Abstract
Cancer cells can use a variety of metabolic substrates to fulfill the bioenergetic and biosynthetic needs of their oncogenic program. Besides bioenergetics, cancer cell metabolism also directly influences genetic, epigenetic and signaling events associated with tumor progression. Many cancer cells are addicted to glutamine, and this addiction is observed in oxidative as well as in glycolytic cells. While both oxidative and bioreductive glutamine metabolism can contribute to cancer progression and glutamine can further serve to generate peptides (including glutathione) and proteins, we report that glutamine promotes the proliferation of cancer cells independently of its use as a metabolic fuel or as a precursor of glutathione. Extracellular glutamine activates transcription factor STAT3, which is necessary and sufficient to mediate the proliferative effects of glutamine in glycolytic and in oxidative cancer cells. Glutamine also activates transcription factors HIF-1, mTOR and c-Myc, but these factors do not mediate the effects of glutamine on cancer cell proliferation. Our findings shed a new light on the anticancer effects of L-asparaginase that possesses glutaminase activity and converts glutamine into glutamate extracellularly. Conversely, cancer resistance to treatments that block glutamine metabolism could arise from glutamine-independent STAT3 re-activation.
Keywords: tumor metabolism, glutamine, HIF-1, mTOR, c-Myc, STAT3
Introduction
Cancer cells can use a variety of metabolic substrates, including glucose, lactate and glutamine, to fuel cataplerotic pathways that ultimately produce ATP and anaplerotic pathways for the biosynthesis of cell constituents.1 When fully processed through glycolysis, glucose generates ATP, NADH, pyruvate and water. Pyruvate can be imported in mitochondria where it is catabolized to generate GTP and electron donors (NADH and FADH2) that fuel oxidative phosphorylation (OXPHOS) for ATP generation at the electron transport chain (ETC). Reducing equivalents of cytosolic NADH can also be transferred to mitochondria (malate-aspartate shuttle) to fuel OXPHOS. Alternatively, several glycolytic intermediates and pyruvate are used for biosynthesis. For example, glucose-6-phosphate can aliment the pentose phosphate pathway (PPP) for nucleotide synthesis and NADPH production for lipogenesis;2 3-phosphoglycerate can fuel the serine pathway for protein and nucleotide synthesis;3 and pyruvate can generate alanine in the cytosol and citrate in mitochondria, from where citrate can be exported for lipogenesis.4 In populations of cancer cells producing most energy through glycolysis, either because of microenvironmental influences such as hypoxia or because they are actively proliferating, glycolysis can also be coupled to lactic fermentation.4 Lactate is released form glycolytic cells in a process facilitated by monocarboxylate transporters (MCTs), primarily MCT4.5,6 It can be recycled by oxidative cancer cells that express inward lactate transporters, essentially MCT1, and lactate dehydrogenase B that catalyzes the oxidation of lactate and NAD+ to pyruvate, NADH and H+.7,8 Thus, pyruvate derived from lactate can be used to generate ATP, alanine and citrate, thereby contributing to cataplerosis and anaplerosis.
Different functions have also been identified for glutamine, and many cancer cell lines are addicted to glutamine.9 Glutamine from liver origin is taken up by cancer cells in a process mainly facilitated by ASC amino-acid transporter 2 (SLC1A5/ASCT2).10 Glutamine can be directly used to generate peptides (including glutathione) and proteins, serve for amino acid exchange through the plasma membrane, or be further processed. Its metabolism, glutaminolysis, involves a 2-step conversion: glutaminases convert glutamine to glutamate (deamidation),11 and glutamate dehydrogenases (GDHs) further convert glutamate to 2-oxoglutarate (deamination).12 Transamination reactions, during which glutamate donates a nitrogen group to generate other amino acids, also generate 2-oxoglutarate.13 In cataplerotic reactions in mitochondria, 2-oxoglutarate oxidation yields malate, GTP, NADH and FADH2.14 Alternatively, 2-oxoglutarate can also fuel bioreductive anaplerotic reactions in the cytosol, which generate citrate for lipid biosynthesis.15
In addition to cataplerotic and anaplerotic metabolism, many metabolic intermediates and several metabolic enzymes can be involved in oncogenic signaling. Signaling pathways modulated by metabolic intermediates are numerous. Modulations can involve posttranslational protein modifications,16 epigenetic regulation,16,17 non-covalent enzyme/transporter modulation,18 as well as direct binding to receptors.19–21 Metabolic enzymes can also directly affect cell signaling and the transcription machinery, as recently reviewed by Lincet and Icard.22 Thus, several functional links exist between metabolic and signaling pathways in cancer cells, and metabolic influences on cell signaling can promote tumor aggressiveness.
In this context, although glutamine metabolism has already been intensively studied in cancer, little is known about direct influences that glutamine could exert as a signaling agent. Because many cancer cell lines are addicted to glutamine independently of their basal metabolic phenotype (glycolytic versus oxidative),23 this study addressed the possibility of a modulation of oncogenic transcription factors by glutamine. We report that glutamine per se activates signal transducer and activator of transcription 3 (STAT3), which promotes cancer cell proliferation.
Results
Glutamine promotes the proliferation of glycolytic and oxidative cancer cells independently of glutaminolysis
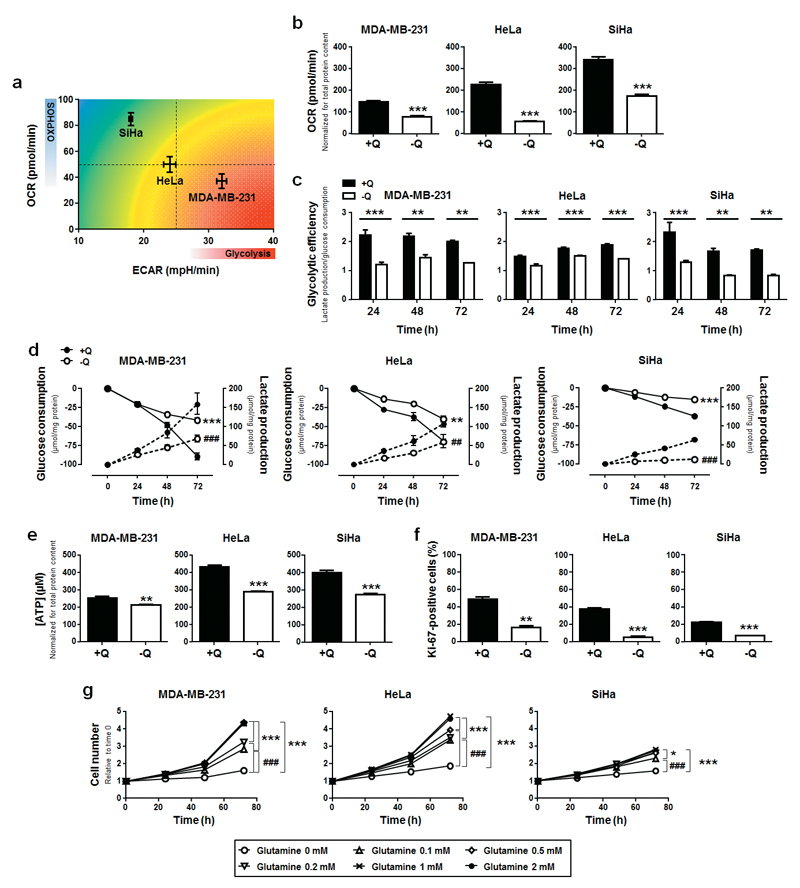
To study the addiction of cancer cells to glutamine, we used human cancer cell lines representing metabolic archetypes. In good agreement with previous characterization,7,8,24–26 in vitro measurements of cellular oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) confirmed that SiHa human cervix cancer cells have a more oxidative basal metabolism (high OCR and low ECAR) than HeLa human cervix cancer cells (intermediate OCR and ECAR), whereas MDA-MB-231 human breast cancer cells were comparatively more glycolytic (low OCR and high ECAR) (Figure 1a). Glutamine deprivation reduced intracellular glutamine concentration in all 3 cell lines, independently of the presence of serum (Figure S1a). Irrespectively of their basal metabolic phenotype, glutamine deprivation also reduced basal OCR (Figure 1b) and the glycolytic efficiency (Figure 1c) of all 3 cell lines. Reduced glycolytic efficiency was due to a simultaneous decrease in glucose uptake and lactate release (Figure 1d). The general depression of oxidative and glycolytic metabolism resulted in a lower capability of the cells to produce ATP (Figure 1e). Glutamine deprivation also strongly reduced their proliferation rate (Ki-67 staining, Figure 1f), rendering the cells almost totally unable to replicate (Figure 1g). Cell proliferation was totally restored when supplying ≥ 1 mM of glutamine. Of note, glutamine deprivation did not trigger cell death, which was evidenced by unaltered caspase-3 activation and PARP cleavage (Figure S1b).
Figure 1. Glutamine deprivation downregulates cancer cell metabolism and proliferation.
(a) The graph shows oxygen consumption rate (OCR, reflecting oxidative phosphorylation [OXPHOS]) on extracellular acidification rate (ECAR, reflecting glycolysis) plotted for MDA-MB-231 (n = 6), HeLa (n = 8) and SiHa (n = 8) cancer cells in complete medium. (b-g) MDA-MB-231, HeLa and SiHa cancer cells were cultured in complete medium containing 2 mM L-glutamine (+Q) or not (-Q). (b) OCR measured with a Seahorse bioenergetic analyzer (***P < 0.005; n = 7 for MDA-MB-213 and HeLa; n = 6 for SiHa). (c) Glycolytic efficiency calculated as the ratio between lactate production and glucose consumption (**P < 0.01, ***P < 0.005; n = 3 for MDA-MB-231 and SiHa: n = 4 for HeLa). (d) Glucose consumption (plain lines) and lactate production (dotted lines) measured using a CMA600 enzymatic analyzer (**P < 0.01, ***P < 0.005 when analyzing glucose consumption; ##P < 0.01, ###P < 0.005 when analyzing lactate production; n = 4). (e) Intracellular ATP content measured using a CellTiter-Glo luminescent assay in MDA-MB-231 (n = 4), HeLa (n = 8) and SiHa (n = 4) cells (**P < 0.01, ***P < 0.005). (f) Cell proliferation measured using Ki-67 staining (**P < 0.01, ***P < 0.005; n = 6). (g) Cell number assessed using a SpectraMax i3 multi-mode microplate reader after treatment with the indicated doses of glutamine (*P < 0.05, ***P < 0.005 compared with media +Q, ###P < 0.005 compared with media -Q; n = 4). (a-g) All quantitative data show means ± SEM.
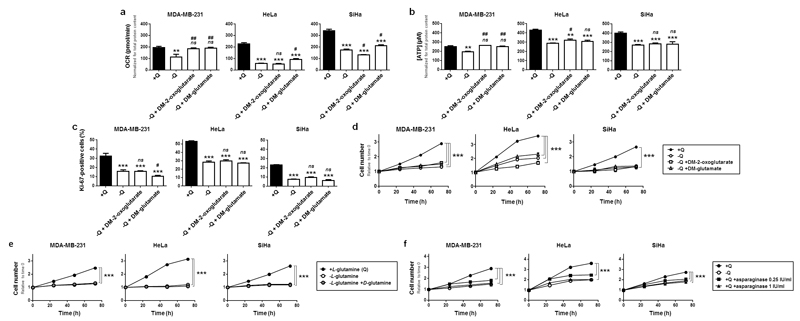
To try to rescue the metabolism and proliferation of glutamine-deprived cancer cells, we provided either glutamate or 2-oxoglutarate, the first two intermediates of glutaminolysis.11–13 To avoid possible transport limitations, cell-permeable precursors dimethyl-glutamate (DM-glutamate, previously shown to regenerate intracellular stores of glutamate and glutathione)27 and dimethyl-2-oxoglutarate (DM-2-oxoglutarate). When used at a concentration of 7 mM, the two compounds did not regenerate glutamine (Figure S2a) but replenished the intracellular pool of glutamate (Figure S2b), a downstream intermediate of glutamine metabolism and a known precursor of 2-oxoglutarate, citrate, succinate and fumarate in glutamine-deprived cancer cells28,29 When used at a low 2 mM concentration, DM-glutamate and DM-2-oxoglutarate restored the OCR (Figure 2a) and ATP production (Figure 2b) of glutamine-deprived MDA-MB-231 cells. However, they did not restore these parameters in HeLa and SiHa cells (Figures 2a-b), and glycolysis was still depressed in MDA-MB-231 cells (Figure S2c). Neither DM-glutamate nor DM-2-oxoglutarate were capable of restoring the proliferation of glutamine-deprived cells (Figures 2c-d), even when the compounds were used at a 7 mM concentration (Figure S2d). The ability of L-glutamine to support cancer cell proliferation was not shared by D-glutamine (Figures 2e and S2e). Furthermore, in the presence of glutamine, the use of L-asparaginase, which converts L-glutamine to L-glutamate extracellularly,30,31 fully recapitulated the antiproliferative effects of glutamine deprivation at a dose of 1 IU/ml (Figure 2f). Together, these data indicated that glutamine can promote cancer cell proliferation independently of its use in oxidative and reductive glutaminolysis.
Figure 2. Dimethyl-glutamate, dimethyl-2-oxoglutarate, D-glutamine and N-acetyl-glucosamine cannot replace L-glutamine to sustain cancer cell proliferation.
(a-f) MDA-MB-231, HeLa and SiHa cancer cells were cultured in complete medium containing 2 mM of L-glutamine (+Q) or not (-Q), and supplemented or not with dimethyl-glutamate (DM-glutamate, 2 mM), dimethyl-2-oxoglutarate (DM-2-oxoglutarate, 2 mM) or D-glutamine (2 mM). (a) OCR was determined using a Seahorse bioenergetic analyzer (**P < 0.01, ***P < 0.005, compared with media +Q; #P < 0.05, ##P < 0.01, ###P < 0.005 compared with medium -Q; ns = not significant; n = 7 for MDA-MB-231 and HeLa; n = 6 for SiHa). (b) Intracellular ATP content measured using a CellTiter-Glo luminescent assay (**P < 0.01, ***P < 0.005, compared with medium +Q, #P < 0.05, ##P < 0.01, ###P < 0.005 compared with medium –Q; ns = not significant; n = 4 for MDA-MB-231 and SiHa; n = 8 for HeLa). (c) Cell proliferation measured using Ki-67 staining (***P < 0.005, compared with medium +Q, #P < 0.05, ###P < 0.005 compared with medium -Q; ns = not significant; n = 4). (d) Cell number assessed using a SpectraMax i3 multi-mode microplate reader (***P < 0.005, compared with medium +Q; n = 6 for MDA-MB-231 and SiHa; n = 4 for HeLa). (e) as in (d) but with cells supplemented with 2 mM of D-glutamine (***P < 0.005 compared with medium +Q; n = 5 for MDA-MB-231 and SiHa; n = 4 for HeLa). (f) as in (d) but with cells supplemented with 0.25 or 1 IU/ml of asparaginase (***P < 0.005 compared with medium +Q; n = 4). (a-f) All quantitative data show means ± SEM.
In addition of being a metabolic fuel, glutamine via glutamate is a precursor of glutathione, a major endogenous antioxidant in cells. However, supplying exogenous glutathione to glutamine-deprived cancer cells27 did not restore their proliferation rate (Figure S2f) and only marginally improved cell number (Figure S2g). Glutamine also fuels the hexosamine pathway, producing N-acetyl-glucosamine for posttranslational protein O-glycosyslation. However, providing N-acetyl-glucosamine under glutamine deprivation32 did not restore cell proliferation (Figure S2h).
Altogether, these first sets of data indicated that none of the major metabolic pathways of glutamine account for its ability to promote cancer cell proliferation. We therefore envisioned that glutamine could act as a signaling agent/modulator to support cancer cell proliferation.
Glutamine independently sustains basal HIF-1 activity and cancer cell proliferation
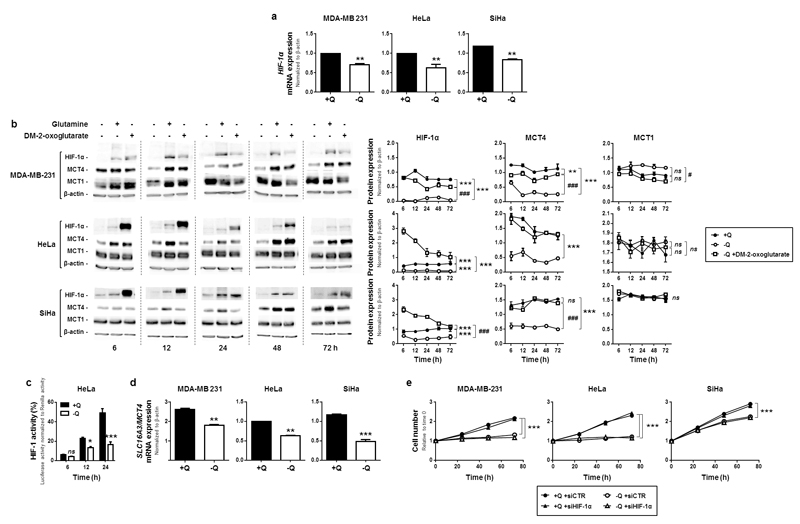
Because glutamine restriction was associated with reduced glycolysis (Figure 1c-d) and glycolysis is important for cell proliferation,26,33 we focused on transcription factor hypoxia-inducible factor-1 (HIF-1) that controls the expression of most glycolytic enzymes and transporters.4,34 HIF-1 is active as a dimer composed of HIF-1α and HIF-1β. While HIF-1β expression is constitutive, HIF-1α expression is finely tuned by transcriptional and posttranscriptional mechanisms.34 We found that glutamine deprivation reduced HIF-1α gene transcription in MDA-MB-231, HeLa and SiHa cancer cells (Figure 3a), which resulted in reduced HIF-1α expression (Figure 3b) and reduced HIF-1 activity (HRE-luciferase reporter assay shown in Figure 3c, where the time-dependent increase in basal HIF-1 activity probably results from hypoxia in the unstirred culture of oxidative HeLa cells).35 Glutamine deprivation reduced the transcription (Figure 3d) and protein expression (Figure 3b) of HIF-1-target MCT45 in the 3 cell lines. Comparatively, despite increased transcription (Figure S3a), the expression of MCT1, which is not under the control of HIF-1,5 was not significantly altered (Figure 3b).
Figure 3. Glutamine controls HIF-1α expression, but HIF-1 does not control the glutamine-dependent proliferation of cancer cells.
(a-d) MDA-MB-231, HeLa and SiHa cancer cells were cultured in complete medium containing 2 mM of L-glutamine (+Q) or not (-Q). (a) HIF-1α mRNA levels were determined after 24 h using qRT-PCR. Data were normalized for β-actin mRNA levels (**P < 0.01; n = 8). (b) Cells in medium -Q were also supplemented or not with dimethyl-2-oxoglutarate (DM-2-oxoglutarate, 2 mM). Representative western blots and graphs depict changes of HIF-1α, MCT4, MCT1 and β-actin expression over time (**P < 0.01, ***P < 0.005, compared with medium +Q; #P < 0.05, ###P < 0.005 compared with medium –Q; ns = not significant; n = 3). (c) HIF-1 activity was measured using a HRE-luciferase reporter in HeLa cells grown in the indicated media for the indicated times (*P < 0.05, ***P < 0.005; ns = not significant; n = 4). (d) SLC16A3/MCT4 mRNA levels were determined using qRT-PCR after 24 h of culture (**P < 0.01, ***P < 0.005; n = 8). (e) Cells were transfected with a control siRNA (siCTR) or a siRNA against HIF-1α (siHIF-1α). Assay media were added 24 h later, and cell number was determined at the indicated times using a SpectraMax i3 multi-mode microplate reader. (***P < 0.005; n = 8 for MDA-MB-231 and SiHa; n = 6 for HeLa). (a-e) All quantitative data show means ± SEM.
To test a causal link between basal HIF-1 activity and glutamine-dependent cancer cell proliferation, we silenced HIF-1α (siHIF-1α) in our model cell lines (target extinction is shown in Figure S3b). Cell proliferation was not affected by siHIF-1α in the presence or absence of glutamine (Figure 3e). These data thus indicated that glutamine exerts independent influences on the proliferation and basal HIF-1 activity of glycolytic and oxidative cancer cells.
That HIF-1 does not control glutamine-dependent cell proliferation was further documented with DM-2-oxoglutarate, which, contrary to endogenous 2-oxoglutarate, activates HIF-1 because it generates high levels of succinate that inhibits HIF prolylhydroxylases (PHDs).36 As expected, DM-2-oxoglutarate inhibited HIF-1 PHDs (ODD-luciferase reporter, Figure S3c), restored HIF-1α protein expression (Figure 3b), the transcriptional activity of HIF-1 (Figure S3d) and MCT4 expression (Figure 3b) in our 3 model cell lines. However, it did not restore cancer cell proliferation upon glutamine deprivation (Figures 2c-d). Thus, in our experimental conditions, cancer cell proliferation was controlled by glutamine independently of HIF-1.
Glutamine deprivation deregulates several oncogenic pathways in cancer cells
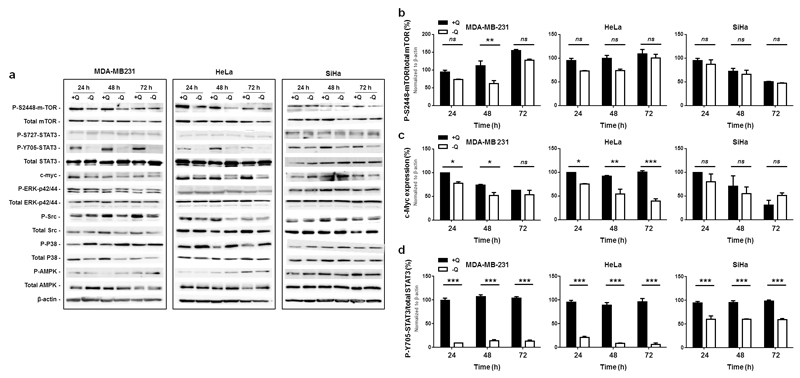
To identify the molecular pathway(s) responsible for the proliferative effects of glutamine, we extended our investigation to several oncogenic pathways known to regulate cancer cell proliferation. We included mammalian target of rapamycin (mTOR) and STAT3 that were previously reported to be under the influence of glutamine: glutamine can activate mTOR through phosphorylation on S2448,37 and mTOR can in turn activate STAT3 by phosphorylation on S72738 (human sequences). Here, western blot analysis revealed that, after 72 h of glutamine starvation, the activities of extracellular signal-regulated kinases (ERKs) (phospho-p42/44 on total p42/44), Src (phospho-Y416-Src/Src), p38 (p-T180/Y182-p38/p38) and AMP-activated protein kinase (AMPK) (phospho-T172-AMPK/AMPK) were only minimally affected (Figure 4a). mTOR phosphorylation on S2448 (p-S2448-mTOR/mTOR) was significantly decreased 48 h after glutamine depletion in MDA-MB-231, but not in HeLa and SiHa cells (Figures 4a&b). Consistently, the phosphorylation of mTOR target p70S6K (p-T389-p70S6K/p70S6K) decreased only in MDA-MB-231 cells (Figure S4a). c-Myc expression was significantly and sustainably decreased in MDA-MB-231 and HeLa, but not in SiHa cells (Figures 4a&c). Comparatively, STAT3 activity (p-Y705-STAT3/STAT3, but not p-S727/STAT3) was highly significantly decreased at all time points in the 3 cell lines (Figures 4a&d).
Figure 4. Glutamine removal deregulates mTOR, c-Myc and STAT3 pathways in cancer cells.
(a-d) MDA-MB-231, HeLa and SiHa cancer cells were cultured for 24, 48 or 72 h in assay medium containing 2 mM of L-glutamine (+Q) or not (-Q). (a) Western blots representative of n = 3 showing the status of several oncogenic pathways. (b) The graph depicts the p-S2448-mTOR/total mTOR ratio (**P < 0.01; ns = not significant; n = 3). (c) The graph depicts c-Myc expression normalized for β-actin (*P < 0.05, **P < 0.01, ***P < 0.005; ns = not significant; n = 3). (d) The graph depicts the p-Y705-STAT3/total STAT3 ratio (***P < 0.005; n = 3). (a-d) All quantitative data show means ± SEM.
STAT3 activation by glutamine controls cancer cell proliferation
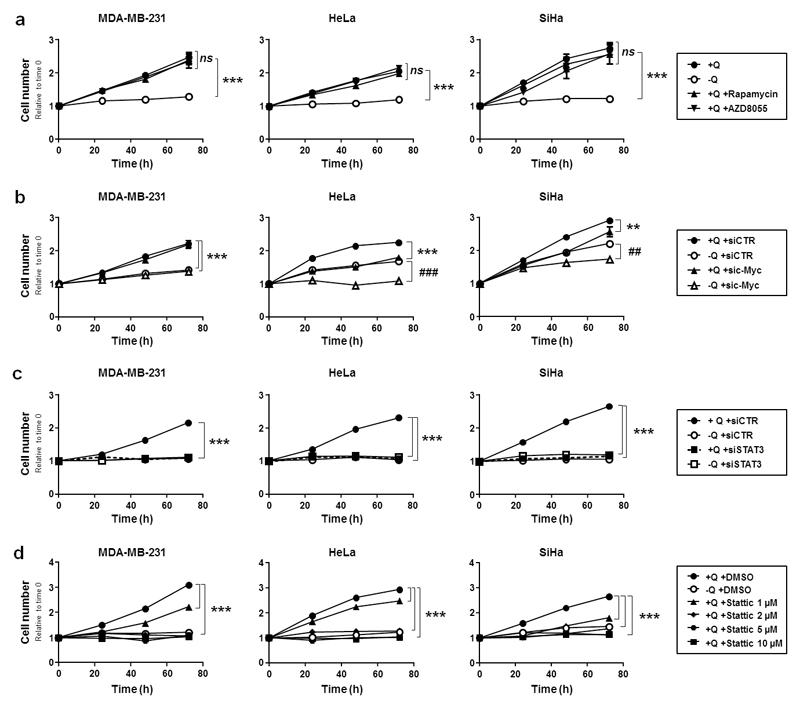
To determine which of these oncogenic pathways controls glutamine-dependent cancer cell proliferation, we targeted mTOR, c-Myc and STAT3 with specific inhibitors and/or siRNAs. Inhibition efficiencies in MDA-MB-231, HeLa and SiHa cells are shown in Figures S4a-d. mTOR inhibition with rapamycin or AZD805539 did not influence the proliferation rate of the cells cultured in the presence of glutamine (Figure 5a). Silencing c-Myc did not affect the proliferation rate of MDA-MB-231 cells (Figure 5b). However, sic-Myc significantly reduced HeLa and SiHa cell proliferation, albeit to the same extent in the presence of glutamine or not. Thus, at the time points that we analyzed, c-Myc and glutamine independently supported cancer cell proliferation. Comparatively, in the presence of glutamine, silencing STAT3 with SmartPool siRNAs (siSTAT3) dramatically repressed the proliferation rate of all 3 cells lines, and the effect was strikingly similar to that seen upon glutamine removal (Figure 5c). Importantly, siSTAT3 exerted no additional antiproliferative effect on the cells when they were glutamine-deprived. Similar to siSTAT3, STAT3 inhibitor Stattic, a specific inhibitor of STAT3 Y705 phosphorylation,40 (Figure S4d) blocked the proliferation of all 3 cell lines in the presence of glutamine (Figure 5d). It did so as efficiently as glutamine removal at doses ≥ 2 µM. These experiments thus unraveled STAT3 as a potential mediator of the effects of glutamine on cancer cells.
Figure 5. STAT3 activation accounts for glutamine-dependent cancer cell proliferation.
(a-d) MDA-MB-231, HeLa and SiHa cancer cells were cultured in complete medium containing 2 mM of L-glutamine (+Q) or not (-Q) during 72 h. Cell number was assessed using a SpectraMax i3 multi-mode microplate reader. (a) The cells were also supplemented with mTOR inhibitors rapamycin (10 nM) or AZD8055 (15 nM) (***P < 0.005, compared with media +Q; ns = non-significant; n = 5). (b) The cells were transfected with a control siRNA (siCTR) or a siRNA against c-Myc (sic-Myc). 24 h later, medium was replaced with medium +Q or medium -Q (**P < 0.01, ***P < 0.005 compared to siCTR +Q; ##P < 0.01, ###P < 0.005 compared to siCTR -Q; n = 8). (c) As in (b) but using SmartPool siRNAs against STAT3 (siSTAT3) (***P < 0.005 compared to siCTR +Q; n = 6 for MDA-MB-231 and HeLa; n = 8 for SiHa). (d) As in (a) but using increasing doses of STAT3 inhibitor Stattic (***P < 0.005 compared to siCTR +Q; n = 6 for MDA-MB-231 and SiHa; n = 5 for HeLa). (a-e) All quantitative data show means ± SEM.
Glutamine activates STAT3 to control cancer cell metabolism and proliferation
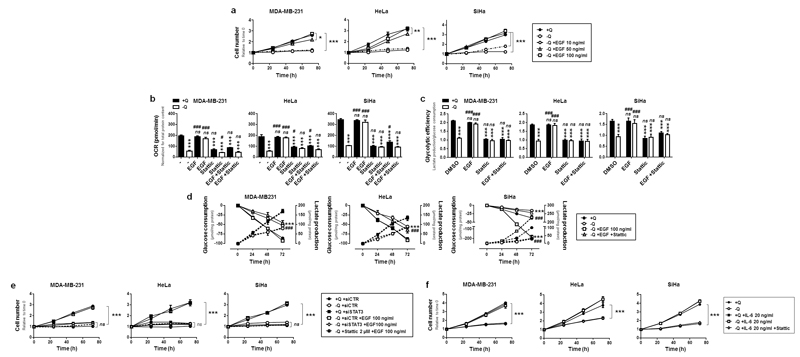
Based on our previous observations, we sought to functionally link glutamine-induced STAT3 activation (Y705 phosphorylation) and cancer cell proliferation. For that aim, we first used epithelial growth factor (EGF), a well-known inducer of STAT3 Y705 phosphorylation41 that did not modify STAT3 S727 phosphorylation in our model cell lines (Figure S5a&b). In the presence of glutamine, EGF did not increase the proliferation rate of MDA-MB-231, HeLa and SiHa cells (Figure S5c). However, in the absence of glutamine, EGF dose-dependently restored proliferation (Figure 6a). It also fully restored OCR (Figure 6b) and glycolytic efficiency (Figure 6c), with no additive effect of glutamine. Both glucose consumption and lactate production were restored (Figure 6d). Most importantly, when EGF was supplied in the absence of glutamine, targeting STAT3 with siSTAT3 or Stattic fully prevented the restoration of cell proliferation (Figure 6e and Figure S5b), OCR (Figure 6b) and glycolytic efficiency (Figure 6c); and EGF did not exert additive effects on glutamine (Figure S5c). Thus, the restoration of proliferative and metabolic parameters by EGF in glutamine-deprived cancer cells was exquisitely dependent on the ability of EGF to activate STAT3 through Y705 phosphorylation.
Figure 6. STAT3 activation restores cancer cell proliferation and metabolism upon glutamine starvation.
(a-f) MDA-MB-231, HeLa and SiHa cancer cells were cultured in complete medium containing 2 mM of L-glutamine (+Q) or not (-Q), and supplemented or not with the indicated amounts of epithelial grow factor (EGF), interleukin-6 (IL-6), and/or with 2 µM of STAT3 inhibitor Stattic. (a) Cell number was assessed using a SpectraMax i3 multi-mode microplate reader at 24, 48 and 72 h (*P < 0.05, **P < 0.001, **P < 0.005 compared with media +Q; n = 6 for MDA-MB-231 and SiHa; n = 4 for HeLa). (b) Where indicated, EGF was used at a dose of 100 ng/ml. OCR was determined in a Seahorse bioenergetic analyzer (***P < 0.005 compared with media +Q; #P < 0.05, ###P < 0.005, compared with media -Q; ns = not significant: n = 8 for MDA-MB-231 and SiHa; n = 6 for HeLa). (c) Glycolytic efficiency calculated as the ratio between lactate production and glucose consumption (***P < 0.005 compared with medium +Q; ###P < 0.005 compared with medium –Q; ns = not significant; n = 4 for MDA-MB-231 and SiHa; n = 3 for HeLa). (d) Glucose consumption (plain lines) and lactate production (dotted lines) measured using a CMA600 enzymatic analyzer (***P < 0.005 when analyzing glucose consumption; ###P < 0.005 when analyzing lactate production; n = 4). (e) The cells were transfected with SmartPool control siRNA (siCTR) or with SmartPool siRNAs against STAT3 (siSTAT3). Media were replaced 24 h later by assay media containing the indicated treatments. Cell number was assessed over time using a SpectraMax i3 multi-mode microplate reader (***P < 0.005 compared to siCTR +Q; ns = not significant compared to siCTR -Q; n = 8). (f) Cells were treated as indicated, and cell number was assessed over time using a SpectraMax i3 multi-mode microplate reader (***P < 0.005 compared with media +Q; n = 8 for MDA-MB-231 and SiHa; n = 6 for HeLa. (a-f) All quantitative data show means ± SEM.
We further verified that STAT3 activation was sufficient to restore the proliferation of glutamine-deprived cancer cells by using interleukin-6 (IL-6), a potent activator of JAK/STAT3 signaling42 (Figure S5d). In the presence of glutamine, IL-6 did not increase the proliferation rate of MDA-MB-231, HeLa and SiHa cells (Figure 6f). However, in the absence of glutamine, IL-6 restored the proliferation of all 3 cancer cell lines, and this effect was inhibited by Stattic. Together, our experiments demonstrated that STAT3 activation is sufficient to restore the proliferation of glutamine-deprived cancer cells.
We next tested whether intracellular glutamine metabolism could mediate the effects of STAT3 on cancer cell proliferation. To definitely exclude a contribution of extracellular glutamine, proliferation rescue experiments were repeated on the cells cultured in glutamine-deprived medium containing dialyzed serum. In these conditions, EGF (Figure S5e) and IL-6 (Figure S5f) still restored cancer cell proliferation in a STAT3-dependent manner. Still, cancer cells deprived of exogenous glutamine can produce intracellular glutamine through protein degradation or de novo synthesis.43,44 However, neither glutaminase inhibitor BPTES (10 µM; Figure S5g) nor SmartPool siRNAs against GDH1 (siGDH; Figure S5h-i), which have previously been functionally validated to inhibit glutamine metabolism,28,45 interfered with the ability of EGF and IL-6 to restore cancer cell proliferation in the absence of extracellular glutamine. In the presence of extracellular glutamine, BPTES and siGDH did not block cancer cell proliferation, further indicating that glutamine metabolism does not account for the STAT3-dependent proliferative effects of glutamine on the cancer cells that we tested.
Finally, we tested whether STAT3 also mediated the effects of glutamine on HIF-1. In the presence of glutamine, Stattic decreased the transcription of HIF-1α (Figure S5j) and MCT4 (Figure S5k), which provided a functional link between STAT3 and basal HIF-1 activity in cancer cells. However, siHIF-1α did not impair the restoration of glutamine-deprived cancer cells proliferation by EGF (Figure S5l). Thus, if on the one hand STAT3 can activate HIF-1, on the other hand HIF-1 is not an effector of STAT3 for the control of glutamine-dependent cancer cell proliferation.
Altogether, our data thus reveal that STAT3 mediates the effects of glutamine on cancer cell proliferation, independently of the need of glutamine to be metabolized and independently of its ability to activate HIF-1.
Discussion
This study aimed to better understand why several cancer cell lines are addicted to glutamine irrespectively of their basal metabolic activities. We report that STAT3 is activated by glutamine and controls cancer cell proliferation independently of the metabolic use of glutamine. We further report that glutamine controls basal HIF-1 activity and cancer cell metabolism, with no direct incidence on cellular proliferation.
In cancer, glutamine is best characterized as a metabolic fuel and as a precursor of glutathione,9 which has led to the general assumption that it exerts most of its protumoral activities as a cataplerotic/anaplerotic substrate (in oxidative and reductive glutaminolysis, as a nitrogen donor and as a substrate of the hexosamine pathway) and as a regulator of the intracellular redox balance (glutathione synthesis and metabolic production of NADPH). These functions of glutamine require its metabolic processing, and did indeed account for the general metabolic depression that we observed in this study using different cancer cell types with different basal metabolic activities (Figures 1 and 2). Accordingly, cell-permeable forms of glutamate and 2-oxoglutarate, which were previously reported to effectively supply glutamate and 2-oxoglutarate intracellularly,27,28 could, to some extent, replace glutamine to support cancer cell metabolism and ATP production. Still, glutamine deprivation had antiproliferative effects that were not rescued by intermediate metabolites of glutamine metabolism (glutamate, 2-oxoglutarate, glutathione and N-acetyl-glucosamine). Proliferation impairment rather primarily depended on the absence of glutamine itself, more precisely on the specific decrease in extracellular L-glutamine availability (Figure S2e). Our further characterization of this paradigm led to the conclusion that, in our experimental conditions, glutamine metabolism does not control glutamine-dependent cancer cell proliferation.
While glutamine can modulate the activity of several transcription factors (including HIF-1, mTOR, c-Myc and STAT3 (this study and references 37,38,46,47)), we found that, among these factors, STAT3 is the only essential mediator of glutamine-dependent cancer cell proliferation. Glutamine activated STAT3 by promoting its phosphorylation on Y705, which was not only necessary for cell proliferation in the presence of glutamine, but also sufficient to restore cell proliferation with EGF and IL-6 in the absence of glutamine (Figure 6e-f and Figure S5b&d). Consequently, Stattic, a specific inhibitor of Y705 STAT3 phosphorylation,40 repressed glutamine-EGF- and IL-6-dependent cancer cell proliferation. Yang et al.46 documented that glutamine can further activate STAT3 through S727 phosphorylation in ovarian cancer cells, which we did not observe in our human breast and cervix cancer cell models. Of note, STAT3 S727-phosphorylation is under the control of mTOR,38 which was barely affected by glutamine deprivation in our experimental models.
STAT3 Y705-phosphorylation is controlled by the EGFR and JAK-STAT pathways.48 At least two possibilities could explain how glutamine activates STAT3. First, one can speculate that glutamine would act intracellularly to activate STAT3, either directly or indirectly following its export in exchange for leucine and other essential amino acids. However, our data argue against this possibility. Indeed, changes in glutamine uptake (use of dialyzed serum in glutamine-free medium) or in glutamine metabolism (use of BPTES, siGDH, DM-glutamate and DM-2-oxoglutarate) did not account for the proliferative effects of STAT3 activation. Thus, it is unlikely that glutamine acts intracellularly to activate STAT3 or to mediate the proliferative effects of STAT3. We therefore consider more likely a second possibility that glutamine could act as a signaling agent/modulator able to activate cell surface receptors that control STAT3. In support of such hypothesis, other metabolites have been previously identified to act on membrane receptors in cancer cells. For example, lactate can activate GPR81 in several cancer cell types,20 glutamate binds to mGluR1 in triple-negative breast cancer cells,49 and the presence of a yet unknown ATP receptor was proposed on colon cancer cells.50 Interestingly, a receptor for L-glutamine, termed Grp, has been identified in bacteria and crystalized in the L-glutamine ligand-bound form.51 No analogue has been disclosed in eukaryotic cells to date, but the possibility of the existence of such receptor in cancer cells certainly deserves further investigation.
Our study does not establish a direct cause-effect relationship between metabolic and proliferation changes in a glutamine-dependent context. However, our data assign a central role to STAT3 in both processes. Indeed, while glutamine starvation downregulated basal oxidative and glycolytic metabolism, STAT3 reactivation by EGF was sufficient to restore both types of metabolism (Figures 6b-d). With respect to aerobic glycolysis, P-Y705-STAT3 was previously reported to activate HIF-1, a master regulator of glycolysis in cancer,34 via Akt and through directly binding to the HIF-1 complex in the cell nucleus.52,53 In addition, as previously observed in melanoma cells exposed to growth factors,54 we report here that, in response to glutamine, STAT3 stimulates the transcription of HIF-1α (Figures S5j). STAT3 can indeed bind to and activate the promoter of the HIF-1α gene.54 HIF-1α, however, was not needed for cancer cell proliferation in the presence of glutamine, did not act as a downstream effector of STAT3 to control cell proliferation, and HIF-1 activation by DM-2-oxoglutarate was not sufficient to restore the proliferation of glutamine-deprived cancer cells. Thus, other regulators of cell metabolism most probably collaborate to couple STAT3-dependent cancer cell proliferation with its associated bioenergetic and biosynthetic needs. Of note, STAT3 can also promote OXPHOS when P-S727-STAT3 relocalizes from the cytosol to the inner mitochondrial membrane where it interacts with Complexes I, II and V of the ETC and with the mitochondrial permeability transition pore, thereby decreasing the production of reactive oxygen species and protecting against cytochrome c release.55 In our experimental models, STAT3 phosphorylation on S727 was detected, but did not change with glutamine availability. This does not preclude a preferential mitochondrial localization of P-S727-STAT3 in the presence of glutamine, but calls for follow-up studies to identify more precisely how STAT3 controls OXPHOS in a glutamine-dependent context.
Our findings could have a significant impact for cancer therapy. Several pharmacological strategies have indeed been developed to starve cancer cells from glutamine, including γ-L-glutamyl-p-nitroanilide (GPNA) that inhibits ASCT2-dependent glutamine uptake,56 BPTES57 that targets glutaminases, and L-asparaginase that has intrinsic glutaminase activity.30 In particular, L-asparaginase is used for the treatment of acute lymphoblastic leukemia, and it blocked cancer cell proliferation to the same extent as glutamine deprivation (Figure 2f). Because the glutaminase activity of L-asparaginase is essential for its cytotoxic activity on leukemic cells,58 the proliferation of which most often requires JAK-STAT3 activation,59 our study suggests that L-asparaginase could inhibit JAK-STAT3 signaling by depleting extracellular glutamine. If confirmed experimentally, this possibility could help to stratify patients for L-asparaginase therapy. Conversely, resistance to drugs inhibiting glutamine uptake and metabolism could depend on growth factors (such as EGF),41 cytokines (such as IL-6)60 and mutations61 that activate receptors responsible for STAT3 Y705-phosphorylation. Because STAT3 can be constitutively activated following mutations61 or stimulated by S727 phosphorylation downstream of the Ras-MAPK pathway,62,63 it would be particularly attractive to test experimentally whether targeting these pathways could increase the response of tumors to inhibitors of glutamine uptake and metabolism.
Conclusively, we report that STAT3 is activated by extracellular glutamine in cancer cells, which directly promotes their proliferation and indirectly sustains the high metabolic activities associated with proliferation. STAT3 activation is independent of glutamine metabolism.
Materials and Methods
Cells and reagents
MDA-MB231 human mammary breast adenocarcinoma (ATCC), HeLa human cervix cancer adenocarcinoma (ATCC) and SiHa human cervix squamous cell carcinoma (ATCC) cells were routinely cultured in DMEM containing 4.5 g/l of glucose and 2 mM of Glutamax, and supplemented with 10% FBS. Assay medium was DMEM without glucose, L-glutamine, phenol red, sodium pyruvate and sodium bicarbonate (D5030, Sigma-Aldrich), reconstituted with 10 mM glucose, 10% FBS, and buffered at pH 7.4 with 3.7 g/L NaHCO3. Cells were grown at 37°C in a humidified 5% CO2 atmosphere. Where indicated, cells were treated with 2 mM of L-glutamine (Invitrogen), 2 mM of dimethyl-L-glutamate (Sigma), 2 mM of dimethyl-2-oxoglutarate (Sigma), 2 mM of D-glutamine (Sigma), 5 mM of reduced glutathione (Merck Millipore),10 nM of rapamycin (Sigma), 15 nM of AZD8055 (Selleckchem), 10-100 ng/ml of recombinant human EGF (PeproTech), 20 ng/ml of recombinant IL-6 (Sigma), 1-10µM of Stattic (Santa Cruz), 10 µM of BPTES or 0.25-1 IU/ml of asparaginase (Sigma). Reagents were dissolved either in DMSO or directly in DMEM according to manufacturer’s indications. When DMSO was used, an equal quantity was also added to control medium. All drugs and reagents were administrated to adherent cells in fresh assay medium.
Metabolic measurements
Oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) were measured using XF Cell Mito Stress Test kit (Seahorse Bioscience) on a Seahorse XF96 bioenergetic analyzer. MDA-MB-231, HeLa and SiHa cells were pre-incubated for 24 h in assay medium. Experiments were performed following manufacturer's instructions. The concentration of glucose and lactate was measured in cell supernatant using specific enzymatic reactions with a CMA600 analyzer (CMA microdialysis) following producer’s instruction. ATP levels were measured using the CellTiter-Glo Luminescent Cell Viability assay from Promega following manufacturer’s instructions on cells that were grown for 24 h in assay medium containing or not L-glutamine (2 mM), dimethyl-L-glutamate (2 mM) or dimethyl-2-oxoglutarate (2 mM). All the above metabolic measurements were normalized for total protein content analyzed at the end of the experiment using the Bio-Rad Protein assay or the Pierce BCA Protein Assay kit (Thermo scientific). The concentration of intracellular glutamine and glutamate was measured in cell lysate using the glutamine/glutamate-Glo Assay kit (courtesy of Promega) following producer’s instruction. Luminescence was measured using a SpectraMax miniMax 300 imaging cytometer, and data were normalized for cell number.
Cell viability and proliferation
Cell number was determined over time from transmitted light images using a SpectraMax miniMax 300 imaging cytometer (Molecular Devices). 5 x 103 cells were plated in black clear bottom 96-well plates (Costar) and treated 24 h later. Cell proliferation was determined using Ki-67 staining. Briefly, cells were cultured for 72 h of culture in assay medium, after which they were immunostained using a mouse monoclonal antibody against Ki-67 (#556003, BD biosciences) and a goat anti-mouse IgG1 secondary antibody (Alexa Fluor 488 conjugate, #A-21121, Invitrogen). Fluorescence of Ki-67-positive cells and total cell number were determined using a SpectraMax miniMax 300 imaging cytometer. Cell proliferation corresponds to the percentage of Ki-67-positive cells on total number analyzed using the SoftMax Pro software.
Western blotting
Western blotting was performed as previously described.8 Briefly, an equal amount of protein extracts were loaded on polyacrylamide gels, separated by SDS-PAGE and then blotted on PVDF membranes. Primary and secondary antibodies are reported in Table S1. Staining was revealed with Amersham Imager 600 (GE Healthcare Life Sciences). Data were analyzed using the Image J software (NIH).
qRT-PCR
Total RNA was isolated from cells using a Maxwell RSC instrument (Promega) following manufacturer’s instructions. qRT-PCR was performed as previously described64 with SYBR green on a ViiA 7417 Real-Time instrument. Primers are reported in Table S2.
HIF-1 and PHD activity
HIF-1 and PHD activities were measured using a dual luciferase kit from Promega with HRE-luciferase (Plasmid #26731, Addgene) as reporter of HIF-1 activity and ODD-luciferase (Plasmid #18965, Addgene) as reporter of PHD activity. pRL Renilla luciferase (Promega) was used as internal control. Cells were co-transfected using the TransIT-2020 transfection reagent (Mirus Bio).
RNA interference
Cells were transfected using Lipofectamine RNAiMAX (Invitrogen) following reverse transfection according to manufacturer’s protocol. siRNAs targeted hHIF-1α 5'-AACUGGACACAGUGUGUUUGA-3' (siHIF-1α) or hc-Myc 5'-GAGAACAGUUGAAACACAA-3' (sic-Myc). ON-TARGETplus SmartPool siRNAs were used to silence hSTAT3 (Dharmacon, #L-003544) and hGLUD1/GDH1 (Dharmacon # L-004032). ON-TARGETplus non-targeting Pool (Dharmacon) or Allstar siRNA (Qiagen) was used as negative control.
Statistics
Data represent means ± SEM. All experiments were repeated at least 3 times independently, and sample size was chosen based on similar experiments in previous publications. SEMs are sometimes smaller than symbols. Similar variance is reflected by small SEMs with equal n between the different conditions of a same experiment. Outliers were identified using Grubb's test. Two-tailed unpaired Student’s t test, one-way ANOVA (Bonferroni post-hoc test) and two-way ANOVA were used where appropriate. P < 0.05 was considered to be statistically significant.
Supplementary Material
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Acknowledgments
Financial Support: This work was supported by a Starting Grant from the European Research Council (ERC No. 243188 TUMETABO), Interuniversity Attraction Pole (IAP) grant #UP7-03 from the Belgian Science Policy Office (Belspo), an Action de Recherche Concertée from the Communauté Française de Belgique (ARC 14/19-058), the Belgian Fonds National de la Recherche Scientifique (F.R.S.-FNRS), the Télévie and the Belgian Fondation contre le Cancer (2012-186). P.S. is a F.R.S.-FNRS Research Associate. A.C. is a Postdoctoral Fellow and M.S. a Research Fellow of the Télévie.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors declare that they have no conflicts of interest.
References
- 1.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci. 2014;39:347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Payen VL, Brisson L, Dewhirst MW, Sonveaux P. Common responses of tumors and wounds to hypoxia. Cancer J. 2015;21:75–87. doi: 10.1097/PPO.0000000000000098. [DOI] [PubMed] [Google Scholar]
- 5.Ullah MS, Davies AJ, Halestrap AP. The plasma membrane lactate transporter MCT4, but not MCT1, is up-regulated by hypoxia through a HIF-1alpha-dependent mechanism. J Biol Chem. 2006;281:9030–9037. doi: 10.1074/jbc.M511397200. [DOI] [PubMed] [Google Scholar]
- 6.Dimmer KS, Friedrich B, Lang F, Deitmer JW, Broer S. The low-affinity monocarboxylate transporter MCT4 is adapted to the export of lactate in highly glycolytic cells. Biochem J. 2000;350(Pt 1):219–227. [PMC free article] [PubMed] [Google Scholar]
- 7.Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Hée VF, Pérez-Escuredo J, Cacace A, Copetti T, Sonveaux P. Lactate does not activate NF-kB in oxidative tumor cells. Front Pharmacol. 2015;6:228. doi: 10.3389/fphar.2015.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mates JM, Segura JA, Martin-Rufian M, Campos-Sandoval JA, Alonso FJ, Marquez J. Glutaminase isoenzymes as key regulators in metabolic and oxidative stress against cancer. Curr Mol Med. 2013;13:514–534. doi: 10.2174/1566524011313040005. [DOI] [PubMed] [Google Scholar]
- 12.Zaganas IV, Kanavouras K, Borompokas N, Arianoglou G, Dimovasili C, Latsoudis H, et al. The odyssey of a young gene: structure-function studies in human glutamate dehydrogenases reveal evolutionary-acquired complex allosteric regulation mechanisms. Neurochem Res. 2014;39:471–486. doi: 10.1007/s11064-014-1251-0. [DOI] [PubMed] [Google Scholar]
- 13.Yang C, Sudderth J, Dang T, Bachoo RM, McDonald JG, DeBerardinis RJ. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 2009;69:7986–7993. doi: 10.1158/0008-5472.CAN-09-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979;254:2669–2676. [PubMed] [Google Scholar]
- 15.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li F, Xu W, Zhao S. Regulatory roles of metabolites in cell signaling networks. J Genet Genomics. 2013;40:367–374. doi: 10.1016/j.jgg.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Chandra V, Hong KM. Effects of deranged metabolism on epigenetic changes in cancer. Arch Pharm Res. 2015;38:321–337. doi: 10.1007/s12272-015-0561-3. [DOI] [PubMed] [Google Scholar]
- 18.Bellance N, Lestienne P, Rossignol R. Mitochondria: from bioenergetics to the metabolic regulation of carcinogenesis. Front Biosci (Landmark Ed) 2009;14:4015–4034. doi: 10.2741/3509. [DOI] [PubMed] [Google Scholar]
- 19.Prickett TD, Samuels Y. Molecular pathways: dysregulated glutamatergic signaling pathways in cancer. Clin Cancer Res. 2012;18:4240–4246. doi: 10.1158/1078-0432.CCR-11-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roland CL, Arumugam T, Deng D, Liu SH, Philip B, Gomez S, et al. Cell surface lactate receptor GPR81 is crucial for cancer cell survival. Cancer Res. 2014;74:5301–5310. doi: 10.1158/0008-5472.CAN-14-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorne JL, Campbell MJ. Nuclear receptors and the Warburg effect in cancer. Int J Cancer. 2015;137:1519–1527. doi: 10.1002/ijc.29012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lincet H, Icard P. How do glycolytic enzymes favour cancer cell proliferation by nonmetabolic functions? Oncogene. 2015;34:3751–3759. doi: 10.1038/onc.2014.320. [DOI] [PubMed] [Google Scholar]
- 23.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Saedeleer CJ, Copetti T, Porporato PE, Verrax J, Feron O, Sonveaux P. Lactate activates HIF-1 in oxidative but not in Warburg-phenotype human tumor cells. PLoS ONE. 2012;7:e46571. doi: 10.1371/journal.pone.0046571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Saedeleer CJ, Porporato PE, Copetti T, Pérez-Escuredo J, Payen VL, Brisson L, et al. Glucose deprivation increases monocarboxylate transporter 1 (MCT1) expression and MCT1-dependent tumor cell migration. Oncogene. 2014;33:4060–4068. doi: 10.1038/onc.2013.454. [DOI] [PubMed] [Google Scholar]
- 26.De Preter G, Neveu MA, Danhier P, Brisson L, Payen VL, Porporato PE, et al. Inhibition of the pentose phosphate pathway by dichloroacetate unravels a missing link between aerobic glycolysis and cancer cell proliferation. Oncotarget. 2016;7:2910–2920. doi: 10.18632/oncotarget.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratnikov B, Aza-Blanc P, Ronai ZA, Smith JW, Osterman AL, et al. Glutamate and asparagine cataplerosis underlie glutamine addiction in melanoma. Oncotarget. 2015;6:7379–7389. doi: 10.18632/oncotarget.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shanware NP, Bray K, Eng CH, Wang F, Follettie M, Myers J, et al. Glutamine deprivation stimulates mTOR-JNK-dependent chemokine secretion. Nat Commun. 2014;5:4900. doi: 10.1038/ncomms5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergstrom J, Furst P, Noree LO, Vinnars E. Intracellular free amino acid concentration in human muscle tissue. J Appl Physiol. 1974;36:693–697. doi: 10.1152/jappl.1974.36.6.693. [DOI] [PubMed] [Google Scholar]
- 30.Hakimi J, Bosmann HB. The intrinsic glutaminase activity of asparaginase: cytotoxicity and cellular resistance. Res Commun Chem Pathol Pharmacol. 1979;26:581–595. [PubMed] [Google Scholar]
- 31.Avramis VI, Tiwari PN. Asparaginase (native ASNase or pegylated ASNase) in the treatment of acute lymphoblastic leukemia. Int J Nanomedicine. 2006;1:241–254. [PMC free article] [PubMed] [Google Scholar]
- 32.Guillaumond F, Leca J, Olivares O, Lavaut MN, Vidal N, Berthezene P, et al. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2013;110:3919–3924. doi: 10.1073/pnas.1219555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Metzen E, Wolff M, Fandrey J, Jelkmann W. Pericellular PO2 and O2 consumption in monolayer cell cultures. Respir Physiol. 1995;100:101–106. doi: 10.1016/0034-5687(94)00125-j. [DOI] [PubMed] [Google Scholar]
- 36.Hou P, Kuo CY, Cheng CT, Liou JP, Ann DK, Chen Q. Intermediary metabolite precursor dimethyl-2-ketoglutarate stabilizes hypoxia-inducible factor-1alpha by inhibiting prolyl-4-hydroxylase PHD2. PLoS ONE. 2014;9:e113865. doi: 10.1371/journal.pone.0113865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yokogami K, Wakisaka S, Avruch J, Reeves SA. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr Biol. 2000;10:47–50. doi: 10.1016/s0960-9822(99)00268-7. [DOI] [PubMed] [Google Scholar]
- 39.Chresta CM, Davies BR, Hickson I, Harding T, Cosulich S, Critchlow SE, et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010;70:288–298. doi: 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- 40.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol. 2006;13:1235–1242. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 41.Zhong Z, Wen Z, Darnell JE., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 42.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 43.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tardito S, Oudin A, Ahmed SU, Fack F, Keunen O, Zheng L, et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat Cell Biol. 2015;17:1556–1568. doi: 10.1038/ncb3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin L, Li D, Alesi GN, Fan J, Kang HB, Lu Z, et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell. 2015;27:257–270. doi: 10.1016/j.ccell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang L, Moss T, Mangala LS, Marini J, Zhao H, Wahlig S, et al. Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Mol Syst Biol. 2014;10:728. doi: 10.1002/msb.20134892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jewell JL, Kim YC, Russell RC, Yu FX, Park HW, Plouffe SW, et al. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science. 2015;347:194–198. doi: 10.1126/science.1259472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siveen KS, Sikka S, Surana R, Dai X, Zhang J, Kumar AP, et al. Targeting the STAT3 signaling pathway in cancer: role of synthetic and natural inhibitors. Biochim Biophys Acta. 2014;1845:136–154. doi: 10.1016/j.bbcan.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 49.Banda M, Speyer CL, Semma SN, Osuala KO, Kounalakis N, Torres Torres KE, et al. Metabotropic glutamate receptor-1 contributes to progression in triple negative breast cancer. PLoS ONE. 2014;9:e81126. doi: 10.1371/journal.pone.0081126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yaguchi T, Saito M, Yasuda Y, Kanno T, Nakano T, Nishizaki T. Higher concentrations of extracellular ATP suppress proliferation of Caco-2 human colonic cancer cells via an unknown receptor involving PKC inhibition. Cell Physiol Biochem. 2010;26:125–134. doi: 10.1159/000320518. [DOI] [PubMed] [Google Scholar]
- 51.Kumarevel T, Nakano N, Ponnuraj K, Gopinath SC, Sakamoto K, Shinkai A, et al. Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding. Nucleic Acids Res. 2008;36:4808–4820. doi: 10.1093/nar/gkn456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Q, Briggs J, Park S, Niu G, Kortylewski M, Zhang S, et al. Targeting Stat3 blocks both HIF-1 and VEGF expression induced by multiple oncogenic growth signaling pathways. Oncogene. 2005;24:5552–5560. doi: 10.1038/sj.onc.1208719. [DOI] [PubMed] [Google Scholar]
- 53.Pawlus MR, Wang L, Hu CJ. STAT3 and HIF1alpha cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells. Oncogene. 2014;33:1670–1679. doi: 10.1038/onc.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niu G, Briggs J, Deng J, Ma Y, Lee H, Kortylewski M, et al. Signal transducer and activator of transcription 3 is required for hypoxia-inducible factor-1alpha RNA expression in both tumor cells and tumor-associated myeloid cells. Mol Cancer Res. 2008;6:1099–1105. doi: 10.1158/1541-7786.MCR-07-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poli V, Camporeale A. STAT3-mediated metabolic reprograming in cellular transformation and implications for drug resistance. Front Oncol. 2015;5:121. doi: 10.3389/fonc.2015.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hassanein M, Hoeksema MD, Shiota M, Qian J, Harris BK, Chen H, et al. SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin Cancer Res. 2013;19:560–570. doi: 10.1158/1078-0432.CCR-12-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson MM, McBryant SJ, Tsukamoto T, Rojas C, Ferraris DV, Hamilton SK, et al. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) Biochem J. 2007;406:407–414. doi: 10.1042/BJ20070039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parmentier JH, Maggi M, Tarasco E, Scotti C, Avramis VI, Mittelman SD. Glutaminase activity determines cytotoxicity of L-asparaginases on most leukemia cell lines. Leuk Res. 2015;39:757–762. doi: 10.1016/j.leukres.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Munoz J, Dhillon N, Janku F, Watowich SS, Hong DS. STAT3 inhibitors: finding a home in lymphoma and leukemia. Oncologist. 2014;19:536–544. doi: 10.1634/theoncologist.2013-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Catlett-Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 61.Groner B. Determinants of the extent and duration of STAT3 signaling. JAKSTAT. 2012;1:211–215. doi: 10.4161/jkst.21469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 63.Gough DJ, Koetz L, Levy DE. The MEK-ERK pathway is necessary for serine phosphorylation of mitochondrial STAT3 and Ras-mediated transformation. PLoS ONE. 2013;8:e83395. doi: 10.1371/journal.pone.0083395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Corbet C, Draoui N, Polet F, Pinto A, Drozak X, Riant O, et al. The SIRT1/HIF2alpha axis drives reductive glutamine metabolism under chronic acidosis and alters tumor response to therapy. Cancer Res. 2014;74:5507–5519. doi: 10.1158/0008-5472.CAN-14-0705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.