Abstract
Chemokine receptors may share common ligands, setting up potential competition for ligand binding, and association of activated receptors with downstream signaling molecules such as β-arrestin. Determining the “winner” of competition for shared effector molecules is essential for understanding integrated functions of chemokine receptor signaling in normal physiology, disease, and response to therapy. We describe a dual-color click beetle luciferase complementation assay for cell-based analysis of interactions of two different chemokine receptors, CXCR4 and ACKR3, with the intracellular scaffolding protein β-arrestin 2. This assay provides real-time quantification of receptor activation and signaling in response to chemokine CXCL12. More broadly, this general imaging strategy can be applied to quantify interactions of any set of two proteins that interact with a common binding partner.
1. INTRODUCTION
Signaling by chemokine receptors, like most other receptors and signal transduction pathways, relies upon regulated formation and dissociation of protein complexes. A single chemokine may bind to two different chemokine receptors, initiating distinct signaling pathways and biologic outputs. Preferential binding of a chemokine ligand to one of two or more competing receptors can determine activation of specific downstream signaling pathways in addition to magnitude and duration of signaling. Inhibiting chemokine binding to one receptor partner may increase availability of the chemokine ligand to signal through another receptor, potentially altering responses to therapy and contributing to drug resistance. Understanding dynamics of signaling by two different chemokine receptors in response to a common chemokine ligand requires analysis of formation and dissociation of complexes of signaling proteins in physiologic environments. While methods such as immunoprecipitation and immunofluorescence can detect association of multiple proteins, such techniques typically are performed at a limited number of fixed time points, precluding real-time analysis, and quantification of signaling.
Protein fragment complementation assays provide a facile approach to detect and quantify protein interactions in signaling pathways in intact cells and animal models, complementing established biochemical assays (Luker & Luker, 2011). Several different protein fragment complementation assays have been developed, including strategies based on fluorescent proteins, metabolic enzymes, and luciferases (Vidi & Watts, 2009). These assays all are based upon splitting a reporter protein into two inactive fragments (amino (N)- and carboxy (C)-termini) that do not or very minimally reassemble spontaneously. N- and C-terminal reporter fragments then are fused to proteins of interest. When fused proteins of interest interact, N- and C-terminal reporter fragments reconstitute a functional reporter protein. Protein fragment complementation assays based on luciferase enzymes provide a particularly powerful approach to quantify dynamics of protein interactions in chemokine signaling. Unlike fluorescence complementation, luciferase complementation does not require maturation time before producing bioluminescence from interacting proteins, and luciferase complementation also is reversible. Luciferase complementation also provides a large dynamic range of signal with low background activity, and the assay format is compatible with moderate- and high-throughput technologies.
Luciferase complementation assays typically have been used to quantify the magnitude and kinetics of interactions between a single pair of proteins fused to N- and C-terminal fragments of luciferases such as firefly, Renilla, or Gaussia (Luker et al., 2012, 2004; Paulmurugan & Gambhir, 2003; Remy & Michnick, 2006). However, these strategies cannot analyze two different proteins competing for interaction with a common protein partner as occurs commonly in nodes of signaling pathways. To accomplish this goal, we have leveraged a recently described dual-color luciferase complementation assay based on green and red spectral variants of click beetle luciferase (Coggins et al., 2014; Villalobos et al., 2010). In the dual-color click beetle luciferase complementation assay, N-terminal fragments of click beetle green and red luciferases, respectively, interact with a C-terminal fragment shared by both N-terminal fragments. N-terminal fragments determine the wavelength of bioluminescence produced by complementation. By using emission filters to separate light from complemented green and red luciferases, investigators can quantify interactions of two different proteins with a shared partner in the same population of cells.
In this chapter, we describe methods we have used to quantify interactions between CXCR4 and ACKR3 (formerly designated CXCR7) with the common intracellular scaffolding protein, β-arrestin 2, in cells that coexpress both receptors. Both CXCR4 and ACKR3 share chemokine CXCL12 as a common ligand, although ACKR3 binds CXCL12 with approximately 10-fold higher affinity (Burns et al., 2006). CXCR4 signals through both G protein and β-arrestin pathways, while ACKR3 biases signaling to β-arrestin-mediated outputs (Rajagopal et al., 2010). Using dual-color click beetle complementation, we demonstrated that CXCL12 preferentially signals through ACKR3 in cells that coexpress this receptor with CXCR4, thereby biasing signaling toward β-arrestin 2 (Coggins et al., 2014). While we describe methods for CXCR4 and ACKR3 interacting with β-arrestin 2, the general approach for dual-color click beetle luciferase complementation can be applied readily to other receptors or protein interactions in chemokine signaling pathways.
2. METHODS
The dual-color click beetle luciferase complementation assay we describe is designed to quantify pair-wise interactions between two proteins, such as receptors CXCR4 and ACKR3, with a common binding partner, β-arrestin 2. Intracellular C-termini of CXCR4 and ACKR3 are fused to N-terminal fragments of click beetle green (CBGN) or click beetle red (CBRN) luciferases. The N-terminal fragments of each enzyme determine the color of bioluminescence produced by complementation with the common C-terminal fragment of click beetle luciferase (CLuc), the latter of which is fused to β-arrestin 2. From previous studies, we had determined that fusing the C-terminus of β-arrestin 2 to CLuc produced optimal induction of signaling in response to chemokine CXCL12 (Luker, Gupta, & Luker, 2008). When designing a new complementation assay, we always check all logical orientations of fusions of CBGN, CBRN, and CLuc with proteins of interest.
The overall workflow of this chapter proceeds from generating fusions between proteins of interest and click beetle luciferase fragments and then describes the protocol for cell-based imaging with these constructs to quantify magnitude and kinetics of protein interactions in chemokine signaling.
2.1 Complementation Reporter Constructs
We describe the overall approach for generating click beetle luciferase complementation reporters for live-cell bioluminescence imaging of chemokine receptor signaling. We refer readers to other standard texts, such as Methods in Molecular Biology, for detailed protocols for molecular biology procedures including PCR and ligations.
2.1.1 Required Materials
cDNA constructs for interacting proteins of interest.
Plasmids for CBGN and CBRN luciferases (Promega) or plasmids with N-terminal (amino acids 1–413) or C-terminal (395–542) fragments of these enzymes. Fragments for click beetle luciferase complementation were selected based on comparable fragments identified for optimal firefly luciferase complementation (Luker et al., 2004; Villalobos et al., 2010). The N-terminal fragment determines the emission spectrum of light from complementation with the common C-terminal fragment.
Vectors with constitutive or inducible (optional) promoters for expression in mammalian cells.
Lentiviral vectors for expressing gene of interest and packaging proteins (optional).
Reagents and equipment for PCR, restriction digests, and DNA ligations.
Reagents to validate expression of fusion proteins, such as antibodies for flow cytometry and/or Western blotting.
2.1.2 Generate Constructs for Click Beetle Luciferase Complementation Fusion Proteins
Design cloning strategy to fuse intracellular C-termini of chemokine receptors to NLuc fragments of CBGN and red luciferases, respectively, and the common CLuc fragment to β-arrestin 2 (Fig. 1). This cloning strategy is based on quantifying interactions of two different receptors, such as CXCR4 and ACKR3, with a common partner protein, such as β-arrestin 2. NLuc fragments must be fused to the C-termini of receptors for complementation with other intracellular targets and pair-wise determinations of each receptor interacting with β-arrestin 2 based on color of bioluminescence. For new complementation assays, we design and test fusions of luciferase fragments positioned at all spatially relevant N- and C-termini of target proteins. The same general approach can be used to analyze interactions of CBGN and red NLuc fusions of any two proteins of interest with a common binding partner fused to CLuc.
We generally add a linker sequence, such as (GGGGS)2, between the protein of interest and luciferase fragments to reduce steric hindrance. A linker is not necessarily required, and other linker sequences and lengths may be optimal for other protein interaction. Investigators may need to test different linker sequences and fusion constructs to optimize ligand-dependent induction of bioluminescence signal (Chichili, Kumar, & Sivaraman, 2013).
Using appropriate molecular biology procedures, generate plasmid constructs with fusion proteins for desired interacting proteins, such as receptors and β-arrestin 2 in this example. We prepare fusion proteins with all logical orientations of NLuc and CLuc fragments on N- and C-termini of target proteins with the goal of optimizing ligand-induced bioluminescence signal above background and negative control interactions (Fig. 2).
Produce plasmids with relevant control constructs for background association of CBGN and CBRN fragments with CLuc. Appropriate control constructs should be matched to spatial localization and size of interacting proteins of interest. Possible controls include mismatched receptors, proteins with mutations in amino acids necessary for protein interaction, and/or other membrane bound and cytosolic proteins.
Verify integrity of plasmids with fusion proteins by DNA sequencing.
If needed, transfer click beetle luciferase fusion proteins to plasmids for constitutive or inducible (optional) mammalian expression. Inducible expression systems, since as tetracycline regulated promoters, can be useful to investigate how different levels of CBGN or CBRN fusions affect signaling. Using vectors with selectable markers, such as drug-resistance genes or coexpressed fluorescent proteins, facilitate identification of transiently and stably transfected cells. To facilitate repeated experiments using cells with well-characterized expression of reporter constructs, we generate stably transduced cells via lentiviral transduction. We commonly use vectors in which the luciferase complementation reporter is coexpressed via an internal ribosomal entry site or P2A site with a drug-resistance marker or fluorescent protein. This strategy improves the ability to obtain a population in which all cells express desired reporter proteins. We work with batch populations of cells to avoid potential artifacts of selecting individual clones on biologic responses of cells.
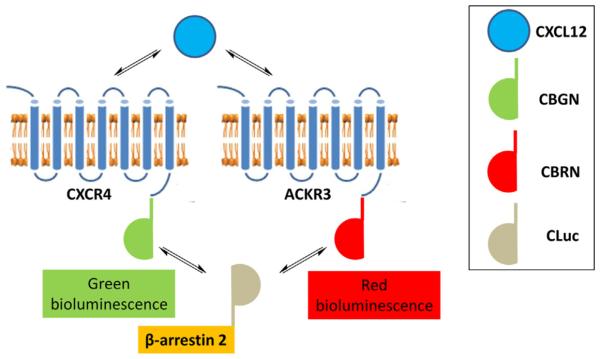
Figure 1.
Dual-color luciferase complementation assay for CXCL12 signaling to β-arrestin 2 via CXCR4 or ACKR3. CXCL12 binding to CXCR4 fused to CBGN drives recruitment of β-arrestin 2-CLuc to produce green (light gray in the print version) bioluminescence, while CXCL12 signaling through ACKR3 fused to CBRN produces red (gray in the print version) bioluminescence upon binding to β-arrestin 2. Measuring bioluminescence from green (light gray in the print version) versus red (gray in the print version) click beetle luciferases provides enables real-time analysis of relative signaling through CXCR4 versus ACKR3 in cells coexpressing both receptors. Figure modified from Wu, Xie, Zhao, Nice, and Huang (2012).
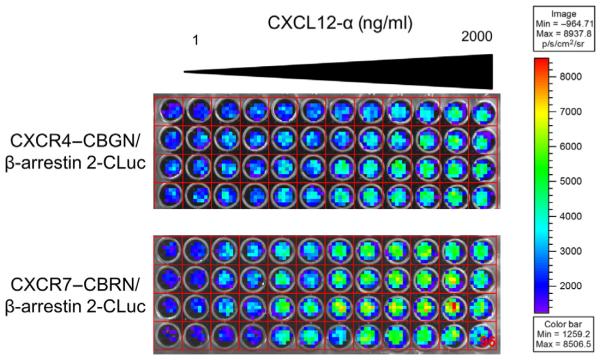
Figure 2.
Cell-based click beetle green and red complementation assay for CXCL12-dependent activation of CXCR4–CBGN and CXCR7 (ACKR3)–CBRN association β-arrestin 2-CLuc. MDA-MB-231 breast cancer cells coexpressing CXCR4–CBGN and β-arrestin 2-CLuc or CXCR7 (ACKR3)–CBRN and β-arrestin 2-CLuc were seeded at 1.5 × 104 cells per well in black wall 96-well plates. We incubated cells with vehicle only (far left column) or increasing concentrations of CXCL12-α (1–2000 ng/ml). The figure shows a representative bioluminescence image of green (top) and red (bottom) bioluminescence obtained 12 and 14 min, respectively, after adding CXCL12. The grid overlay is used to quantify photons per well by region-of-interest analysis. Bioluminescence is depicted as a pseudocolor display with red and blue defining high and low values for photon flux.
2.2 Dual-Color Bioluminescence Live-Cell Imaging
2.2.1 Required Materials
Supplies for culturing mammalian cells, including media, sterile culture flasks/dishes, and temperature controlled incubators.
HEK-293T cells or other cell line that can be transfected readily for initial testing of constructs and preparation of recombinant viral vectors.
Established cell lines or primary cells of interest for specific biologic question(s).
Tissue culture treated sterile multiwell plates (96- or 384-well) with black sides, clear bottom, and lid (corning or other vendor). Black sides prevent light generated from one well from contaminating light produced by an adjacent well.
Sterile pipette tips (low adherence coating preferred) for dispensing reagents.
Standard and multichannel pipettes.
Sterile phosphate-buffered saline (PBS) solution.
Chemokine ligand for receptor(s) of interest prepared as a stock solution. For chemokine CXCL12 (R&D Systems or other vendor), we prepare a 50 μg/μl stock in PBS with 0.2% Probumin brand BSA (Millipore) stored at −20 °C or lower. It is important to select a BSA carrier that does not contain proteases that can degrade CXCL12 or other chemokine.
d-luciferin (Promega or other vendor) prepared as sterile-filtered stock solution at 15 mg/ml in PBS and stored at −20 °C.
10. Bioluminescence imaging instrument with high sensitivity and software for data quantification and analysis (IVIS, Perkin-Elmer, or similar equipment).
2.2.2 Live-Cell Imaging
When developing an assay for new interacting proteins, we first test all orientations (N-terminal and C-terminal fusions as relevant) of CBGN and red NLuc fusions paired with the common interacting CLuc fusion by transient transfection in 293T cells. Investigators may use other cell lines that transfect readily. We also include appropriate control fusion constructs in these tests. The objectives of the initial assays with transient transfections are to (1) identify optimal orientations of fusion proteins for imaging based on greatest ligand-dependent induction of signal above control and (2) verify expression of fusion proteins by means such as Western blotting or flow cytometry. These data inform optimal orientations of NLuc and CLuc fusion proteins to use for stable cell lines. The basic assay protocol detailed in subsequent steps is the same for transiently transfected or stable cell lines.
Plate cells at a density of 1 × 104–2 × 104 cells per well in 100 μl complete growth medium with serum in black walled, clear bottom 96-well plates for tissue culture. Since complementation of click beetle luciferase fragments produces less bioluminescence than intact CBGN or red, we typically use 96-well rather than 384-well plates for cell-based assays. This approach allows us to use shorter acquisition times in each emission channel and improves resolution for kinetics of signaling. For 384-well plate assays, we use 3 × 103–5 × 103 cells per well. We typically culture cells for two days in 96-well plates before assays.
On the day of the assay, gently aspirate medium from wells and use a multichannel pipette to add 50 μl per well of phenol red free DMEM medium with 0.2% probumin 30 min before imaging. The added medium should be equilibrated to 37 °C in a tissue culture incubator before adding to cells. We use phenol red free DMEM to reduce absorption of emitted bioluminescence. The 0.2% probumin serves as a carrier for added chemokine, such as CXCL12, and provides a brief period of serum starvation prior to assays. We typically add any inhibitors at desired concentrations during this 30 min incubation. Return cells to the 37 °C incubator.
To produce consistent, physiologic signaling responses, it is essential to minimize cooling of cells in 96-well plates during medium exchanges and addition of reagents. When we remove cells from the tissue culture incubator, we place plates on an insulated surface, such as the lid of a Styrofoam box or absorbent bench pad, rather than directly on a counter top. We also work with plates on a counter immediately adjacent to the tissue culture incubator. Adding reagents at room temperature rather than prewarmed to 37 °C slows reaction kinetics and increases variability among experiments.
Using a multichannel pipette, add 7 μl per well of D-luciferin from a 15 mg/ml stock and incubate at 37 °C for 5 min. Adding D-luciferin begins the bioluminescence reaction from any preformed protein complexes, so it is necessary to add this and subsequent reagents as rapidly as possible.
Immediately before imaging, add 14 μl per well of phenol red free DMEM with 0.2% probumin and desired concentrations of chemokine ligand, such as CXCL12. For CXCL12, we typically use 0–2000 ng/ml. We typically use 4–6 replicate wells per experimental condition. It is important to have a set of wells treated with only vehicle control to use for normalizing imaging data (see Section 2.3).
Immediately place plate in bioluminescence imaging instrument at the smallest field of view that will include all samples of interest. For interactions of CXCR4 and ACKR3 with β-arrestin 2, we acquire 20 images with large binning and 2 min exposure, alternating between 530–550 and 690–710 nm emission filters (IVIS Spectrum, Perkin-Elmer). Imaging parameters should be programmed in advance of placing the plate into the instrument to minimize delays in beginning imaging. Acquisition times will vary for other interacting proteins depending on the amount of bioluminescence produced by complementation. Total duration of imaging also will vary based on kinetics of a signaling pathway of interest. As alternatives to wavelengths listed above, filters that collect light <550 nm (green) and >680 nm (red) provide acceptable separation of CBGN and red bioluminescence.
Cells may be collected at the end of the assay for subsequent analysis by Western blotting or other biochemical assays as needed.
2.3 Data Analysis
Quantify bioluminescence in each well over time by region-of-interest analysis software with the imaging instrument. For IVIS instruments, analyze imaging signal as photon flux rather than counts to correct for any differences in image acquisition time among different experiments. Any wells with saturated pixels cannot be quantified accurately and should be excluded from analysis. Subsequent studies should use shorter image acquisition times to avoid this problem.
Normalize ligand-dependent changes in bioluminescence over time (without or with inhibitors) to corresponding signal from untreated controls at the same time point to account for any changes in bioluminescence due to availability of D-luciferin substrate.
ACKNOWLEDGMENT
This work was supported by NIH Grants R01CA170198 and R01CA196018.
REFERENCES
- Burns J, Summers B, Wang Y, Melikian A, Berahovich R, Miao Z, et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. The Journal of Experimental Medicine. 2006;203:2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichili V, Kumar V, Sivaraman J. Linkers in the structural biology of protein–protein interactions. Protein Science. 2013;22:153–157. doi: 10.1002/pro.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggins N, Trakimas D, Chang S, Ehrlich A, Ray P, Luker K, et al. CXCR7 controls competition for recruitment of beta-arrestin 2 in cells expressing both CXCR4 and CXCR7. PLoS One. 2014;9:e98328. doi: 10.1371/journal.pone.0098328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker K, Gupta M, Luker G. Imaging CXCR4 signaling with firefly luciferase complementation. Analytical Chemistry. 2008;80:5565–5573. doi: 10.1021/ac8005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker G, Luker K. Luciferase protein complementation assays for bioluminescence imaging of cells and mice. Methods in Molecular Biology. 2011;680:29–43. doi: 10.1007/978-1-60761-901-7_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker K, Mihalko L, Schmidt B, Lewin S, Ray P, Shcherbo D, et al. In vivo imaging of ligand receptor binding with Gaussia luciferase complementation. Nature Medicine. 2012;18:172–177. doi: 10.1038/nm.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker K, Smith M, Luker G, Gammon S, Piwnica-Worms H, Piwnica-Worms D. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulmurugan R, Gambhir S. Monitoring protein-protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Analytical Chemistry. 2003;75:1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal S, Kim J, Ahn S, Craig S, Lam C, Gerard N, et al. Beta-arrestin- but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:628–632. doi: 10.1073/pnas.0912852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy I, Michnick S. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nature Methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- Vidi P, Watts V. Fluorescent and bioluminescent protein-fragment complementation assays in the study of G protein-coupled receptor oligomerization and signaling. Molecular Pharmacology. 2009;75:733–739. doi: 10.1124/mol.108.053819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalobos V, Naik S, Bruinsma M, Dothager R, Pan M, Samrakindi M, et al. Dual-color click beetle luciferase heteroprotein fragment complementation assays. Chemistry & Biology. 2010;17:1018–1029. doi: 10.1016/j.chembiol.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Xie N, Zhao X, Nice E, Huang C. Dissection of aberrant GPCR signaling in tumorigenesis—A systems biology approach. Cancer Genomics Proteomics. 2012;9:37–50. [PubMed] [Google Scholar]