Abstract
Query Integrator System (QIS) is a database mediator framework intended to address robust data integration from continuously changing heterogeneous data sources in the biosciences. Currently in the advanced prototype stage, it is being used on a production basis to integrate data from neuroscience databases developed for the SenseLab project at Yale University with external neuroscience and genomics databases. The QIS framework uses standard technologies and is intended to be deployable by administrators with a moderate level of technological expertise: It comes with various tools, such as interfaces for the design of distributed queries. The QIS architecture is based on a set of distributed network-based servers, data source servers, integration servers, and ontology servers, that exchange metadata as well as mappings of both metadata and data elements to elements in an ontology. Metadata version difference determination coupled with decomposition of stored queries is used as the basis for partial query recovery when the schema of data sources alters.
A major long-term goal of the national Human Brain Project (HBP),1 a loosely knit consortium of neuroscience researchers, is interoperability between the “federation” of databases produced by its members. The challenge is to devise a robust approach that, among other things, enhances the ability to answer complex research questions. At the most basic level, database interoperation means querying multiple databases in a single logical operation to retrieve data of interest from each.
This paper outlines barriers to interoperability of bioscience databases, summarizes previous interoperation approaches, and then describes Query Integrator System (QIS), a system developed to allow multidatabase network-based queries in the biosciences. QIS is based on a distributed architecture and is designed to facilitate maintenance of query integrity as the underlying database schemas evolve over time. QIS has been developed in the context of SenseLab,2,3 an ongoing neuroinformatics project at Yale University supported by the HBP. We discuss its current status, plans for future work, and lessons learned.
Background
Barriers to Federation of Bioscience Databases
Considerable technical expertise is required to set up an effective federated-query infrastructure. Bioscience database schemas evolve significantly and rapidly because of scientific progress, changing research goals, and system redesign as better ways of representing data are discovered. For performance and safety reasons, participating databases typically do not support unrestricted Structured Query Language (SQL) queries. Instead, predefined, parameterized queries provide commonly requested results. Here, alterations in database structure may cause predefined queries to “break.”
In a database federation, especially one involving international collaborations, different groups may use subtly or overtly different names to refer to the same class of data (synonymy). Conversely, different groups may use the same name in subtly different ways (polysemy). These differences in meaning, if not documented explicitly, complicate interoperation. To address these issues, one needs shared controlled vocabulary support. Specifically, both data and metadata (“data that describe other data”) must be mapped to concepts in controlled vocabularies. Such curator-intensive mapping efforts yield only modest benefit if existing standard vocabularies provide insufficient domain coverage or if the field progresses faster than corresponding curatorial efforts at vocabulary enhancement. It therefore may be necessary to create federation-specific (“local”) vocabularies and to devise mechanisms to facilitate the identification of candidates for new concepts within individual databases at both the metadata and data level.
Individual participating schemas must also be accompanied by detailed textual annotations. Such annotations must be far more extensive and lucid than documentation developed for internal purposes because they must provide considerable overview for researchers who are unfamiliar with a particular group's interests and experimental methodology. Annotations must often exist at two levels: metadata and data. Creating uniformly high-quality annotations is time-consuming, and, even when they exist, they may not be enough. Kans and Oullette4 state: “As good as annotations can be, they will never surpass a published article in fully representing large amounts of biology. It is therefore imperative to ensure the proper link between a research publication and the primary data it will cite.”
In addition, federated search mechanisms must appropriately exclude data that are still preliminary and not available for public access beyond the research group creating an individual database. Some of the work required to establish federated search mechanisms (such as describing the semantics of data carefully and defining which data/metadata are public) is unavoidable no matter what approach is used. A robust framework, however, can minimize other barriers to effective retrieval from federated databases.
Existing Approaches for Database Federation
The first step in data interoperability involves data source access. The simplest method is based on downloadable files, which are impractical for large, volatile datasets. Remote (direct) database access through vendor-independent standards such as ODBC (Open Database Connectivity) carries implications such as increased resource administration, security risks, and institutional firewall restrictions. For data sources exposed only as Web pages, the limitation of HTML (interleaving content with formatting) makes content extraction tedious, nonscalable, and fragile as data proliferate and Web page cosmetics change without notice. Second-generation Web sites, such as National Center for Biotechnology Information's Entrez and the current version of SenseLab, circumvent this problem by allowing users to get data as extended markup language (XML) (“pure” content) using programmable ad hoc query interfaces. The use of site-specific XML data alone, however, is not intended to address federation across multiple sites.
A mechanism to relate contents in federated databases uses the global schema approach based on an agreed-on (and infrequently revised) standard definition of domain-specific data types and classes and their interrelationships. Anyone who uses this standard (or a subset) must not deviate from it. Microsoft's “BizTalk”5 E-commerce specification exemplifies this approach, which is appropriate in situations in which the organization of domain knowledge does not change very fast. Its broad applicability in rapidly evolving areas such as bioscience seems doubtful.
In contrast to global schema-based systems, mediator systems allow a single query to be translated into the language recognized by heterogeneous databases, extracting their information and integrating the results in a single dataset. One type of mediator uses a single repository that stores both the schema description (metadata) as well as data of every included database. This approach, often used within a single geographically dispersed organization, has the advantage that queries against the integrated data run relatively fast because one does not attempt to do “joins” of geographically separated tables over the Internet. However, it becomes enormously more complicated in the presence of highly heterogeneous schemas that are maintained autonomously and may change often; the extremely close coordination required between master and satellite sites may not be feasible in relatively loose research federations.
Other mediators6,7,8 limit themselves to metadata exchange and leave the data in their original databases and format. Here, queries are described in a common language and are translated into the specific data source syntax, and results are converted into a common output format. Some of these systems are commercially available, e.g., IBM's DiscoveryLink7 and Genetic Exchange's discoveryHub6,9; they are limited with respect to the ranges of data sources accessed. Further, they do not expressly address the issue of schema evolution. Although they are valuable for dealing with data source connectivity in domains where the individual logical schemas evolve very slowly, if at all, the applicability of these systems to federations where continual schema evolution is the rule is doubtful. We contend that, in the bioscience domain, one must address heterogeneous data mediation and schema adaptation together to achieve robust evolvable data integration.
Current Research in Schema Evolution and Database Federation
From the vast literature, we focus only on papers dealing with problems directly related to this paper's theme. McBrien and Poulovassilis10 treat a database schema as a graph structure, defining changes in terms of operations (such as addition and removal) on the graph's nodes and edges. This paper does not differentiate between nodes representing tables and nodes representing a table's columns; also, transformations such as column datatype changes are not considered. The treatments by Ram and Shankaranarayanan11 and Li and Tari12 are much more comprehensive. The former tries to automate the transformation of one schema to another by storing the atomic changes in computable form. The second associates schema evolution with version management, identifying the former as an application of the latter and defining “versioning algebra” in terms of operations on schema elements.
Determination of differences between two schemas, or between two versions of a schema, is the inverse of the schema-evolution problem. Kim and Seo13 devised a taxonomy of schema differences at the class/table, attribute/column, and domain/data type levels. Sheth and Kashyap14 have modified this taxonomy to emphasize semantic differences.
In a loose federation, however, individual schemas tend to evolve in a far more unplanned and nonorderly fashion than conceived in the above-mentioned papers. Specifically, individual groups' system architects may freely alter their schemas as needed but may communicate details of such changes to the federation much later. In such scenarios, it is desirable to minimize and/or streamline communication efforts by enabling discovery of schema version differences by the federation mediator.
Structural differences between schema versions (addition or removal of tables/columns, changes in column properties, and changes in table relationships) can be discovered in a fairly straightforward manner using existing database connectivity technology. The database administrator must only allow a mediator program read-only access to the database. However, the meaning of the differences in terms of the domain, or evolving domain-specific needs, cannot generally be inferred automatically, even if detailed annotations are provided because of the generally unstructured and narrative nature of the latter.
Previous Efforts in Bioinformatics
The TAMBIS (Transparent Access to Multiple Bioinformatics Information Sources)15,16 project creates a bioinformatics domain ontology using the GRAIL Description Logic Language17, mapping concepts to existing information sources. Queries against that ontology access individual sources in a user-transparent manner. Although this approach is novel, scalability and expressivity are a concern. The TAMBIS team uses custom function libraries for each information source, which provide a function-based view of the source. Although its underlying query language, Collection Programming Language,18 supports issuing of SQL commands that are passed untranslated to a database, it is well known that function-based perspectives of data that are essentially tabular are not appropriate when one wishes to execute the equivalent of SQL statements that join several tables arbitrarily. Further, the depth and quality of the TAMBIS ontology, or its overlap with existing bioinformatics ontologies such as Gene Ontology (http://www.geneontology.org), are also difficult to evaluate because the ontology contents are not contributed currently to a source such as the Unified Medical Language System.19
The PQL structured language (PQL) approach of Mork et al.20,21 relies on metadata describing the entities and relationships (“links”) between entities in a federated schema plus additional metadata on intended semantics or judgments about database curation quality to discover multiple ways of answering loosely defined queries, such as finding all proteins “closely related” to a disease. The PQL query language resembles the XQuery language used to query XML documents. It provides access to several data sources, including nonstructured textual data such as Online Mendelian Inheritance in Man.22 PQL's metadata appear to be created manually by the system developers: Certainly, there is no other way to create metadata about topics such as curation quality. This approach is interesting and valid. It is geared, however, to discovering ways to answer loosely defined queries rather than performing consistency checking on existing queries that are well defined, which is the focus of our work.
System Design Objectives
The objectives underlying the creation of the QIS were as follows:
to integrate data sources within the HBP that use technologies and approaches not supported by commercial mediator systems, e.g., our own Entity-Attribute-Value, with Classes and Relationships (EAV/CR) approach23,24 and the “common data model for neuroscience”25
to devise a scalable approach that can work in a loosely coupled database federation that explicitly addresses the issue of schema evolution within the participating databases
to devise robust mechanisms for metadata exchange
to address the “federated query fragility problem” by devising mechanisms that use differences in metadata versions to facilitate automatic detection of schema evolution and assess their impact on existing stored queries and to use such impact assessment, where possible, for partial or complete recovery of stored queries when target schemas have altered
to support interoperation with a straightforward separation between public and private data
to facilitate recording of system semantics through an infrastructure that integrates ontologies and detailed text annotations with federation and allows sites to contribute candidate concepts to an ontology development group in a seamless fashion
to devise a low-cost and lightweight system that requires a relatively modest infrastructure that researchers with limited informatics skills can operate and that can eventually be distributed as an open source
QIS addresses the following issues: data source connectivity, heterogeneous schema mapping, common query formulation, query adaptation, and data delivery. QIS also features simplified deployment, easy maintenance, enforced security, firewall independency, alert systems, and domain independency.
System Description
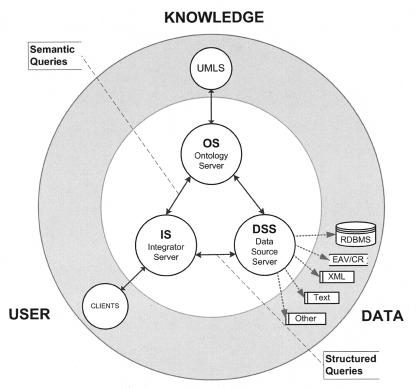
QIS belongs to the class of mediator systems that limit themselves to metadata exchange. It uses a distributed architecture that is composed of three main functional units: integrator servers (ISs), data source servers (DSSs), and the ontology server (OS). These units form the system's middle tier, connecting “data consumers” (client applications requesting query execution) with “data providers” (back-end data sources containing the data) and knowledge sources (Ontologies) (▶). All servers use a database management system (DBMS) (currently Microsoft SQL Server) in addition to a Web server (Microsoft Internet Information Server). The annotated schemas of each unit are described in Appendix 1 (available as an online data supplement at www.jamia.org).
Figure 1.
Query Integration System—architectural overview. The QIS architecture is based on three middle-tier servers. The data source server (DSS) connects to disparate supported data sources. The Integrator Server (IS) stores, coordinates query execution, and returns query results to Web applications. The Ontology Server (OS) maps data sources' metadata and data elements to concepts in standard vocabularies. EAV/CR = Entity-Attribute-Value, with Classes and Relationships; RDBMS = Rational Database Management System; XML = extended markup language; UMLS = Unified Medical Language System.
DSSs provide access to various data sources within a single group (or cooperating groups within an institution). In addition to traditional relational databases and EAV/CR databases that are built on top of relational technology, they also access XML files and flat files (spreadsheets, text). DSS administrators add definitions of data sources to the DSS through a Web interface.
Schema capture is the process of capturing metadata about a database (e.g., its table, column, and relationship definitions) into structured form. This process is partly automated through connectivity technologies that query a database's system data dictionary, or an XML schema. However, the captured metadata need to be manually enriched by mapping schema elements, where possible, to elements in ontologies to assist automatic schema discovery and by detailed textual annotations to provide semantic overview.
Each DSS itself is like a “metadatabase” that accesses one or more individual databases at a site. The administrators of individual databases must define the subset of the data and metadata within their own schema that is “public.” This is done through a three-step process for each data source:
A special account with restricted privileges is created for the DSS. This account cannot alter data and can access only a limited set of tables or views.
For tables containing both public and private data, an extra Boolean column is added to indicate whether the row in question is publicly accessible, with a default of “No/False” so rows that are to be made public must be manually set.
The administrator creates views that define the subset of public columns/tables. Where the views use tables with both public and private data, the view must specify a filter that the “public” flag must be “True.” The DSS account is now given permission to these views.
For non–account-oriented data, such as XML or text files, which must be accessed in their entirety, the DSS stores the URL of the source.
ISs store “public” metadata from DSSs as well as queries that access single or multiple data sources. They allow building of queries against the DSSs through a graphical user interface. QIS is primarily intended to allow other Web-based applications to execute predefined queries on the IS through Web-service26 mechanisms. That is, the IS operates “behind the scenes,” and the federation's end users connect to such an application rather than to the IS directly. IS administrators perform tasks such as registering new DSSs and registering individuals with domain expertise who can design queries.
An OS maintains an integrated schema, plus content, of one or more controlled vocabularies used within the federation. Alternatively, it may provide a gateway to relate these vocabularies to standardized content maintained elsewhere, or it may replicate such content. Parts of the OS schema are recorded redundantly at the IS level.
The OS supports mapping of elements in individual data sources to vocabulary elements by curators who specialize in ontology development. More important, once this information is replicated on the DSS, mappings at both the metadata and data levels are also forwarded to the IS. The OS can now act as an information source map (ISM).27 Such information makes it possible to ask questions of varying granularity, such as “which data sources contain information on neurons” (where mappings are likely to exist at the metadata level) and “which data sources contain information on cerebellar Purkinje cells” (where mappings will likely exist at the data level). The mapping to specific schema elements in a data source allows assisted query composition against that data source that would actually return the desired data.
Data and metadata elements from DSSs that are candidates for local concept creation are exported to the OS in a facilitated fashion, as discussed later. Other potential services envisaged for the OS are term translation and unit conversion. The OS also lets ontology curators collaborate with DSS curators to jointly define new “local” (federation-specific) concepts: This is necessary when existing ontologies offer insufficient coverage. This infrastructure can also be used to submit new, curated concepts to a standard vocabulary for inclusion in a new release.
Communication between the various QIS nodes is XML encoded and HTTP delivered to support communication through network firewalls. Asynchronous processing is supported using customized queuing services. Other software technologies used by the system are Microsoft Active Data Objects for data and schema access, Extended Markup Language Document Object Model, and SAX (Simple Application Programming Interface for XML) for dataset manipulation, and Scalable Vector Graphics28 for standardized entity relationship (ER) diagram generation.
System Features
Dealing with Schema Evolution
For robustness of the federated query infrastructure, changes to the physical or conceptual schema at the individual data sources must be propagated efficiently to the IS. The DSS performs periodic automated schema extraction from its individual data sources and computes a schema version difference by comparing the new schema with the old. This computation uses the principle underlying the well-known diff algorithm,29 the basis of source-code control systems.30 Diff is an example of an algorithm that determines the “edit distance”31 between two objects (text files, DNA sequences), where change is defined as the series of additions, deletions, or replacements required to transform one object to the other. In QIS, changes are computed first at the aggregate (class/table/view) level and then at the atomic/column level.
Not all replacements can be inferred automatically. For example, changes in data type, length, and precision of a column are inferred reliably, but the less common renaming of a column/table appears as a combination of an addition and a deletion, and it is typically up to a curator to note that the two differences can be merged into one: Algorithms that try to infer replacements based on synonymy between old and new names are not guaranteed to work reliably for tables or columns whose names may be abbreviated or use characters such as underscores. In the case of EAV/CR data sources, automatic schema evolution is facilitated due to the metadata identification's preservation regardless of element renaming.
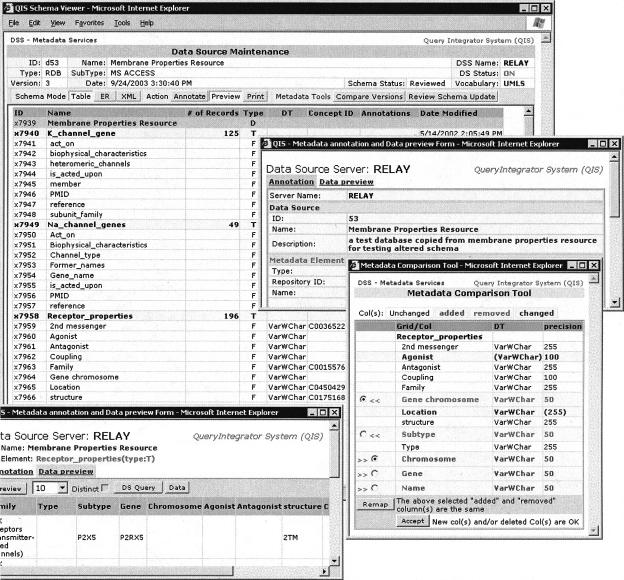
The DSS computes the “deltas” (differences) between the old and new schemas. The discovery of deltas is used to notify the responsible DSS curators. The deltas can then be annotated and/or curated to identify replacements. ▶ shows a screenshot of the curation/annotation interface. Replacements result in a version increment of the affected element; deletions result in a version change of the parent element. In general, all ancestors' versions are increased by one for the changed elements, and deltas contain information about the elements whose version differs. Obsolete metadata entries are moved to a metadata “history” table.
Figure 2.
The metadata maintenance toolset. In the rear window, the schema viewer shows the Membrane Properties Resource metadata description in tabular form. This information can alternatively be shown as an Entity relationship diagram or extended markup language. Selection of the “Receptor_properties” grid shows the detailed metadata annotation: description, concept identification, and semantic relationship (only for columns). The administrator can preview the underlying data to clarify the content. The front window shows the versioning update tool showing the differences (deltas) between previous and current versions with controls to resolve them.
The curated deltas are sent from the DSS to the IS. At the IS, they are used to update (synchronize) the metadata for that data source. Although metadata updates are intended to be largely automatic, periodic reports on the delta audit trail can be the basis for a dialog between the IS curators and DSS curators. This may happen if, for example, the annotation of particular elements is insufficient for the IS curators' understanding. Note that the deltas only identify structural differences; attributing meaning to these changes in terms of the domain is beyond their scope. That is why human annotations are necessary. Automatic delta identification, however, is critically important because identifying differences and presenting these to a human expert facilitate their comprehensive annotation.
Composing Queries: Preserving Integrity of Federated Queries
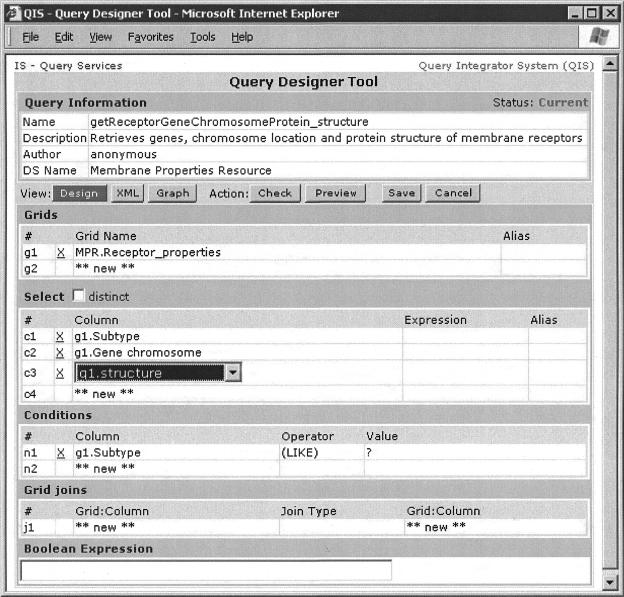
The IS, as a query repository, provides a tool (▶) to support the creation, maintenance, storage, and execution of data queries. To create federated queries, one first creates several single data source queries and then combines them into a query of queries by specifying intersection, union, or difference operations.
Figure 3.
Query designer tool at the integrator server showing information about the “getReceptorGeneChromosomeProtein_structure” query. This particular query extracts information from the “Receptor_properties” table in the Membrane Properties Resource (MPR) database. The extracted information includes the following fields: subtype, gene chromosome, and structure. This is a parameterized query that requires a specific text string pattern to fit the subtype field. Note that the tool refers to grid and column rather than tables and fields that are specific to Rational Database. This tool interacts with the user in three different ways: visual query by example (current), extended markup language, and entity-relationship graph. The query can be checked for correctness, executed in preview mode, and saved. The Graphic User Interface is based on a drill-down metaphor in which users pick one or more tables/sets from one or more data sources and then select columns/attributes from these. (When making a selection, choices are dynamically presented as pull-down menus, as shown for item #c3). Constraints are then specified for columns/attributes of interest.
A QIS query description derives from SQL-like languages but is represented in XML to facilitate syntax validation and future feature extensibility. The query is basically decomposed (“preparsed”) into its constituent elements, represented in terms of their metadata repository “unique identifiers.” Further, for atomic/column elements in a query, the IS records whether the element is part of the output (i.e., to be displayed), whether it is used in the equivalent of a “join” to bridge between two tables, and whether it is part of a query criterion/filter (in SQL parlance, part of the WHERE clause). Further details of the query language are beyond the scope of this paper but are available at http://ycmi.med.yale.edu/qis/qis_main.htm#Query.
Because this information is stored and indexed by metadata identifiers, delta computation determines which queries are affected by a change in a particular element. The authors of the affected queries are now alerted, so that the query may be fixed manually. However, the severity of this impact can also be determined automatically as described below. In many cases, even before fixing, “self-repairing” mechanisms can be activated dynamically so that the query can still run, returning partial or complete results.
Atomic elements (columns) used in joins will typically break the query irreparably if they are missing in a new schema version because the column's omission would cause a pathological Cartesian join.
Changes in data type are tolerable if the join condition is modified dynamically by a type-conversion function so as to temporarily restore the old data type. Query performance, however, may be impaired because this step can be computationally expensive.
Changes in length or precision of a column will not affect a query.
Atomic elements used in a query filter will, if removed, make the query less selective. In the worst case, one may return an entire table instead of the desired rows: This is generally unacceptable. Data type changes in such elements can be compensated temporarily by dynamic type conversion.
Atomic elements that are only displayed have the least impact. Such elements can simply be dropped from the query and replaced with placeholders indicating that they are now missing. Similarly, if the query is part of a union query that reaches out to multiple data sources or even multiple DSSs, data from other (unaltered) sources can still be returned.
If a table has changed by addition of new columns, queries do not need to be altered. However, query authors using that table are informed about the new columns, so that these can be used if necessary. Queries on single data sources that return all the columns in a table/view can automatically take advantage of the new columns.
Delta determination is ordinarily a batch process, and a user may execute a federated query against a changed schema before delta computation, and the query may fail. The DSSs will, however, sense this failure through standard error detection mechanisms. The failure triggers delta computation and subsequent metadata update. If possible, the query will be automatically reissued, relying on the self-repairing mechanisms described above to return partial or complete results, albeit after some delay. (This scenario is illustrated later.)
Pilot Implementation: Neuroscience and Genomics
The QIS components have been built on the Microsoft Windows platform using a Web server (MS Internet Information Server), the MS SQL server DBMS (although our database access code is vendor neutral), and the Microsoft .NET platform. Communication between servers is XML based. We have implemented a demonstration QIS-Client Web site that displays the information provided by one IS (at http://ycmi-hbp.med.yale.edu/QISClient-IIS/M1506.asp). The site invokes sample queries stored in an IS and displays code samples demonstrating how the system can be used from other Web sites.
Neuroscience
These examples are “behind-the-scenes” queries used in production SenseLab. (The SenseLab application acts as a “client” of QIS.)
The first query combines data from SenseLab's CellPropDB, which stores experimental data on neuronal cell properties, and the membrane properties inventory resource (MPIR), a standalone MS-Access database independently developed and maintained by a Yale researcher, which stores different information on neuronal membrane data. The CellPropDB query extracts receptor and ion channels information for a user-specified cell type, which is the parameter to this query. The MPIR query extracts gene information associated with a list of receptors and ion channels (the parameter to the second query). The join yields to the genes expressed in a particular cell. This example is in use by the production version of SenseLab. For the thalamic relay neuron's information, go to http://senselab.med.yale.edu/senselab/CellPropDB/GeneData.asp?cellid=262.
The second query integrates data from CellPropDB and the University of Southern California's Brain Architecture Management System (BAMS)32 (http://brancusi.usc.edu/bkms), to which we have restricted access as part of a collaboration. A query of brain structure (such as the hippocampus) from CellPropDB is augmented with neuroscience nomenclature provided by BAMS. To view this query, go to http://senselab.med.yale.edu/senselab/CellPropDB/cpdbRegions.asp?sr=1, and follow the hyperlink under “hippocampus” in the third column. Here, the program code uses the QIS application-programming interface to execute a specific query (of BAMS), and merge its results into a local (SenseLab) result set.
Genomics
The “Genomics-Microarray” query example from the demonstration QIS-Client Web site accesses three genomic databases, maintained by different Yale groups:
the Yale Microarray Database (YMD), a large repository of institutionally generated experimental information
a local gene annotation database (GAD) that contains curated genomic data on approximately 70,000 genes from the National Center for Biotechnology Information's LocusLink and Unigene datasets
a Yale installation of the well-known Gene Ontology (GO) database, which is curated by the Gene Ontology consortium (http://www.geneontology.org/GO.doc.html)
The query is intended to get microarray experiment results for any genes with cytokine activity. It takes about 2 minutes to run and operates as follows. All gene ontology accession numbers where the descriptions containing “cytokine” are pulled from GO. GAD is then queried to fetch the GenInfo ID, gene symbol, and gene name for these accession numbers. YMD is finally queried to fetch summaries of microarray experiments that were indexed by these GenInfo IDs.
Query Adaptation
This example is based on an actual case from the SenseLab project. The membrane properties resource database records, for each receptor molecule, the gene from which it is derived and its chromosomal cytogenetic location. The latter was originally expressed in the string form in which it is typically recorded in the literature, e.g., “11q12-q13.” (The hyphen indicates a region of uncertainty within two cytogenetic bands.) We decided to partition the location information into its three components: chromosome, upper (short-arm, p-terminal) location extent, and lower (long-arm, q-terminal) location extent. In the above example, the values of the three fields would be 11, q12, and q13. The extents can be converted to numeric fractions, which allow searching by location range as well as generation of graphics (“ideograms”) showing uncertainty regions.
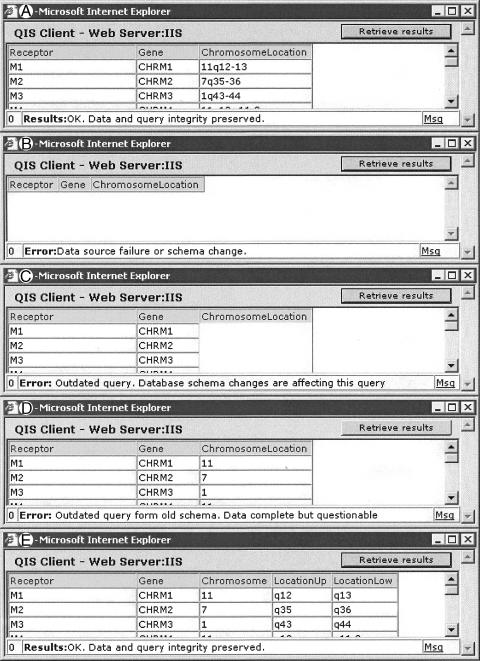
▶ shows the results for the query “show me the genes and chromosomal location information for all muscarinic receptors” with the original database structure. ▶ shows the query after the structure has been altered—the query has failed—with an error message about the query being out of date. As result, the DSS initiates an automated metadata refresh: The system now realizes that the “ChromosomeLocation” field is missing and shows only Gene information (▶). (Note that partial results are still returned rather than the query failing completely; these assist troubleshooting by the query designer.)
Figure 4.
Query adaptation: this query asks for genes and chromosome location information for all muscarinic membrane receptors. (A) The query runs against the original schema of a particular data source server and returns intended results. (B) The schema has changed; the result field “chromosome location” is partitioned into three fields (chromosome and upper and lower chromosomal location extents). When the query runs again, it fails, triggering an automatic metadata refresh at the integrator server. (C) Partial recovery: gene information is returned, but chromosome location is not. (D) Initial exploration and use of data preview show that the “chromosome” field returns only the initial part of the location information. (E) Incorporating the additional two fields returns the original information. Only a user with query-creation privileges sees the screens in B through D (which allow troubleshooting and query repair). To users without such privileges, an error message would simply state that the query is obsolete and that an administrator has been notified.
▶ shows the result of human intervention. ▶ shows the result after preliminary exploration, where the field “Chromosome” replaces “Chromosome Location.” It can be seen that the results returned lack cytogenetic information. ▶ shows the result of a correct query rewrite, where three fields replace the initial field.
OS Operation
The OS currently hosts a replicated copy of the National Library of Medicine's Unified Medical Language System (UMLS). Approximately a third of the concepts (metadata + data) in SenseLab have been mapped to UMLS concepts in batch mode using a tailored version of an algorithm originally described,33 with results manually verified.
The OS currently provides an important function for SenseLab, which, although residing within a single physical database, is divided into a number of “virtual” databases or portals to provide direct access to data of interest to a variety of neuroscience communities (e.g., neuronal modelers, olfactory receptor researchers). Although this division is convenient for the regular visitor of SenseLab, it is a barrier to the new user who wants to directly access objects of interest without first having to know in which virtual database they might lie. Further, neuroscience has numerous synonyms (“5-HT,” “serotonin,” “5-hydroxytryptamine” are the same molecule, as are “norepinephrine” and “noradrenaline”). It is desirable to use UMLS's synonymy information to facilitate query expansion during searching.
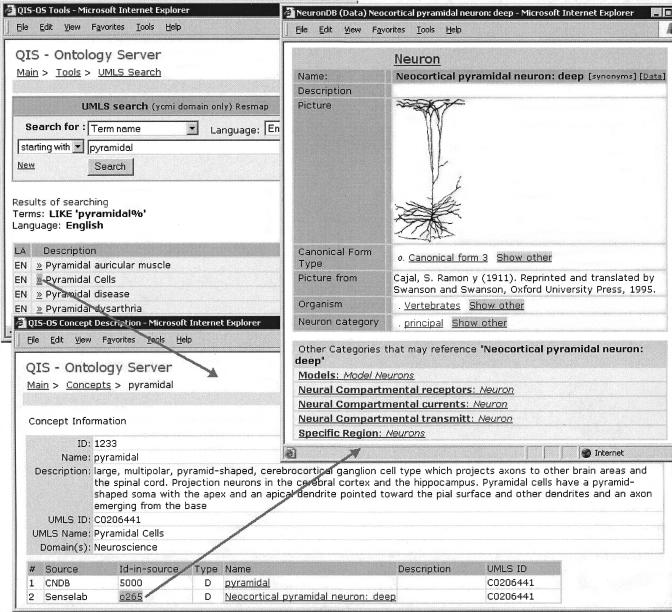
We therefore allow search of UMLS terms based on partial phrases that the user enters: UMLS terms matching the query are returned; when the user selects the term of interest, the details of the matching concept(s)s are returned (note: some terms are ambiguous and map to more than one concept). Any mapped objects in our local databases are also returned. Associated hyperlinks lead the user to detailed information on each object. In ▶, the user has searched the UMLS for terms beginning with “pyramidal.” A list of matching terms is returned: Clicking on “Pyramidal Cells” shows details (taken from UMLS's MRDEF table) as well as local objects mapped to that concept. The resulting page shows the pyramidal cell found in two databases in the federation: the Cortical Neuron Database (located at the Gardner Laboratory at Cornell University) and SenseLab. Clicking on the hyperlink associated with the second row then takes the user to details of the neocortical pyramidal neuron within SenseLab. Other features of the OS include finding common concepts between a pair of federated databases and vocabulary creation by promoting concepts within those databases not available in UMLS.
Figure 5.
Using the ontology server. The user searches the Unified Medical Language System (UMLS) for terms beginning with “pyramidal.” A list of matching terms is returned (EN in the figure indicates that the term is in English). Clicking on Pyramidal Cells shows details (taken from UMLS's MRDEF table) as well as local objects mapped to that concept. Clicking on the hyperlink associated with the second row (o265 is the unique internal identification of the object) takes the user to details of the neocortical pyramidal neuron within SenseLab. One may then inspect additional information on the neuron, such as the receptors and currents associated with individual compartments in the neuron, by clicking on further hyperlinks (details not shown). This query involves all three types of servers: the ontology server provides access to the UMLS and also replicates the mapping of objects in local databases to UMLS concepts: the integrator server actually mediates the query that fetches information from SenseLab (by making a request of SenseLab's data source).
The OS is described in depth, with an online tutorial, at the following URL: http://ycmi.med.yale.edu/QIS/components.htm#OS/.
Current Status
Individual parts of the QIS framework are already being used on a production basis in SenseLab, which receives an average of 3,000 hits per day, excluding Web-bots. The DSS, IS, and OS are currently all housed on a single CPU in separate physical databases. Therefore, there is currently no need for a protocol for the communication of deltas between the different server units. In the geographically separated server scenario ultimately envisaged, however, such a protocol will be necessary.
Query features: Some query aggregate functions and subqueries are allowed in some data sources that implicitly support them. Because many databases implement them differently, we allow their use in a limited fashion, resembling pass-through queries in ODBC.
Client application interfacing: The system currently provides XML-HTTP requests. We currently do not support the Simple Object Access Protocol (SOAP) because QIS is still in too early a stage to merit the creation of specific contractual interface names and parameters. Supporting evolution of the underlying database schemas will require SOAP meta-interfaces rather than interfaces that hard code the current view of domain knowledge. To avoid burdening the reader with these technical details, sample code and documentation can be found at http://ycmi.med.yale.edu/QIS/interfacing.htm/.
Query optimization: Little to no optimization of multidatabase queries is currently performed. The query designer must specify the order of operations on individual data sources, such as in the genomics example. Only multidatabase queries in which the composite query is explicitly designated as a union operation can be optimized by having each of the component queries run in parallel. Caching of specific intermediate data sets, based on query usage statistics, may also improve performance. QIS performance depends on several factors: query execution, data transference, and multidata-source query processing. Query execution can take from a few seconds to minutes. For this reason, all processing is asynchronous, and the system's tracing mechanisms inform the client about elapsed events. As in all distributed queries, performance is influenced by network bandwidth, CPU performance, data storage mechanisms, and RAM availability.
Integrity and Security: To protect access to restricted functions such as query authoring and metadata annotation, both IS and DSS use log in–based access control. Client applications can be restricted with passwords or to specific network addresses. For sensitive data, encryption is implemented using secure socket layer and server certificates.
Discussion
Many mainstream DBMSs are excessively fragile even when dealing with a single (nonfederated) database. A well-known example is the Oracle DBMS: Adding a new column to a table causes all views defined on that table (which use all of that table's columns) to be rendered invalid; any operations accessing this view will fail. These views must be manually “recompiled,” a tedious process requiring identifying the numerous views in the database that are bad and then fixing these, typically by text editing. The use of such technology by itself in a distributed database scenario requires considerable augmentation. A more robust dependency model could facilitate consistency maintenance. The research contribution of QIS is in the development of an explicit dependency model in the context of federated schemas.
Issues of Scalability
We provide both theoretical reasons and benchmarks to argue that the QIS architecture, which is based on metadata exchange between the three kinds of servers, is highly scalable.
In any database, metadata are typically a small fraction of data. Thus, a table may contain millions of rows, but its metadata describe a fixed, small number of columns in that table.
Metadata are significantly less volatile than data; that is, although changes in schema definition are common in a scientific database, schema changes do not occur every day.
Schema capture and delta computation are distributed over several DSSs, each of which is concerned only with its registered data sources. Only the deltas propagate between the servers. Based on a preliminary version of an XML-based protocol that we are devising for delta propagation, we have estimated that the encoding of a single column change in a table should not take more than 300 bytes (including the XML tags).
Mapping of data/metadata elements in data sources to controlled-vocabulary concepts is manually intensive. This type of task is well known in clinical informatics settings and is a critical part of system integration efforts. The process is performed against local copies of standard vocabularies (such as UMLS) and is not communication bandwidth intensive. The time/bandwidth required to transmit information about mapped concepts to an OS is negligible in comparison.
Benchmarks
The schema capture/delta computation process takes less than 2 seconds against all of SenseLab (76 classes modeled in EAV/CR, 238 attributes across all classes). We have also arranged for the Yale DSS to access, as a data source, the BAMS database at the University of Southern California (USC). This MySQL database, which uses a conventional relational database structure, has a total of 22 tables and 144 columns. To prevent contention for resources with internal users, BAMS deliberately “throttles” queries from non-USC users (i.e., runs them with low priority). Live schema capture/delta computation of BAMS from the Yale DSS takes approximately 20 seconds using a dual Pentium Xeon 2 GHz with 1 GB RAM that also hosts several other databases.
Future Work
Local ontology development is currently in its infancy within the HBP. We intend to provide extensive infrastructure support for the development and maintenance of local, domain-specific vocabularies. We are also implementing “semantic queries” in which a user can identify elements of interest in the ontology, and the system can compose appropriate queries against data sources in an automated or assisted fashion. The specifications of such queries can be saved for reuse, so that even if there are currently very few data of interest to a specific query within the federation, the same query may return more results when rerun in future as the contents of the federated databases expand.
We need to devise efficient protocols for delta transmission. There are several kinds of deltas: metadata differences between DSS schema versions (transmitted between DSS and IS), ontology version differences (between OS and IS), and changes in ontological mappings at the metadata and data levels (between DSS and OS).
We also plan to improve query responsiveness. Nonparameterized queries of relatively static data can be scheduled to run periodically, and their results cached on ISs to avoid unnecessary reprocessing. This solution is particularly useful to automatically populate client tables containing information from multiple data sources (genomic, ontological, or publication data).
Lessons Learned
Optimizing the Use of Metadata: Metadata improve the understandability of a database's contents. Current database engines provide limited metadata annotation facilities. Such limitations hinder the ability to formulate distributed queries. For the DSS alone, these metadata are being used to generate real-time data previews from a particular table or column. Data preview minimizes the number of iterations required for correct query formulation. The use of explicit relationships in data sources that implicitly do not support them also improves the understanding of the database structure.
Ontology Mapping: The use of ontological annotations improves on ordinary metadata descriptions, facilitating the localization of data of interest. One contribution of this paper is in integrating ontology-based approaches with federated query technology. We believe that National Library of Medicine's idea of the “Information Sources Map,” which was emphasized during the early years of UMLS development, is one that needs to be resurrected and fleshed out. Rather than simply indicating that a particular database contains information about a particular topic of interest, the mapping of vocabulary terms to actual metaschema/data elements goes a step further in fetching contextual information. The problem of “meaning” of data has often been somewhat ignored in the computer science literature on database federation, which has often discussed unrealistic scenarios such as mapping elements in different databases to each other based on their names. In the future, ontological mappings could play a crucial role in mediated ontological queries (queries based on ontologies that are translated to structured queries in ontologically annotated database schemas).
-
Query Adaptation by Decomposition and Versioning: The second contribution of this paper is the use of versioning approaches and decomposition of queries into their atomic components to achieve the goals of metadata refresh and the offline flagging of queries depending on altered metadata elements well before they are actually executed. Schema synchronization and versioning allow rapid determination of which queries are affected by a change to a specific element and the extent to which automated recovery mechanisms can operate successfully.
Current systems often break without any indication or reason of their failure. More important, many mainstream DBMSs are excessively fragile even when dealing with a single (nonfederated) database. With a more robust dependency model, consistency maintenance could be performed automatically.
We are continuing to accumulate experience with QIS, and releasing the framework through open-source mechanisms will help us evolve it based on the needs of a variety of groups that choose to experiment with it.
Supplementary Material
Supported by NIH grants P01 DC04732, G08 LM05583, and U01 ES10867.
The authors thank David Tuck of the Yale Department of Pathology, Kei Cheung of the Yale Center of Medical Informatics, and Mihail Bota at the University of Southern California, part of the BAMS group.
The existing QIS code base will be made freely available on request to the first author.
References
- 1.Koslow S, Huerta M. Neuroinformatics: An Overview of the Human Brain Project. Mahwah, NJ: Lawrence Erlbaum Associates, 1997.
- 2.Miller PL, Nadkarni P, Singer M, Marenco L, Hines M, Shepherd G. Integration of multidisciplinary sensory data: a pilot model of the human brain project approach. J Am Med Inform Assoc. 2001;8:34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shepherd GM, Healy MD, Singer MS, et al. Senselab: a project in multidisciplinary, multilevel sensory integration. In: Koslow SH, Huerta MF, editors. Neuroinformatics: An Overview of the Human Brain Project. Mahwah, NJ: Lawrence Erlbaum Associates, 1997, pp 21–56.
- 4.Kans JA, Ouellette BFF. Submitting DNA sequences to the databases. In: Baxevanis AD, Ouellette BFF, editors. Bioinformatics: A Practical Guide to the Analysis of Genes and Proteins. New York: John Wiley & Sons, 1998.
- 5.Li P. BizTalk Server Developer's Guide. New York: Osborne/McGraw-Hill, 2002.
- 6.Genetic Exchange Inc. Exploiting the life science data explosion to speed new drug discovery. Turn Massive Amounts of Data into Gems of Knowledge Using discoveryHub. Available at: http://www.geneticxchange.com/v3/product/whitepapers/WPexplosion.pdf. Accessed Dec 17, 2002.
- 7.Hass LM, Schwarz PM, Kodali P, Kotlar E, Rice JE, Swope WC. DiscoveryLink: A system for Integrated Access to Life Science Data Sources. IBM Syst J. 2001;40:489. [Google Scholar]
- 8.Josifovski V, Risch T. Query decomposition for a distributed object-oriented mediator system. Dist Parallel Databases. 2002;11:307–36. [Google Scholar]
- 9.Chung SY, Wong L. Kleisli: a new tool for data integration in biology. Trends Biotechnol. 1999;17:351–5. [DOI] [PubMed] [Google Scholar]
- 10.McBrien P, Poulovassilis A. Schema evolution in heterogeneous database architectures, a schema transformation approach. In: Pidduck AB, editor. 14th International Conference on Advanced Information Systems Engineering (CAiSE 2002). Berlin: Springer-Verlag, 2002, pp 484–99.
- 11.Ram S, Shankaranarayanan G. Dynamically Managing Schema Changes in a HDE—Pitfalls and Possibilities. 2003. Available at: http://smgnet.bu.edu/smgnet/css/staff/pub/GetFile.CFM/Shankar,_G_11.pdf?did=229&Filename=Shankar,_G_11.pdf. Accessed Jan 25, 2004.
- 12.Li X, Tari Z. Class versioning for schema evolution. In: Proceedings of the Australian Database Conference (ADC), Perth, Australia, 1998, pp 117–28.
- 13.Kim W, Seo J. Classifying schematic and data heterogeneity in multidatabase systems. IEEE Comput. 1991;24:12–8. [Google Scholar]
- 14.Sheth AP, Kashyap V. So far (schematically) yet so near (semantically). In: Proceedings of the International Federation for Information Processing (IFIP) Working Group on Database Semantics Conference on Interoperable Database Systems (DS-5) 1992, pp 283–312.
- 15.Paton NW, Stevens R, Baker PG, Goble CA, Bechhofer S, Brass A. Query processing in the TAMBIS Bioinformatics Source Integration System. In: Proceedings of the 11th International Conference on Scientific and Statistical Databases (SSDBM), 1999. Los Alamitos, CA: IEEE Press, 1999, pp 138–47.
- 16.Stevens R, Baker P, Bechhofer S, et al. TAMBIS: Transparent access to multiple bioinformatics information sources. Bioinformatics. 2000;16:184–6. [DOI] [PubMed] [Google Scholar]
- 17.Rector AL, Bechhofer S, Goble CA, Horrocks I, Nowlan WA, Solomon WD. The GRAIL concept modeling language for medical terminology. Artif Intell Med. 1997;9:139–71. [DOI] [PubMed] [Google Scholar]
- 18.Wong LS. The Collection Programming Language. 1996. Available at: http://citeseer.ist.psu.edu/wong96collection.html/. Accessed Apr 20, 2004.
- 19.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993;32:281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mork P, Halevy A, Tarczy-Hornoch P. A model for data integration systems of biomedical data applied to online genetic databases. AMIA Fall Symp. 2001:473–7. [PMC free article] [PubMed]
- 21.Mork P, Shaker R, Halevy A, Tarczy-Hornoch P. PQL: a declarative query language over dynamic biological schemata. AMIA Fall Symp. 2002:533–7. [PMC free article] [PubMed]
- 22.OMIM. Online Mendelian Inheritance in Man. 2002. Available at: http://www.ncbi.nlm.nih.gov/omim/. Accessed Nov 10, 2002.
- 23.Nadkarni PM. Management of evolving map data: data structures and algorithms based on the framework map. Genomics. 1995;30:565–73. [DOI] [PubMed] [Google Scholar]
- 24.Marenco L, Tosches N, Crasto C, Shepherd G, Miller PL, Nadkarni PM. Achieving evolvable Web-database bioscience applications using the EAV/CR framework: recent advances. J Am Med Inform Assoc. 2003;10:444–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardner D, Knuth KH, Abato M, et al. Common data model for neuroscience data and data model exchange. J Am Med Inform Assoc. 2001;8:17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaye D. Loosely Coupled: The Missing Pieces of Web Services. Kentfield, CA: RDS Press, 2003.
- 27.Masys D. An evaluation of the source selection elements of the prototype UMLS Information Sources Map. In: Proceedings of the Annual Symposium on Computer Applications in Medical Care. 1992:295–8. [PMC free article] [PubMed]
- 28.Andersson O, Armstrong P, Axelsson H, et al. Scalable Vector Graphics (SVG) 1.1 Specification. 2003. Available at: http://www.w3.org/TR/2003/REC-SVG11-20030114/. Accessed Jul 7, 2003.
- 29.Haertel M, Hayes D, Stallman R, Tower L, Eggert P. GNU DIFF. In. 2.7 ed. Cambridge, MA: Free Software Foundation, 1992. Program and documentation available at: ftp://prep.ai.mit.edu/pub/gnu/diffutils-2.7.
- 30.Krinke J, Zeller A. Linux/Unix Programming Toolset: Version Control, Construction, Testing, and Debugging. New York: John Wiley & Sons, 2001.
- 31.Sankoff DE, Kruskal JB. Time Warps, String Edits, and Macromolecules: The Theory and Practice of Sequence Comparison. Reading, MA: Pearson Addison-Wesley, 1983.
- 32.Bota M, Dong HW, Swanson LW. From gene networks to brain networks. Nat Neurosci. 2003;6(8):795–9. [DOI] [PubMed] [Google Scholar]
- 33.Nadkarni PM, Chen RS, Brandt CA. UMLS concept indexing for production databases: a feasibility study. J Am Med Inform Assoc. 2001;8:80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.