Abstract
Cardiovascular disease has been the biggest killer in the United States for decades, with almost a million new cases each year. Even though mammalian rodent neonatal cardiomyocytes show proliferative potential for up to 5 days, adult cardiomyocytes lose this ability. Insufficient cardiomyocyte proliferation is one of the major reasons for the lack of regeneration of myocardial tissue, post injury. Several studies have looked at the mechanisms responsible for the arrest in proliferation at an adult stage. Following up on a recent study by Eulalio et al’s study on functional screening of 875 miRNAs for neonatal cardiomyocyte proliferation, we recently identified several miRNAs that induce proliferation in naturally senescent adult cardiomyocytes. Additional studies by Mahmood et al 2013 have identified Meis1 as the major regulator of cardiomyocyte cell cycle. In our present study we have identified three of the adult cardiomyocyte proliferation inducing miRNAs to have binding sites on the 3’UTR of Meis1 gene by in-silico analysis and luciferase assay. Additionally we found these miRNAs; miR-548c-3p, miR-509-3p, and miR-23b-3p to induce significant proliferation in adult cardiomyocytes through translational inhibition of Meis1. We found a significant increase in the number of ACMs with each miRNA, in combination, and with siRNA mediated inhibition of Meis1 gene. We confirmed that these microRNAs, through inhibition of Meis1, affect its downstream targets and thereby regulate cell-cycle progression. Further investigating of the mechanism of action of these miRNAs can identify other treatment options for abnormalities associated with the lack of cardiac regeneration post myocardial injury.
Keywords: miR-548c, miR-509, miR-23b, Meis1, Adult Cardiomyocytes, Cardiomyocyte Proliferation, Cardiac regeneration
Introduction
In the United States alone there are close to a million new cases of heart attacks each year and cardiovascular diseases (CVDs) are responsible for over 315 billion dollars (US) of direct and indirect associated cost [1]. CVDs have been the biggest killer in the United States for several decades, and this year one in every three deaths was associated with it [1]. With obesity rates at an all-time high, CVDs do not show signs of slowing down thereby resulting in high rates of mortality. Interestingly, recent reports have shown several approaches of regenerating myocardial tissue post injury and one such study by Porello et al. [2] has shown a complete regeneration of neonatal mice hearts post myocardial injury. However, unlike neonatal cardiomyocytes, inducing proliferation of adult cardiomyocytes (ACMs) is challenging as they are known to be in a senescent state [3, 4]. While lower organisms like newt and zebrafish have been reported to regenerate lost myocardium [5, 6] this phenomenon is almost absent in an adult mammalian heart.
Since their discovery, microRNAs have been studied in almost every disease type and have shown very promising therapeutic effects in many cases [7, 8]. MicroRNAs are 18-22nt long double stranded RNAs which are known canonically to bind to the 3’UTR of their target gene and inhibit its expression. While transcribed in the nucleus and cleaved by the drosha complex, microRNAs are exported to the cytoplasm by exportin and cleaved into a double stranded duplex by the dicer complex. Moreover, canonically one strand (guide) of the double stranded duplex forms a complex with Ago2 (RISC) while the other strand (passenger) is degraded. This RISC complex can then repress translation leading to inhibition of target gene function [9, 10]. While there have been other pathways proposed for the processing of microRNAs, this is the canonical mode of action for microRNA mediated inhibition of gene expression [11].
In our previous work we screened several miRNAs that can induce proliferation in ACMs and amongst them miR-548c-3p, miR-509-3p, and miR-23b-3p have direct binding sites in the 3’UTR region of Meis1 [12, 13].
Meis1 belongs to the TALE (three amino acid loop extension) family of homeodomain transcription factor and is known for its role in cell differentiation. Moreover, levels of Meis1 have been reported to increase with age, along with its role in cardiomyocyte cell cycle regulation [14]. Inhibition of Meis1 showed an increase in proliferation and eventually cardiac regeneration [14]. Here we identified miRNAs (miR-548c-3p, miR-509-3p, and miR-23b-3p) that can regulate Meis1 and bring naturally senescent ACMs back into cell cycle thereby inducing proliferation in otherwise senescent cells. In addition, we show regulation of cell-cycle genes with both miRNAs and siRNA mediated knockdown of Meis1 gene indicating these miRNAs are up-stream regulators of Meis1.
Material and Methods
Ethics statement
All protocols were approved by the Institutional Animal Care and Use Committee of the University of Cincinnati, College of Medicine (IACUC). All experiments performed follow the guidelines of the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH).
Isolation of rat ventricular cardiomyocytes
Hearts from 10-12 weeks old adult rats were used for cell isolation using Langendroff-perfusion method (Bell RM, 2011). Briefly, hearts were removed from anesthetized rats and perfused with Krebs-Hanseleit bicarbonate buffer. At 10 and 15 minutes post perfusion 12.5μl and 25μl of 0.1M CaCl2 was added to the buffer solution (circulating through the heart), respectively to reach a final concentration of 0.1mM. At this point, the hearts were finely minced and passed through a 100μm filter to remove non-cardiomyocytes. Flow through was spun at 300 rpm for 3-5 minutes at room temperature. Cardiomyocytes were suspended in 10% bovine serum albumin (BSA, Sigma MO) in a 1:1 ratio with Kresbs-Hanseleit bicarbonate buffer. Fifty, 75, and 100μl of CaCl2 were added at 5 minute intervals to a final concentration of 1mM.
Cardiomyocytes were plated on laminin coated plates (Life Technologies, CA), in DMEM media containing 5% fetal bovine serum (Corning, Life Technologies, CA) and 1% penicillin-streptomycin (Life Technologies, CA) and cultured in 5% CO2 in a humidified incubator at 370 C.
Transfection
Cardiomyocytes isolated from rat hearts were transfected 24 hours post plating. C-elegans miR-67 (Cel-miR-67) was used as control miRNA. Cardiomyocytes were transfected with 50nM of miRNA mimic (Dharmacon, GE) using lipofectamine RNAiMAX (Life Technologies, CA) according to the manufacturer’s protocol.
Luciferase assay
293T cells were transfected with the luciferase construct containing the 3’UTR of Meis1. MiRNAs were transfected simultaneously (as described above) and media was harvested 48 hours post transfection for analysis of luciferase activity according to the manufacturer’s recommended protocol (GeneCopoeia).
Immuno-staining
Cells were fixed with 4% paraformaldehyde for 15 mins and permeabalized with 0.5% Triton X-100 in PBS for 20 minutes. Cells were then stained with primary antibody (in CAS-Block, Life Technologies) overnight at 40C. Antibodies used included: rabbit monoclonal antibody against Troponin-I (Santa Cruz Biotechnology sc-15368), histone H3 phosphorylation at serine 10 (Cell Signaling 9706S). EdU staining was performed using Click-iT EdU Alexa Fluor Imaging Kit (Invitrogen, CA) according to manufacturer’s protocol.
EdU images were taken at 10X magnification using an OLYMPUS microscope. pH3 images were taken at 20X magnification.
Western blotting
Western blot analysis was performed by separating proteins on 10% sodium dodecyl sulfate-polyacrylamide gels (Invitrogen). Protein was transferred onto a PVDF membrane and blocked using 5% milk (Bio-Rad, CA), followed by probing with respective primary and secondary antibodies. Primary antibodies used: Rabbit anti-cyclin-dependent Kinase (CDK2) at 1:1000 dilution (Cell Signaling, MA), rabbit anti-Bax at 1:1000 dilution (Cell Signaling, MA), rabbit anti-β-actin at 1:1000 dilution (Cell Signaling, MA), goat-anti-Meis1/2 at 1:1000 dilution (Santa Cruz, CA). Secondary antibodies were used at 1:5000 dilution (Cell Signaling, MA). In addition Wes ProteinSimple (Protein Simple, CA), a new capillary based method which is acquiring wide acceptance, was also used in some cases to perform protein analysis [15, 16].
In-silico analysis
Binding sites for individual miRNAs were confirmed using TargetScan-prediction of microRNA targets [13]. DAVID Functional Annotation Tool [17, 18] was used to determine pathways regulated by individual miRNAs.
Statistics
Sample size in each experiment was ≥ 3. Student’s t-test was used for statistical analysis and significance calculation. P<0.05 was considered to be statistically significant. All quantifications and cell counting were blinded and counted by at least two individuals.
Results
MiR-548c-3p, miR-509-3p, and miR-23b-3p bind to the 3’UTR of Meis1 and inhibit its expression
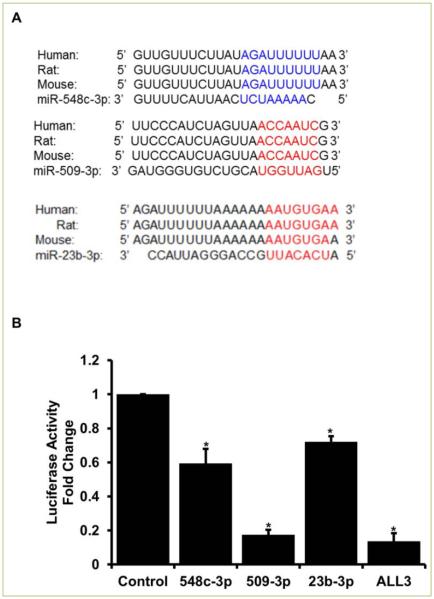
Based on our previous work [12] we aimed to elucidate a mechanistic understanding of miRNAs inducing proliferation in naturally senescent ACMs. Three of the 10 miRNAs identified to promote proliferation of ACMs had a binding site on the 3’ untranslated region (UTR) of the Meis1 gene as determined by in-silico analysis. Interestingly, all three miRNAs have binding sites that are conserved amongst species (human, mouse, and rat) as shown in Figure 1A. We next performed luciferase assay to validate these binding sites. As shown in Figure 1B, all the three miRNAs individually, as well as together, confer a significant inhibition of Meis1 luciferase activity. The next step was to determine if these miRNAs can induce a significant proliferation individually and if simultaneous transfection can induce a higher rate of proliferation in ACMs.
Figure 1. miR-548c-3p, miR-509-3p, and miR-23b-3p bind to the 3’UTR and inhibit the expression of Meis1 gene.
A. Using in-silico approach binding sites were identified in the 3’UTR of Meis1 gene for all three miRNAs. B. Luciferase assay using the 3’UTR of Meis1 was used to validate respective miRNA binding. Student t-test was performed to determine significance *p<0.05. ALL3= all three miRNAs together at 50nM concentration each.
MiR-548c-3p, miR-509-3p, and miR-23b-3p promote proliferation of ACMs
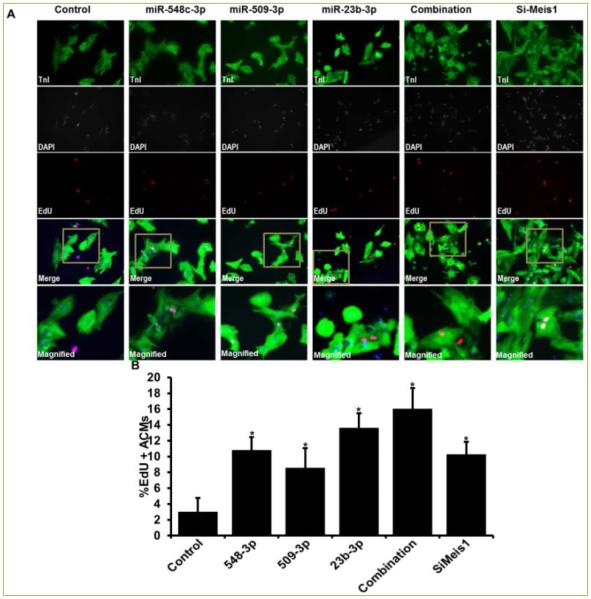
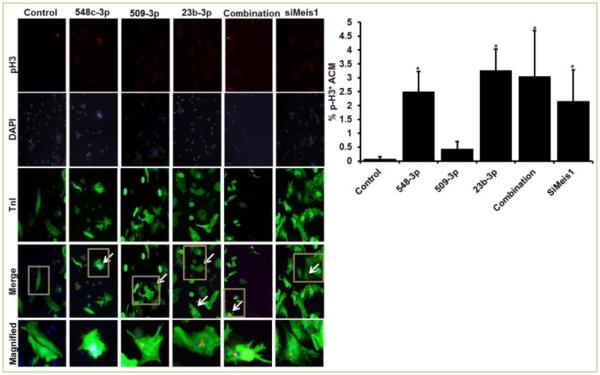
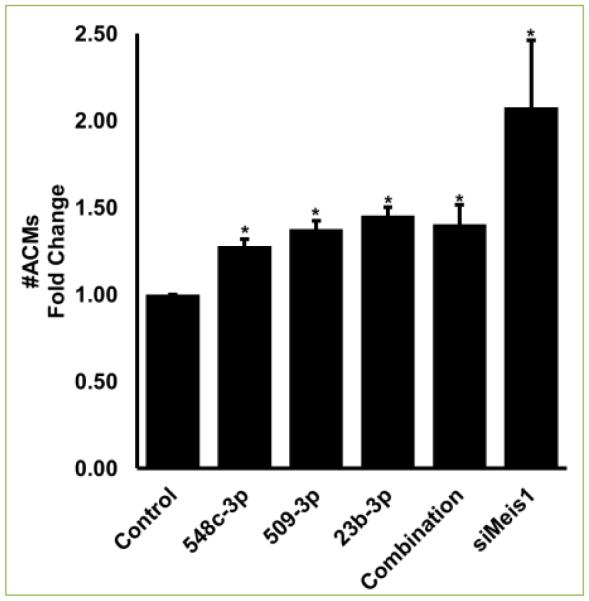
ACMs transfected with miR-548c-3p, miR-509-3p, and miR-23b induced significant proliferation of 10.83%, 8.57%, and 13.64%, respectively. Interestingly, all three miRNAs together induced a robust proliferation of 16.04% while siRNA mediated silencing of Meis1 gene (siMeis1) showed 10.30% EdU positive ACMs (Figure 2). Since EdU indicated ACMs positive for DNA synthesis, we next confirmed if DNA synthesis also led to mitosis in these ACMs. To measure mitosis we counted phospho-histone H3 (pH3) positive ACMs by immunostaining. As shown in Figure 3, miR-548c-3p and miR-23b induced significant increase in pH3 positive ACMs 2.51% and 3.27%, respectively. However, miR-509-3p only showed 0.44% pH3 positive cells (while control showed 0.08%). Moreover, all three miRNAs together induced over 3.1%, while si-Meis1 showed 2.16% mitotic ACMs (Figure 3). As a result of increased proliferation a significant increase in cell number was observed on day 7 post transfection, with individual miRNAs, combination of all three, and siMeis1 (Figure 4). These results confirmed that these miRNAs can inhibit the expression of Meis1 and bring senescent ACMs back into cell cycle.
Figure 2. miR-548c-3p, miR-509-3p, and miR-23b-3p promote proliferation of ACM.
A. Representative Fluorescence images of ACMs transfected with cel-miR-67(control), miR-548c-3p, miR-509-3p, miR-23b, and all three miRNAs together and stained for cardiac marker TnI (green), nuclear stain DAPI (blue) and DNA synthesis marker Edu (Red); B. Quantitative analysis of percentage EdU positive ACMs. Meis1 siRNA = siMeis1. Student t-test was performed to determine significance *p<0.05. Combination= all three miRNAs together at 50nM concentration each.
Figure 3. miRNA-548c-3p, miRNA-509-3p, and miRNA-23b promote mitosis in ACMs.
A. Representative fluorescence images of ACMs transfected with cel-miR-67(control), miR-548c-3p, miR-509-3p, miR-23b, and all three miRNAs together and stained for TnI (green), DAPI (blue) and pH3 (Red); B. Quantitative analysis of percentage pH3 positive ACMs. Meis1 siRNA = siMeis1. Student t-test performed to determine significance.*p<0.05. Combination= all three miRNAs together at 50nM concentration each.
Figure 4. miRNA-548c-3p, miRNA-509-3p, and miRNA-23b transfection increases number of ACMs.
Fold change in the number of ACM on day 6 after transfection in various groups when compared to ACMs in control transfected group. Student t-test performed to determine significance. *p<0.05. Combination= all three miRNAs together at 50nM concentration each.
MiR-548c-3p, miR-509-3p, and miR-23b-3p regulate cell-cycle by inhibiting Meis1
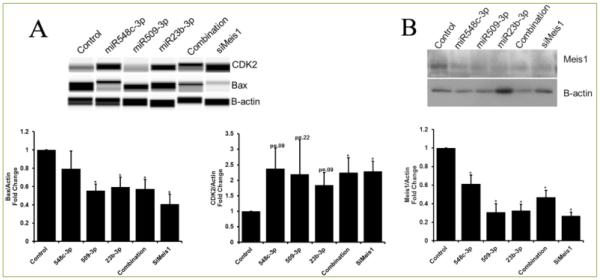
As shown previously by Mahmoud, et. al., inhibition of Meis1 regulates cell cycle genes. Here we confirmed that the inhibition of Meis1 by each of these miRNAs individually and combination was similar to that observed by siRNA mediated inhibition of Meis1 (siMeis1) expression. Additionally, Bax (negative regulator of cell-cycle/survival) was down-regulated by individual miRNAs, miRNA combination, and siMeis1. Moreover, Cyclin-dependent Kinase 2(CDK2) was significantly upregulated by combination of all three miRNAs and siMeis1; however this effect was slight but not significant with any of the individual miRNA alone (Figure 5). This could be due insufficient silencing and other compensatory mechanisms and targets of miRNAs. Our data confirmed that these miRNAs work through Meis1 and have the same downstream targets as Meis1 thus forcing naturally senescent adult cardiomyocytes back into cell cycle.
Figure 5. Cell-cycle genes are regulated by miRNA-548c-3p, miRNA-509-3p, and miRNA-23b.
A) Protein expression of cell-cycle regulators, CDK2 and Bax, in the presence of miR548c-3p, miR-509-3p, miR-23b-3p, combination (all three miRNAs), and siMeis1, shows differential expression. B) Meis1 protein expression was significantly reduced with individual miRNAs, combination, and with siMeis1. Student t-test was performed to determine significance *p<0.05.
Multiple cellular pathways and processes are regulated by these miRNAs
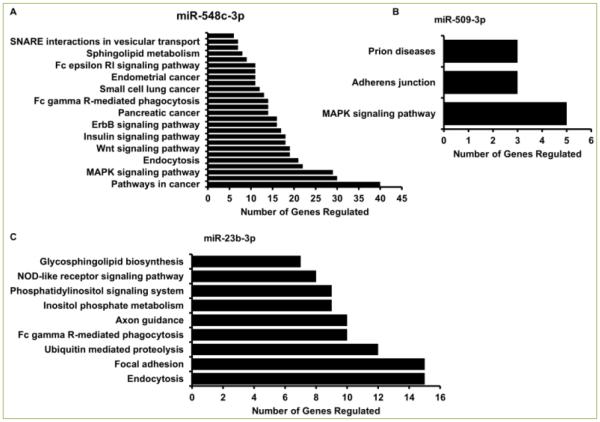
Given the multi-target action of miRNAs, we performed a comprehensive in-silico meta-analysis to elucidate pathways and cellular processes regulated by these miRNAs. As shown in Figure 6A each of these miRNAs regulated several pathways. To name a few, miR-548c-3p regulates 40 genes related to cancer, 29 genes in the MAPK signaling pathway, and 19 genes in the cell cycle and Wnt signaling pathway, and can play a key role in a plethora of biological processes. According to the database available through DAVID Functional Annotation Tool [17, 18] genes related to seven different types of cancers have been previously reported to be regulated by miR-548c-3p. While miR-509-3p was seen to be involved in Prion diseases, adherens junction, and the MAPK signaling pathway (Figure 6B), miR-23b-3p regulates over 10 genes in Ubiquitin mediated proteolysis, focal adhesion, and endocytosis (Figure 6C). Together, these miRNAs by in-silico analysis are shown to regulate thousands of genes that can go on to regulate almost every pathway in an organism.
Figure 6. Multiple cellular pathways are regulated by these miRNAs.
In silico analysis using DAVID Bioinformatics Resource shows cellular pathways and processes regulated by miR-548c-3p (A), miR-509-3p (B), and miR-23b-3p(C) along with number of genes regulated in each pathway.
Discussion
Our results have shown that exogenous transfection of miRNAs (miR-548c-3p, miR-509-3p, and miR-23b-3p) can induce significant proliferation of naturally senescent ACMs. We show that freshly isolated ACMs from 10 week old rats can be induced to proliferate with just a single transfection of miRNA mimic. Moreover, to better understand the mechanism we have shown that mRNA translation of Meis1 was inhibited by these miRNAs and even more so when added simultaneously. While less than 1% of ACMs are reported to proliferate in their lifetime [3] here we reported over 16% EdU positive (DNA synthesis) ACMs and over 3% ACMs undergoing mitosis following specific miRNA transfection. Recent studies have shown CMs from neonatal and adult mice and rats to proliferate [6] and even regenerate lost tissue completely [3, 19-21], this study shows a robust increase in proliferation in ACMs by miRNA mediated inhibition of Meis1 expression. Our luciferase data confirmed the mRNA inhibition of Meis1 gene and confirmed the previous reports that inhibition of Meis1 can depress the cell cycle repression of ACMs. Following siRNA mediated silencing of Meis1, over 10% ACMs were positive for Edu, while over 2% were undergoing active mitosis. Increased DNA synthesis and mitosis lead to an increase in total number of ACMs in each condition and up to double in siMeis1 condition. In addition, we showed downregulation of Meis1 protein expression by miR-548c-3p, miR-509-3p, and miR-23b-3p (and all three miRNAs together), similar to that of siRNA mediated inhibition of Meis1. We observed a significant reduction of negative cell-cycle regulator, Bax, with individual miRNAs as well as a combination of all three. However, only slight increase in positive cell cycle regulator, CDK2, was observed with individual miRNAs but this effect was significantly higher with the combination of all three miRNAs and siMeis1. We attribute this to imperfect binding of miRNAs on the 3’UTR and other compensatory mechanism that may make up for the inhibition of genes. However, this is a question of much concern and future research. Nonetheless, we have to keep in mind the multi-targeted effects of miRNA mediated inhibition of genes and other cell-cycle regulators that could be altered.
While we performed a gene specific approach to determine the effect of these miRNAs in inducing proliferation and division of senescent ACMs, we showed the multi-targeting side of miRNAs mode of action. Since miRNAs bind with a match of 5-6 base-pairs, this allows for a single miRNA targeting a plethora of genes and pathways. As shown in Figure 6, miR-548c-3p alone has been reported to regulate seven different forms of cancers and over a thousand genes. We performed a meta-analysis of genes and pathways regulated by these miRNAs and found genes in almost every pathways being possibly regulated by just these three miRNAs. However these targets were not experimentally validated and one of the best approaches to identify the actual targets is to do a miRNA-pull down assay followed by RNA-seq. While miRNAs usually silence translation of target genes, there have been reports that miRNAs can control other miRNAs [11] and this is an area of great interest in the small-RNA research community.
In conclusion, this research holds a very high and promising therapeutic potential for using miRNAs as a therapeutic option, following in-vivo studies in rodents along with larger animal models. Therapeutic miRNA delivery to promote endogenous ACM proliferation is promising when compared to cell therapy since the majority of transplanted cells are known to be lost within a short period, while raising concerns about the tumorigenic nature of transplanting exogenous cells [22]. Conversely, miRNAs are readily available, and are easy to administer which makes this approach a highly relevant and an achievable one. Although, there is a need to validate the possible targets of these miRNAs in cardiac tissue, this work provides an understanding of miRNA targets and how multiple miRNAs can targets a specific gene which in this case is an established cell-cycle regulator Meis1.
Acknowledgement
We would like to thank the department of Pathology and Laboratory Medicine at the University Of Cincinnati College Of Medicine.
This work is supported by National Institute of Health (NIH) grant R01HL-106190 to RPHA. The University Research Council (URC) Graduate Student Research Fellowship, University of Cincinnati to RP. U.S. Health Resource and Services Administration Award (HRSA), D34HP16299 to LBJ.
Footnotes
Author Contributions
Conceived and designed experiments: RP and RPHA. Performed the experiments: RP, YY, and LJ. Analyzed the data: RP, YY, LJ, and RPHA. Wrote the paper: RP, LJ, and RPHA.
References
- 1.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, et al. Heart disease and stroke statistics--2015 update: A report from the american heart association. Circulation. 2015;131:e29–322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 2.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimura W, Xiao F, Canseco DC, Murlidhar S, Thet S, Zhang HM, et al. Hypoxia fate mapping identifies cycling cardiomyocytes in the adult heart. Nature. 2015;523:226–230. doi: 10.1038/nature14582. [DOI] [PubMed] [Google Scholar]
- 4.Puente BN, Kimura W, Muralidhar SA, Moon J, Amatruda JF, Phelps KL, et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell. 2014;157:565–579. doi: 10.1016/j.cell.2014.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabé-Heider F, Walsh S, Zupicich J, Alkass K, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009 Apr 3;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zucchigna S, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 7.Lee R, Feinbaum R, Ambros V. A short history of a short RNA. Cell. 2004;116:S89–92. doi: 10.1016/s0092-8674(04)00035-2. 1 p following S96. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Cannell IG, Kong YW, Bushell M. How do microRNAs regulate gene expression? Biochem Soc Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- 10.Aida Martinez-Sanchezemail, Murphy Chris L. MicroRNA target Identification—Experimental approaches. Biology. 2013;2:189–205. doi: 10.3390/biology2010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matkovich SJ, Hu Y, Dorn GW., 2nd Regulation of cardiac microRNAs by cardiac microRNAs. Circ Res. 2013;113:62–71. doi: 10.1161/CIRCRESAHA.113.300975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandey R, Ahmed RP. MicroRNAs inducing proliferation of quiescent adult cardiomyocytes. Cardiovascular Regenerative Medicine. 2015;2 doi: 10.14800/crm.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 14.Mahmoud AI, Kocabas F, Muralidhar SA, Kimura W, Koura AS, Thet S, et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497:249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao B, Sanders MJ, Carmena D, Bright NJ, Haire LF, Underwood E, et al. Structural basis of AMPK regulation by small molecule activators. Nat Commun. 2013;4:3017. doi: 10.1038/ncomms4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris VM. Protein detection by simple western analysis. Methods Mol Biol. 2015;1312:465–468. doi: 10.1007/978-1-4939-2694-7_47. [DOI] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Mahmoud AI, Canseco D, Xiao F, Sadek HA. Cardiomyocyte cell cycle: Meis-ing something? Cell Cycle. 2014;13:1057–1058. doi: 10.4161/cc.28379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canseco DC, Kimura W, Garg S, Mukherjee S, Bhattacharya S, Abdisalaam S, et al. Human ventricular unloading induces cardiomyocyte proliferation. J Am Coll Cardiol. 2015;65:892–900. doi: 10.1016/j.jacc.2014.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puente BN, Kimura W, Muralidhar SA, Moon J, Amatruda JF, Phelps KL, et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell. 2014;157:565–579. doi: 10.1016/j.cell.2014.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldthwaite CA., Jr. Mending a broken heart: Stem cells and cardiac repair. 2007 [Google Scholar]