Abstract
The functionality of natural biopolymers has inspired significant effort to develop sequence-defined synthetic polymers for applications including molecular recognition, self-assembly, and catalysis. Conjugation of synthetic materials to biomacromolecules has played an increasingly important role in drug delivery and biomaterials. Here we develop the controlled synthesis of novel oligomers from hydroxyproline-based building blocks and conjugate these materials to siRNA. Hydroxyproline-based monomers enable incorporation of broad structural diversity into defined polymer chains. Using a perfluorocarbon purification handle, we are able to purify diverse oligomers with a single solid phase extraction method. We show the efficiency of synthesis by building 14 unique trimers and 4 hexamers from 6 diverse building blocks. We adapt this method to parallel synthesis of hundreds of materials in 96-well plates. This strategy provides a platform for library screening of modified biomolecules.
Keywords: sequence-defined polymers, siRNA conjugate, fluorous synthesis, bioconjugate
Sequence-defined polymers play many roles in biological systems. The enormous structural diversity of biopolymers such as proteins and nucleic acids enables their many functions including catalysis, self-assembly, molecular recognition, and information storage. The potential of these sequenced biopolymers has inspired interest in developing synthetic polymers with precisely defined monomer sequences for applications in biomaterials as well as for the tuning of bulk material properties.[1] Strategies for controlling monomer sequence in synthetic polymers include controlled radical polymerizations,[2–6] DNA-templated synthesis,[7] and sequential or iterative synthesis.[8–12] Sequential synthesis, in which each monomer is added to a polymer chain individually, offers complete control over polymer sequence and enables the incorporation of a variety of diverse properties. Sequential synthesis methods are typically carried out on a solid support and include peptoid synthesis via the submonomer method,[8,9] aminolysis of thiolactones,[10] the Passerini[11] and Biginelli[13] multicomponent reactions, triazene-based polymers[14], poly(alkoxyamine amide)s[15], and poly(amidoamine)s.[16,17] While solid-supported synthesis allows for facile purification, solution-phase synthesis conditions are desired because they provide homogenous reaction kinetics and allow for direct analysis of reaction mixtures without cleavage of materials from the solid support.[18,19]
Sequence-defined polymers provide precise control of composition, and thereby chemical and material properties, making them attractive for various applications that can benefit from parallel synthesis and library screening, such as nucleic acid delivery,[20,21] biomaterials,[22–25] and drug discovery.[26–28] The combination of synthetic materials and biologics through bioconjugation has also played an increasingly important role in the development of novel therapeutics.[29–31] The integration of biomolecular conjugation with library screening may be particularly useful in the development of novel bioconjugates. Due to this potential to impact multiple fields, novel and scalable approaches to generating sequence-controlled polymers are needed.
Here we present a unique strategy for the synthesis of sequence-defined oligomers from building blocks based on hydroxyproline using only methods that are adaptable to high-throughput synthesis. We hypothesized that hydroxyproline, modified to create diverse monomer building blocks, could be sequentially linked to form novel polyurethanes while minimizing the use of protecting group chemistry. Our method allows for the efficient incorporation of monomers that span a diverse structural space. Monomer coupling reactions consistently proceeded with nearly quantitative conversion and oligomers were recovered with excellent purity. In place of a solid support, we employed a fluorous protecting group as a purification handle, providing the advantage of solution-phase kinetics while allowing for facile purification via solid phase extraction. Fluorous solid phase extraction (FSPE) exploits the interaction of perfluorocarbon-tagged molecules with a perfluorocarbon-modified silica matrix, allowing for facile separation of fluorous-tagged material from untagged materials. This strategy was adaptable to parallel synthesis in a multi-well plate format, as reaction conditions and purification methods were universal despite varying oligomer structure.
This hydroxyproline-based monomer structure was chosen for its ability to form stable polymer chains while incorporating a variety of side chain functionalities. Linear monomer backbone structures were also explored, but cyclization of these monomers or of polymer chains was observed to compete with monomer coupling, especially when basic side chains were included (Supporting Information). Such cyclization side reactions are observed in other stepwise polymer synthesis methods; for example, diketopiperazines and aspartimides are known to form during peptide synthesis.32 The cyclic structure of monomers based on hydroxyproline prevents this uncontrolled cyclization and favors formation of the desired product. The only side reaction observed during synthesis of hydroxyproline-based polyurethanes was hydrolysis of the active intermediate resulting in incomplete coupling.
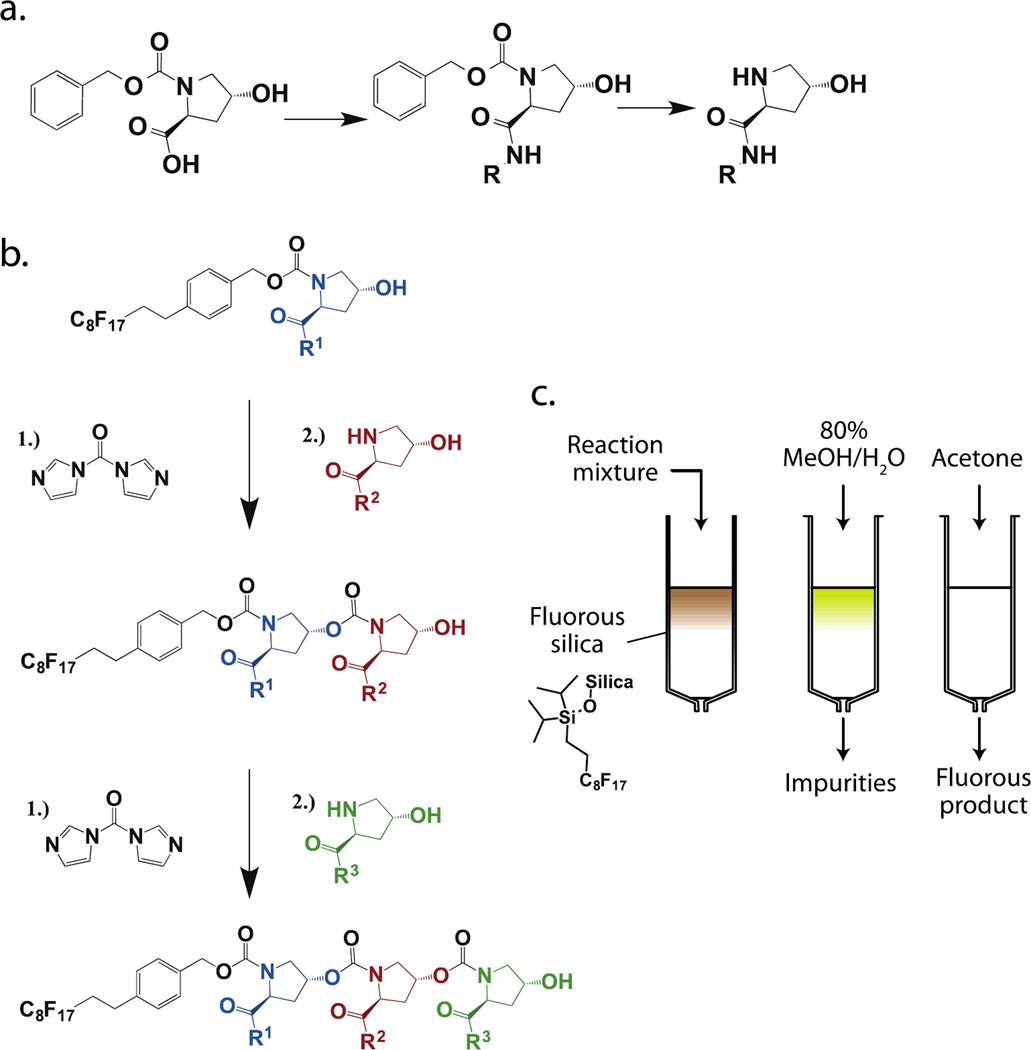
Monomers were synthesized by amidation of the carboxylic acid on CbZ-protected hydroxyproline, followed by deprotection of the amine (Figure 1a). Fluorous-CbZ-protected starting monomers were synthesized by coupling of fluorous-CbZ-NHS ester to hydroxyproline, followed by amidation of the carboxylic acid. Monomer coupling conditions were chosen to drive complete conversion of the fluorous-tagged monomer or polymer chain. The hydroxyl group of the starting monomer was activated by addition of excess carbonyldiimidazole (CDI). The free hydroxyl was completely converted to the activated imidazole-N-carboxylic ester after one hour as observed by LCMS. Excess reagent was quenched and the intermediate was dried thoroughly to avoid hydrolysis of the imidazole-N-carboxylic ester. The second monomer was then added in excess and reacted overnight to yield the carbamate product (Figure 1b).
Figure 1. Synthesis and purification of hydroxyproline-based oligocarbamates.
a.) Monomer synthesis by amidation of hydroxyproline b.) Monomer coupling using carbonyldiimidazole (CDI) c.) Purification by fluorous solid phase extraction (FSPE)
A fluorous protecting group at the end of each polymer chain allowed for rapid purification of oligomers by solid phase extraction (SPE) through fluorous silica (Figure 1c). After each monomer addition the reaction mixture was loaded onto a fluorous silica matrix and washed with an aqueous solvent, removing excess monomer and any impurities that lack a fluorous tag. The fluorous product was then eluted with a fluorophilic solvent such as acetone or tetrahydrofuran. This method afforded efficient separation of oligomers from excess reagents for a wide range of monomer and oligomer properties. Fluorous tags provided the benefit of facile purification afforded by solid-phase synthesis, but with the advantage of solution-phase reaction kinetics and direct analysis of materials without the need for cleavage of materials from a solid support. This method can be readily adapted for high-throughput purification, as fluorous columns are available in 96-well plate format.
To test the robustness and versatility of this method we synthesized a set of 6 diverse monomers and then combined these into 14 different trimers. We chose a monomer set that encompasses broad chemical space, including side chains of varying hydrophobicity, size, and charge in order to test the ability of this approach to generate structurally diverse materials. Reactions were monitored by LCMS and monomer coupling consistently proceeded with high conversion (Figure 2, Supporting Information). Minimal impurities were observed, and the most prevalent impurity was unreacted starting material (Supporting Information). This was due to hydrolysis of the imidazole-N-carboxylic ester intermediate, as formation of the intermediate proceeded reliably with complete conversion. This hydrolysis was the only side reaction observed, and hydrolysis was eliminated by thorough drying of all reagents and by controlling the exposure of the intermediate to moisture. Purity was optimal when the intermediate was dried down within 30 minutes of quenching and used within two hours.
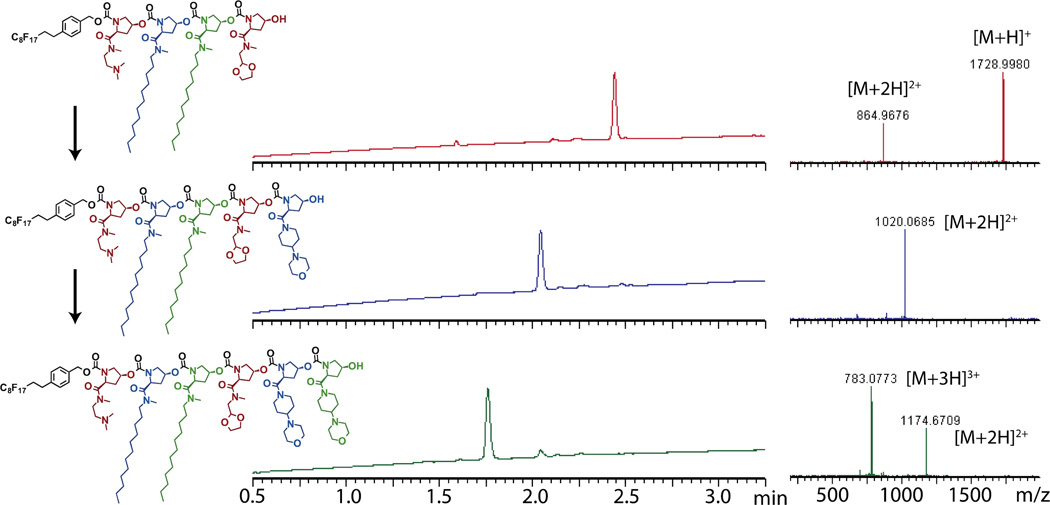
Figure 2. LCMS analysis of oligomer synthesis.
LC traces of tetramer, pentamer, and hexamer show UV absorbance at 214 nm. Mass spectra show ions contributing to the corresponding peak in the MS trace. Calculated masses: 1727.82 (tetramer), 2036.99 (pentamer), 2346.15 (hexamer). Purity of hexamer = 91% with the major observed impurity being unreacted pentamer.
For the 14 trimers synthesized, product purity was consistently high (averaging 94% for dimers and 92% for trimers), and recovered yield was variable, averaging 72% per step (Tables 1 and 2). We observed reduced yield for the most hydrophilic oligomers, which was due to partial elution of product in the aqueous wash during Fluorous SPE. Reducing the amount of washing improved recovery in subsequent steps without reducing purity. While not attempted here, we hypothesize that yields with more hydrophilic monomers could be further improved by using purification tags with longer perfluorocarbon chains.
Table 1. Purity and yield of dimers.
Purity (%)/Yield (%)
| R1\R2 | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 | 100/48 | 95/54 | ||||
| 2 | 90/55 | 97/76 | ||||
| 3 | 96/80 | 80/68 | ||||
| 4 | 100/90 | 90/94 | 87/86 | |||
| 5 | 100/48 | 97/85 | ||||
| 6 | 100/68 |
Table 2. Purity and yield of trimers.
Purity (%)/Yield (%)
| R1R2\R3 | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 12 | 71/36 | |||||
| 14 | 100/48 | 89/14 | ||||
| 21 | 73/83 | 78/34 | ||||
| 25 | 100/67 | |||||
| 34 | 100/64 | |||||
| 36 | 90/54 | |||||
| 42 | 100/82 | |||||
| 44 | 100/87 | |||||
| 46 | 91/58 | |||||
| 53 | 100/97 | |||||
| 54 | 99/86 | |||||
| 63 | 95/60 |
To evaluate the utility of this method for synthesizing longer chains, we selected a subset of the trimers, which exhibited varying hydrophilicity and charge, and extended these chains to hexamers (Figure 2). As before, monomer coupling proceeded smoothly, often with complete conversion. Incomplete conversion due to hydrolysis of the intermediate was again the only side reaction observed (Figure 2). Incompletely reacted oligomers are retained by the fluorous SPE purification, results in the loss of a monomer from the sequence as polymerization is continued. To retain purity at higher degrees of polymerization, the moisture exposure of intermediates must be tightly controlled. Recovered yield averaged 80% per step, an improvement over the average yield observed during trimer synthesis. This improvement was due to adjustments made to the purification process – the amount of aqueous washing was reduced by half, which improved yields without sacrificing purity.
Using these methods, we were able to produce defined polymers with significant chemical diversity. Since the reaction and purification conditions require few steps and are functional with various structures, we believe this synthetic approach maybe appealing for parallel synthesis and screening applications. To evaluate the utility of this method for high-throughput parallel synthesis, we next synthesized and purified a library of 216 unique trimers entirely in 96-well plate format. An automated liquid handler was programmed to combine solutions of starting monomer and CDI in a dry nitrogen environment. After CDI quenching, solvents were evaporated using a Genevac system. Plates were then returned to the liquid handling robot, which was programmed to add solutions of the next monomer. Purification was carried out by fluorous solid phase extraction on 96-well fluorous plates according to the manufacturer’s protocol.
LCMS analysis of selected wells showed that oligomers were successfully synthesized in high-throughput format (Supporting Information). Fluorous SPE in 96-well plates was able to separate fluorous-tagged oligomers from excess reagents, as only fluorous-tagged products were detected in LCMS analysis of purified samples (Supporting Information). Purity of trimers synthesized high-throughput was lower than it was for those synthesized individually (average purity = 92% for classical synthesis, 71% for high-throughput synthesis, Supporting Information), with the major source of impurities being hydrolysis of the imidazole-N-carboxylic ester. This increase in hydrolysis may be due to the increased drying time required to evaporate the larger volume of solvent after CDI quenching (up to 3 h was required for this workup, compared to 1 h in pilot experiments). In prior experiments we observed increased hydrolysis when the intermediate was handled for more than 2 hours, and we reason that reducing this workup time may improve purity in high throughput synthesis. This could be achieved by switching to a more volatile solvent, such as tetrahydrofuran, for the CDI activation steps of the synthesis. Oligomers were weighed after one additional reaction and purification step (addition of azido-PEG as discussed below), and the overall recovered yield of trimer-azides averaged 52% with a standard deviation of 11.5% (Supporting Information). The fluorous purification tag was removed from the trimer-azide library by heating in trifluoroacetic acid (Supporting Information).
One potential application for such libraries of sequence-defined oligomers may be in nucleic acid delivery, where combinatorial library screens have played an important role in developing novel materials. We reasoned that hydroxyproline-based oligomers might enable the expansion of these combinatorial studies to include siRNA conjugate delivery materials. For this purpose, we next explored whether our materials could be conjugated to siRNA for library screening applications. Using the same CDI chemistry, we attached a short polyethylene glycol with an azide functionality to a subset of five selected trimers (Supporting Information). After purification by FSPE, the fluorous protecting group was removed from each of the trimer-azides by heating in trifluoroacetic acid (Supporting Information).
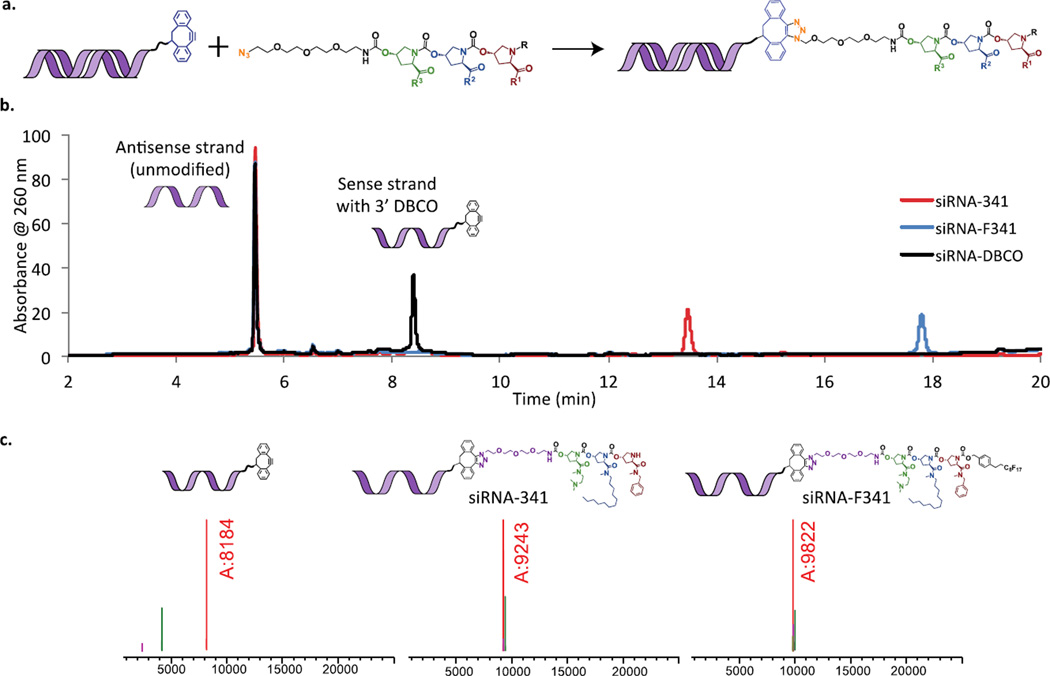
The azide-modified trimers were conjugated to siRNA bearing a dibenzocyclooctyne (DBCO) group the 3’ end the sense strand, which allows for facile “click” coupling via 1,3-dipolar cycloaddition without the need for a copper catalyst.[33] Both fluorous-protected trimers and deprotected trimers conjugated efficiently to the siRNA using a solvent system of 30% acetonitrile in phosphate-buffered saline (PBS). Excess azide was removed from the mixture by ethanol precipitation of the nucleic acid (Supporting Information). Conjugation efficiency was analyzed by LCMS (Figure 4). sConjugation consistently proceeded essentially to completion, based on the shift in elution time in the LC trace. Average recovery of siRNA conjugate material after ethanol precipitation was 95.5% (Supporting Information).
Figure 4. Conjugation of hydroxyproline-based oligomers to siRNA.
a.) Azide-modified trimers, with and without fluorous tags, were conjugated to dibenzocyclooctyne-modified siRNA and resulting conjugates were purified by ethanol precipitation. b.) LCMS analysis of siRNA conjugates: LC trace. siRNA-341 = DBCO-modified siRNA conjugated to trimer 341 (structure shown in Figure 5c.) siRNA-F341 = siRNA conjugated to fluorous-protected trimer 341 (structure shown in Figure 5c.) siRNA-DBCO = unmodified siRNA. c.) Observed masses corresponding to single-stranded siRNA conjugates shown. Calculated masses: 9243 (siRNA-341), 9823 (siRNA-F341).
In conclusion, we have shown that sequence-controlled oligomers can be efficiently synthesized from hydroxyproline building blocks using only methods that are readily adaptable to parallel synthesis in multiwell plates. We demonstrate that these materials are able to incorporate a wide variety of monomer properties with precise control and excellent purity. We further demonstrate that these materials can be conjugated to biomacromolecules in a manner amenable to high-throughput screening applications. Our method for the synthesis of novel hydroxyproline-based materials affords precise control over polymer structure using simple chemistry and mild reaction conditions. These materials may benefit applications that require precise control over polymer structure, such as self-assembly and molecular recognition, as well as applications that require the exploration of vast structural space, such as library screening. We anticipate that these materials may find utility in applications such as protein modification, delivery of biologics, and other biomaterial applications.
Supplementary Material
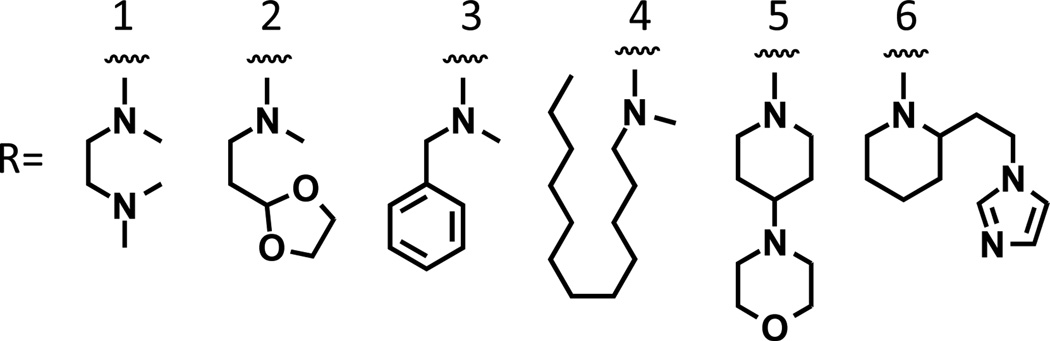
Figure 3. Structures of monomer side chains.
A set of 14 unique trimers was synthesized from six starting monomers. For naming purposes in Tables 1 and 2, monomer side chains are assigned a number 1–6.
Acknowledgments
This work was supported by the National Insitutes of Health (grant EB000244), the Koch Institute Support (core grant P30-CA14051) from the National Cancer Institute, and Alnylam Pharmaceuticals. We thank Ramesh Indrakanti of Alnylam for his assistance in analysis of siRNA conjugates.
References
- 1.Lutz JF, Ouchi M, Liu DR, Sawamoto M. Science. 2013;341:6146. doi: 10.1126/science.1238149. [DOI] [PubMed] [Google Scholar]
- 2.Pfeifer S, Lutz JF. J. Am. Chem. Soc. 2007;129:9542. doi: 10.1021/ja0717616. [DOI] [PubMed] [Google Scholar]
- 3.Ida S, Terashima T, Ouchi M, Sawamoto M. J. Am. Chem. Soc. 2009;131:10808. doi: 10.1021/ja9031314. [DOI] [PubMed] [Google Scholar]
- 4.Satoh K, Matsuda M, Nagai K, Kamigaito M. J. Am. Chem. Soc. 2010;132:10003. doi: 10.1021/ja1042353. [DOI] [PubMed] [Google Scholar]
- 5.Hibi Y, Ouchi M, Sawamoto M. Angew. Chem., Int. Ed. 2011;50:7434. doi: 10.1002/anie.201103007. [DOI] [PubMed] [Google Scholar]
- 6.Chan-Seng D, Zamfir M, Lutz JF. Angew. Chem., Int. Ed. 2012;51:12254. doi: 10.1002/anie.201206371. [DOI] [PubMed] [Google Scholar]
- 7.Niu J, Hili R, Liu DR. Nat. Chem. 2013;5:282. doi: 10.1038/nchem.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuckermann RN, Kerr JM, Kent SBH, Moos WH. J. Am. Chem. Soc. 1992;114:10646. [Google Scholar]
- 9.Olivos HJ, Alluri PG, Reddy MM, Salony D, Kodadek T. Org. Lett. 2002;4:4057. doi: 10.1021/ol0267578. [DOI] [PubMed] [Google Scholar]
- 10.Espeel P, et al. Angew. Chem., Int. Ed. 2013;52:13261. doi: 10.1002/anie.201307439. [DOI] [PubMed] [Google Scholar]
- 11.Solleder SC, Meier M. Angew. Chem., Int. Ed. 2014;53:711. doi: 10.1002/anie.201308960. [DOI] [PubMed] [Google Scholar]
- 12.Porel M, Alabi C. J. Am. Chem. Soc. 2014;136:13162. doi: 10.1021/ja507262t. [DOI] [PubMed] [Google Scholar]
- 13.Boukis AC, Llevot A, Meier MAR. Macromol. Rapid Commun. 2016;37:643. doi: 10.1002/marc.201500717. [DOI] [PubMed] [Google Scholar]
- 14.Grate JW, Mo K, Daily MD. Angew. Chemie. 2016;128:3993. doi: 10.1002/anie.201509864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy RK, et al. Nat. Commun. 2015;6:7237. doi: 10.1038/ncomms8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartmann L, Krause E, Antonietti M, Börner HG. Biomacromolecules. 2006;7:1239. doi: 10.1021/bm050884k. [DOI] [PubMed] [Google Scholar]
- 17.Hartmann L, Häfele S, Peschka-Süss R, Antonietti M, Börner HG. Chem. - A Eur. J. 2008;14:2025. doi: 10.1002/chem.200701223. [DOI] [PubMed] [Google Scholar]
- 18.Tzschucke CC, et al. Angew. Chem., Int. Ed. 2002;41:3964. doi: 10.1002/1521-3773(20021104)41:21<3964::AID-ANIE3964>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Pfeifer S, Zarafshani Z, Badi N, Lutz JF. J. Am. Chem. Soc. 2009;131:9195. doi: 10.1021/ja903635y. [DOI] [PubMed] [Google Scholar]
- 20.Akinc A, et al. Nat. Biotechnol. 2008;26:561. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whiteheada K, et al. Nat. Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Algahtani MS, et al. J. Control. Release. 2014;190:115–126. doi: 10.1016/j.jconrel.2014.06.045. [DOI] [PubMed] [Google Scholar]
- 23.Gu M, et al. Biomaterials. 2013;34:6133. doi: 10.1016/j.biomaterials.2013.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang F, Mei Y, Langer R, Anderson DG. Comb. Chem. High Throughput Screen. 2009;12:554. doi: 10.2174/138620709788681916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hook AL, et al. Adv. Mater. 2013;25:2542. doi: 10.1002/adma.201204936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timmerman P, et al. J. Biol. Chem. 2009;284:34126. doi: 10.1074/jbc.M109.041459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lian W, Upadhyaya P, Rhodes C, Liu Y, Pei D. J. Am. Chem. Soc. 2013;135:11990. doi: 10.1021/ja405106u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D, Li Y, Zhang Y, Liu Y, Shi G. Comb. Chem. High Throughput Screen. 2012;15:232. doi: 10.2174/138620712799218626. [DOI] [PubMed] [Google Scholar]
- 29.Kanasty R, Dorkin JR, Vegas A, Anderson D. Nat. Mater. 2013;12:967. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 30.Leal M, et al. Ann. N. Y. Acad. Sci. 2014;1321:41. doi: 10.1111/nyas.12499. [DOI] [PubMed] [Google Scholar]
- 31.Duncan R, Vicent MJ. Adv. Drug Delivery Rev. 2013;65:60. doi: 10.1016/j.addr.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 32.D’Hondt M, et al. J. Pharm. Biomed. Anal. 2014;101:2. doi: 10.1016/j.jpba.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 33.Jayaprakash KN, et al. Org. Lett. 2010;12:10. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.