Abstract
The authors have developed the Genetics Home Reference, a consumer resource that addresses the health implications of the Human Genome Project. The research results made possible by the Human Genome Project are being made available increasingly in scientific databases on the Internet, but, because of the often highly technical nature of these databases, they are not readily accessible to the lay public. The authors' goal is to provide a bridge between the clinical questions of the public and the richness of the data emanating from the Human Genome Project. The Genetics Home Reference currently focuses on single gene or polygenic conditions that are also topics on MEDLINEplus, the National Library of Medicine's primary consumer health site. As knowledge of genetics expands, the interrelationships between genes and diseases will continue to unfold, and the site will reflect these developments.
People are increasingly seeking health information online1,2,3,4 from the tens of thousands of Web sites that disseminate health information. However, many consumers are concerned about the credibility and reliability of health-related Web sites5 as well as about the readability and accessibility levels of materials on these sites even when they are otherwise reliable.6 For example, a recent survey of patients' use of the Internet for genetics-related material found that one of the primary reasons that patients and their families turn to the Internet is to find information that is expressed in “layperson's” terms.7 In many cases, however, when they do visit genetics-related sites, they find them confusing and hard to understand.
We have developed the Genetics Home Reference,8 a consumer resource that addresses the health implications of the Human Genome Project and makes them more readily accessible to the lay public. The Genetics Home Reference provides basic information, in a question and answer format, on the nature of genes and how they give rise to various conditions and diseases. For each condition, the site provides basic information about it, how common it is, what the genetic causes and inheritance mechanisms are, and, importantly, where the user can go to find additional information. For each gene, the site provides information about the normal function of the gene, its chromosomal location, and what conditions are associated with mutations or regulatory function changes in the gene. Each description includes a glossary, a list of alternative names for the gene or condition being described, and the sources for the information consulted for each description. In addition, each condition or gene description links directly to pertinent information available on a variety of other resources, including MEDLINEplus,9 ClinicalTrials.gov,10 PubMed,11 Gene Tests,12 Gene Reviews,12 LocusLink,13 and Online Mendelian Inheritance in Man.14
As a further guide to users of the Genetics Home Reference, we have developed a resource called Help Me Understand Genetics, which explains, together with diagrams and other visuals, some basic concepts in genetics. This resource offers, for example, easy-to-understand explanations of DNA, genes, proteins, chromosomes, and how genes control the growth and division of cells. The resource also has sections on the nature of genetic disorders, genetic consultation and testing, gene therapy, and genomic research.
In this report, we focus on the motivation for developing the Genetics Home Reference and the ways in which it differs from some other information resources that focus on inherited disorders. We discuss the principles that guided our design and implementation of the system including the underlying knowledge structures. We conclude with the lessons we have learned and some of the challenges we expect to face in the future.
Background
Before developing the Genetics Home Reference, we conducted some preliminary investigations to get a sense of the type of information that was available and that might be of interest to users of such a site. We first reviewed all queries from the help desk logs that were submitted to National Library of Medicine's (NLM) National Center for Biotechnology Information (NCBI) during a one-week time period in January 2002. For those queries that were specifically related to questions about genetic disorders, we analyzed the nature of the questions to see what type of information was being sought. We then investigated the available resources for answering such questions.
We found that, although there were several very high quality resources available, they were not straightforward for members of the public to find, navigate, or use.15 This is not surprising, since many of the resources have been designed specifically for biologists, geneticists, or practicing clinicians. For example, GeneCards was developed to provide working biologists with ready access to distributed information that supports the functional analysis of genes.16 It integrates a subset of the information stored in major data sources dealing with human genes and their products. Some of the many resources from which GeneCards draws its information include the HUGO gene nomenclature,17 the SWISS PROT protein database,18 the GENBANK sequence database,19 and the Human Gene Mutation Database.20 Another example of a rich resource of information on human genes and genetic disorders is the Online Mendelian Inheritance in Man (OMIM).21 OMIM is intended for use by physicians, professionals concerned with genetic disorders, genetics researchers, and advanced students in science and medicine. GeneTests,12 and its related database, GeneReviews, is a site intended primarily for practicing medical geneticists, but it also contains a range of educational materials to help health care providers understand the appropriate use of genetic counseling and testing.22,23,24 GeneTests contains information on laboratory tests for inherited diseases, including directories of U.S. and international genetics clinics, and GeneReviews contains summaries relating genetic testing information to diagnosis, management, and genetic counseling of specific inherited diseases.
Our background work indicated that (1) the public is actively trying to find information about inherited disorders from both consumer health sites and from biological databases; (2) the paths through the World Wide Web to find such information usually do not proceed directly from consumer health sites to the specific genetic information related to health risks or genes; (3) considerable high-quality, publicly accessible information is available; and, importantly, (4) models exist in the genetics scientific community to bring together multiple pieces of information from other resources for a specific audience. Because we believe the public has difficulty accessing the available systems and the information they contain, we focused on the development of a new system, which would be accessible to the lay public.
Design Objectives
The development of the Genetics Home Reference has been guided by three main principles. Each of these principles is related to the primary goal of creating a resource that would be accessible to the lay public and sensitive to their issues. The first principle was to link the site directly to consumer health resources so that it would be easy for the public to find and understand the resources. The second principle was to interrelate and integrate existing information resources, including the full spectrum from consumer health resources to those used by the clinical and scientific community. The third principle was to apply informatics methods to enhance the sustainability of the knowledge base and allow the project to expand gracefully to the anticipated size of the types of health implications of the Human Genome Project. The following section discusses how these principles have been followed in the development of the Genetics Home Reference.
The overall sensitivity to consumer health issues led to the list of health implications being simply called “conditions” rather than diseases or disorders. This decision was made for two reasons. First, the consequences of genetic mutations include both healthy states and disease states. Second, some conditions that are anomalous (i.e., hearing loss) should not be labeled as disease states.
Design Principle 1: Make the Site Easy for the Public to Access with Understandable Content
The first principle led to direct links to primary topics in MEDLINEplus, search-based links to ClinicalTrials.gov, and links to patient support group sites. The content needed to be structured around a set of specific questions related to clearly defined entities (either health conditions or genes) that were related to each other in clearly defined ways. A list of synonyms and alternate names was essential to help avoid confusion between entities, leading to the use of the Unified Medical Language System (UMLS)25 with special consideration for gene and protein names. The content needed to be written in a straightforward fashion and the technical terms defined. Explanatory material needed to be available so that basic concepts could be reviewed as often as needed.
This led us to the use of terms and definitions available in the UMLS but augmented for specific dictionary terms from several other sources including GeneReviews/GeneTests, the NIH Office of Rare Diseases,26 the Department of Energy's Human Genome Project,27 and the National Cancer Institute.28 This also led us to develop an online resource called Help Me Understand Genetics that provides a basic set of information about genetics and inherited conditions. This resource is organized into short segments that define and illustrate basic concepts in genetics and then provide Web links to more detailed information about the topic.
We carried out two informal studies to help with the site layout and level of presentation of the content. The first was an early presentation of some proposed material to a group of first-year college students. This helped determine which types of items needed to be included in the glossaries and also whether the content was generally understandable by the group. We later asked a group of online users to give us feedback on the ease of navigation through the site, the helpfulness of the information, the understandability of the information, overall impressions, and specific comments about link placement and searching options.
Design Principle 2: Interrelate and Integrate Existing Resources Extending from Consumer Health Resources to Clinical and Scientific Resources
With the second principle, we developed a system that bridges diverse communities and information sources. Bridging diverse communities involves interpreting and contextualizing those complex concepts used by researchers to a format easily understood by a less-technical lay public. Additionally, we link to more technical resources than those usually handled by consumer health sites, including the OMIM, GeneTests, LocusLink, and others containing high-quality information about genes and genetics conditions.
Design Principle 3: Create an Informatics-based Knowledge Resource that Would Enhance the Sustainability of the Project
The third principle arose from the requirements imposed by the first and second principles and the ultimate size of the project. We discuss here three aspects of this informatics principle:
1. A Naming Convention for Health Conditions, Subconditions, and Genes
We use the specific guidelines within the medical genetics profession29 when naming the health conditions and list the other names as synonyms. An example is the decision to use the name Alzheimer Disease instead of Alzheimer's Disease, since the American Medical Association and American College of Medical Genetics both suggest avoiding the possessive form of eponyms. We also use the official gene name as declared by the Human Genome Nomenclature Committee but use the protein name in the written discussion because it is generally more readable.
Many of the health conditions discussed in Genetics Home Reference have multiple genetically determined subtypes. We include those subtypes that have a specific genetic basis found in more than one family and do not include those where the subtype has not been established genetically. This facilitates a complete data model with a relationship between a specific health condition and a change in a specific gene, together with specific literature references. We, therefore, include only four genetically determined subtypes of Alzheimer Disease, because only four genes have been definitively related to Alzheimer Disease, even though there are more subtypes suspected on the basis of family and population studies.
2. A Data Model to Relate the Health Conditions and Genes to Each Other
The relationships among genes and conditions are a many-to-many relationship with conditions being related to multiple genes and genes being related to multiple conditions. This implies first a network of gene–condition entities. Further, there are multiple types of relationships. This is not only a causal relationship in which a mutation in a specific place in a specific gene always causes a specific disorder but also includes relationships with lesser degrees of specificity or probability. For this reason, we have four different gene–condition relationships in the current model with the full anticipation that we will expand this list of relationships in the future. The four occur when:
Mutations in the gene cause the disorder, (e.g., the delta F508 mutation in the CFTR gene causes the disease cystic fibrosis); or
Mutations in the gene increase the risk of the specific condition, (e.g., mutations in the ATM gene in the heterozygous condition increase the risk for a woman to develop breast cancer); or
Mutations in the gene (or other changes such as in the regulatory regions) are associated with the health condition, (i.e., changes in the HRAS gene have been associated with the development of bladder cancer); or
Mutations in the gene modify the course of the disease process (i.e., changes in the SMN2 gene modify the course of spinal muscular atrophy).
Each gene can be related to multiple conditions in all of the above ways.
Various conditions and subtypes of conditions are related to each other. The conditions in our initial project were chosen to represent genetically fairly well-characterized health conditions. To date, these conditions can be related to each other in parent–child relationships, which may involve a series of such parent–child relationships. We anticipate representing more complex relationships as we expand to include multifactored conditions.
The genes have relationships with other genes, with the knowledge of these rapidly expanding. Among these relationships is membership in a gene family, membership in a common biochemical pathway, functional interactions with other genes, and modifiers of the function of other genes. Within the Genetics Home Reference, we decided to include such relationships within “gene groups.” These groupings are meant to facilitate retrieval of genes related to each other and to conditions within the Genetics Home Reference. Each gene can be related to multiple gene groupings.
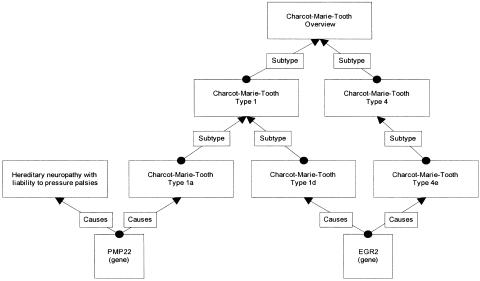
The overall data model between entities is a directed acyclic graph in which an entity can have more than one parent and more than one relationship. The overall management of these relationships within the Genetics Home Reference content manager is represented by a visual “content map” that links all related conditions and genes including condition subtypes and the various types of linking relationships. ▶ illustrates a partial content map for Charcot-Marie-Tooth (CMT) Disease showing two of the four CMT subtypes and two of 15 genes. We illustrate the nested parent–child subtype relationship for the CMT conditions. The PMP22 gene causes two separate health conditions (hereditary neuropathy with liability to pressure palsies and CMT type 1a).
Figure 1.
Partial content map for Charcot-Marie-Tooth disease. Charcot-Marie-Tooth disease shows the complex relationships between health conditions and genes. This figure only shows two of the fifteen genes and two of the disease subtypes. Geneticists use subtype relationships to distinguish between different inheritance patterns, different genetic causality, and different course or symptoms of the disease. This diagram shows that a health condition may be related to another health condition and to many genes; the genes may be related to many health conditions and may also be related to other genes.
The EGR2 gene causes two different subtypes (CMT 1d and CMT 4e) of CMT. The gene family relationship is not shown on the diagram: the PMP22 gene is part of the epithelial membrane gene family, whereas the EGR2 gene is part of the early growth response gene family.
Our work on the relationships between entities builds on the classification hierarchy developed by Edwards and Tarczy-Hornoch30 for use in GeneTests/GeneReviews. They use a directed graph with nodes that are either genes or disorders. We have built on this work to include multiple relationships between genes or disorders, to relate the disorders to each other via external knowledge sources as well as to relate the genes to each other within gene groups and also via external knowledge sources.
3. A Method to Relate the Content to External Knowledge Sources
We use two terminologies included in the UMLS to ground the Genetics Home Reference content to external classifications of both health conditions and genes. Because we link to other NLM consumer sites, MeSH31 was the logical choice for the hierarchy of health conditions. Linking to MeSH allows us to cross-reference the Genetics Home Reference to both MEDLINEplus and to ClinicalTrials.gov. This further facilitates ongoing discussions with the MeSH team about genetics issues. We use the Gene Ontology (GO)32 to create a browsable hierarchy for the genes. The GO presents a technical classification of gene function that is not very consumer oriented but is widely used in the scientific community. Further, the GO has recently been added to the UMLS and thus provides the opportunity for additional knowledge source linkages in the future. We use the top levels of the GO for browsing by gene. An example of this browse function using the GO is demonstrated by the list of genes that are involved in biological processes relating to behavior. The list includes the causal genes for Huntington Disease, Alzheimer Disease, and Lesch-Nyhan Syndrome. This GO browsing is parallel to the decision to use the top level MeSH hierarchy for browsing by conditions. Similarly, the user can browse the health conditions for those that affect mental health and behavior.
System Description
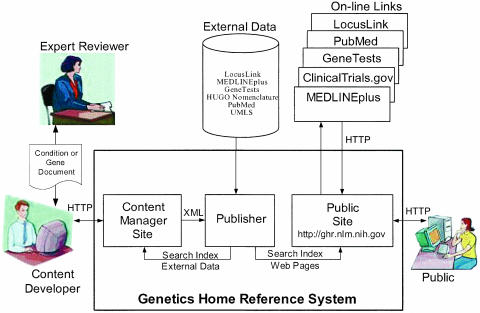
Our system provides support for collecting and maintaining content while responding to continual changes in data structure and supporting a stable public Web site. A modular architecture within an overall content manager system, shown in ▶, suits these needs. The content manager houses separate components for content development and public presentation, linked by a publishing component. A small in-house team of content developers selects the relevant data and augments it with other information to create the Genetics Home Reference content. The content is reviewed and revised by external medical genetics experts before it is released to the public site. Additional content manager components allow us to interrelate, index, and cross-reference the information with data from other high-quality, publicly available sources and publish it on the public site. The architecture is adapted from NLM's ClinicalTrials.gov,33 although substantial modifications were needed for the specific nature of the Genetics Home Reference project. We will concentrate our discussion on those items that are unique to the Genetics Home Reference system. We have included the data model in Appendices 1 and 2.
Figure 2.
Genetics Home Reference System architecture. The Genetics Home Reference system architecture is composed of a content manager system linked through a publisher to the public site. The system automatically collates available information, support data entry, and expert review of content, and presents a lay person's format to the public.
The content manager has been customized to extract and preprocess data from the major human data sources of the Human Genome Project, including LocusLink, OMIM, GeneTests, the Human Gene Nomenclature Committee (HGNC), and the GO. LocusLink data are filtered and sorted to identify the records that we automatically present as search suggestions on the public site. These data sources are linked to MEDLINEplus, ClinicalTrials.gov, and PubMed via search-based links and via MeSH-indexed content links. The HGNC data are the source of the “nomeid” that permits unambiguous linkage to other technical data sources.
The content manager has semi-automated techniques to handle the voluminous and ever-changing data of the Human Genome Project. Three types of automation are of note: (1) decoding, (2) automatic suggestions, and (3) change detection. First, advanced terminology or notation is automatically decoded into consumer-friendly words. For example, our system automatically decodes the standard description of the location of a gene. Thus, the location code for the APP gene, 21q21, is translated into “the APP gene is located on the long arm of chromosome 21 at position 21.” We follow up with a link to the “Help Me Understand Genetics” section that describes how to understand gene location codes. Second, the content manager searches available information to find suggestions on possibly related content. For example, when a gene record is first created, the system searches the LocusLink and HGNC data to find possibly related information. The suggestions are presented to the content developer who selects the best suggestions, searches for other suggestions, or chooses to ignore the suggestion. This assists in quickly finding the official gene name, location code, possible synonyms, LocusLink identifier, possible links to OMIM, possible links to GeneTests, and the SWISS-Prot ID. Third, the content manager periodically searches these multiple data sources to detect changes relating to the Genetics Home Reference data elements. The system presents notice of each detected change to the content developers. For example, when HGNC changes a gene symbol, the system detects the change and notifies the content developers who decide if or when the change warrants an update to the public site.
Status Report
The Genetics Home Reference became operational in May 2003 with about a dozen health conditions and genes. Content has been added steadily since then and usage has grown accordingly. ▶ shows the set of MEDLINEplus topics currently linked to the Genetics Home Reference content (about 100 conditions and 150 genes). We give some trends and observations over the first year of operation. The “Help Me Understand Genetics” section is heavily used and has many positive user comments. The browse and search functions to find content have approximately equal usage, with more usage of the browse function for conditions than for genes. In general, the health conditions are viewed more frequently than the genes; however, the gene pages are viewed quite frequently overall, helping to dispel the notion that the information about the genes would not be of interest to a consumer audience. The most frequently viewed glossary concepts include not only a combination of genetics terms (autosomal recessive, mutation, chromosome, etc.) but also more general medical terms (hemoglobin, autism, symptom). Most users access the site via search engines or from a MEDLINEplus topic.
Table 1.
MEDLINEplus Topics Linked to Genetics Home Reference Content (June 2004)
| • Adrenal gland disorders | • Genetic disorders |
| • Alpha-1 antitrypsin deficiency | • Genetic testing |
| • Alzheimer's caregivers | • Head and brain malformations |
| • Alzheimer's disease | • Hearing disorders and deafness |
| • Amyotrophic lateral sclerosis | • Hearing problems in children |
| • Anemia | • Hemochromatosis |
| • Ataxia telangiectasia | • Hemophilia |
| • Birth defects | • Huntington's disease |
| • Bladder cancer | • Infertility |
| • Bleeding disorders | • Kidney disease |
| • Bone diseases | • Leukodystrophies |
| • Bone marrow diseases | • Liver diseases |
| • Brain diseases | • Male breast cancer |
| • Breast cancer | • Male genital disorders |
| • Cancer | • Marfan syndrome |
| • Cardiomyopathy | • Metabolic disorders |
| • Carpal tunnel syndrome | • Muscular dystrophy |
| • Charcot-Marie-Tooth disease | • Neurofibromatosis |
| • Cleft lip and palate | • Neurologic diseases |
| • Connective tissue disorders | • Neuromuscular diseases |
| • Creutzfeldt-Jakob disease | • Osteogenesis imperfecta |
| • Cystic fibrosis | • Peripheral nerve disorders |
| • Degenerative nerve disease | • Phenylketonuria |
| • Dementia | • Porphyria |
| • Developmental disabilities | • Pulmonary hypertension |
| • Dwarfism | • Retinal disorders |
| • Ehlers-Danlos syndrome | • Sickle cell anemia |
| • Eye cancer | • Skin pigmentation disorders |
| • Facial injuries and disorders | • Spinal muscular atrophy |
| • Fragile X syndrome | • Stroke |
| • Gaucher's disease | • Tay-Sachs disease |
| • Genes and gene therapy | • Thyroid diseases |
| • Genetic brain disorders | • Tuberous sclerosis |
| • Genetic counseling | • Wilson's disease |
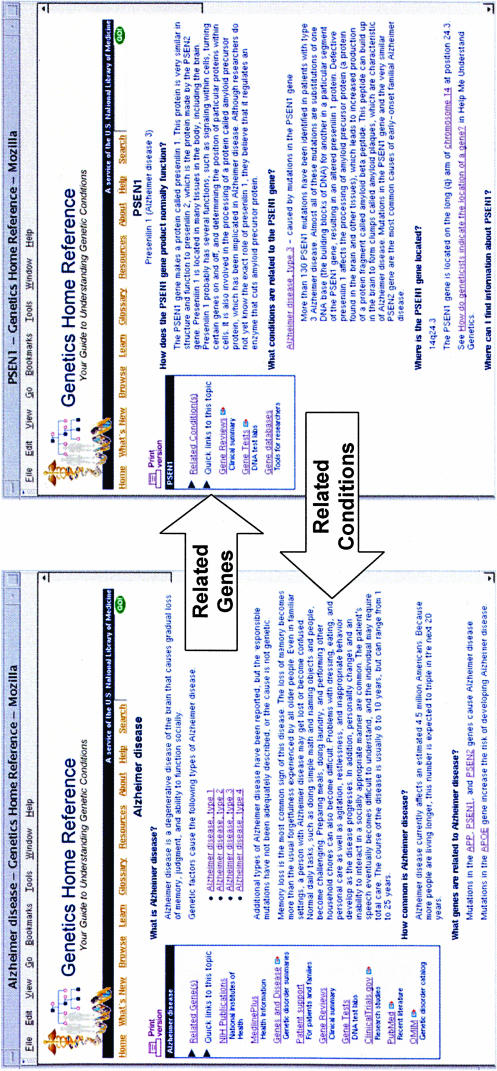
The system concentrates on the relationship between health conditions and the genes that are related to them. ▶ shows a partial view of the Alzheimer disease presentation from the Genetics Home Reference public site and illustrates the linkage between the health condition summary and the gene summary. The body of each condition or gene summary is displayed in a question and answer form. The left column displays links to specific pages on selected sites that present information on the selected topic. Also, the left column displays the link that connects related conditions and genes. Each condition or gene page links to a page listing the sources used to write the content. Each condition or gene page has links to glossary entry information as well as “Help Me Understand Genetics” topics that may aid in understanding the content.
Figure 3.
Health condition and gene pages from Genetics Home Reference. The Genetics Home Reference interlinks health conditions and genes to facilitate understanding of known gene-condition relationships. This example of Alzheimer disease shows the linkage to one of the genes and also the links to other information resources to assist in pursuing the topics more fully.
Discussion
We have addressed numerous challenges in the development of the Genetics Home Reference, but many others remain. We have built on the experiences and techniques of other researchers in similar domains. We worked closely with the technical team from ClinicalTrials.gov and have reused the architecture and a range of tools and code. We extended the GeneTests/GeneReviews classification hierarchy for genes and health conditions to include additional relationships and other entities. We plan to utilize other UMLS knowledge sources (including the recent addition of Gene Ontology to the UMLS) to further structure the content and the external links.
We struggle to strike the right balance in the Genetics Home Reference. This balance needs first to stay focused on providing information for a consumer audience but in an extremely technical area with great depth and detail. Our philosophy is to provide a learning resource for families with inherited conditions in which the individuals can find information at varying levels of complexity but will be able to come back multiple times over time and learn more. We strive to have some information at an introductory level but do not shrink from providing links to technical resources that a more advanced user would appreciate. This balance is also provided for health professionals who need to learn quickly about these rare health conditions and their genetic basis and to be able to discuss the information with family members. We are bridging multiple communities: consumers, health care providers, researchers, and students; such inter-community communications are difficult.
We are cognizant of the difference between providing information and providing advice. We have provided several links to other agencies that provide more assistance on specific topics (e.g., the Genetic and Rare Disease Information Center) and to Web sites to search for a genetics counselor or a genetics clinic located near the searcher to ask about personal conditions. Providing information on genetic disorders involves such potentially controversial issues as cloning, gene therapy, stem cell research, and prenatal diagnosis. The wording of the content as well as the wording of the links to other information resources is carefully considered.
The largest challenge is scaling the content to keep up with the constant changes in the underlying biological databases and their anticipated growth. Several of our efforts in the system design are in anticipation of these challenges. Thus, we believe that it is essential to structure the content internally and to relate this content to other knowledge sources. We are building on our initial efforts of discrepancy detection in our source databases to be able to detect important changes in content on a regular basis. Currently, there are more than 1,600 genes linked with specific human health conditions* and the rate of new discoveries is accelerating. Our current system needs to expand to cover paradigms other than single gene disorders. We are already expanding to include selected chromosome conditions. Future work with pharmacogenetics, immunogenetics, and environmental susceptibilities will undoubtedly require additions to the knowledge model and underlying data sources. We need to have automated tools to augment a small team of content experts as we proceed into more aspects of health implications of the Human Genome Project.
Conclusion
We have developed a system called the Genetics Home Reference to begin to answer a consumer's question, “How is genetics related to my health condition?” Our goal is to present to the public the health implications of the research evolving from the Human Genome Project. The first phase is to focus on topics that are in MEDLINEplus that have major single gene involvement, and then to eventually cover all genes that are implicated in human health conditions. The major informatics aspects of the system are its underlying data model to link health conditions and genes, the explicit linkage to structured knowledge sources, and the resulting bridging of the consumer health and scientific domains. The next phases will include working on the tools to allow us to find, retrieve, manipulate, and present this content in a manner the public can use. We are involved in ongoing evaluation studies to determine how we might improve its accessibility to the public.
Appendix 1. Health Condition Data Elements
| Public Site Presentation | Data Element | Data Type |
|---|---|---|
| What is condition name? | Name | Free text |
| Description | Free text | |
| Symptoms | Free text | |
| Subtype Condition Name | Free text | |
| How common is condition name? | Statistics | Free text |
| What genes are related to condition name? | Mutation Gene Symbol | From HUGO nomenclature or LocusLink |
| Mutation Type | Choose from: | |
| Causes | ||
| Predisposes | ||
| Associated | ||
| Etiology | Free text | |
| How do people inherit condition name? | Inheritance | Derived, editable free text |
| Default values for: | ||
| Unknown pattern | ||
| Autosomal dominant | ||
| Autosomal recessive | ||
| X-Linked | ||
| and additional text: | ||
| 1 affected parent | ||
| Somatic | ||
| Homozygous | ||
| Predispose | ||
| New mutation | ||
| Inherited or new mutation | ||
| Anticipation | ||
| Carrier | ||
| Environment | ||
| Sometimes Unknown | ||
| X-linked recessive | ||
| X-linked dominant | ||
| Where can I find information about condition name? | URL | Free text |
| Title | Free text | |
| What other names do people use for condition name? | Synonym | Name: free text |
| What glossary definitions help with understanding condition name? | Glossary concept or synonym | Link to glossary |
| What sources were used to construct this information? | URL | Free text |
| Title | Free text |
Appendix 2. Gene Data Elements
| Public Site Presentation | Data Element | Data Type |
|---|---|---|
| Official Symbol | HGNC Symbol | Free text |
| Official Name | HGNC Name | Free text |
| How does the gene product normally function? | Function | Free text |
| Gene group gene symbol and name | Free text | |
| What conditions are related to the gene? | Mutation condition name | Free text |
| Mutation type | Choose from: | |
| Causes | ||
| Predisposes | ||
| Associated | ||
| Modifies | ||
| Mutation description | Free text | |
| Where can I find information about the gene? | URL | Free text |
| Title | Free text | |
| Where is the gene located? | Location symbol | Free text |
| Location description | Derived, editable free text | |
| What other names do people use for the gene or gene products? | Synonym | Free text |
| What glossary definitions help with understanding the gene? | Glossary concept or synonym | Link to glossary |
| What sources were used to construct this information? | URL | Free text |
| Title | Free text |
The authors thank the other members of the Genetics Home Reference team (M Cheh, SLH Davenport, E Dorfman, CM Fomous, J Gillen, N Ide, R Loane, R Logan, SM Morrison, DM Mucci, P Wolfe, K Zeng) as well as the expert reviewers.
Based on a search in June 2004 of the LocusLink database (http://www.locuslink.ncbi.nlm.nih.gov_) with the query Disease_known AND Has_seq for the human species.
References
- 1.Cline RJ, Haynes K. Consumer health information seeking on the Internet: the state of the art. Health Educ Res. 2001;16:671–92. [DOI] [PubMed] [Google Scholar]
- 2.Fox S, Fallows D. Internet Health Resources, 2003. Washington, DC: Pew Foundation Pew Internet and American Life Project.
- 3.Gupte CM, Hassan AN, McDermott ID, Thomas RD. The internet—friend or foe? A questionnaire study of orthopaedic out-patients. Ann R Coll Surg Engl. 2002;84:187–92. [PMC free article] [PubMed] [Google Scholar]
- 4.Rokade A, Kapoor PKD, Rao S, Rokade V, Reddy KTV, Kumar BN. Has the internet overtaken other traditional sources of health information? Questionnaire survey of patients attending ENT outpatient clinics. Clin Otolaryngol. 2002;27:526–8. [DOI] [PubMed] [Google Scholar]
- 5.Eysenbach G, Powell J, Kuss OSE. Empirical studies assessing the quality of health information for consumers on the world wide web: a systematic review. JAMA. 2002;287:2691–700. [DOI] [PubMed] [Google Scholar]
- 6.Rudd RE, Moeykens BA, Colton TC. Health and literacy: a review of medical and public health literature. Annual Review of Adult Learning and Literacy. San Francisco, CA: Jossey-Bass, 1999, pp 158–99.
- 7.Taylor MR, Alman A, Manchester DK. Use of the Internet by patients and their families to obtain genetics-related information [comment]. Mayo Clin Proc. 2001;76(8):772–6. [DOI] [PubMed] [Google Scholar]
- 8.U.S. National Institutes of Health, National Library of Medicine. Genetics Home Reference. [Online] Available at http://ghr.nlm.nih.gov. Accessed June 25, 2004.
- 9.U.S. National Institutes of Health, National Library of Medicine. MEDLINEplus. [Online] Available at http://www.medlineplus.gov. Accessed June 25, 2004.
- 10.U.S. National Institutes of Health, National Library of Medicine. ClinicalTrials.gov. [Online] Available at http://clinicaltrials.gov/. Accessed June 25, 2004.
- 11.U.S. National Institutes of Health, National Library of Medicine. PubMed. [Online] Available at http://www.ncbi.nlm.nih.gov.PubMed. Accessed June 25, 2004.
- 12.University of Washington, Seattle (with funding from U.S. National Institutes of Health, National Library of Medicine, National Human Genome Research Institute). GeneTests. [Online] Available at http://www.genetests.org. Accessed June 25, 2004.
- 13.U.S. National Institutes of Health, National Library of Medicine, National Center for Biotechnology Information. LocusLink (changing to Entrez Gene soon). [Online] Available at http://www.ncbi.nlm.nih.gov/LocusLink. Accessed June 25, 2004.
- 14.U.S. National Institutes of Health, National Library of Medicine, National Center for Biotechnology Information and Johns Hopkins University. Online Mendelian Inheritance in Man (OMIM). [Online] Available at http://www.ncbi.nlm.nih.gov/Omim/. Accessed June 25, 2004.
- 15.Mitchell J, McCray A, Bodenreider O. From phenotype to genotype: issues in navigating the available information resources. Methods Inf Med. 2003;42:557–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Safran M, Solomon I, Shmueli O, et al. GeneCards 2002: towards a complete, object-oriented, human gene compendium. Bioinformatics. 2002;18:1542–3. [DOI] [PubMed] [Google Scholar]
- 17.U.S. National Institutes of Health, National Library of Medicine, National Center for Biotechnology Information, University College London, UK Medical Research Council, and Wellcome Trust. HUGO Human Genome Nomenclature Committee (HGNC). [Online] Available at http://www.gene.ucl.ac.uk/nomenclature/. Accessed June 25, 2004.
- 18.Swiss Institute of Bioinformatics and European Bioinformatics Institute. Swiss-Prot: Curated Protein Sequence Database. [Online] Available at http://us.expasy.org.sprot/. Accessed June 25, 2004.
- 19.U.S. National Institutes of Health, National Library of Medicine, National Center for Biotechnology Information. GenBank. [Online] Available at http://www.ncbi.nih.gov/GenBank/. Accessed June 25, 2004.
- 20.University of Wales College of Medicine, Institute of Medical Genetics in Cardiff. Human Gene Mutation Database (HGMD). [Online] Available at http://archive.uwcm.ac.uk.uwcm/mg/hgmd0.html. Accessed June 25, 2004.
- 21.Hamosh A, Scott AF, Amberger J, Bocchini C, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledge base of human genes and genetic disorders. Nucleic Acids Res. 2002;1:52–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pagon RA, Tarczy-Hornoch P, Baskin PK, et al. GeneTests-GeneClinics: genetic testing information for a growing audience. Hum Mutat. 2002;19:501–9. [DOI] [PubMed] [Google Scholar]
- 23.Tarczy-Hornoch P, Covington ML, Edwards J, Shannon P, Fuller S, Pagon RA. Creation and maintenance of helix, a Web based database of medical genetics laboratories, to serve the needs of the genetics community. Proceedings / AMIA Annual Symposium. 1998:341–5. [PMC free article] [PubMed]
- 24.Tarczy-Hornoch P, Shannon P, Baskin P, Espeseth M, Pagon RA. GeneClinics: a hybrid text/data electronic publishing model using XML applied to clinical genetic testing. J Am Med Inform Assoc. 2000;7:267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.U.S. National Institutes of Health, Office of Rare Diseases. Rare Diseases Information. [Online] Available at http://rarediseases.info.nih.gov. Accessed June 25, 2004.
- 27.U.S. Department of Energy, Office of Science. Human Genome Project. [Online] Available at http://ornl.gov/TechResources/Human_Genome. Accessed June 25, 2004.
- 28.U.S. National Institutes of Health, National Cancer Institute. NCI Dictionary of Cancer Terms. [Online] Available at http://www.nci.nih.gov/dictionary. Accessed June 25, 2004.
- 29.McKusick VA. Mendelian Inheritance in Man. Baltimore, MD: Johns Hopkins University Press, 1996.
- 30.Edwards J, Tarczy-Hornoch P. Implementation of a classification hierarchy for the GeneTests/GeneClinics genetic testing databases. Proceedings/AMIA Annual Symposium. 2002:235–9. [PMC free article] [PubMed]
- 31.U.S. National Institutes of Health, National Library of Medicine. Medical Subject Headings (MeSH). [Online] Available at http://www.nlm.nih.gov/mesh/meshhome.html. Accessed June 25, 2004.
- 32.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCray AT, Ide NC. Design and implementation of a national clinical trials registry. J Am Med Inform Assoc. 2000;7:313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]