Abstract
Oxidatively damaged proteins accumulate with age in almost all cell types and tissues. The activity of chaperone-mediated autophagy (CMA), a selective pathway for the degradation of cytosolic proteins in lysosomes, decreases with age. We have analyzed the possible participation of CMA in the removal of oxidized proteins in rat liver and cultured mouse fibroblasts. Added to the fact that CMA substrates, when oxidized, are more efficiently internalized into lysosomes, we have found a constitutive activation of CMA during oxidative stress. Oxidation-induced activation of CMA correlates with higher levels of several components of the lysosomal translocation complex, but in particular of the lumenal chaperone, required for substrate uptake, and of the lysosomal membrane protein (lamp) type 2a, previously identified as a receptor for this pathway. In contrast with the well characterized mechanism of CMA activation during nutritional stress, which does not require de novo synthesis of the receptor, oxidation-induced activation of CMA is attained through transcriptional up-regulation of lamp2a. We conclude that CMA is activated during oxidative stress and that the higher activity of this pathway under these conditions, along with the higher susceptibility of the oxidized proteins to be taken up by lysosomes, both contribute to the efficient removal of oxidized proteins.
INTRODUCTION
Accumulation of oxidized protein is a common feature of aged cells (Dunlop et al., 2002; Grune and Davies, 1997; Grune et al., 2001, 2002b; Stadtman, 2001). Many physiological and pathological processes lead to the generation of free radicals and consequent damage of intracellular components, including proteins. In most of these oxidative events, damaged proteins are removed from the cell through degradation by proteases (Dunlop et al., 2002). The activity of different intracellular proteolytic systems decreases with age (Terman, 1995; Cuervo and Dice, 1998a; Friguet et al., 2000; Carrard et al., 2002; Donati et al., 2001; Merker et al., 2001; Friguet, 2002; Keller et al., 2002; Ward, 2002), and this impaired activity is considered responsible for the deficient removal of oxidized proteins in old organisms (Grune, 2000; Merker and Grune, 2000; Dunlop et al., 2002; Szweda et al., 2002).
The susceptibility of oxidized proteins to proteases changes with the duration and degree of oxidative damage (Dunlop et al., 2002; Mehlhase and Grune, 2002). Thus, mild oxidation induces partial protein unfolding and facilitates proteolytic cleavage (Grune et al., 1995, 1997). However, persistent or extensive oxidative damage usually promotes protein aggregation, due to the exposure of patches of hydrophobic amino acids. Once a protein aggregates, it becomes less susceptible to proteolytic cleavage (Hoff et al., 1993; Grune et al., 1997; Davies, 2001; Demasi and Davies, 2003). Kinetics of degradation of oxidized proteins in vitro have been analyzed using different types of proteases (Merker and Grune, 2000; Dunlop et al., 2002; Szweda et al., 2002). One of the most extensively analyzed protease in this respect has been the proteasome. Although most of the studies linking the proteasome to the degradation of oxidized proteins have been carried out in vitro (Rivett, 1985; Davies, 2001), recent evidence supports the participation of the proteasome in the removal of oxidized proteins in vivo also (Hosler et al., 2003; Shringarpure et al., 2003). Thus, treatment of culture cells with proteasome inhibitors or antisense oligonucleotides against essential proteasome subunits results in accumulation of oxidized proteins inside cells and diminished rates of cell survival during oxidative stress. The degradation of most oxidized proteins by the proteasome is ATP and ubiquitin independent (Shringarpure et al., 2003), suggesting that the exposure of hydrophobic regions in the surface of the protein acts as recognition signal for this protease.
The lysosomal system, the other major proteolytic system in cells, has not been often considered as a possible candidate for the removal of oxidized proteins because of its lack of selectivity. In mammalian cells three different main mechanisms contribute to the degradation of intracellular components inside lysosomes (autophagy) (Dice, 2000; Reggiori and Klionsky, 2002; Wang and Klionsky, 2003; Cuervo, 2004a,b). Two of those mechanisms, macroautophagy and microautophagy, result in the degradation of complete regions of the cytosol, including organelles, in the lysosomal lumen. Although degradation of organelles by these autophagic pathways can be selective (i.e., specific removal of damaged mitochondria sparing normal functioning ones) (Lemasters et al., 2002), there is no evidence supporting selectivity for the degradation of soluble cytosolic proteins. In contrast, the main characteristic of a third form of autophagy, chaperone-mediated autophagy (CMA), is its selectivity regarding the substrates (cytosolic proteins) degraded through this pathway (Cuervo and Dice, 1998b; Dice, 2000; Dice et al., 2003).
CMA is a generalized form of autophagy present in most cell types and tissues (Massey et al., 2004). All the CMA substrate proteins described to date are soluble cytosolic proteins containing a targeting motif biochemically related to the pentapeptide KFERQ (Chiang and Dice, 1988; Dice et al., 2003). This motif, present in ∼30% of the proteins in the cytosol, is recognized by a cytosolic chaperone, the heat shock cognate protein of 73 kDa (cyt-hsc70) (Chiang and Dice, 1988; Chiang et al., 1989). The interaction with this chaperone, modulated by the hsc70 cochaperones (Agarraberes and Dice, 2001), targets the substrate to the lysosomal membrane, where it interacts with the lysosomal membrane protein (lamp) type 2a (Cuervo and Dice, 1996). Substrates need to be unfolded before translocation into the lysosomal lumen, and several cytosolic chaperones associated to the lysosomal membrane have been proposed to assist in the unfolding (Agarraberes and Dice, 2001). Translocation of the substrate requires the presence of a variant of hsc70, lyshsc70, in the lysosomal lumen (Agarraberes et al., 1997; Cuervo et al., 1997) and is followed by the rapid proteolysis of the substrate by resident lysosomal proteases (half-life in the lysosomal lumen of 5–10 min).
Although some basal level of CMA activity is probably present in most cells, nutritional stress has been shown to maximally activate this pathway (Wing et al., 1991; Cuervo et al., 1995). Activation during nutrient deprivation is associated with higher levels of lys-hsc70 in the lysosomal lumen and of lamp2a at the lysosomal membrane (Cuervo et al., 1995; Agarraberes et al., 1997; Cuervo and Dice, 2000b). Because the interaction of substrate proteins with lamp2a is a limiting step for this pathway, changes in levels of lamp2a at the lysosomal membrane modulate CMA activity (Cuervo and Dice, 2000b,c). Interestingly, all the conditions known to activate CMA, up-regulation of the lysosomal levels of lamp2a does not require de novo synthesis of the protein, but instead it is attained through down-regulation of its degradation and by relocation of a fraction of the protein from the lysosomal lumen into the membrane (Cuervo and Dice, 2000b).
In addition to starvation, activation of CMA has also been observed in rat liver and kidney after exposure to gasoline derivatives (Cuervo et al., 1999); in fibroblasts from galactosialidosis patients, which lack the protective protein/cathepsin A (Cuervo et al., 2003); and in cells overexpressing lamp2a (Cuervo and Dice, 1996). CMA activity is reduced during renal tubular cell growth (Franch et al., 2001) and in aging (Cuervo and Dice, 2000a). The decrease in CMA activity in old cells, known to accumulate oxidized proteins (Cuervo and Dice, 2000a), combined with the fact that activation of CMA during toxic exposure results in the selective degradation of a protein altered by the toxic compound (Cuervo et al., 1999), and with our finding that in the presence of antioxidants degradation of the inhibitor of the nuclear factor κB by CMA decreases (Cuervo et al., 1998), prompted us to analyze a possible role of CMA in the removal of oxidized proteins from cells. We show in this work that oxidation of CMA substrates facilitates their translocation into lysosomes for degradation via CMA and also that CMA itself is activated during oxidative stress. Unexpectedly, the oxidative stress-mediated activation of CMA is attained through a novel mechanism, different from the previous well-characterized activation of CMA in response to nutritional stress.
MATERIALS AND METHODS
Animals and Cells
Male Wistar rats (200–250 g) were fed ad libitum or starved for 20 or 48 h before sacrifice. An age-controlled rat strain (Fisher 344) was used for the study of age-related changes and 3-, 12-, and 22-mo-old rats were compared. Mild oxidative stress was induced in rats with two single i.p. injections of paraquat (40 mg/100 g body weight) separated by 24 h. Lysosomal isolation was carried out 24 h after the last injection. Mouse fibroblasts (NIH3T3) were from the American Type Culture Collection (Manassas, VA). Cells were maintained in DMEM (Sigma-Aldrich, St. Louis, MO) in the presence of 10% newborn calf serum. To deprive cells from serum, plates were extensively washed with Hanks' balanced salt solution (Invitrogen, Carlsbad, CA), and fresh medium without serum was added. Mild oxidative stress was induced in culture cells by supplementation of the culture medium with 100 μM H2O2 or 40 μM paraquat for 1 h. Assays were carried out 12–24 h after removing the oxidizing compound.
Chemicals
Sources of chemicals and antibodies were as described previously (Cuervo and Dice, 1996; Cuervo et al., 1997; Cuervo and Dice, 2000a; Martin et al., 2002). The antibodies against the cytosolic tail of rat and mouse lamp2a and lamp2c were prepared in our laboratory (Cuervo and Dice, 1996; Zhang, Bandhyopadhyay, Kiffin, Massey, and Cuervo, unpublished data). The antibody against mouse lamp1 (1D4B) was from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA), and antibodies against hsp90 (SPA-845), hsp72 (SPA-810), hsp40 (SPA-400), hip (SPA-766), and Bag-1 (AAM-400E) were from Stressgene Biotechnologies (Victoria, BC, Canada). Carbonyl groups in oxidized proteins were detected using the OxyBlot oxidized protein detection kit from Chemicon International (Temecula, CA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ribonuclease A (RNase A) were radiolabeled with [14C]formaldehyde by reductive methylation as described previously (Jentoft and Dearborn, 1983). Cytosolic proteins from mouse fibroblasts in culture were metabolically radiolabeled by incubation with [14C]leucine (2 μCi/ml) at 37°C for 2 d (Cuervo et al., 1997). Cells were disrupted by nitrogen cavitation, and the cytosolic fraction was the supernatant of a centrifugation at 100,000 × g for 1 h at 4°C. Oxidized radiolabeled cytosolic proteins were prepared by the same procedure from cells treated with different prooxidants for 24 h before isolation.
Isolation of Lysosomal Fractions
Rat liver lysosomes were isolated from a light mitochondrial-lysosomal fraction in a discontinuous metrizamide density gradient (Wattiaux et al., 1978). Lysosomal fractions with different activities for CMA were separated as described previously (Cuervo et al., 1997). Lysosomes from cultured cells were isolated as described previously (Storrie and Madden, 1990). Preparations with more than 10% broken lysosomes, measured as β-hexosaminidase latency, were discarded. Lysosomal matrices and membranes were isolated after hypotonic shock (Ohsumi et al., 1983).
Uptake and Degradation of Proteins by Isolated Lysosomes
GAPDH or RNase A were incubated in MOPS buffer (10 mM 3-(N-morpholino)propanesulfonic acid [MOPS] pH 7.3, 0.3 M sucrose) with chymostatin-treated lysosomes as described previously (Cuervo et al., 1997). Transport was measured after proteinase K treatment of the samples, SDS-PAGE, and immunoblot as the amount of GAPDH associated to lysosomes resistant to the protease. In another group of experiments, to overcome any possible changes in the proteolytic susceptibility of the oxidized proteins to proteinase K, substrates were incubated with chymostatin-treated or untreated lysosomes, and uptake was calculated as the difference between the amount of substrate associated to lysosomes (chymostatin-treated lysosomes) and the amount of substrate bound to their membrane (untreated lysosomes) (Salvador et al., 2000). Degradation of [14C]GAPDH (260 nM; 1.5 × 108 dpm/nmol), [14C]RNase A (150 nM; 0.9 × 108 dpm/nmol), or a pool of radiolabeled cytosolic proteins (500 dpm/μg) by intact lysosomes or lysosomal matrices was measured as described previously (Terlecky and Dice, 1993; Cuervo et al., 1997). After incubation for 30 min at 37°C in MOPS/dithiothreitol (DTT) buffer (10 mM MOPS pH 7.3, 0.3 M sucrose, 5.4 μM cystein, and 1 mM DTT) (for intact lysosomes) or in 0.03 M sucrose (for lysosomal matrices), samples were precipitated in 10% trichloroacetic acid and filtered through a multiscreen assay system (Millipore, Bedford, MA) using a 0.45 μm pore filter. Radioactivity in the flow through and in the filter was converted to disintegrations per minute in a WinSpectral 1414 liquid scintillation analyzer (PerkinElmer Life and Analytical Sciences, Boston, MA) by correcting for quenching using an external standard. Proteolysis was expressed as the percentage of the initial acid-insoluble radioactivity (protein) transformed into acid-soluble radioactivity (amino acids and small peptides) at the end of the incubation.
Rates of degradation of lamp2a in the isolated membranes were followed by immunoblot with a specific antibody against the cytosolic tail of lamp2a as described previously (Cuervo and Dice, 2000b). Briefly, isolated lysosomal membranes were incubated in MOPS buffer at 37°C, and at different times aliquots were removed and subjected to SDS-PAGE and immunoblot for lamp2a.
GAPDH Oxidation
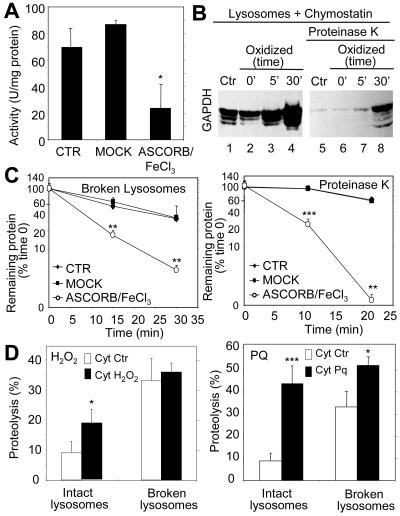
Metal-catalyzed oxidation of rabbit muscle d-glyceraldehyde-3-phosphate dehydrogenase EC 1.2.1.12 (Roche Diagnostics, Indianapolis, IN) was carried out as described previously (Rivett and Levine, 1990). GAPDH (10 mg of protein) was dialyzed against 50 volumes of HEPES buffer (50 mM HEPES, pH 7.2, 100 mM KCl, 10 mM MgCl2) supplemented or not (Mock) with 25 mM ascorbic acid and 0.1 mM FeCl3 at 37°C. The oxidative reaction was stopped adding 1 mM EDTA, and the samples were dialyzed against HEPES buffer for 24 h with four changes. The two first changes were supplemented with 1 mM EDTA. The efficiency of oxidation was monitored by measuring the enzymatic activity of GAPDH after being exposed to the oxidant mixture for different periods of time. As described in Figure 2, incubation of GAPDH with the oxidant mixture for 30 min resulted in 70% decrease of its enzymatic activity.
Figure 2.
Oxidation of CMA substrates facilitates their uptake by lysosomes. (A) GAPDH was exposed to an ascorbic acid/iron oxidizing reaction for 30 min as described under Materials and Methods. The remaining specific enzymatic activity of untreated GAPDH (CTR) and of GAPDH exposed to the oxidant mixture (ASCORB/FeCl3) or to the dialysis buffer only (MOCK) is shown. Values are mean ± SE of two different preparations with triplicate measurements. (B) Unmodified GAPDH (Ctr) or GAPDH exposed to the oxidant mixture for the indicated periods of time was incubated for 30 min under standard conditions with intact chymostatin-treated liver lysosomes isolated from 20-h starved rats. Where indicated, samples were treated with proteinase K to remove GAPDH bound to the cytosolic side of the lysosomal membrane. At the end of the incubation, lysosomes were collected by centrifugation and subjected to SDS-PAGE and immunoblot for GAPDH. (C) The different forms of GAPDH (50 μg) described in A were incubated with lysosomal enzymes (25 μg of protein of broken lysosomes; pH 4.5) (left) or proteinase K (0.1 μg) (right) at 37 or 0°C, respectively. At the indicated times aliquots were taken and subjected to SDS-PAGE and Coomassie Blue staining. Values are the mean + SE of the densitometric values for GAPDH from three different experiments. (D) Mouse fibroblasts were metabolically labeled with [3H]leucine for 48 h. A group of fibroblasts was exposed to 100 μM H2O2 (left) or 40 μM paraquat (Pq) for 1 h during the labeling. After cavitation, cytosolic fractions (Cyt) from untreated (Ctr) and H2O2 or paraquat-treated fibroblasts were prepared and incubated for 30 min at 37°C with intact rat liver lysosomes (25 μg of protein) or with lysosomes disrupted by a hypotonic shock (broken lysosomes; 15 μg of protein) under standard conditions. At the end of the incubation proteolysis was calculated as the amount of acid precipitable radioactivity (protein) transformed in acid soluble (amino acids and small peptides). Values are the mean + SE of six different experiments (*p < 0.05, **p < 0.01, ***p < 0.001).
Immunocytochemical Staining
Immunofluorescence studies of cultured cells were performed following conventional procedures (Cuervo and Dice, 2000c). Cells grown on coverslips until confluent and kept in the presence or absence of serum for 20 h were fixed with a 3% formaldehyde solution, blocked, and then incubated with the primary and corresponding fluorescein isothiocyanate or Cy5-conjugated secondary antibodies as described previously (Cuervo and Dice, 2000c). Mounting medium contained 4,6-diamidino-2-phenylindole staining to highlight the cellular nucleus. Images were acquired with an Axiovert 200 fluorescence microscope (Carl Zeiss, Thornwood, NY) and subjected to deconvolution with the manufacturer's software. Colocalization was determined using MetaMorph (Universal Imaging, Downingtown, PA). All digital microscopic images were prepared using Adobe Photoshop 6.0 software (Adobe Systems, Mountain View, CA).
mRNA Quantification
Total RNA was extracted from rat livers using the RNeasy protect mini kit (QIAGEN, Valencia, CA) following the manufacturer's indications and stored at -80°C until use. The first strand cDNA was synthesized from 1 μg of the total RNA with the SuperScript II RNase H Reverse Transcriptase (Invitrogen) and oligo-(dT)12–18 primer. A region of the exon 5 of lamp2a and of actin were amplified with specific primers (lamp2a, 5′-GTCTCAAGCGCCATCATACT-3′ [forward] and 5′-TCCAAGGAGTCTGTCTTAAGTAGC-3′ [reverse]; actin, 5′-AAGGACTCCTATAGTGGGTGACGA-3′ [forward] and 5′-ATCTTCTCCATGTCGTCCCAGTTG-3′ [reverse]) by using the SYBR green polymerase chain reaction (PCR) kit (PE Biosystems, Warrington, United Kingdom). Amplification of the lamp2a and actin DNA products (120 and 108 bp, respectively) was measured in real time in a light cycler (SmartCycler; Cepheid, Sunnyvale, CA). For both genes, the presence of a single amplified product was verified by agarose gel electrophoresis, and by analysis of the melting curves of the reverse transcription-PCR reaction. The expression levels of lamp2a in different samples were normalized with respect of those of actin in the same samples. Differences between samples were calculated based in their differences of the cycle numbers to reach a certain fluorescence intensity level. Because the size of the amplified fragments was very similar we did not need to correct for fragment length. Standard PCR was carried out with the same primers but increasing dilutions of the cDNA pool. PCR products were subjected to electrophoresis in 1.2% agarose gels and densitometry. The densitometric intensity of the lamp2a products was normalized with respect to the intensity of the actin products for the same sample.
General Methods
Protein concentration was determined by the Lowry method (Lowry et al., 1951) by using bovine serum albumin as a standard. Lysosomal enzymatic activities were measured as reported previously (Storrie and Madden, 1990). After SDS-PAGE (Laemmli, 1970) and immunoblotting (Towbin et al., 1979), the proteins recognized by the specific antibodies were visualized by chemiluminescence methods (Renaissance; PerkinElmer Life and Analytical Sciences). Oxidized proteins were visualized after derivatization with DNPH by immunoblot with an antibody against the DNP moiety following the manufacturer's recommendation. Densitometric quantification of the immunoblotted membranes and stained gels was done with an Image Analyzer System (S1800; Inotech, Sunnyvale, CA). Student's t test was used for statistical analyses.
RESULTS
Oxidized Proteins in Lysosomes
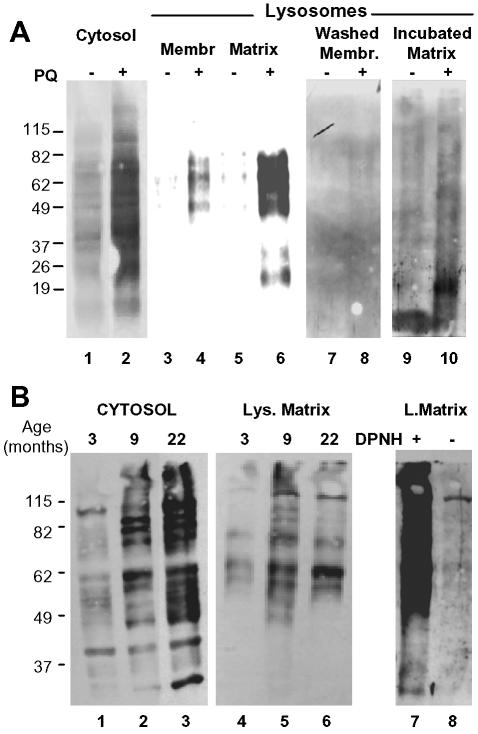
To test the contribution that the lysosomal system, and in particular CMA, might play in the removal of oxidized cytosolic proteins, we first analyzed the presence of those proteins inside lysosomes after oxidative stress. We induced mild oxidative stress in rats by treatment with bleomycin, 1,1′-dimethyl-4,4′-bipyridinium dichloride (paraquat), a superoxide-generating drug with well-characterized prooxidant effects (Haly, 1979). To verify the oxidative effect of paraquat in rat liver we compared the content of carbonyl-containing proteins in the cytosol of untreated and paraquat-treated rats (Figure 1A, lanes 1 and 2). Although some oxidized proteins can be observed in the cytosol of untreated rats, we found a significant increase in the content of these modified proteins in the cytosol of rats injected with a sublethal dose of paraquat for two consecutive days. Lysosomes active for CMA can be isolated from rat liver based on their high content of the lumenal chaperone lys-hsc70 required for substrate uptake (Cuervo et al., 1997). When we isolated this particular lysosomal population from treated and untreated rats and separated their membranes from their lumenal content, we found carbonyl-containing proteins in both the membrane and lumenal fraction of the lysosomes from paraquat-treated rats (Figure 1A, compare lanes lanes 3 and 5 to lanes 4 and 6). Washing the lysosomal membranes with 1 M NaCl released most of the oxidized proteins (Figure 1A, lanes 7 and 8), suggesting that they were likely cytosolic proteins associated to the membrane, rather than the result of oxidation of integral lysosomal membrane proteins. Likewise, when we incubated the lysosomal matrices at 37°C before electrophoresis to promote the degradation of nonlysosomal proteins in the lysosomal lumen, the level of oxidized proteins in this fraction decreased drastically (Figure 1A, lanes 9 and 10). Note that the new lower molecular weight bands detected in the matrices of paraquat-treated animals after the incubation at 37°C could be proteolytic products of the proteins detected in the freshly isolated matrices. These results support that the oxidized proteins detected inside the lysosomes were most likely nonlysosomal proteins delivered there to be degraded. Interestingly, despite of their cytosolic origin, the electrophoretic pattern of the oxidized proteins associated with the lysosomal fraction differs from the ones in the cytosol. Because these differences are also observed in the membrane associated proteins, we do not think that they are due to distinct susceptibility of some of the oxidized proteins to proteases; instead we consider it an indication that a subset of oxidized proteins is selectively taken up by lysosomes.
Figure 1.
Oxidized proteins in lysosomes active for CMA. (A) Cytosol (20 μg of protein) and lysosomal membranes and matrices (10 μg of protein) separated after hypotonic shock from lysosomes enriched in hsc70 isolated from livers of untreated or paraquat (PQ)-treated rats were derivatized and subjected to SDS-PAGE and immunoblot against 2,4-dinitrophenylhydrazine (DPNH) moieties. To show the protein pattern in all lanes, exposure of lanes 1 and 2 was 5 times shorter than for the other lanes. Lanes 7 and 8 are duplicate samples of lanes 3 and 4, in which membranes were washed with 1 M NaCl before derivatization. Lanes 9 and 10 are duplicate samples of lanes 5 and 6 in which the matrices were incubated for 20 min at 37°C before derivatization. No bands were detected in the same samples when derivatization was omitted (our unpublished data). (B) Cytosol (40 μg of protein) and lysosomal matrices (20 μg of protein) were isolated from livers of 3-, 9- and 22-mo-old rats. Content of oxidized proteins in these fractions was analyzed as in A. Exposure of lanes 1–3 was 3 times shorter than for lanes 4–6. Lanes 7 and 8 show the same sample than lane 5 derivatized or not with DPNH and immunobloted as the others. The film is overexposed (8 times lane 5 exposure) to show the low content of protein bands nonspecifically recognized by the antibody.
As reported previously, even in the absence of treatment with a prooxidant agent, oxidized proteins can be detected in normal liver and their content increases with age (Figure 1B). When we analyzed the association of oxidized proteins to lysosomes from livers of 3- and 9-mo-old rats, we found an increase in the content of oxidized proteins in the lysosomal lumen with age, similar to the one observed in the cytosol (Figure 1B, compare lanes 1 and 2 and 4 and 5). In contrast, in the oldest animals (22 mo old), for which we have previously described a severe decrease in CMA activity (Cuervo and Dice, 2000a), despite the higher content of oxidized proteins in the cytosolic fraction, in proportion, the amount of oxidized proteins detected in lysosomes was lower than in the 9-mo-old animals (Figure 1B, compare lanes 5 and 6). Similar assays to the ones described above confirmed that the oxidized proteins were of extralysosomal origin and could be degraded by the lysosomal proteases (our unpublished data). The last panel of Figure 1B shows the specificity of the antibody used in these studies against the carbonyl groups, which in nonderivatized samples only weakly cross-reacted with a single protein (compare signal in derivatized (lane 7) and nonderivatized samples (lane 8). Although macroautophagy, a nonselective form of autophagy, also decreases with age (Terman, 1995; Donati et al., 2001), the fact that the lysosomal population analyzed has been shown to be very active for CMA and that only a subset of oxidized proteins was detected in lysosomes encouraged us to analyze the role of CMA in the removal of oxidized proteins.
Oxidation of CMA Substrates
We first analyzed whether the oxidation of known CMA substrates facilitated their degradation by CMA. We oxidized GAPDH, a well-characterized CMA substrate (Aniento et al., 1993), by incubating this protein with an ascorbic acid/iron oxidizing mixture. After 30 min of incubation with this mixture, ∼70% of GAPDH activity was lost (Figure 2A), and carbonyl groups were easily detectable in the purified protein (our unpublished data). We have previously optimized an in vitro system that allows the analysis of binding and uptake of CMA substrates by isolated lysosomes (Aniento et al., 1993; Cuervo et al., 1997, 1999). When we compared the association to lysosomes of untreated GAPDH or of GAPDH exposed for increasing periods of time to the oxidizing mixture, we found an increase in both the amount of protein bound to the lysosomal membrane and translocated into the lysosomal lumen (proteinase K resistant) with longer oxidation times of GAPDH (Figure 2B). Oxidation of ovalbumin, a protein that does not contain a KFERQ-like motif and consequently is not a CMA substrate, did not increase its association to lysosomes (our unpublished data). The higher amount of GAPDH detected in the lysosomal lumen could also result from an increased resistance of the oxidized protein to degradation, either by the lysosomal proteases or by proteinase K, which removes the protein associated to the cytosolic side of the lysosomal membrane. However, as shown in Figure 2C, the oxidized protein was even more readily degraded by both the pool of proteases located in the lysosomal lumen and by exogenously added proteinase K.
An increase in lysosomal translocation of oxidized proteins was also evident when instead of using a specific CMA substrate, we compared the degradation of a pool of oxidized cytosolic proteins, in which ∼30% of them are possible CMA substrates (contain KFERQ-related motifs) (Figure 2D). Radiolabeled cytosolic proteins isolated from fibroblasts exposed to H2O2 (Figure 2D, left) or paraquat (Figure 2D, right) were degraded faster than cytosolic proteins from untreated fibroblasts when incubated with intact rat liver lysosomes. Because these differences were greatly reduced when the lysosomal membranes were disrupted before the incubation with the cytosolic proteins, giving free access of the proteases to the substrates, we conclude that, as shown for GAPDH, increased degradation of the oxidized proteins resulted from their higher rates of translocation via CMA into lysosomes. These results support that oxidation of proteins that are substrates for CMA facilitates their degradation through this pathway.
Activation of CMA during Oxidative Stress
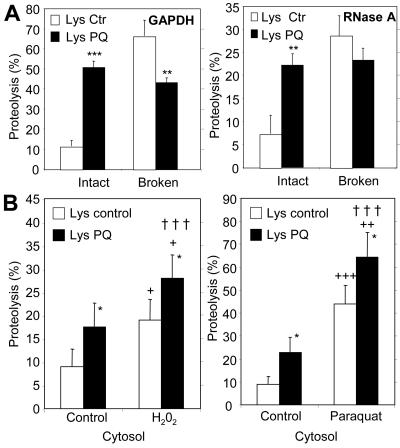
Independently of the effect of oxidation on CMA substrates, which makes them more susceptible to degradation by this pathway, we found a constitutive activation of CMA during oxidative stress. We isolated lysosomes from rats treated with a sublethal dose of paraquat for two consecutive days to generate a mild oxidative stress response. Under these conditions, we did not find significant differences with control in lysosomal membrane stability assessed by measuring β-hexosaminidase latency (Supplemental Figure 1) (Storrie and Madden, 1990) and extralysosomal proteolytic activity (Aniento et al., 1993). Lysosomes isolated from rats exposed to paraquat degraded larger amounts of unmodified GAPDH than lysosomes isolated from untreated animals (Figure 3A, left). Similar results were observed for RNase A, another well-characterized CMA substrate (Terlecky and Dice, 1993; Cuervo et al., 1994) (Figure 3A, right). In both cases, the increase in substrate degradation was no longer observed if the lysosomal membrane was disrupted. In fact, the degradation of both substrates by lysosomal proteases was slightly lower in the paraquat-treated animals. This decrease could result from a decrease in the activity of some lysosomal proteases during oxidative stress, as reported previously (O'Neil et al., 1997; Carr, 2001; Crabb et al., 2002). In any case, the higher ability to degrade substrates of the intact lysosomes from paraquat-treated animals was thus a consequence of increased binding/uptake rather than of higher proteolysis in the lysosomal lumen.
Figure 3.
Additive effect of mild oxidative stress on CMA substrate proteins and on lysosomes. (A) Degradation of [14C]GAPDH (left) and [14C]RNase A (right) by lysosomes isolated from rats treated with paraquat as described under Materials and Methods. Incubations were carried out as described in Fig. 2D. Values are the mean + SE of five to eight different experiments (*p < 0.05, **p < 0.01, ***p < 0.001). (B) Degradation of radiolabeled cytosolic proteins prepared as in Figure 2D from untreated fibroblasts (control) or from fibroblasts exposed to H2O2 (left) or paraquat (right), by intact liver lysosomes from untreated rats (Lysosomes control) or from rats treated with paraquat (Lysosomes PQ) as described under Materials and Methods. Proteolysis was calculated as in Figure 2D. Values are mean + SE of six different experiments. *, differences compared with lysosomes control; +, differences compared with cytosol control; †, differences comparing lysosomes and cytosol control, to lysosomes and cytosol treated (*p < 0.05, **p < 0.01, ***p < 0.001).
We then determined whether oxidative stress could increase CMA-mediated degradation of proteins by acting on both the substrates, increasing their ability to be taken up by the lysosomes, and on the lysosomes, increasing their ability to take up substrates. For that purpose, we verified first that, as described above for GAPDH and RNase A, cytosolic proteins from untreated fibroblasts were more readily degraded by intact lysosomes from paraquat-treated rats than by lysosomes from untreated rats (Figure 3B). Then, we analyzed the effect of combining lysosomes and cytosolic proteins that had been both exposed to the prooxidant. When we incubated the lysosomes from paraquat-treated rats with the cytosolic proteins isolated from fibroblasts treated with H2O2 (Figure 3B, left) or with paraquat (Figure 3B, right), we found that these proteins were degraded even faster. We obtained similar results using lysosomes isolated from H2O2 treated fibroblasts (our unpublished data). Based on this additive effect, we conclude that not only oxidation-induced changes in the substrate proteins, but also in the lysosomal compartment, are responsible for the higher rates of CMA observed during oxidative stress.
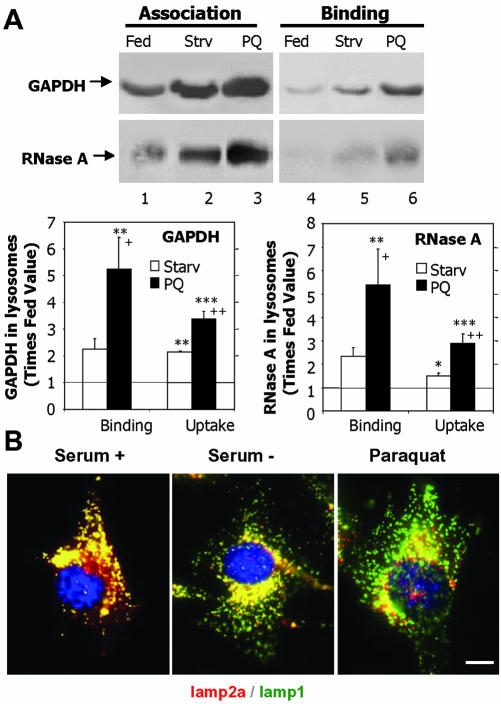
We further analyzed the activation of CMA during oxidative stress by separately analyzing binding and uptake of unmodified substrate proteins by lysosomes isolated from paraquat-treated rats. In these studies, to eliminate any possible differences in the ability of exogenous proteases to remove the protein bound to the cytosolic side of the lysosomal membrane, we compared instead the amount of substrate associated to lysosomes previously treated or not with a strong cocktail of protease inhibitors. In the absence of protease inhibitors the substrate that translocates inside the lysosomal lumen is rapidly degraded by the lysosomal proteases (half-life of proteins in the lysosomal lumen is 5–7 min; Aniento et al., 1993). Consequently, the protein associated to the lysosomes at the end of the incubation represents the protein bound to the cytosolic side of the lysosomal membrane (Salvador et al., 2000). In contrast, if lysosomal proteases are inhibited, the protein recovered associated to lysosomes corresponds to both protein bound and internalized into the lysosomal lumen. As described previously, the amount of internalized substrate can then be calculated by subtracting the protein bound from the amount of protein associated (Salvador et al., 2000). Consistent with our previous results, the amount of GAPDH and RNase A bound to the lysosomal membrane was significantly higher (4- to 5-fold increase) in lysosomes from paraquat-treated rats (Figure 4A). A two- to threefold increase was also detected in the uptake of both substrates by these lysosomes, when calculated as described above (Figure 4A, bottom). We have previously shown that CMA is activated under starvation conditions (Cuervo et al., 1995). When we compared both stimuli, starvation and paraquat, we found that treatment with the prooxidant agent resulted in stronger activation of CMA. Levels of binding and uptake of both substrates were significantly higher in lysosomes isolated from fed rats treated with paraquat than in lysosomes from 48-h starved animals. To determine whether the observed increase in CMA during paraquat treatment could be due, at least in part, to some degree of starvation in this group of animals, we measured food-intake in the animals exposed to paraquat. However, we did not find significant differences in food consumption between the paraquat-treated and untreated animals when maintained in ad libitum conditions, suggesting that activation of CMA after paraquat treatment was not due to starvation, but to the oxidative stress.
Figure 4.
Activation of CMA during mild oxidative stress. (A) Association and binding of GAPDH and RNase A to intact lysosomes from untreated fed rats, 48-h starved rats (Strv) or fed rats treated with paraquat (PQ). Lysosomes in lanes 1–3 were preincubated with chymostatin to prevent protein degradation. Incubations were carried out in an isotonic buffer, as described under Materials and Methods, and levels of associated proteins were determined by immunoblot of the lysosomes collected by centrifugation. Bottom: densitometric quantification (mean + SE) of 4–6 immunoblots similar to the ones shown here. Values are expressed as the fold of increase in binding and in uptake (association-binding) for the lysosomes from starved or paraquat-treated rats, compared with lysosomes from untreated fed animals. *, differences compared with fed animals; +, differences compared with starved animals (*p < 0.1, **p < 0.05, ***p < 0.01) (B) Immunofluorescence for lamp2a (red) and lamp1 (green) in mouse fibroblasts after removing the serum from the culture medium (serum-) or after treatment with paraquat in cells maintained in serum supplemented medium. The merged image of both fluorochrome channels is shown. Bar, 50 μm.
Activation of CMA is associated with the relocalization toward the perinuclear region of the lysosomes with higher CMA activity (those with higher levels of lys-hsc70 and lamp2a) (Agarraberes et al., 1997; Cuervo and Dice, 2000c). As shown in Figure 4B, in culture fibroblasts maintained in the presence of serum, lamp2a colocalized with other lysosomal membrane proteins (lamp1 is shown here), displaying a typical vesicular punctated pattern. When serum was removed from the culture medium, the lamp2a-enriched lysosomes preferentially localized in the perinuclear region. Treatment with paraquat displayed a similar perinuclear pattern for lamp2a even when the cells were maintained in the presence of serum (Figure 4B, right).
These results are consistent with a constitutive activation of CMA during oxidative stress in the two experimental models in which CMA has been better characterized, rat liver and fibroblasts in culture (Terlecky and Dice, 1993; Cuervo et al., 1997).
Oxidative Stress-induced Changes in Lysosomes Active for CMA
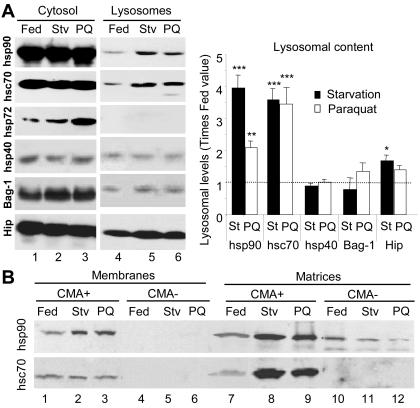
Several cytosolic chaperones associate with the lysosomal membrane and are required for CMA substrate binding/uptake into lysosomes (Agarraberes and Dice, 2001). Although the specific function of each of these cochaperones in CMA remains unknown, studies with human fibroblasts in culture have revealed that the levels of some of them are up-regulated when CMA is activated by removal of serum, whereas others remain unchanged (Agarraberes and Dice, 2001). In lysosomes isolated from livers of rats starved for 48 h, in addition to the previously reported increase in lys-hsc70, we found higher levels of hsp90, and, in less extent, of Hip, compared with lysosomes from control-fed animals (Figure 5A). Treatment with paraquat (in fed rats) resulted in similar increase in the lysosomal content of lyshsc70 and of Hip, but only a discrete increase in the content of hsp90, when compared with untreated animals. None of the treatments modified the levels of other associated chaperones (hsp40 or Bag-1), nor did they result in lysosomal association of the inducible form of the hsp70 family, hsp72, which was however elevated in the cytosol after paraquat treatment (Figure 5A). Because the function of hsp90 in lysosomes is not known, it is not clear why the increase in its lysosomal levels is not as pronounced as the one observed during starvation, but because rates of uptake are still up-regulated under these conditions, it points to a nonlimiting role for hsp90 in the translocation complex.
Figure 5.
Changes in lysosome-associated chaperones during mild oxidative stress. (A) Cytosol and lysosomes (75 μg of protein) isolated from normally fed, 48-h starved rats (Stv), or fed rats treated with paraquat (PQ) as labeled were subjected to SDS-PAGE and immunoblot for the indicated proteins. Right, densitometric quantification (mean + SE) of the chaperones in the lysosomal fraction in four immunoblots similar to the ones shown here. Levels in lysosomes from fed animals were given an arbitrary value of 1 (dotted line). *, differences compared with fed animals (*p < 0.1, **p < 0.05, ***p < 0.01). (B) Lysosomes with high (CMA+) and low (CMA-) activity for CMA (Cuervo et al., 1997) were isolated from the same groups of animals as detailed in A. After hypotonic shock and centrifugation, membranes and matrices (50 μg of protein) were separated and subjected to SDS-PAGE and immunoblot for hsc90 (top) or hsc70 (bottom).
Two components have been shown to be rate-limiting in the uptake of CMA substrates by lysosomes: the chaperone in the lysosomal lumen (lys-hsc70) (Cuervo et al., 1997; Agarraberes et al., 1997) and the receptor at the lysosomal membrane (lamp2a) (Cuervo and Dice, 2000b,c). We have previously reported in rat liver the presence of two lysosomal populations with similar morphological and enzymatic characteristics but different CMA activity (Cuervo et al., 1997). The main difference found, so far, between these two groups of lysosomes is the enrichment of lys-hsc70 in the lumen of the lysosomes with higher CMA activity (Cuervo et al., 1997). These two groups of lysosomes are normally purified together (they contribute 75 and 25% high activity and low activity, respectively, to the fraction that we used in these studies), but can be physically separated by further density and differential centrifugation (Cuervo et al., 1997). The lysosomes with lower CMA activity can become more active under specific conditions, such as prolonged (88-h) starvation or aging, which correlates with an increase in their lumenal levels of lys-hsc70 (Cuervo et al., 1997; Cuervo and Dice, 2000a). Because we found higher levels of lyshsc70 in the lysosomes from paraquat-treated animals, we separated these two lysosomal subpopulations to determine whether there was a net increase in lys-hsc70 per lysosome, or it resulted from enrichment of the less active lysosomes in lys-hsc70. As shown in Figure 5B, after treatment with paraquat there were no significant changes in the content of lys-hsc70 in the membranes or matrices of the less active group. Accordingly, the ability of this group of lysosomes to selectively take up CMA substrates did not change in paraquat-treated rats (our unpublished data). The percentage of hsc70-enriched lysosomes, determined as recovery of hexosaminidase activity in this fraction, was similar in untreated and paraquat-treated rats (our unpublished data). For the more active group of lysosomes, as observed during starvation, levels of lys-hsc70 at the lysosomal membrane remained constant, whereas levels of lys-hsc70 in the lysosomal lumen increased approximately fourfold. Separation of the lysosomal membranes and matrices also revealed that hsp90 increased in both compartments during starvation and after paraquat treatment (Figure 5B, top), but the increase in the lumen of lysosomes from paraquat-treated rats was smaller than in starved animals. These differences in the luminal content of hsp90 could explain why, when working with total lysosomes (Figure 5A), we found that paraquat induced a lower increase in lysosomal levels of hsp90 than starvation. Whether this hsp90 in the lysosomal lumen is functional, or whether it is only internalized to be degraded (amino acid sequence analysis of hsc90 revealed the presence of two KFERQ-related motifs) remains unknown.
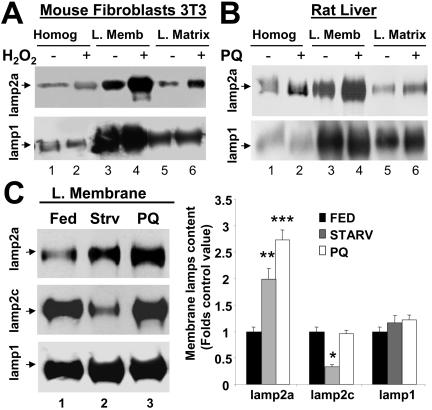
Levels of lamp2a at the lysosomal membrane directly correlate with CMA activity (Cuervo and Dice, 2000b). Lysosomes isolated from mouse fibroblasts treated with H2O2 (Figure 6A) or from livers of rats exposed to paraquat (Figure 6B) had higher levels of lamp2a at their membrane than lysosomes isolated from the corresponding untreated controls. This increase seems selective for lamp2a, because levels of other lysosomal membrane proteins (lamp1 shown here) remained unchanged. In agreement with the binding and uptake data (Figure 4A), the increase of lamp2a in the lysosomal membrane induced by paraquat was higher than the one induced by starvation (Figure 6C). For the two other lysosomal membrane proteins analyzed, levels of lamp1 remained unchanged in both conditions, and levels of lamp2c were not affected by the treatment with paraquat, but decreased significantly during starvation. Whether this decrease in lamp2c content is related to the increase in lamp2a or it happens independently is currently under investigation.
Figure 6.
Lamp2a levels at the lysosomal membrane increase during mild oxidative stress. (A and B) Immunoblot for lamp2a and lamp1 of homogenates (50 μg of protein), lysosomal membranes (L. Memb) and lysosomal matrices (L. Matrix) (10 μg of protein) isolated from cultured mice fibroblasts, exposed or not to 100 μM H2O2 (A) or from liver of rats treated or not with paraquat (PQ). (C) Imunoblot for lamp2a, lamp2c, and lamp1 of lysosomal membranes isolated from livers of fed rats, 48-h starved (Strv) rats, or fed rats treated with paraquat (PQ). Right, densitometric quantification of six to eight immunoblots as the ones shown here. Values are expressed as fold of the values in fed untreated animals and are the mean + SE of six different experiments. Levels in lysosomes from fed animals were given an arbitrary value of 1. *, differences compared with fed animals (*p < 0.05, **p < 0.01, ***p < 0.001).
A Novel Mechanism for Activation of CMA during Oxidative Stress
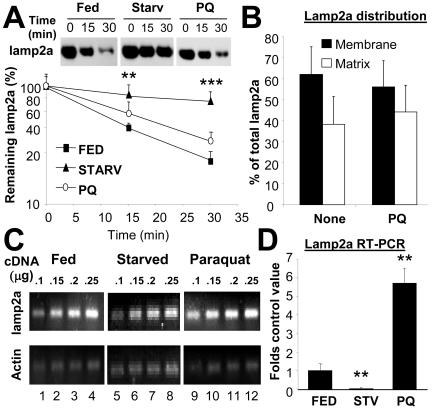
We have previously shown that CMA activity can be modulated by changes in the levels of lamp2a at the lysosomal membrane (Cuervo and Dice, 2000b). In fact, overexpression of lamp2a results in higher rates of CMA proportional to the increase in the receptor protein (Cuervo and Dice, 1996, 2000c). During starvation, the most extensively studied stimuli for CMA, the increase of lamp2a at the lysosomal membrane does not result from de novo synthesis of the protein. Instead, a decrease in the degradation rate of lamp2a, along with the relocation of part of the lamp2a resident in the lysosomal lumen toward the lysosomal membrane account for most of the lamp2a increase in the membrane (Cuervo and Dice, 1996, 2000c). To determine whether similar mechanisms were responsible for the increased levels of lamp2a during oxidative stress, we first compared the rates of degradation of lamp2a in membranes of lysosomes from rats fed, starved, or treated with paraquat. In contrast with the significant reduction in lamp2a degradation induced by starvation, degradation of lamp2a was only slightly slower in rats treated with paraquat than in untreated fed rats (Figure 7A). Likewise, the distribution of lamp2a between membrane and matrix was not significantly affected by the treatment with paraquat, suggesting that the increased levels of lamp2a in the membrane were not the result of recruitment of the lamp2a normally present in the lumen (Figure 7B).
Figure 7.
A novel mechanism for activation of CMA during mild oxidative stress. (A) Degradation of lamp2a in isolated membranes from normally fed rats, 48-h starved rats, or fed rats treated with paraquat (PQ), as indicated under Materials and Methods. Degradation of lamp2a was followed by immunoblot of the lysosomal membranes with an antibody specific against its cytosolic tail, at different times of the incubation. Values are the mean + SD of the densitometric quantification of immunoblots from four different experiments. A representative immunoblot is shown in the top insets. (B) Distribution of lamp2a between the lysosomal membrane and the matrix in lysosomes isolated from normally fed rats, treated or not with PQ. Values are mean + SE of the densitometric quantification of four immunoblots similar to the one shown in Figure 6B. (C) A 120-nucleotide fragment of lamp2a and a 108-nucleotide fragment of actin were amplified, through PCR, from increasing concentrations of total mRNA isolated from the livers of normally fed, 48-h starved, or paraquat-treated fed rats. The electrophoretic pattern of the products in a 2% agarose gel is shown. (D) Semiquantitative real-time PCR was used to compare mRNA expression levels for lamp2a in the same samples as detailed in C. Values were corrected for actin amplification in each samples and are expressed as fold increase compared to mRNA lamp2a values in fed untreated rats (that was given an arbitrary value of 1). Values are mean + SD for four different experiments. *, differences compared with fed animals (**p < 0.01, ***p < 0.001).
Contrary to starved animals, levels of lamp2a mRNA in the liver of rats treated with paraquat were significantly higher than in untreated rats. We used both a dilution-based quantitative PCR method (Figure 7C) and real-time PCR (Figure 7D) to compare levels of lamp2a mRNA in livers from fed, starved, or paraquat (fed)-treated rats. Treatment with paraquat induced an increase in lamp2a mRNA levels of approximately sixfold in fed animals, whereas starvation resulted in a slight decrease in lamp2a mRNA levels. Therefore, contrary to starvation, where the increase in lamp2a levels did not require new protein to be synthesized, most of the increase in lamp2a levels detected in paraquat-treated animals was a consequence of de novo synthesis of the protein.
We conclude that activation of CMA is part of the normal oxidative stress response and it contributes to the selective removal of oxidized proteins from the cytosol. The higher rates of CMA observed under these conditions are the combined result of an increase in the susceptibility of the proteins to be taken up and degraded by lysosomes, and an enhanced ability of the lysosomes for substrate uptake. Unexpectedly, during mild oxidative stress, activation of CMA is mediated by a novel mechanism different from the previously characterized activation of CMA during nutritional stress.
DISCUSSION
Our results provide for the first time evidence for the participation of lysosomes in the removal of oxidized proteins during mild oxidative stress through CMA. This novel role for CMA is supported by the fact that 1) oxidized proteins can be detected in the lumen of lysosomes active for CMA (Figure 1); 2) in conditions with declined CMA, such as aging, the amount of oxidized proteins translocated into lysosomes is reduced (Figure 1B); 3) oxidized CMA substrates bind and are taken up more efficiently by isolated lysosomes than their unmodified forms (Figure 2); 4) lysosomes from cells or rats exposed to prooxidants display higher rates of binding and uptake of CMA substrate proteins (Figures 3 and 4); and 5) blockage of CMA in cultured cells increases their susceptibility to prooxidant compounds and decreases their viability (Massey, Kiffin, and Cuervo, unpublished data). Activation of CMA during oxidative-stress is attained through the up-regulation of specific components of the lysosomal translocation complex (lysosomal chaperones and the lysosomal membrane receptor) (Figures 5 and 6). Interestingly, although these changes are similar to the ones described when CMA is activated by nutritional stress, the mechanism involved in the up-regulation is different. Therefore, this finding suggests that stress-mediated activation of CMA varies depending on the nature of the stress.
Most of the previous studies on the lysosomal role during oxidative stress have focused on the contribution of this subcellular compartment to the oxidative damage, rather than on a possible protective role. In fact, the destabilizing effects of severe oxidative stress (>350 μM hydrogen peroxide) on lysosomes have been well documented (Brunk et al., 1995; Ollinger and Brunk, 1995), showing that the release of lysosomal enzymes plays a major role in intracellular damage and in cell death under these conditions. Lower concentrations of hydrogen peroxide (250 μM for 30 min) still cause leakage of some lysosomal enzymes, although the lysosomal damage is reversible (Brunk et al., 1995; Ollinger and Brunk, 1995). In our study, we did not find changes in the stability of the lysosomal membrane, assayed either as release of specific enzymes or with a more sensitive proteolytic method. It is possible that the lower doses of hydrogen peroxide and paraquat used in our study (100 μM and 40 mg/100 g body weight, respectively) were enough to originate cytosolic protein damage, but they did not alter the lysosomal compartment. Other possibility is that the initial lysosomal damage still takes place. but due to its reversibility under these conditions (Brunk et al., 1995; Ollinger and Brunk, 1995), it is not longer detectable by the time the lysosomes are isolated (24 h after the second injection). In addition, because most of the studies regarding lysosomal stability and oxidative stress have been performed in intact cells, analyzing the leakage of fluorescent probes from the lysosomal compartment, we cannot discard that different lysosomal populations might be differently affected by prooxidants. In our assays, the cell fractionation method used has been optimized to isolate a subset of intracellular lysosomes active for CMA (10–15% of total) (Cuervo et al., 1997). Particular characteristics of the CMA-active lysosomes, such as, for example, a lower concentration of iron in their lumen, could explain their higher resistance to prooxidants. Our studies on CMA-active lysosomes in old animals also support their unique characteristics. Thus, although accumulation of lipofuscin in lysosomes is a commonly used biomarker of aging, CMA-active lysosomes from old rat livers rarely accumulate this pigment (Cuervo and Dice, 2000a).
The ability of proteasomes to degrade oxidized proteins has been previously well documented in vitro (Rivett, 1985; Davies, 2001), and more recently in vivo (Grune et al., 2002a; Hosler et al., 2003; Shringarpure et al., 2003). Most of the studies assessing oxidation-induced changes in lysosomal proteolytic behavior have focused in the analysis of the enzymatic activity of cathepsins, the lysosomal proteases. Moderate oxidative stress does not significantly change the activity of most cathepsins, whereas more severe oxidizing conditions result in increased or decreased cathepsin activity depending on the cellular conditions (Sitte et al., 2000a,b). This lack of correlation between the intracellular accumulation of oxidized proteins and the activity of lysosomal enzymes sets the basis for arguments against the lysosomal participation in removal of oxidized proteins (Sitte et al., 2000a). However, the activity of the lysosomal proteases is a nonlimiting step for any of the forms of autophagy. The intralysosomal concentration of cathepsins is such that, once substrates reach the lysosomal lumen, they are rapidly degraded. This makes the delivery of substrates the limiting step. Bergamini and colleagues were one of the first groups to point out that, if the degradation of proteins by lysosomes, instead of the enzymatic activity of lysosomal proteases, is considered, there is a good inverse correlation between lysosomal proteolysis and intracellular content of oxidized proteins (Vittorini et al., 1999). Our current study also supports a good correlation between the intracellular content of oxidized proteins and the delivery of substrate to lysosomes via CMA (binding/uptake). We have previously shown that, in the group of lysosomes active for CMA, the activity of most lysosomal enzymes does not change significantly with age, and yet rates of CMA are lower in aged cells (Cuervo and Dice, 2000a). The impaired ability of this group of lysosomes to take up substrates in older animals could explain why there is a lower content of oxidized proteins in their lumen, despite the higher level of oxidized cytosolic proteins (Figure 1B). It is unlikely that the lower levels of oxidized proteins detected in the lysosomal lumen of old rats, result from faster degradation of these proteins inside lysosomes, because even when we inhibited lysosomal degradation in rats, before lysosomal isolation (by i.p. of leupeptin; our unpublished results), we still found lower content of oxidized proteins in the lysosomes from the oldest animals.
The contribution of proteasomes to the removal of oxidized proteins in vivo has been demonstrated by analyzing the consequences of blocking its catalytic activity in oxidized protein removal (Grune et al., 2002a; Hosler et al., 2003; Shringarpure et al., 2003). This approach, however, cannot be used to evaluate the contribution of autophagy in this process. Even if the lysosomal proteases are inhibited (either with specific protease inhibitors or by raising the intralysosomal pH), we do not expect to find changes in the cytosolic content of oxidized proteins, because only their proteolysis, but not their translocation into the lysosomal lumen, would be blocked. The accumulation of undegraded products in the lysosomal lumen can be tolerated for a long time without affecting their translocation ability. An alternative approach would be to directly inhibit the delivery of the substrates. No chemical inhibitors are available to block CMA. We have recently succeeded in blocking substrate translocation by using RNA interference against the lysosomal receptor in cultured fibroblasts (Massey, Kiffin, and Cuervo, unpublished data). As mentioned before, these cells have a lower resistance to oxidative stress. However, even in this system, it is difficult to asses lysosomal versus proteasome contribution, because blockage of CMA results in changes in the composition of the 20S/26S proteasomes and, consequently, in its proteolytic activities (Massey, Kiffin, and Cuervo, unpublished data). Likewise, chronic inhibition of proteasomes, similar to the one that occurs in aging, alters the ability of the cells to activate autophagy in response to stress (Ding et al., 2003). Although an experimental challenge, this cross-talking among the different proteolytic systems offers perhaps a more physiological view of what is really happening in the cells during oxidative stress. In the same way that the relative contribution of proteasomes and lysosomes to total protein degradation varies depending on the cell type and on the cellular conditions (Fuertes et al., 2003), participation of these two systems in the removal of oxidized proteins is probably also dynamic. In fact, there are now numerous examples of proteins that can be degraded by more than one proteolytic pathway (Cuervo et al., 1998; Lenk et al., 1999). Future studies should be directed at understanding how those fluctuations are regulated and to identify possible ways of stimulating one system to compensate for the failure of another.
Whereas during severe oxidative stress, protein aggregation and cross-linking are common events, mild oxidative stress has been shown to generate excellent proteolytic substrates (Grune et al., 1995; Gomes-Marcondes and Tisdale, 2002). Our work supports that in the case of CMA, mild oxidation of CMA substrates facilitates not only their degradation by lysosomal proteases but also their binding/translocation across the lysosomal membrane (Figure 4A). Although the exact mechanism behind this facilitated uptake is still elusive, we hypothesize that protein unfolding, likely to occur during oxidation (Imai et al., 2003), could accelerate uptake, by diminishing the time required for substrate unfolding before translocation. Oxidized substrates were taken up faster by lysosomes even when we did not add cytosolic hsc70 in the translocation cocktail (our unpublished data). However, because the isolated lysosomes contain significant amounts of hsc70 in the cytosolic side of the membrane, we cannot discard that partial unfolding might make the KFERQ-motif more accessible for the interaction with hsc70 and facilitate in this way unfolding/translocation of the substrate. Our unpublished data showing that manipulations, such as partial denaturation of CMA substrates or extensive truncation, increase their rates of uptake into lysosomes via CMA (Salvador, Aguado, Cuervo, and Knecht, unpublished data), also support the proposed facilitating role of partial protein unfolding under mild oxidative conditions. Whether the activation of CMA under these conditions is a response to the presence of these partially unfolded proteins in the cytosol, or it is mediated by other cytosolic components generated during the oxidative stress remains to be elucidated.
An interesting remaining question is how the cells decide whether an oxidized protein should be degraded by the proteasome or in the lysosomal compartment. About 30% of cytosolic proteins contain a KFERQ-motif, suggesting that, in theory, this would be the subset of proteins degraded through CMA during mild oxidative stress (Figure 1A). However, a very intriguing idea proposed by Gracy et al., 1998 is that some amino acid modifications, such as deamidation and oxidation, could result in the generation of KFERQ-like containing motifs in proteins previously lacking this sequence. As other targeting motifs, the KFERQ-like motif is degenerate, resulting from the combination of a basic, a hydrophobic and an acid residue with a fourth basic or hydrophobic residue, flanked on either side by a glutamine. Oxidation for example of a histidine to aspartic acid could provide the acid residue necessary to complete a KFERQ-like motif. On the other hand, the same process could eliminate KFERQ-like motifs in proteins that normally are substrates for this pathway, resulting in their impaired degradation in conditions such as aging. This modification of the targeting motif, added to the fact that CMA activity decreases with age, could explain the lower content of oxidized proteins in the lumen of CMA active lysosomes in old rats, despite the higher levels of oxidized proteins in their cytosol. Our laboratory is currently trying to identify changes in the targeting motif of CMA substrates with age using a shot-gun proteomic approach.
Particularly exciting is the finding that although activation of CMA during oxidative and nutritional stress have the same consequences on the levels of the lysosomal translocation components, up-regulation, at least for the lysosomal receptor, obeys different mechanisms. During starvation CMA is activated to supply the cells with amino acids required for the synthesis of essential proteins. Under these conditions, de novo synthesis of lamp2a to increase CMA rates is probably not a reasonable option due to the shortage of amino acids. Instead, more conservative mechanisms (down-regulation of lamp2a degradation and intralysosomal relocation) are adopted (Cuervo and Dice, 2000b). If oxidative damage takes place under normal nutritional conditions, as the supply of amino acids would not be compromised, de novo synthesis of lamp2a is up-regulated to activate CMA (Figure 7C). De novo synthesis might be advantageous under these conditions, because it provides a faster mechanism for CMA activation.
In conclusion, we have identified for the first time a role of CMA as part of the oxidative stress response. Because CMA activity is severely impaired during aging, we hypothesize that part of the accumulation of oxidized proteins observed in old organisms may result from the malfunctioning of CMA. Future efforts aimed to restore normal CMA activity in old cells would help to understand the relevance of the cross-talk among different proteolytic pathways, and the compensatory mechanisms activated when the activity of one or more of these pathways is compromised.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Fernando Macian and the other members of our laboratory for valuable suggestions and comments. This work was supported by the National Institutes of Health/National Institute on Aging grant AG-021904, by an Ellison Medical Foundation Research Award (to A.M.C.), and by the Ministerio de Ciencia y Tecnología of Spain grants BMC2001-0816 and SAF2002-00206 and Fondo de Investigación Sanitaria grant RGDMG031212 (to E.K.). R.K. is a National Institutes of Health/National Institute on Aging postbachelor fellow.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–06–0477. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–06–0477.
Abbreviations used: CMA, chaperone-mediated autophagy; GAPDH, glyceraldehide-3-phosphate dehydrogenase; hsc70, heat shock cognate protein of 70 kDa; lamp, lysosomal associated membrane protein; MOPS, 3-(N-morpholino)propanesulfonic acid; RNase A, ribonuclease A.
The online version of this article contains supplementary material accessible through http://www.molbiolcell.org.
References
- Agarraberes, F.A., and Dice, J.F. (2001). A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J. Cell Sci. 114, 2491-2499. [DOI] [PubMed] [Google Scholar]
- Agarraberes, F., Terlecky, S.R., and Dice, J.F. (1997). An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J. Cell Biol. 137, 825-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aniento, F., Roche, E., Cuervo, A.M., and Knecht, E. (1993). Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J. Biol. Chem. 268, 10463-10470. [PubMed] [Google Scholar]
- Brunk, U.T., Zhang, H., Dalen, H., and Ollinger, K. (1995). Exposure of cells to nonlethal concentrations of hydrogen peroxide induces degeneration-repair mechanisms involving lysosomal destabilization. Free Radic. Biol. Med. 19, 813-822. [DOI] [PubMed] [Google Scholar]
- Carr, A.C. (2001). Hypochlorous acid-modified low-density lipoprotein inactivates the lysosomal protease cathepsin B: protection by ascorbic and lipoic acids. Redox Rep. 6, 343-349. [DOI] [PubMed] [Google Scholar]
- Carrard, G., Bulteau, A.L., Petropoulos, I., and Friguet, B. (2002). Impairment of proteasome structure and function in aging. Int. J. Biochem. Cell Biol. 34, 1461-1474. [DOI] [PubMed] [Google Scholar]
- Chiang, H.L., and Dice, J.F. (1988). Peptide sequences that target proteins for enhanced degradation during serum withdrawal. J. Biol. Chem. 262, 6797-6805. [PubMed] [Google Scholar]
- Chiang, H.L., Terlecky, S.R., Plant, C.P., and Dice, J.F. (1989). A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 246, 382-385. [DOI] [PubMed] [Google Scholar]
- Crabb, J.W., O'Neil, J., Miyagi, M., West, K., and Hoff, H.F. (2002). Hydroxynonenal inactivates cathepsin B by forming Michael adducts with active site residues. Protein Sci. 11, 831-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo, A.M. (2004a). Autophagy: in sickness and in health. Trends Cell Biol. 14, 70-77. [DOI] [PubMed] [Google Scholar]
- Cuervo, A.M. (2004b). Autophagy: many paths to the same end. Mol. Cell. Biochem. 263, 55-72 [DOI] [PubMed] [Google Scholar]
- Cuervo, A.M., and Dice, J.F. (1996). A receptor for the selective uptake and degradation of proteins by lysosomes. Science 273, 501-503. [DOI] [PubMed] [Google Scholar]
- Cuervo, A.M., and Dice, J.F. (1998a). How do intracellular proteolytic systems change with age? Front. Biosci. 3, 25-43. [DOI] [PubMed] [Google Scholar]
- Cuervo, A.M., and Dice, J.F. (1998b). Lysosomes, a meeting point of proteins, chaperones, and proteases. J. Mol. Med. 76, 6-12. [DOI] [PubMed] [Google Scholar]
- Cuervo, A.M., and Dice, J.F. (2000a). Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 275, 31505-31513. [DOI] [PubMed] [Google Scholar]
- Cuervo, A.M., and Dice, J.F. (2000b). Regulation of lamp2a levels in the lysosomal membrane. Traffic 1, 570-583. [DOI] [PubMed] [Google Scholar]
- Cuervo, A.M., and Dice, J.F. (2000c). Unique properties of lamp2a compared to other lamp2 isoforms. J. Cell Sci. 113, 4441-4450. [DOI] [PubMed] [Google Scholar]
- Cuervo, A.M., Dice, J.F., and Knecht, E. (1997). A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J. Biol. Chem. 272, 5606-5615. [DOI] [PubMed] [Google Scholar]
- Cuervo, A.M., Hildebrand, H., Bomhard, E.M., and Dice, J.F. (1999). Direct lysosomal uptake of alpha2-microglobulin contributes to chemically induced nephropathy. Kidney Int. 55, 529-545. [DOI] [PubMed] [Google Scholar]
- Cuervo, A.M., Hu, W., Lim, B., and Dice, J.F. (1998). IkB is a substrate for a selective pathway of lysosomal proteolysis. Mol. Biol. Cell 9, 1995-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo, A.M., Knecht, E., Terlecky, S.R., and Dice, J.F. (1995). Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am. J. Physiol. 269, C1200-C1208. [DOI] [PubMed] [Google Scholar]
- Cuervo, A. M., Mann, L., Bonten, E., d'Azzo, A., and Dice, J. (2003). Cathepsin A regulates chaperone-mediated autophagy through cleavage of the lysosomal receptor. EMBO J. 22, 12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo, A.M., Terlecky, S.R., Dice, J.F., and Knecht, E. (1994). Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J. Biol. Chem. 269, 26374-26380. [PubMed] [Google Scholar]
- Davies, K.J. (2001). Degradation of oxidized proteins by the 20S proteasome. Biochimie 83, 301-310. [DOI] [PubMed] [Google Scholar]
- Demasi, M., and Davies, K.J. (2003). Proteasome inhibitors induce intracellular protein aggregation and cell death by an oxygen-dependent mechanism. FEBS Lett. 542, 89-94. [DOI] [PubMed] [Google Scholar]
- Dice, J., Finn, P., Majeski, A., Mesieres, N., and Cuervo, A. (2003). Chaperone-mediated autophagy. In: Autophagy, ed. D.J. Klionsky, Georgetown, TX: Landes Bioscience, 158-177.
- Dice, J.F. (2000). Lysosomal Pathways of Protein Degradation, Austin, TX: Landes Bioscience.
- Ding, Q., Dimayuga, E., Martin, S., Bruce-Keller, A.J., Nukala, V., Cuervo, A.M., and Keller, J.N. (2003). Characterization of chronic low-level proteasome inhibition on neural homeostasis. J. Neurochem. 86, 489-497. [DOI] [PubMed] [Google Scholar]
- Donati, A., Cavallini, G., Paradiso, C., Vittorini, S., Pollera, M., Gori, Z., and Bergamini, E. (2001). Age-related changes in the autophagic proteolysis of rat isolated liver cells: effects of antiaging dietary restrictions. J. Gerontol. 56, B375-B383. [DOI] [PubMed] [Google Scholar]
- Dunlop, R.A., Rodgers, K.J., and Dean, R.T. (2002). Recent developments in the intracellular degradation of oxidized proteins. Free Radic. Biol. Med. 33, 894-906. [DOI] [PubMed] [Google Scholar]
- Franch, H.A., Sooparb, S., Du, J., and Brown, N.S. (2001). A mechanism regulating proteolysis of specific proteins during renal tubular cell growth. J. Biol. Chem. 276, 19126-19131. [DOI] [PubMed] [Google Scholar]
- Friguet, B. (2002). Protein repair and degradation during aging. Sci. World J. 2, 248-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friguet, B., Bulteau, A.L., Chondrogianni, N., Conconi, M., and Petropoulos, I. (2000). Protein degradation by the proteasome and its implications in aging. Ann. N.Y. Acad. Sci. 908, 143-154. [DOI] [PubMed] [Google Scholar]
- Fuertes, G., Martin de Llano, J., Villarroya, A., Rivett, A.J., and Knecht, E. (2003). Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem. J. 375, 75-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes-Marcondes, M., and Tisdale, M. (2002). Induction of protein catabolism and the ubiquitin-proteasome pathway by mild oxidative stress. Cancer Lett. 180, 69-74. [DOI] [PubMed] [Google Scholar]
- Gracy, R., Talent, J., and Zvaigzne, A. (1998). Molecular wear and tear leads to terminal marking and the unstable isoforms of aging. J. Exp. Zool. 282, 18-27. [PubMed] [Google Scholar]
- Grune, T. (2000). Oxidative stress, aging and the proteasomal system. Biogerontology 1, 31-40. [DOI] [PubMed] [Google Scholar]
- Grune, T., and Davies, K.J. (1997). Breakdown of oxidized proteins as a part of secondary antioxidant defenses in mammalian cells. Biofactors 6, 165-172. [DOI] [PubMed] [Google Scholar]
- Grune, T., Klotz, L.O., Gieche, J., Rudeck, M., and Sies, H. (2001). Protein oxidation and proteolysis by the nonradical oxidants singlet oxygen or peroxynitrite. Free Radic. Biol. Med. 30, 1243-1253. [DOI] [PubMed] [Google Scholar]
- Grune, T., Reinheckel, T., and Davies, K.J. (1997). Degradation of oxidized proteins in mammalian cells. FASEB J. 11, 526-534. [PubMed] [Google Scholar]
- Grune, T., Reinheckel, T., Joshi, M., and Davies, K.J. (1995). Proteolysis in cultured liver epithelial cells during oxidative stress. Role of the multicatalytic proteinase complex, proteasome. J. Biol. Chem. 270, 2344-2351. [DOI] [PubMed] [Google Scholar]
- Grune, T., Reinheckel, T., Li, R., North, J.A., and Davies, K.J. (2002a). Proteasome-dependent turnover of protein disulfide isomerase in oxidatively stressed cells. Arch. Biochem. Biophys. 397, 407-413. [DOI] [PubMed] [Google Scholar]
- Grune, T., Stolzing, A., Jakstadt, M., McLaren, J.S., Speakman, J.R., and Szweda, P.A. (2002b). Proteolysis, free radicals, and aging. Free Radic. Biol. Med. 33, 259-265. [DOI] [PubMed] [Google Scholar]
- Haly, T. (1979). Review of the toxicity of paraquat. Clin. Toxicol. 14, 1-46. [DOI] [PubMed] [Google Scholar]
- Hoff, H.F., Zyromski, N., Armstrong, D., and O'Neil, J. (1993). Aggregation as well as chemical modification of LDL during oxidation is responsible for poor processing in macrophages. J Lipid Res. 34, 1919-1929. [PubMed] [Google Scholar]
- Hosler, M.R., Wang-Su, S.T., and Wagner, B.J. (2003). Targeted disruption of specific steps of the ubiquitin-proteasome pathway by oxidation in lens epithelial cells. Int. J Biochem. Cell Biol. 35, 685-697. [DOI] [PubMed] [Google Scholar]
- Imai, J., Yashiroda, H., Maruya, M., Yahara, I., and Tanaka, K. (2003). Proteasomes and molecular chaperones: cellular machinery responsible for folding and destruction of unfolded proteins. Cell Cycle 2, 585-590. [PubMed] [Google Scholar]
- Jentoft, N., and Dearborn, D. (1983). Protein labeling by reductive alkylation. Methods Enzymol. 91, 570-579. [DOI] [PubMed] [Google Scholar]
- Keller, J.N., Gee, J., and Ding, Q. (2002). The proteasome in brain aging. Age Res Rev. 1, 279-293. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. (1970). Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- Lemasters, J.J., Qian, T., He, L., Kim, J.S., Elmore, S.P., Cascio, W.E., and Brenner, D.A. (2002). Role of mitochondrial inner membrane permeabilization in necrotic cell death, apoptosis, and autophagy. Antioxid. Redox Signal. 4, 769-781. [DOI] [PubMed] [Google Scholar]
- Lenk, S.E., Susan, P.P., Hickson, I., Jasionowski, T., and Dunn, W.A., Jr. (1999). Ubiquitinated aldolase B accumulates during starvation-induced lysosomal proteolysis. J. Cell Physiol. 178, 17-27. [DOI] [PubMed] [Google Scholar]
- Lowry, O.H., Rosebrough, N.J., Farr, A.L., and Randall, R.J. (1951). Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265-275. [PubMed] [Google Scholar]
- Martin, A., Joseph, J.A., and Cuervo, A.M. (2002). Stimulatory effect of vitamin C on autophagy in glial cells. J. Neurochem. 82, 538-549. [DOI] [PubMed] [Google Scholar]
- Massey, A., Kiffin, R., and Cuervo, A.M. (2004). Pathophysiology of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 36, 2420-2434. [DOI] [PubMed] [Google Scholar]
- Mehlhase, J., and Grune, T. (2002). Proteolytic response to oxidative stress in mammalian cells. Biol. Chem. 383, 559-567. [DOI] [PubMed] [Google Scholar]
- Merker, K., and Grune, T. (2000). Proteolysis of oxidised proteins and cellular senescence. Exp. Gerontol. 35, 779-786. [DOI] [PubMed] [Google Scholar]
- Merker, K., Stolzing, A., and Grune, T. (2001). Proteolysis, caloric restriction and aging. Mech. Ageing Dev. 122, 595-615. [DOI] [PubMed] [Google Scholar]
- Ohsumi, Y., Ishikawa, T., and Kato, K. (1983). A rapid and simplified method for the preparation of lysosomal membranes from rat liver. J. Biochem. 93, 547-556. [PubMed] [Google Scholar]
- Ollinger, K., and Brunk, U.T. (1995). Cellular injury induced by oxidative stress is mediated through lysosomal damage. Free Radic. Biol. Med. 19, 565-574. [DOI] [PubMed] [Google Scholar]
- O'Neil, J., Hoppe, G., Sayre, L.M., and Hoff, H.F. (1997). Inactivation of cathepsin B by oxidized LDL involves complex formation induced by binding of putative reactive sites exposed at low pH to thiols on the enzyme. Free Radic. Biol. Med. 23, 215-225. [DOI] [PubMed] [Google Scholar]
- Reggiori, F., and Klionsky, D.J. (2002). Autophagy in the eukaryotic cell. Eukaryot. Cell 1, 11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivett, A., and Levine, R. (1990). Metal-catalyzed oxidation of Escherichia coli glutamine synthetase: correlation of structural and functional changes. Arch. Biochem. Biophys. 278, 26-34. [DOI] [PubMed] [Google Scholar]
- Rivett, A.J. (1985). Preferential degradation of the oxidatively modified form of glutamine synthetase by intracellular mammalian proteases. J. Biol. Chem. 260, 300-305. [PubMed] [Google Scholar]
- Salvador, N., Aguado, C., Horst, M., and Knecht, E. (2000). Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J. Biol. Chem. 275, 27447-27456. [DOI] [PubMed] [Google Scholar]
- Shringarpure, R., Grune, T., Mehlhase, J., and Davies, K.J. (2003). Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J. Biol. Chem. 278, 311-318. [DOI] [PubMed] [Google Scholar]
- Sitte, N., Merker, K., Von Zglinicki, T., Davies, K.J., and Grune, T. (2000a). Protein oxidation and degradation during cellular senescence of human BJ fibroblasts: part II–aging of nondividing cells. FASEB J. 14, 2503-2510. [DOI] [PubMed] [Google Scholar]
- Sitte, N., Merker, K., Von Zglinicki, T., Grune, T., and Davies, K.J. (2000b). Protein oxidation and degradation during cellular senescence of human BJ fibroblasts: part I–effects of proliferative senescence. FASEB J. 14, 2495-2502. [DOI] [PubMed] [Google Scholar]
- Stadtman, E.R. (2001). Protein oxidation in aging and age-related diseases. Ann. N.Y. Acad. Sci. 928, 22-38. [DOI] [PubMed] [Google Scholar]
- Storrie, B., and Madden, E. (1990). Isolation of subcellular organelles. Methods Enzymol. 182, 203-225. [DOI] [PubMed] [Google Scholar]
- Szweda, P.A., Friguet, B., and Szweda, L.I. (2002). Proteolysis, free radicals, and aging. Free Radic. Biol. Med. 33, 29-36. [DOI] [PubMed] [Google Scholar]
- Terlecky, S., and Dice, J.F. (1993). Polypeptide import and degradation by isolated lysosomes. J. Biol. Chem. 268, 23490-23495. [PubMed] [Google Scholar]
- Terman, A. (1995). The effect of age on formation and elimination of autophagic vacuoles in mouse hepatocytes. Gerontology 41 (suppl 2), 319-326. [DOI] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures and some applications. Proc. Natl. Acad. Sci. USA 76, 4350-4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittorini, S., Paradiso, C., Donati, A., Cavallini, G., Masini, M., Gori, Z., Pollera, M., and Bergamini, E. (1999). The age-related accumulation of protein carbonyl in rat liver correlates with the age-related decline in liver proteolytic activities. J. Gerontol. A Biol. Sci. Med. Sci. 54, B318-B323. [DOI] [PubMed] [Google Scholar]
- Wang, C.W., and Klionsky, D.J. (2003). The molecular mechanism of autophagy. Mol. Med. 9, 65-76. [PMC free article] [PubMed] [Google Scholar]
- Ward, W.F. (2002). Protein degradation in the aging organism. Prog. Mol. Subcell. Biol. 29, 35-42. [DOI] [PubMed] [Google Scholar]
- Wattiaux, R., Wattiaux-De Coninck, S., Ronveaux-Dupal, M.F., and Dubois, F. (1978). Isolation of rat liver lysosomes by isopycnic centrifugation in a metrizamide gradient. J. Cell Biol. 78, 349-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing, S.S., Chiang, H.L., Goldberg, A.L., and Dice, J.F. (1991). Proteins containing peptide sequences related to KFERQ are selectively depleted in liver and heart, but not skeletal muscle, of fasted rats. Biochem. J. 275, 165-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.