host cells in vivo were more tolerant to both INH and RIF than those recovered from resting host cells. C57BL/6J mice were infected with mCherry-expressing M. tuberculosis Erdman for 21 days, and M. tuberculosis-containing myeloid cells with different immune activation status isolated from lung tissue using flow cytometry. (A) CD11b+ mCherry+ CD80high cells (activated population) and CD11b+ mCherry+ CD80low cells (resting population) were sorted according to the depicted gating strategies. (B–C) Isolated cells were established in culture and subjected to treatment with 1 µg/ml INH or RIF, or an equivalent volume of DMSO. Following 24 h drug treatment, bacterial survival was determined by CFU enumeration (B), and the percentage of M. tuberculosis surviving drug treatment quantified by normalizing bacterial load in drug-treated samples against that in DMSO-treated samples (C). This figure is modified from the work of Lui et al, (34).
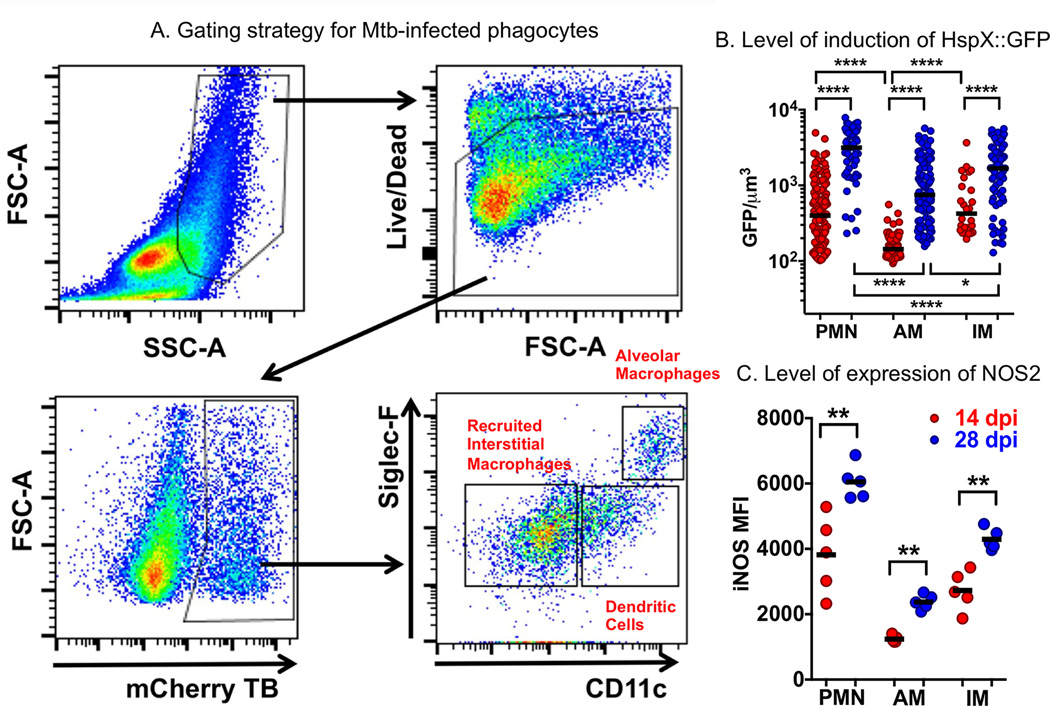
We are currently probing the phagocyte subsets with greater resolution to determine the bacterial fitness in the different host cell populations. Similarly to the reported findings of Srivastava and colleagues (1, 33), we detect M. tuberculosis in alveolar macrophages, neutrophils, recruited interstitial macrophages and monocyte-derived dendritic cells. The relative number of bacteria in the different phagocyte subsets is extremely dynamic during the first 4 weeks of infection as the mouse transitions from a naïve host to one with an acquired immune response. Some preliminary analysis of the dynamic nature of this interaction is illustrated in Figure 4 (Huang and Russell, unpublished data). We show analysis of the single cell suspensions isolated from infected mouse lung tissue. The flow cytometric analysis involves the identification of live cells, that are infected with reporter strains of M. tuberculosis, in this case the hspX’::GFP, smyc’::mCherry strain. Gating on the mCherry-positive cells these can be subjected to preliminary assignment as alveolar macrophages, monocyte-derived dendritic cells, and recruited interstitial macrophages on the basis of the levels of surface expression of SiglecF and CD11c. The cells are fixed for further analysis to correlate the bacteria’s stress response status (hspX’::GFP) with the level of expression of the host nitric oxide synthase (NOS2). The level of induction of GFP expression in M. tuberculosis increases between the 14 and 28 day time points, which is consistent with the increased expression of NOS2 in the different cell subsets. The level of expression of NOS2 is lowest in the alveolar macrophages, as is the level of induction of GFP in those M. tuberculosis. These data suggest that the alveolar macrophage population represents a less stressful host cell environment than the other phagocyte populations shown here. But a more extensive analysis of the hypothesis that these different phagocyte populations present differing degrees of permissiveness or control of bacterial growth is in progress. This figure is intended to show the experimental approaches behind this rationale, and not provide a definitive answer.
Figure 4. Examination of the bacterial stress reporter strain hspX’::GFP, smyc’::mCherry Erdman M. tuberculosis at the level of different host phagocyte populations in murine lung infection model.
A. The flow cytometry gating strategy for the identification of M. tuberculosis-infected phagocyte subsets from infected mouse lung showing preliminary identification of alveolar macrophages, recruited interstitial macrophages, and monocyte-derived dendritic cell populations. B. The level of expression of NO-driven GFP under regulation of the hspX promoter identifying those phagocytes that induce the highest level of bacterial stress, and how the stress intensifies from 14 to 28 days post-infection. The levels of induction of expression of GFP indicate that the most stressful host cells appear to be neutrophils, and the least stressful, alveolar macrophages. C. Labeling of the host cells with antibody against NOS2 demonstrates the direct correlation between expression levels of the host nitric oxide synthase, NOS2, and levels of expression of the bacterial stress response reporter hspX’::GFP, shown in panel B.
Nonetheless, what these reporter strain studies have shown us is that the environment(s) within the host phagocytes are heterogeneous, both spatially and temporally, and that this heterogeneity has fundamental consequences on bacterial fitness and growth, and ultimately on the progression of the infection to active disease.
Chemical genetics of M. tuberculosis infection in macrophages
Intracellular pathogens such as M. tuberculosis have evolved to specifically survive within their respective host cells and these pathogens are inextricably linked to the unique constraints of the various cells they infect. With this in mind we conducted an unbiased chemical screen designed to identify small molecules that specifically target pathways required for M. tuberculosis replication in macrophages. In collaboration with Vertex Pharmaceuticals we screened a collection of ~340k small molecules and identified 1,359 compounds that inhibit M. tuberculosis replication in macrophages. Characterization of these hits revealed that only ~50% of the 300 most potent compounds (IC50 values < 5.0 µM in the macrophage assay) displayed equivalent inhibitory activity when M. tuberculosis was grown in standard Middlebrook 7H9 OADC medium. Unfortunately the majority of these “universally active” compounds have previously been described in the literature. These compounds were likely discovered because of their ability to inhibit M. tuberculosis growth in standard media formulations. This class of compounds represents the classic type of antibiotics that inhibit M. tuberculosis growth and target essential bacterial pathways regardless of the growth conditions.
The remaining ~50% of the most potent compounds from the macrophage assay displayed no inhibitory activity against M. tuberculosis in 7H9 OADC media indicating that this “macrophage active” group of compounds likely target some aspect of M. tuberculosis physiology that is required for replication in macrophages (36). To begin to understand the mechanism of action for these “macrophage active” compounds we evaluated the compounds against M. tuberculosis cultured in conditions thought to mimic the intra-phagosomal environment. We tested these compounds when M. tuberculosis was exposed to in vitro pH and NO stresses or during growth in media that contained either fatty acids or cholesterol as carbon sources. To our surprise, we found that ~50% of the ~150 “macrophage active” compounds are cholesterol dependent and inhibit M. tuberculosis when the bacteria are grown in cholesterol media. Based on this observation we hypothesized that some of these compounds directly target aspects of cholesterol metabolism or propionyl-CoA assimilation. To identify compounds involved in these pathways we counter screened all of the 1,359 hits to discover specific inhibitors of enzymes involved in propionyl-CoA assimilation through the methylcitrate cycle (MCC) or required for cholesterol breakdown. To do this, we focused on compounds that rescue M. tuberculosis growth from a cholesterol-dependent intoxication in a M. tuberculosis ΔIcl1 mutant. In this assay compounds that inhibit key bottleneck enzymes involved in cholesterol breakdown or propionyl-CoA assimilation reverse toxicity in the M. tuberculosis ΔIcl1 mutant and allow the bacteria to grow in the presence of cholesterol (36). Using this method we have now discovered ~20 inhibitors that specifically block these metabolic pathways in M. tuberculosis and these compounds have IC50 values between 5–20 µM in macrophages.
The rescue approach using the M. tuberculosis ΔIcl1 mutant did not reveal any putative targets for our most potent cholesterol dependent, “macrophage active” compounds. To understand the mechanism of action for these compounds we used transcriptional profiling to characterize M. tuberculosis’s response to these compounds in cholesterol media. By this approach we discovered 7 structurally unrelated compounds that prevent M. tuberculosis from metabolizing cholesterol as indicated by the strong down regulation of the KstR regulon (36). To further understand these compounds we next screened a M. tuberculosis transposon library and isolated mutants resistant to these compounds and we found that inactivating Rv1625c/cya conferred resistance to all 7 of these compounds. The protein, Rv1625c/cya is a well-characterized adenylyl cyclase that forms cAMP from ATP (37, 38). We have now confirmed that this group of compounds directly stimulates Rv1625c/cya to overproduce cAMP. These compounds are unusual in that they inhibit cholesterol utilization and induce very high levels of cAMP in M. tuberculosis, which can potentially perturb multiple different pathways (39–45). Additionally, the high levels of cAMP produced by the M. tuberculosis in response to these compounds may modulate the TNF-α production in the infected cell by activating cytosolic sensors of cAMP in the infected macrophage (46).
Together, these data are consistent with previous observations (47–51) indicating that cholesterol is an important nutrient for M. tuberculosis and that pathways associated with the utilization of this molecule are required for growth in macrophages (52–55). This work further demonstrates that the cholesterol utilization pathways are “druggable” and tractable as targets for drug discovery. However, these observations do raise some new and puzzling questions such as: (i) why do other nutrients, such as fatty acids, not substitute for cholesterol in supporting M. tuberculosis growth in macrophages, (ii) what is the link between cAMP and cholesterol metabolism, and (iii) is there an equivalent to carbon catabolite repression in M. tuberculosis that is activated in macrophages to govern M. tuberculosis’s utilization of host-derived nutrients? In recent years our field has gained new insight into these aspects of M. tuberculosis physiology and pathogenesis and here we incorporate these findings with our own observations and current understanding of M. tuberculosis metabolism in macrophages.
Lipid utilization by M. tuberculosis in macrophages
Although M. tuberculosis likely utilizes a many different nutrients in vivo and during infection in macrophages the bacterium appears to preferentially utilize host-derived lipids (fatty acids and cholesterol) as carbon sources to produce energy and fuel biosynthetic pathways. The notion that M. tuberculosis preferentially metabolizes host lipids during infection began with classic experiments reported by Segal and Bloch in 1954. These studies demonstrated that ex vivo respiration rates of M. tuberculosis isolated from mouse lung tissues was stimulated by fatty acid substrates but not carbohydrates (56). Consistent with the idea that lipids are important nutrients the early genome annotation efforts revealed that M. tuberculosis has an expanded repertoire of genes associated with lipid metabolism and β-oxidation, which exceeded 250 genes (57). Although several of the M. tuberculosis genes originally annotated as having a β-oxidation functions are now known to have anabolic activities (58, 59) or act in multimeric complexes to oxidize lipids (60, 61) the M. tuberculosis genome still encodes more genes involved in lipid metabolism and β-oxidation relative to other bacteria. Another critical pathway that is required when bacteria metabolize lipids is the anaplerotic glyoxylate cycle and a key enzyme in this pathway is isocitrate lyase (Icl). Early gene expression studies demonstrated that Icl1 is strongly induced by M. tuberculosis during infection in macrophages (62–64) and that the Icl1 gene is required for M. tuberculosis pathogenesis in vivo (53). A series of elegant experiments from John McKinney’s lab established that M. tuberculosis expresses two Icl homologs (Icl1/Icl2) and these enzymes play critical roles lipid metabolism in the glyoxylate cycle and the methyl citrate cycle (MCC) (55, 65, 66). Early transcriptional profiling experiments of M. tuberculosis during infection in macrophages reported by Schnappinger et al. found that numerous genes associated with lipid metabolism were induced in M. tuberculosis (67). Importantly, over one half of the genes associated with metabolic adaptations in this report are now known to be involved in cholesterol metabolism (FadD19, FadE27, EchA19, FadA5, FadD3, FadE28, FadE29, FadE31, and FadA6) and/or propionyl-CoA assimilation through the MCC (Icl1 and PrpC) in M. tuberculosis (68–70). More recent transcriptional studies on M. tuberculosis in macrophages demonstrated that genes involved in propionyl-CoA assimilation, lipid break down, and cholesterol metabolic genes are induced in M. tuberculosis across a 14 day infection period in macrophages (24). Lastly, the growth of M. tuberculosis on lipid-based substrates during infection in macrophages would also necessitate that M. tuberculosis synthesizes carbohydrates de novo through gluconeogenesis. Recent studies have confirmed that PckA, a key bottleneck enzyme in gluconeogenesis is required for optimal Mb growth in macrophages (71). In total, these observations continue to support the idea that M. tuberculosis relies heavily on lipids (fatty acids and cholesterol) as a carbon sources and that the flux of propionyl-CoA through the MCC represents a critical axis in M. tuberculosis central metabolism during infection in macrophages.
Fatty acids
M. tuberculosis can utilize exogenously acquired fatty acids as: (i) substrates for β-oxidation to produce energy, (ii) acyl-primers for polyketide lipid synthesis, or (iii) can be directly assimilated into phospholipids and/or TAG (59, 72). The fatty acid oxidation pathways in M. tuberculosis still remain largely uncharacterized and the lipid substrates for the various β-oxidation enzymes poorly defined. In addition to catabolism, fatty acids can be used as acyl-AMP primers for the biosynthesis of polyketide lipids PDIM, poly-acylated trehaloses (PATS), and sulfolipid (SL) (73). Interestingly, the fate of fatty acids in M. tuberculosis can be impacted when the bacteria metabolizes cholesterol. M. tuberculosis produce more PDIM and SL when the bacterium metabolizes cholesterol and this is also observed during infection in mouse tissues (52, 74, 75). During growth on cholesterol M. tuberculosis also synthesizes a modified form PDIM and SL lipids that contain additional methyl-branching which consistent with the assimilation of more methylmalonyl-CoA (derived from propionyl-CoA) into these lipids (52, 74, 75). Thus, metabolizing cholesterol induces the production of methyl-branched polyketide lipids, which requires a pool of available fatty acids as acyl-primers. Metabolic labeling experiments with infected macrophages were used to track exogenously added 14C-labeled fatty acids into PDIM and TAG (28) and we demonstrated that the growth defect of M. tuberculosis ΔIcl1 mutant in macrophages can be overcome by stimulating foam cell formation through the provision of excess fatty acid to the host macrophages (28). This is consistent with the idea that during infection fatty acids can be shunted into the anabolism of methyl-branched lipids and TAG to act as a sink for the assimilation of excess propionyl-CoA during infection (76). Fatty acids can also be assimilated directly into cell membrane phospholipids or converted into TAG. Phospholipids are required to maintain cytoplasmic membrane integrity while TAG is classically thought to function as a carbon storage molecule that can be turned over when nutrients are limiting (72, 77). Cytosolic accumulation of TAG also negatively regulates M. tuberculosis growth and exporting TAG from the bacterial cytosol appears to be required for optimal growth of M. tuberculosis in macrophages (78, 79). In addition to fueling biosynthesis and energy production, lipid metabolic pathways can influence the physiology of M. tuberculosis during infection. For example, TAG synthesis can also reduce M. tuberculosis growth rate and antibiotic sensitivity by reducing acetyl-CoA availability (80). The balance of fatty acid catabolism and anabolism can also influence cytosolic redox homeostasis during infection in macrophages (76).
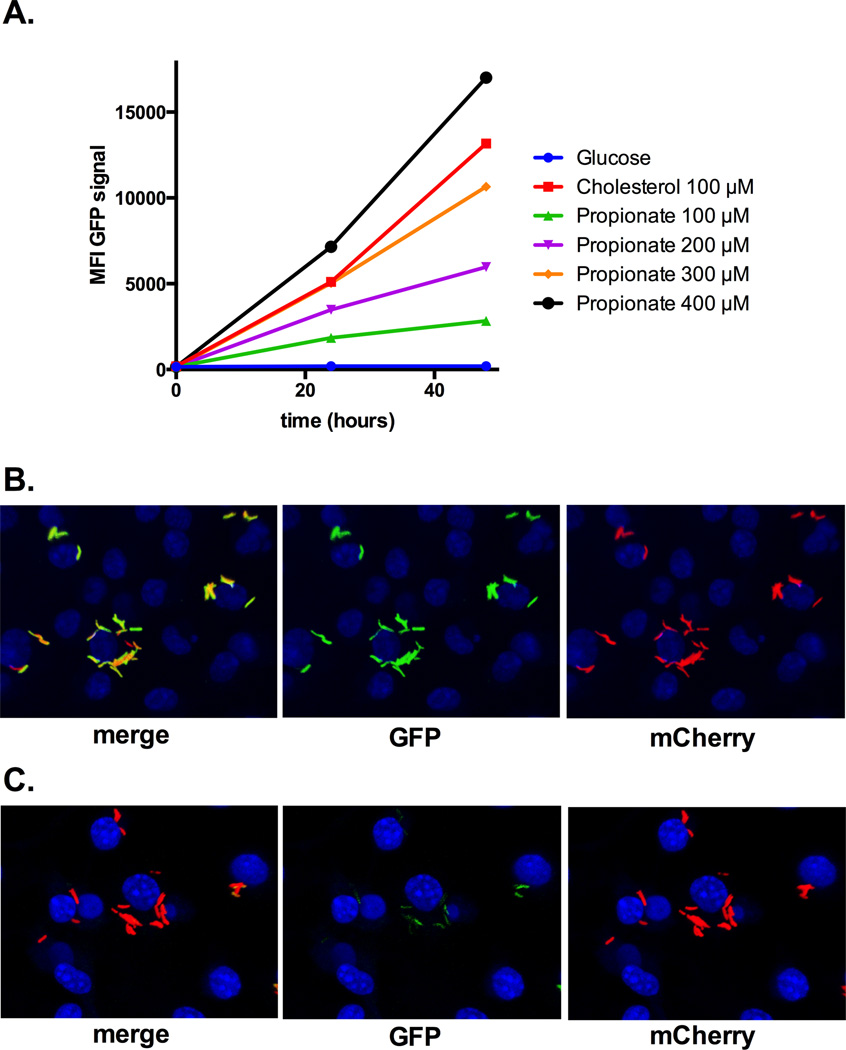
Cholesterol
Cholesterol has been repeatedly shown to be required for optimal growth and persistence of M. tuberculosis during infection (36, 47–49, 51, 81). Mechanisms involved in cholesterol import, side chain degradation, and early ring cleavage events have been partially characterized in M. tuberculosis (49, 52, 81–95). The current model is that when cholesterol is broken down by M. tuberculosis, the 2- and 3-carbon intermediates acetyl-CoA, propionyl-CoA, and pyruvate are generated. The 3-carbon intermediate, propionyl-CoA, plays an important role in M. tuberculosis physiology and recent studies have indicated that cholesterol is the major source of propionyl-CoA during infection in macrophages. The MCC genes (prpC and prpD) are induced by propionate and cholesterol indicating that the transcriptional induction of prpC and prpD is activated by propionate or propionyl-CoA (52, 96). Griffin et al. demonstrated deleting the Mce4 cholesterol transporter in M. tuberculosis could reverse propionyl-CoA toxicity induced by the Icl1 inhibitor, 3-nitropropionate (3-NP) during infection in macrophages (52). It is now understood that 3-NP inhibits the pure M. tuberculosis Icl1 enzyme (55) but also inhibits the M. tuberculosis succinate dehydrogenase in intact cells (97). Since the prpD promoter is responsive to cholesterol-derived propionate or propionyl-CoA we fused the promoter region of prpD to GFP and made cholesterol reporter strain in M. tuberculosis prpD’::GFP, smyc’::mCherry. This reporter strain responds to cholesterol and stoichiometric amounts of propionate, Figure 5. Importantly, the GFP signal in this reporter strain is strongly induced during infection in macrophages and an inhibitor of M. tuberculosis cholesterol metabolism completely abolishes the GFP signal Figure 5. Together these observations support the idea that cholesterol is the major source of propionyl-CoA for M. tuberculosis during infection. Additionally, it is likely that other sources of propionyl-CoA such as branched chain amino acids, or odd chain fatty acids are minor contributors to the bacterial pools o propionyl-CoA during infection in macrophages. Whether the requirement for cholesterol for optimal growth in macrophages reflects the abundance, availability of the molecule, or a preference for the catabolic intermediates released from the molecule remains unclear. Given that M. tuberculosis has the capacity to simultaneously metabolize simple substrates (98) it is puzzling why other nutrients such as fatty acids do not fully substitute for cholesterol during infection. Since the cholesterol catabolism produces only 2- and 3-carbon intermediates this molecule may fulfill a dedicated role in M. tuberculosis metabolism. Fatty acids are more versatile substrates in metabolism and can be β-oxidized, stored, or used for biosynthesis. Perhaps M. tuberculosis uses cholesterol to fuel central metabolism and signals the bacterium to shunt fatty acids away from oxidation pathways during infection. This might explain why genetically and chemically inactivating cholesterol metabolism negatively impacts bacterial growth in macrophages despite the presence of other nutrients such as fatty acids. Clearly this is a poorly understood area of M. tuberculosis physiology and perhaps a potential therapeutic intervention could be developed that disrupts these complex pathways.
Figure 5. The prpD’::GFP reporter strain responds to cholesterol during infection.
A GFP expression is induced with propionate and cholesterol. The GFP mean fluorescent intensity (MFI) was determined from 10,000 bacteria by flow cytometery. B resting bone marrow derived macrophages were infected with the M. tuberculosis prpD’::GFP reporter strain for 24 hours. Within macrophages the prpD’::GFP reporter is active. C treating macrophages were infected with the M. tuberculosis prpD’::GFP reporter strain with an inhibitor abolishes reporter signal. Nuclei are stained with DAPI and the images are courtesy of Kaley Wilburn.
The role of ICL and the MCC
It has been repeatedly demonstrated that the M. tuberculosis Icl enzymes are required for optimal bacterial growth in macrophages. Complicating our understanding of this growth phenotype is that M. tuberculosis expresses two Icl enzymes termed Icl1 and Icl2 (65). These enzymes are also bifunctional and catalyze the isocitrate lyase reaction in the glyoxylate cycle and the methyl-isocitrate lyase reaction in the MCC that are required for the assimilation of acetyl-CoA and propionyl-CoA, respectively. Recently, Eoh and Rhee reported a metabolomic-based characterization of the effects of blocking propionyl-CoA and acetyl-CoA assimilation in an M. tuberculosis Icl1/Icl2 mutant. This work demonstrated that when the M. tuberculosis Icl1/Icl2 mutant is provided acetate or propionate inactivation of the methyl-isocitrate lyase activity is primarily responsible for bactericidal effect of the Icl1/Icl2 mutation which depletes central metabolic intermediates, blocks gluconeogenesis, and perturbs multiple downstream aspects of cellular homeostasis (99). These studies required defined growth conditions and carbon sources to carefully quantify the isotopically labeled substrates which would make these experiments difficult to conduct with intracellular bacteria. That said, our studies with intracellular M. tuberculosis also indicate that the methyl-isocitrate lyase activity is principally responsible for the growth defect of the M. tuberculosis ΔIcl1 mutant. It is established that vitamin-B12 activates the methylmalonyl pathway and can reroute propionyl-CoA into central metabolism bypassing the MCC (100). Adding vitamin-B12 to macrophages infected with M. tuberculosis ΔIcl1 mutant restores growth of the mutant to wild type levels (28). This vitamin-B12 dependent growth restoration could be interpreted as simply opening an additional anaplerotic pathway that fuels central metabolism however, we do not think this is the case because chemically or genetically inactivating PrpC in a M. tuberculosis ΔIcl1 background also rescues growth of the bacteria to wild type levels in 7H9 OADC containing cholesterol and in macrophages (36). Because this chemical and genetic rescue occurs without seemingly opening any anaplerotic route for propionyl-CoA it is likely that the “dead ended” MCC pathway in the M. tuberculosis ΔIcl1 mutant allows the accumulation of a toxic intermediate metabolite of the MCC and is responsible for the growth defect of this mutant.
Lipid acquisition from the host cell
The mechanisms required for the bacterial acquisition of host-derived lipids is another poorly understood aspect of M. tuberculosis pathogenesis and physiology. Microscopy-based studies indicate that phagosomes containing M. tuberculosis can interact with lipid-loaded droplets in foamy macrophages (101, 102) and exogenously added radiolabeled fatty acids can be incorporated by M. tuberculosis during intracellular growth (28, 72). However, it remains unclear if the fatty acids were acquired from host lipid droplets, membrane bilayers, or other sources. One possibility is that M. tuberculosis acquires cholesterol and fatty acids from serum-derived lipoproteins that traffic within the macrophage endocytic network (103). Serum lipoproteins can be internalized by macrophages using the same scavenger receptor as the M. tuberculosis bacillus (104).
On the bacterial side, the mycobacterial Mce4 cholesterol transporter complex is involved in the assimilation of host cholesterol during infection (50). Interestingly, a mutant lacking the permease subunit of the Mce4 transporter can grow on cholesterol as a sole carbon source after a lag period and the mutant can partially metabolize the substrate (50). This suggests that cholesterol transport is redundant or a compensatory pathway exists in M. tuberculosis to assimilate either the intact sterol or some form of the molecule. In preliminary studies we have observed a ~50% reduction in GFP expression using our prpD’::GFP, smyc’::mCherry reporter in a M. tuberculosis ΔMce4 mutant during infection in macrophages (data not shown). Together these observations indicate that Mce4 is clearly involved in cholesterol assimilation but during infection some form of the sterol may still be metabolized to propionyl-CoA in the Mce4 mutant. Despite the important role fatty acids play in M. tuberculosis physiology our understanding of how fatty acids are transported across the M. tuberculosis cell envelope remains uncharacterized.
Manipulating the host cell for nutritional purposes
The immune response mounted against M. tuberculosis also perturbs the host cell metabolism to seemingly favor M. tuberculosis survival. For example, chronic inflammation observed in human tuberculosis leads to the accumulation of lipid-loaded foamy macrophages that are infected with M. tuberculosis and the development of these cells is associated with transcriptional and metabolic reprogramming of cells within the granuloma (101, 105). One mechanism involved in remodeling the macrophage involves M. tuberculosis activation of peroxisome proliferator-activated receptors (PPARs) (106). PPARs are a family of transcription factors that respond to fatty acid metabolites and during M. tuberculosis infection increases PPARγ expression in a toll-like receptor 2 dependent manner, leading to foam cell formation (106–108). Conversely, antagonists of PPARγ down regulate the lipid content of infected macrophages which has a negative effect on intracellular growth of M. tuberculosis (107–109). Many of the mechanisms involved in foam cell formation remain uncharacterized however a recent report suggested that M. tuberculosis infection may stimulate PPARγ to increase the expression of CD36 through testicular receptor 4 (110). CD36 is a low-density lipoprotein receptor that is involved in uptake of serum-derived lipoproteins and this may increase the lipid concentration and drive foam cell formation. Another mechanism that has recently been described by Singh et al demonstrates that M. tuberculosis infection induces glycolysis in infected macrophages, which can drive foam cell formation (111). Specifically, M. tuberculosis-directed dysregulation of the anti-lipolytic G-Protein-coupled receptor GPR109A in infected cells leads to increased lipid droplet accumulation and a permissive bacterial growth environment.
Conclusion
The interaction between M. tuberculosis and its host cell is highly complex and extremely intimate. Were it not for the disease, one might regard this interaction at the cellular level as an almost symbiotic one. The metabolic activity and physiology of both cells are shaped by this co-existence. We believe that where this appreciation has greatest significance is in the field of drug discovery. Evolution rewards efficiency, and recent data from many groups discussed in this chapter indicate that M. tuberculosis has evolved to utilize the environmental cues within its host to control large genetic responses or regulons. However these regulons may represent chinks in the bacterium’s armor because they can constrain M. tuberculosis’s metabolic plasticity. The prime example is how the presence of cholesterol within the host cell appears to limit the ability of M. tuberculosis to utilize or assimilate other carbon sources (36). And that is the reason for this title of this book chapter. We believe firmly that to understand the physiology of M. tuberculosis, and to identify new drug targets, it is imperative that the bacterium be interrogated within the context of its host cell. The M. tuberculosis-infected macrophage truly is the “minimal unit of infection”.
Acknowledgments
The authors would like to acknowledge the support of the US Public Health Services support through the National institutes of Health awards AI118582, AI067027 and HL055936 (DGR) and AI099569 and AI119122 (BCV). DGR is also grateful for the support of the Bill and Melinda Gates Foundation.
Literature Cited
- 1.Srivastava S, Ernst JD, Desvignes L. Beyond macrophages: the diversity of mononuclear cells in tuberculosis. Immunol Rev. 2014;262:179–192. doi: 10.1111/imr.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leemans JC, Juffermans NP, Florquin S, van Rooijen N, Vervoordeldonk MJ, Verbon A, van Deventer SJ, van der Poll T. Depletion of alveolar macrophages exerts protective effects in pulmonary tuberculosis in mice. J Immunol. 2001;166:4604–4611. doi: 10.4049/jimmunol.166.7.4604. [DOI] [PubMed] [Google Scholar]
- 3.Leemans JC, Thepen T, Weijer S, Florquin S, van Rooijen N, van de Winkel JG, van der Poll T. Macrophages play a dual role during pulmonary tuberculosis in mice. J Infect Dis. 2005;191:65–74. doi: 10.1086/426395. [DOI] [PubMed] [Google Scholar]
- 4.Antonelli LR, Gigliotti Rothfuchs A, Goncalves R, Roffe E, Cheever AW, Bafica A, Salazar AM, Feng CG, Sher A. Intranasal Poly-IC treatment exacerbates tuberculosis in mice through the pulmonary recruitment of a pathogen-permissive monocyte/macrophage population. J Clin Invest. 2010;120:1674–1682. doi: 10.1172/JCI40817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorhoi A, Yeremeev V, Nouailles G, Weiner J, 3rd, Jorg S, Heinemann E, Oberbeck-Muller D, Knaul JK, Vogelzang A, Reece ST, Hahnke K, Mollenkopf HJ, Brinkmann V, Kaufmann SH. Type I IFN signaling triggers immunopathology in tuberculosis-susceptible mice by modulating lung phagocyte dynamics. Eur J Immunol. 2014;44:2380–2393. doi: 10.1002/eji.201344219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mattila JT, Ojo OO, Kepka-Lenhart D, Marino S, Kim JH, Eum SY, Via LE, Barry CE, 3rd, Klein E, Kirschner DE, Morris SM, Jr, Lin PL, Flynn JL. Microenvironments in tuberculous granulomas are delineated by distinct populations of macrophage subsets and expression of nitric oxide synthase and arginase isoforms. J Immunol. 2013;191:773–784. doi: 10.4049/jimmunol.1300113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marino S, Cilfone NA, Mattila JT, Linderman JJ, Flynn JL, Kirschner DE. Macrophage polarization drives granuloma outcome during Mycobacterium tuberculosis infection. Infect Immun. 2015;83:324–338. doi: 10.1128/IAI.02494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gideon HP, Phuah J, Myers AJ, Bryson BD, Rodgers MA, Coleman MT, Maiello P, Rutledge T, Marino S, Fortune SM, Kirschner DE, Lin PL, Flynn JL. Variability in tuberculosis granuloma T cell responses exists, but a balance of pro- and anti-inflammatory cytokines is associated with sterilization. PLoS Pathog. 2015;11:e1004603. doi: 10.1371/journal.ppat.1004603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan S, Russell DG. Trans-species communication in the Mycobacterium tuberculosis-infected macrophage. Immunol Rev. 2015;264:233–248. doi: 10.1111/imr.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaible UE, Sturgill-Koszycki S, Schlesinger PH, Russell DG. Cytokine activation leads to acidification and increases maturation of Mycobacterium avium-containing phagosomes in murine macrophages. J Immunol. 1998;160:1290–1296. [PubMed] [Google Scholar]
- 11.Via LE, Fratti RA, McFalone M, Pagan-Ramos E, Deretic D, Deretic V. Effects of cytokines on mycobacterial phagosome maturation. J Cell Sci. 1998;111(Pt 7):897–905. doi: 10.1242/jcs.111.7.897. [DOI] [PubMed] [Google Scholar]
- 12.Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 14.MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci U S A. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonough KA, Kress Y, Bloom BR. Pathogenesis of tuberculosis: interaction of Mycobacterium tuberculosis with macrophages. Infect Immun. 1993;61:2763–2773. doi: 10.1128/iai.61.7.2763-2773.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myrvik QN, Leake ES, Wright MJ. Disruption of phagosomal membranes of normal alveolar macrophages by the H37Rv strain of Mycobacterium tuberculosis. A correlate of virulence. Am Rev Respir Dis. 1984;129:322–328. [PubMed] [Google Scholar]
- 17.Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, Enninga J. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012;8:e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 19.Russell DG. The ins and outs of the Mycobacterium tuberculosis-containing vacuole. Cell Microbiol. 2016 doi: 10.1111/cmi.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment. The Journal of experimental medicine. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohde KH, Abramovitch RB, Russell DG. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe. 2007;2:352–364. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Rohde KH, Veiga DF, Caldwell S, Balazsi G, Russell DG. Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS Pathog. 2012;8:e1002769. doi: 10.1371/journal.ppat.1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abramovitch RB, Rohde KH, Hsu FF, Russell DG. aprABC: a Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol Microbiol. 2011;80:678–694. doi: 10.1111/j.1365-2958.2011.07601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohde KH, Veiga DF, Caldwell S, Balazsi G, Russell DG. Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gill WP, Harik NS, Whiddon MR, Liao RP, Mittler JE, Sherman DR. A replication clock for Mycobacterium tuberculosis. Nat Med. 2009;15:211–214. doi: 10.1038/nm.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Homolka S, Niemann S, Russell DG, Rohde KH. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog. 2010;6:e1000988. doi: 10.1371/journal.ppat.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohde K, Yates RM, Purdy GE, Russell DG. Mycobacterium tuberculosis and the environment within the phagosome. Immunol Rev. 2007;219:37–54. doi: 10.1111/j.1600-065X.2007.00547.x. [DOI] [PubMed] [Google Scholar]
- 28.Lee W, VanderVen BC, Fahey RJ, Russell DG. Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J Biol Chem. 2013;288:6788–6800. doi: 10.1074/jbc.M112.445056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan S, Sukumar N, Abramovitch RB, Parish T, Russell DG. Mycobacterium tuberculosis responds to chloride and pH as synergistic cues to the immune status of its host cell. PLoS Pathog. 2013;9:e1003282. doi: 10.1371/journal.ppat.1003282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, Riley LW, Schnappinger D. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Research. 2005;33:e21–e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sukumar N, Tan S, Aldridge BB, Russell DG. Exploitation of Mycobacterium tuberculosis reporter strains to probe the impact of vaccination at sites of infection. PLoS Pathog. 2014;10:e1004394. doi: 10.1371/journal.ppat.1004394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 2008;133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf AJ, Linas B, Trevejo-Nunez GJ, Kincaid E, Tamura T, Takatsu K, Ernst JD. Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo. J Immunol. 2007;179:2509–2519. doi: 10.4049/jimmunol.179.4.2509. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Tan S, Huang L, Abramovitch RB, Rohde KH, Zimmerman MD, Chen C, Dartois V, VanderVen BC, Russell DG. Immune activation of the host cell induces drug tolerance in Mycobacterium tuberculosis both in vitro and in vivo. J Exp Med. 2016;213:809–825. doi: 10.1084/jem.20151248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manina G, Dhar N, McKinney JD. Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe. 2015;17:32–46. doi: 10.1016/j.chom.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 36.VanderVen BC, Fahey RJ, Lee W, Liu Y, Abramovitch RB, Memmott C, Crowe AM, Eltis LD, Perola E, Deininger DD, Wang T, Locher CP, Russell DG. Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium's metabolism is constrained by the intracellular environment. PLoS Pathog. 2015;11:e1004679. doi: 10.1371/journal.ppat.1004679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo YL, Seebacher T, Kurz U, Linder JU, Schultz JE. Adenylyl cyclase Rv1625c of Mycobacterium tuberculosis: a progenitor of mammalian adenylyl cyclases. Embo j. 2001;20:3667–3675. doi: 10.1093/emboj/20.14.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ketkar AD, Shenoy AR, Ramagopal UA, Visweswariah SS, Suguna K. A structural basis for the role of nucleotide specifying residues in regulating the oligomerization of the Rv1625c adenylyl cyclase from M. tuberculosis. J Mol Biol. 2006;356:904–916. doi: 10.1016/j.jmb.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 39.Agarwal N, Lamichhane G, Gupta R, Nolan S, Bishai WR. Cyclic AMP intoxication of macrophages by a Mycobacterium tuberculosis adenylate cyclase. Nature. 2009;460:98–102. doi: 10.1038/nature08123. [DOI] [PubMed] [Google Scholar]
- 40.Choudhary E, Bishai W, Agarwal N. Expression of a subset of heat stress induced genes of mycobacterium tuberculosis is regulated by 3',5'-cyclic AMP. PLoS One. 2014;9:e89759. doi: 10.1371/journal.pone.0089759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahramanoglou C, Cortes T, Matange N, Hunt DM, Visweswariah SS, Young DB, Buxton RS. Genomic mapping of cAMP receptor protein (CRP Mt) in Mycobacterium tuberculosis: relation to transcriptional start sites and the role of CRPMt as a transcription factor. Nucleic Acids Res. 2014;42:8320–8329. doi: 10.1093/nar/gku548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knapp GS, Lyubetskaya A, Peterson MW, Gomes AL, Ma Z, Galagan JE, McDonough KA. Role of intragenic binding of cAMP responsive protein (CRP) in regulation of the succinate dehydrogenase genes Rv0249c-Rv0247c in TB complex mycobacteria. Nucleic Acids Res. 2015;43:5377–5393. doi: 10.1093/nar/gkv420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee HJ, Lang PT, Fortune SM, Sassetti CM, Alber T. Cyclic AMP regulation of protein lysine acetylation in Mycobacterium tuberculosis. Nat Struct Mol Biol. 2012;19:811–818. doi: 10.1038/nsmb.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shleeva M, Goncharenko A, Kudykina Y, Young D, Young M, Kaprelyants A. Cyclic AMP-dependent resuscitation of dormant Mycobacteria by exogenous free fatty acids. PLoS One. 2013;8:e82914. doi: 10.1371/journal.pone.0082914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu H, Hegde SS, Blanchard JS. Reversible acetylation and inactivation of Mycobacterium tuberculosis acetyl-CoA synthetase is dependent on cAMP. Biochemistry. 2011;50:5883–5892. doi: 10.1021/bi200156t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aronoff DM, Canetti C, Serezani CH, Luo M, Peters-Golden M. Cutting edge: macrophage inhibition by cyclic AMP (cAMP): differential roles of protein kinase A and exchange protein directly activated by cAMP-1. Journal of immunology (Baltimore, Md. : 1950) 2005;174:595–599. doi: 10.4049/jimmunol.174.2.595. [DOI] [PubMed] [Google Scholar]
- 47.Chang JC, Harik NS, Liao RP, Sherman DR. Identification of Mycobacterial Genes That Alter Growth and Pathology in Macrophages and in Mice. Journal of Infectious Diseases. 2007;196:788–795. doi: 10.1086/520089. [DOI] [PubMed] [Google Scholar]
- 48.Hu Y, van der Geize R, Besra GS, Gurcha SS, Liu A, Rohde M, Singh M, Coates A. 3-Ketosteroid 9alpha-hydroxylase is an essential factor in the pathogenesis of Mycobacterium tuberculosis. Mol Microbiol. 2010;75:107–121. doi: 10.1111/j.1365-2958.2009.06957.x. [DOI] [PubMed] [Google Scholar]
- 49.Nesbitt NM, Yang X, Fontan P, Kolesnikova I, Smith I, Sampson NS, Dubnau E. A Thiolase of Mycobacterium tuberculosis Is Required for Virulence and Production of Androstenedione and Androstadienedione from Cholesterol. Infect. Immun. 2010;78:275–282. doi: 10.1128/IAI.00893-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proceedings of the National Academy of Sciences. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van der Geize R, Yam K, Heuser T, Wilbrink MH, Hara H, Anderton MC, Sim E, Dijkhuizen L, Davies JE, Mohn WW, Eltis LD. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proceedings of the National Academy of Sciences. 2007;104:1947–1952. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Griffin JE, Pandey AK, Gilmore SA, Mizrahi V, McKinney JD, Bertozzi CR, Sassetti CM. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem Biol. 2012;19:218–227. doi: 10.1016/j.chembiol.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McKinney JD, Honer zu Bentrup K, Munoz-Elias EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Jr, Russell DG. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 54.McKinney JD, zu Bentrup KH, Munoz-Elias EJ, Miczak A, Chen B, Chan W-T, Swenson D, Sacchettini JC, Jacobs WR, Russell DG. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 55.Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bloch H, Segal W. Biochemical differentiation of Mycobacterium tuberculosis grown in vivo and in vitro. J Bacteriol. 1956;72:132–141. doi: 10.1128/jb.72.2.132-141.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 58.Krithika R, Marathe U, Saxena P, Ansari MZ, Mohanty D, Gokhale RS. A genetic locus required for iron acquisition in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:2069–2074. doi: 10.1073/pnas.0507924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trivedi OA, Arora P, Sridharan V, Tickoo R, Mohanty D, Gokhale RS. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature. 2004;428:441–445. doi: 10.1038/nature02384. [DOI] [PubMed] [Google Scholar]
- 60.Yang M, Guja KE, Thomas ST, Garcia-Diaz M, Sampson NS. A distinct MaoC-like enoyl-CoA hydratase architecture mediates cholesterol catabolism in Mycobacterium tuberculosis. ACS Chem Biol. 2014;9:2632–2645. doi: 10.1021/cb500232h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang M, Lu R, Guja KE, Wipperman MF, St Clair JR, Bonds AC, Garcia-Diaz M, Sampson NS. Unraveling Cholesterol Catabolism in Mycobacterium tuberculosis: ChsE4-ChsE5 alpha2beta2 Acyl-CoA Dehydrogenase Initiates beta-Oxidation of 3-Oxo-cholest-4-en-26-oyl CoA. ACS Infect Dis. 2015;1:110–125. doi: 10.1021/id500033m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Graham JE, Clark-Curtiss JE. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS) Proc Natl Acad Sci U S A. 1999;96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Honer Zu Bentrup K, Miczak A, Swenson DL, Russell DG. Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J Bacteriol. 1999;181:7161–7167. doi: 10.1128/jb.181.23.7161-7167.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sturgill-Koszycki S, Haddix PL, Russell DG. The interaction between Mycobacterium and the macrophage analyzed by two-dimensional polyacrylamide gel electrophoresis. Electrophoresis. 1997;18:2558–2565. doi: 10.1002/elps.1150181411. [DOI] [PubMed] [Google Scholar]
- 65.Gould TA, van de Langemheen H, Munoz-Elias EJ, McKinney JD, Sacchettini JC. Dual role of isocitrate lyase 1 in the glyoxylate and methylcitrate cycles in Mycobacterium tuberculosis. Mol Microbiol. 2006;61:940–947. doi: 10.1111/j.1365-2958.2006.05297.x. [DOI] [PubMed] [Google Scholar]
- 66.Muñoz-Elías EJ, Upton AM, Cherian J, McKinney JD. Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Molecular Microbiology. 2006;60:1109–1122. doi: 10.1111/j.1365-2958.2006.05155.x. [DOI] [PubMed] [Google Scholar]
- 67.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment. The Journal of Experimental Medicine. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ouellet H, Johnston JB, de Montellano PR. Cholesterol catabolism as a therapeutic target in Mycobacterium tuberculosis. Trends Microbiol. 2011;19:530–539. doi: 10.1016/j.tim.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wipperman MF, Sampson NS, Thomas ST. Pathogen roid rage: cholesterol utilization by Mycobacterium tuberculosis. Crit Rev Biochem Mol Biol. 2014;49:269–293. doi: 10.3109/10409238.2014.895700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yam KC, Okamoto S, Roberts JN, Eltis LD. Adventures in Rhodococcus - from steroids to explosives. Can J Microbiol. 2011;57:155–168. doi: 10.1139/W10-115. [DOI] [PubMed] [Google Scholar]
- 71.Marrero J, Rhee KY, Schnappinger D, Pethe K, Ehrt S. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc Natl Acad Sci U S A. 2010;107:9819–9824. doi: 10.1073/pnas.1000715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog. 2011;7:e1002093. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quadri LE. Biosynthesis of mycobacterial lipids by polyketide synthases and beyond. Crit Rev Biochem Mol Biol. 2014;49:179–211. doi: 10.3109/10409238.2014.896859. [DOI] [PubMed] [Google Scholar]
- 74.Jain M, Petzold CJ, Schelle MW, Leavell MD, Mougous JD, Bertozzi CR, Leary JA, Cox JS. Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. Proc Natl Acad Sci U S A. 2007;104:5133–5138. doi: 10.1073/pnas.0610634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang X, Nesbitt NM, Dubnau E, Smith I, Sampson NS. Cholesterol metabolism increases the metabolic pool of propionate in Mycobacterium tuberculosis. Biochemistry. 2009;48:3819–3821. doi: 10.1021/bi9005418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn AJ. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 2009;5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Daniel J, Deb C, Dubey VS, Sirakova TD, Abomoelak B, Morbidoni HR, Kolattukudy PE. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol. 2004;186:5017–5030. doi: 10.1128/JB.186.15.5017-5030.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gaur RL, Ren K, Blumenthal A, Bhamidi S, Gonzalez-Nilo FD, Jackson M, Zare RN, Ehrt S, Ernst JD, Banaei N. LprG-mediated surface expression of lipoarabinomannan is essential for virulence of Mycobacterium tuberculosis. PLoS Pathog. 2014;10:e1004376. doi: 10.1371/journal.ppat.1004376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martinot AJ, Farrow M, Bai L, Layre E, Cheng TY, Tsai JH, Iqbal J, Annand JW, Sullivan ZA, Hussain MM, Sacchettini J, Moody DB, Seeliger JC, Rubin EJ. Mycobacterial Metabolic Syndrome: LprG and Rv1410 Regulate Triacylglyceride Levels, Growth Rate and Virulence in Mycobacterium tuberculosis. PLoS Pathog. 2016;12:e1005351. doi: 10.1371/journal.ppat.1005351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baek SH, Li AH, Sassetti CM. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS biology. 2011;9:e1001065. doi: 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yam KC, D'Angelo I, Kalscheuer R, Zhu H, Wang JX, Snieckus V, Ly LH, Converse PJ, Jacobs WR, Jr, Strynadka N, Eltis LD. Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000344. doi: 10.1371/journal.ppat.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Capyk JK, Casabon I, Gruninger R, Strynadka NC, Eltis LD. Activity of 3-ketosteroid 9alpha-hydroxylase (KshAB) indicates cholesterol side chain and ring degradation occur simultaneously in Mycobacterium tuberculosis. J Biol Chem. 2011;286:40717–40724. doi: 10.1074/jbc.M111.289975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Capyk JK, D'Angelo I, Strynadka NC, Eltis LD. Characterization of 3-ketosteroid 9{alpha}-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis. J Biol Chem. 2009;284:9937–9946. doi: 10.1074/jbc.M900719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Capyk JK, Kalscheuer R, Stewart GR, Liu J, Kwon H, Zhao R, Okamoto S, Jacobs WR, Jr, Eltis LD, Mohn WW. Mycobacterial cytochrome p450 125 (cyp125) catalyzes the terminal hydroxylation of c27 steroids. J Biol Chem. 2009;284:35534–35542. doi: 10.1074/jbc.M109.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Casabon I, Crowe AM, Liu J, Eltis LD. FadD3 is an acyl-CoA synthetase that initiates catabolism of cholesterol rings C and D in actinobacteria. Mol Microbiol. 2013;87:269–283. doi: 10.1111/mmi.12095. [DOI] [PubMed] [Google Scholar]
- 86.Casabon I, Swain K, Crowe AM, Eltis LD, Mohn WW. Actinobacterial acyl coenzyme A synthetases involved in steroid side-chain catabolism. J Bacteriol. 2014;196:579–587. doi: 10.1128/JB.01012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Casabon I, Zhu SH, Otani H, Liu J, Mohn WW, Eltis LD. Regulation of the KstR2 regulon of Mycobacterium tuberculosis by a cholesterol catabolite. Mol Microbiol. 2013;89:1201–1212. doi: 10.1111/mmi.12340. [DOI] [PubMed] [Google Scholar]
- 88.Dresen C, Lin LY, D'Angelo I, Tocheva EI, Strynadka N, Eltis LD. A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism. J Biol Chem. 2010;285:22264–22275. doi: 10.1074/jbc.M109.099028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Driscoll MD, McLean KJ, Levy C, Mast N, Pikuleva IA, Lafite P, Rigby SE, Leys D, Munro AW. Structural and biochemical characterization of Mycobacterium tuberculosis CYP142: evidence for multiple cholesterol 27-hydroxylase activities in a human pathogen. J Biol Chem. 2010;285:38270–38282. doi: 10.1074/jbc.M110.164293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Frank DJ, Madrona Y, Ortiz de Montellano PR. Cholesterol ester oxidation by mycobacterial cytochrome P450. J Biol Chem. 2014;289:30417–30425. doi: 10.1074/jbc.M114.602771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Griffin JE, Gawronski JD, DeJesus MA, Ioerger TR, Akerley BJ, Sassetti CM. High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lack NA, Yam KC, Lowe ED, Horsman GP, Owen RL, Sim E, Eltis LD. Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism. J Biol Chem. 2010;285:434–443. doi: 10.1074/jbc.M109.058081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ouellet H, Guan S, Johnston JB, Chow ED, Kells PM, Burlingame AL, Cox JS, Podust LM, De Montellano PRO. Mycobacterium tuberculosis CYP125A1, a steroid C27 monooxygenase that detoxifies intracellularly generated cholest-4-en-3-one. Molecular Microbiology. 2010;77:730–742. doi: 10.1111/j.1365-2958.2010.07243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thomas ST, VanderVen BC, Sherman DR, Russell DG, Sampson NS. Pathway Profiling in Mycobacterium tuberculosis: elucidation of a cholesterol-derived catabolite and the enzymes that catalyze its metabolism. Journal of Biological Chemistry. 2011;286:43668–43678. doi: 10.1074/jbc.M111.313643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang X, Nesbitt NM, Dubnau E, Smith I, Sampson NS. Cholesterol Metabolism Increases the Metabolic Pool of Propionate in Mycobacterium tuberculosis. Biochemistry. 2009;48:3819–3821. doi: 10.1021/bi9005418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Masiewicz P, Brzostek A, Wolanski M, Dziadek J, Zakrzewska-Czerwinska J. A novel role of the PrpR as a transcription factor involved in the regulation of methylcitrate pathway in Mycobacterium tuberculosis. PLoS One. 2012;7:e43651. doi: 10.1371/journal.pone.0043651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eoh H, Rhee KY. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2013;110:6554–6559. doi: 10.1073/pnas.1219375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.de Carvalho LP, Fischer SM, Marrero J, Nathan C, Ehrt S, Rhee KY. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chemistry & biology. 2010;17:1122–1131. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 99.Eoh H, Rhee KY. Methylcitrate cycle defines the bactericidal essentiality of isocitrate lyase for survival of Mycobacterium tuberculosis on fatty acids. Proc Natl Acad Sci U S A. 2014;111:4976–4981. doi: 10.1073/pnas.1400390111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Savvi S, Warner DF, Kana BD, McKinney JD, Mizrahi V, Dawes SS. Functional characterization of a vitamin B12-dependent methylmalonyl pathway in Mycobacterium tuberculosis: implications for propionate metabolism during growth on fatty acids. J Bacteriol. 2008;190:3886–3895. doi: 10.1128/JB.01767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim MJ, Wainwright HC, Locketz M, Bekker LG, Walther GB, Dittrich C, Visser A, Wang W, Hsu FF, Wiehart U, Tsenova L, Kaplan G, Russell DG. Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMBO Mol Med. 2010;2:258–274. doi: 10.1002/emmm.201000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Peyron P, Vaubourgeix J, Poquet Y, Levillain F, Botanch C, Bardou F, Daffe M, Emile JF, Marchou B, Cardona PJ, de Chastellier C, Altare F. Foamy macrophages from tuberculous patients' granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 2008;4:e1000204. doi: 10.1371/journal.ppat.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Graham A, Allen AM. Mitochondrial function and regulation of macrophage sterol metabolism and inflammatory responses. World J Cardiol. 2015;7:277–286. doi: 10.4330/wjc.v7.i5.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Philips JA, Rubin EJ, Perrimon N. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science. 2005;309:1251–1253. doi: 10.1126/science.1116006. [DOI] [PubMed] [Google Scholar]
- 105.Russell DG, Cardona P-J, Kim M-J, Allain S, Altare F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat Immunol. 2009;10:943–948. doi: 10.1038/ni.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Almeida PE, Silva AR, Maya-Monteiro CM, Torocsik D, D'Avila H, Dezso B, Magalhaes KG, Castro-Faria-Neto HC, Nagy L, Bozza PT. Mycobacterium bovis bacillus Calmette-Guerin infection induces TLR2-dependent peroxisome proliferator-activated receptor gamma expression and activation: functions in inflammation, lipid metabolism, and pathogenesis. J Immunol. 2009;183:1337–1345. doi: 10.4049/jimmunol.0900365. [DOI] [PubMed] [Google Scholar]
- 107.Almeida PE, Carneiro AB, Silva AR, Bozza PT. PPARgamma Expression and Function in Mycobacterial Infection: Roles in Lipid Metabolism, Immunity, and Bacterial Killing. PPAR Res. 2012;2012:383829. doi: 10.1155/2012/383829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rajaram MV, Brooks MN, Morris JD, Torrelles JB, Azad AK, Schlesinger LS. Mycobacterium tuberculosis activates human macrophage peroxisome proliferator-activated receptor gamma linking mannose receptor recognition to regulation of immune responses. J Immunol. 2010;185:929–942. doi: 10.4049/jimmunol.1000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Salamon H, Bruiners N, Lakehal K, Shi L, Ravi J, Yamaguchi KD, Pine R, Gennaro ML. Cutting edge: Vitamin D regulates lipid metabolism in Mycobacterium tuberculosis infection. J Immunol. 2014;193:30–34. doi: 10.4049/jimmunol.1400736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mahajan S, Dkhar HK, Chandra V, Dave S, Nanduri R, Janmeja AK, Agrewala JN, Gupta P. Mycobacterium tuberculosis modulates macrophage lipid-sensing nuclear receptors PPARgamma and TR4 for survival. J Immunol. 2012;188:5593–5603. doi: 10.4049/jimmunol.1103038. [DOI] [PubMed] [Google Scholar]
- 111.Singh V, Jamwal S, Jain R, Verma P, Gokhale R, Rao KV. Mycobacterium tuberculosis-driven targeted recalibration of macrophage lipid homeostasis promotes the foamy phenotype. Cell Host Microbe. 2012;12:669–681. doi: 10.1016/j.chom.2012.09.012. [DOI] [PubMed] [Google Scholar]