Abstract
Aberrant secreted proteins can be destroyed by ER-associated protein degradation (ERAD), and a prominent, medically relevant ERAD substrate is the cystic fibrosis transmembrane conductance regulator (CFTR). To better define the chaperone requirements during CFTR maturation, the protein was expressed in yeast. Because Hsp70 function impacts CFTR biogenesis in yeast and mammals, we first sought ER-associated Hsp40 cochaperones involved in CFTR maturation. Ydj1p and Hlj1p enhanced Hsp70 ATP hydrolysis but CFTR degradation was slowed only in yeast mutated for both YDJ1 and HLJ1, suggesting functional redundancy. In contrast, CFTR degradation was accelerated in an Hsp90 mutant strain, suggesting that Hsp90 preserves CFTR in a folded state, and consistent with this hypothesis, Hsp90 maintained the solubility of an aggregation-prone domain (NBD1) in CFTR. Soluble ERAD substrate degradation was unaffected in the Hsp90 or the Ydj1p/Hlj1p mutants, and surprisingly CFTR degradation was unaffected in yeast mutated for Hsp90 cochaperones. These results indicate that Hsp90, but not the Hsp90 complex, maintains CFTR structural integrity, whereas Ydj1p/Hlj1p catalyze CFTR degradation.
INTRODUCTION
Mutated proteins that are unstable or fold slowly can accumulate in the ER, aggregate, and/or induce apoptosis (Thomas et al., 1995; Kaufman, 1999). However, eukaryotic cells have evolved the ability to identify and degrade these aberrant proteins by a pathway termed ER-associated protein degradation (ERAD). After their identification ERAD substrates are “retro-translocated” (or “dislocated”) to the cytosol, ubiquitinated, and targeted to the 26S proteasome for degradation (Ellgaard et al., 1999; Romisch, 1999; Tsai et al., 2002; Kostova and Wolf, 2003; McCracken and Brodsky, 2003). How ERAD substrates are selected is not completely clear, but a family of proteins, known as molecular chaperones, are involved in this process. Several molecular chaperones bind short, linear arrays of amino acids enriched for hydrophobic residues that normally represent buried regions in folded proteins. As a result chaperones can retain mis-folded proteins in solution and facilitate their refolding and/or retro-translocation.
Previous work indicated unique chaperone requirements for the degradation of soluble and integral membrane proteins in yeast (for review see Fewell et al., 2001). For example, the ER lumenal Hsp70 molecular chaperone, BiP (Kar2p in yeast) is required for the efficient degradation of CPY* (Plemper et al., 1997) and pro-α-factor (Brodsky et al., 1999), which are soluble substrates, but is dispensable for the degradation of some mis-folded yeast membrane proteins (Plemper et al., 1998; Zhang et al., 2001; Taxis et al., 2003). In contrast, cytoplasmic Hsp70 is required for the proteolysis of several ER membrane proteins but is dispensable for CPY* and pro-α-factor degradation (Hill and Cooper, 2000; Zhang et al., 2001). Because the ATPase activity of Hsp70s—and thus their ability to trap polypeptide substrates—is enhanced by interaction with specific Hsp40 cochaperones (“J-domain”–containing proteins), it was anticipated that lumenal and cytoplasmic Hsp40s would also be required for the degradation of soluble and integral membrane ERAD substrates. Indeed, two Hsp40 homologues in the ER lumen, known as Scj1p and Jem1p, function redundantly to facilitate the BiP-dependent solubilization and retro-translocation of soluble ERAD substrates (Nishikawa et al., 2001). However, the cytoplasmic Hsp40s that cooperate with the yeast Hsp70, Ssa1p, to promote the degradation of integral membrane substrates have not been conclusively identified.
Wild-type proteins can also be ERAD substrates, and one prominent example is the cystic fibrosis transmembrane conductance regulator (CFTR): ∼80% of wild-type CFTR is degraded by ERAD (Jensen et al., 1995; Ward et al., 1995). Mutations in CFTR can cause cystic fibrosis (CF), the most prevalent fatal inherited disease in Caucasians, and the most common disease-causing allele in CFTR is the deletion of a phenylalanine at position 508 (ΔF508). The effect of this mutation is that ∼100% of the protein is degraded (Cheng et al., 1990). Not surprisingly, several cytoplasmic molecular chaperones, including Hsp70, Hsp40s, and Hsp90, and the ER lumenal quality control lectin, calnexin, interact with immature wild-type and ΔF508 CFTR (Yang et al., 1993; Pind et al., 1994; Loo et al., 1998; Meacham et al., 1999; Farinha et al., 2002; Zhang et al., 2002). It is thought that the interaction of Hsp70 and possibly Hsp90 targets CFTR to an E3 ubiuqitin ligase, CHIP, whose activity is required for subsequent proteasome-mediated degradation (Höhfeld et al., 2001; Meacham et al., 2001).
To date, it has been difficult to ascertain whether the interactions between specific chaperones and CFTR result from their attempts to fold the protein and/or to target it for degradation. This fact is highlighted by conflicting studies when the Hsp90 chaperone is disabled and CFTR biogenesis is measured. Hsp90 is one of the most abundant proteins in the eukaryotic cytoplasm, constituting up to ∼2% of total protein in nonstressed cells and is required for the folding of a diverse set of client proteins including transcription factors, steroid hormone receptors, and protein tryosine kinases (Buchner, 1996; Caplan, 1999; Young et al., 2001). Hsp90 action requires the participation of several cochaperones, which constitute the Hsp90 complex and include Hsp70, Hsp40, Hop, Hip, cis-trans prolyl isomerases (cyclophilins), and p23. When CFTR biogenesis was examined in BHK and CHO cells treated with geldanamycin (GA), an Hsp90 inhibitor, enhanced CFTR degradation was observed (Loo et al., 1998); however, extended GA treatment might exert pleiotropic effects, including activation of heat shock factor (Zou et al., 1998) and up-regulation of ER chaperones (Lawson et al., 1998). In contrast, CFTR was stabilized when GA was added to a reaction containing dog pancreas microsomes into which CFTR was inserted after in vitro transcription/translation (Fuller and Cuthbert, 2000). In neither study was the effect of Hsp90 cochaperones on CFTR degradation investigated.
To better define the roles of molecular chaperones on CFTR biogenesis, and more generally to elucidate how membrane proteins are targeted for ERAD, we expressed CFTR in the budding yeast Saccharomyces cerevisiae because of the genetic tools available in this organism and because the trafficking and degradation pathways and participating chaperones are conserved between yeast and humans. We showed previously that CFTR is a bona fide ERAD substrate in yeast as it is in mammals and that Hsp70 facilitates degradation because CFTR stabilization was observed in yeast containing a rapid-onset, temperature-sensitive SSA1 mutant allele (Zhang et al., 2001). Yeast expression of CFTR has been used by others to purify the protein and obtain single-channel recordings (Huang et al., 1996), to demonstrate that the COPII machinery sorts ERAD substrates to a degradative ER sub-compartment (Fu and Sztul, 2003) and to further delineate the requirements for its degradation (Kiser et al., 2001; Gnann et al., 2004). For example, a recent report indicates that CFTR degradation is attenuated in cdc48, ufd1, and htm1/mnl1 mutant yeast, suggesting a role for the cytosolic Cdc48p-Ufd1p-Npl4p complex and an ER degradation enhancing α-mannosidase-like protein (EDEM) in CFTR quality control (Gnann et al., 2004).
In this report, we have again used the yeast system and find that Hsp90, but not the Hsp90 complex, helps maintain CFTR in a folded state. We have also identified two ER-associated, but cytoplasmically disposed Hsp40 cochaperones, Ydj1p and Hlj1p, which function redundantly to facilitate CFTR degradation. Finally, because Hsp90 and Ydj1p/Hlj1p are dispensable for the turnover of a soluble ERAD substrate, our results further define the chaperone requirements for the degradation of membrane and soluble ERAD substrates.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
Yeast strains (Table 1) were grown at 26°C unless indicated otherwise and standard methods for growth, preparation of media, and transformation of yeast cultures were used (Adams et al., 1997). The hlj1Δ/ydj1-151 and the hlj1Δ mutant strain were constructed using PCR-based gene disruption as described previously (Longtine et al., 1998). In brief, the hlj1Δ/ydj1-151 strain was created by transformation of strain ACY17b with pMET25-GFP-HLJ1 (Beilharz et al., 2003) followed by direct transformation with an hlj1::TRP1 disruption cassette, generated by PCR using template vector pFA6a-TRP1 and the primer sequences (forward-GCAATATAACTTGTGGAAGGTCTATAATAGGAAGAACCGGATCCCCGGGTTAATTAA; reverse-CGTTGGAAAATAATAAAAAAATGACAGTAAAAATATTGTGTGAATTCGAGCTCGTTTAAAC).
Table 1.
Yeast strains used in this study
| Strain name | Genotype | Source |
|---|---|---|
| p82a | MAT a, ade2-1, leu2-3, 112, his3-11, 15, trp1-1, ura3-1, can1-100, hsc82::LEU2, hsp82::LEU2, pTGPD-HSP82 | Nathan and Lindquist, 1995 |
| G313N | MAT a, ade2-1, leu2-3, 112, his3-11, 15, trp1-1, ura3-1, can1-100, hsc82::LEU2, hsp82::LEU2, pTGPD-HSP82-G313N | Nathan and Lindquist, 1995 |
| STI1/SBA1 | MAT a, his3, leu2, met15, ura3 | Invitrogen, Carlsbad, CA |
| sti1Δ | MAT a, his3, leu2, met15, ura3, sti1::KANR | Invitrogen, Carlsbad, CA |
| sba1Δ | MAT a, his3, leu2, met15, ura3, sba1::KANR | Invitrogen, Carlsbad, CA |
| W3031b | MAT α, ade2, his3, leu2, ura3, trp1, can1-100 | Shirayama et al., 1993 |
| ACY17b | MAT α, ade2, his3, leu2, ura3, trp1, can1-100, ydj1-2::HIS3, ydj1-151::LEU2 | Caplan et al., 1992 |
| E0020 | MAT α, ura3, leu2, his3, trp1, sse1::HIS3 | Shirayama et al., 1993 |
| HLJ1/YDJ1 | MAT a, ade2, his3, leu2, ura3, trp1 | This study |
| hlj1Δ | MAT a, ade2, his3, leu2, ura3, trp1, Δhlj1::HIS3 | This study |
| hlj1Δ/ydj1-151 | MAT α, ade2, his3, leu2, ura3, trp1, can1-100 ydj1-2::HIS3, ydj1-151::LEU2, hlj1::TRP1 | This study |
| STI1/SSE1 | MAT a, GAL2, his2-11, 15, leu2-3, 112, lys1, lys2, trp1Δ1, ura3-52 | Nicolet and Craig, 1989 |
| sti1Δ/sse1Δ | MAT a, GAL2, his2-11, 15, leu2-3, 112, lys1, lys2, trp1Δ1, ura3-52 sti1::HIS3, sse1::KANR | Liu et al., 1999 |
Transformants were selected by plating on minimal medium lacking tryptophan and were counterselected for the URA3-marked pMET25-GFP-HLJ1 plasmid by plating on minimal medium supplemented with amino acids, uracil and 5-fluoro-orotic acid. The resulting strain failed to grow on minimal medium lacking uracil, but grew on medium lacking histidine and tryptophan, confirming the double mutant had been constructed. The hlj1Δ strain was constructed by transformation of the isogenic wild-type strain with a hlj1::HIS3 disruption cassette generated by PCR using template vector pFA6a-HIS3MX6 and the same primers listed above.
ERAD Assays
Yeast strains expressing HA-CFTR were grown to logarithmic phase (OD600 = 0.4–0.8) at 26°C in synthetic complete medium lacking uracil, but supplemented with glucose to a final concentration of 2% (SC-ura), and protein synthesis was stopped by the addition of cycloheximide to a final concentration of 50 μg/ml. Cells were shifted to 37°C and 2.0–2.5 ODs of cells were removed at the indicated time points. The cells were washed, and total protein was precipitated (Zhang et al., 2002). Proteins were resolved on either 10 or 12.5% SDS-polyacrylamide gels, transferred to nitrocellulose, and probed with mouse monoclonal anti-HA antibody (12CA5, Roche Molecular Biochemicals, Indianapolis, IN) and polyclonal anti-Sec61p (Stirling et al., 1992). The signals were quantified using 125I-secondary antibody and phosphorImage analysis (Fuji Medical Systems, Stamford, CT) or horseradish peroxidase-conjugated secondary antiserum and a Kodak 440CF Image Station and the associated Kodak 1D (V. 3.6) software (Rochester, NY). The degradation of a mis-folded, HA-tagged form of carboxypeptidase Y (HA-CPY*) was measured by 35S metabolic labeling/pulse-chase analysis as described (Zhang et al., 2001).
Protein Purifications
The following proteins were purified as described: Ssa1p (McClellan and Brodsky, 2000), Ydj1p (Cyr et al., 1992), NBD1 (G404-L644) of CFTR (Qu and Thomas, 1996), and Sba1p (Fang et al., 1998). Yeast Hsc90 (Hsc82p) was isolated using a modified protocol provided by Dr. David Toft (Mayo Clinic, Rochester, MN) from yeast strain ECUpep4 (Jakob et al., 1995) with deletions in the chromosomal HSC82 and HSP82 genes and that encodes HSC82 on a 2-μm plasmid. ECUpep4 cells were grown to logarithmic phase (OD600 = 0.8–1.0) in YPD (1% yeast extract, 2% peptone, and 2% dextrose) at 26°C, and the cells were harvested and frozen in liquid nitrogen. The cell pellets were thawed, resuspended in 3–5 volumes of buffer 1 (20 mM Tris-HCl, pH 7.5, 4 mM EDTA, 1 mM DTT) supplemented with protease inhibitors (1 mM PMSF, 1 μg/ml leupeptin, 0.5 μg/ml pepstatin A), and subjected to glass bead lysis by vigorous agitation. Unbroken cells were pelleted by centrifugation at ∼2800 × g for 10 min at 4°C, and cell membranes were pelleted by centrifugation at ∼48,000 × g for 40 min at 4°C. The cleared lysate was applied to a DE52 column equilibrated in buffer 1, and the column was washed with 2 volumes of buffer 1 and with 2 volumes of buffer 1 containing 50 mM KCl. Bound protein was eluted with a gradient of buffer up to 1 M KCl, and fractions were collected and peak Hsc82p-containing fractions, as assessed by SDS-PAGE and Coomassie brilliant blue staining, were pooled and dialyzed against buffer 2 (20 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM EDTA, 1 mM DTT, 10% glycerol) overnight at 4°C. The dialysate was loaded onto a high-performance Q-Sepharose column equilibrated in buffer 2, and the column was washed with buffer 2 and then buffer 2 containing 50 mM KCl (flow rate ∼1.0 ml/min) at 4°C. Bound protein was eluted with a gradient up to 1 M KCl. Peak Hsc82p-containing fractions were pooled, diluted to ∼1.5 mg/ml protein, and dialyzed against buffer 3 (20 mM Tris-HCl, pH 7.5, 50 mM KCl, 0.1 mM EDTA, 1 mM DTT, 10% glyercol). The final protein concentration was determined using the Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, CA) with BSA as the standard, and the final purity of Hsc82p was determined to be ∼80%.
To purify the Hlj1p J-domain, a DNA fragment containing the N-terminal 259 base pairs of HLJ1 was subcloned into pQE60 to add a hexahistidine C-terminal tag, and subsequently this fragment was subcloned into pGEX-KG at the BamHI and HindIII restriction sites in order to attach an N-terminal GST tag. The resulting GST-Hlj1p-6His–tagged protein (containing the J-domain) was expressed in M15 Escherichia coli and a 100-ml culture was grown in LB + KAN (25 μg/ml) to an OD600 ∼2.4. A total of 10 ml of the culture was diluted into 1 l of LB + KAN (25 μg/ml) and grown to OD600 ∼ 0.2 at 37°C, and then expression was induced by the addition of IPTG to a final concentration of 0.2 mM. Cells were incubated for ∼7 h at 37°C until an OD600 of ∼0.8 was reached and were harvested and frozen at -80°C. The cells were thawed and resuspended in 10 ml of Buffer 88 (20 mM HEPES, pH 6.8, 150 mM KOAc, 5 mM MgOAc, 250 mM sorbitol) containing 0.1% Triton X-100 and 1 mM EDTA, and protease inhibitors (1 mM PMSF, 1 μg/ml leupeptin, 0.5 μg/ml pepstatin A), and were broken using a sonic dismembrator (Fisher Scientific, Pittsburgh, PA). The lysate was cleared by centrifugation at 10,000 rpm for 10 min at 4°C in an SS34 rotor (Sorvall, Newton, CT), and the lysate was added to a 1 ml, ∼50% slurry of gluthanione-agarose beads (Sigma, St. Louis, MO) and incubated for ∼2 h with rotation at 4°C. The gluthanione beads were washed (10 min, 4°C) with 25 ml of buffer 88 supplemented with 0.1% Triton X-100, 1 mM EDTA, and protease inhibitors, followed by a second wash with the same buffer supplemented with 1 M KCl, and finally washed with buffer 88 plus 0.1% Triton X-100. The bound Hlj1p was eluted by two 1-ml washes of 50 mM Tris, pH 8, 5 mM reduced glutathione. Protein concentration was assessed as described above. Only the GST-Hlj1p-6His fusion protein was evident on a Coomassie Brilliant Blue–stained SDS-PAGE gel.
Biochemical Assays
Sba1p pull-down assays using purified hexahistidine-tagged Sba1 and Ni-NTA resin were performed essentially as described (Fang et al., 1998). Ssa1p-ATP complex formation and single-turnover ATPase assays were performed as published by incubating preformed Ssa1p-[α32P]ATP complex with the indicated protein (Hlj1p-J-domain-GST chimera, Ydj1p, and GST) at a final concentration of 0.2 μM (Sullivan et al., 2000).
For luciferase aggregation assays, firefly luciferase (Sigma) at an initial concentration of ∼0.65 μM was preincubated in the presence or absence of ∼20 μM purified Hsc82p in 150 μl of refolding buffer (10 mM MOPS/KOH, pH 7.2, 50 mM KCl, 3 mM MgCl2, 3 mM ATP, 2 mM DTT) for 20 min at 25°C before 500 μl of refolding buffer was added at 45°C to yield a final concentration of 0.15 μM for luciferase and 2.4 μM for Hsc82p. Aggregation was measured by light scattering at a wavelength of 320 nm at 45°C in a 14DS UV-VIS-IR spectrophotometer (AVIV, Lakewood, NJ).
The ability of Hsc82p to prevent the aggregation of NBD1 was investigated as described (Strickland et al., 1997). Our NBD1 construct spans amino acids G404 to L644 from helix H1b to helix H9, which includes the F1-type core ATP-binding subdomain (Lewis et al., 2004). This permits the measurement of early folding intermediates (Strickland et al., 1997). The assay was performed using the purified hexahistidine-tagged NBD1 diluted ∼100-fold out of 6 M guanidine-HCl buffer, 20 mM HEPES, pH 7.5, and into 650 μl of refolding buffer (100 mM Tris-HCl, pH 7.4, 0.385 M l-arginine, 10 mM DTT, 200 mM KCl, 20 mM MgCl2) to a final concentration of 2 μM. Protein aggregation was measured over time at a wavelength of 400 nm at 37°C in a 14DS UV-VIS-IR spectrophotometer (AVIV) in the absence or presence of the indicated concentrations of Hsc82p and other indicated reagents. Results of aggregation experiments were plotted as the relative amount of aggregation, normalized to the NBD1 control at 10 min, vs. time, and fitted to a single exponential using KaleidaGraph software version 3.0.4 (Abelbeck Software, Reading, PA) to determine initial rates. Macbecin I/II was kindly provided by the Drug Synthesis and Chemistry Branch in the Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, at the National Cancer Institute.
For the carbonate extraction and protease sensitivity assays, microsomal membrane fractions were prepared by differential centrifugation as previously described (Burri et al., 2003). Proteins were extracted from membranes by resuspension in 100 mM Na2CO3 and incubated for 30 min on ice with intermittent agitation on a Vortex mixer (Fujiki et al., 1982). Soluble and insoluble proteins were separated by centrifugation at 100,000 × g. Trypsin treatments were performed in 100 μl of lysis buffer (250 mM sorbitol, 150 mM potassium acetate, 5 mM magnesium acetate, 0.5 mM PMSF, 1.2 μg/ml leupeptin, 0.75 μg/ml antipain, 0.25 μg/ml chymostatin, 1 μg/ml pepstatin, 50 mM HEPES, pH 6.8) containing 1.5 μg trypsin and incubated on ice for 30 min, before a 10-fold excess of soybean trypsin inhibitor, or TCA was added to stop the reaction (Beilharz et al., 1998).
RESULTS
Hsp40 Cochaperones Function Redundantly during CFTR, But Not CPY* Degradation
We previously reported that a cytoplasmic Hsp70 in yeast, Ssa1p, facilitates CFTR degradation (Zhang et al., 2001) and in mammalian cells Hsc70 cooperates with Hdj2, an Hsp40 homologue, during CFTR biogenesis (Meacham et al., 1999). Ydj1p is the yeast Hdj2p homologue, is tethered to the ER membrane via a farnesyl moiety and interacts with Ssa1p based on genetic and biochemical studies (Caplan et al., 1992; Cyr et al., 1992; Becker et al., 1996). However, CFTR degradation was unaffected in yeast containing a temperature-sensitive allele of YDJ1 (Zhang et al., 2001), suggesting either that there are inherent differences between CFTR biogenesis in yeast and mammals or that more than one functionally redundant Hsp40 in yeast cooperates with Ssa1p to facilitate CFTR degradation.
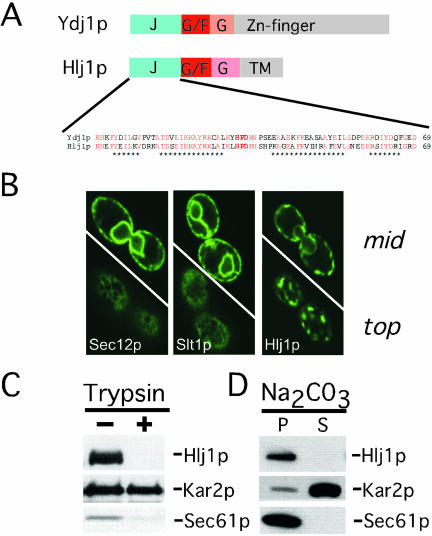
There are >20 J-domain–containing proteins in yeast and at least 14 of these reside in the cytoplasm (Costanzo et al., 2001; Walsh et al., 2004). Recently, an Hsp40 homologue, Hlj1p, was revealed in a search for tail-anchored membrane proteins (Beilharz et al., 2003). The N-terminal domain of Hlj1p is 58% identical to the J-domain of Ydj1p (Figure 1A) and the Hlj1p J-domain was predicted to reside in the cytosol (Beilharz et al., 2003). To test this prediction, we expressed GFP-tagged forms of Hlj1p, the tail-anchored SNARE Slt1p (Burri and Lithgow, 2004) and Sec12p, an integral ER membrane protein required for COPII-mediated vesicle budding (Barlowe, 2003). Confocal fluorescence microscopy was performed and the ER residence of Sec12p, Hlj1p, and Slt1p were apparent (Figure 1B). Trypsin digestion of isolated ER microsomes showed that Hlj1p is accessible to exogenous trypsin, as is the polytopic ER membrane protein Sec61p (Figure 1C). In contrast, Kar2p/BiP was protease resistant and protected from trypsin by the microsomal membrane. Moreover, neither Hlj1p nor Sec61p were solubilized by carbonate (Figure 1D). Together, these results indicate that Hlj1p is tethered to the ER membrane via a C-terminal anchor and, like Ydj1p, contains a cytoplasmically oriented J-domain.
Figure 1.
Hlj1p is a tail-anchored, integral membrane protein localized to the ER membrane. (A) Alignment of Hlj1p with Ydj1p. J, J-domain; G/F, glycine/phenylalanine-rich region; G, glycine-rich region; TM, transmembrane domain. The sequences of the J-domains are depicted and asterisks denote regions corresponding to the four predicted alpha-helices. Amino acids shaded red are identical between Hlj1p and Ydj1p and the functionally essential HPD motif is boldfaced. (B) Yeast expressing GFP-Sec12p (an integral ER membrane protein; Nakano et al., 1988), GFP-Slt1p, a C-terminally anchored protein (Beilharz et al., 2003; Burri et al., 2003) or GFP-Hlj1p were analyzed by confocal fluorescence microscopy and Z-sections are shown from midway through representative cells (mid) and through the periphery of the same cells (top). (C) A crude microsomal membrane fraction was isolated from yeast expressing GFP-Hlj1 and was incubated in the presence (+) or absence (-) of trypsin. The presence of Hlj1p, the lumenal protein Kar2p/BiP, and the polytopic ER membrane protein Sec61p were detected by immunoblot analysis. (D) A crude microsomal membrane fraction was isolated from yeast cells expressing GFP-Hlj1, treated with 0.1 M Na2CO3, and centrifuged to separate soluble proteins (S) from insoluble material (P). Immunoblots for the integral membrane ER protein Sec61p and the lumenal protein Kar2p were used to control for carbonate treatment.
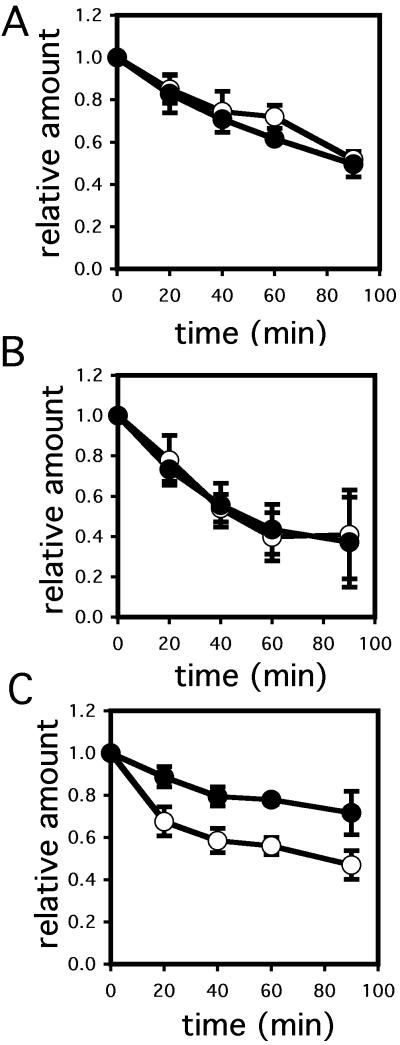
To test whether Hlj1p is involved in CFTR degradation, we expressed an HA-epitope–tagged form of CFTR under the control of a constitutive promoter in an hlj1 deletion mutant and in an isogenic wild-type yeast strain and performed cycloheximide chase analyses (see Materials and Methods). We found that the rates of CFTR degradation were identical (Figure 2A). Consistent with previous results (Zhang et al., 2001), we also found that CFTR degradation was unaffected in the ydj1-151 strain compared with an isogenic wild-type (Figure 2B). However, in a hlj1Δ/ydj1-151 double mutant CFTR degradation was slowed relative to the wild-type strain (Figure 2C). The degradation of Ste6p*, a mis-folded yeast ABC transporter and ERAD substrate, is also attenuated in a hlj1Δ/ydj1-151 double mutant (Huyer et al., 2004). These data suggest that Ydj1p and Hlj1p function redundantly to facilitate the degradation of CFTR and at least one other integral membrane ERAD substrate.
Figure 2.
CFTR degradation is attenuated in an hlj1/ydj1 double mutant, but not in yeast mutated individually for each gene. Wild-type and mutant yeast strains expressing CFTR were subjected to cycloheximide chase analysis as described in Materials and Methods, and the amount of CFTR at time zero was set to 1.0. (A) The relative amounts of CFTR remaining in wild-type (○) and hlj1Δ (•) yeast strains over time were plotted. (B) The relative amounts of CFTR remaining in wild-type (○) and ydj1-151 (•) yeast, and (C) in wild-type (○) and hlj1Δ/ydj1-151 mutant yeast (•) over time were plotted. In each panel the data represent the means of three independent experiments ± SEM.
Unique chaperone requirements for the ERAD of soluble vs. integral membrane proteins have been observed (Fewell et al., 2001). To examine this distinction further, we measured the degradation of CPY*, a soluble ERAD substrate (Hiller et al., 1996), in the hlj1Δ/ydj1-151 double mutant and in the isogenic wild-type strains by pulse-chase analysis. We found nearly identical rates of degradation in the two strains (Figure 3), indicating that Ydj1p and Hlj1p are dispensable for the degradation of CPY*.
Figure 3.
CPY* degradation is unaffected in the ydj1-151/hlj1Δ mutant yeast strain. CPY* degradation was assessed by pulse-chase analysis as described in Materials and Methods in wild-type (○) and in ydj1-151/hlj1Δ mutant (•) yeast. Data represent the means of three independent experiments ± SEM.
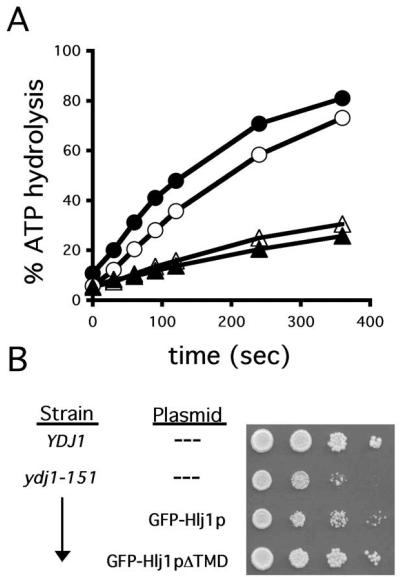
On the basis of data presented in Figure 2, we predicted that the effect of Ydj1p/Hlj1p during CFTR degradation is through their interaction with Ssa1p, the Hsp70 that in turn catalyzes CFTR degradation in yeast (Zhang et al., 2001). The functional interaction between J-domain–containing proteins and their cognate Hsp70s is best examined by measurements of Hsp70 ATP hydrolysis in the presence and absence of an Hsp40/J-domain–containing protein. For example, a GST fusion protein containing the J-domain of Sec63p, a membrane protein whose J-domain faces the ER lumen, stimulates BiP's ATPase activity in vitro (Corsi and Schekman, 1997). To examine whether Hlj1p functionally interacts with Ssa1p, we similarly constructed and purified a GST-tagged fusion protein that contains the Hlj1p J-domain and incubated the purified protein with preformed [α-32P]ATP-Ssa1p complex. Hlj1p stimulated Ssa1p in this single-turnover ATPase assay to a similar extent as equimolar amounts of Ydj1p (Figure 4A), indicating that both Hlj1p and Ydj1p interact with the yeast cytoplasmic Hsp70. Further evidence to support this proposition is shown in Figure 4B in which the abilities of full-length Hlj1p (GFP-Hlj1p) and a form of Hlj1p lacking the transmembrane domain (GFP-Hlj1pΔTMD) to suppress a slow-growth phenotype of ydj1-151 mutant cells were examined. We observed that the viability of the ydj1-151 mutant increased when Hlj1p was expressed, suggesting that Hlj1p partially supplants Ydj1p function. We also noted in this experiment that the soluble Hlj1p derivative is more effective at improving the growth of the mutant strain than full-length Hlj1p, suggesting that overexpression of the membrane anchor might be somewhat toxic; consistent with this hypothesis, we have observed toxicity derived from the overexpression of other stable, wild-type ER membrane proteins in yeast (our unpublished observations).
Figure 4.
Hlj1p and Ydj1p functions partially overlap. (A) The ATPase activity of Ssa1p is enhanced by purified Ydj1p and by a fusion protein containing the Hlj1p J-domain. A preformed [α-32P]ATP-Ssa1p complex was incubated for the indicated times with equimolar amounts of the Hlj1p J-domain fusion protein (•), Ydj1p (○), GST (▴), or buffer (▵) at 30°C, and the extent of ATP hydrolysis was assessed as described in Materials and Methods. (B) Wild-type and ydj1-151 yeast either lacking or containing a MET17-driven GFP-Hlj1p or GFP-Hlj1pΔTMD expression construct were grown overnight and serial dilutions were plated on selective medium containing 500 μM methionine at 30°C for 2 d. GFP-Hlj1p and GFP-Hlj1pΔTMD expression was confirmed by fluorescence microscopy (unpublished data).
Mutations in Hsp90 Enhance CFTR Degradation But Have No Effect on CPY* Turnover
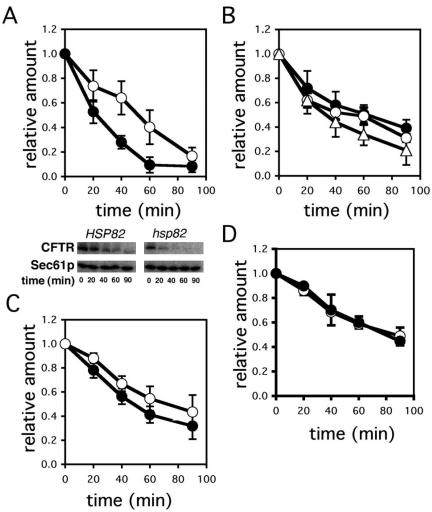
The role of the Hsp90 molecular chaperone during CFTR maturation in mammals is controversial (Loo et al., 1998; Fuller and Cuthbert, 2000; see Introduction). To better define the action of Hsp90 during CFTR biogenesis we expressed CFTR in yeast deleted for the genes encoding the constitutive (HSC82) and heat-inducible (HSP82) Hsp90s. Yeast Hsp82 and Hsc82p are ∼97% identical at the amino acid level and are functionally interchangeable, but at least one homologue must be expressed to maintain viability (Borkovich et al., 1989). Therefore, the wild-type strain for this experiment contains a plasmid-borne copy of HSP82, and the mutant strain contains a temperature-sensitive allele, hsp82ts (G313N). The mutant protein is extremely unstable when cells are shifted to the nonpermissive temperature of 37°C and is rapidly degraded (Bohen and Yamamoto, 1993; Fliss et al., 2000). This results in the equivalent of a null phenotype immediately after temperature shift. After cells were grown at a permissive temperature, CFTR degradation in these strains was monitored at 37°C by cycloheximide chase analysis. As shown in Figure 5A, the rate of CFTR degradation was significantly higher in the mutant strain. Similar results were obtained when CFTR degradation in the G170D hsp82 mutant was examined (unpublished data), which also rapidly loses activity at the nonpermissive temperature (Nathan and Lindquist, 1995). Because only the immature, unfolded form of CFTR is an ERAD substrate (Gelman et al., 2002), these data suggest that Hsp90 is important to maintain the stability of CFTR.
Figure 5.
CFTR degradation is accelerated in an Hsp90 mutant yeast strain, but not in yeast strains mutated for Hsp90 cochaperones. Yeast strains expressing CFTR were subjected to a cycloheximide chase and immunoblot analysis as described in Materials and Methods. (A) The degradation of CFTR in wild-type (○) and hsp82 (•) strains is plotted as the relative amount of CFTR remaining over time. Data represent the means of four independent experiments ± SEM. Bottom: representative Western blot. Sec61p is an ER integral protein and serves as a loading control. (B) The degradation of CFTR in wild-type (○), sti1Δ (•), and sba1Δ (▵) yeast strains is plotted as the relative amount of CFTR remaining over time. Data represent the means of 3–4 independent experiments ± SEM (C) CFTR degradation in wild-type (○) and sse1Δ (•) yeast strains is plotted as the relative amount of CFTR over time. Data represent the means of five independent experiments ± SEM (D) CFTR degradation in wild-type (○) and sti1Δ/sse1Δ (•) yeast strains is plotted as the relative amount of CFTR over time. Data represent the means of two independent experiments ± range.
Hsp90 associates with several cochaperones to form a macromolecular complex required for the folding and activation of select client proteins (Caplan, 1999; Richter and Buchner, 2001; Young et al., 2001). Although the role of Hsp90 in mammalian cells can be assessed using ansamycin antibiotics, examining the functions of Hsp90 cochaperones is more challenging. In yeast, however, mutations in Hsp90 cochaperones are readily available. We therefore examined the roles of three well-defined Hsp90 cochaperones (Sti1p, Sba1p, Sse1p) on CFTR stability in yeast. Sti1p is the yeast Hop homologue, and both Hsp70 and Hsp90 can dock onto the TPR domains of Sti1p (Johnson et al., 1998). Sba1p is the p23 homologue that stabilizes Hsp90 substrate binding (Fang et al., 1998). Sse1p is a yeast Hsp110 homologue that resides in the Hsp90 complex (Liu et al., 1999; Goeckeler et al., 2002). Deletion of the genes encoding each of these factors compromises Hsp90 complex-mediated processes in yeast (Chang et al., 1997; Fang et al., 1998; Liu et al., 1999; Cox and Miller, 2002). When CFTR degradation was examined in isogenic wild-type strains and in sti1Δ, sba1Δ, or sse1Δ yeast, we detected no statistically significant differences in the rates of CFTR degradation in the mutants compared with wild-type yeast (Figure 5, B and C). Furthermore, CFTR degradation was unaltered in an sti1Δ/sse1Δ double mutant (Figure 5D) that has a severe growth defect at 37°C (Liu et al., 1999), indicating that the accelerated degradation observed in the hsp82 mutant was not simply the result of shifting temperature-sensitive cells to the nonpermissive temperature. Together, these results demonstrate that Hsp90, but not the Hsp90 complex, participates in CFTR biogenesis in yeast. Moreover, the action of Hsp90 during CFTR biogenesis is not via an indirect effect on cellular signaling pathways because the same signaling pathways are compromised in strains mutated for the cochaperones.
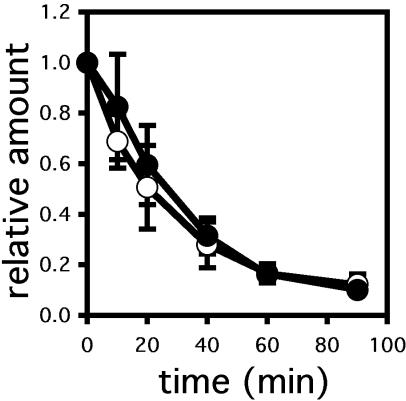
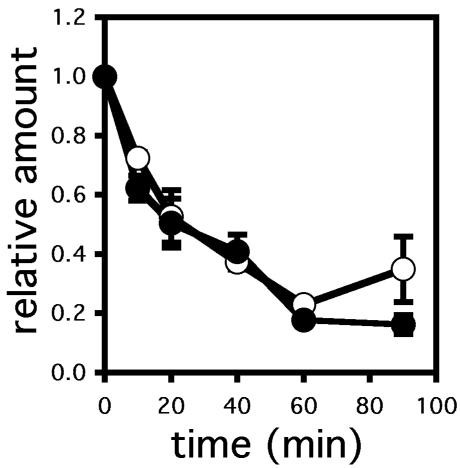
As discussed above, distinct chaperones requirements for the degradation of soluble and membrane proteins have been noted, and this distinction was supported further by the data presented in Figure 3. Therefore, we examined CPY* degradation in the hsp82 mutant and in the isogenic wild-type strain but found that there was no statistically significant difference in the rate or extent of degradation (Figure 6). We conclude that the ERAD of CPY* is Hsp90 independent.
Figure 6.
CPY* degradation is unaffected in the hsp82 mutant yeast strain. CPY* degradation was assessed by pulse-chase analysis as described in Materials and Methods in wild-type (○) and Hsp90 mutant (•) yeast. Data represent the means of three independent experiments ± SEM.
Yeast Hsp90 Prevents the Aggregation of the First Nucleotide-binding Domain of CFTR
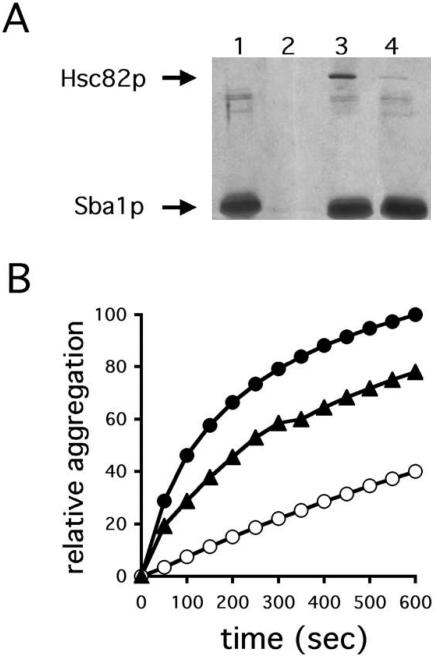
The data presented in Figure 5 suggest that Hsp90 helps protect CFTR from degradation and might therefore be important to maintain CFTR in its folded conformation. In general, a loss of structural integrity can be accompanied by protein aggregation, and it is well known that the first nucleotide-binding domain (NBD1) in CFTR is aggregation prone; moreover, the low efficiency or slow rate of NBD1 folding directly determines the efficacy of CFTR maturation, and maintaining NBD1 solubility prevents the formation of off-pathway aggregates (Qu and Thomas, 1996; Strickland et al., 1997; Zhang et al., 1998). To test directly whether yeast Hsp90 maintains NBD1 solubility, we isolated Hsc82p (see Materials and Methods). First, to confirm that the highly enriched protein was active, we examined the chaperone's ability to suppress the aggregation of firefly luciferase because mammalian Hsp90 was previously shown to slow the aggregation of this substrate (Wiech et al., 1992). Luciferase aggregation was suppressed by ∼65% when a 16:1 M ratio of Hsc82p to luciferase was used (our unpublished observations), consistent with previous data (Minami et al., 2001).
Next, we examined whether Hsc82p associated with the Sba1p cochaperone (Fang et al., 1998). Proficient interaction between Hsc82p and Sba1p was observed by pull-down assay, and importantly the degree of association decreased about sixfold in the presence of Macbecin (Figure 7A), an ansamycin antibiotic that inhibits yeast Hsp90 function both in vivo and in vitro (Bohen, 1998; Fang et al., 1998; Donze and Picard, 1999; Liu et al., 1999). Finally, we assessed Hsc82p prevention of NBD1 aggregation at a 5:1 M ratio and found that aggregation was suppressed by ∼60% (Figure 7B), indicating that yeast Hsp90 maintains NBD1 in solution. The ability of Hsp82p to maintain NBD1 in solution was reduced somewhat if Macbecin was added to a final concentration of 50 μM (our unpublished observations). This partial effect may be due to the fact that Hsp90 contains two polypeptide binding sites, only one of which is sensitive to ansamycin antibiotics (Young et al., 1997; Scheibel et al., 1998). These data are also consistent with previous work in which a partial effect of ansamycin antibiotics on Hsp90-dependent activities was noted (Minami et al., 2001). In any event, these results suggest that Hsp90 stabilizes CFTR by binding NBD1.
Figure 7.
Yeast Hsp90 binds the Sba1p/p23 cochaperone and suppresses the aggregation of NBD1 early-folding intermediates. (A) Hexa-histidine–tagged Sba1p was prebound to nickel-linked resin and then incubated in either the absence (lane 1) or presence (lanes 2–4) of highly enriched Hsc82p. Bound protein was eluted, resolved by SDS-PAGE and visualized by silver staining. The reaction shown in lane 2 lacks prebound Sba1p and the reaction shown in lane 4 was supplemented with Macbecin to a final concentration of 50 μM. (B) CFTR-NBD1 was diluted out of denaturant into refolding buffer at 37°C in the absence (•) or presence of Hsp90 at a 2.5:1 (▴) and 5:1 (○) molar ratio, and light scattering was measured as described in Materials and Methods. Data obtained at 50-s intervals are shown.
DISCUSSION
Our continued analysis of CFTR degradation in yeast, described in this report, has uncovered several novel aspects of molecular chaperone function and of the ERAD pathway for integral membrane proteins, which we address individually in the following sections.
First, we have identified the Hsp40 homologues in yeast that facilitate CFTR degradation—most likely in conjunction with an Hsp70, Ssa1p—and suggest that Hlj1p and Ydj1p function redundantly. Both chaperones enhance Ssa1p ATPase activity and the extent of CFTR stabilization in the hlj1/ydj1 mutant strain (Figure 2C) is similar to that observed in the ssa1 mutant (Zhang et al., 2001). These data are reminiscent of the reported functional redundancy and interactions between BiP and the ER lumenal Hsp40 chaperones, Scj1p and Jem1p, during the ERAD of soluble proteins (Nishikawa et al., 2001). The functional redundancy displayed by Ydj1p and Hlj1p is not limited to CFTR turnover because yeast mutated for the genes encoding these cochaperones also exhibit slowed degradation of Ste6p* (Huyer et al., 2004), another integral membrane ERAD substrate. Other recent studies have also hinted at a role for Hlj1p in protein quality control. For example, hlj1Δ mutants grew poorly when they express a Huntingtin fragment (HD53Q; Willingham et al., 2003) and the degradation of a synthetic, integral membrane ERAD substrate was mildly suppressed in an hlj1Δ strain (Taxis et al., 2003). The functional overlap of YDJ1 and HLJ1 reported here might explain the weak phenotype previously observed in the hlj1Δ mutant.
Second, we suggest that the yeast Hsp90 chaperone, Hsp82p, is required to maintain the folded state of CFTR because hsp82 mutant yeast degrade CFTR faster than isogenic wild-type yeast and because Hsp82p prevents NBD1 aggregation, a domain whose folding is critical for CFTR maturation (Qu and Thomas, 1996; Qu et al., 1997; Zhang et al., 1998). A significant prevention of aggregation was achieved at an Hsp82p:NBD1 M ratio of 5:1, an amount that is not unreasonable given the high concentration of cellular Hsp90 (Buchner, 1996). The more rapid degradation of CFTR in Hsp90 mutant yeast is not due to the well-characterized impact of the Hsp90 complex on cellular signaling pathways because no effect on CFTR degradation was observed in yeast mutated for Hsp90 cochaperones either individually (Sti1p, Sba1p, Sse1p) or in combination (Sti1p/Sse1p). We therefore conclude that CFTR is one of several cellular proteins that require Hsp90 for efficient folding, a list that includes p53, Src and steroid hormone receptors (Richter and Buchner, 2001). In contrast, it is important to note that Hsp90 facilitates the ERAD of apolipoprotein B (Gusarova et al., 2001) and an insulin receptor mutant (Imamura et al., 1998). These data indicate that some substrates utilize Hsp90 for protection, whereas other substrates engage Hsp90 en route to degradation. In some cases, Hsp90 is involved in both events, acting first to promote folding, and then if folding cannot proceed, targeting the substrate to the proteasome (Schneider et al., 1996). A role for Hsp90 in degradation is also supported by connections between Hsp90 and the ubiquitin-proteasome machinery: For example, Hsp90 binds to the 19S cap of the yeast proteasome (Verma et al., 2000), and in mammals Hsp90 function is linked to CHIP, an E3 ubiquitin ligase (Connell et al., 2001).
Third, we found that Hsp90 cochaperones do not impact CFTR biogenesis in yeast. To our knowledge, this is the first investigation of the relative contributions of Hsp90 vs. Hsp90 complex members in membrane protein biogenesis. In contrast, a recent study demonstrated that deletion of individual Hsp90 cochaperones had differential effects on the activity of the yeast MAP kinase, Ste11p (Lee et al., 2004), suggesting that interactions between client proteins and Hsp90/Hsp90 cochaperones are likely to be complex. In the best-characterized example, an Hsp90 folding pathway has been proposed based on in-depth studies of the progesterone and estrogen receptor folding pathways. Two distinct Hsp90 complexes are evident in this pathway: an early complex containing Hop (Sti1p), Hsp40, and Hsp70, and a mature complex containing p23 (Sba1p) and cyclophilins. The transition from the early to the late complex involves conformational changes in Hsp90 upon ATP binding and hydrolysis, and upon p23 binding (Smith, 1998; Pratt and Toft, 2003). It is unknown, however, whether other Hsp90 subcomplexes exist and how additional Hsp90 cochaperones impact this pathway. We therefore cannot rule out the possibility that a novel Hsp90 subcomplex might be important for CFTR biogenesis in yeast.
We also cannot rule out the possibility that Hsp90 cochaperones might be required for CFTR biogenesis in mammals. Formally, the ability of GA to induce more rapid degradation of CFTR in mammalian cells (Loo et al., 1998) might have occurred through direct inhibition of Hsp90 function or through an effect on Hsp90 complex maturation. In fact, it was noted in the published study that p23 associates with CFTR. Until individual Hsp90 cochaperones can be disabled in mammalian cells, this issue cannot be resolved.
Fourth, our data further define the unique chaperone requirements for the degradation of soluble and integral membrane proteins in the ER. Ssa1p impacts the ERAD of integral membrane substrates (Hill and Cooper, 2000; Zhang et al., 2001) and the Ydj1p/Hlj1p pair (this study) acts similarly. Precisely how these chaperones facilitate membrane protein turnover is not completely clear. In mammals the Hsp70-Hsp40 complex is directly linked to the ubiquitin-proteasome degradation machinery through its association with CHIP (Meacham et al., 2001). An Hsp70 cochaperone, BAG-1, might augment Hsp70-catalyzed degradation by transferring substrates from Hsp70 to the proteasome; BAG-1 is an Hsp70 nucleotide exchange factor that promotes substrate release, contains a ubiquitin-like element, and binds to the proteasome (Höhfeld, 1998; Höhfeld et al., 2001). In yeast the cytoplasmic Hsp70-Hsp40 chaperone complex (Ssa1p-Ydj1p/Hlj1p) might similarly link the selection of integral membrane ERAD substrates to one or more of the E3 ligases that play a role in ERAD (Bays et al., 2001; Deak and Wolf, 2001; Swanson et al., 2001). Besides the unique chaperone requirements for the degradation of membrane and soluble proteins, further distinctions in the selection of ERAD substrates were recently uncovered from Ng and colleagues, and it will be interesting to determine how chaperones impact the degradation of substrates through the described ERAD-C and ERAD-L pathways (Vashist and Ng, 2004). Future research efforts will seek to identify how the chaperone machinery is coupled to the action of the ubiquitin-proteasome system during the selection of CFTR and other membrane proteins.
Acknowledgments
We thank Drs. P. Thomas, A. Caplan, D. Toft, S. Lindquist, D. Thiele, R. Duda, J. Buchner, and the members of the Brodsky Laboratory for reagents, strains, and helpful discussions. We also thank Yinchern Law for excellent work in the early stages of the project. This study was supported by grant MCB-0110331 from the National Science Foundation and by grant DK-60835 from the National Institutes of Health to J.L.B., and by a grant from the Australian Research Council to T.L.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–07–0584. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–07–0584.
References
- Adams, A., Gottschling, D., Kaiser, C., and Stearns, T. (1997). Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Barlowe, C. (2003). Signals for COPII-dependent export from the ER: what's the ticket out? Trends Cell Biol. 13, 295-300. [DOI] [PubMed] [Google Scholar]
- Bays, N.W., Gardner, R.G., Seelig, L.P., Joazeiro, C.A., and Hampton, R.Y. (2001). Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 3, 24-29. [DOI] [PubMed] [Google Scholar]
- Becker, J., Walter, W., Yan, W., and Craig, E.A. (1996). Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 16, 4378-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz, T., Egan, B., Silver, P.A., Hofmann, K., and Lithgow, T. (2003). Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J. Biol. Chem. 278, 8219-8223. [DOI] [PubMed] [Google Scholar]
- Beilharz, T., Suzuki, C.K., and Lithgow, T. (1998). A toxic fusion protein accumulating between the mitochondrial membranes inhibits protein assembly in vivo. J. Biol. Chem. 273, 35268-35272. [DOI] [PubMed] [Google Scholar]
- Bohen, S.P. (1998). Genetic and biochemical analysis of p23 and ansamycin antibiotics in the function of Hsp90-dependent signaling proteins. Mol. Cell. Biol. 18, 3330-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohen, S.P., and Yamamoto, K.R. (1993). Isolation of Hsp90 mutants by screening for decreased steroid receptor function. Proc. Natl. Acad. Sci. USA 90, 11424-11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich, K.A., Farrelly, F.W., Finkelstein, D.B., Taulien, J., and Lindquist, S. (1989). hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9, 3919-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, J.L., Werner, E.D., Dubas, M.E., Goeckeler, J.L., Kruse, K.B., and McCracken, A.A. (1999). The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 274, 3453-3460. [DOI] [PubMed] [Google Scholar]
- Buchner, J. (1996). Supervising the fold: functional principles of molecular chaperones. FASEB J. 10, 10-19. [PubMed] [Google Scholar]
- Burri, L., and Lithgow, T. (2004). A complete set of SNAREs in yeast. Traffic 5, 45-52. [DOI] [PubMed] [Google Scholar]
- Burri, L., Varlamov, O., Doege, C.A., Hofmann, K., Beilharz, T., Rothman, J.E., Sollner, T.H., and Lithgow, T. (2003). A SNARE required for retrograde transport to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 100, 9873-9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan, A.J. (1999). Hsp90's secrets unfold: new insights from structural and functional studies. Trends Cell Biol. 9, 262-268. [DOI] [PubMed] [Google Scholar]
- Caplan, A.J., Cyr, D.M., and Douglas, M.G. (1992). YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell 71, 1143-1155. [DOI] [PubMed] [Google Scholar]
- Chang, H.C., Nathan, D.F., and Lindquist, S. (1997). In vivo analysis of the Hsp90 cochaperone Sti1 (p60). Mol. Cell. Biol. 17, 318-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S.H., Gregory, R.J., Marshall, J., Paul, S., Souza, D.W., White, G.A., O'Riordan, C.R., and Smith, A.E. (1990). Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell 63, 827-834. [DOI] [PubMed] [Google Scholar]
- Connell, P., Ballinger, C.A., Jiang, J., Wu, Y., Thompson, L.J., Höhfeld, J., and Patterson, C. (2001). The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3, 93-96. [DOI] [PubMed] [Google Scholar]
- Corsi, A.K., and Schekman, R. (1997). The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. J. Cell Biol. 137, 1483-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo, M.C. et al. (2001). YPD, PombePD and WormPD: model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res. 29, 75-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, M.B., and Miller, C.A., 3rd. (2002). The p23 co-chaperone facilitates dioxin receptor signaling in a yeast model system. Toxicol. Lett. 129, 13-21. [DOI] [PubMed] [Google Scholar]
- Cyr, D.M., Lu, X., and Douglas, M.G. (1992). Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J. Biol. Chem. 267, 20927-20931. [PubMed] [Google Scholar]
- Deak, P.M., and Wolf, D.H. (2001). Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J. Biol. Chem. 276, 10663-10669. [DOI] [PubMed] [Google Scholar]
- Donze, O., and Picard, D. (1999). Hsp90 binds and regulates Gcn2, the ligand-inducible kinase of the alpha subunit of eukaryotic translation initiation factor 2. Mol. Cell. Biol. 19, 8422-8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard, L., Molinari, M., and Helenius, A. (1999). Setting the standards: quality control in the secretory pathway. Science 286, 1882-1888. [DOI] [PubMed] [Google Scholar]
- Fang, Y., Fliss, A.E., Rao, J., and Caplan, A.J. (1998). SBA1 encodes a yeast hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol. Cell. Biol. 18, 3727-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinha, C.M., Nogueira, P., Mendes, F., Penque, D., and Amaral, M.D. (2002). The human DnaJ homologue (Hdj)-1/heat-shock protein (Hsp) 40 co-chaperone is required for the in vivo stabilization of the cystic fibrosis transmembrane conductance regulator by Hsp70. Biochem. J. 366, 797-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewell, S.W., Travers, K.J., Weissman, J.S., and Brodsky, J.L. (2001). The action of molecular chaperones in the early secretory pathway. Annu. Rev. Genet. 35, 149-191. [DOI] [PubMed] [Google Scholar]
- Fliss AE, Benzeno S, Rao J, Caplan AJ. (2000). Control of estrogen receptor ligand binding by Hsp90. J. Steroid. Biochem. Mol. Biol. 72, 223-230. [DOI] [PubMed] [Google Scholar]
- Fu, L., and Sztul, E. (2003). Traffic-independent function of the Sar1p/COPII machinery in proteasomal sorting of the cystic fibrosis transmembrane conductance regulator. J. Cell Biol. 160, 157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki, Y., Hubbard, A.L., Fowler, S., and Lazarow, P.B. (1982). Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93, 97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, W., and Cuthbert, A.W. (2000). Post-translational disruption of the delta F508 cystic fibrosis transmembrane conductance regulator (CFTR)-molecular chaperone complex with geldanamycin stabilizes delta F508 CFTR in the rabbit reticulocyte lysate. J. Biol. Chem. 275, 37462-37468. [DOI] [PubMed] [Google Scholar]
- Gelman, M.S., Kannegaard, E.S., and Kopito, R.R. (2002). A principal role for the proteasome in endoplasmic reticulum-associated degradation of misfolded intracellular cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 277, 11709-11714. [DOI] [PubMed] [Google Scholar]
- Gnann, A., Riordan, J.R., and Wolf, D.H. (2004). CFTR degradation depends on the lectins Htm1p/EDEM and the Cdc48 protein complex in yeast. Mol. Biol. Cell 15, 4125-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeckeler, J.L., Stephens, A., Lee, P., Caplan, A.J., and Brodsky, J.L. (2002). Overexpression of yeast Hsp110 homolog Sse1p suppresses ydj1-151 thermo-sensitivity and restores Hsp90-dependent activity. Mol. Biol. Cell 13, 2760-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarova, V., Caplan, A.J., Brodsky, J.L., and Fisher, E.A. (2001). Apoprotein B degradation is promoted by the molecular chaperones hsp90 and hsp70. J. Biol. Chem. 276, 24891-24900. [DOI] [PubMed] [Google Scholar]
- Hill, K., and Cooper, A.A. (2000). Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 19, 550-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller, M.M., Finger, A., Schweiger, M., and Wolf, D.H. (1996). ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science 273, 1725-1728. [DOI] [PubMed] [Google Scholar]
- Höhfeld, J. (1998). Regulation of the heat shock conjugate Hsc70 in the mammalian cell: the characterization of the anti-apoptotic protein BAG-1 provides novel insights. Biol. Chem. 379, 269-274. [PubMed] [Google Scholar]
- Höhfeld, J., Cyr, D.M., and Patterson, C. (2001). From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2, 885-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, P., Stroffekova, K., Cuppoletti, J., Mahanty, S.K., and Scarborough, G.A. (1996). Functional expression of the cystic fibrosis transmembrane conductance regulator in yeast. Biochim. Biophys. Acta 1281, 80-90. [DOI] [PubMed] [Google Scholar]
- Huyer, G., Piluek, W. F., Fansler, Z., Kreft, S., Hochstrasser, M., Brodsky, J.L., and Michaelis, S. (2004). Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multi-spanning membrane protein and a soluble lumenal protein. J. Biol. Chem. 279, 38369-38378. [DOI] [PubMed] [Google Scholar]
- Imamura, T. et al. (1998). Involvement of heat shock protein 90 in the degradation of mutant insulin receptors by the proteasome. J. Biol. Chem. 273, 11183-11188. [DOI] [PubMed] [Google Scholar]
- Jakob, U., Meyer, I., Bugl, H., Andre, S., Bardwell, J.C., and Buchner, J. (1995). Structural organization of procaryotic and eucaryotic Hsp90. Influence of divalent cations on structure and function. J. Biol. Chem. 270, 14412-14419. [DOI] [PubMed] [Google Scholar]
- Jensen, T.J., Loo, M.A., Pind, S., Williams, D.B., Goldberg, A.L., and Riordan, J.R. (1995). Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell 83, 129-135. [DOI] [PubMed] [Google Scholar]
- Johnson, B.D., Schumacher, R.J., Ross, E.D., and Toft, D.O. (1998). Hop modulates Hsp70/Hsp90 interactions in protein folding. J. Biol. Chem. 273, 3679-3686. [DOI] [PubMed] [Google Scholar]
- Kaufman, R.J. (1999). Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13, 1211-1233. [DOI] [PubMed] [Google Scholar]
- Kiser, G.L., Gentzsch, M., Kloser, A.K., Balzi, E., Wolf, D.H., Goffeau, A., and Riordan, J.R. (2001). Expression and degradation of the cystic fibrosis transmembrane conductance regulator in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 390, 195-205. [DOI] [PubMed] [Google Scholar]
- Kostova, Z., and Wolf, D.H. (2003). For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 22, 2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson, B., Brewer, J.W., and Hendershot, L.M. (1998). Geldanamycin, an hsp90/GRP94-binding drug, induces increased transcription of endoplasmic reticulum (ER) chaperones via the ER stress pathway. J. Cell. Physiol. 174, 170-178. [DOI] [PubMed] [Google Scholar]
- Lee, P., Shabbir, A., Cardozo, C., and Caplan, A.J. (2004). Sti1 and Cdc37 can stabilize Hsp90 in chaperone complexes with a protein kinase. Mol. Biol. Cell 15, 1785-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, H.A. et al. (2004). Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 23, 282-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.D., Morano, K.A., and Thiele, D.J. (1999). The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J. Biol. Chem. 274, 26654-26660. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A., 3rd, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Loo, M.A., Jensen, T.J., Cui, L., Hou, Y., Chang, X.B., and Riordan, J.R. (1998). Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J. 17, 6879-6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan, A.J., and Brodsky, J.L. (2000). Mutation of the ATP-binding pocket of SSA1 indicates that a functional interaction between Ssa1p and Ydj1p is required for post-translational translocation into the yeast endoplasmic reticulum. Genetics 156, 501-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken, A.A., and Brodsky, J.L. (2003). Evolving questions and paradigm shifts in endoplasmic-reticulum-associated degradation (ERAD). Bioessays 25, 868-877. [DOI] [PubMed] [Google Scholar]
- Meacham, G.C., Lu, Z., King, S., Sorscher, E., Tousson, A., and Cyr, D.M. (1999). The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 18, 1492-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham, G.C., Patterson, C., Zhang, W., Younger, J.M., and Cyr, D.M. (2001). The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3, 100-105. [DOI] [PubMed] [Google Scholar]
- Minami, M., Nakamura, M., Emori, Y., and Minami, Y. (2001). Both the N- and C-terminal chaperone sites of Hsp90 participate in protein refolding. Eur. J. Biochem. 268, 2520-2524. [DOI] [PubMed] [Google Scholar]
- Nakano, A., Brada, D., and Schekman, R. (1988). A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J. Cell Biol. 107, 851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan, D.F., and Lindquist, S. (1995). Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol. Cell. Biol. 15, 3917-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolet, C.M., and Craig, E.A. (1989). Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol. Cell. Biol. 9, 3638-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa, S.I., Fewell, S.W., Kato, Y., Brodsky, J.L., and Endo, T. (2001). Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 153, 1061-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pind, S., Riordan, J.R., and Williams, D.B. (1994). Participation of the endoplasmic reticulum chaperone calnexin (p88, IP90) in the biogenesis of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 269, 12784-12788. [PubMed] [Google Scholar]
- Plemper, R.K., Bohmler, S., Bordallo, J., Sommer, T., and Wolf, D.H. (1997). Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388, 891-895. [DOI] [PubMed] [Google Scholar]
- Plemper, R.K., Egner, R., Kuchler, K., and Wolf, D.H. (1998). Endoplasmic reticulum degradation of a mutated ATP-binding cassette transporter Pdr5 proceeds in a concerted action of Sec61 and the proteasome. J. Biol. Chem. 273, 32848-32856. [DOI] [PubMed] [Google Scholar]
- Pratt, W.B., and Toft, D.O. (2003). Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 228, 111-133. [DOI] [PubMed] [Google Scholar]
- Qu, B.H., Strickland, E.H., and Thomas, P.J. (1997). Localization and suppression of a kinetic defect in cystic fibrosis transmembrane conductance regulator folding. J. Biol. Chem. 272, 15739-15744. [DOI] [PubMed] [Google Scholar]
- Qu, B.H., and Thomas, P.J. (1996). Alteration of the cystic fibrosis transmembrane conductance regulator folding pathway. J. Biol. Chem. 271, 7261-7264. [DOI] [PubMed] [Google Scholar]
- Richter, K., and Buchner, J. (2001). Hsp 90, chaperoning signal transduction. J. Cell. Physiol. 188, 281-290. [DOI] [PubMed] [Google Scholar]
- Romisch, K. (1999). Surfing the Sec61 channel: bidirectional protein translocation across the ER membrane. J. Cell Sci. 112, 4185-4191. [DOI] [PubMed] [Google Scholar]
- Scheibel, T., Weikl, T., and Buchner, J. (1998). Two chaperone sites in Hsp90 differing in substrate specificity and ATP dependence. Proc. Natl. Acad. Sci. USA 95, 1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C., Sepp-Lorenzino, L., Nimmesgern, E., Ouerfelli, O., Danishefsky, S., Rosen, N., and Hartl, F.U. (1996). Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90. Proc. Natl. Acad. Sci. USA 93, 14536-14541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama, M., Kawakami, K., Matsui, Y., Tanaka, K., and Toh-e, A. (1993). MSI3, a multicopy suppressor of mutants hyperactivated in the RAS-cAMP pathway, encodes a novel HSP70 protein of Saccharomyces cerevisiae. Mol. Gen. Genet. 240, 323-332. [DOI] [PubMed] [Google Scholar]
- Smith, D.F. (1998). Sequence motifs shared between chaperone components participating in the assembly of progesterone receptor complexes. Biol. Chem. 379, 283-288. [DOI] [PubMed] [Google Scholar]
- Stirling, C.J., Rothblatt, J., Hosobuchi, M., Deshaies, R., and Schekman, R. (1992). Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol. Biol. Cell 3, 129-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland, E., Qu, B.H., Millen, L., and Thomas, P.J. (1997). The molecular chaperone Hsc70 assists the in vitro folding of the N-terminal nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 272, 25421-25424. [DOI] [PubMed] [Google Scholar]
- Sullivan, C.S., Tremblay, J.D., Fewell, S.W., Lewis, J.A., Brodsky, J.L., and Pipas, J.M. (2000). Species-specific elements in the large T-antigen J domain are required for cellular transformation and DNA replication by simian virus 40. Mol. Cell. Biol. 20, 5749-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, R., Locher, M., and Hochstrasser, M. (2001). A conserved ubiquitin ligase of the nuclear envelop/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev. 15, 2660-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis, C., Hitt, R., Park, S.H., Deak, P.M., Kostova, Z., and Wolf, D.H. (2003). Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J. Biol. Chem. 278, 35903-35913. [DOI] [PubMed] [Google Scholar]
- Thomas, P.J., Qu, B.H., and Pedersen, P.L. (1995). Defective protein folding as a basis of human disease. Trends Biochem. Sci. 20, 456-459. [DOI] [PubMed] [Google Scholar]
- Tsai, B., Ye, Y., and Rapoport, T.A. (2002). Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell. Biol. 3, 246-255. [DOI] [PubMed] [Google Scholar]
- Vashist, S., and Ng, D.T. (2004). Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 165, 41-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, R., Chen, S., Feldman, R., Schieltz, D., Yates, J., Dohmen, J., and Deshaies, R.J. (2000). Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11, 3425-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, P., Bursac, D., Law, Y., Cyr, D., and Lithgow, T. (2004). The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5, 567-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, C.L., Omura, S., and Kopito, R.R. (1995). Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83, 121-127. [DOI] [PubMed] [Google Scholar]
- Wiech, H., Buchner, J., Zimmermann, R., and Jakob, U. (1992). Hsp90 chaperones protein folding in vitro. Nature 358, 169-170. [DOI] [PubMed] [Google Scholar]
- Willingham, S., Outeiro, T.F., DeVit, M.J., Lindquist, S.L., and Muchowski, P.J. (2003). Yeast genes that enhance the toxicity of a mutant huntingtin fragment or alpha-synuclein. Science 302, 1769-1772. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Janich, S., Cohn, J.A., and Wilson, J.M. (1993). The common variant of cystic fibrosis transmembrane conductance regulator is recognized by hsp70 and degraded in a pre-Golgi nonlysosomal compartment. Proc. Natl. Acad. Sci. USA 90, 9480-9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J.C., Moarefi, I., and Hartl, F.U. (2001). Hsp 90, a specialized but essential protein-folding tool. J. Cell Biol. 154, 267-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J.C., Schneider, C., and Hartl, F.U. (1997). In vitro evidence that hsp90 contains two independent chaperone sites. FEBS Lett. 418, 139-143. [DOI] [PubMed] [Google Scholar]
- Zhang, F., Kartner, N., and Lukacs, G.L. (1998). Limited proteolysis as a probe for arrested conformational maturation of delta F508 CFTR. Nat. Struct. Biol. 5, 180-183. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Michaelis, S., and Brodsky, J.L. (2002). CFTR expression and ER-associated degradation in yeast. Methods Mol. Med. 70, 257-265. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Nijbroek, G., Sullivan, M.L., McCracken, A.A., Watkins, S.C., Michaelis, S., and Brodsky, J.L. (2001). Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell 12, 1303-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J., Guo, Y., Guettouche, T., Smith, D.F., and Voellmy, R. (1998). Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94, 471-480. [DOI] [PubMed] [Google Scholar]