Abstract
Background & Aims
Microbes may increase susceptibility to inflammatory bowel disease (IBD) by producing bioactive metabolites that affect immune activity and epithelial function. We undertook a family based study to identify microbial and metabolic features of IBD that may represent a predisease risk state when found in healthy first-degree relatives.
Methods
Twenty-one families with pediatric IBD were recruited, comprising 26 Crohn’s disease patients in clinical remission, 10 ulcerative colitis patients in clinical remission, and 54 healthy siblings/parents. Fecal samples were collected for 16S ribosomal RNA gene sequencing, untargeted liquid chromatography–mass spectrometry metabolomics, and calprotectin measurement. Individuals were grouped into microbial and metabolomics states using Dirichlet multinomial models. Multivariate models were used to identify microbes and metabolites associated with these states.
Results
Individuals were classified into 2 microbial community types. One was associated with IBD but irrespective of disease status, had lower microbial diversity, and characteristic shifts in microbial composition including increased Enterobacteriaceae, consistent with dysbiosis. This microbial community type was associated similarly with IBD and reduced microbial diversity in an independent pediatric cohort. Individuals also clustered bioinformatically into 2 subsets with shared fecal metabolomics signatures. One metabotype was associated with IBD and was characterized by increased bile acids, taurine, and tryptophan. The IBD-associated microbial and metabolomics states were highly correlated, suggesting that they represented an integrated ecosystem. Healthy relatives with the IBD-associated microbial community type had an increased incidence of elevated fecal calprotectin.
Conclusions
Healthy first-degree relatives can have dysbiosis associated with an altered intestinal metabolome that may signify a predisease microbial susceptibility state or subclinical inflammation. Longitudinal prospective studies are required to determine whether these individuals have a clinically significant increased risk for developing IBD.
Keywords: Microbiome, Metabolomics, Inflammatory Bowel Disease, Family Cohort
Abbreviations used in this paper: AUC, area under the curve; CD, Crohn’s disease; IBD, inflammatory bowel disease; LC/MS, liquid chromatography/mass spectrometry; OTU, operational taxonomic unit; PCoA, principal coordinates analysis; PCR, polymerase chain reaction; rRNA, ribosomal RNA; ToFMS, time-of-flight mass spectrometry; UC, ulcerative colitis; UPLC, ultra-performance liquid chromatography
Summary.
Healthy first-degree relatives of pediatric inflammatory bowel disease patients can have a fecal microbial and metabolomics profile seen in many patients with quiescent inflammatory bowel disease. This potential predisease state was associated with an increased likelihood of increased fecal calprotectin levels.
The inflammatory bowel diseases (IBDs)—comprising Crohn’s disease (CD) and ulcerative colitis (UC)—are chronic inflammatory diseases with a growing prevalence worldwide.1 IBD is believed to arise from a combination of genetic susceptibility and environmental factors that trigger an inappropriate mucosal inflammatory response.2 A pathogenic role for the intestinal microbiome at the junction of genetics and environment is supported by the resistance of germ-free mice to experimental colitis models and the transmissibility of colitis by microbiota derived from mice with genetic defects in mucosal immunity.3, 4, 5 IBD patients have reduced microbial diversity and alterations in the composition and function of the intestinal microbiome compared with healthy controls.6, 7, 8, 9, 10 Founder effects and diet are important environmental factors in microbial composition, and are associated with IBD disease risk.11 These findings support the hypothesis that IBD involves a dysbiosis arising from a combination of environmental and genetic perturbations of the mucosal habitat controlling microbial composition.12, 13
It remains unclear which differentially abundant microbes in IBD patients are instigators of disease rather than bystanders responding to the altered environment of the inflamed intestine. An important prospect for resolution is the expanding recognition of bioactive microbial products that profoundly can affect mucosal immune responses and epithelial homeostasis.14 For instance, IBD patients have reduced fecal concentrations of short-chain fatty acids such as butyrate that limit inflammatory responses in colitis models.15, 16, 17 This may be attributable to shifts in the microbiome including reduced abundance of the prominent butyrate-producer Faecalibacterium prausnitzii.8, 9, 18 Administration of F prausnitzii or its culture supernatant ameliorated experimental colitis; however, treatment of mice with butyrate at the concentration in culture supernatant did not affect colitis severity.18 This underscores the need to more fully characterize the IBD-associated metabolome and the scope of microbial products relevant to disease pathogenesis.
If IBD develops as a consequence of a host response to a proinflammatory microbiome and its associated microbial products, as has been reported in animal models, IBD patients would be predicted to have harbored dysbiosis before the development of disease. We performed a family-based study of the microbiome and metabolome of pediatric IBD patients and their unaffected first-degree relatives, who are at heightened risk for developing IBD. We hypothesized that unaffected siblings and parents—previously reported to have increased fecal calprotectin levels compared with healthy unrelated controls—may carry a predisease microbial risk state owing to shared genetic and environmental factors with the IBD proband.19, 20 We found that individuals in the family cohort grouped into 1 of 2 states based on fecal microbial and metabolomics profiles, which could be defined bioinformatically as OTU types and metabotypes (multidimensional community clusters characterized by differences in the abundance of signature taxa or metabolites, respectively).21 These OTU types and metabotypes were correlated highly with one another, and with disease status. We propose that in families at risk for IBD, a subset of healthy individuals harbor a stable intestinal microbial/metabolomic state that may confer increased susceptibility to IBD.
Materials and Methods
Cohort Recruitment and Sample Collection
Twenty-one CD and UC probands younger than the age of 18 were recruited from the Pediatric IBD Center at the Cedars-Sinai Medical Center. All patients were in clinical remission at the time of collection as evidenced by a Harvey–Bradshaw Index score of less than 5 for CD patients or a partial Mayo score of less than 2 for UC patients. Family members of these probands were recruited and assessed by detailed questionnaire for history of IBD. Six siblings and 9 parents were known to have IBD. Non-IBD family members were validated by lack of symptoms or medications suggestive of undiagnosed IBD. Participants were provided with a toilet hat, sample containers, and cold packs for home collection. Freshly defecated feces were frozen immediately at the clinic or at home and then brought to the clinic on cold packs for storage at -80°C. Frozen fecal samples were ground with a mortar and pestle in the presence of liquid nitrogen and then aliquoted for microbiome and metabolome analysis. Fecal calprotectin was measured from frozen aliquots of stool by an enzyme-linked immunosorbent assay performed by Quest Diagnostics (Los Angeles, CA). This research was approved by the Cedars-Sinai Medical Center Institutional Review Board under IRB 3766. All authors had access to the study data and reviewed and approved the final manuscript.
16S Ribosomal RNA Gene Sequencing
Genomic DNA was extracted using the PowerSoil DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA) with a 30-second, beat-beating step using a Mini-Beadbeater-16 (BioSpec Products, Bartlesville, OK).22 Polymerase chain reaction amplification of bacterial 16S ribosomal RNA (rRNA) genes was performed using extracted genomic DNA as the template. The 100-μL reactions contained 50 mmol/L Tris (pH 8.3), 500 μg/mL bovine serum albumin, 2.5 mmol/L MgCl2, 250 μmol/L of each of the 4 deoxynucleotide triphosphates, 400 nmol/L of each primer, 4 μL of DNA template, and 2.5 U JumpStart Taq DNA polymerase (Sigma-Aldrich, St. Louis, MO). The polymerase chain reaction (PCR) primers (F515/R806) targeted the V4 hypervariable region of the 16S rRNA gene, with the reverse primers including a 12-bp Golay barcode.23 Thermal cycling was performed in an MJ Research PTC-200 (Bio-Rad, Inc, Hercules, CA) with the following parameters: 94°C for 5 minutes; 35 cycles of 94°C for 20 seconds, 50°C for 20 seconds, and 72°C for 30 seconds; and 72°C for 5 minutes. PCR products were purified using the MinElute 96 UF PCR Purification Kit (Qiagen, Valencia, CA). DNA sequencing was performed using an Illumina HiSeq 2000 (Illumina, Inc, San Diego, CA). Clusters were created using template concentrations of 1.9 pmol/L and PhiX at 65 K/mm2. Sequencing primers targeted 101 base pair reads of the 5’ end of the amplicons and 7 base pair barcode reads.23 Reads were filtered using the following parameters: minimum Q-score, - 20; maximum number of consecutive low-quality base calls allowed before truncating, - 3; and maximum number of N characters allowed, - 0. All filtered V4 reads had a length of 101 bp. The number of sequences per sample ranged from 17,946 (an outlier; the second lowest sequence depth was 478,168) to 848,638, with a mean of 620,720. Raw 16S rRNA sequence data were deposited under National Center for Biotechnology Information BioProject ID PRJNA324147. Operational taxonomic units (OTUs) were picked in QIIME 1.8.0 against the May 2013 version of the Greengenes database (http://greengenes.secondgenome.com), prefiltered at 97% identity.24 The number of reads assigned to 97% OTUs ranged from 445,985 to 805,639, excluding the lowest depth sample (16,941).
OTUs with only a single assigned read were removed, leaving 7967 OTUs. α Diversity was assessed in QIIME using Faith’s phylogenetic diversity, Chao1, and the Shannon index with data rarefied to 445,975 sequences. Statistical significance was assessed using the Mann–Whitney U test. β Diversity was assessed on unrarefied data using the square root of the Jensen–Shannon divergence (calculated in R with a script available at http://enterotype.embl.de/enterotypes.html) and on data rarefied to 445,975 sequences using Bray–Curtis and unweighted UniFrac (calculated in QIIME). Principal coordinates analysis was used to visualize β diversity. Adonis with 100,000 permutations was used to assess statistical significance of differences in β diversity.25
Ultra-Performance Liquid Chromatography/Time-of-Flight Mass Spectrometry Untargeted Metabolomics
A total of 50 mg of each fecal sample was dried, resuspended in 150 μL of Optima liquid chromatography/mass spectrometry (LC/MS) grade water on ice, subjected to heat shock in a 37°C water bath for 90 seconds, then chilled on ice. One microliter from each sample was removed for protein concentration measurement. Six hundred microliters of chilled methanol containing internal standards for lipidomics and metabolomics was added to each sample and incubated on ice for 15 minutes as previously described.26 An equal volume of chloroform then was added to the mixture. The samples then were centrifuged at 16,000g for 10 minutes. The organic and aqueous phases were separated carefully, followed by addition of 600 μL of chilled acetonitrile for overnight incubation at -20°C. The samples then were centrifuged at maximum speed at room temperature for 10 minutes. Supernatants were combined and transferred to new glass tubes, dried under a gentle stream of N2, and resuspended in 100 μL of solvent A for LC/MS.
The MS analysis was performed by injecting 2-μL aliquots of each sample into a reverse-phase 50 × 2.1 mm H-class Ultra-Performance Liquid Chromatography (UPLC) Acquity 1.7-μmol/L BEH C18 column (Waters Corp, Milford, MA, USA), coupled to a time of flight mass spectrometry (ToFMS). The mobile phase consisted of water and 0.1% formic acid (solvent A), 100% acetonitrile (solvent B), and isopropanol/acetonitrile (90:10, vol:vol) with 10 mmol/L ammonium formate (solvent C). The Xevo G2-S mass spectrometer (Waters Corp) was operated in the positive (ESI+) and negative (ESI-) electrospray ionization modes scanning a 50- to 1200-m/z range. The following 13-minute gradient was used: 95%/5% solvent A/solvent B at 0.45 mL/min for 8 minutes, 2%/98% solvent A/solvent B for 1 minute, 2%/98% solvent B/solvent C for 1.5 minutes, 50%/50% solvent A/solvent B for 1.5 minutes, and 95%/5% solvent A/solvent B for the remaining half a minute. The lock-spray consisted of leucine-enkephalin (556.2771 [M+H]+ and 554.2615 [M-H]-). The MS data were acquired in centroid mode and processed using MassLynx software (Waters Corp) to construct a data matrix consisting of the retention time, m/z, and intensity (via the peak area normalized to protein concentration) for each ion. A total of 3206 ions were detected in the positive mode and 1420 ions were detected in the negative mode.
Our in-house statistical analysis program was used to putatively identify ions, using the Human Metabolome Database, LipidMaps, the Kyoto Encyclopedia of Genes and Genomes database, and BioCyc. The m/z values were used to putatively assign IDs to the ions by neutral mass elucidation, which was accomplished by considering the possible adducts (H+, Na+, and/or NH4+ in the ESI+ mode; and H- and Cl- in the ESI- mode). The masses then were compared with the exact mass of small molecules in the databases, from which putative metabolites were identified with a mass error of 20 ppm or less. MS/MS validation was performed by comparing the fragmentation pattern of each metabolite and lipid of interest against either that of its pure chemical form or its published MS/MS spectra in Metlin or our in-house database.27 Validation was performed for 123 spectral features, of which 37 were validated successfully.
Community-Type Analysis
Individuals were grouped into OTU types and metabotypes by applying Dirichlet multinomial mixture models to 16S sequence data and UPLC-ToFMS metabolomics data.28 This approach has been reported to outperform partitioning around medoids for clustering individuals by their microbiome in simulation studies and using Human Microbiome Project data.29 Modeling was performed using the get.communitytype function in Mothur v.1.36.1 with default parameters for the minimum (5) and maximum (10) number of partitions tested.30 Model fit for each possible number of OTU types or metabotypes was estimated using the Laplace approximation. The exponent of the difference in model fit serves as an estimate of the probability of one model over another.
Differential Abundance Testing
Nonrarefied 16S rRNA sequence data and UPLC-ToFMS metabolomics data were filtered to remove OTUs and spectral features present in less than 10% of samples. The sample with 17,946 16S rRNA sequences was excluded. The resulting filtered data sets were analyzed using DESeq2 (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html). This algorithm performs normalization using size factors estimated by the median-of-ratios method, uses an empiric Bayesian approach to shrink dispersion, and fits the data to negative binomial models.31 Covariates included sex, Jewish ancestry, current anti–tumor necrosis factor therapy, mode of delivery, family group, IBD diagnosis, and OTU type/metabotype. P values for variables in the linear models (eg, IBD status) were converted to q values to correct for multiple hypothesis testing. This was implemented with the R script q value (https://github.com/jdstorey/qvalue), which for each P value estimates the false-discovery rate based on the distribution of P values.32 Q values less than 0.05 were considered significant. Hierarchical clustering of metabolite abundance data and visualization with heat maps were performed in GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E). Rows and columns were clustered by Pearson correlation coefficient with the average linkage method.
Random Forest Classifiers
Random forest classifiers to predict OTU type, metabotype, and CD were created in R using the randomForest package (https://cran.r-project.org/web/packages/randomForest) with 1001 trees and mtry = 2.33 Features were inputted into the algorithm if they were associated significantly with the trait in multivariate DESeq2 models and were present in at least 30% of samples. The accuracy of random forest classifiers was estimated using 10-fold cross-validation. The OTU-type classifier trained on the family cohort was used to predict the OTU type of fecal samples in the RISK cohort (a previously published pediatric Crohn’s disease inception cohort).9 Subjects with recent exposure to antibiotics were excluded. The RISK cohort data were kindly supplied by the authors of that study (Gevers and Huttenhower) as a count table of 97% OTUs picked against the Greengenes database of OTUs, allowing for matching of OTUs with those in the random forest classifier. The OTU-type classifier was constructed from 57 OTUs that were differentially abundant by OTU type (as visualized in Figure 1E), present in at least 30% of samples in the family cohort, and present in at least 30% of samples in the RISK cohort after both data sets were rarefied to 20,000 sequences/samples to adjust for different sequence depths.9
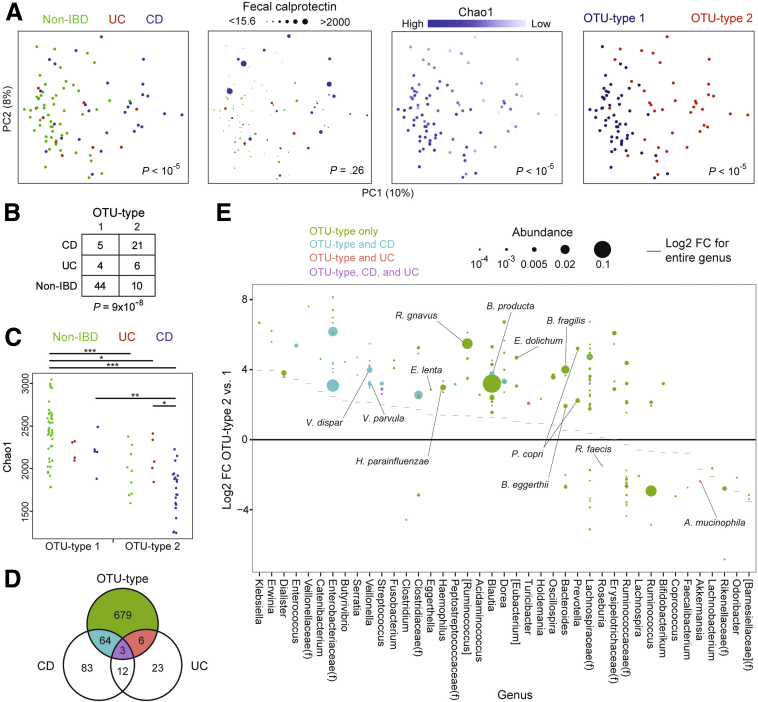
Figure 1.
A low-diversity microbial community type is associated with IBD but also is present in healthy relatives. (A) PCoA plots were used to visualize differences in microbial composition across samples (β diversity) as measured by the square root of the Jensen–Shannon divergence. Dots represent fecal samples colored by IBD status (left 2 panels), Chao1 (a measure of microbial richness), or OTU type (determined using Dirichlet multinomial mixture models). Dot size corresponds to fecal calprotectin level in the second panel from the left. P values for the association of the indicated variable (IBD status, calprotectin, Chao1, and OTU type) with microbial composition were calculated using Adonis. (B) Contingency table of IBD status by OTU type. P value was calculated using the Fisher exact test. (C) Chao1 by IBD status and OTU type. P values were calculated using the Mann–Whitney U test. *P < .05, **P < .005, ***P < .0001. (D) Venn diagram indicating differential OTUs for CD (vs non-IBD), UC (vs non-IBD), and OTU type (2 vs 1) in multivariate DESeq2 models including sex, Jewish ancestry, current anti–tumor necrosis factor therapy, mode of delivery, family group, IBD diagnosis, and OTU type as covariates. (E) OTUs with a statistically significant difference in abundance between OTU types 2 and 1 in multivariate DESeq2 models are shown. Effect size is represented as the log2 fold-change (FC) between OTU types 2 and 1. Dot size is proportional to the abundance of that OTU after normalization in DESeq2. Only OTUs with a mean normalized abundance greater than 10-4 are included. Color signifies whether the OTU also is differentially abundant in CD, UC, or both. The genus of each OTU is shown on the horizontal axis, ordered by the log2 FC between OTU types 2 and 1 of the genus as a whole. OTUs are grouped into families (f) where indicated if they did not have an assigned genus. OTUs putatively identified at the species level in the Greengenes database are annotated.
Inter’omic Analysis
Similarity between the microbiome and metabolomics data sets was assessed by Procrustes superimposition of principal coordinates constructed from distance matrices calculated using the square root of the Jensen–Shannon divergence.34 Significance of the Procrustes analysis was determined using 10,000 Monte Carlo permutations in QIIME. An inter’omic network was constructed in Cytoscape 3.2.1 (http://cytoscape.org) from OTUs and spectral features that had a random forest importance score greater than 2 in classifiers for OTU type and metabotype. Spearman correlations were calculated for all OTU–OTU, metabolite–metabolite, and OTU–metabolite pairs using the residuals of DESeq2 multivariate models incorporating IBD status, OTU type or metabotype, sex, current anti–tumor necrosis factor therapy, Jewish ethnicity, mode of delivery, and family. P values were computed for Spearman correlation coefficients based on the asymptotic t approximation and then were converted to q values in R. Correlation pairs with a q value less than 0.05 represented edges in the network.
Logistic Regression
Multivariate and univariate logistic regression were performed in R using the glm function. Multivariate models for IBD that included parent/child status and Jewish ethnicity had improved fit by the likelihood ratio test relative to models without these covariates.
Results
A Low-Diversity IBD-Associated Microbial Community Type Is Present In Healthy First-Degree Relatives of Pediatric IBD Patients
We recruited a cohort of 21 families with pediatric IBD, consisting of 36 IBD patients (21 probands, 6 siblings, and 9 parents) and 54 healthy first-degree relatives (Table 1). The IBD subjects included 26 with CD and 10 with UC. All were in clinical remission to reduce confounding of microbial and metabolomics analyses by the effects of active disease. Twenty of 22 of the probands and their siblings with CD had small intestinal involvement; of these, 14 also had colonic involvement. All but 5 of the IBD subjects were on medication, including 21 on a tumor necrosis factor inhibitor (with or without immunomodulators; ie, methotrexate or 6-mercaptopurine), 2 exclusively on an immunomodulator, and 9 on mesalamine therapy. The probands and their siblings underwent an extensive characterization of their neonatal and early life medical history. No differences were seen between children with IBD and their healthy siblings in mode of delivery (vaginal vs Cesarean), birth order, gestational age, birth weight, perinatal disease, maternal antibiotic use predelivery, maternal age, and early life antibiotic treatment. The cohort was predominantly of European ancestry, with the majority self-identifying as Jewish (60 individuals). Two parents were of non-European ancestry (1 Asian, 1 Hispanic), accounting for 4 mixed-race children in the cohort.
Table 1.
Cohort Demographics
| Probands | Siblings with IBD | Healthy siblings | Parents | |
|---|---|---|---|---|
| N | 21 | 6 | 21 | 42 |
| Crohn's disease | 17 | 5 | 4 | |
| Ulcerative colitis | 4 | 1 | 5 | |
| Sex | ||||
| Male | 14 | 5 | 10 | 21 |
| Female | 7 | 1 | 11 | 21 |
| Race/ethnicity | ||||
| Caucasian | 19 | 6 | 19 | |
| Jewish | 13 | 4 | 15 | |
| Mixed | 2 | 0 | 2 | |
| Age at diagnosis, y | 10 | 18.4 | ||
| Age at sampling, y | 13.6 | 19.7 | 12.7 | |
| Crohn's disease location | ||||
| Small intestine | 15 | 5 | ||
| Colon involvement | 10 | 4 | ||
| UGI involvement | 6 | 1 | ||
| Perianal disease | 7 | 1 | ||
| Colon only | 2 | 0 | ||
| IBD medications at sampling | ||||
| Anti-TNF | 16 | 3 | 2 | |
| Methotrexatea | 10 | 1 | 0 | |
| 6-MP/azathioprinea | 2 | 0 | 0 | |
| Mesalamine | 6 | 1 | 2 | |
| None | 1 | 1 | 3 | |
| Birth order | 1.7 | 1.3 | 1.9 | |
| Mode of delivery | ||||
| Vaginal | 16 | 4 | 15 | |
| Cesarean | 5 | 2 | 6 | |
| Gestational age, wkb | 0.2 | −0.3 | 0.0 | |
| Birth weight, lb | 7.4 | 6.8 | 7.0 | |
| Perinatal disease | 8 | 2 | 3 | |
| Anemia | 3 | 0 | 1 | |
| Preterm labor | 1 | 2 | 0 | |
| Preeclampsia | 1 | 0 | 0 | |
| Other | 3 | 0 | 2 | |
| Antibiotics predelivery | 3 | 1 | 3 | |
| Maternal age at delivery, y | 33.9 | 32 | 33.4 | |
| Exclusive breastfeedingc | 20 | 4 | 17 | |
| ≥2 antibiotics during first year | 4 | 1 | 4 |
NOTE. There were no statistically significant differences in metadata between children with IBD and healthy siblings.
6-MP, 6-mercaptopurine; TNF, tumor necrosis factor; UGI, upper gastrointestinal.
Immunomodulators (methotrexate, 6-mercaptopurine) were combined with anti–tumor necrosis factor therapy in all except 2 patients.
Gestational age was expressed as weeks from full-term.
Exclusive breastfeeding was for at least 4 months after birth.
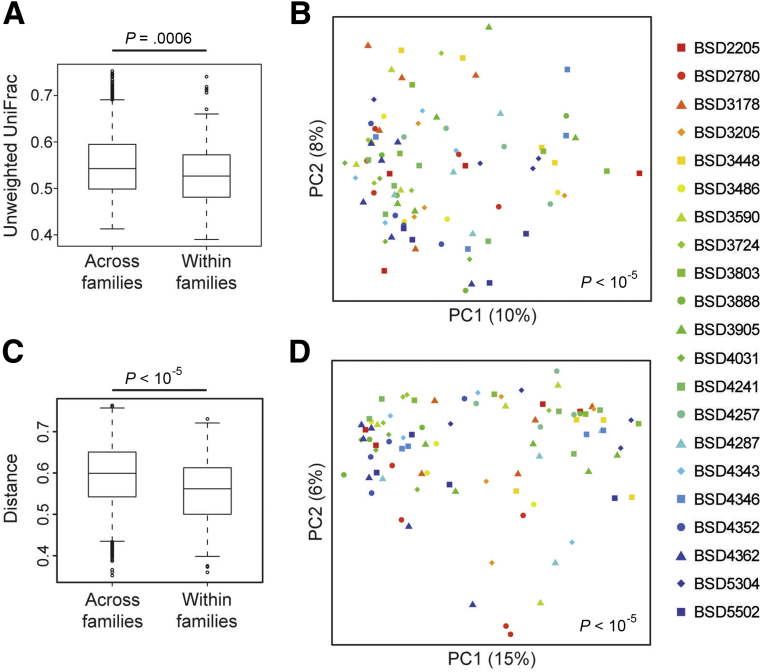
High-depth 16S rRNA sequencing was performed on fecal samples (mean, 620,720 reads per sample) to detect low-abundance taxa that may be critical for IBD susceptibility. Consistent with previous studies, CD patients had lower microbial diversity than both UC patients and healthy individuals as measured by 3 metrics: Chao1 (richness), Faith’s phylogenetic diversity, and the Shannon index (evenness) (CD vs non-IBD, P < .0001 for all measures; CD vs UC, P < .05–.005). Differences in microbial composition across samples were visualized using principal coordinate analysis (PCoA). Samples formed a gradient by IBD status and microbial diversity, both of which were highly significantly associated with overall composition using Adonis, a nonparametric method of analysis of variance (Figure 1A). These findings were confirmed using Bray–Curtis or unweighted UniFrac distances (data not shown). CD patients and healthy relatives mostly occupied distinct regions of the PCoA space but had substantial overlap; UC patients were interspersed throughout. A subset of Crohn’s patients in this cohort had increased fecal calprotectin levels to greater than 2000 μg/g despite being in clinical remission, but calprotectin was not associated with overall microbial composition. Microbial composition was found to vary across families (Adonis, P < 10-5) (Figure 2A and B).
Figure 2.
Microbial composition and metabolomics vary across families. (A) Box plots showing unweighted UniFrac distances for all pairwise combinations of individuals across families or within the same family were generated in QIIME using the function make_distance_boxplots. Lower values indicate a greater similarity of microbial communities. Increased similarity when comparing pairs of samples within families relative to the similarity of pairs of samples from different families implies an effect of family on microbial composition. This approach has been used in studies comparing monozygotic and dizygotic twins to show a role for genes in regulating microbial composition.51 Statistical significance was calculated in QIIME using t tests with 100,000 Monte Carlo simulations. (B) PCoA plot showing microbial composition divided by family (each symbol/color combination represents 1 of the 21 families in this cohort). P value was calculated using Adonis. (C) Box plots showing root square Jensen–Shannon divergence (distance) of the fecal metabolome for all pairwise combinations of individuals across and within IBD groups or families. Statistical significance was calculated using t tests with 100,000 Monte Carlo simulations. (D) PCoA plot showing metabolomics data by family.
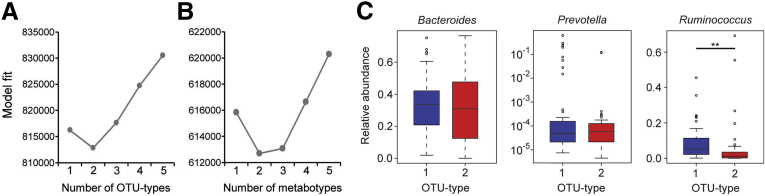
It has been reported that the human fecal microbiome can be divided into discrete community types.21, 29 To investigate whether variation in microbial composition in this cohort could be represented by microbial community types, we classified individuals using a conservative, rigorous method based on Dirichlet multinomial mixture modeling of OTUs at the 97% sequence identity threshold, roughly corresponding to species.28 This model best fit the data when the cohort was divided into 2 groups that we termed OTU types, which were detectable at a sequence depth of 10,000 reads or more per sample (Figure 3A, and data not shown). These OTU types did not correspond to the 3 enterotypes reported in a genus-level analysis of European, American, and Japanese fecal microbiota that were differentiated by abundance of Bacteroides, Prevotella, and Ruminococcus (Figure 3C).21 OTU type 2 was associated strongly with IBD, particularly CD (Figure 1B). Nineteen of the 21 families included at least 1 individual with IBD who carried OTU type 2, indicating that the association of this microbial community state with IBD was not a family-specific trait. Although most healthy relatives had OTU type 1, 10 carried OTU type 2. In PCoA plots, OTU type 1 encompassed a region with high microbial diversity whereas OTU type 2 mapped to a lower-diversity region (Figure 1A). The difference in microbial α diversity between OTU types was observed in separate comparisons of CD subjects and healthy relatives, and confirmed using phylogenetic diversity and the Shannon index (data not shown). However, within the same OTU type, differences in microbial diversity between CD and non-IBD did not reach significance (Figure 1C). Hence, OTU type 2 represents a low-diversity microbial state independent of IBD status. Two families contained 5 of 10 of the healthy relatives with OTU type 2, although the relationship between OTU type and family among healthy relatives did not reach statistical significance (Fisher exact test, P = .08) (Table 2).
Figure 3.
Dirichlet multinomial mixture models support the presence of 2 OTU types and 2 metabotypes. (A and B) The model fit is shown for the given number of (A) OTU types and (B) metabotypes estimated from Dirichlet multinomial mixture models using the Laplace approximation. The exponent of the difference in model fit is the estimated probability of one model over another. (C) No statistically significant difference in Bacteroides or Prevotella abundance was observed between the 2 OTU types in our cohort. Ruminococcus was increased in OTU type 1 but this was not the exclusive driver of OTU type because 2 individuals with OTU type 2 had greater Ruminococcus levels than all individuals with OTU type 1. **P < .005.
Table 2.
Metabotypes and OTU Types of Healthy First-Degree Relatives Are Associated With Family
| Family | OTU type |
Metabotype |
||
|---|---|---|---|---|
| 1 | 2 | 1 | 2 | |
| BSD2205 | 2 | 0 | 2 | 0 |
| BSD2780 | 1 | 0 | 1 | 0 |
| BSD3178 | 2 | 1 | 1 | 2 |
| BSD3205 | 2 | 0 | 0 | 2 |
| BSD3448 | 0 | 3 | 0 | 3 |
| BSD3486 | 2 | 0 | 2 | 0 |
| BSD3590 | 2 | 0 | 1 | 1 |
| BSD3724 | 3 | 0 | 3 | 0 |
| BSD3803 | 2 | 0 | 1 | 1 |
| BSD3888 | 3 | 0 | 3 | 0 |
| BSD3905 | 3 | 0 | 2 | 1 |
| BSD4031 | 1 | 1 | 2 | 0 |
| BSD4241 | 4 | 0 | 2 | 2 |
| BSD4257 | 1 | 2 | 1 | 2 |
| BSD4287 | 2 | 0 | 1 | 1 |
| BSD4343 | 2 | 1 | 3 | 0 |
| BSD4346 | 2 | 0 | 2 | 0 |
| BSD4352 | 2 | 0 | 2 | 0 |
| BSD4362 | 4 | 1 | 5 | 0 |
| BSD5304 | 1 | 1 | 1 | 1 |
| BSD5502 | 3 | 0 | 3 | 0 |
| Total | 44 | 10 | 38 | 16 |
| P value | .08 | .03 | ||
NOTE. Contingency tables are shown for OTU type and metabotype of the 54 non-IBD individuals across the 21 families in this cohort. P values were calculated using the Fisher exact test. Two families are highlighted (BSD4257, BSD3448) that contain at least 2 individuals with the IBD-associated OTU type and metabotype.
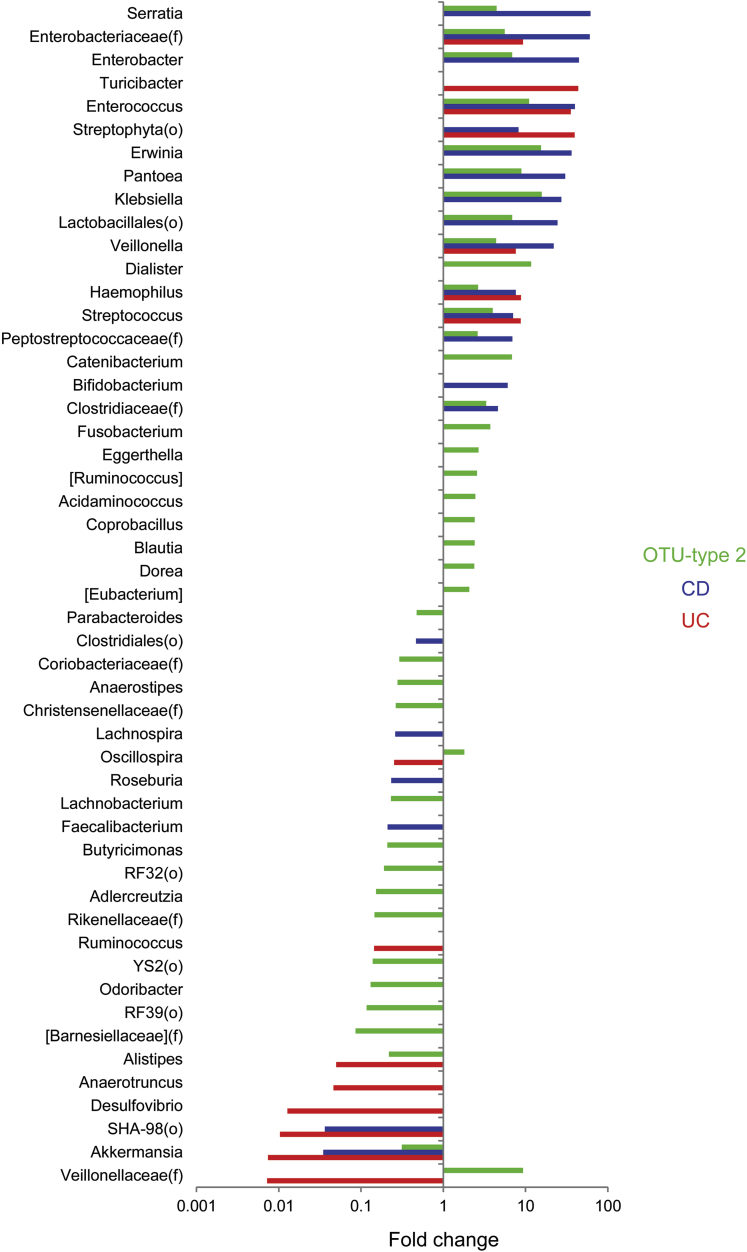
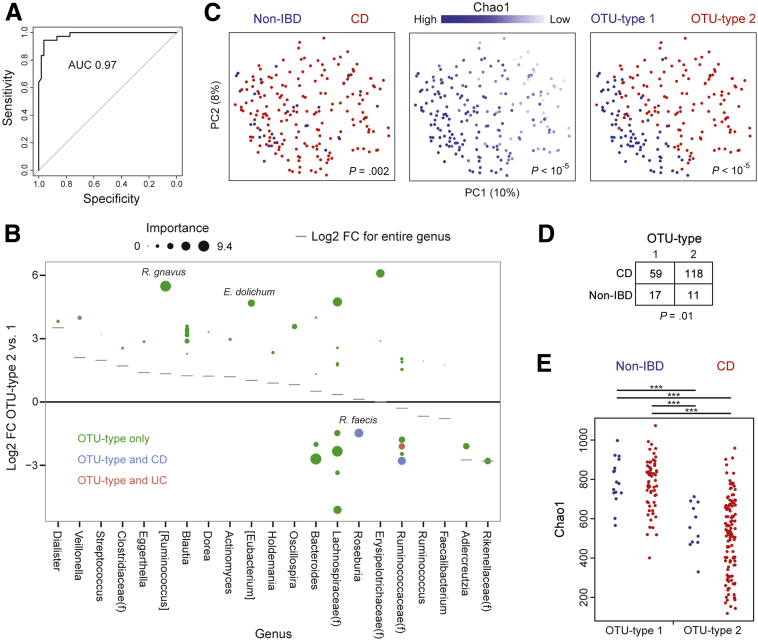
We then identified taxa associated with IBD status and OTU type using DESeq2, an algorithm that uses an empiric Bayesian approach to shrink dispersion and fits the data to negative binomial models. At the genus level, microbial community type was associated with shifts in the abundance of 40 microbial genera compared with 21 for CD and 15 for UC (Figure 4). Similarly, far more OTUs were differentially abundant between the 2 OTU types (752) than between non-IBD individuals and either CD (162) or UC (44) patients (Figure 1D and Supplementary Table 1). Rarefaction analysis showed that the number of differentially abundant microbial taxa increased with greater sequence depth, whereas α and β diversity comparisons were largely unaffected by sequence depth greater than 3000 (data not shown). OTU type 2 was enriched in abundant members of the Blautia (Ruminococcus), and Bacteroides genera and depleted in an abundant member of the Ruminococcus genus (Figure 1E). Some OTUs, including Veillonella dispar, Veillonella parvula, Blautia producta, Haemophilus parainfluenzae, and multiple Enterobacteriaceae, were associated independently with both OTU type and CD. The CD-specific microbial signature included increased Bifidibacterium adolescentis and Parabacteroides distasonis and decreased F prausnitzii and Bacteroides fragilis. OTU type 2, CD, and UC were associated with decreased Akkermansia mucinophila (Figures 1E and 4).
Figure 4.
Microbial genera associated with IBD status and OTU type. Fold-change is shown for genera associated with CD (vs non-IBD), UC (vs non-IBD), or OTU type (2 vs 1) with q less than .05 in multivariate DESeq2 models including sex, Jewish ancestry, current anti–tumor necrosis factor therapy, mode of delivery, family group, IBD diagnosis, and OTU type as covariates. Some taxa represent unclassified members of the indicated family (f) or order (o).
Prediction of Microbial Community Types in an Independent Pediatric CD Cohort
We then investigated whether the 2 OTU types described in this cohort could be mapped to the RISK cohort, consisting of new-onset pediatric Crohn’s disease patients and children presenting with gastrointestinal symptoms but without evidence of inflammatory disease.9 We first created a random forest classifier for microbial community type using the family cohort data. This classifier was highly accurate in 10-fold cross-validation, with an area under the curve (AUC) of the receiver operating characteristic curve of 0.97 (Figure 5A). The relative contribution of OTUs to the classifier was expressed as importance scores, which measure the loss in accuracy of the random forest classifier when that feature is permutated randomly. The OTUs that had the greatest impact on the random forest classifier for OTU types were [Ruminococcus] gnavus (increased in OTU type 2), an unidentified Bacteroides (increased in OTU type 1), 2 unidentified members of the Lachnospiraceae family (1 associated with each OTU type), and Roseburia faecis (increased in OTU type 1), of which only R faecis was associated independently with IBD (Figure 5B). The classifier then was applied to stool samples in the RISK cohort from patients (n = 177) and controls (n = 28) to identify OTU types in this independent cohort. These stool samples formed a gradient by microbial diversity similar to that of the stool samples from the family cohort (Figures 1A and 5C). As in the family cohort, predicted OTU type 2 in the RISK cohort was associated with CD as well as with lower microbial diversity irrespective of disease status (Figure 5D and E).
Figure 5.
Microbial community types were predicted in an independent cohort of pediatric CD patients using a random forest classifier. (A) A classifier was created to predict OTU type in the family cohort using 57 OTUs that were differentially abundant in DESeq2 models and were present in at least 30% of samples. The receiver operating characteristic curve of this classifier based on 10-fold cross-validation is shown. (B) Log2 FC between OTU types 2 and 1 in the family cohort for OTUs included in the random forest classifier. Size is proportional to the importance score of the OTU in the classifier, which measures the loss in accuracy of the classifier when the OTU is permutated randomly. (C) PCoA plots visualizing microbial composition in the RISK cohort of pediatric CD patients and controls with gastrointestinal symptoms but no evidence of inflammatory disease. Samples are colored by IBD status, Chao1, or OTU type predicted from the classifier shown in panel A. P values were calculated using Adonis. (D) Contingency table of IBD status by predicted OTU type in the RISK cohort. P value was calculated using the Fisher exact test. (E) Chao1 was determined by IBD status and predicted OTU type in the RISK cohort. P values were calculated using the Mann–Whitney U test. ***P < .0001.
Intestinal Metabotypes Are Associated With IBD
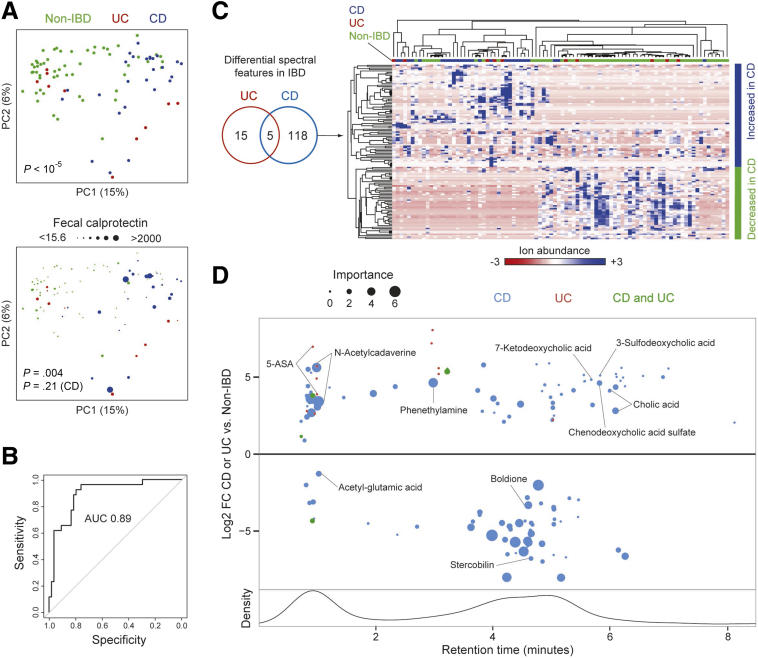
We performed untargeted UPLC/ToFMS metabolomics to further investigate the functional consequences of altered microbial composition in our family cohort. Our pipeline detected 4626 spectral features across 2 acquisition modes. Differences across samples in the intensities of these spectral features were visualized by PCoA. Healthy relatives and CD patients mostly separated to opposite ends of the PCoA space with an area of overlap between the two, although UC patients were found in both regions (Figure 6A). Metabolomics signatures also varied by family (Adonis, P < 10-5), showing that shared genetic and/or environmental factors within families influenced the composition of the metabolome (Figure 2C and D). A random forests classifier was trained on the metabolomics data to determine the discriminative power of fecal metabolites to predict CD from non-IBD. Differences in metabolomics profiles between CD and non-IBD were sufficient for the classifier to have an AUC of 0.89 in 10-fold cross-validation (Figure 6B). We then used multivariate models to identify specific spectral features associated with CD or UC, adjusting for covariates including sex, anti–tumor necrosis factor therapy, Jewish ethnicity, mode of delivery, and calprotectin. CD patients had altered abundance of 123 spectral features whereas UC patients showed comparatively little change: 20 spectral features, of which 5 were shared with CD and 2 were validated as mesalamine (a common treatment for UC in this cohort) (Figure 6C). Hierarchical clustering of differential spectral features in CD showed that most Crohn’s patients and a subset of healthy individuals grouped together whereas other non-IBD individuals had a distinct profile. CD-associated metabolites included amino acid derivatives (phenylethylamine and N-acetylcadaverine) and bile acids (cholic acid, 7-ketodeoxycholic acid, chenodeoxycholic acid sulfate, and 3-sulfodeoxycholic acid) (Figure 6D). Metabolites that were increased in non-IBD individuals included a heme degradation product (stercobilin), a glutamate derivative (acetyl-glutamic acid), and a product of steroid catabolism (boldione).
Figure 6.
Metabolomic features of CD also were seen in a subset of healthy relatives. (A) PCoA plots were used to visualize differences in metabolomics profiles across samples as measured by the square root of the Jensen–Shannon divergence. Dots represent fecal samples, colored by IBD status. Dot size corresponds to fecal calprotectin level in the lower panel. P values were calculated using Adonis. Separate calculations were made for the whole cohort and for just the CD subset because increased calprotectin was seen primarily in a subset of CD. (B) A random forest classifier was created to distinguish CD from non-IBD using fecal metabolomics profiles. The receiver operating characteristic curve of this classifier based on 10-fold cross-validation is shown. (C) Venn diagram of the differential spectral features for CD or UC vs non-IBD in multivariate DESeq2 models including sex, Jewish ancestry, current anti–tumor necrosis factor therapy, mode of delivery, family group, and IBD diagnosis as covariates. The heat map shows intensity values for the 123 spectral features with differential abundance in CD vs non-IBD. Intensity is represented on a color scale spanning 3 standard deviations above and below the mean across all samples. Samples and spectral features were clustered hierarchically by Pearson correlation coefficient with the average linkage method. IBD status of samples is indicated by colored boxes above the heat map. (D) Spectral features differentially abundant in CD or UC (as summarized in the Venn diagram in panel C) are shown, arranged by retention time. Effect size is represented as the log2 FC in CD or UC compared with non-IBD in multivariate DESeq2 models. Features associated with both CD and UC are plotted by the log2 FC in CD vs non-IBD. Size is proportional to the importance score of the feature in the random forest classifier for CD. The names of validated metabolites are shown. Lower panel: the density of detected spectral features by retention time on the UPLC column.
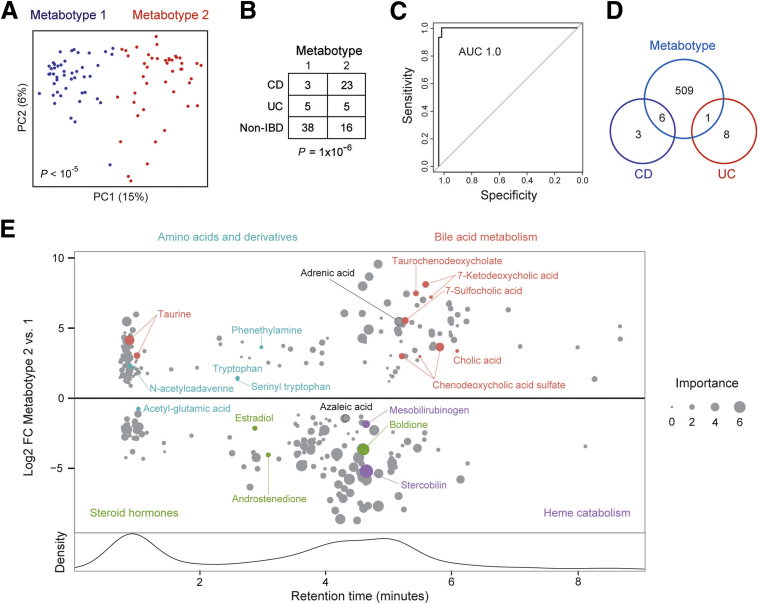
The grouping of some non-IBD individuals with CD patients by hierarchical clustering suggested the existence of metabotypes in this cohort analogous to the 2 OTU types. Dirichlet multinomial mixture models supported 2 metabotypes (Figure 3B). PCoA plots showed separation of individuals in the cohort by metabotype and a notable overlap of metabotype 1 with non-IBD individuals (Figures 6A and 7A). As with OTU type, metabotype was found to be correlated highly with IBD status (Fisher exact test, P = 1 × 10-6) and also was correlated with family (P = .03) (Figure 7B and Table 2). A random forest classifier for metabotype was trained with an AUC of 1.0, indicative of the robust difference between the 2 metabotypes (Figure 7C). We then used multivariate models that included metabotype to identify IBD-associated spectral features independent of metabotype (Figure 7D and Supplementary Data File 2). CD and UC were found to have a limited metabolic signature with only 9 differential features each, whereas metabotype was associated with 516 spectral features. This suggested that metabotype accounted for much of the variation in the metabolome that had been attributed to disease status without considering metabotype. Metabotype 2 was characterized by increased levels of the same bile acids and amino acid derivatives previously attributed to CD as well as taurine, tryptophan, serinyl tryptophan, and an omega-6 fatty acid (adrenic acid) (Figure 7E). Metabotype 1 had increased levels of steroid hormones (estradiol, androstenedione, and boldione), products of heme catabolism (stercobilin and mesobilirubinogen), azaleic acid, and acetyl-glutamic acid. Estradiol and androstenedione abundance was higher in females than in males, but in both sexes increased levels of these hormones were seen in metabotype 1 (data not shown).
Figure 7.
Fecal metabotypes are associated with IBD status. (A) PCoA plots of the metabolomics profile of fecal samples colored by metabotype (determined using Dirichlet multinomial mixture models). (B) Contingency table of IBD status by metabotype. The P value was calculated using the Fisher exact test. (C) A random forest classifier was created to predict metabotype. The receiver operating characteristic (ROC) curve of this classifier based on 10-fold cross-validation is shown. (D) Venn diagram showing the number of differential spectral features for metabotypes 2 vs 1, CD vs non-IBD, and UC vs non-IBD in multivariate DESeq2 models including sex, Jewish ancestry, current anti–tumor necrosis factor therapy, mode of delivery, family group, IBD diagnosis, and metabotype as covariates. (E) Metabotype-associated spectral features are shown, plotted by retention time. Effect size is represented as the log2 FC of metabotypes 2 vs 1 in multivariate DESeq2 models. Size is proportional to the importance score of the feature in the random forest classifier for metabotype. The names of validated metabolites are shown, color-coded by metabolite category.
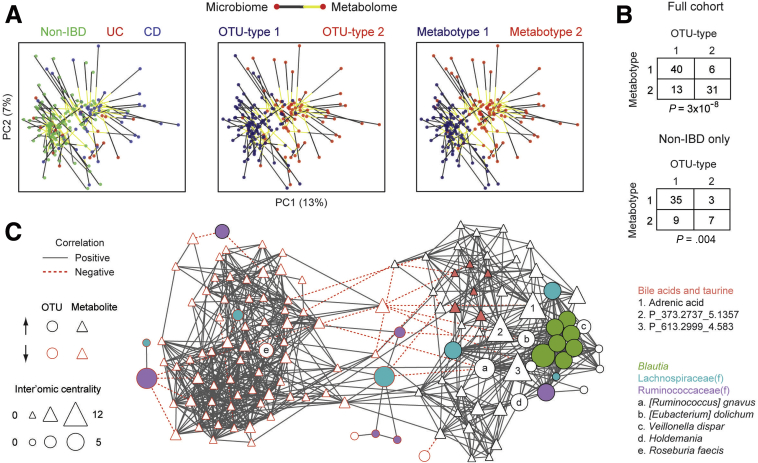
OTU Type and Metabotype Are Highly Correlated
The similarity between OTU types and metabotypes suggested a functional relationship between the microbiome and metabolome. This was visualized using Procrustes to rotate and rescale the metabolomics PCoA data to allow it to be superimposed onto the microbiome PCoA data. The 2 data sets showed a high degree of concordance that was statistically significant in Monte Carlo simulations with a P value less than .0001 (Figure 8A). Superimposed microbiome and metabolomics data were separated not only by IBD status but also by OTU type and metabotype. Contingency table analysis showed a strong association between OTU types and metabotypes (Fisher exact test, P = 3 × 10-8) that was present even when considering only the healthy relatives in this cohort (P = .004) (Figure 8B).
Figure 8.
Microbe–metabolite interactions between the IBD-associated microbial community type and metabotype. (A) The PCoA plot for the metabolome (visualized in Figure 6A) was rotated and rescaled using Procrustes, then superimposed on the PCoA plot for the microbiome (visualized in Figure 1A). Points represent 16S rRNA and UPLC/ToFMS data, color-coded by IBD status, OTU type, or metabotype. Each line connects the microbial and metabolomics data from one individual. (B) Contingency table of OTU type and metabotype for the full cohort and only non-IBD individuals. P values were calculated using the Fisher exact test. (C) An inter’omic network was constructed with nodes representing OTU-type–associated microbes and metabotype-associated spectral features with an importance score greater than 2 in random forest classifiers. Microbe (circle) and metabolite (triangle) nodes are outlined in red if they are decreased in OTU type 2 or metabotype 2, respectively. Node size reflects inter’omic degree centrality—the number of connections to nodes of the opposite data type (ie, microbe–metabolite pairs). Edges represent statistically significant correlations (q < 0.05) between microbe–microbe, metabolite–metabolite, or microbe–metabolite pairs. These correlations were made using residuals from multivariate DESeq2 models adjusting for sex, Jewish ancestry, current anti–tumor necrosis factor therapy, mode of delivery, family group, IBD diagnosis, and OTU type/metabotype. This approach highlights correlations that cannot be explained by these factors. Selected microbes and metabolites are indicated by fill color, numbers, or letters.
We next sought to investigate the individual microbe–metabolite associations that underpin the relationship between microbial community type and metabotype. An inter’omic network was constructed using all OTU-type–associated microbes and metabotype-associated metabolites with an importance score of 2 (ie, 2 standard deviations above having no effect on classification) (Figure 8C). Edges in this network represented statistically significant correlations between 2 microbes, 2 metabolites, or a microbe and metabolite after adjustment for sex, Jewish ancestry, current anti–tumor necrosis factor therapy, mode of delivery, family group, IBD diagnosis, and OTU type/metabotype. Inter’omic degree centrality—the number of edges to nodes of the opposite data type (ie, microbe–metabolite correlations)—was calculated to highlight microbes associated with the largest number of metabolites and vice versa. Three metabolites were found to have a high inter’omic centrality, including adrenic acid and 2 unidentified metabolites, as a result of association with a group of co-correlated Blautia OTUs, [Ruminococcus] gnavus, [Eubacterium] dolichum, Veillonella dispar, and Holdemania. The metabotype 2 associated bile acids were highly co-correlated in the network. Two bile acids, 7-sulfocholic acid and chenodeoxycholic acid sulfate, were associated with a Lachnospiraceae OTU enriched in OTU type 2.
Increased Incidence of Elevated Fecal Calprotectin in Healthy First-Degree Relatives With the IBD-Associated Microbial Community Type
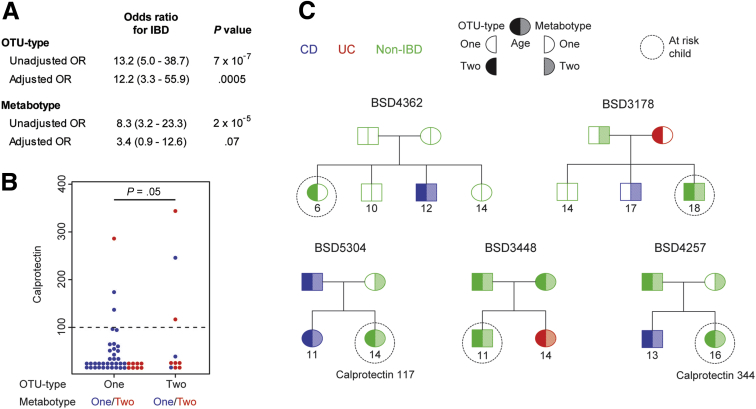
We then evaluated the relative association of OTU type and metabotype with IBD in multivariate logistic regression models. OTU type and metabotype each had a statistically significant correlation with IBD in univariate logistic regression with odds ratios of 13.2 and 8.3, respectively, although only OTU type retained significance in multivariate logistic regression models that included OTU type, metabotype, parent/child status, and Jewish ethnicity (Figure 9A). We investigated whether OTU type was associated with an increased risk of subclinical intestinal inflammation as evidenced by increased fecal calprotectin.35 The majority of samples had calprotectin levels below the limit of detection. We selected 100 μg/g as a cut-off value for increased fecal calprotectin based on a meta-analysis of the use of calprotectin to differentiate IBD from healthy controls or irritable bowel syndrome.36 Only 3 of 44 healthy relatives (7%) with OTU type 1 and 3 of 10 relatives (30%) with OTU type 2 had increased calprotectin levels (Figure 9B). This corresponded to an odds ratio of 5.9 (confidence interval, 0.9–37.9) for the association of OTU type 2 with increased calprotectin level, with a P value of .05. Five of 21 healthy siblings carried the IBD-associated OTU type, of whom 4 also had the IBD-associated metabotype (Figure 9C).
Figure 9.
Healthy first-degree relatives with the IBD-associated microbial community type are at higher risk for increased fecal calprotectin. (A) Unadjusted odds ratios (ORs) and 95% confidence intervals were calculated for IBD risk using univariate logistic regression. Adjusted ORs were obtained from a multivariate logistic regression model incorporating OTU type, metabotype, parent/child status, and Jewish ethnicity. (B) Dot plot showing fecal calprotectin in the 54 healthy first-degree relatives, stratified by OTU type. The lowest 2 rows of dots represent samples below or near the limit of detection of 15.6 μg/g. The P value was calculated from univariate logistic regression for increased calprotectin level (cut-off value is indicated by the dashed line). (C) Pedigrees are shown for 5 families with healthy children who carry an IBD-associated OTU type. Two of these children had an increased fecal calprotectin level.
Discussion
Although there has been much focus on the microbiota associated with disease activity in IBD, a separate and experimentally challenging concept is the possibility of distinctive basal states of the intestinal microbiome that confer disease susceptibility. We found that variation in the microbiome and metabolome among disease-quiescent IBD patients and their first-degree relatives can be represented by 2 intestinal OTU types and metabotypes. The existence of an IBD-associated OTU type and metabotype in healthy relatives suggests that dysbiosis with its associated metabolic products may be a pre-existing trait that precedes the acquisition of disease. The bacterial taxa characterizing the IBD-associated OTU type included multiple OTUs identified as Enterobacteriaceae and 2 putatively identified as Prevotella copri. This parallels studies of Toll-like receptor 5 knockout mice and NLRP6 inflammasome knockout mice showing that colitis susceptibility was associated with blooms of Enterobacteriaceae and Prevotellaceae, respectively.5, 37 Although unaffected family members were not evaluated endoscopically for IBD, the study design included clinical assessment at recruitment and periodic monitoring for acquisition of clinical disease. Healthy individuals with the IBD-associated OTU type and metabotype were found predominantly in several families. Such states may have arisen owing to shared founder effects, dietary factors, and genetic variants between patients and some of their family members that influence host–microbiome homeostasis.11, 12, 38
The metabotypes identified in this cohort may be driven by shifts in bacterial metabolism of host-derived factors such as bile acids due to altered microbial community structure, explaining the close correlation of metabotype with OTU type. Metabotype 2 showed increased levels of cholic acid (a primary bile acid) and a sulfated derivative (7-sulfocholic acid), as well as taurine-conjugated and sulfated derivatives of the other primary bile acid, chenodeoxycholic acid. These findings parallel a study that reported increased conjugated and sulfated bile acids in the feces of CD patients attributed to reduced capacity of the CD microbiome to deconjugate and desulfate bile acids.39 The increased levels of cholic acid may reflect reduced conversion to other bile acid metabolites. The other validated bile acid associated with metabotype 2, 7-ketodeoxycholic acid, has not been reported previously to be increased in CD or UC patients and may reflect altered bacterial metabolism of deoxycholic acid (a secondary bile acid).40, 41 The bile acid shifts in metabotype 2 likely are relevant to the development of dysbiosis and IBD given the considerable evidence that bile acid signaling through farnesoid X receptor and TGR5 is critical for maintaining the epithelial barrier and promoting antibacterial defenses.42, 43, 44 There also are recent data indicating that bile acids including cholic acid can induce enterochromaffin cells to secrete serotonin, a neurotransmitter shown to promote experimental colitis.45, 46 This could be potentiated by the increased levels in metabotype 2 of tryptophan, an essential amino acid that is the precursor to serotonin and that also can be metabolized by microbes into immunomodulatory metabolites including kynurenines and an aryl hydrocarbon receptor ligand, indole-3-aldehyde.47, 48 The possible causal link between OTU type and metabotype, with its associated alteration of bile acid metabolism, warrants mechanistic investigation in models of germ-free mice colonized with feces of each OTU type/metabotype. Interestingly, 19 of 90 subjects had discordant OTU type and metabotype, suggesting that other factors besides the microbiome such as diet also could influence the metabolome.49
A strength of this study was that its central finding (a disease-related microbial community type detectable in healthy subjects) also was observed in the RISK cohort, which consisted of more than 200 unrelated pediatric patients and controls from a large network of pediatric gastroenterology clinics across North America.9 At least 2 prior publications have reported that IBD patients cluster into 2 groups by microbiome composition.6, 10 In an early Sanger sequencing study of ileal and colonic surgical specimens, OTU presence/absence was used to group non-IBD subjects and the majority of IBD patients into 1 cluster while a subset of IBD patients formed a second cluster with increased Proteobacteria and Actinobacteria OTUs. The second cluster was associated with abscess, suggesting that it was driven by CD patients with penetrating disease. A second study used genus level fecal microbiome data to group CD patients with active disease into near and far clusters to reflect their closeness to the microbiome of healthy controls.10 The far cluster was characterized by lower microbial diversity and differential abundance of 30 genera, of which 7 had concordant differences between the 2 OTU types defined here (Figure 4). The present study builds on these observations by showing that microbial community types found in IBD patients also are present in healthy relatives. Furthermore, microbial community types were associated with metabolomics profiles and subclinical intestinal inflammation, indicating functional relevance of this community structure. A limitation of this study was the small number of high-risk families and their lack of ethnic and racial diversity. However, we are not aware of other published family based or longitudinal cohorts analyzed for microbiome and metabolites. Their availability will be important to validate the present observations and refine them in broader populations that include other racial or ethnic groups.
The microbial state defined by OTU type 2 was associated not just with diagnosed IBD but also with a potential subclinical disease state defined by increased fecal calprotectin in healthy first-degree relatives. Nevertheless, most individuals with the IBD-associated OTU type and metabotype had normal calprotectin level. This suggests that these microbial changes are not simply the result of subclinical inflammation, although we cannot rule out that OTU type and metabotype are more sensitive biomarkers for low-grade inflammation than calprotectin. We speculate that healthy individuals with an IBD-associated microbial community type and metabotype have a predisease state that increases the risk for the future development of both subclinical and overt IBD. Validating this interpretation will require prospective longitudinal studies to assess the incidence of IBD in individuals stratified by OTU type, metabotype, and calprotectin, and to assess the stability of OTU types and metabotypes across time and diet. Analytically, it also will require bioinformatically fastidious methods to refine and simplify the definition of microbial community types that robustly are preserved between independent cohorts and data sets, and illuminate and resolve the methodologic and ecologic questions raised by prior studies of intestinal microbiome clustering.50 Future cohort studies would need to incorporate known genetic and epidemiologic risk factors to determine the risk for IBD attributable to OTU type and metabotype after adjusting for these other factors. In addition, studies of germ-free mice colonized with feces from healthy relatives stratified by OTU type and metabotype are warranted to investigate whether these microbial states influence susceptibility to experimental models of IBD. If validated, prospective identification of healthy individuals with a high-risk microbiome/metabolome creates the opportunity to prevent disease development by targeting the dysbiosis and/or its metabolic consequences in the intestine.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was funded by The Helmsley Charitable Trust, The Crohn’s and Colitis Foundation of America, the Fineberg Foundation, United States Public Health Service (PO1DK046763), National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (T32DK07180-39 to J.P.J., DK062413 and DK046763-19 to D.P.B.M.), National Institutes of Health/National Institute of Allergy and Infectious Diseases (AI067068 to D.P.B.M.), National Institutes of Health/Agency for Healthcare Research and Quality (HS021747 to D.P.B.M.), the National Institutes of Health/National Center for Advancing Translational Science (UL1TR000124), and the Cedars-Sinai F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute. D.P.B.M. was also supported by the European Union and The Joshua L. and Lisa Z. Greer Chair in Inflammatory Bowel Disease Genetics.
Contributor Information
Jonathan Braun, Email: jbraun@mednet.ucla.edu.
Marla Dubinsky, Email: Marla.dubinsky@mssm.edu.
Supplementary Material
References
- 1.Ng S.C., Bernstein C.N., Vatn M.H. Geographical variability and environmental risk factors in inflammatory bowel disease. Gut. 2013;62:630–649. doi: 10.1136/gutjnl-2012-303661. [DOI] [PubMed] [Google Scholar]
- 2.McGovern D.P., Kugathasan S., Cho J.H. Genetics of inflammatory bowel diseases. Gastroenterology. 2015;149:1163–1176 e2. doi: 10.1053/j.gastro.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sellon R.K., Tonkonogy S., Schultz M. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garrett W.S., Lord G.M., Punit S. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elinav E., Strowig T., Kau A.L. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frank D.N., St Amand A.L., Feldman R.A. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willing B.P., Dicksved J., Halfvarson J. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854 e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 8.Morgan X.C., Tickle T.L., Sokol H. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gevers D., Kugathasan S., Denson L.A. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis J.D., Chen E.Z., Baldassano R.N. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's disease. Cell Host Microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee D., Albenberg L., Compher C. Diet in the pathogenesis and treatment of inflammatory bowel diseases. Gastroenterology. 2015;148:1087–1106. doi: 10.1053/j.gastro.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs J.P., Braun J. Immune and genetic gardening of the intestinal microbiome. FEBS Lett. 2014;588:4102–4111. doi: 10.1016/j.febslet.2014.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham D.B., Xavier R.J. From genetics of inflammatory bowel disease towards mechanistic insights. Trends Immunol. 2013;34:371–378. doi: 10.1016/j.it.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorrestein P.C., Mazmanian S.K., Knight R. Finding the missing links among metabolites, microbes, and the host. Immunity. 2014;40:824–832. doi: 10.1016/j.immuni.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furusawa Y., Obata Y., Fukuda S. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 16.Marchesi J.R., Holmes E., Khan F. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- 17.Bjerrum J.T., Wang Y., Hao F. Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn's disease and healthy individuals. Metabolomics. 2015;11:122–133. doi: 10.1007/s11306-014-0677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokol H., Pigneur B., Watterlot L. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thjodleifsson B., Sigthorsson G., Cariglia N. Subclinical intestinal inflammation: an inherited abnormality in Crohn's disease relatives? Gastroenterology. 2003;124:1728–1737. doi: 10.1016/s0016-5085(03)00383-4. [DOI] [PubMed] [Google Scholar]
- 20.Montalto M., Curigliano V., Santoro L. Fecal calprotectin in first-degree relatives of patients with ulcerative colitis. Am J Gastroenterol. 2007;102:132–136. doi: 10.1111/j.1572-0241.2006.00884.x. [DOI] [PubMed] [Google Scholar]
- 21.Arumugam M., Raes J., Pelletier E. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong M., Jacobs J.P., McHardy I.H. Sampling of intestinal microbiota and targeted amplification of bacterial 16S rRNA genes for microbial ecologic analysis. Curr Protoc Immunol. 2014;107:7.41.1–7.41.11. doi: 10.1002/0471142735.im0741s107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso J.G., Lauber C.L., Walters W.A. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caporaso J.G., Kuczynski J., Stombaugh J. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McArdle B.H., Anderson M.J. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001;82:290–297. [Google Scholar]
- 26.Goudarzi M., Weber W.M., Mak T.D. Metabolomic and lipidomic analysis of serum from mice exposed to an internal emitter, cesium-137, using a shotgun LC-MS(E) approach. J Proteome Res. 2015;14:374–384. doi: 10.1021/pr500913n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith C.A., O'Maille G., Want E.J. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 28.Holmes I., Harris K., Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One. 2012;7:e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding T., Schloss P.D. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schloss P.D., Westcott S.L., Ryabin T. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 34.McHardy I.H., Goudarzi M., Tong M. Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome. 2013;1:17. doi: 10.1186/2049-2618-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sands B.E. Biomarkers of inflammation in inflammatory bowel disease. Gastroenterology. 2015;149:1275–1285 e2. doi: 10.1053/j.gastro.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 36.von Roon A.C., Karamountzos L., Purkayastha S. Diagnostic precision of fecal calprotectin for inflammatory bowel disease and colorectal malignancy. Am J Gastroenterol. 2007;102:803–813. doi: 10.1111/j.1572-0241.2007.01126.x. [DOI] [PubMed] [Google Scholar]
- 37.Carvalho F.A., Koren O., Goodrich J.K. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J., Linnenbrink M., Kunzel S. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proc Natl Acad Sci U S A. 2014;111:E2703–E2710. doi: 10.1073/pnas.1402342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duboc H., Rajca S., Rainteau D. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–539. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- 40.Jansson J., Willing B., Lucio M. Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS One. 2009;4:e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Gall G., Noor S.O., Ridgway K. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res. 2011;10:4208–4218. doi: 10.1021/pr2003598. [DOI] [PubMed] [Google Scholar]
- 42.Inagaki T., Moschetta A., Lee Y.K. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cipriani S., Mencarelli A., Chini M.G. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS One. 2011;6:e25637. doi: 10.1371/journal.pone.0025637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vavassori P., Mencarelli A., Renga B. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol. 2009;183:6251–6261. doi: 10.4049/jimmunol.0803978. [DOI] [PubMed] [Google Scholar]
- 45.Yano J.M., Yu K., Donaldson G.P. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghia J.E., Li N., Wang H. Serotonin has a key role in pathogenesis of experimental colitis. Gastroenterology. 2009;137:1649–1660. doi: 10.1053/j.gastro.2009.08.041. [DOI] [PubMed] [Google Scholar]
- 47.Vujkovic-Cvijin I., Dunham R.M., Iwai S. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med. 2013;5:193ra91. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zelante T., Iannitti R.G., Cunha C. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 49.Thorburn A.N., Macia L., Mackay C.R. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity. 2014;40:833–842. doi: 10.1016/j.immuni.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 50.Koren O., Knights D., Gonzalez A. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodrich J.K., Waters J.L., Poole A.C. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.