Abstract
The ATR checkpoint kinase coordinates cellular responses to DNA replication stress. Budding yeast contain three activators of Mec1 (the ATR orthologue); however, only TOPBP1 is known to activate ATR in vertebrates. We identified ETAA1 as a replication stress response protein in two proteomic screens. ETAA1-deficient cells accumulate double-strand breaks, sister chromatid exchanges, and other hallmarks of genome instability. They are also hyper-sensitive to replication stress and have increased frequencies of replication fork collapse. ETAA1 contains two RPA-interaction motifs that localize ETAA1 to stalled replication forks. It also interacts with several DNA damage response proteins including the BLM/TOP3α/RMI1/RMI2 and ATR/ATRIP complexes. It binds ATR/ATRIP directly using a motif with sequence similarity to the TOPBP1-ATR activation domain; and like TOPBP1, ETAA1 acts as a direct ATR activator. ETAA1 functions in parallel to the TOPBP1/RAD9/HUS1/RAD1 pathway to regulate ATR and maintain genome stability. Thus, vertebrate cells contain at least two ATR activating proteins.
DNA replication is challenged by difficult to replicate sequences, DNA damage, and collisions with transcriptional machinery. DNA damage response (DDR) pathways respond to replication stress to maintain genome stability, and DDR defects cause developmental disorders and cancer1,2.
Replication protein A (RPA) binds and protects single-stranded DNA (ssDNA) at stalled replication forks3. It also recruits DDR proteins such as ATRIP which is part of the ATR/ATRIP checkpoint kinase complex4–6. ATR is then activated by a direct interaction with TOPBP17, which requires RHINO, and the RAD9/RAD1/HUS1 (911) and MRE11/RAD50/NBS1 (MRN) complexes for its ATR-activating function8–10. In budding yeast, there are at least three Mec1ATR-activating proteins including Dpb11 (a TOPBP1 orthologue), Ddc1, and Dna211–14. As yet, TOPBP1 is the only known ATR activator in mammals.
A second RPA interacting protein at stalled forks is the BLM helicase15. Mutations in BLM cause Bloom syndrome16, and BLM-deficient cells suffer from chromosomal abnormalities such as an increase in sister chromatid exchanges (SCEs)17,18. BLM functions with Topoisomerase IIIα, RMI1, and RMI2 (BTR complex) to generate non-cross over products during recombination19–24. TOPBP1 interacts with BLM and regulates its ability to prevent SCEs through a mechanism reported to be independent of its ATR activating function25,26.
ETAA1 is an uncharacterized protein that derives its name from a study of Ewing tumor antigens27. Additionally, ETAA1 was identified as an ATM/ATR substrate28, and genome wide association studies found single nucleotide polymorphisms at the ETAA1 locus increase pancreatic cancer risk29,30. We find that ETAA1 is a replication stress response protein that localizes to stalled forks via a direct interaction with RPA. It also interacts with several other DDR proteins including ATR/ATRIP and the BTR complexes. ETAA1 maintains genome integrity by activating ATR using a motif that has sequence similarity to the TOPBP1 ATR-activation domain (AAD). Furthermore, ETAA1 acts in a distinct pathway from TOPBP1.
Results
ETAA1 is an RPA-interacting protein that localizes to stalled forks
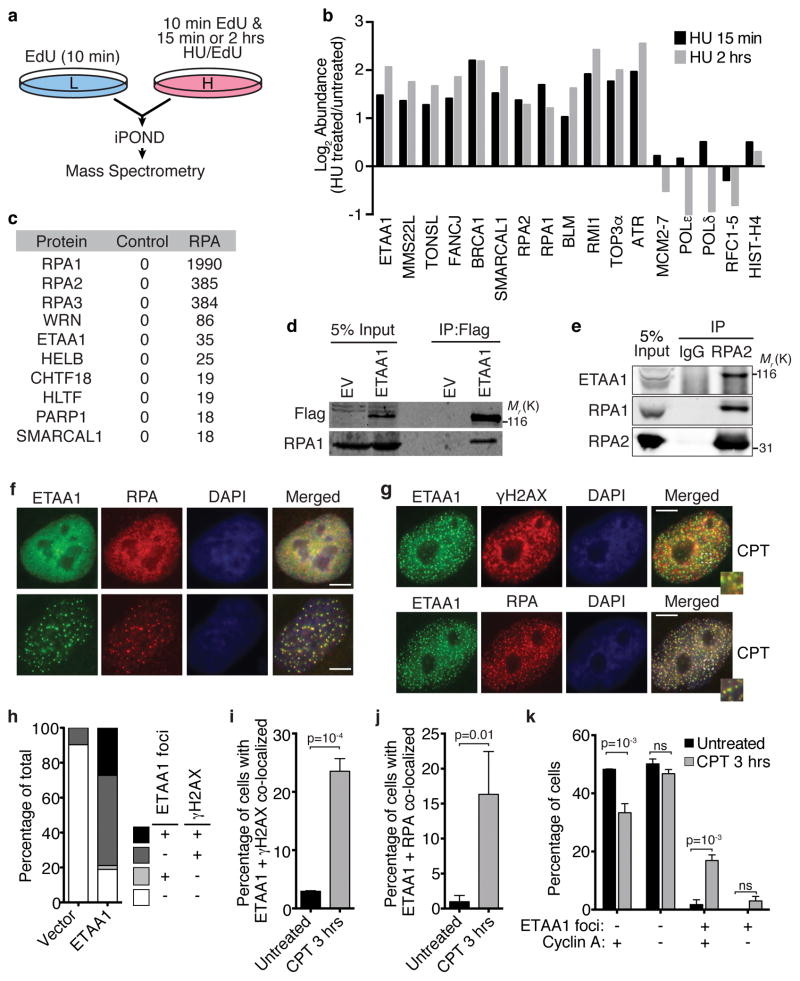
We recently conducted a proteomic screen utilizing iPOND (isolation of Proteins On Nascent DNA) combined with quantitative mass spectrometry to identify proteins enriched at stalled replication forks31. Samples treated with hydroxyurea (HU) for 15 minutes or two hours were compared to untreated cells (Fig. 1a). 72 proteins are significantly enriched at the HU-stalled forks compared to elongating forks at these times31. These include known DDR proteins like ATR, RPA, BLM, SMARCAL1, BRCA1, FANCJ, MMS22L, and TONSL as well as ETAA1 (Fig. 1b).
Figure 1. ETAA1 is enriched at stalled replication forks and interacts with RPA.
(a,b) HEK293T cells grown in heavy isotope media and incubated with EdU and HU were compared to EdU-labeled cells grown in light isotope media. Replication fork proteins were isolated and detected by iPOND and mass spectrometry. (b) The log2 of the average abundance ratio for selected proteins or complexes is depicted. The full iPOND-MS dataset is presented elsewhere31. (c) Flag-RPA1 was immunopurified from HEK293T nuclear extracts and interacting proteins were identified by mass spectrometry. The table indicates the number of peptides identified for each protein. The control sample was an immunopurification from untransfected cells. The mass spectrometry experiment was performed once. (d) HEK293T cells were transfected with a Flag-ETAA1 or empty expression vector (EV), and nuclear extracts used for immunoprecipitation with Flag antibodies. Immunoprecipitates were immunoblotted with the indicated antibodies after separation by SDS-PAGE. Representative blots from one of five independent experiments are shown. (e) Nuclear extracts from HEK293T cells were used for immunoprecipitation with RPA2 or control IgG antibodies followed by immunoblotting. Representative blots from one of two independent experiments are shown. (f–h) U2OS cells were transiently transfected with a Flag-ETAA1 expression vector and stained and imaged for Flag-ETAA1, RPA, and γH2AX. Scale bars are 5 μm. In (g) cells were treated with 100 nM camptothecin (CPT) for 3 hrs. (i–k) Stable cell lines expressing GFP-Flag-ETAA1 were sorted by flow cytometry to select the 10% of cells expressing the lowest levels, stained for Flag-ETAA1, RPA, γH2AX, and cyclin A as indicated, and scored for focal co-localization before and after treatment with 100 nM CPT. Error bars are SEM from n=3 experiments; student’s, two-tailed, unpaired t-test. Unprocessed original scans of blots in d and e are shown in Supplementary Fig. 8 and source data for i–k are available in Supplemental Table 1.
A proteomic screen to identify RPA-interacting proteins also identified ETAA1 (Fig. 1c). We validated the interaction by co-immunoprecipitation of RPA with Flag-ETAA1 (Fig. 1d). Additionally, RPA2 co-immunoprecipitates with endogenous ETAA1 (Fig. 1e).
While a previous study identified ETAA1 on the cell surface or in the cytoplasm27, we found ETAA1 is localized exclusively in the nucleus. Overexpressed Flag-ETAA1 localizes to intranuclear foci in approximately 25% of cells and is diffusely pan-nuclear in others (Fig. 1f). When it is localized to foci, ETAA1 co-localizes with RPA (Fig. 1f). Overexpressed ETAA1 is also localized in foci when cells are treated with agents that cause replication stress including camptothecin (CPT), cisplatin (CISP), and hydroxyurea (HU) (Fig. 1g, Supplemental Fig. 1a). These ETAA1 foci also co-localize with RPA and partially co-localize with γH2AX.
The untreated cells overexpressing ETAA1 with focal localization almost invariably contained elevated γH2AX levels, and many of the cells with pan-nuclear ETAA1 also contain elevated γH2AX suggesting that ETAA1 overexpression stimulates DNA damage signaling (Fig. 1h). To better assess how ETAA1 localizes without overexpression, we generated stable cell lines by lentiviral infection and used fluorescence activated cell sorting to select for the 10% of cells with the lowest GFP-ETAA1 levels. In over 95% of these cells, ETAA1 is localized diffusely throughout the nucleoplasm, but after treatment with CPT, it localizes to nuclear foci that also contain RPA and γH2AX (Fig. 1i,j). Most cells with ETAA1 foci also contained cyclin A indicating they are in S or G2 phase (Fig. 1k). We conclude that ETAA1 overexpression induces DNA damage signaling and ETAA1 focal accumulation, but that when it is expressed at lower levels it is primarily recruited to replication foci in response to stress.
ETAA1 binds two RPA domains to recruit it to damaged forks
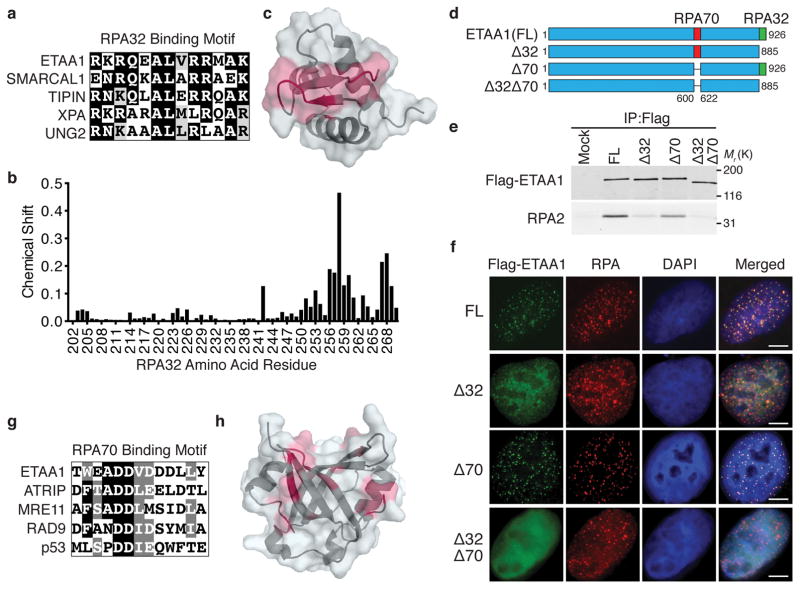
To test whether the interaction with RPA recruits ETAA1 to stalled forks, we first examined a series of ETAA1 fragments for their ability to co-immunoprecipitate RPA. RPA binding is largely restricted to an ETAA1 fragment containing amino acids 571–926 (Supplemental Fig. 1b,c). Sequence alignments identified an evolutionarily conserved motif within this fragment consisting of amino acids 900–912 that closely resembles the RPA32C binding motif of other RPA32C-interacting proteins including SMARCAL1 and TIPIN32,33 (Fig. 2a). An NMR chemical shift perturbation approach demonstrated that this ETAA1 motif binds directly to the same surface of RPA32C as previously observed for other RPA32C-interacting proteins33,34 (Fig. 2b,c).
Figure 2. ETAA1 interacts with RPA through the RPA32C and RPA70N domains.
(a) Sequence alignment of ETAA1900–912 with the RPA32C binding motif of other RPA32C-interacting proteins. (b) Plot of RPA32C chemical shift perturbations induced by the binding of the ETAA1 peptide calculated from 15N-1H HSQC NMR spectra of 15N-labeled RPA32C obtained in the absence and presence of peptide. (c) Map of RPA32C residues with chemical shift perturbations greater than one standard deviation (SD) above the mean (red) on the structure of RPA32C (PDBID 4OU0). (d) Schematic diagram of ETAA1 mutants examined in e and f. (e) Nuclear extract from HEK293T cells mock transfected or transfected with ETAA1 expression constructs were used for immunoprecipitation with Flag antibodies; FL, full length. Immunoprecipitates were immunoblotted with Flag or RPA2 antibodies. (f) U2OS cells were transfected with the indicated ETAA1 expression vectors and treated with 100 nM CPT for 3 hrs prior to examining ETAA1 and RPA localization. Scale bars are 5 μm. (g) Sequence alignment of ETAA1599–611 with the RPA70N-interaction motif of other RPA70N-interacting proteins. (h) Map of RPA70N residues with chemical shift perturbations greater than one SD above the mean (red) on the structure of RPA70N (PDBID 2B29) calculated from NMR 15N-1H HSQC spectra of 15N-labeled RPA70N obtained in the absence and presence of ETAA1 peptide. Unprocessed original scans of blots in e are shown in Supplementary Fig. 8. All panels are representative of two experiments.
Deletion of the ETAA1 RPA32C interaction motif (ETAA1Δ32) greatly reduced, but did not eliminate its ability to associate with RPA and localize to foci (Fig. 2d–f, Supplemental Fig. 1f). Knockdown of RPA70 in cells expressing ETAA1Δ32 abrogated this residual localization suggesting an additional RPA-interaction surface (Supplemental Fig. 1g). Indeed, fragments of ETAA1 containing either residues 600–678 or 574–724 co-immunoprecipitate RPA; whereas ETAA1 fragments containing residues 2–569, 623–885 or 623–724 do not (Supplemental Fig. 1b,d,e), thereby narrowing the interacting motif to amino acids 600–623. This region has sequence homology to the RPA70N-interacting peptides of ATRIP, MRE11, RAD9, and p5335 (Fig. 2g). NMR chemical shift mapping with this ETAA1 peptide indicates that it directly binds the basic cleft in RPA70N (Fig. 2h). Deletion of this motif in ETAA1 (ETAA1Δ70) caused a slight reduction in RPA co-immunoprecipitation, and modest impairment in localization to RPA foci (Fig. 2e,f, and Supplemental Fig. 1f). Deletion or mutation of both RPA binding motifs largely abolished RPA co-immunoprecipitation and eliminated focal accumulation (Fig. 2e,f, and Supplemental Fig. 1f,h). Thus, ETAA1 interacts with both the 70N and 32C domains of RPA, and these interactions recruit ETAA1 to stalled replication forks.
ETAA1 is a replication stress response protein
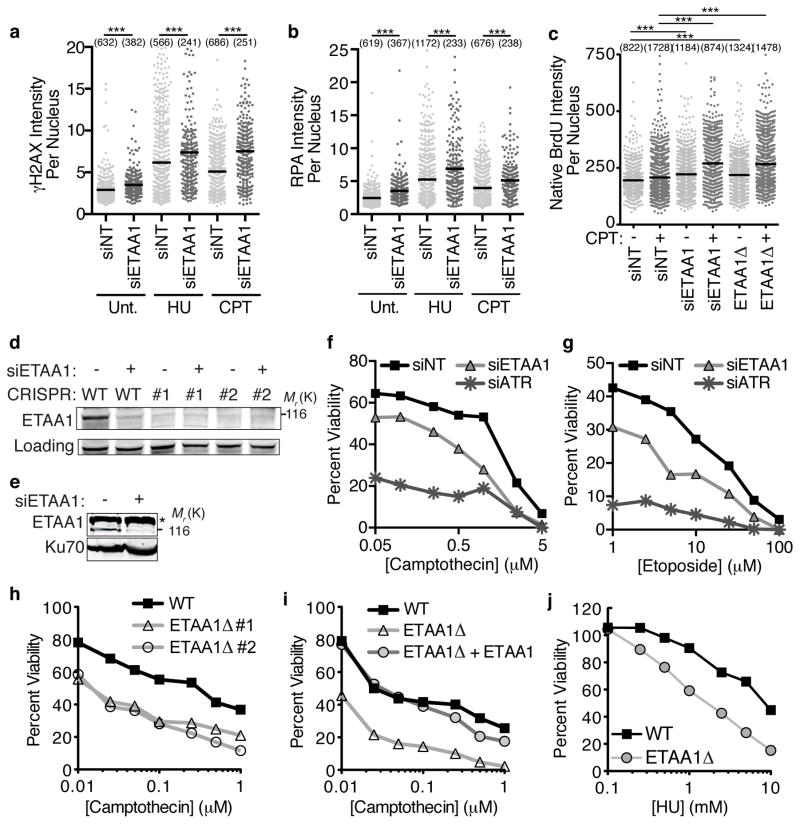
To determine if ETAA1 has an essential function in the replication stress response, we examined the consequences of ETAA1 gene silencing. Even in untreated U2OS cells, ETAA1 knockdown caused an increase in the appearance of DNA damage markers including increased γH2AX and chromatin-associated RPA (Fig. 3a,b). These differences were more pronounced in cells challenged with either HU or CPT, and happened in multiple cancer cell lines including HeLa, H157, and BT549 (Supplemental Fig. 2a,b). The increase in chromatin-associated RPA suggested that there may be additional ssDNA in ETAA1-deficient cells. Indeed, staining with BrdU antibodies in non-denaturing conditions confirmed this ssDNA increase (Fig. 3c).
Figure 3. Loss of ETAA1 results in increased DNA damage and sensitivity to DNA damaging agents.
(a,b) U2OS cells transfected with non-targeting (NT) or ETAA1 siRNAs were left untreated (Unt), or treated with 2 mM HU or 100 nM CPT for 3 hrs. Soluble proteins were extracted with detergent prior to fixation. γH2AX and RPA intensity were quantified by immunofluorescence imaging. (c) U2OS cells transfected with siRNA or ETAA1Δ U2OS cells were labeled with BrdU for 24 hrs and then treated with CPT for 3 hrs as indicated. Cells were fixed and stained with BrdU antibodies in non-denaturing conditions to measure ssDNA levels. In a–c the intensity of each nucleus and mean intensity from a representative experiment of at least two independent experiments is shown... Significance was determined by the Mann-Whitney test. ***p<0.001 The numbers avove each sample indicates the n value, which represents the number of nuclei imaged. (d) Immunoblot to confirm ETAA1 siRNA knockdown and gene deletion. A cross-reacting protein that migrates at a similar position as ETAA1 is observed in some ETAA1 immunoblots. (e) Nuclear extracts were prepared from U2OS cells transfected with pooled ETAA1 siRNAs and ETAA1 was detected by immunoblotting after SDS-PAGE. Star denotes cross-reacting protein. (f–g) U2OS cells were transfected with NT, ETAA1, or ATR siRNAs and treated with CPT or etoposide for 24 hrs. Viability compared to untreated cells was measured 72 hrs after initial addition of drug. Untreated cell viability was set at 100%. (h–j) Wild-type or ETAA1Δ U2OS cells were treated with CPT or HU for 24 hrs and viability was measured as in f and g. In i ETAA1Δ cells stably expressing wild type ETAA1 were also examined. In all viability assays, the mean viability from three technical replicates of a representative experiment is graphed. Three biological replicates were completed for all panels except h, which was repeated twice. Unprocessed original scans of blots in d are shown in Supplementary Fig. 8 and source data for a,c,b,f,g,h,i, and j are in Supplemental Table 1.
ETAA1 knockdown resulted in hypersensitivity to CPT as well as etoposide (Fig. 3d–g). This phenotype is not due to off-target effects since multiple siRNAs cause hypersensitivity (Supplemental Fig. 2c). Furthermore, we generated ETAA1Δ cells using CRISPR-Cas9 and again found that two independent knockout cell lines were hypersensitive to CPT and contain elevated ssDNA levels (Fig. 3c,d,h). Additionally, expression of a wild-type ETAA1 cDNA in ETAA1Δ cells was able to complement this defect (Fig. 3i). ETAA1-deficient cells are also hypersensitive to HU (Fig. 3j). Hypersensitivity is not limited to ETAA1-deficient U2OS cells as knockdown of ETAA1 in most other cell types also caused hypersensitivity to both CPT and HU (Supplemental Fig. 2 d–k). However, we did not observe increased sensitivity to ionizing radiation, cisplatin, or the PARP inhibitors Olaparib or BMN673 in ETAA1-deficient cells (Supplemental Fig. 2l–o).
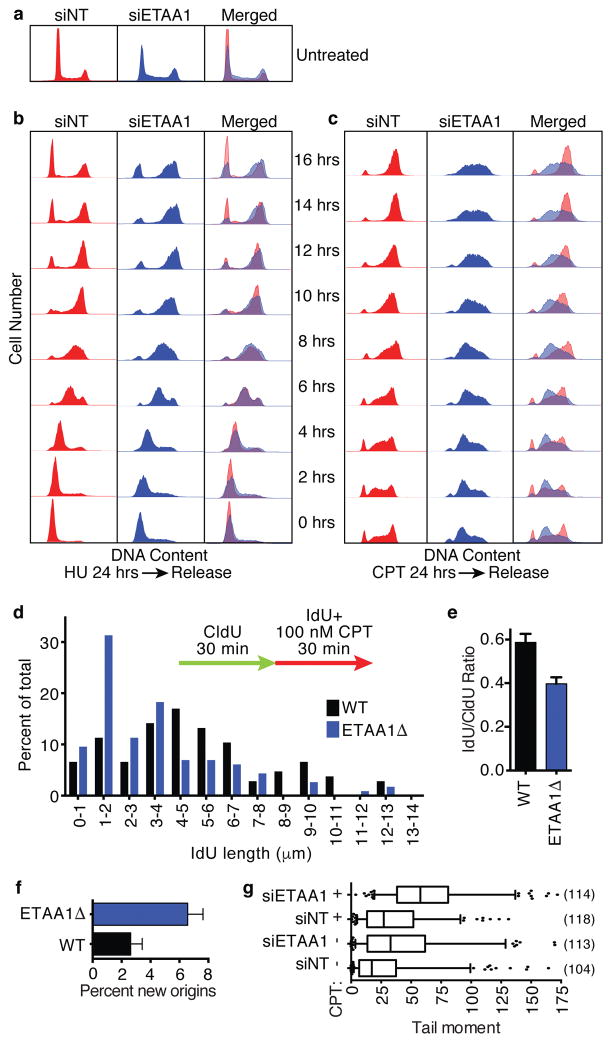
In the absence of added genotoxic stress, ETAA1-deficient cell populations exhibited slightly higher percentages of cells with greater than 2n DNA content compared to controls (Fig. 4a) consistent with some difficulty in DNA replication. After an HU challenge, control cells rapidly resume DNA synthesis and complete the cell division cycle by 16 hrs. ETAA1 knockdown resulted in a slightly slower recovery with fewer cells able to complete the cell division cycle (Fig. 4b). The differences between control and ETAA1-deficient cells were even more pronounced upon treatment with CPT. These cells accumulated in early to mid S-phase, and were largely unable to complete DNA synthesis after removing CPT (Fig. 4c).
Figure 4. ETAA1 is needed to recover from replication stress.
(a–c) U2OS cells were transfected with non-targeting or ETAA1 siRNAs and left untreated (a) or challenged with 2 mM HU (b) or 100 nM CPT (c) for 24 hrs. After 24 hrs, drug was removed and samples were collected every 2 hrs for 16 hrs. Collected cells were fixed, stained with propidium iodide, and DNA content was measured by flow cytometry. Data is representative of two experiments. (d–e) Wild-type or ETAA1 knockout cells were labeled with CldU and IdU and treated with 100 nM CPT as indicated during the second labeling period. DNA fibers stretched on a microscope slide were stained with IdU and CldU antibodies, imaged, and the lengths of fiber tracks measured. n=107 and 116 fibers for WT and ETAA1Δ respectively. One of two biological replicates is shown. (f) The percent of new origins (red only fibers) were also quantitated. n=500 fibers for WT and 508 fibers for ETAA1Δ. (g) U2OS cells transfected with non-targeting or ETAA1 siRNAs and left untreated or treated with 1 μM CPT for 1 hr were subjected to a neutral comet assay to measure double-strand breaks. The box depicts 25–75%, whiskers are 5–95%, and the line is the median value. The numbers of comets measured (n values) from one of two independent experiments are indicated. Source data for d,e,f and g is in Supplemental Table 1.
To confirm that ETAA1-deficient cells have difficulty in DNA replication in response to replication stress, we performed DNA fiber labeling experiments. Elongation rates in unchallenged ETAA1Δ and control U2OS cells are similar (0.21±0.01 μm/min and 0.20±0.01 μm/min respectively). ETAA1Δ cells treated with CPT exhibit significant shortening of replication track lengths compared to controls consistent with increased fork collapse (Fig. 4d,e). We also observed an increase in origin firing in ETAA1Δ cells (Fig. 4f). Furthermore, neutral comet assays indicate that ETAA1-deficient cells contain elevated levels of double-strand breaks with and without added replication stress (Fig. 4g). Thus, we conclude that ETAA1 is needed to maintain replication fork stability.
ETAA1 interacts with multiple DDR including ATR and BLM
ETAA1 lacks any predicted domain structure other than a potential coiled-coil motif. To determine if it exerts its genome maintenance functions through protein-protein interactions we immunopurified Flag-ETAA1 and identified ETAA1-interacting proteins by mass spectrometry. As expected, all three subunits of RPA were observed in the ETAA1 immunopurifications (Supplemental Fig 3a). In addition, ETAA1 complexes contain many proteins that act at damaged replication forks including all four subunits of the BTR complex, both subunits of the ATR/ATRIP checkpoint kinase complex, BRCA1, BRCA2, HLTF, FANCM and FANCJ.
We validated that overexpressed ETAA1 interacts with BLM, HLTF, BRCA1, BRCA2 and ATR/ATRIP in co-immunoprecipitation experiments (Supplemental Fig. 3b,c). In contrast, we did not observe FANCD2 in the ETAA1 immunoprecipitates. ETAA1 co-fractionates with BLM, TOP3α, and RMI1 over a size exclusion column (Supplemental Fig. 3d). Some ATR, RPA2 and HLTF also co-fractionate in these high molecular weight complexes. Thus, ETAA1 likely participates in one or more large DDR protein complexes.
ETAA1 activates ATR
Since ETAA1 interacts with ATR and ATRIP we next asked whether it participates in the ATR signaling pathway by examining ATR substrate phosphorylation. We observe a modest decrease in RPA phosphorylation in U2OS cells transfected with ETAA1 siRNA (Fig. 5a). This difference cannot be explained by decreased RPA association with damaged replication forks in ETAA1-deficient cells since there is actually an increase in chromatin-associated RPA (Fig. 3b). Quantitation of multiple experiments confirmed the differences in RPA phosphorylation in two clones of ETAA1Δ HEK293T cells (Fig. 5b,c). Decreased RPA phosphorylation following ETAA1 knockdown was also observed in HeLa, HCT116, H157, BT549, and A549 cells (Supplemental Fig. 4). In contrast to RPA, ATR-dependent CHK1 phosphorylation was largely unaffected by ETAA1 inactivation (Fig. 5a,d and Supplemental Fig. 4). There is a change in ETAA1 protein migration and detection on immunoblots following treatment with CPT consistent with it being an ATM/ATR substrate (Supplemental Fig. 4)28.
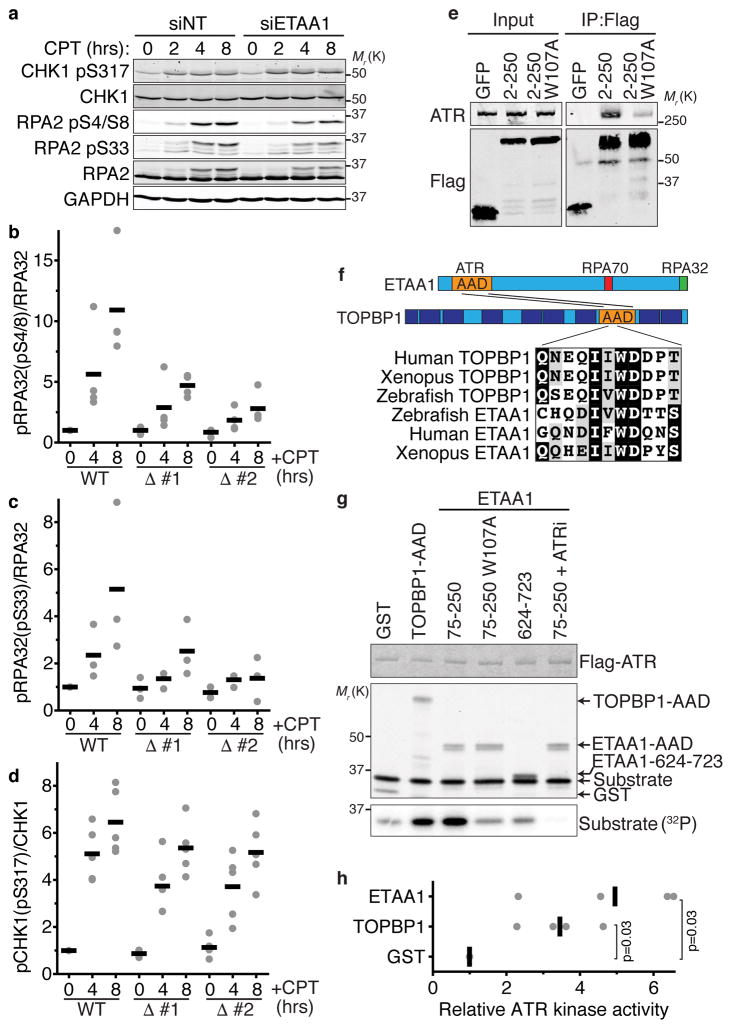
Figure 5. ETAA1 activates ATR.
(a) U2OS cells transfected with non-targeting or ETAA1 siRNAs or (b–d) wild-type and two ETAA1Δ HEK293T cell clones were treated with 100 nM CPT for 2, 4, or 8 hrs. Cell lysates were separated by SDS-PAGE and immunoblotted with the indicated antibodies. The blots in a are representative of two experiments. (b–d) The amount of phosphorylated versus total RPA and CHK1 from n=4, 3, and 5 experiments in b, c, and d respectively is shown. Black bars are the mean. (e) Immunoprecipitates of GFP-Flag-NLS-tagged ETAA1 fragments expressed in HEK293T cells were immunoblotted for Flag and ATR. Representative blots from one of two independent experiments are shown. (f) Schematic diagram of the ETAA1 protein indicating the ATR-interacting domain and evolutionarily conserved sequence similarity with the TOPBP1-AAD. (g) Purified ATR/ATRIP complexes were incubated with GST-TOPBP1 or ETAA1 proteins purified from E. coli, substrate, and γ-32P-ATP. The kinase reactions were separated by SDS-PAGE prior to immunoblotting or quantitating substrate phosphorylation by phosphorimaging. ATRi, ATR inhibitor. (h) ATR kinase activity relative to the control reaction of n=4 independent experiments is graphed. Significance was calculated with the Mann-Whitney test. Unprocessed original scans of blots in a, e and g are shown in Supplementary Fig. 8, and source data for b, c, d, and h is provided in Supplemental Table 1.
To understand whether the defective RPA phosphorylation in ETAA1-deficient cells is due to a defect in ATR regulation, we mapped the ATR-binding motif in ETAA1 to an N-terminal region (Fig. 5e). Sequence analysis identified a highly evolutionarily conserved tryptophan (residue 107) accompanied by a short region of sequence similarity to the ATR-activation domain (AAD) of TOPBP1 within this region (Fig. 5f). In TOPBP1 this tryptophan is essential to bind and activate ATR7. Mutation of ETAA1 W107 to alanine reduced the ability of ETAA1 to co-immunoprecipitate ATR (Fig. 5e).
Based on the similarity to TOPBP1 and the reduction in RPA phosphorylation in ETAA1-deficient cells, we considered the possibility that ETAA1 acts as a direct ATR activator. Indeed, like the TOPBP1 AAD, an ETAA1 fragment containing amino acids 75–250 purified from E. coli strongly activates ATR in vitro (Fig. 5g,h). This activation requires W107, other fragments of ETAA1 do not stimulate ATR, and an ATR inhibitor eliminates the kinase activity indicating specificity. Thus, ETAA1 contains an AAD within amino acids 75–250, and ETAA1 is a direct ATR activator like TOPBP1.
We hypothesized that the increased DNA damage signaling that we observed in cells overexpressing ETAA1 (Fig. 1) could be due to ectopic activation of ATR like what happens upon TOPBP1-AAD overexpression7,36,37. It is also possible that ETAA1 overexpression could cause damage by interfering with RPA function yielding an RPA-exhaustion-like phenotype38. To test these ideas, we overexpressed various ETAA1 proteins and measured γH2AX. The high level of γH2AX induced by wild-type ETAA1 is reduced but not eliminated by mutation of the RPA-interacting motifs (Supplemental Fig. 5a,c,e). These proteins are expressed at least 10-fold higher than the stable ETAA1 expressing cell lines that lack spontaneous ETAA1 foci (Supplemental Fig. 5f). A fragment of ETAA1 missing the ETAA1 AAD (aa251–926) does not cause γH2AX despite its ability to localize to RPA-foci (Supplemental Fig. 5b–d).
The ETAA1 AAD by itself is sufficient to induce γH2AX when highly overexpressed and the level of γH2AX induced is correlated with its expression level (Supplemental Fig. 5b,c,e,g). In contrast, the ETAA1 AAD containing the W107A mutation largely does not induce damage signaling (Supplemental Fig. 5b,c,h). Thus, we conclude that overexpression of ETAA1 promotes DNA damage signaling by binding both RPA and ATR, and high levels of expression of just the ETAA1 AAD is sufficient to activate ATR.
ETAA1 requires its RPA-interaction and ATR-activation domains to maintain genome stability
We next tested whether the ETAA1 replication stress response functions are dependent on its ability to bind RPA and activate ATR. First, we complemented the ETAA1Δ cells with wild-type or RPA binding mutant GFP-ETAA1 cDNA. Cells were sorted to select for the 10% of cells with the lowest expression of GFP-ETAA1 proteins (Supplemental Fig. 6a,b). While wild-type ETAA1 fully complements the CPT hypersensitivity of ETAA1Δ cells, the ETAA1-ΔRPA expressing cells remain modestly hyper-sensitive (Supplemental Fig. 6c). Furthermore, ETAA1Δ cells also exhibited significantly higher levels of genome instability as measured by micronuclei formation, which could be rescued by wild-type but not RPA-binding defective ETAA1 (Supplemental Fig. 6d). Thus, we conclude that RPA binding is needed for ETAA1 to maintain genome stability.
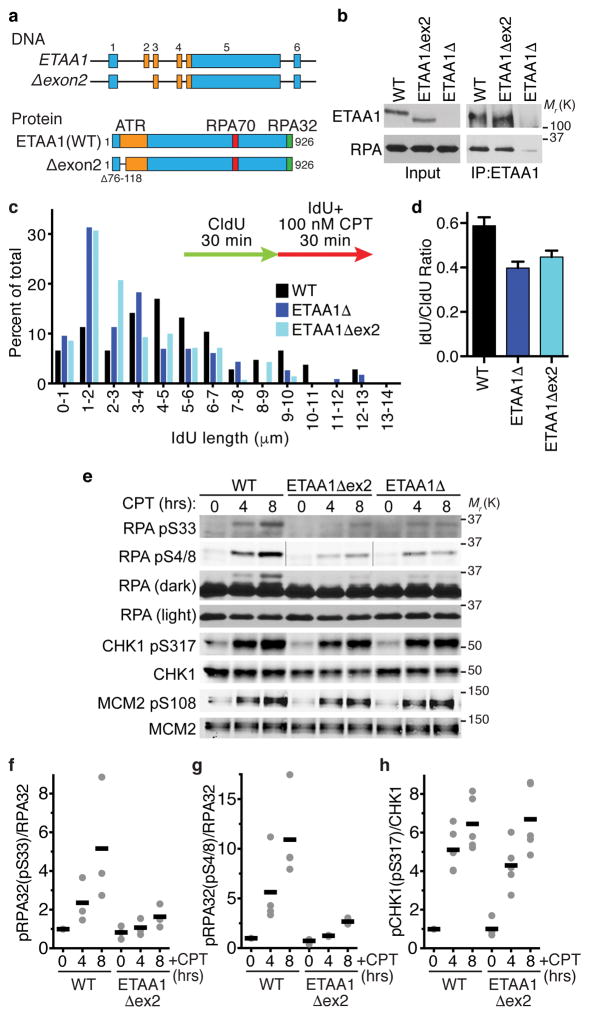
To examine the cellular functions of the ETAA1 AAD and avoid problems caused by ETAA1 overexpression we devised a strategy to delete the AAD in the endogenous ETAA1 gene locus. A portion of the AAD including W107 is encoded by ETAA1 exon 2. Cas9-mediated deletion of exon 2 using two guide RNAs spanning the 5’ intron-exon boundary results in the production of an ETAA1 mRNA in which exon 1 splices to exon 3. This mutation maintains the open reading frame and results in the expression of an ETAA1Δexon2 protein missing residues 76–118 that remains capable of binding RPA (Fig. 6a,b).
Figure 6. The ETAA1 ATR activation domain is needed to maintain fork stability and promote ATR signaling.
(a) Schematic of the ETAA1Δexon2 gene and protein. (b) ETAA1 immunoprecipitates from wild-type, ETAA1Δ, and ETAA1Δexon2 cells were immunoblotted for ETAA1 and RPA. Data are representative of two experiments. (c–d) Cells were labeled with CldU and IdU and treated with 100 nM CPT during the second labeling period. DNA fibers stretched on a microscope slide were stained with IdU and CldU antibodies, imaged, and the lengths of fiber tracks measured. n=107, 116, and 105 fibers for WT, ETAA1Δ, and ETAA1Δexon2 respectively. One of two biological replicates is shown. (e–h) Wild-type, ETAA1Δ, and ETAA1Δexon2 HEK293T cells were treated with 100 nM CPT for 0, 4, or 8 hrs. Cell lysates were immunoblotted for phosphorylated and total RPA and CHK1. Black bars are the mean from n=3, 4, and 5 experiments in f, g, and h respectively. The wild-type and ETAA1Δ data presented in panels c and d of this figure are the same as in figure 4 since the wild-type, ETAA1Δ, and ETAA1exon2 cells were compared in the same experiments. Unprocessed original scans of blots in b and e are shown in Supplementary Fig. 8, and source data for c,d,f,g, and h are provided in Supplemental Table 1
ETAA1Δexon2 cells show a very similar reduction in DNA replication track lengths as the ETAA1Δ cells in response to a CPT challenge (Fig. 6c,d). They also exhibit a reduction in RPA phosphorylation (Fig. 6e,f,g), although there is no obvious defect in CHK1 or MCM2 phosphorylation (Fig. 6e,h).
ETAA1-deficient cells exhibit elevated levels of SCEs
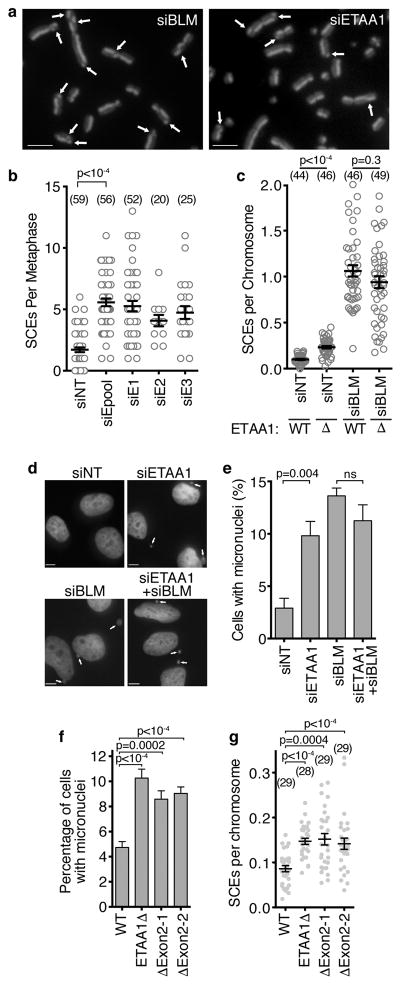
Many of the other proteins that ETAA1 associates with are involved in recombination-based repair mechanisms. However, we did not observe a significant hypersensitivity of ETAA1-deficient cells to ionizing radiation (Supplemental Fig. 2l) or PARP inhibition (Supplemental Fig.2n,o). The interaction of ETAA1 with both HLTF and the BTR complex was of interest since the budding yeast orthologue of HLTF (RAD5) promotes template switching, while the BLM orthologue (SGS1) helps to dissolve these repair intermediates to prevent crossovers39. The human BTR complex also promotes non-crossover repair outcomes during replication fork repair20,23,40. Thus, a defining characteristic of BLM-deficient cells is a striking increase in SCEs. Indeed, BLM silencing increases SCEs by approximately 10-fold while silencing ETAA1 resulted in a 2.5-fold increase (Fig. 7a,b). ETAA1Δ cells also exhibited an increased frequency of SCEs compared to controls (Fig. 7c). The frequency of SCEs in ETAA1Δ and wild-type cells transfected with BLM siRNA were not different suggesting an epistatic relationship (Fig. 7c). Both ETAA1 and BLM-deficiency also yield increased levels of micronuclei, and again ETAA1 and BLM are epistatic for this phenotype (Fig. 7d,e). ETAA1Δexon2 cell lines also exhibit elevated levels of micronuclei and SCEs similar to the ETAA1Δ cells indicating that ETAA1 must be able to activate ATR to prevent genetic instability (Fig. 7f,g).
Figure 7. ETAA1 deficient cells have elevated sister chromatid exchanges and genetic instability.
(a–b) SCEs were imaged and scored after ETAA1 knockdown. Scale bars are 5 μm. Four eperiments were performed for siNT and siEpool and one experiment was completed for the other siRNAs. Mean and SEM from a representative experiment is shown and significance calculated by ANOVA with a Dunnett multiple comparison post-test. The number of metaphases analyzed (n value) is indicated. (c) SCEs were scored in wild-type or ETAA1Δ U2OS cells transfected with non-targeting or BLM siRNAs. Data are representative from three independent experiments and significance was calculated with a Mann-Whitney test. The number of metaphases (n value) is indicated. (d,e) Micronuclei were imaged and scored in U2OS cells transfected with non-targeting, ETAA1, or BLM siRNAs as indicated. Data are mean and SD of n=3 independent experiments. Scale bar is 5 μm. (f) Micronuclei and (g) SCEs were scored in wild-type, ETAA1Δ, and two independent ETAA1Δexon2 U2OS cell clones. Mean, SEM, and number of metaphases analyzed (n value) is presented. Significance in e, f and g was determined by ANOVA with a Dunnett multiple comparison post-test. Source data for b,c,e,f, and g are provided in Supplemental Table 1.
ETAA1 and TOPBP1 function in distinct pathways
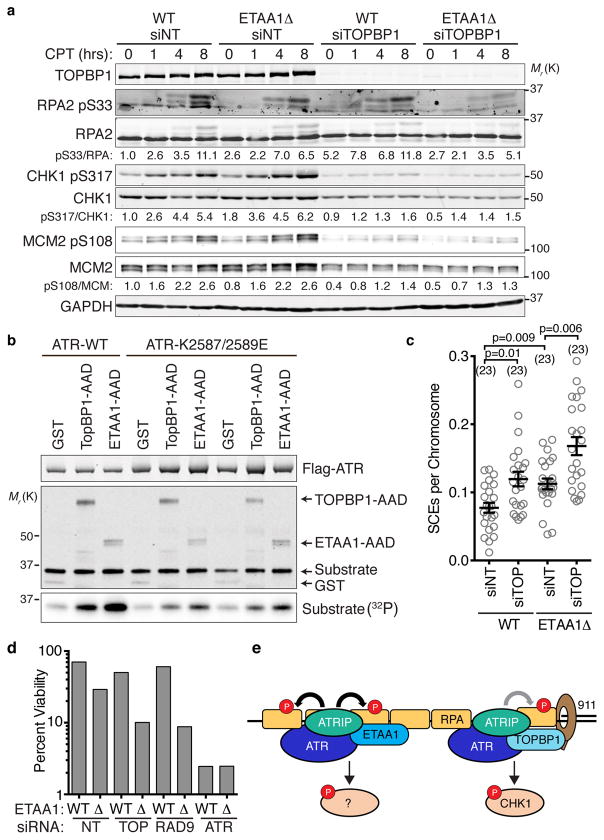
ETAA1 is similar to TOPBP1 in that both activate ATR, complex with BLM, and prevent SCEs. Thus, we considered whether they function in the same or distinct pathways. Unlike ETAA1 deficiency, TOPBP1 knockdown has a strong affect on ATR-dependent CHK1 phosphorylation (Fig. 8a). However, we consistently observe that RPA phosphorylation is primarily dependent on ETAA1 and only modestly affected by TOPBP1 knockdown. We also did not observe TOPBP1 in our ETAA1 purifications suggesting they function in distinct pathways.
Figure 8. ETAA1 and TOPBP1 act in separate pathways to regulate ATR and maintain genome stability.
(a) Wild-type or ETAA1Δ U2OS cells were transfected with non-targeting or TOPBP1 siRNAs, treated with 100 nM CPT for the indicated times, lysed, and immunoblotted with the indicated antibodies. Blots are representative of two experiments. (b) Purified wild-type or mutant ATR/ATRIP complexes were incubated with GST or the AAD domains of TOPBP1 or ETAA1 in the presence of substrate and γ-32P-ATP. The kinase reactions were separated by SDS-PAGE prior to immunoblotting or quantitating substrate phosphorylation by phosphorimaging. Two independent replicates of the mutant ATR kinase reactions are shown. (c) SCEs in wild-type or ETAA1Δ U2OS cells transfected with non-targeting or TOPBP1 siRNAs were quantitated. Error bars are SEM and significance was calculated with a Mann-Whitney test. The number of metaphases scored (n value) is indicated. (d) Cells transfected with non-targeting, TOPBP1, RAD9, or ATR siRNAs were challenged with 10 nM CPT for 24 hrs and viability was determined by clonogenic assay. Untreated viability was set at 100%. Bars indicate the mean of three technical replicates from a representative experiment of two independent experiments (e) Simplified model of ATR activation by ETAA1 and TOPBP1. The 911 complex stabilizes TOPBP1 at stalled forks and assists in the TOPBP1-dependent activation pathway. ETAA1 is recruited directly by RPA and functions independently of 911 and TOPBP1 to activate ATR. Unprocessed original scans of blots in a and b are shown in Supplementary Fig. 8, and source data for c and d are provided in Supplemental Table 1.
TOPBP1 binding and ATR activation requires ATR amino acids between the kinase and FATC domains called the PIKK regulatory domain (PRD)41. Mutations in the PRD do not interfere with the basal activity of ATR, but they greatly reduce the ability of TOPBP1 to activate ATR41. Like TOPBP1, ETAA1-dependent ATR activation is greatly diminished by ATR PRD mutations (Fig. 8b). This result and the similarity of the AAD motifs suggest that ETAA1 and TOPBP1 utilize a similar mechanism to activate ATR.
ETAA1 requires its AAD to prevent SCEs (Fig. 7g), while TOPBP1 is reported to regulate SCEs independently of its AAD25,26. TOPBP1 knockdown yields a similar increase in SCEs as ETAA1-deficiency and TOPBP1 knockdown in ETAA1Δ cells further increases SCE frequency above that of either TOPBP1 or ETAA1-deficiency alone again consistent with operation in distinct pathways (Fig. 8c). Previous studies have made conflicting conclusions about whether TOPBP1 regulation of BLM is through changing its stability25,26. We did not observe large changes in BLM protein levels when we knocked down TOPBP1, and ETAA1 deficiency also does not alter BLM protein levels (Supplemental Fig. 7).
Finally, if ETAA1 and TOPBP1 pathways are distinct we would expect loss of ETAA1 to be synthetically lethal with TOPBP1 or 911 deficiency. Indeed, knockdown of TOPBP1 or RAD9 resulted in decreased survival of ETAA1Δ cells following a challenge with CPT (Fig. 8d). Taken together, these data indicate that ETAA1 and TOPBP1 function in distinct pathways to activate ATR signaling and maintain genome stability.
Discussion
Previous studies identified three ATR (Mec1) activators in budding yeast (Dpb11, Ddc1, and Dna2)12–14. We now report that human cells contain at least two ATR activators, TOPBP1 and ETAA1. This conclusion is consistent with that of the accompanying paper that independently identified ETAA1 as an ATR activator42. TOPBP1 is the Dpb11 functional orthologue; however, ETAA1 does not resemble any of the yeast ATR activating proteins outside of the key residues needed to activate ATR.
TOPBP1 and ETAA1 act in separate pathways to maintain genome integrity and regulate ATR. The ability of ETAA1 to bind directly to RPA distinguishes it from TOPBP1, which requires the MRN and 911 complexes for its recruitment and ATR activating function10,43. 911 is only loaded on DNA gaps when a free 5’ end is available at the ssDNA-dsDNA junction. Thus, extensive ssDNA generation may not generate more 911-TOPBP1-ATR signaling complexes. We propose that ETAA1 helps to propagate ATR activation along stretches of ssDNA since it can bind RPA and function independently of 911 (Fig. 8e).
RPA is particularly dependent on ETAA1 for phosphorylation; whereas other ATR substrates like CHK1 and MCM2 are more dependent on TOPBP1. CHK1 phosphorylation requires the replisome component CLASPIN44, and MCM2 is part of the replicative helicase. Thus, it is possible that the proximity to the replisome where it may be more likely to have a 5’ DNA junction to load 911/TOPBP1 could determine ETAA1 vs. TOPBP1 dependency. The ability of TOPBP1 and ETAA1 to interact with DDR proteins or their relative level of expression, which differs considerably across cell types, may be additional levels of substrate selection.
In conclusion, ETAA1 is a replication stress response protein needed to maintain genome stability. ETAA1 complexes with multiple DDR proteins and one mechanism of ETAA1 action in the replication stress response is as a direct ATR activator. The requirement for ETAA1 to maintain genome stability could be why polymorphisms in the ETAA1 locus increase the risk of pancreatic cancer29,30.
Supplementary Material
Acknowledgments
The research was supported primarily by R01GM116616 to D.C with additional support from R01CA102729 and the Vanderbilt-Ingram Cancer Center. NMR experiments were supported by R01GM65484 and P01CA092584 to W.J.C. T.E.B. is supported by training grant T32CA009582-28. We thank Rong Guo for performing the initial Flag-RPA1 immunopurifications and W. Hayes McDonald in the Vanderbilt Proteomics Laboratory for performing the mass spectrometry.
Footnotes
Author contributions
T.E.B, J.W.L, G.K, C.C., H.D., G.G.G., and D.C. performed most of the experiments. M.D.F. and R.P. performed the NMR experiments with supervision from W.J.C. T.E.B. and D.C. conceived of the project and wrote the manuscript. D.C. supervised the project.
Competing financial interests
The authors have no competing financial interests.
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–37. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–8. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 5.Ball HL, Myers JS, Cortez D. ATRIP Binding to Replication Protein A-Single-stranded DNA Promotes ATR – ATRIP Localization but Is Dispensable for Chk1. Phosphorylation. 2005;16:2372–2381. doi: 10.1091/mbc.E04-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortez D, et al. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–6. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 7.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–55. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 8.Lindsey-Boltz LA, Kemp MG, Capp C, Sancar A. RHINO forms a stoichiometric complex with the 9-1-1 checkpoint clamp and mediates ATR-Chk1 signaling. Cell Cycle. 2015;14:99–108. doi: 10.4161/15384101.2014.967076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotta-Ramusino C, et al. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science. 2011;332:1313–7. doi: 10.1126/science.1203430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duursma AM, Driscoll R, Elias JE, Cimprich KA. A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol Cell. 2013;50:116–22. doi: 10.1016/j.molcel.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mordes DA, Nam EA, Cortez D. Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci U S A. 2008;105:18730–4. doi: 10.1073/pnas.0806621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navadgi-Patil VM, Burgers PM. The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol Cell. 2009;36:743–53. doi: 10.1016/j.molcel.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navadgi-Patil VM, Burgers PM. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem. 2008;283:35853–9. doi: 10.1074/jbc.M807435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Burgers PM. Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev. 2013;27:313–21. doi: 10.1101/gad.204750.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brosh RM, et al. Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J Biol Chem. 2000;275:23500–8. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 16.Ellis NA, et al. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 17.Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc Natl Acad Sci U S A. 1974;71:4508–12. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem. 2014;83:519–52. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plank JL, Wu J, Hsieh TS. Topoisomerase IIIalpha and Bloom’s helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc Natl Acad Sci U S A. 2006;103:11118–23. doi: 10.1073/pnas.0604873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J Biol Chem. 2006;281:13861–4. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- 21.Singh TR, et al. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22:2856–68. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–4. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 23.Wu L, et al. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci U S A. 2006;103:4068–73. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu D, Guo R, Sobeck A. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes …. 2008:2843–2855. doi: 10.1101/gad.1708608.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blackford AN, et al. TopBP1 interacts with BLM to maintain genome stability but is dispensable for preventing BLM degradation. Mol Cell. 2015;57:1133–41. doi: 10.1016/j.molcel.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Chen J, Gong Z. TopBP1 controls BLM protein level to maintain genome stability. Mol Cell. 2013;52:667–78. doi: 10.1016/j.molcel.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borowski A, et al. Structure and function of ETAA16: a novel cell surface antigen in Ewing’s tumours. Cancer Immunol Immunother. 2006;55:363–74. doi: 10.1007/s00262-005-0017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 29.Childs EJ, et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat Genet. 2015;47:911–916. doi: 10.1038/ng.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu C, et al. Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat Genet. 2012;44:62–6. doi: 10.1038/ng.1020. [DOI] [PubMed] [Google Scholar]
- 31.Dungrawala H, et al. The Replication Checkpoint Prevents Two Types of Fork Collapse without Regulating Replisome Stability. Mol Cell. 2015;59:998–1010. doi: 10.1016/j.molcel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bansbach CE, Bétous R, Lovejoy CA, Glick GG, Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 2009;23:2405–2414. doi: 10.1101/gad.1839909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mer G, et al. Structural Basis for the Recognition of DNA Repair Proteins UNG2, XPA, and RAD52 by Replication Factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 34.Feldkamp MD, Mason AC, Eichman BF, Chazin WJ. Structural analysis of replication protein A recruitment of the DNA damage response protein SMARCAL1. Biochemistry. 2014;53:3052–61. doi: 10.1021/bi500252w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu X, et al. The basic cleft of RPA70N binds multiple checkpoint proteins, including RAD9, to regulate ATR signaling. Mol Cell Biol. 2008;28:7345–53. doi: 10.1128/MCB.01079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toledo LI, Murga M, Gutierrez-Martinez P, Soria R, Fernandez-Capetillo O. ATR signaling can drive cells into senescence in the absence of DNA breaks. Genes Dev. 2008;22:297–302. doi: 10.1101/gad.452308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ball HL, et al. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol Cell Biol. 2007;27:3367–77. doi: 10.1128/MCB.02238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toledo LI, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 39.Putnam CD, Hayes TK, Kolodner RD. Post-replication repair suppresses duplication-mediated genome instability. PLoS Genet. 2010;6:e1000933. doi: 10.1371/journal.pgen.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bussen W, Raynard S, Busygina V, Singh AK, Sung P. Holliday junction processing activity of the BLM-Topo IIIalpha-BLAP75 complex. J Biol Chem. 2007;282:31484–92. doi: 10.1074/jbc.M706116200. [DOI] [PubMed] [Google Scholar]
- 41.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–89. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haahr Peter, Hoffmann Saskia, Tollenaere Maxim AX, Ho Teresa, Toledo Luis Ignacio, Matthias Simon Bekker-Jensen, Markus Raschle NM. Nat Cell Biol. 2016. Activation of the ATR kinase by the RPA-binding protein ETAA1. [DOI] [PubMed] [Google Scholar]
- 43.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K-i, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumagai A, Dunphy WG. Claspin, a Novel Protein Required for the Activation of Chk1 during a DNA Replication Checkpoint Response in Xenopus Egg Extracts. Mol Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.