Abstract
Proteomic detection of non-annotated microproteins indicates the translation of hundreds of small open reading frames in human cells, but whether these microproteins are functional is unknown. Here, we report the discovery and characterization of a 7-kilodalton human microprotein we named non-annotated P-body dissociating polypeptide (NoBody). NoBody interacts with mRNA decapping proteins, which remove the 5’ cap from mRNAs to promote 5’-3’ decay. Decapping proteins participate in mRNA turnover and nonsense mediated decay (NMD). NoBody localizes to mRNA decay-associated RNA-protein granules called P-bodies. Modulation of NoBody levels reveals that its abundance is anti-correlated with cellular P-body numbers and alters the steady-state levels of a cellular NMD substrate. These results implicate NoBody as a novel component of the mRNA decapping complex and demonstrate potential functionality of a newly discovered microprotein.
Introduction
Recent advances in genomics and mass spectrometry-based proteomics have revealed that traditional computational genome annotation algorithms. For example, ribosome footprinting coupled with deep sequencing (Ribo-Seq) revealed thousands of putative non-annotated protein-coding smORFs in eukaryotic genomes1,2. Likewise, modified proteomics strategies have identified hundreds to thousands of translated human smORFs through direct detection of expressed peptides and small proteins (microproteins)3,4. These findings demonstrate that microprotein-encoding smORFs constitute a significant fraction of genomes5,6. However, it is unclear how many of these newly discovered smORFs encode functional microproteins.
While the generality of microprotein function remains uncertain, functional fly, mouse and human microproteins have been described5. For example, the sarcolamban and myoregulin microproteins regulate muscle function in flies and mice, respectively, by binding to a calcium channel7,8. The human microprotein MOTS-C9 regulates metabolic homeostasis, and human MRI-210 modulates non-homologous end-joining DNA repair,in vitro. These precedents suggest that continued elucidation of the functions of small ORFs may reveal novel regulatory roles of microproteins. However, because sequence conservation of smORFs is low relative to known protein-coding genes3,11, homology-based functional characterization is difficult.
Most of the eukaryotic microproteins that have been characterized to date exert their functions through microprotein-protein interactions, suggesting that this property can be leveraged when investigating microproteins of unknown function. In this report, we describe application of functional proteomics to identify the interaction partners of a conserved ~7-kilodalton mammalian microprotein, which we call NoBody (non-annotated P-body dissociating polypeptide) based on its cellular function (vide infra). Through this approach, we demonstrate that NoBody interacts with mRNA decapping proteins, which are the molecular components of the first enzymatic step in the 5’-to-3’ mRNA decay pathway. NoBody pulls down and covalently cross-links to the purified protein enhancer of decapping 4 (EDC4), an activator of mRNA decapping. The 5’-to-3’ mRNA decay pathway regulates turnover of normal cellular mRNAs, as well as transcripts targeted for decay via AU-rich element (ARE) recognition and nonsense-mediated decay (NMD)12, and the molecular components of this pathway localize to cytoplasmic ribonucleoprotein granules called P-bodies13. Remarkably, manipulation of NoBody expression levels reveals that they are anti-correlated with P-body numbers, with P-bodies largely absent from cells over-expressing NoBody. NoBody levels are also anti-correlated with the steady-state concentration of an NMD substrate mRNA. Taken together, our studies reveal that NoBody interacts with decapping proteins to modulate P-body numbers in cells, demonstrating that NoBody may represent a new functional human microprotein.
Results
A non-coding gene produces the NoBody microprotein
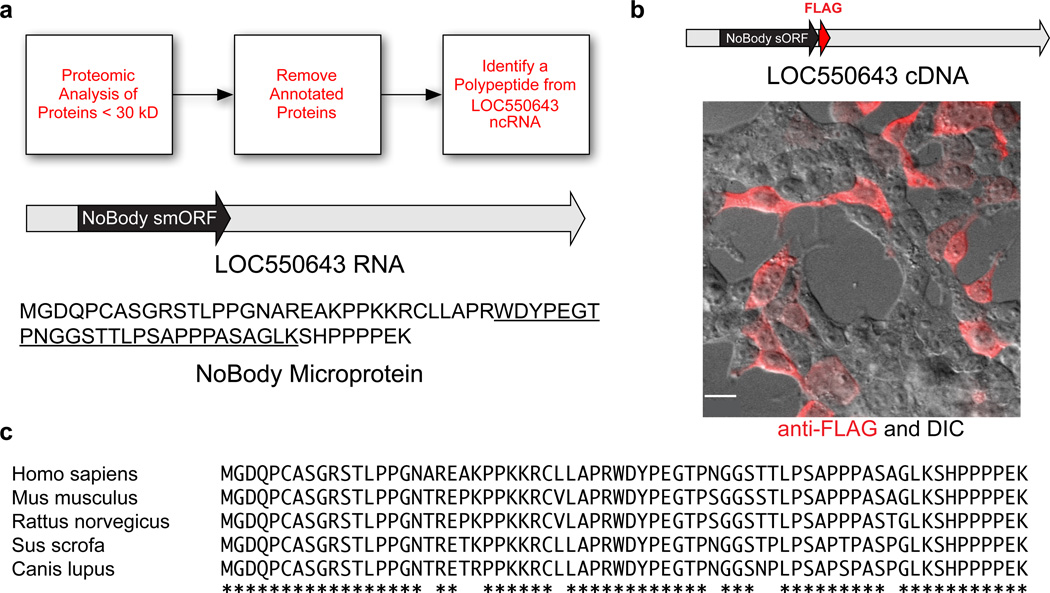
Using proteomics, we detected an unreported sORF-encoded polypeptide that is translated from the LINC01420/LOC550643 RNA (Fig. 1a), which is currently annotated in the NCBI database as non-coding. Our detection of this polypeptide in K562 cells is, to our knowledge, the first experimental evidence that this gene is translated (Supplementary Results, Supplementary Fig. 1a). NoBody is also detected in HEK293T and MDA-MB-231 cells (Supplementary Fig. 1b,c), indicating expression in several human cell lines from different tissues of origin. An expression construct comprising the full-length LINC01420 transcript with an epitope tag appended to the 3’ end of the putative sORF generated robust expression of NoBody in transfected human cells, demonstrating that the NoBody sORF is translated (Fig. 1b).
Figure 1. The LOC550643/LINC01420 gene encodes the NoBody peptide in a short open reading frame (sORF).
(a) K562 and HEK293T cellular peptides were enriched and subjected to multidimensional LC-MS proteomics. Peptide mass spectra were searched against a custom protein database obtained from the 3-frame translation of RNA-Seq data from these cell lines. Annotated peptides were removed by BLAST search to afford a list of non-annotated peptides. This workflow led to the discovery of a tryptic peptide (underlined sequence) derived from a polypeptide translated from a sORF (black) in the LOC550643 RNA transcript (gray). The polypeptide is hereafter referred to as NoBody. (b) Transfection of an expression construct corresponding to the annotated full-length LOC550643 cDNA sequence (gray), with an epitope tag (red) at the C-terminus of the putative short ORF (black) into HEK293T cells resulted in expression of NoBody (red anti-FLAG immunofluorescence image superimposed on differential interference contrast (DIC) image). Scale bar, 20 µm. (c) ClustalW2 alignment of full-length NoBody polypeptide sequence from a variety of mammals. Amino acid identity is indicated by asterisks.
A translated nucleotide BLAST (tBLASTn) search of the NCBI NR database (Fig. 1c), revealed that NoBody exhibits sequence conservation with computationally predicted small mammalian proteins of similar length, but not with any larger proteins or domains of known function. A web-based nucleotide BLAST search of the Drosophila melanogaster genome18 with relaxed parameters (expect value 100) returned no hits to any valid open reading frames; similarly, web-based BLAT search of the zebrafish genome19–21 also returned no hits. This may suggest that NoBody is specific to the mammalian lineage, though it does not rule out the existence of distant homologs. However, the high sequence conservation of NoBody among mammals supports a functional role for this protein.
The LINC01420 gene is located on the X chromosome (Supplementary Fig. 2a). Analysis of the gene structure reveals three exons separated by two introns, which are removed in the mature mRNA (Supplementary Fig. 2b,c)22, as well as alternative splice donor sites. The NoBody coding sequence is the first (5’) ORF in the transcript. LINC01420 expression is in the first or second highest gene expression quantile in most human tissues based on publicly available tissue-specific RNA-Seq data (Supplementary Fig. 2d), demonstrating that it is highly and ubiquitously expressed23. A Mus musculus homolog, 2210013O21Rik, is encoded on the mouse X chromosome at position +7612154249216.0022 and, while intron lengths and annotated alternative splice sites vary between human and mouse, the overall exon structure is conserved (Supplementary Fig. 3).
NoBody interacts with mRNA decapping proteins
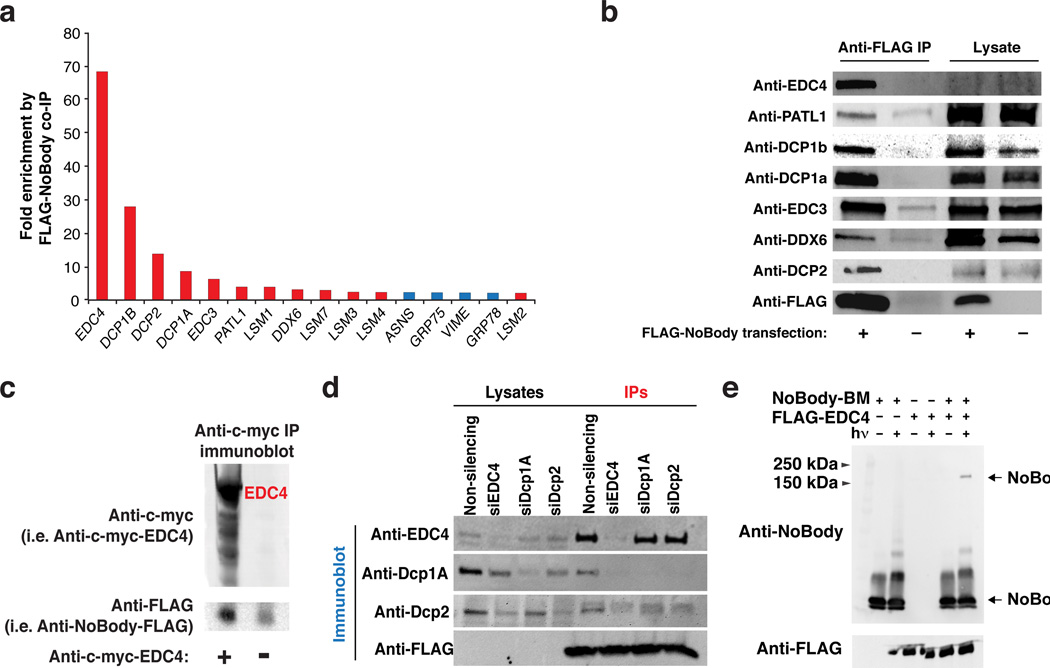
Though the sequence of NoBody is conserved in mammals, it is not homologous to any functional proteins, and is predicted to have little to no secondary structure. Because the function of NoBody could not be predicted based on sequence or structural conservation, we turned to functional proteomics. We immunoprecipitated FLAG-tagged NoBody from transiently transfected HEK293T cells, and quantified proteins specifically enriched over a vector-only immunoprecipitation control via semi-quantitative proteomics. After excluding common contaminants24, we found that only twelve proteins were specifically enriched >2-fold by NoBody, all of which are components of mRNA decapping and 5’-to-3’ mRNA decay complexes (Fig. 2a, Supplementary Data Set 1). Western blotting of co-immunoprecipitates of NoBody confirmed that it enriches proteins with established roles in mRNA decapping, including enhancer of decapping proteins 3 and 4 (EDC3 and EDC4), two orthologs of decapping protein 1 (Dcp1A and Dcp1B), and decapping protein 2 (Dcp2), as well as additional 5’-to-3’ decay factors (Fig. 2b). Reciprocal co-immunoprecipitation of FLAG-NoBody with myc-tagged EDC4 and Dcp1A confirmed its co-purification with these proteins (Fig. 2c, Supplementary Fig. 5). The highly specific enrichment of the decapping complex proteins, as quantified by mass spectrometry and lack of enrichment of background proteins, demonstrates that NoBody must be associated with the decapping complex.
Figure 2. NoBody enriches a complex of proteins involved in mRNA decapping and cross-links to EDC4.
(a) Quantitative proteomics of FLAG-NoBody immunoprecipitates from HEK293T lysates identified putative NoBody interaction partners (N = 1) (see Supplementary Data 1 for total list). Red bars correspond to proteins with a role in mRNA decapping. (b) Confirmation of NoBody-interaction partners by anti-FLAG immunoprecipitation and Western blotting (N = 3). (c) Anti-c-myc immunoprecipitation of c-myc-EDC4 and c-myc-Dcp1A from HEK293T cells co-expressing FLAG-NoBody (cells expressing NoBody-FLAG alone as a negative control) followed by anti-FLAG immunoblotting demonstrates that FLAG-NoBody is enriched by EDC4. (d) HEK293T cells were transfected with either non-targeting siRNA, or siRNA targeting EDC4, Dcp1A, or Dcp2. 48 hours later, cells were lysed and recombinantly expressed and purified FLAG-NoBody was added. Anti-FLAG immunoprecipitation (IP) isolated NoBody-interacting complexes. (e) NoBody was labeled with the photo-cross-linker 4-(Maleimido)benzophenone (NoBody-BP) and irradiated at 365 nm in the presence or absence of purified FLAG-EDC4. In the presence of EDC4, a higher molecular weight cross-linked product is observed at a molecular weight that would correspond to a 1:1 complex between EDC4 and NoBody.
EDC4 was the most abundant protein enriched by NoBody, based on semi-quantitative spectral counting, suggesting that it could be the direct interaction partner of NoBody (Supplementary Data Set 1). In order to test this hypothesis, we investigated interaction of NoBody with the decapping complex in cell lysates depleted of individual candidate interaction partners (Fig. 2d). We used siRNA to silence cellular expression of several candidate interaction partners: EDC4, Dcp1A, and Dcp2. Purified, recombinant FLAG-NoBody was then added to the cell lysates and immunoprecipitated to identify interaction partners. While we were unable to achieve complete silencing for Dcp1A and Dcp2, their expression levels and co-purification with NoBody were reduced relative to the non-silenced control. Under these conditions, even though the levels of Dcp1A and Dcp2 were depleted, EDC4 still copurified with NoBody. On the other hand, when EDC4 was silenced, none of the other decapping proteins co-purified with NoBody. These data indicate Nobody interacts with the decapping complex via direct interactions with EDC4, though does not completely rule out interactions with other proteins.
We then demonstrated that NoBody can pull down EDC4 in vitro (Supplementary Fig. 4). GST (glutathione S-transferase)-tagged NoBody was expressed and purified from E. coli, and FLAG-tagged EDC4 was immunopurified from HEK293T cells. FLAG-EDC4 was specifically retained on a GST column only in the presence of GST-NoBody, consistent with an interaction between these proteins. However, while EDC4 appears pure by silver staining, we cannot rule out the trace presence of other proteins, which may also interact with NoBody.
We therefore utilized covalent photo-cross-linking to probe a direct molecular contact between NoBody and EDC4 both in isolation and in cell lysates. First, purified NoBody was chemically labeled with a benzophenone photo-cross-linker and irradiated in the presence of immunopurified EDC4. If benzophenone labeled NoBody (NoBody-BP) directly contacts EDC4, then irradiation should result in a covalent bond between NoBody and EDC4. Probing with a rabbit polyclonal antibody we raised to NoBody (see Online Methods) revealed that addition of EDC4 to NoBody-BP, followed by irradiation, results in the formation of a NoBody-reactive species that migrates at a molecular weight consistent with a complex of the two proteins (Fig. 2e). To further support a direct interaction between NoBody and EDC4 in cells, we synthesized a modified NoBody22–41 peptide, with an N-terminal rhodamine and a C-terminal benzophenone (Rh-NoBody22–41-BP). Again, addition of Rh-NoBody22–41-BP to HEK293 lysates overexpressing EDC4 resulted in a light-dependent Rh-NoBody22–41-BP labeling of a protein consistent with the size of EDC4 (Supplementary Fig. 7). The addition of excess biotin-NoBody22–41-BP selectively decreased the labeling of this band without affecting the intensity of background bands, demonstrating that it is specific (Supplementary Fig. 6). Combined, these in-cell and in vitro experiments are consistent with a direct interaction between NoBody and EDC4, though we cannot exclude additional interactions with other proteins.
Sequence determinants of NoBody-EDC4 co-purification
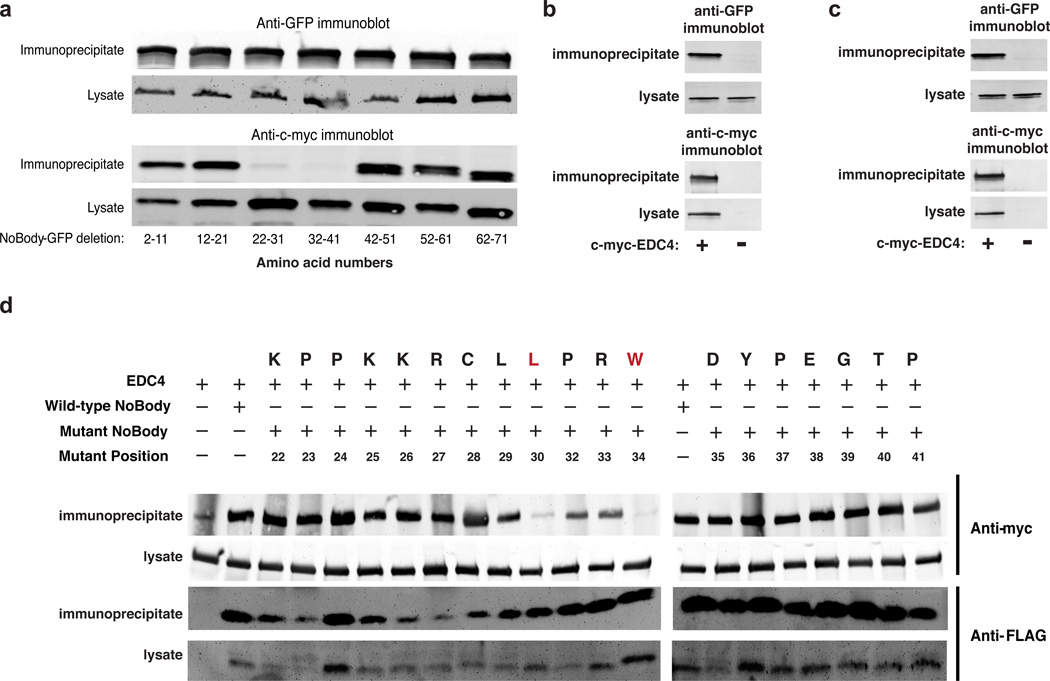
A series of deletion mutants of NoBody were generated to identify the region of NoBody necessary for co-purification with EDC4. Each of these constructs was fused to enhanced green fluorescent protein (EGFP) in order to equalize expression of all deletion mutants after transient transfection (Fig. 3a). Only NoBody-EGFP deletion mutants that lacked amino acids 22–41 (Δ22–31 and Δ32–41) did not co-precipitate with co-transfected myc-EDC4 (Fig. 3a). We found that amino acids 22–41 of NoBody are sufficient for EDC4 association by attaching these amino acids to the N- and C-termini of EGFP. Both the N- and C-terminal fusions of amino acids 22–41 of NoBody to EGFP co-precipitated with myc-EDC4 (Fig. 3b,c), while EGFP alone did not (Supplementary Fig. 7).
Figure 3. Sequence dependence of NoBody-EDC4 co-precipitation.
(a) In order to determine the region of the NoBody peptide responsible for interacting with EDC4, a series of 10-amino-acid deletions spanning the length of NoBody was prepared and fused to EGFP to equalize their expression. The constructs were co-transfected with c-myc-EDC4 in HEK293T cells, then lysates were subjected to anti-myc immunoprecipitation followed by anti-GFP Western blotting. 5% of each lysate and 25% of each immunoprecipitate was loaded in each lane. (b,c) The 20-amino-acid fragment of NoBody required for EDC4 co-precipitation (NoBody(22–41)) was fused to the N- and C-termini of EGFP to assay its sufficiency for interaction with EDC4. The NoBody(22–41)-EGFP fusions were co-transfected into HEK293T cells with c-myc-EDC4, then lysates subjected to anti-c-myc immunoprecipitation followed by anti-GFP Western blotting. A negative control (d) was performed with non-fused EGFP alone. 2–5 % of lysate and 40–50 % of each immunoprecipitate was loaded in each lane. (e) Alanine scanning mutagenesis of full-length FLAG-tagged NoBody peptide between NoBody amino acid residues 22 to 41 was performed to identify residues essential for the interaction with EDC4. These constructs were co-expressed with c-myc-EDC4 in HEK293T cells and subjected to anti-FLAG immunoprecipitation, followed by anti-c-myc Western blotting to assess EDC4 interaction.
Next, to identify specific residues required for the NoBody co-precipitation with EDC4, we performed an alanine scan between amino acids 22–41 of FLAG-NoBody. Immunoprecipitation of FLAG-NoBody alanine mutants revealed two key residues, Leu30 and Trp34, that impair co-precipitation of NoBody with myc-EDC4 (Fig. 3d). These data demonstrate the high sequence-specificity of the association of NoBody and EDC4, consistent with a selective molecular interaction.
NoBody levels are anti-correlated with P-body numbers
The proteins involved in mRNA decapping and 5’-to-3’ mRNA decay localize to RNA-protein (RNP) granules called mRNA processing bodies, or P-bodies, in all eukaryotic cells25. Previously characterized mRNA decapping proteins are known to affect P-body numbers and/or morphology when over-expressed or silenced26. For example, silencing of EDC4 eliminates P-bodies, while over-expression of EDC4 increases P-body numbers and size17,27. Conversely, silencing of Dcp2 increases P-body number and size17,27. We therefore examined the effect of NoBody expression level on P-bodies.
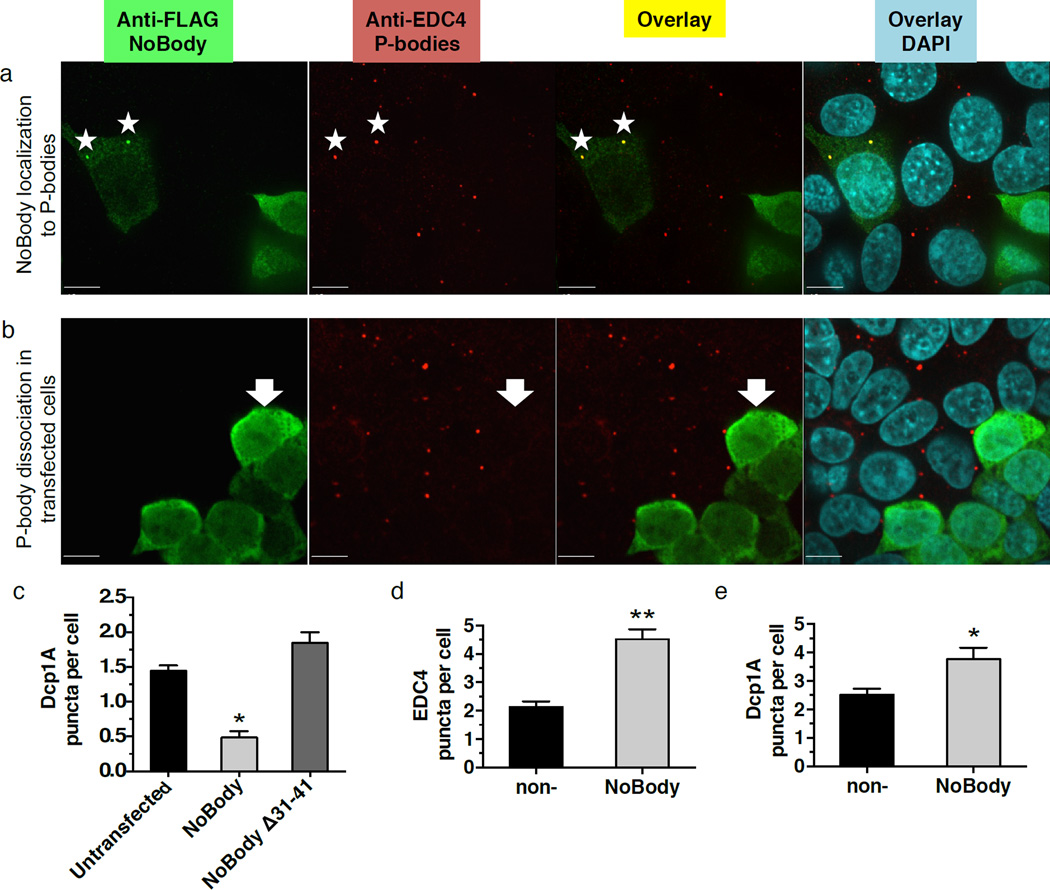
Transiently transfected P-body proteins can localize primarily to the cytoplasm if overexpressed17. Therefore, we expressed the NoBody coding sequence using lentiviral transduction, which we demonstrated permits the P-body proteins Dcp2 and Rck1 to primarily co-localize with endogenous P-body foci (Supplementary Fig. 8). Endogenous EDC4 and Dcp1A were used as P-body markers, and formed foci consistent with P-bodies in untransfected cells (Fig. 4a, Supplementary Fig. 8–10). Surprisingly, cells expressing NoBody do not have detectable P-bodies (Fig. 4b, Supplementary Fig. 9a, 10a). P-body dispersal requires residues 31–41 of NoBody, indicating that the interaction between NoBody and EDC4 is involved (Fig. 4c). P-body dispersal by NoBody is generalizable to three additional cell lines (Supplementary Fig. 11), demonstrating that it is not cell line-specific.
Figure 4. NoBody localizes to P-bodies and its expression level is inversely correlated with P-body numbers.
HEK293T cells were transfected with FLAG-NoBody via lentiviral infection at low viral titer. After fixation cells were stained with antibodies detecting a P-body marker (Dcp1A, red) and FLAG-NoBody (green), then counterstained with DAPI to visualize the nucleus (blue). In the merged image, NoBody/Dcp1A co-localization appears yellow. (a) In a minority of transfected cells (~10%), NoBody forms puncta that co-localize with P-bodies (starred). (b) In a majority of transfected cells (~90%), over-expression of NoBody leads to an absence of macroscopically detectable P-bodies (arrow). (c) HEK293T cells were transfected with FLAG-NoBody or the non-EDC4-interacting mutant FLAG-NoBodyΔ32–41. Endogenous P-bodies were visualized with anti-Dcp1A. Four fields of view, including the representative images, were used to quantitate average P-bodies per cell, representing >377 cells, in each average. Data represent mean values +/− S.E.M. Data represent mean values +/− S.E.M, and significance was evaluated with ANOVA (d–e) HEK293T cells were transfected with non- or NoBody-silencing siRNA, then fixed and endogenous P-bodies were detected using either anti-EDC4 (d) or anti-Dcp1A (e) immunofluorescence. For quantitation of P-bodies per cell, >6 fields of view were analyzed, totaling >400 cells for each measurement. Data represent mean values +/− S.E.M, and significance was evaluated with a two-tailed t-test. Scale bars, 10 µm.
Previous studies have indicated that proteins readily exchange between P-bodies and the cytosol28. In line with this conclusion, the level of endogenous Dcp1A was identical by Western blot between control cells and NoBody infected cells (Supplementary Fig. 9b), confirming that P-bodies, and their constituent proteins, are being dispersed or solubilized rather than degraded.
We utilized another P-body reporter protein, GFP-Dcp2, to further examine this phenomenon. Cells expressing GFP-Dcp2 have normal P-bodies, and GFP-Dcp2 forms foci that co-localize with endogenous EDC4, indicating that retroviral transduction of cells does not affect P-bodies in and of itself (Supplementary Fig. 12). Co-expression of GFP-Dcp2 and NoBody results in loss of GFP-Dcp2 puncta as well as endogenous P-bodies (Supplementary Fig. 12). The dispersal of GFP-Dcp2 from puncta to a diffuse localization again demonstrates that NoBody causes P-body dispersal rather than degradation.
Finally, we showed that P-body dispersal requires the NoBody peptide, not the LINC01420 mRNA. While all imaging and functional experiments in this report were conducted using expression constructs harboring only the NoBody coding sequence, not the full-length cDNA (except Figure 1b, see Methods), it is possible that this small fragment of the transcript could be active at the RNA level and interact with P-bodies. We therefore generated a lentiviral vector with the NoBody start codon deleted. Transduction of cells with this construct deficient for translation failed to elicit a change in P-bodies, demonstrating the peptide itself is required for this phenotype (Supplementary Fig. 13).
Conversely, silencing endogenous NoBody expression increased the number of P-bodies per cell. We observed a 1.5- to 2-fold increase in P-bodies per cell when NoBody was silenced (Fig. 4d,e). This effect is not caused by a non-specific effect of siRNA-mediated gene silencing, because silencing an unrelated gene (GAPDH) causes no change in P-body numbers (Supplementary Fig. 14). Taken together, our results are consistent with an inverse correlation of the cellular concentration of the NoBody microprotein with P-body numbers that depends on its interaction with the mRNA-decapping complex.
NoBody localizes to P-bodies
Since over-expression of NoBody results in dissolution of P-bodies, we reasoned that low levels of NoBody expression might result in a subpopulation of cells where the NoBody expression is low enough to detect but the NoBody concentration is not high enough to affect P-body structure. Indeed, lentiviral transfection of FLAG-NoBody with low viral titers resulted in a population of cells (~10% of total) where NoBody co-localized with the P-body marker EDC4 (Fig. 4a). Cells exhibiting higher NoBody expression did not show this co-localization and NoBody was found to be mostly cytoplasmic (Fig. 4b). This suggests that NoBody localizes to P-bodies at low expression levels.
NoBody is anti-correlated with levels of an NMD substrate
NMD is a pathway of RNA quality control that targets premature termination codon-containing mRNAs for rapid degradation to prevent production of truncated protein products29,30. While NMD has been reported to promote RNA degradation via SMG6-mediated endonucleolysis followed by both 5’-to-3’ and 3’-to-5’ decay of the resulting fragments, the decay of some NMD substrates is also sensitive to mRNA decapping. In support of a role for decapping in NMD, silencing of Dcp2 has been reported to inhibit NMD31,32, and Upf1 interacts with human Dcp1A and Dcp233. While it is unclear if the decapping complex is a catalytic component of the NMD pathway, or if its role is indirect, at least some NMD substrates in some cell lines are protected by loss of Dcp2. We therefore sought to determine whether NoBody affects the cellular levels of an endogenous cellular NMD substrate.
We chose the Calu-6 cell line for our assay, which harbors a well-characterized mutant p53 gene containing a premature termination codon34. The Calu-6 p53 transcript is a potent NMD substrate35, and inhibition of NMD increases steady-state levels of mutant p53 mRNA in this cell line36,37. This reporter gene has been utilized to study the function of the upstream NMD regulator SMG-135,38 and to develop novel small-molecule inhibitors of NMD36,37. We therefore perturbed levels of cellular NoBody and measured changes in steady-state p53 mRNA abundance via quantitative RT-PCR with normalization to beta-actin. We note, however, that the Calu-6 p53 gene has not previously been shown to be a Dcp2 substrate.
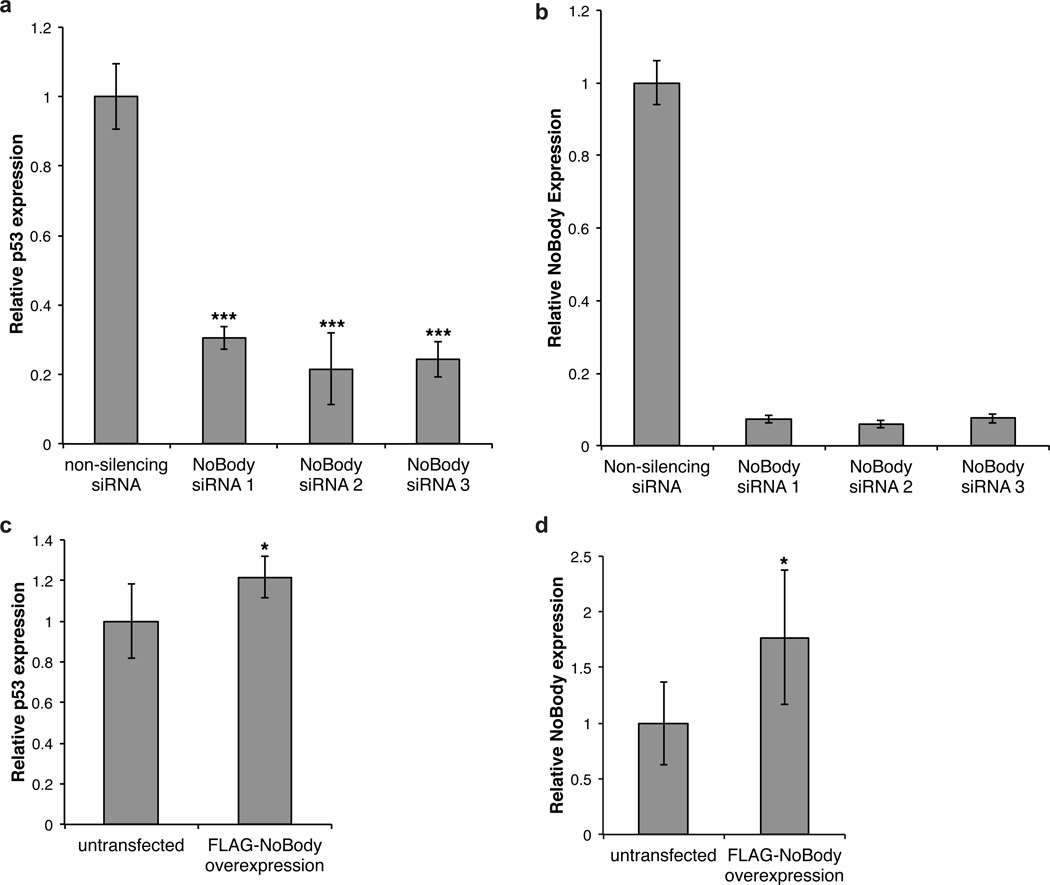
We first confirmed that this assay is responsive to small-molecule NMD inhibitors39; as previously reported, cycloheximide, actinomycin D, and caffeine all increase p53 mRNA (relative to a reference gene, beta-actin) ~4–16-fold over vehicle (Supplementary Fig. 15). Next, we transfected Calu-6 cells with two different siRNA sequences targeting NoBody, each of which afforded >80% silencing, and observed a nearly two-fold decrease in p53 levels relative to a non-silencing control (Fig. 5a,b) Conversely, weak over-expression of lentivirally transfected NoBody (75% above endogenous NoBody levels) led to a small but statistically significant 22% increase in p53 mRNA relative to untransfected cells (Fig. 5c,d). These results demonstrate that the cellular concentration of NoBody is inversely linked to steady-state abundance of an NMD substrate in Calu-6 cells, though the effect of NoBody on the decay rate of this mRNA has not been established, and by extension the mechanism by which its abundance is increased in the absence of NoBody remains to be elucidated.
Figure 5. Perturbation of NoBody expression modulates levels of a nonsense-mediated decay substrate in cells.
(a) Calu-6 cells were transfected with 2 independent siRNA sequences targeting NoBody for RNAi-mediated silencing, or with a non-targeting siRNA as a negative control. RNA was isolated 48 hours later, and levels of NoBody and p53 mRNA were determined by quantitative RT-PCR with reference to beta-actin. Data represent the mean of six biological replicates (**, p<0.01, ***, p<0.001). (b) NoBody silencing in Calu-6 was measured by quantitative RT-PCR with reference to beta-actin. (c) Calu-6 cells were lentivirally transfected for over-expression of the FLAG-NoBody coding sequence; untransfected cells served as a control. RNA was isolated 48 hours later, and levels of NoBody and p53 mRNA were determined by quantitative RT-PCR with reference to beta-actin. Data represent the mean of six biological replicates (*, p<0.05). (d) NoBody overexpression after lentiviral transfection in Calu-6 (b) was measured by quantitative RT-PCR with reference to beta-actin. Data represent mean values +/− standard deviation, and significance was evaluated with two-tailed t-test.
Discussion
Our search for functional microproteins led to us to the identification of NoBody as a conserved component of the mRNA decapping complex that cross-links to EDC4. The first report describing EDC4 postulated the existence of an uncharacterized “small subunit” of this protein17, which is consistent with the size and EDC4 binding activity of NoBody. The discovery of NoBody provides new opportunities to investigate this protein and its contribution to mRNA decapping. For example, whether NoBody is a constitutive and stoichiometric EDC4 binding partner, or a dynamic regulator of EDC4 activity, is a key question that will require elucidation of the absolute cellular concentrations of these proteins.
The observation that P-body numbers change upon modulation of NoBody expression is consistent with similar changes in P-body numbers and morphologies reported when over-expressing and silencing expression of known 5’-to-3’ mRNA decay factors. Whether P-body numbers change in our experiments due to direct structural perturbation of P-bodies, or as a secondary result of an increase or decrease in the rate of enzymatic mRNA degradation40, is unclear. Therefore, the observation of the inverse correlation of P-body numbers to NoBody expression is best interpreted as evidence for the involvement of NoBody in the decapping complex. However, NoBody is, to our knowledge, the smallest protein thus far described that perturbs P-body numbers, and therefore may prove valuable in future studies of P-body structure and function. Our observation that loss of NoBody causes a decrease in the cellular levels of an NMD substrate is most consistent with negative regulation of mRNA decay in cells by NoBody, though the exact mechanism of this regulation will require additional assays (e.g. kinetic measurements of mRNA decay and biochemical mRNA decapping assays), to fully elucidate the molecular function and mechanism of NoBody. At minimum, our results suggest that NoBody expression is inversely coupled to the expression level of a mutant oncogene.
Rapid advancements in technology have driven our ability to dig deep into the unexplored sequence space of the human genome and proteome. It is now possible to use RNA deep sequencing data to create databases of all potential coding sequences in cells and tissues. Coupled to ribosome-sequencing1,2 (Ribo-Seq) or proteomics, these data have identified previously non-annotated short protein-coding genes. The total number of these newly identified microproteins in human cells has now reached the thousands, but their contribution to the functional proteome is only beginning to be explored. Our study demonstrates that a newly described microprotein, despite its small size, can perturb cellular RNP granules and gene expression, suggesting that the continued discovery and functional characterization of microproteins stands to provide new insights into important cellular processes.
Online Methods
Data Analysis
Statistics, two-sided t-test or ANOVA, were performed using Excel or Prism, and equal variance between samples being compared was established using an F-test.
Cell Lines
COS7, HEK293T, K562, HeLa, Calu-6 were purchased from ATCC and early-passage stocks were in order to ensure cell line provenance and sterility. The BGC-823 cell line was purchased from the Istituto Nazionale per la Ricerca sul Cancro (Genova, Italy) and early-passage stocks were used in order to ensure cell line provenance and sterility.
Antibodies
Primary antibodies for both Western blotting and immunofluorescence include: mouse monoclonal anti-FLAG (Sigma, F3165); rabbit polyclonal anti-Dcp1a (Sigma, D5444); rabbit polyclonal anti-PatL1 (Abcam, ab124257); rabbit polyclonal anti-EDC4 (Sigma, SAB4200114); rabbit polyclonal anti-Dcp1b (Novus Biologicals, NBP1-82018); rabbit polyclonal anti-Dcp2 (Novus Biologicals, NBP1-41070); rabbit monoclonal anti-DDX6X (Abcam, ab174277); rabbit polyclonal anti-EDC3 (Abcam, ab168851); anti-c-Myc (VWR, 95040-930, this antibody is no longer available at VWR); and rabbit monoclonal anti-GFP (Abcam, ab183734). Secondary antibodies for fluorescent Western blotting were goat anti-mouse IR dye 800 (LI-COR, 925-32210) and goat anti-rabbit IR dye 680 (LI-COR, 925-68071). Secondary antibodies for chemiluminescent Western blotting were goat anti-rabbit peroxidase conjugate (Rockland, 611-103-122) and goat anti-mouse peroxidase conjugate (Rockland, 610-1319). Secondary antibodies for immunofluorescence are goat anti-mouse Alexa Fluor 568 (Life Technologies, A-11004), and goat anti-rabbit Alexa Fluor 488 (Life Technologies, A-11008), 568 (Life Technologies, A-11011), and 647 (Life Technologies, A-21245). Immunoprecipitation was performed with the following antibody beads: anti FLAG M2 affinity gel (Sigma, A2220), anti-myc tag agarose beads (MBL International Corp., M047-8). The NoBody antibody was generated by GenScript and validated as described in Supplementary Figure 16.
Cloning and genetic constructs
A cDNA clone for LOC550643 was obtained from Open Biosystems. The NoBody coding sequence was subcloned with an N-terminal FLAG epitope tag into pcDNA3. Constructs comprising the FLAG-NoBody coding sequence only were used for all experiments except Figure 1. A separate construct encoding the entirety of the NCBI cDNA sequence for LOC550643 was synthesized by Genscript, with a FLAG epitope tag at the 3’ end of the NoBody coding sequence, then subcloned into pcDNA3. This full-length cDNA clone was only used to generate Figure 1; all other experiments utilized expression constructs harboring the NoBody coding sequence only.
A cDNA clone encoding EDC4 was obtained from Open Biosystems; N-terminal myc and FLAG epitope tag fusions were created in this construct using inverse PCR. Deletion mutants of NoBody were also generated by inverse PCR. The GFP fusion to NoBody was constructed in pcDNA3 using restriction cloning. GFP-Dcp1A and GFP-Dcp2 were obtained from Addgene as a gift from Eliza Izurralde41. Retroviral constructs were subcloned into the pFCPGW plasmid.
For bacterial expression, the NoBody coding sequence was cloned into BamHI and XhoI sites of the pGEX-6-P3 vector (GE Life Sciences). This construct yields a fusion protein consisting of Glutathione S-Transferase (GST) followed by NoBody with an amino-terminal FLAG tag. A PreScission protease cleavage site between the GST tag and NoBody peptide facilitated removal of the GST tag after purification. An identical construct that lacked the FLAG tag was generated by deletion PCR.
Cell culture and transfection
HEK293T and HeLa cells were cultured in DMEM supplemented with 10% fetal bovine serum, penicillin, and streptomycin. Cells were maintained in a 5% CO2 atmosphere at 37 °C. Plasmid transfection was performed with Lipofectamine 2000 and Opti-MEM according to the manufacturer’s instructions. siRNA specific for human LOC550643, GAPDH, and a non-silencing siRNA were obtained from Qiagen and transfected using Dharmafect 1 or 2 according to the manufacturer’s instructions. For immunoprecipitation, cells were lysed 24 hours post-transfection; for immunofluorescence imaging, cells transfected with plasmids or siRNA were assayed 24 or 48 hours post-transfection, respectively. Lentivirally infected cells were assayed 48 hours post-infection.
siRNA and qRT-PCR primers
siRNA and qRT-PCR primer product numbers and providers utilized in this study are provided in Supplementary Table 1 and 2.
Peptidomics and LOC550643 SEP identification
The peptidomics experiments in which NoBody was detected in K562, H293T and MDA-MB-231 cells were previously reported3,42. NoBody was excluded from the originally reported list of SEPs because predicted proteins in the non-redundant protein database (NCBI) were filtered out.
Confirmation of NoBody expression from LOC550643/LINC01420 cDNA clone via immunofluorescence
HEK 293T or HeLa cells were grown to 80% confluency on no. 1.5 glass coverslips in 48-well plates. Cells were transfected with a mammalian expression plasmid (pcDNA3) into which a complete cDNA clone corresponding to LOC550643/LINC01420 and putatively encoding the NoBody peptide had been inserted, with a FLAG epitope tag appended at the 3’-end of the putative coding sequence by inverse PCR. Transfections were carried out with Lipofectamine 2000 according to the manufacturer’s instructions. 24 hours post-transfection, cells were fixed with 4% formaldehyde in phosphate buffered saline, permeabilized with methanol at −20°C, and blocked with fluorescence blocking buffer (Rockland) for 1 hour at 4°C. Cells were stained with mouse anti-FLAG M2 antibody at a 1:1000 dilution in Rockland blocking buffer overnight at 4°C, followed by 3 phosphate-buffered saline (PBS) washes. Goat anti-mouse Alexa Fluor 568 was applied at a 1:1000 dilution in Rockland buffer for 1 hour at room temperature, then washed 3× with PBS. Cells were post-fixed with 4% formaldehyde in PBS, then imaged by confocal microscopy.
Confocal microscopy
Coverslips were inverted and imaged in PBS in MatTek imaging dishes. Confocal imaging was performed on a Zeiss AxioObserver inverted confocal microscope with 40 ×, 63 ×, or 100 × oil immersion objective, Yokogawa spinning disk confocal head, Quad-band notch dichroic mirror (405/488/568/647), and 405 (diode), 491 (DPSS), 561 (DPSS), and 640 nm (diode) lasers (all 50 mW). Hoechst was imaged using 405 laser excitation, 445/40 emission; Alexa Fluor 488/GFP was imaged using 491 laser excitation, 528/38 emission; Alexa Fluor 568 was imaged using 561 laser excitation, 617/73 emission; and Alexa Fluor 647 was imaged using 640 laser excitation, 700/75 emission. Differential interference contrast (DIC) images were also collected. All image collection and analysis were performed using Slidebook software (Intelligent Imaging Innovations).
Database search
A translated nucleotide BLAST (tBLASTn) search of the NCBI NR nucleotide database43 was performed with standard parameters (max target sequences 100, expect threshold 10, word size 3) to assess NoBody conservation. Conserved proteins were aligned with Clustal Omega using standard parameters. In order to directly probe the zebrafish genome for similar sequences, the NoBody sequence was searched against the zebrafish genome19 using the BLAT algorithm20 via the UCSC genome browser21,44. Both NoBody DNA and protein sequences were used as the query sequence under default search parameters. The FlyBase web interface was used to probe the Drosophila melanogaster genome for sequences similar to the NoBody protein coding sequence via tBLASTn search with relaxed parameters (no low complexity filter, expect value 100).
Co-immunoprecipitation and proteomics
HEK 293T cells were transfected with the FLAG-tagged NoBody coding sequence (coding sequence only) in a mammalian expression plasmid (pcDNA3) or empty pcDNA3 as a negative control. Transfections were carried out using Lipofectamine 2000 and 10 µg DNA per 10 cm dish of cells. 24 hours post-transfection, cells were harvested and lysed using Tris-buffered saline (TBS) with 1% Triton X-100 and Roche Complete protease inhibitor cocktail tablets. 400 µL lysis buffer was used per pellet. Cells were lysed on ice for 20 min followed by centrifugation at 14000 rpm, 4°C, 15 min. Lysate samples were saved for analysis of loading. A 50 µL aliquot of anti-FLAG agarose beads (clone M2, Sigma) was washed with 1 mL TBS-T, collected by centrifugation for 1 min at 3000 rpm, then suspended in the cell lysate supernatant. Bead suspensions were rotated at 4°C for 1 hour, then washed 3 times with TBS-T. Elution was in 30 µL of 3× FLAG peptide (Sigma), at a final concentration of 100 µg/mL in TBS-T at 4°C for 1 hour. Beads were removed by centrifugation and the entire supernatant was loaded on an SDS-PAGE gel.
For proteomics, immunoprecipitates were separated by SDS-PAGE with Coomassie stain and imaged. For quantitation via spectral counting, the gel was cut straight across with a clean razor into 10 equal-size slices for both the positive and negative control.
Protein-containing gel slices were digested with trypsin overnight. The resulting peptide mixtures were extracted from the gel and run directly on an Orbitrap Velos instrument (Thermo Fisher Scientific) with 90-minute liquid chromatography and tandem mass spectrometry (LC-MS/MS) using a standard TOP20 method procedure. Briefly, MS1 m/z regions for 395 –1600 m/z ions were collected at 60K resolving power and used to trigger MS/MS in the ion trap for the top 20 most abundant ions. Active dynamic exclusion of 500 ions for 90 secs was used during the LC-MS/MS method. Peptides were eluted with 300 nL/min flow rate using a NanoAcquity pump (Waters). Samples were trapped for 15 minutes with flow rate of 2 µL/min on a trapping column 100-micron ID packed for 5 cm in-house with 5 µm Magic C18 AQ beads (Waters) and eluted with a gradient to 20 cm 75-micron ID analytical column (New Objective) packed in-house with 3 µm Magic C18 AQ beads (Waters).
Mass spectra were analyzed using our in-house Proteome Browser System against uniprot_human database. Carbamidomethylated cysteines were set as a fixed modification, with oxidation of methionine and N-terminal acetylation as variable modifications. A mass deviation of 20 ppm was set for MS1 peaks and 0.6 Da was set as maximum allowed for MS/MS peaks and a maximum of two missed cleavages were allowed. Maximum false discovery rates were set to 0.01 both on peptide and protein levels. Minimum required peptide length was five amino acids.
Protein quantitation was accomplished via spectral counting, where the number of total peptides observed for each identified protein was taken as the total spectral counts and compared for the IP vs. negative control sample. All proteins elevated >10-fold relative to the negative control and present at >20 spectral counts in the IP sample were considered candidates for confirmation by Western blotting. The data represents a single run (i.e. n = 1).
Western blotting confirmation of interaction partners
Interaction candidates identified by proteomics were subsequently confirmed by co-immunoprecipitation and Western blotting with antibodies against the endogenous proteins. Transfections of HEK293T cells and immunoprecipitations were carried out as described above (see “Co-immunoprecipitation and proteomics”). 1% of cell lysate (5µL/400µL) and 15% of the immunoprecipitate was loaded onto three replicate gels for analysis with multiple antibodies. Lysates and IP samples were mixed with protein loading buffer, boiled, and separated on 4–20% Tris/glycine SDS-PAGE gels (BioRad). Proteins were transferred to Immobilon-FL PVDF (Millipore), for fluorescence imaging, or nitrocellulose membranes, for chemiluminescence imaging, for 2 h at 400 mA. Immunoblots were blocked with Rockland fluorescent blocking buffer, then probed with primary antibodies at a 1:1000 dilution in the same buffer for ~2 hours at 4°C. The membrane was washed three times with TBS-T. For chemiluminescence imaging, secondary antibodies were applied at a dilution of 1:10,000 in 3% bovine serum albumin in TBS-T, then washed 3× with TBS-T prior to development with Clarity ECL Western Blotting Substrate (Bio-Rad) and imaged using a UVP ChemiDoc-It2 Imaging System with Visionworks Software.
Reciprocal immunoprecipitation
HEK293T cells were transfected with the FLAG-tagged NoBody coding sequence in pcDNA3, alone as a negative control, or co-transfected with myc-tagged EDC4 in pCMV-Sport6 or myc-tagged Dcp1A in pcDNA3. 24 hours post-transfection, cells were lysed and immunoprecipitations were carried out as described above, but using anti-myc agarose beads (MBL International Corp). 50% of each immunoprecipitate was loaded onto a gel and transferred to membranes for Western blotting as described above. Primary antibodies used were rabbit anti-myc and mouse anti-FLAG M2, and secondary antibodies were goat anti-mouse IR dye 800 and goat anti-rabbit IR dye 680. For fluorescence imaging, secondary antibodies were applied at a dilution of 1:4000 in Rockland fluorescent blocking buffer. After a final 3× TBS-T wash, infrared fluorescence imaging was performed on a LICOR Odyssey instrument.
Co-immunoprecipitation after siRNA-mediated decapping protein silencing
HEK293T cells were grown to 50% confluency in T25 flasks, then transfected with non-silencing siRNA, or pools of siRNA targeting Dcp1A, Dcp2, or EDC4 using Dharmafect I according to the manufacturer’s instructions. 48 hours after transfection, cells were harvested and lysed as described above in 400 µL lysis buffer. 9 µg purified FLAG-NoBody protein (see below) was added to each lysate. 50 µg anti-FLAG agarose beads were washed and added to the lysates, and rotated for 4h at 4°C. Beads were washed 4× with TBS-T, then elution was performed by boiling in 40 µL SDS-PAGE loading buffer for 10 min. 5% of each lysate and 50% of each immunoprecipitate was separated on a 4–20% SDS-PAGE gel, then transferred to nitrocellulose membranes for Western blotting and chemiluminescence imaging as described above.
Deletion and mutation analysis of NoBody interaction interface
To identify the portion of the NoBody sequence that interacts with EDC4, cells were transfected with (a) myc-EDC4 in pCMVSport6 and one of a series of 10-amino-acid deletions of NoBody fused to GFP in pcDNA3, (b) myc-EDC4 pCMVSport6 and residues 22–41 of NoBody fused to the C-terminus of GFP in pcDNA3, or the NoBody construct alone; (c) myc-EDC4 pCMVSport6 and residues 22–41 of NoBody fused to the N-terminus of GFP in pcDNA3, or the NoBody construct alone; or (d) myc-EDC4 in pCMVSport6 and GFP-pcDNA3, or GFP-pcDNA3 alone. Cells were lysed and immunoprecipitations with anti-c-myc agarose beads was performed as described above. For parts B and C, 5% of lysates and 50% of immunoprecipitates were loaded onto 4–20% SDS-PAGE gels; for part D, 2% of lysates and 40% of immunoprecipitates were loaded. For parts b and c, proteins were transferred to PVDF for fluorescence Western blotting; for part d, proteins were transferred to nitrocellulose for chemiluminescent Western blotting as described above. Primary antibodies used were rabbit anti-GFP and rabbit anti-myc.
To refine the NoBody interaction analysis, alanine scanning mutagenesis was performed. HEK293T cells in 6-well plates were transfected with myc-EDC4-pCMVSport6 alone or co-transfected with myc-EDC4-pCMVSport6 and the FLAG-NoBody coding sequence in pcDNA3 as negative and positive controls, respectively. Each additional well was transfected with myc-EDC4-pCMVSport6 in combination with the coding sequence for a single alanine point mutant of FLAG-tagged NoBody from residues 22–41 (except A31, which is already an alanine residue). We note that NoBody numbering corresponds to the endogenous NoBody coding sequence and does not include the FLAG tag. 24 hours after transfection, cells were lysed in 200 µL lysis buffer and anti-FLAG immunoprecipitations were performed as described. 10% of lysate and 50% of immunoprecipitate was loaded on 4–20% gels, with positive and negative controls included on each gel for reference and comparison, and transferred to nitrocellulose membranes for chemiluminescent Western blotting as described above. Primary antibodies used were mouse anti-FLAG M2 and rabbit anti-myc. If samples that were to be compared had to be run on separate gels due to well number limitations, the corresponding membranes were developed and imaged simultaneously.
NoBody purification
For protein expression, the NoBody coding sequence, with a N-terminal glutathione S-transferase (GST) fusion tag, with or without a FLAG epitope tag on the N-terminus of NoBody, was expressed in E. coli BL21 Gold (DE3) competent cells. Cells were lysed by sonication in PBS buffer (10 mM phosphate buffer, 2.7 mM KCl, 137 mM NaCl, pH 7.4) containing 0.1 % Triton X-100, 0.15 mg/mL lysozyme, 60 µg/mL DNAse (Roche) and 1 tablet of Complete mini EDTA-free protease inhibitor (Roche). Sonication was carried out on ice, using 10 second pulses with intervals of 20 seconds for a total pulse time of 2.5 minutes at 45 % amplitude using a Model 120 sonic dismembrator (Fisher Scientific). Broken cells were separated by centrifugation at 20,000 × g and the supernatant containing NoBody was loaded on a glutathione agarose column (Pierce) at 4 °C. Following incubation of lysate with the matrix for 30 minutes, the matrix was washed extensively with PBS buffer. On-column cleavage of the GST tag was done by adding 20U of GST-PreScission protease (GE LifeSciences) to the NoBody-bound GST matrix at 4 °C overnight with rotation. The NoBody peptide was further purified using an Amicon Ultra-15 centrifugal filter unit (EMD Millipore) with a molecular weight cut-off of 30,000 Daltons. The flow-through from this step was concentrated by a filter unit with a cut-off of 3,000 Daltons so that NoBody was retained. FLAG-NoBody, FLAG-NoBody(W34A), and NoBody without a FLAG tag were also expressed and purified in this manner.
NoBody-EDC4 GST pull-down in vitro
To probe complex formation between EDC4 and NoBody in vitro, 0.6 µM FLAG-EDC4 was incubated with 3 µM GST-FLAG-NoBody on ice for 15 minutes in PBS buffer. The reactions were then added separately to microfuge tubes containing 25 µl of settled Glutathione agarose (Thermo Fisher Scientific) that had been washed and equilibrated in cold PBS buffer. To allow for binding of GST to the resin, tubes were incubated at 4 °C on a rotator for 30 minutes and centrifuged at 1000 × g for 1 minute at 4 °C. The resin was washed three times with 500 µl of cold PBS each time and finally with 50 µl of PBS. Tubes were centrifuged at 1000 × g for 1 minute. To elute the proteins, 50 µl of Glutathione containing buffer (10 mM reduced glutathione in 50 mM Tris pH 8.0, 150 mM NaCl) was added to the resin and incubated on a rotator at 4 °C for 30 minutes. The suspensions were transferred to BioSpin columns (BioRad) and centrifuged at 1000 × g at 4 °C for 1 minute to separate the agarose beads from the solution. At each step, the flow through, final wash and elution samples were collected. 5 µl of 5× SDS loading dye was added to 50 µl of samples and 10 µl was loaded per lane on a 4–20 % SDS gel. Following SDS PAGE, western blotting was performed using anti-FLAG antibodies as described in a previous section of this manuscript. In parallel, 10 µl of each sample was loaded on a 10 % SDS gel, followed by silver staining.
Photo-cross-linking in vitro
Purified NoBody was treated with a twofold molar excess of (tris(2-carboxyethyl)phosphine) TCEP-HCl at 4 °C for 1 hour, to ensure disulfide bond reduction followed by treatment with a tenfold molar excess of 4-(maleimido)benzophenone (Santa Cruz Biotechnology) in the dark at 4 °C overnight, on a rotator. The reaction was quenched by adding dithiothreitol (DTT) at a ten-fold molar excess above the cross-linker concentration. Unreacted cross-linker was removed using a Biospin 6 column (BioRad). Photo-cross-linking reactions were carried out in a microtiter plate held on ice. The final concentrations of FLAG-EDC4 and benzophenone-NoBody were 1 µM and 50 µM respectively in 20 mM sodium phosphate, 50 mM NaCl, pH 7.1. For photo-cross-linking, samples were irradiated with a UV lamp (UVP) at 365 nm for 20 mins. Samples were subjected to SDS PAGE followed by western blotting with an antibody to the NoBody peptide or FLAG tag to visualize EDC4.
Photo-cross-linking in cell lysates
HEK293 cells were transiently transfected with EDC4-myc-pCMV-Sport6 using Lipofectamine 2000. Cells were harvested after 20 hours of transfection, lysed in RIPA buffer supplemented with protease inhibitor tablets. Cell debris was spun down at 20,000 × g for 20 min at 4°C. Total cell lysate (both transfected and non-transfected) was diluted to 1 mg/mL in PBS. 300 µL of the lysate was used for each reaction. Purified Rhodamine-NoBody(22–41)-benzophenone (BP) and Biotin-NoBody(22–41)-BP peptides were purchased from Peptide 2.0. Both peptides were dissolved in DMSO to make a 10mM stock. Rhodamine-NoBody(22–41)-BP was used at 25 uM concentration, Biotin-NoBody(22–41)-BP peptides were used at 100 uM to compete for binding. Sample mixtures were incubated for 1 hour by rotating at 4°C, followed by UV crosslinking (Stratalinker 1800, 365 nm) for 60 min while maintaining samples on ice. Reactions were quenched by directly adding 4× SDS loading dye to each sample, and boiled for 5 min. 30 µl of each sample was loaded on a 4–12% Bis-Tris gel and run in MES SDS running buffer at 200 V for 20 min. The gel was imaged using Typhoon Imager 8600: Gain 550, 50 micron, and 532ex/580em. The same gel was stained in InstantBlue to show the total protein loading. The gel was imaged using Odyssey CLx, IR700 gray scale.
Lentivirus production and infection
Lentivirus was produced as previously described45. Briefly, HEK 293T cells were transfected with construct in pFCPGW, along with pVSV-G and pdelta8.91, and growth media replaced 5 hours later. 48 hours post-transfection, media containing viruses was harvested, filtered through 0.45-µm filter, aliquots were made and flash-frozen.
APEX-Dcp2 and APEX-Rck1 imaging
APEX is a peroxidase reporter protein useful for protein imaging and proteomics, and was used as a fusion tag in these experiments. APEX-Dcp2 and Rck1 also bear a FLAG epitope tag fusion. Lentiviral particles were prepared for transduction of FLAG-APEX-Dcp2 and FLAG-APEX-Rck1 transgenes, and added to HEK 293T cells grown to 75% confluency on fibronectin-coated glass coverslips in 48-well plates. Lentivirus was titered from 0.1 µL to 50 µL in each well (total growth media volume, 250 µL) to control expression level, and concentrations affording detectable protein exhibiting the desired localization were selected for imaging. Cells were fixed as described above, then blocked with Rockland fluorescence blocking buffer up to overnight at 4°C. Staining was performed with rabbit anti-EDC4 (to detect endogenous P-bodies) and mouse anti-FLAG (to detect the transgenes) at a 1:1000 dilution in Rockland fluorescence blocking buffer overnight at 4°C, then washed 3× with PBS. Secondary antibodies were goat anti-mouse Alexa Fluor 488 and goat anti-rabbit Alexa Fluor 568. Nuclei were counterstained with DAPI. Imaging was performed as described above.
NoBody over-expression and imaging
HEK293T cells at 75% confluency were transduced with lentiviral particles containing the FLAG-NoBody coding sequence (coding sequence only) in a lentiviral expression vector. For a negative control experiment, identical HEK293T cells were transfected with FLAG-NoBody ∆31–41, a non-interacting mutant. Lentiviral particle concentration was titered as described above. For the batches of virus utilized in these figures, approximately 10 µL of lentiviral particle suspension per well of cells was optimal. 36 hours after infection, cells were fixed and blocked as described above. Cells were stained with 1:1000 dilution of anti-FLAG (to detect NoBody), anti-EDC4, and/or anti-Dcp1A antibody to detect endogenous P-bodies, followed by fluorescent secondary antibodies and DAPI as described above and in the figures. If quantitation was performed, cells were manually counted in the DAPI (nucleus) channel, and P-bodies were counted in the EDC4 and/or Dcp1A channels using Slidebook software for automated image detection. For individual fields of view, the average number of P-bodies per cell was calculated as the total number of P-bodies divided by the number of nuclei detected. Multiple fields of view were then analyzed to determine global averages and standard deviation as described in the figure legend. Analysis of NoBody over-expression and imaging in COS7, HeLa, and BGC-823 was performed identically.
NoBody silencing and imaging
HEK293T cells were grown on glass coverslips in a 48-well plate to 50% confluency, then transfected with a pool of 4 siRNAs (Qiagen) against LOC550643 using Dharmafect I according to the manufacturer’s instructions. Negative controls included a non-targeting siRNA, and a pool of 4 siRNAs targeting GAPDH, an unrelated gene. 48 hours after transfection, cells were fixed as described above, blocked, and immunostained with anti-EDC4 or anti-Dcp1A primary antibodies, fluorescent secondary antibodies, and DAPI as described above. Cells were manually counted in the DAPI (nucleus) channel, and P-bodies were counted in the EDC4 and/or Dcp1A channels using Slidebook software for automated image detection. For individual fields of view, the average number of P-bodies per cell was calculated as the total number of P-bodies divided by the number of nuclei detected. Multiple fields of view were then analyzed to determine global averages and standard deviation as described in the figure legend.
NoBody and GFP-Dcp2 imaging
HEK293T cells were grown to 75% confluency on coverslips in 48-well plates. Cells were lentivirally transfected with GFP-Dcp2 alone or in tandem with FLAG-NoBody coding sequence (lentiviruses were mixed prior to adding to cells) as described above. Virus was titered to achieve GFP-Dcp2 co-localization with endogenous P-bodies. 48 hours after transfection, cells were fixed and stained as described above.
Dcp1A expression levels after NoBody over-expression
For transient transfection, HEK293T cells were grown to 75% confluency in a 6-well plate, then transfected with the FLAG-NoBody coding sequence in pcDNA3, or empty vector as a negative control, using Lipofectamine 2000 and Opti-MEM according to the manufacturer’s instructions. 24 hours post-transfection, cells were harvested and lysed using Tris-buffered saline (TBS) with 1% Triton X-100 and Roche Complete protease inhibitor cocktail tablets. 100 µL lysis buffer was used per pellet. Cells were lysed on ice for 20 min followed by centrifugation at 14000 rpm, 4°C, 15 min. Supernatant was mixed with 50 µL of 3× SDS-PAGE loading buffer, then boiled and 30 µL were loaded on a 4–20% SDS-PAGE gel. For lentiviral transfection, HEK293T cells were grown to 75% confluency and transfected with a concentration of lentivirus utilized for cellular imaging, or mock transfection was used as a negative control. 48 hours after transfection, cells were lysed and lysates were loaded onto the same SDS-PAGE gel as described. Western blotting was performed as described above, using anti-FLAG, anti-Dcp1A, and anti-alpha tubulin primary antibodies and fluorescent secondary antibodies, followed by IR fluorescence detection on an Odyssey imager, all as described above.
Non-translatable NoBody imaging
HEK293T cells were grown to 50% confluency on coverslips in 48-well plates, then transfected with lentiviruses encoding the FLAG-NoBody coding sequence, or the FLAG-NoBody coding sequence lacking its start codon. 48 hours after transfection, cells were fixed, immunostained, and imaged. Cells and P-bodies were counted and quantified as described above.
EDC4 immunopurification
FLAG-tagged EDC4 in pCMVSport6 was transfected into HEK293T cells using 10 µg DNA per T25 flask of cells. 10 flasks were used for each purification. 24 hours post-transfection, cells were harvested and lysed using tris-buffered saline (TBS) with 1% Triton X-100 (TBS-T), Roche Complete protease inhibitor cocktail tablets, and a phosphatase inhibitor tablet PhosSTOP (Sigma). 400 µL lysis buffer was used per pellet. Cells were lysed on ice for 20 min followed by centrifugation at 18000 × g at 4°C, 15 min. Lysate samples were saved for analysis of loading. A 50 µL aliquot of anti-FLAG agarose beads (clone M2, Sigma) was washed with 1 mL TBS-T, collected by centrifugation for 1 min at 1000 × g, then suspended in the cell lysate supernatant. Bead suspensions were rotated at 4°C for 2 hours, then washed 3 times with TBS-T, followed by extensive TBS washes. Elution was in 50 µL of 3× FLAG peptide (Sigma), at a final concentration of 1 mg/mL in TBS at 4°C for 1 hour. Beads were removed by centrifugation, excess FLAG peptide was removed using a Bio-Spin 6 column (BioRad), previously equilibrated in TBS. The protein concentration of FLAG-EDC4 eluted from the spin column was determined by measuring the absorbance at 280 nm.
Cellular nonsense-mediated decay
Calu-6 cells, which contain a premature termination codon mutation in the p53 gene, were grown to 60–80% confluency in 6-well plates. For small molecule-mediated NMD silencing, cells were treated with water as vehicle control for caffeine, or DMSO as vehicle control for wortmannin and cycloheximide. Experimental cells were treated with 10 mM caffeine in water, 100 µg/mL cycloheximide, 10 µM wortmannin) in growth media for 4 hours in the 37C incubator. After incubation, cells were washed 3 times with DPBS and RNA was isolated for analysis. For this control experiment, two biological replicates were prepared, and triplicate technical replicates were performed for each.
For gene silencing, cells were transfected with Qiagen Flexitube Gene Solution siRNAs using Dharmafect I (GE Healthcare) according to the manufacturer’s instructions. For overexpression, cells were lentivirally transfected with FLAG-NoBody-pFCPGW which encodes only the FLAG-tagged NoBody coding sequence. 48 hours after transfection, cells were washed 3 times with DPBS and RNA was isolated for analysis. Three biological replicates were prepared and analyzed for each condition in this experiment.
For overexpression, 50% confluent Calu-6 cells were transfected with a 1:10 dilution of FLAG-NoBody-pFCPGW lentiviral particle-containing media in the growth media. 48 hours after transfection, cells were washed 3 times with DPBS and RNA was isolated for analysis. Six biological replicates were prepared and analyzed for each condition in this experiment.
Total RNA was isolated using the Qiagen RNeasy kit and genomic DNA was removed via DNase I digest. Reverse transcription was performed with the iScript cDNA synthesis kit (BioRad), and qPCR with gene-specific primers (Qiagen; see Supplementary Table 2) was performed with iTaq Universal SYBR Green Supermix (BioRad) on a BioRad CFX96 qPCR System. Data were analyzed with a delta-delta Cq method with reference to beta-actin for normalization, and Student’s t-test (two-tailed) was applied to assess significance. Data represent mean and standard deviation.
Full-size Western blots are shown in Supplementary Note.
Supplementary Material
Acknowledgments
We thank Professor Alice Ting for the generous use of her Zeiss AxioObserver inverted confocal microscope. This study was supported by a George E. Hewitt Foundation for medical research Postdoctoral Fellowship (Q.C.), NIH (R01 GM102491, A.S), the NCI Cancer Center Support Grant P30 (CA014195 MASS core, A.S.), The Leona M. and Harry B. Helmsley Charitable Trust grant (#2012-PG-MED002, A.S.), and Dr. Frederick Paulsen Chair/Ferring Pharmaceuticals (A.S.). S. A. S. was supported by Yale University West Campus start-up funds, an American Cancer Society Institutional Research Grant Individual Award for New Investigators from the Yale Cancer Center (IRG-58-012-57) and a NIH Ruth L. Kirchstein postdoctoral fellowship (1F32GM099408). N.G.D. was supported by an Anderson Endowed Postdoctoral Fellowship in the Biological Sciences.
Footnotes
Author contributions. S.A.S. and A.S. conceived the project. N.G.D., J.M., L.W., Q.C., K.H.L, S.A.S. and A.S. designed the experiments. S.A.S. conducted the majority of the experiments and data analyses, including the identification of NoBody, the discovery of NoBody-binding partners, cellular imaging, and NMD studies. N.G.D. performed NoBody purification and in vitro EDC4 co-purification and cross-linking. J.M. generated NoBody expression constructs, and J.M. and Q.C. carried out the cellular EDC4-NoBody crosslinking experiments. L.W. assisted with identification of NoBody binding partners and co-immunoprecipitation experiments. K.H.L. carried out cellular imaging. E.O.C. and J.L.A. provided advice and experimental help with NMD studies. B.A.B. provided proteomics assistance. N.G.D., A.S. and S.A.S. wrote the manuscript with input from all authors.
Competing financial interests. The authors declare no competing financial interest.
References
- 1.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slavoff SA, et al. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nature chemical biology. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanderperre B, et al. Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PloS one. 2013;8:e70698. doi: 10.1371/journal.pone.0070698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saghatelian A, Couso JP. Discovery and characterization of smORF-encoded bioactive polypeptides. Nature chemical biology. 2015;11:909–916. doi: 10.1038/nchembio.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Storz G, Wolf YI, Ramamurthi KS. Small proteins can no longer be ignored. Annual review of biochemistry. 2014;83:753–777. doi: 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magny EG, et al. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013;341:1116–1120. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- 8.Anderson DM, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee C, et al. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 2015;21:443–454. doi: 10.1016/j.cmet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slavoff SA, Heo J, Budnik BA, Hanakahi LA, Saghatelian A. A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining. The Journal of biological chemistry. 2014;289:10950–10957. doi: 10.1074/jbc.C113.533968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvunis AR, et al. Proto-genes and de novo gene birth. Nature. 2012;487:370–374. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fillman C, Lykke-Andersen J. RNA decapping inside and outside of processing bodies. Current opinion in cell biology. 2005;17:326–331. doi: 10.1016/j.ceb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coller J, Parker R. Eukaryotic mRNA decapping. Annual review of biochemistry. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Molecular cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 18.dos Santos G, et al. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic acids research. 2015;43:D690–D697. doi: 10.1093/nar/gku1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howe K, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kent WJ. BLAT--the BLAST-like alignment tool. Genome research. 2002;12:656–664. doi: 10.1101/gr.229202. Article published online before March 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent WJ, et al. The human genome browser at UCSC. Genome research. 2002;12:996–1006. doi: 10.1101/gr.229102. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome biology. 2006;7(Suppl 1):S12, 11–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng X, et al. Tissue-specific transcriptome sequencing analysis expands the non-human primate reference transcriptome resource (NHPRTR) Nucleic acids research. 2015;43:D737–D742. doi: 10.1093/nar/gku1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellacheruvu D, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nature methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Molecular cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 26.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nature reviews. Molecular cell biology. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 27.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Molecular and cellular biology. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aizer A, et al. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Molecular biology of the cell. 2008;19:4154–4166. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Current opinion in cell biology. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Popp MW, Maquat LE. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet. 2013;47:139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Molecular cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Song M, Kiledjian M. Differential utilization of decapping enzymes in mammalian mRNA decay pathways. RNA. 2011;17:419–428. doi: 10.1261/rna.2439811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Molecular and cellular biology. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehman TA, et al. p53 mutations, ras mutations, and p53-heat shock 70 protein complexes in human lung carcinoma cell lines. Cancer Res. 1991;51:4090–4096. [PubMed] [Google Scholar]
- 35.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes & development. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin L, et al. Identification and characterization of small molecules that inhibit nonsense-mediated RNA decay and suppress nonsense p53 mutations. Cancer Res. 2014;74:3104–3113. doi: 10.1158/0008-5472.CAN-13-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nickless A, et al. Intracellular calcium regulates nonsense-mediated mRNA decay. Nat Med. 2014;20:961–966. doi: 10.1038/nm.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brumbaugh KM, et al. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Molecular cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Keeling KM, et al. Attenuation of nonsense-mediated mRNA decay enhances in vivo nonsense suppression. PloS one. 2013;8:e60478. doi: 10.1371/journal.pone.0060478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Molecular cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braun JE, et al. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5' exonucleolytic degradation. Nature structural & molecular biology. 2012;19:1324–1331. doi: 10.1038/nsmb.2413. [DOI] [PubMed] [Google Scholar]
- 42.Ma J, et al. The Discovery of Human sORF-Encoded Polypeptides (SEPs) in Cell Lines and Tissue. Journal of proteome research. 2014 doi: 10.1021/pr401280w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson M, et al. NCBI BLAST: a better web interface. Nucleic acids research. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karolchik D, et al. The UCSC Genome Browser database: 2014 update. Nucleic acids research. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nature protocols. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.