Abstract
The brown citrus aphid, Toxoptera citricida (Kirkaldy), is considered the primary vector of citrus tristeza virus, a severe pathogen which causes losses to citrus industries worldwide. The alate (winged) form of this aphid can readily fly long distances with the wind, thus spreading citrus tristeza virus in citrus growing regions. To better understand the biology of the brown citrus aphid and the emergence of genes expressed during wing development, we undertook a large-scale 5′ end sequencing project of cDNA clones from alate aphids. Similar large-scale expressed sequence tag (EST) sequencing projects from other insects have provided a vehicle for answering biological questions relating to development and physiology. Although there is a growing database in GenBank of ESTs from insects, most are from Drosophila melanogaster and Anopheles gambiae, with relatively few specifically derived from aphids. However, important morphogenetic processes are exclusively associated with piercing-sucking insect development and sap feeding insect metabolism. In this paper, we describe the first public data set of ESTs from the brown citrus aphid, T. citricida. The cDNA library was derived from alate adults due to their significance in spreading viruses (e.g., citrus tristeza virus). Over 5180 cDNA clones were sequenced, resulting in 4263 high-quality ESTs. Contig alignment of these ESTs resulted in 2124 total assembled sequences, including both contiguous sequences and singlets. Approximately 33% of the ESTs currently have no significant match in either the non-redundant protein or nucleic acid databases. Sequences returning matches with an E-value of ≤ −10 using BLASTX, BLASTN, or TBLASTX were annotated based on their putative molecular function and biological process using the Gene Ontology classification system. These data will aid research efforts in the identification of important genes within insects, specifically aphids and other sap feeding insects within the Order Hemiptera.
The sequence data described in this paper have been submitted to Genbank's dbEST under the following accession numbers.: CB814527-CB814982, CB832665-CB833296, CB854878-CB855147, CB909714-CB910020, CB936196-CB936346, CD449954-CD450759.
| Abbreviation: | |
|---|---|
| EST | expressed sequence tag |
Keywords: Aphididae, cDNA, EST, Gene expression, Hemiptera, Development, Toxoptera
Introduction
The brown citrus aphid, Toxoptera citricida (Kirkaldy), is one of the most devastating pests of citrus, causing extensive crop losses worldwide. Feeding by this aphid alone can cause severe damage to citrus. However, it poses an even greater threat to citrus because of its efficient transmission of citrus tristeza closterovirus (Fasulo and Halbert, 1993).
Since the brown citrus aphid genomic sequence is not available, expressed sequence tags (ESTs) derived from single-pass sequencing of cDNA clones prepared from the brown citrus aphid provide an invaluable resource for the identification of genes associated with the biology of the alate adult life stage. In the past, cloning of genes encoding enzymes of specific biochemical pathways by single-pass sequencing of cDNA clones has been a very successful strategy, particularly when the cDNA libraries have been prepared from tissues with high activity for the respective enzymes (Coyle-Thompson and Banerjee 1993; Newman et al., 1994; Blaxter et al., 1996; Cooke et al., 1996; Rounsley et al., 1996). This enables investigators to isolate genes derived from specific tissues and/or life stages for more detailed study, which may include developing efficient biocontrol methods.
Additionally, ESTs and their accompanying cDNAs, provide the means to construct glass or nylon based arrays that can be used for transcript profiling on a genome-wide scale (DeRisi et al., 1997; Ruan et al., 1998; Egger et al., 2002). A careful bioinformatic analysis identifying life stage-specific ESTs is a prerequisite in order to obtain a comprehensive and representative set of cDNAs for gene expression studies by arrays (Loftus et al., 1999). Given that there are only a small number of insect ESTs in public databases it was essential to build a life-stage specific library derived from aphids so that analysis of metabolism and development on a genome-wide scale could be accomplished. Even without subsequent array analysis, a relatively large number of ESTs from a specific life stage can provide clues toward the expression of specific genes important to the functions expressly connected with that life stage (Rafalski et al., 1998; Arbeitman et al., 2002). In most cases and within statistical limitations, the abundance of a specific cDNA in the EST collection is a measure of gene expression (Audic and Claverie, 1997). This technique, referred to as a “digital or electronic northern”, has been utilized in several similar studies to gauge relative gene expression in various tissues. The data sets are available at GenBank, dbEST under the following accession numbers.: CB814527-CB814982, CB832665-CB833296, CB854878-CB855147, CB909714-CB910020, CB936196-CB936346, CD449954-CD450759.
Materials and Methods
Aphid rearing and collection
Alate brown citrus aphids, Toxoptera citricida, were obtained from a healthy colony maintained by WB Hunter at the USDA, ARS, U.S. Horticultural Research Laboratory, Ft. Pierce, FL. The founders were collected from a single collection site in Orlando, Florida. The colony was reared under continuous asexual reproduction for a period of 3 years on sweet orange, Madam vinous, seedlings in screen cages contained in an insectary, and held at 25°C, 16 L: 8 D. Plants free of insecticide and bearing new flush were cycled into cages on a weekly basis. Aphids and their host plants were surveyed biweekly for any incidence of contaminating insect species (e.g., mites, parasitoids, fungus gnats, shore flies, etc.). High-density aphid populations produced alate aphids that were collected by aspiration within two days of emergence. All alates were collected from the top of the cage so as to avoid sample contamination with other developmental forms or host plant tissue. Upon collection, alates were immediately submerged into liquid nitrogen prior to total RNA isolation. Approximately 50–100 alates were placed into 95% ethanol and stored at −80°C to be used as voucher specimens.
cDNA library construction
Approximately 4500 1–2 day old alate aphids were used in the construction of an expression library. Whole aphids were ground in liquid nitrogen and total RNA extracted using guanidinium salt-phenol-chloroform procedure as described by Strommer et al. (1993). Poly(A)+ RNA was purified using two rounds of selection on oligo dT magnetic beads according to the manufacturer's instructions (Dynal, www.dynal.no). A directional cDNA library was constructed in Lambda Uni-ZAP® XR Vector using Stratagene's ZAP-cDNA Synthesis Kit (Stratagene, www.stratagene.com). The resulting DNA was packaged into lambda particles using Gigapack® III Gold Packaging Extract (Stratagene). An amplified library was generated with a titer of 1.0 × 109 plaque-forming units per mL. Mass excision of the amplified library was carried out using Ex-Assist® helper phage (Stratagene). An aliquot of the excised, amplified library was used for infecting XL1-Blue MRF' cells and subsequently plated on LB agar containing 100 µg/mL ampicillin. Bacterial clones containing excised pBluescript SK(+) phagemids were recovered by random colony selection.
Sequencing of clones
pBluescript SK(+) phagemids were grown overnight at 37°C and 240 rpm in 96-deep well culture plates containing 1.7 mL of LB broth, supplemented with 100 µg/mL ampicillin. Archived stocks were prepared from the cell cultures using 75 µl of a LB-amp, glycerol mixture and 75 µl of cells. These archived stocks are held at the Horticultural Research Laboratory where they are kept in an ultra low temperature freezer set at −80°C. Plasmid DNA was extracted using the Qiagen 9600 liquid handling robot and the QIAprep 96 Turbo miniprep kit according to the recommended protocol (QIAGEN, www.quigen.com).
Sequencing reactions were performed using the ABI PRISM® BigDye™ Primer Cycle Sequencing Kit (Applied Biosystems, home.appliedbiosystems.com) along with a universal T3 primer. Reactions were prepared in 96-well format using the Biomek2000™ liquid handling robot (Beckman Coulter, www.beckman.com). Sequencing reaction products were precipitated with 70% isopropanol, resuspended in 15 µL sterile water and loaded onto an ABI 3700 DNA Analyzer (Applied Biosystems).
Computer analysis
Base confidence scores were designated using TraceTuner® (Paracel, www.paracel.com). Low-quality bases (confidence score <20) were trimmed from both ends of sequences. Quality trimming, vector trimming and sequence fragment alignments were executed using Sequencher® software (Gene Codes, www.genecodes.com). Contaminating sequences such as rRNA and mitochondrial DNA were identified using BLASTN and were excluded from analysis along with sequences less than 100 nucleotides in length after both vector and quality trimming. Additional ESTs that corresponded to vector contaminants were removed from the dataset. To estimate the number of genes represented in the library and the redundancy of specific genes, ESTs were assembled into contigs using Sequencher®. Contig assembly parameters that were set using a minimum overlap of 50 bases and 95% identity match.
Functional annotation of ESTs
Putative sequence identity was determined based on BLAST similarity searches using the National Center for Biotechnology Information (NCBI) BLAST server (http://www.ncbi.nlm.nih.gov) with comparisons made to both non redundant nucleic acid (BLASTN) and protein (BLASTX) databases. ESTs that had no significant similarity to any publicly available sequences using BLASTN and BLASTX were then screened individually using TBLASTX.
The top 5 hits for each assembled sequence were then formatted using an in-house parsing program that allowed for direct import into a Microsoft Excel® spreadsheet for further analysis. Sequence matches with E-value scores ≤ −10 were considered significant and were categorized according to the Gene Ontology (GO) classification system based on annotation of the 5 ‘best hit’ matches in BLASTX searches. All D. melanogaster matches were cataloged using FlyBase (www.flybase.org). Those sequences without a D. melanogaster hit were annotated using AmiGO (www.geneontology.org).
Results and Discussion
Generation and assembly of adult alate ESTs
An initial 5180 clones were sequenced from the 5′ end. These sequences were trimmed of vector and low-quality sequence and filtered for minimum length (100 bp), producing 4267 high-quality ESTs of 481 bp average length. These ESTs were analyzed with the Sequencher® assembly program to identify those that represent redundant transcripts. ESTs were assembled into 468 contiguous sequences (contigs) with 1656 ESTs remaining as singlets, suggesting a 61% redundancy. Thus, the combined set of contigs and singlets included 2124 sequences (hereafter referred to as ‘assembled sequences’), putatively representing different transcripts. Only 22 contig sequences contained more than 10 ESTs.
EST quality analysis and sequence survey
Of the 2124 assembled sequences analyzed, 993 (representing 2132 ESTs) were similar to known protein sequences in the non-redundant protein database (BLASTX; E ≤ −10). Seven of these assembled sequences, representing 13 ESTs, were identified by BLASTX as contaminating vector sequences and were removed from the dataset.
Because some genes encode RNAs rather than proteins, it was necessary to run BLASTN against our dataset. Eight assembled sequences were identified as ribosomal and 2 were identified as mitochondrial DNA, representing 582 and 65 ESTs respectively, and were removed from the dataset. Although the number of ribosomal sequences appears inflated, it has been shown that several non-coding RNAs, such as rRNA, have mRNA-like modifications, such as polyadenylation and splicing. Because this EST dataset was derived from a cDNA library that was enriched for poly(A+) RNA, it is reasonable to assume that some non-coding RNAs should be present (MacIntosh et al., 2001). An additional 76 ESTs were identified as either rRNA or mitochondrial using TBLASTX, leaving 2031 assembled sequences used in subsequent functional analyses.
Of the initial 2124 assembled sequences (representing 4267 ESTs), 1045 (representing 1412 ESTs) showed no significant similarity (E>−10) to any publicly available sequence using BLASTX, BLASTN, or TBLASTX. This result suggests that a large percentage (∼33%) of the ESTs sequenced here are novel. However, this estimation of potential unique sequences within the cDNA library is most likely to be an overestimation due to several factors, such as computer alignment parameters and low quality internal sequences (White et al., 2000). Moreover, assembled sequences may have lacked an open reading frame because they were too short causing ESTs to consist mostly or entirely of a noncoding region (e.g., 3′ untranslated region) (Whitfield et al., 2002).
Functional annotation of ESTs
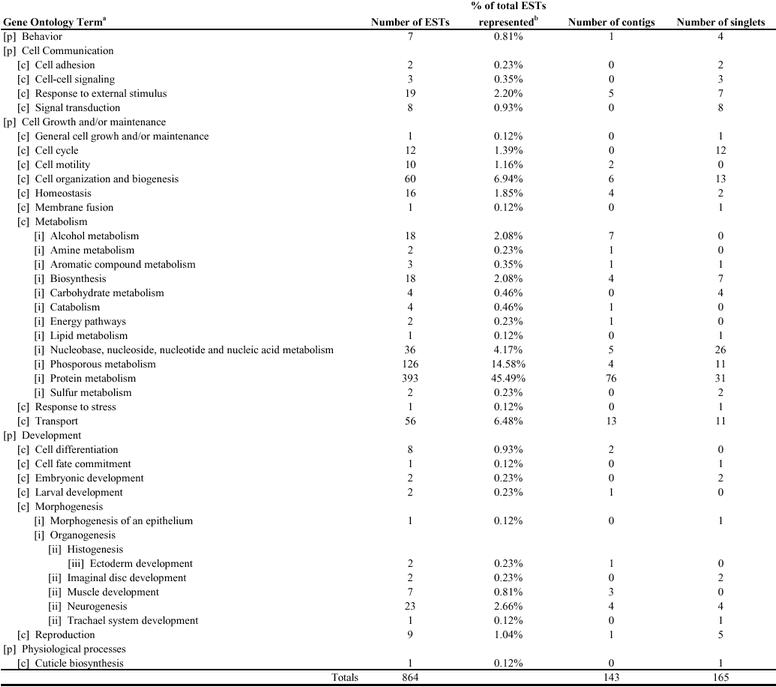
Each Toxoptera citricida assembled sequence was tentatively assigned Gene Ontology classification based on annotation of the top 5 “best hit” matches (E ≤ −10) using BLASTX. Nearly all of these were characterized with respect to the functionally annotated genes in D. melanogaster using FlyBase. Of the 993 sequences demonstrating similarity to known protein sequences, 332 (33%) of these were of unknown molecular function and 685 (69%) were of unknown biological process. Tables 1 and 2 summarize assignments of Toxoptera sequences to major molecular functions and biological processes, respectively.
Table 1.
Molecular Function
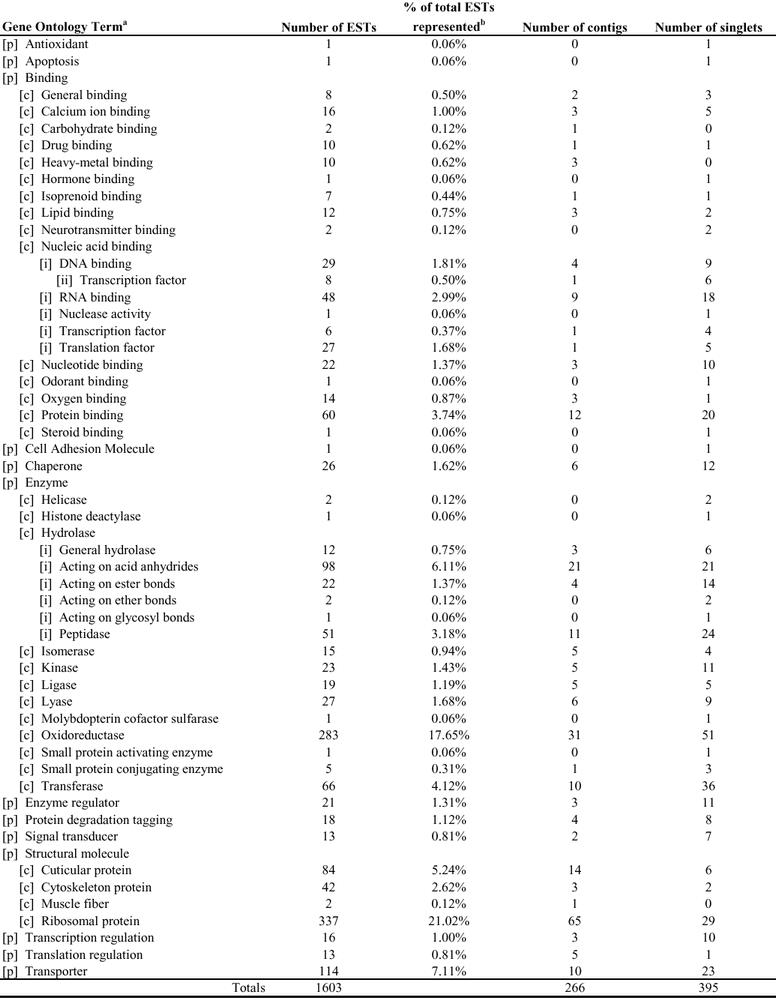
Table 2.
Biological Process
Genes of interest within the EST dataset
The BLASTX results provide useful information regarding the homology of proteins that may be critical for insect cellular communication and development. Table 3 lists sequences of the brown citrus aphid that match to D. melanogaster genes implicated in signal transduction, cell differentiation, cell fate commitment, embryonic and larval development, morphogenesis, reproduction and cuticle biosynthesis. Typically, genes involved in early development would not be present in cDNA libraries derived from adult tissues. However, many aphid species are composed entirely of viviparous parthenogenetic females. These insects telescopic generations as embryogenesis occurs in un-born daughters, producing up to three generations developing within an adult individual (Sabater et al., 2001). Therefore, genes involved in the development of several life stages may be represented simultaneously in this analysis.
Table 3.
Genes of interest in the Alate BrCA EST dataset
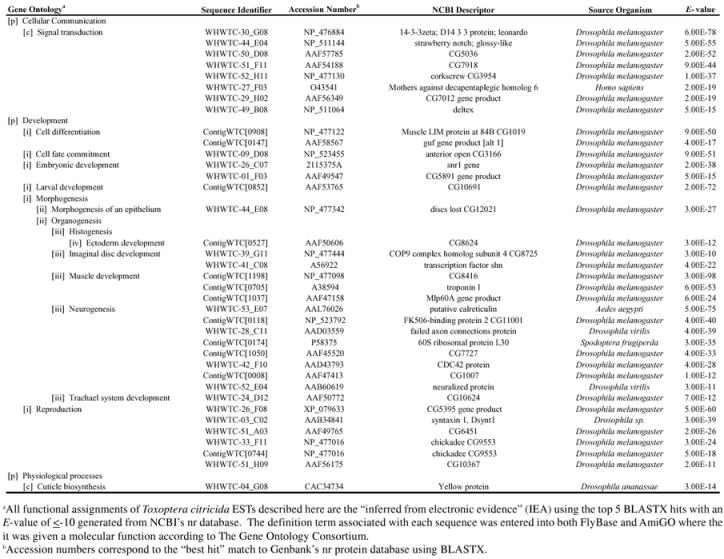
For the purposes of this paper, brown citrus aphid sequences were grouped into distinct gene ontology classifications. However, it is important to recognize that many of these gene products act in concert with one another to control cell fate determination which, in turn, drives morphological changes such as eye, leg, and wing development (e.g., the Notch pathway) (Coyle-Thompson and Banerjee, 1993; Baonza et al., 2000).
Conclusions
We have provided a large data set of ESTs from the alate brown citrus aphid and have begun to analyze this valuable resource. The analysis of this data set is continually evolving and some of the conclusions may have to be revised as more advanced bioinformatic tools become available. Being the first EST data set for the brown citrus aphid, its preliminary examination clearly shows that it is substantially different from the aphid EST data set currently available to the public. For the most part, there is considerable congruence between conventional biochemistry regarding insect metabolism and the number of ESTs encoding metabolic enzymes. This data set provides the first experimental access to these genes and the basis for more in-depth molecular and genomic analysis. Moreover, it identifies genes that are critical in the physiology, reproduction, development, and wing morphogenesis of aphids. Genetic information is crucial to advancing our understanding of aphid biology, and will play a major role in the development of future non-chemical, gene-based control strategies against these insect pests.
Acknowledgments
We thank J. Mozoruk for helpful discussions and review of the manuscript. Special thanks to L.E. Hunnicutt for data analysis, annotation of data, and assistance in manuscript construction and review.
Footnotes
The use or mention of a trademark or proprietary product does not constitute an endorsement, guarantee, or warranty of the product by the U.S. Department of Agriculture and does not imply its approval to the exclusion of other suitable products.
References
- Arbeitman MN, Furlong EEM, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Research. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Blaxter ML, Raghavan N, Ghosh I, Guiliano D, Lu W, Williams SA, Slatko B, Scott AL. Genes expressed in Brugia malayi infective third stage larvae. Molecular and Biochemical Parasitology. 1996;77:77–93. doi: 10.1016/0166-6851(96)02571-6. [DOI] [PubMed] [Google Scholar]
- Baonza A, de Celis JF, García-Bellido A. Relationships between extramacrochaetae and Notch signaling in Drosophila wing development. Development. 2000;127:2383–2393. doi: 10.1242/dev.127.11.2383. [DOI] [PubMed] [Google Scholar]
- Cooke R, Raynal M, Laudie M, Grellet F, Delseny M, Morris PC, Guerrier D, Giraudat J, Quigley F, Clabault G, Li YF, Mache R, Krivitzky M, Gy IJ, Kreis M, Lecharny A, Parmentier Y, Marbach J, Fleck J, Clement B, Phillips G, Herve C, Bardet C, Tremousaygue D, Hofte H. Further progress towards a catalogue of all Arabidopsis genes: Analysis of a set of 5,000 non-redundant ESTs. Plant Journal. 1996;9:101–124. doi: 10.1046/j.1365-313x.1996.09010101.x. [DOI] [PubMed] [Google Scholar]
- Coyle-Thompson CA, Banerjee U. The strawberry notch gene functions with Notch in common developmental pathways. Development. 1993;119:377–395. doi: 10.1242/dev.119.2.377. [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Egger B, Leemans R, Loop R, Kammermeier L, Fan Y, Radimerski T, Strahm MC, Certa U, Reichert H. Gliogenesis in Drosophila: Genome-wide analysis of downstream genes of glial cells missing in the embryonic nervous system. Development. 2002;129:3295–3309. doi: 10.1242/dev.129.14.3295. [DOI] [PubMed] [Google Scholar]
- Fasulo TR, Halbert SE. Aphid Pests of Citrus. ENY-811, Entomology and Nematology Department, Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida 1993 [Google Scholar]
- Loftus SK, Chen Y, Gooden G, Ryan JF, Birznieks G, Hilliard M, Baxevanis AD, Bittner M, Meltzer P, Trent J, Pavan W. Informatic selection of a neural crestmelanocyte cDNA set for microarray analysis. Proceeding of the National Academy of Sciences USA. 1999;96:9277–9280. doi: 10.1073/pnas.96.16.9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntosh GC, Wilkerson C, Green PJ. Identification and Analysis of Arabidopsis Expressed Sequence Tags Characteristic on Non-Coding RNAs. Plant Physiology. 2001;127:765–776. [PMC free article] [PubMed] [Google Scholar]
- Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Sommerville S, Thomashow M. Genes galore: A summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiology. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski JA, Hanafey M, Miao GH, Ching A, Lee JM, Dolan M, Tingey S. New experimental and computational approaches to the analysis of gene expression. Acta Biochimica Polonica. 1998;45:929–934. [PubMed] [Google Scholar]
- Rounsley SD, Glodek A, Sutton G, Adams MD, Somerville CR, Venter JC, Kerlavage AR. The construction of Arabidopsis expressed sequence tag assemblies: A new resource to facilitate gene identification. Plant Physiology. 1996;112:1177–1183. doi: 10.1104/pp.112.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y, Gilmore J, Conner T. Towards Arabidopsis genome analysis: Monitoring expression profiles of 1,400 genes using cDNA microarrays. Plant Journal. 1998;15:821–833. doi: 10.1046/j.1365-313x.1998.00254.x. [DOI] [PubMed] [Google Scholar]
- Sabater B, van Ham RCHJ, Martínez-Torres D, Silva F, Latorre A, Moya A. Molecular evolution of aphids and their primary (Buchnera sp.) and secondary endosymbionts: Implications for the role of symbiosis in insect evolution. Interciencia. 2001;26(10):508–512. [Google Scholar]
- Strommer JN, Gregerson R, and Vayda M. 1993 Isolation and characterization of plant mRNA. Methods in Plant Molecular Biology and Biotechnology. 49–65. [Google Scholar]
- White JA, Todd J, Newman T, Focks N, Girke T, de Ilarduya OM, Jaworski JG, Ohlrogge JB, Benning C. A new set of Arabidopsis expressed sequence tags from developing seeds. The metabolic pathway from carbohydrates to seed oil. Plant Physiology. 2000;124:1582–1594. doi: 10.1104/pp.124.4.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield CW, Band MR, Bonaldo MF, Kumar CG, Liu L, Pardinas JR, Robertson HM, Soares MB, Robinson GE. Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Research. 2002;12:555–566. doi: 10.1101/gr.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]


