Abstract
Invariant natural killer T (iNKT) cells are innate-like lymphocytes that protect against infection, autoimmune disease and cancer. However, little is known about epigenetic regulation of iNKT cell development. Here, we show that the H3K27me3 histone demethylase UTX is an essential cell-intrinsic factor that controls an iNKT lineage-specific gene expression program and epigenetic landscape in a demethylase activity dependent manner. UTX-deficient iNKT cells exhibited impaired expression of iNKT signature genes due to a decrease in activation-associated H3K4me3 and an increase in repressive H3K27me3 marks within the promoters that UTX occupies. We identified JunB as a novel regulator of iNKT development and show that target gene expression of both JunB and iNKT master transcription factor PLZF was UTX-dependent. We determined iNKT super-enhancers and demonstrated that UTX-mediated regulation of super-enhancer accessibility was a key mechanism for iNKT lineage commitment. These findings reveal how UTX regulates iNKT cell development through multiple epigenetic mechanisms.
Invariant natural killer T (iNKT) cells are a subset of T lymphocytes with a limited T cell antigen receptor (TCR) repertoire that recognizes lipid antigens presented by CD1d molecules on the surface of antigen-presenting cells1,2. The prototypical lipid antigen is the glycosphingolipid α-galactosylceramide (α-GalCer) derived from a marine sponge3, which can be loaded onto CD1d tetramers for the detection of iNKT cells through binding to their invariant TCR. Following antigen recognition, iNKT cells respond rapidly in an innate-like fashion and secrete inflammatory cytokines, including interferon-γ (IFN-γ) and interleukin-4 (IL-4), in copious amounts1,2. This early response influences the outcome of downstream immune reactions and endows iNKT cells with regulatory properties. Due to the diversity of their effector functions, iNKT cells are involved in many pathological processes. Accordingly, they are important in host defense against infections, prevent autoimmune disorders, and protect against cancer1,4.
Deciphering molecular mechanisms that control iNKT lineage specification is essential for developing potential therapeutic applications that target iNKT cells. Commitment to the iNKT lineage involves positive selection of iNKT cell precursors by CD4+CD8+ double-positive (DP) thymocytes that express CD1d-glycolipid complexes5. After positive selection, TCR signaling activates the calcineurin-NFAT pathway that induces expression of transcription factor Egr2 (ref.6). Following its induction, Egr2 activates the iNKT master regulator PLZF (encoded by Zbtb16) and the IL-2–IL-15 receptor common β subunit (CD122, encoded by Il2rb) that lead to subsequent steps of iNKT cell development, comprising cytokine expression and proliferation in response to the IL-15–CD122 axis7–10. T-bet (encoded by Tbx21) is a key transcription factor that regulates iNKT cell differentiation and acquisition of NK cell traits during terminal maturation. After egress from the thymus, iNKT cells mostly reside in the liver and spleen to perform effector functions11. Although recent reports contribute to an understanding how key transcription factors establish iNKT cell identity in a stepwise process12,13, it is unclear how a multitude of transcription factors is orchestrated within an epigenetic framework that controls lineage-specific gene expression in iNKT cells.
During development, cell fate determination relies on the activation of cell type-restricted transcription factors that act at promoters and enhancers of genes. This requires epigenetic programming to ensure the establishment of a proper chromatin organization. Stretch-enhancer elements (often called super-enhancers) play a critical role in the control of cell identity14,15. Dynamic regulation of the histone methylation state at promoters or enhancers by histone-modifying enzymes is a key epigenetic mechanism that impacts lineage-specific gene expression16–18. The polycomb repressive complex 2 (PRC2) catalyzes the trimethylation of histone H3 on lysine 27 (H3K27) that is associated with poised or repressed states of promoters and enhancers17,19,20. By contrast, demethylation of H3K27me3 by the histone demethylases UTX (ubiquitously transcribed tetratricopeptide repeat, X chromosome, encoded by Kdm6a) and JMJD3 (Jumonji C domain-containing protein 3, encoded by Kdm6b) correlates with active chromatin states that facilitate gene expression21–25. UTX and JMJD3 serve important roles in early development21–23,26, epigenetic reprogramming27, cellular differentiation24,28–30, and cancer31,32. The role of UTX in different T lymphocyte subsets has been addressed recently33–35, however, the underlying mechanisms remained to be elucidated.
In this study, we assessed the epigenetic mechanisms through which UTX controls the development of iNKT cells. UTX deficiency resulted in downregulation of iNKT signature genes, including Tbx21, Il2rb, and Klrd1, and blocked iNKT cell development. We demonstrated that H3K27me3 abundance was increased and H3K4me3 abundance was decreased around the promoters of downregulated signature genes in UTX-deficient iNKT cells. We found that UTX partners with the iNKT master regulator PLZF, and UTX-deficient iNKT cells failed to activate PLZF target genes and harbored increased H3K27me3 around their gene promoters. Moreover, we identified a novel role for the AP-1 transcription factor JunB in iNKT cell generation. By uncovering the super-enhancer landscape of iNKT cells, we demonstrated that UTX is required for the accessibility of the super-enhancers that mediate iNKT lineage specification. Thus, we found that UTX engages multiple gene regulatory mechanisms to facilitate lineage-specific gene expression and development of iNKT cells.
RESULTS
UTX is required for iNKT cell development
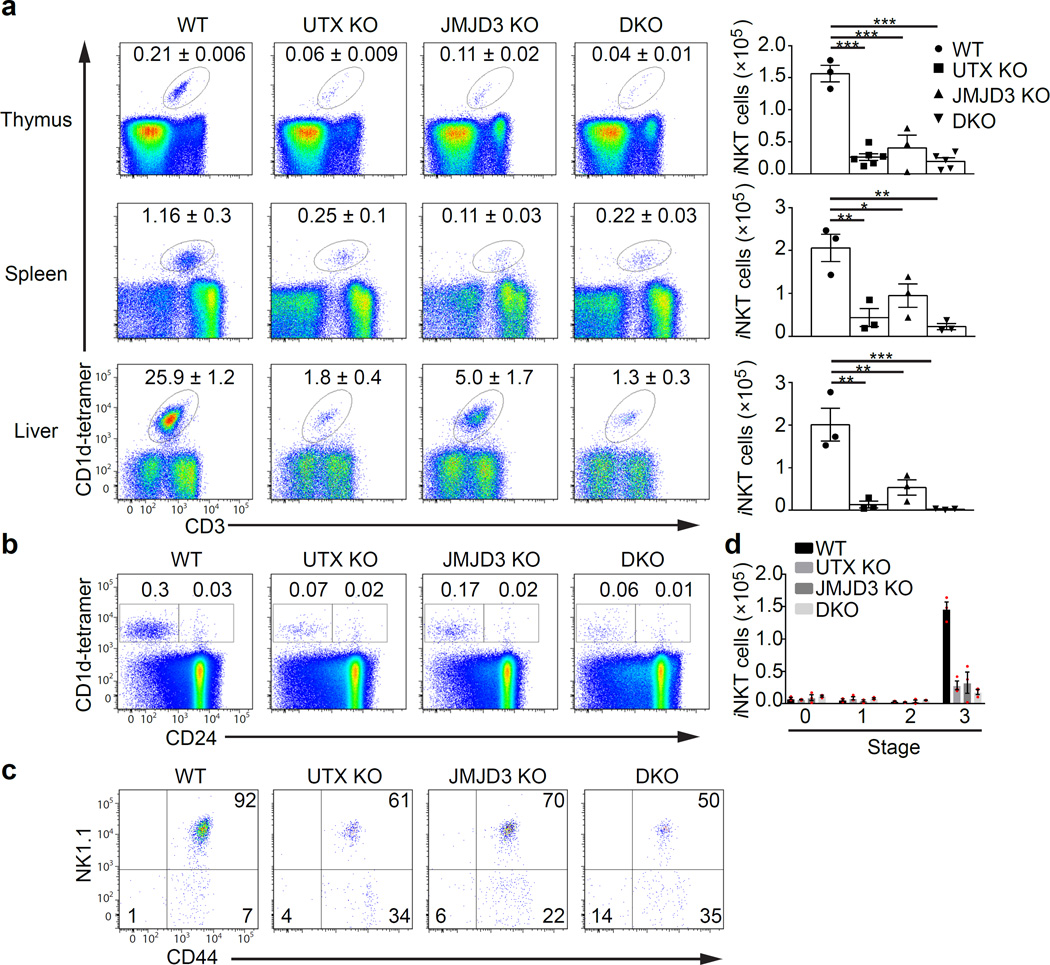
Initially, we determined requirements for UTX (Kdm6a) and JMJD3 (Kdm6b) broadly in the hematopoietic system. For this purpose, we generated conditional UTX-deficient (UTX-KO) and JMJD3-deficient (JMJD3-KO) mice prior to interbreeding with a Vav-Cre allele for gene inactivation in the adult hematopoietic system (Supplementary Fig. 1a–e). We observed that iNKT cell numbers were strikingly decreased in thymus, spleen, and liver of UTX-KO or JMJD3-KO mice as compared to wild-type controls, with a more pronounced effect in UTX deficiency (Fig. 1a). UTX and JMJD3 double-knockout (DKO) animals had a similar phenotype to UTX-KO, suggesting JMJD3 ablation had no additive effects. We confirmed the deletion of Utx and Jmjd3 transcripts in blood, DP thymocytes, and iNKT cells of knockout animals by qPCR (Supplementary Fig. 1f). Using molecular markers to distinguish the stages of iNKT cell development36, analysis of the remaining thymic iNKT cells in UTX- or JMJD3-KO mice revealed a maturation block that prevented iNKT cells from fully entering into stage 3 with a relative accumulation in stages 1 and 2 (Fig. 1b–d). These results indicate that UTX and JMJD3 play a selective role in iNKT cells during blood cell differentiation. Since the iNKT cell phenotype proved to be predominant in UTX deficiency, we focused subsequent analyses on UTX.
Figure 1.
H3K27 demethylases are essential for iNKT cell maturation and development. (a) Frequency of iNKT cells in UTX KO (n = 5 mice), JMJD3 KO (n = 3 mice), or UTX/JMJD3 double-deficient (DKO, n = 5 mice) mice. iNKT cells were detected by staining with anti-CD3 and α-GalCer-loaded CD1d-tetramers (CD1d-tetramer) using flow cytometry. Right panel shows absolute cell numbers of iNKT cells in the different experimental groups. (b–d) Thymic iNKT cells from experimental groups (n = 4 per group) as described in (a) were analyzed for their maturation stages using flow cytometry. (b) Flow cytometry analysis of iNKT cells at stage 0 (CD24+CD1d-tetramer+) and stage 1–3 (CD24−CD1d-tetramer+). Percentages of iNKT cells are depicted in the dot plots. (c) Thymic iNKT cells were gated on CD24−tetramer+ iNKT cells and additionally analyzed for CD44 and NK1.1 expression. Stage 1: CD44−NK1.1−, stage 2: CD44+NK1.1−, stage 3: CD44+NK1.1+. (d) Absolute cell numbers of thymic iNKT cells from the experiment described in (b,c). *P < 0.05; **P < 0.01; ***P < 0.001 (one-way analysis of variance (ANOVA) and multiple comparisons). Each symbol represents an individual mouse; data are mean ± s.e.m from three independent experiments (a–d)
Next, we assessed the influence of UTX loss on the functional properties of iNKT cells. After stimulation with α-GalCer, IFN-γ production was reduced in thymic iNKT cells of UTX-deficient mice compared to wild-type, reflecting a lack of fully mature cells (Supplementary Fig. 2a). However, synthesis of IL-4 in the thymus and IFN-γ production of the few remaining liver iNKT cells in UTX-KO mice was unaltered (Supplementary Fig. 2a). A recent classification, though not mutually exclusive to the maturation model, describes iNKT cell development by the subtypes NKT1, NKT2, and NKT17, based on their cytokine profile and specific transcription factors37. We observed a significant reduction of T-bet-dependent NKT1 cells in UTX-deficient mice compared to wild-type (Supplementary Fig. 2b). To exclude that the impaired development of iNKT cells was a consequence of failure in antigen presentation or the rearrangement of the Vα14-Jα18 TCR, we demonstrated equivalent CD1d expression and abundance of Vα14-Jα18 transcripts between UTX-deficient and wild-type DP thymocytes (Supplementary Fig. 2c,d). Thus, UTX does not primarily affect the function, but rather the development of iNKT cells.
UTX controls iNKT lineage-specific gene expression
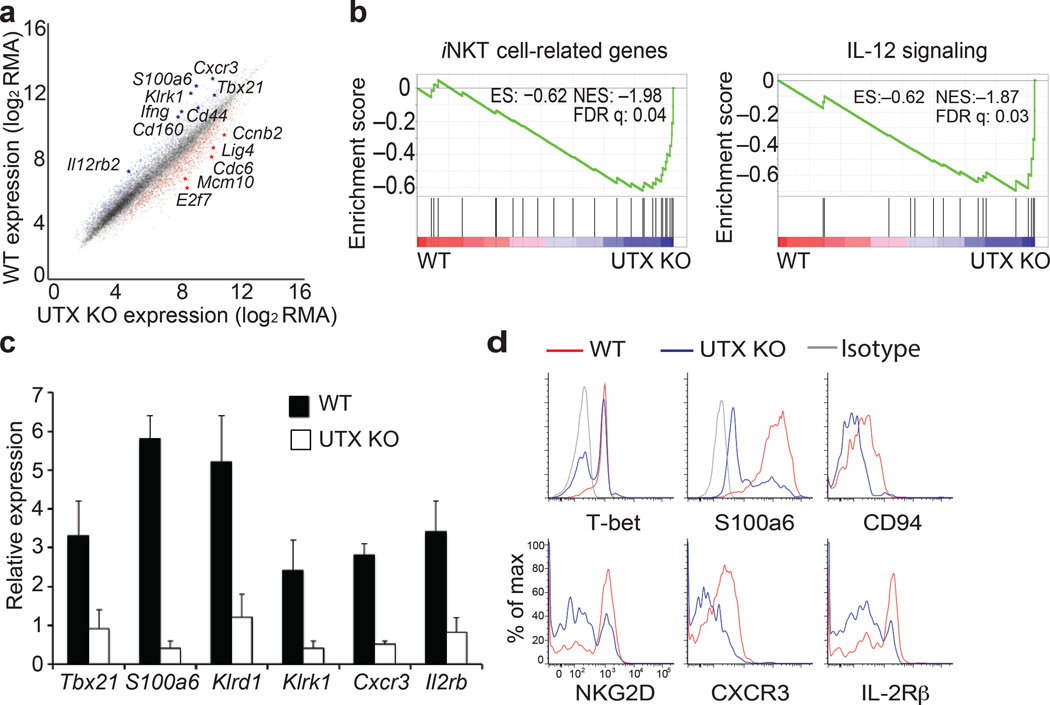
To elucidate the molecular mechanism by which UTX contributes to iNKT cell development, we sorted thymic iNKT cells from UTX-deficient or wild-type mice and performed gene expression analysis (Supplementary Fig. 3a). Compared to wild-type, downregulated genes in UTX-KO iNKT cells consisted of genes that are induced during iNKT cell maturation and included the critical transcription factor T-bet (Tbx21)11, NK cell receptor NKG2D (Klrk1), chemokine and cytokine receptors Cxcr3 and CD122 (Il2rb)9, as well as cytokine IFN-γ (Ifng), and calcium regulator Calcyclin (S100a6)13 (Fig. 2a). Gene set enrichment analysis (GSEA) of downregulated genes in UTX-deficient iNKT cells revealed enrichment for genes involved in iNKT cell differentiation as well as signaling pathways, including IL-12 and NFAT that have been previously reported to play important roles in iNKT cells6,13 (Fig. 2b, Supplementary Fig. 3b). We validated the gene expression of a subset of UTX-dependent downregulated signature genes by qPCR and observed marked downregulation of Tbx21, S100a6, the NK cell receptors Klrd1 and Klrk1, Cxcr3, and Il2rb in UTX-deficient thymic iNKT cells (Fig. 2c). Correspondingly, iNKT cells from UTX-KO mice expressed less protein of those gene products (Fig. 2d). By contrast, UTX deficiency resulted in upregulation of genes involved in cell cycle, DNA replication, and DNA repair pathways (Supplementary Fig. 3c). We confirmed loss of Utx transcripts and UTX-dependent downregulation of signature genes in different stages of UTX-KO iNKT cells (Supplementary Fig. 3d,e). These findings indicate that iNKT cell development requires a UTX-mediated lineage-specific gene expression program.
Figure 2.
UTX is required for lineage-specific expression of signature genes in iNKT cell development. (a–c) Gene expression profiles of UTX-deficient (UTX KO) or wild-type (WT) thymic iNKT cells. (a) Scatter plot shows up- or downregulated genes in UTX KO iNKT cells. RMA; Robust Multi-array Average (normalized fluorescence units of the probes on array). Data are from two experiments with five mice (WT) or four mice (KO). (b) Gene Set Enrichment Analysis (GSEA) of downregulated genes in UTX KO iNKT cells. ES, enriched score; NES, normalized ES; FDR, false discovery rate. (c) Expression of downregulated genes in UTX KO iNKT cells were validated by quantitative RT-PCR. Relative expression values are depicted normalized to Actin. Data are mean ± s.d. from five independent experiments. (d) Flow cytometry analysis of thymic iNKT cells from WT or UTX-deficient mice, staining for the gene products of the iNKT cell expression signature. Analysis is gated on CD3+CD1d-tetramer+ cells, and histograms depict the fluorescence intensity of the proteins indicated. Data are representative of three independent experiments.
UTX regulates the epigenetic landscape of iNKT cells
UTX-mediated H3K27 demethylation and concomitant catalysis of H3K4 trimethylation by MLL2 around the promoters correlates with active gene expression23. Therefore, we hypothesized that during iNKT cell development, transcriptional activation of iNKT lineage-specific genes involves UTX-dependent chromatin regulation. To address this hypothesis, we examined the epigenetic landscape of iNKT cells from UTX-KO or wild-type thymi by genome-wide ChIP-Seq analysis for H3K4me3 and H3K27me3 marks, correlating with activation or repression, respectively. We applied model-based analysis of ChIP-Seq (MACS2) to identify distinct peaks in each condition. Although genome-wide H3K4me3 peaks were largely shared between wild-type and UTX-KO iNKT cells (Fig. 3a), genome-wide H3K27me3 peaks were greatly increased in the absence of UTX (Fig. 3b), consistent with the role of UTX in H3K27me3 demethylation. In addition to this accumulation in global H3K27me3 content in UTX-KO iNKT cells (9,645, Fig. 3b), we observed that some regions exhibited wild-type-specific H3K27me3 peaks (4,250, Fig. 3b). While chromosomal distributions of wild-type- and UTX-KO-specific H3K27me3 peaks were similar (Supplementary Fig. 4a,b), wild-type-specific H3K27me3 peaks were more abundant in distal intergenic regions compared to UTX-KO-specific peaks (33.5% WT vs. 24.7% UTX-KO, Supplementary Fig. 4c,d). Notably, we detected increased abundance of UTX-KO-specific H3K27me3 peaks in proximal promoters compared to wild-type-specific peaks (10.4% WT vs. 13.4% UTX-KO, Supplementary Fig. 4c,d). Genomic Regions Enrichment of Annotations Tool (GREAT) analysis demonstrated that regions with a loss of H3K27me3 peaks in UTX-KO iNKT cells were enriched for bivalent genes that harbor both H3K27me3 and H3K4me3 in their promoters, as well as genes that are involved in erythrocyte development (Supplementary Fig. 5a). Interestingly, regions that gained H3K27me3 peaks in UTX deficiency were enriched for genes that require MLL for their transcription, as well as genes induced in memory T cells (Supplementary Fig. 5b).
Figure 3.
UTX regulates the chromatin landscape of iNKT cells. (a,b) Genome-wide distribution of H3K4me3 (active) or H3K27me3 (repressive) histone marks on UTX-deficient (KO) or wild-type (WT) iNKT cells. Data are from two independent experiments. The Venn diagrams depict the numbers of genome-wide peaks of the histone marks H3K4me3 (a), and H3K27me3 (b), in WT or KO iNKT cells. (c–e) Chromatin landscape of WT or KO iNKT cells around gene promoters. 0 marks transcription start site (TSS). Shown is the average abundance of H3K27me3 and H3K4me3 depicted as z-score in reads per million (rpm). Average profiles are grouped including (c) all iNKT cell genes, or (d) downregulated genes (H3K4me3: P < 1 × 10−5, H3K27me3: P < 1 × 10−5), or (e) upregulated genes (H3K4me3: P = 0.16, H3K27me3: P = 9 × 10−4), with WT in blue and KO in green. (f–i) Integrating gene expression with chromatin state. Downregulated genes in KO iNKT cells were further subdivided into four clusters (C1–C4) dependent on their histone mark pattern. Gene expression is shown as logarithmic fold change (log2FC) in a whisker plot. The box extends from the lower to upper quartile values of the data, with a line at the median. The whiskers extend from the box 1.5 inter-quartile range on each side. Flier points are data points outside the whiskers. Average abundance of H3K27me3 and H3K4me3 around gene promoters in the different clusters is depicted as z-score in reads per million (rpm). C1–4 = clusters 1 to 4. P-values indicate statistical significance based on permutation test where 100,000 permutations were used to calculate the distribution of the difference between two average profiles.
Since UTX-mediated alterations of histone marks around promoters impact gene expression, we interrogated the average histone mark abundance at iNKT cell gene promoters (Fig. 3c–e). In UTX-KO iNKT cells, we observed a significant accumulation of H3K27me3 around the transcription start sites (TSS, indicated as 0 in the graph) and the promoters of downregulated genes (Fig. 3d, Supplementary Fig. 5c, P < 1 × 10−5, permutation test). Moreover, we observed a marked reduction of H3K4me3 in the downregulated genes of UTX-KO iNKT cells (Fig. 3d, P < 1 × 10−5, permutation test). While there was a less significant accumulation of H3K27me3 around upregulated gene promoters of UTX-KO iNKT cells compared to downregulated genes (Supplementary Fig. 5d, P = 9 × 10−4), there was no notable change in H3K4me3 abundance (Fig. 3e, P = 0.16). GREAT analysis of the promoter regions of downregulated genes revealed enrichment for genes involved in iNKT cell development (Supplementary Fig. 5e), while analysis of upregulated genes demonstrated association with cell cycle and DNA repair pathways (Supplementary Fig. 5f). These results suggest that UTX controls iNKT development by regulating the chromatin landscape around the promoters of downregulated genes that are involved in iNKT cell differentiation.
iNKT signature gene promoters are regulated by UTX
To uncover different patterns of chromatin state and explore promoters that exhibit significant UTX-dependent chromatin regulation, we identified clusters based on the distribution of H3K4me3 and H3K27me3 around the promoters of downregulated genes (Fig. 3f–i, Supplementary Fig. 6a) and upregulated genes (Supplementary Fig. 6b,c). For downregulated genes, cluster 1 contained gene promoters with similar abundance of H3K4me3 in both wild-type and UTX-KO iNKT cells and a modest increase in H3K27me3 in the absence of UTX (Fig. 3f). Cluster 2 consists of gene promoters with very low amounts of H3K4me3 and comparable abundance of H3K27me3 in both wild-type and UTX-KO iNKT cells (Fig. 3g). Interestingly, promoters in cluster 3 (Fig. 3h) and cluster 4 (Fig. 3i) had a striking reduction in H3K4me3 content and accumulation of H3K27me3, especially around the TSS (P < 1 × 10−5, permutation test), suggesting that these promoters are affected by UTX-dependent chromatin regulation. Integrating gene expression data with chromatin state revealed that genes most downregulated in expression are grouped within cluster 3, highlighting that UTX-mediated removal of H3K27me3 around these promoters is critical for activation of transcription (Fig. 3h). We assessed the identity of these promoters in cluster 3 and cluster 4 (Supplementary Table 1) that exhibited significant UTX-dependent chromatin regulation by GREAT analysis. We found that immune response genes and signature genes of iNKT cell differentiation were significantly enriched in cluster 3 (Klr family, Cxcr3, S100a6) and cluster 4 (Tbx21) (Supplementary Fig. 6d). Cluster analysis of upregulated genes showed that abundance of H3K4me3 and H3K27me3 was largely comparable between UTX-KO and wild-type iNKT cells (Supplementary Fig. 6b,c). These results underscore that UTX deficiency specifically affects the chromatin state around the promoters of a subset of downregulated genes, which constitutes the iNKT cell signature in cluster 3 and cluster 4, without a robust effect on the chromatin state of other genes.
UTX binds to iNKT signature gene promoters
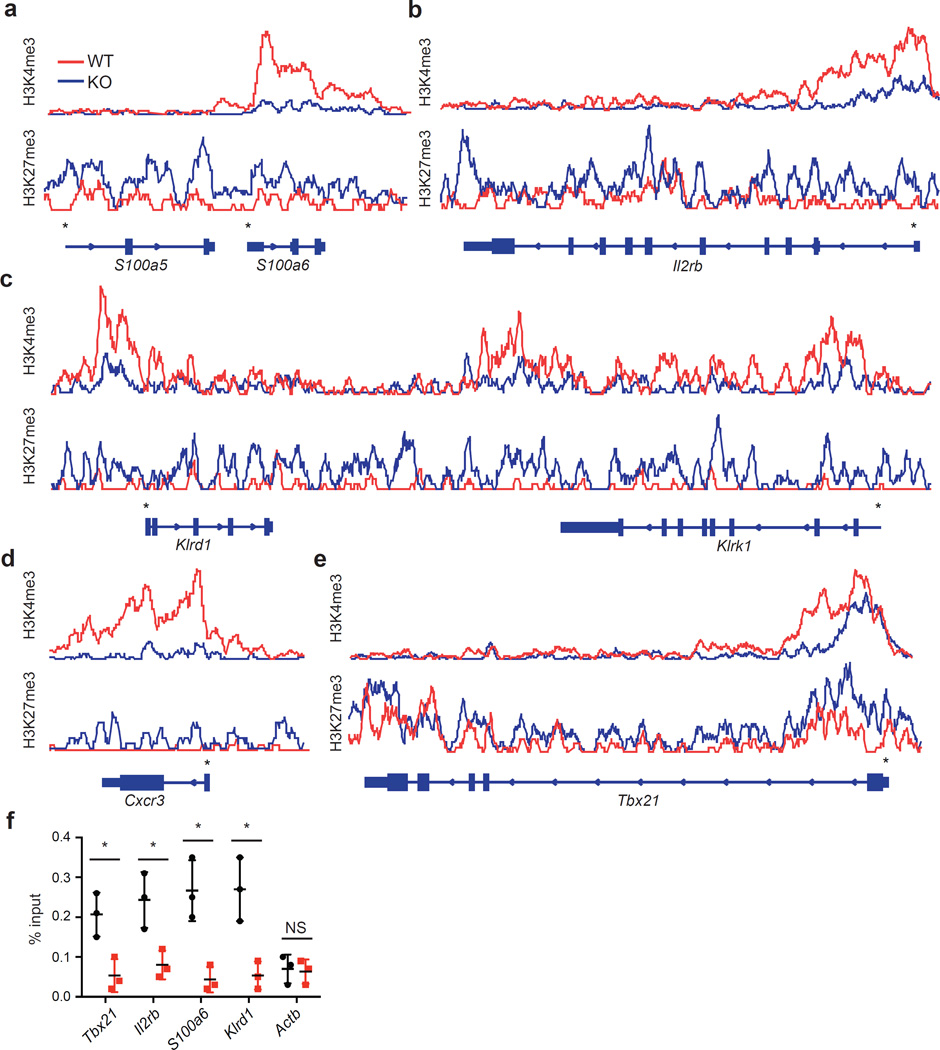
To assess specific alterations in H3K4me3 and H3K27me3 content around the UTX-dependent gene promoters, we generated overlay tracks of ChIP-Seq data from wild-type and UTX-KO iNKT cells using Integrative Genomics Viewer (IGV). We found a marked reduction in H3K4me3 abundance and an accumulation of H3K27me3 around the promoters of signature genes S100a6, Il2rb, Klrd1, Klrk1, Cxcr3, and Tbx21 in UTX-KO iNKT cells (Fig. 4a–e). To investigate whether UTX is physically recruited to these promoters, we performed UTX ChIP-PCR on sorted wild-type iNKT cells. Compared to the housekeeping gene Actin (Actb) which was not regulated by UTX based on gene expression and ChIP-Seq data, we detected significant occupancy of UTX at the promoter regions of the representative genes Tbx21, Il2rb, S100a6, and Klrd1 (Fig. 4f). These results suggest that UTX directly controls the epigenetic landscape around the promoters of iNKT lineage-specific genes to facilitate their transcription.
Figure 4.
UTX occupies iNKT signature gene promoters that exhibit UTX-dependent chromatin regulation. (a–e) ChIP-Seq overlay tracks of H3K27me3 and H3K4me3 marks for representative iNKT cell signature genes from UTX-deficient (KO) or wild-type (WT) iNKT cells (Data from Fig. 3). Gene structure and direction of transcription is depicted below the tracks. Gene promoters are indicated with an asterisk (*). (f) Assessment of UTX occupancy around signature gene promoters by ChIP-PCR. Shown is the relative enrichment of promoter sequences of iNKT cell signature genes by UTX ChIP compared to isotype control IgG. Actin (Actb) was used as negative control. *P < 0.05, using unpaired t-test. Data are mean ± s.d. from three independent experiments.
UTX enzyme activity is required for iNKT development
Although the data above strongly implicate a critical role of UTX in regulating iNKT lineage-specific gene expression and development, it is important to assess whether UTX directly acts within iNKT cells. A peculiarity of iNKT cells is that they originate from and concomitantly are selected by DP thymocytes38. This feature allows the use of mixed bone marrow chimeric mice to distinguish whether a gene defect is intrinsic to iNKT cells, or if extrinsic antigen presentation and selection by CD1d-expressing DP thymocytes is responsible for an observed phenotype. We transferred bone marrow cells from wild-type and UTX-deficient mice in a 1:1 ratio into Rag2−/− animals, and subsequently determined the contribution of each donor bone marrow to the overall iNKT cell pool in recipient hosts. In the thymus, the population of stage 0–1 iNKT cells of UTX-deficient origin predominated over wild-type cells, which was due to relative accumulation of immature iNKT cells in UTX deficiency (Supplementary Fig. 7a,b). Accordingly, the majority of iNKT cells at stage 3 were of wild-type origin, indicating again a block of maturation in the absence of UTX (Supplementary Fig. 7a, b). In the liver, most iNKT cells were wild-type and few UTX-deficient cells contributed to the peripheral iNKT cell pool (Supplementary Fig. 7c). Conventional T cells displayed a balanced mixed chimerism, highlighting a specific effect of UTX in iNKT cells (Supplementary Fig. 7c). Thus, the block in maturation reflects an intrinsic defect in iNKT cells imposed by loss of UTX, which cannot be rescued by the presence of wild-type DP thymocytes.
Next, we asked whether the downregulated gene expression program and developmental block of iNKT cells could be rescued by in vivo reconstitution with UTX, and whether its demethylase activity is required. Using lentiviral transduction of bone marrow cells followed by transplantation, UTX-deficient bone marrow transduced with an empty virus failed to produce a proper population of iNKT cells in the thymus (Fig. 5a). Importantly, UTX-KO bone marrow reconstituted with UTX exhibited substantial iNKT cell development, whereas reconstitution with an enzyme-dead mutant failed to generate a sizeable iNKT cell population (Fig. 5a). Upon analysis of the different maturation stages of iNKT cells, we observed that the few UTX-KO iNKT cells that developed from bone marrows transduced with empty virus or enzyme-dead mutant UTX could not fully mature to stage 3 (Fig. 5b). In sharp contrast, UTX reconstitution facilitated full development of iNKT cells in the thymus (Fig. 5b). We found that liver iNKT cells were virtually lacking in the UTX-KO group transduced with empty virus. UTX reconstitution rescued this phenotype by engendering a sizable liver iNKT cell population, whereas only a very minor iNKT cell fraction developed in the presence of enzyme-dead UTX (Fig. 5c,d). We demonstrated that Utx gene expression was comparable between full-length and enzyme-mutant UTX reconstituted mice, excluding different reconstitution efficiencies (Fig. 5e). In parallel, we analyzed the gene expression of iNKT cells from thymi of reconstituted mice. Notably, reconstitution with full-length UTX rescued expression of the iNKT cell signature, including Tbx21, Klrd1, and Cxcr3 (Fig. 5e). By contrast, enzyme-dead UTX failed to rescue signature gene expression, although an enzyme-independent contribution could be observed for Tbx21 (Fig. 5e). Altogether, these data demonstrate that the enzymatic demethylase function of UTX is essential for the proper generation of iNKT cells by establishing the iNKT gene expression program.
Figure 5.
Demethylase activity of UTX is required for generation of iNKT cells. Bone marrow cells from wild-type (WT) mice transduced with lentivirus containing empty vector (empty), or UTX-deficient bone marrow (UTX KO) transduced with empty vector (empty), or full-length UTX (UTX fl), or enzyme-dead UTX (UTX ed), were transferred to Rag2−/− hosts. Twelve weeks later, recipient mice were analyzed for frequency and maturation of iNKT cells using flow cytometry. (a) Percentages of thymic iNKT cells are depicted in the bar graph. (b) Thymic iNKT cell maturation was assessed by measuring CD44 and NK1.1 expression gated on CD1d-tetramer+ cells, and is shown in the bar graph according to stages 1–3. (c,d) Liver iNKT cells were detected as CD3+CD1d-tetramer+ cells, and their percentages are depicted in the dot plots (c) and the bar graph (d). (e) Thymic iNKT cells from reconstituted mice were analyzed for UTX and signature gene expression by quantitative RT-PCR, and relative expression values were normalized to Actin. *P < 0.05, **P < 0.01, ***P < 0.001; NS, not significant, using one-way ANOVA and multiple comparisons. Data are mean ± s.e.m from two independent experiments with four mice per group.
JunB is a novel regulator of iNKT development
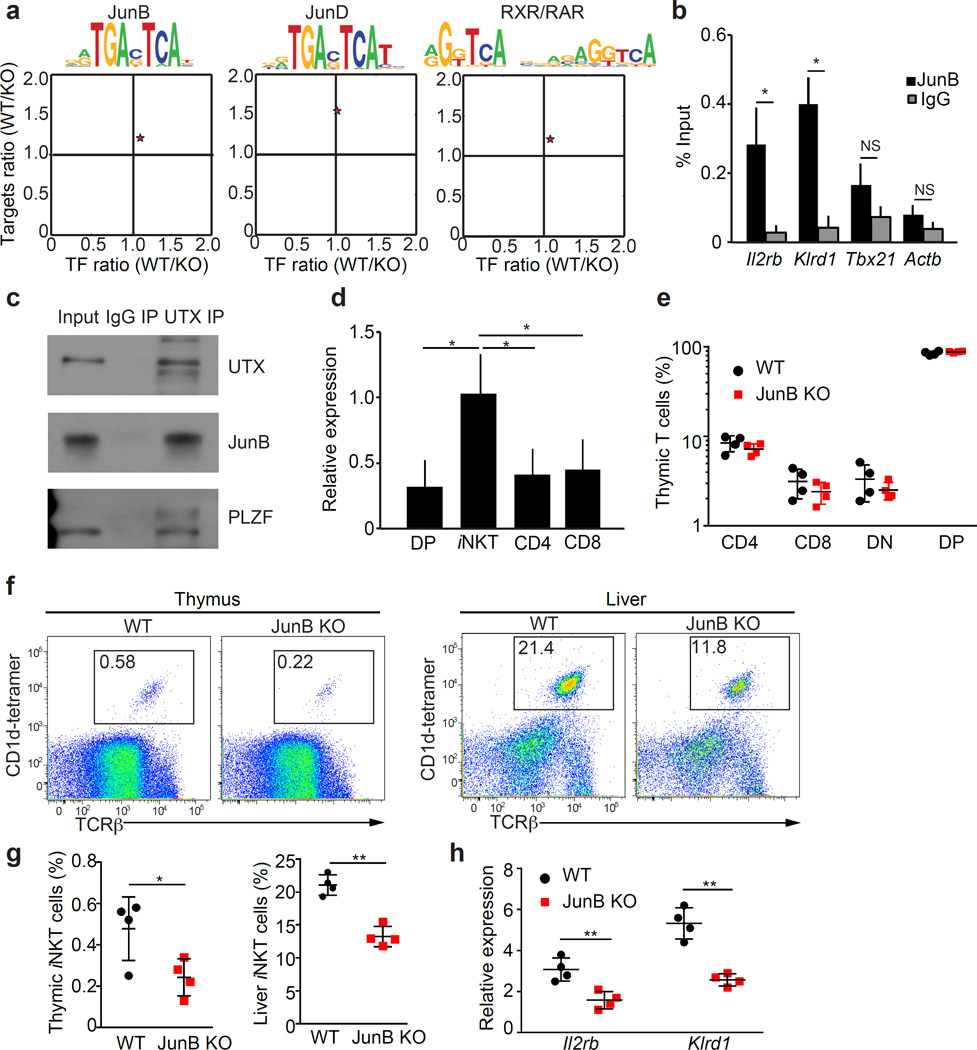
To gain further mechanistic insights how UTX regulates gene expression in iNKT cells, we performed motif enrichment analysis (using Haystack, please see Methods) and identified potential transcription factors that work together with UTX on iNKT signature gene promoters. We found significant enrichment of the target motifs for the AP-1 transcription factors JunB and JunD in cluster 3, as well as RAR/RXR in cluster 4, around the promoters of genes that showed UTX-dependent chromatin and transcriptional regulation (Fig. 6a). To test whether JunB directly regulates UTX-dependent iNKT cell genes, we performed JunB ChIP-PCR. JunB bound the promoters of signature genes from cluster 3 that were enriched for JunB motifs (Il2rb and Klrd1) (Fig. 6b, Supplementary Table 2). To assess whether UTX cooperates with this putative iNKT transcription factor, we performed immunoprecipitation using iNKT cell lysates and demonstrated that UTX interacts with JunB (Fig. 6c). Notably, UTX was also able to specifically bind the iNKT cell transcription factor PLZF (Fig. 6c). Furthermore, JunB expression was preferentially induced in iNKT cells compared to other thymocyte subsets, highlighting a potential role of JunB in iNKT cell generation (Fig. 6d). To test this hypothesis, we analyzed JunB-deficient (JunB-KO) mice and found that the frequencies of conventional CD4+ and CD8+ T cells, as well as DN and DP thymocytes were similar in JunB-KO animals compared to wild-type (Fig. 6e). However, the frequency of thymic and peripheral iNKT cells was significantly reduced in JunB-KO mice (Fig. 6f,g). To assess whether JunB regulates the transcription of UTX-dependent iNKT signature genes in cluster 3, we performed qPCR using sorted thymic iNKT cells from WT and JunB-KO mice. We found that signature genes Il2rb and Klrd1 (both gene promoters are bound by JunB, Fig. 6b) are significantly downregulated in JunB-KO iNKT cells (Fig. 6h). Taken together, these results show that UTX interacts with transcription factors, such as JunB, to establish lineage-specific gene expression in iNKT cells.
Figure 6.
Transcription factor JunB partners with UTX to regulate iNKT cell signature and development. (a) AP-1 transcription factor target motifs are enriched among downregulated genes in UTX-deficient iNKT cells. For motif enrichment, the analysis tool Haystack was used (P-values; JunB: 0.006, JunD: 0.006, RXR/RAR: 0.005). Depicted are target motif sequences and the expression ratios of transcription factors (TF) and their targets between wild-type (WT) and UTX KO (KO). (b) JunB ChIP-PCR analysis for UTX-dependent gene promoters in thymic iNKT cells. Shown is the relative enrichment of promoter sequences of UTX-dependent iNKT cell signature genes after JunB-ChIP compared to the isotype control (IgG). Actin (Actb) was used as a negative control. (c) Molecular interaction between UTX, JunB, and PLZF in iNKT cells as assessed by immunoprecipitation (IP) for UTX, compared to isotype control (IgG) (10% input). Data are representative of three independent experiments. (d) JunB expression in thymic CD4, CD8, double-positive (DP), and iNKT cells was analyzed by quantitative RT-PCR. Relative expression values were normalized to Actin. (e) Percentages of thymic CD4, CD8, double-negative (DN), and double-positive (DP) lymphocytes in WT and JunB KO mice. (f,g) Frequency of thymic and liver iNKT cells in WT and JunB KO mice was assessed using flow cytometry and depicted in representative dot plots (left panels). (h) Reduced expression levels of UTX-dependent iNKT signature genes in JunB KO thymic iNKT cells were analyzed by quantitative RT-PCR. Relative expression values were normalized to Actin. *P < 0.05, **P < 0.01; NS, not significant, using unpaired t-test. Data are mean ± s.e.m from three independent experiments (b–d) or two experiments with four mice per group (e–h).
UTX-deficient iNKT cells fail to activate PLZF target genes
In light of physical association of UTX with PLZF, we asked whether UTX regulates PLZF-mediated activation of gene expression in iNKT cells. By utilizing a PLZF ChIP-Seq data set that defined PLZF-activated genes in iNKT cells39, we determined that loss of UTX in iNKT cells leads to impaired activation of PLZF target gene expression (Fig. 7a). PLZF-activated target genes were significantly downregulated in UTX-KO compared to randomly selected genes (Fig. 7b). To assess whether this downregulation is accompanied by promoter accumulation of H3K27me3 in UTX-KO iNKT cells, we compared average abundance of H3K27me3 around PLZF-activated target gene promoters and randomly selected gene promoters. We found that H3K27me3 content around the promoters of PLZF-activated genes, but not random genes, is significantly increased in UTX-KO iNKT cells (Fig. 7c). Overlay tracks for PLZF target genes including Il18r1, Il12rb1 and Eya2 demonstrate a clear UTX-dependent accumulation of H3K27me3 and a concomitant reduction in H3K4me3 around the promoter regions that PLZF occupies (Fig. 7d–f). Accordingly, we confirmed that the expression of Il18r1, Il12rb1 and Eya2 was significantly reduced in UTX-KO iNKT cells (Fig. 7g). These data indicate that UTX controls the epigenetic landscape and transcription of PLZF-activated genes.
Figure 7.
UTX deficiency impairs activation of PLZF target genes in iNKT cells. (a) Heatmap of PLZF-activated genes illustrates reduction in gene expression in UTX KO iNKT cells. (b) Gene expression log2 ratios (WT/UTX KO) of PLZF-activated genes compared to random genes. (c) Accumulation of H3K27me3 marks around PLZF-activated gene promoters in UTX KO iNKT cells. Depicted are average levels of H3K27me3 abundance around PLZF-activated gene promoters (± 1kb of TSS) compared to randomly picked promoters in WT or UTX KO iNKT cells. Data are represented with whisker plots (n = 17 regions per group). The box extends from the lower to upper quartile values of the data, with a line at the median. The whiskers extend from the box 1.5 inter-quartile range on each side. Flier points are data points outside the whiskers. NS, not significant; *P < 0.05, using Mann-Whitney U test (b,c). (d–f) Representative tracks demonstrating loss of H3K4me3 and gain of H3K27me3 around the PLZF-activated genes: Il18r1 (d), Il12rb1 (e), Eya2 (f). Gene structure and direction of transcription is depicted below the tracks. Gene promoters are indicated with an asterisk (*). (g) Reduced expression levels of PLZF-activated genes in UTX KO thymic iNKT cells were confirmed by quantitative RT-PCR. Relative expression values were normalized to Actin. *P < 0.05, using unpaired t-test. Data are mean ± s.d. from three independent experiments.
UTX facilitates accessibility of iNKT super-enhancers
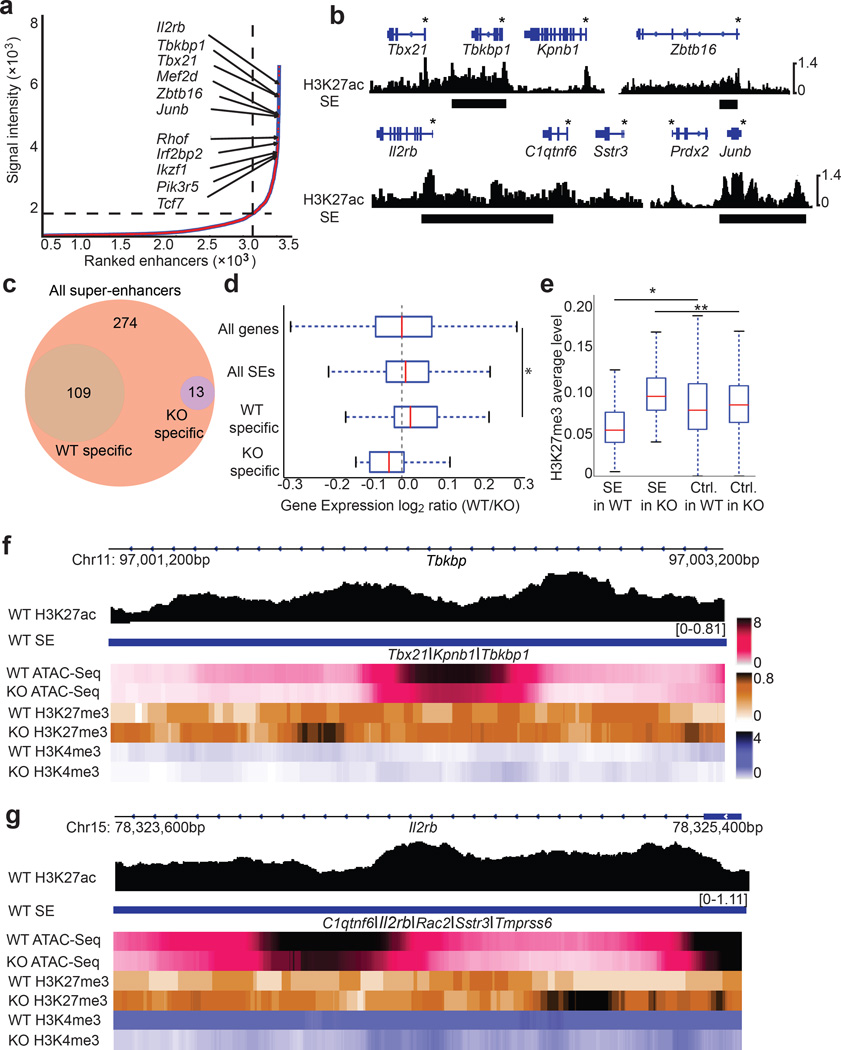
Because super-enhancers bestow lineage specificity14,15, and iNKT cells are vulnerable to loss of UTX, we hypothesized that another mechanism by which UTX controls iNKT lineage commitment is through regulation of super-enhancer accessibility. First, we uncovered the super-enhancer landscape of iNKT cells by performing genome-wide H3K27ac ChIP-Seq and defined super-enhancers as large enhancer elements with highly abundant H3K27ac14,15. We identified 396 super-enhancers that include elements proximal to genes encoding known regulators of the iNKT cell lineage, such as Tbx21, Zbtb16, and Il2rb, as well as putative novel regulators, including the transcription factor Junb (Fig. 8a,b, Supplementary Fig. 8a,b, Supplementary Table 3). To reveal potential pathways that are associated with the genes proximal to the super-enhancers identified in iNKT cells, we performed GREAT analysis of the super-enhancer elements. We found that the AP-1 pathway was significantly enriched in our analysis, corroborating JunB as a crucial novel regulator of the iNKT cell lineage (Supplementary Fig. 8c).
Figure 8.
UTX facilitates super-enhancer accessibility in iNKT cells. (a) Ranking of super-enhancers (SE) in iNKT cells. SEs were ranked based on H3K27ac signal intensity using the ROSE algorithm. (b) H3K27ac ChIP-Seq tracks of representative iNKT cell SEs are shown. Gene bodies are depicted above the tracks, and SEs are marked as black bars. Gene promoters are indicated with an asterisk (*). (c) Chromatin accessibility of SEs identified in (a) was analyzed using ATAC-Seq. The Venn diagram depicts numbers of accessible WT SEs with specific WT−, or KO ATAC-Seq peaks in iNKT cells. Data are from two independent experiments. (d) Expression of genes nearby UTX-dependent SEs is downregulated in KO iNKT cells. Gene expression log2 ratios (WT/KO) of all genes (n = 20,628), genes nearby all defined iNKT cell SEs (n = 396), or iNKT SEs with WT-specific accessibility (UTX-dependent, n = 109), or iNKT SEs with KO-specific accessibility (n = 13) are shown. Data is represented with a whisker plot. *P = 0.002, using Mann-Whitney U test (e) Accumulation of H3K27me3 marks around SE regions in KO iNKT cells. Depicted are average levels of H3K27me3 abundance in the vicinity of the defined SE regions in (a) compared to randomly picked control regions (C) in WT or KO iNKT cells. n = 396 regions per group. Data are represented with a whisker plot. *P = 5.9 × 10−10, **P = 1.9 × 10−14 using Mann-Whitney U test. (f,g) Representative tracks demonstrating loss of accessibility and gain of H3K27me3 around the defined SE regions for T-bet (Tbx21) (f) and Il2rb (g). Depicted are the ChIP-Seq tracks of H3K27ac and heat maps of ATAC-Seq, H3K27me3, and H3K4me3 in WT or KO iNKT cells around the SEs for Tbx21 and Il2rb.
To determine whether iNKT super-enhancer accessibility is affected in the absence of UTX, we performed an assay for transposase-accessible chromatin using sequencing (ATAC-Seq)40 that captures accessible chromatin regions in sorted wild-type and UTX-KO iNKT cells. Amongst 396 super-enhancers we identified, 109 super-enhancers had wild-type-specific ATAC-Seq peaks and therefore lost accessibility in UTX-KO iNKT cells (Fig. 8c, Supplementary Table 4). We found that genes nearby super-enhancers that showed UTX-dependent accessibility are downregulated in UTX-KO by comparing log2 gene expression ratios (WT/KO) of these genes to all genes (P = 0.002) (Fig. 8d).
To interrogate whether this loss in accessibility of super-enhancers and reduced transcription of nearby genes is accompanied by accumulation of H3K27me3 in UTX deficiency, we compared average H3K27me3 abundance around the defined iNKT super-enhancer regions to randomly picked control regions in UTX-KO iNKT cells. We detected significant accumulation of H3K27me3 (P = 1.9 × 10−14) around super-enhancer regions in UTX-KO (Fig. 8e). Super-enhancer regions in wild-type iNKT cells exhibited less H3K27me3 abundance compared to randomly picked control regions (P = 5.9 × 10−10), which is consistent with the notion that active enhancers are devoid of H3K27me317,41. Control regions in wild-type and UTX-KO iNKT cells had similar H3K27me3 levels (P = 0.14) (Fig. 8e). Notably, super-enhancers that showed UTX-dependent accessibility are nearby important iNKT cell regulators, such as Tbx21 (Fig. 8f, Supplementary Fig. 8a), and Il2rb (Fig. 8g, Supplementary Fig. 8b). Furthermore, GREAT analysis demonstrated that genes involved in lymphocyte differentiation and IL-2 signaling are enriched around these regions (Supplementary Fig. 8d). To determine the transcription factors that bind these super-enhancers, we performed motif analysis and found enrichment of the transcription factors RelA and Bhlhe40 (Supplementary Fig. 8e), which were recently shown to cooperate with T-bet to control iNKT cell functions42. Although a small fraction of super-enhancers (n = 13) gained accessibility in UTX-KO iNKT cells (Fig. 8c, Supplementary Table 4), we could not detect any gene set enrichment for these regions (Supplementary Fig. 8d). Overall, these results suggest that UTX regulates accessibility of super-enhancers that establish iNKT cell identity.
DISCUSSION
Discoveries of key transcription factors8,9,11 and gene expression programs13 contribute to an understanding of the iNKT cell lineage. In this context, a recent study showed the diversity of gene programs in different iNKT cell subsets based on RNA-Seq transcriptomics and the description of enhancer elements43. However, the epigenetic mechanisms that govern iNKT cell identity are undefined. Here, we demonstrate a selective, cell-intrinsic, and catalytic role of the H3K27me3 demethylase UTX in regulating the epigenetic landscape and lineage-specific gene expression of iNKT cells. Our data and a previous study35 reveal a requirement for the H3K27me3 demethylases UTX and JMJD3 in iNKT cell development. We found that this requirement is due to UTX-dependent regulation of iNKT signature gene expression. UTX-deficient iNKT cells exhibited a developmental block and failed to induce expression of transcription factors and signaling molecules involved in iNKT cell terminal maturation, such as T-bet (Tbx21)11 and CD122 (Il2rb)9,10.
Epigenetic regulation of gene expression by UTX has been studied primarily in the context of demethylation of repressive H3K27me3 marks. Moreover, UTX associates with MLL2, which catalyzes trimethylation of H3K4 at the promoters in order to facilitate transcription of lineage-specifying genes21,23,29. Accordingly, in iNKT cells, UTX-dependent signature gene promoters, such as those in Tbx21 and Il2rb, accumulated H3K27me3 and harbored reduced H3K4me3 levels in the absence of UTX.
We found that gene promoters that exhibit UTX-dependent chromatin and transcriptional regulation are bound by the AP-1 transcription factor JunB. A previous report that JunB is part of a gene network preferentially upregulated in iNKT cells compared to NK cells and conventional T cells is consistent with our findings13. While indirect evidence through overexpression or deficiency of the negative regulator BATF shows that AP-1 activity promotes generation of iNKT cells44,45, lack of the Fos family member Fra2 leads to increased iNKT cell numbers46. Interestingly, JunB/AP1 directly controls expression of Ifng, a hallmark cytokine released by mature iNKT cells47. Our studies using JunB-deficient mice reveal a significant reduction of thymic as well as peripheral iNKT cell numbers. Taken together, our data indicate that JunB is a novel regulator that influences iNKT cell development.
UTX can function both dependent and independent of its demethylase enzyme activity29,48. We found that iNKT cell development requires UTX demethylase activity, highlighting a critical role for the removal of H3K27me3 marks from iNKT signature gene promoters. Interestingly, iNKT cells lacking components of PRC2, which is responsible for H3K27me3 deposition, exhibit impaired maturation and increased accumulation in the thymus and spleen35,49. It is proposed that iNKT cell development involves transition from a poised PLZF (Zbtb16) promoter in DP thymocytes, harboring both H3K27me3 and H3K4me3 marks, to an active state in iNKT cells, characterized by H3K4me3 and devoid of H3K27me335. Although we did not observe significant accumulation of H3K27me3 around the PLZF (Zbtb16) promoter and a robust decrease in PLZF expression in UTX-deficient iNKT cells, we demonstrated that UTX regulates the expression and epigenetic landscape of downstream PLZF-activated genes in iNKT cells.
Recent studies identified super-enhancers demarcated by elevated H3K27ac levels as a hallmark of key genes associated with cell identity and genetic risk of disease14,15,50. Our study defines for the first time the iNKT cell super-enhancer landscape and demonstrates that signature genes, including Zbtb16, Tbx21, and Il2rb, exhibit super-enhancer elements. In addition, we uncovered numerous novel regulators of iNKT cell identity, providing an invaluable resource for understanding epigenetic control of iNKT lineage specification and disease-associated genes in iNKT cells.
UTX interacts with chromatin regulators like SWI/SNF and the MLL complex that can impact enhancer activation23,29,48. Since dynamic regulation of H3K27me3 and H3K27ac content determines enhancer activity17,41, another mechanism for UTX-mediated gene regulation raised by our study involves control of lineage-specific super-enhancer accessibility by UTX in iNKT cells. Accessibility of super-enhancers nearby downregulated signature genes, such as Tbx21 and Il2rb, was lost in UTX-deficient iNKT cells. This finding suggests that proper maturation of iNKT cells requires a dual mechanism that involves UTX-mediated regulation of the epigenetic landscape around both promoters and enhancers of signature genes. Taken together, our data offer a new perspective on transcriptional control of iNKT cell development and delineate multiple mechanisms that UTX engages to regulate the lineage-specific gene expression program of iNKT cells.
Methods
Experimental mice
Targeting strategy for generating Utx (Kdm6a)f/f or Jmjd3 (Kdm6b)f/f mice is shown in Supplementary Fig. 1. Briefly, Jmjd3f/f embryonic stem cells (ESCs) were generated by flanking exons 17 and 19 of the Jmjd3 locus with loxP sites. These exons encode the catalytic JmjC domain. The sequence-verified targeting vector was linearized and electroporated into CJ9 ES cells (129sv background). Homologous recombination was assessed by Southern blot analysis by digesting genomic DNA with NheI (5′ probe) or KpnI (3′ probe), using specific external probes. Utxf/f ESCs were obtained from EUCOMM. After sequence and genotype verification, karyotypically normal ESC clones were injected into blastocysts to generate chimera. Mice were backcrossed for at least 10 generations to C57BL/6 background. Additionally, UTX- and JMJD3-deficient animals were crossed to generate doubly deficient mice. Wild-type control mice were derived from littermates. Vav-Cre, CD45.1-congenic, and Rag2−/− mice were purchased from The Jackson Laboratory. JunB-KO (Junbfl/flMORE-Cre) mice were generated as previously described51. Sex- and age-matched animals between 8 and 12 weeks of age were used for experiments. To estimate proper number of animals, preliminary experiments were performed. Mice were allocated at random to experimental groups. Mouse studies were performed in a non-blinded fashion. The Institutional Animal Care and Use Committee (IACUC) of Boston Children’s Hospital approved all animal experiments.
Reagents
Phorbol myristate acetate (PMA), ionomycin, fetal bovine serum (FBS), and phosphate-buffered saline (PBS) were purchased from Sigma. Percoll was obtained from GE Healthcare Life Sciences. Viability Dyes (eFluor-780 and eFluor-450) were purchased from eBioscience. RPMI media 1640 and Propidium Iodide (PI) were purchased from Life Technologies. α-Galactosylceramide (PBS57, α-GalCer) was obtained from Enzo Life Sciences. Brefeldin A was acquired from BioLegend.
Antibodies
For flow cytometry, cells were stained with following antibodies: anti-CD3ε (145-2C11), anti-CD8 (53-6.7), anti-CD4 (GK1.5), anti-CD24 (M1/69), anti-CD45.1 (A20), anti-CD45.2 (104), anti-B220 (RA3-6B2), anti-NKG2D (CX5), anti-IL-2Rβ (TM-β1), anti-CD94 (18d3), anti-IFN-γ (XMG1.2), anti-IL-4 (11B11), and anti-T-bet (4B10) were purchased from BioLegend. Anti-NK1.1 (PK136), anti-TCRβ (H57-597), anti-CD44 (IM7), anti-IgG1 (A85-1), anti-CD1d (1B1), and anti-CD90.2 (53-2.1) were obtained from BD Biosciences. Anti-IL-17 (ebio17B7) and anti-Rorγt (AFKJS-9) were purchased from eBioscience. Anti-PLZF (D-9) was obtained from Santa Cruz Biotechnology. Anti-CXCR3 (220803) was purchased from R&D Systems. Anti-S100a6 (EPNCIR121) was obtained from Abcam. Anti-CD16/CD32 (2.4G2) was acquired from Bio-Xcell (Malaysia).
Flow Cytometry
Cells were stained with antibodies against surface makers in flow cytometry buffer (PBS supplemented with 0.5% bovine serum albumin) on ice for 30 min. Subsequently, cells were washed and analyzed using a FACSCanto™ II flow cytometer (BD Biosciences). Propidium iodide was added just before flow cytometry analysis to exclude dead cells.
Tetramer analysis
For staining of iNKT cells, Alexa647- or Phycoerythrin (PE)-conjugated α-GalCer-loaded CD1d tetramers (shortly called CD1d tetramer) were obtained from the Tetramer Core Facility of the NIH. For blocking non-specific binding of immunoglobulin to Fc receptors, cells were incubated with anti-CD16/CD32 in flow cytometry buffer for 10 min on ice. After incubation, cells were stained with fluorescently conjugated CD1d tetramer and viability dye for 40 min on ice. To perform molecular studies, CD1d tetramer-positive cells were sorted using FACSARIA III (BD Biosciences).
Intracellular staining for cytokines and transcription factors
To stain cytokines intracellularly, cells were fixed and permeabilized with BD Cytofix/Cytoperm followed by staining with fluorescently labeled antibodies against IFN-γ, IL-4, or IL-17 in Perm wash buffer (BD Biosciences). To analyze expression of transcription factors in iNKT cells, thymocytes were stained with fluorescently conjugated CD1d tetramer, followed by fixation/permeabilization using the Foxp3/transcription factor staining buffer set (eBioscience). Subsequently, cells were incubated with purified anti-PLZF, followed by staining with fluorescently labeled anti-mouse IgG1. Thereafter, cells were stained with antibodies against T-bet and Rorγt.
Isolation of liver lymphocytes
To isolate liver mononuclear cells, livers were perfused with PBS through the portal vein, excised, and minced through a metal mesh. Tissue homogenates were collected in 50 ml of media (RPMI supplemented with 2.5% FBS) and incubated for 15 min, prior to taking the supernatant. Cells were washed twice, suspended in medium containing 33% Percoll, and centrifuged at 500 g at 20 °C without break. Thereafter, cell pellets were collected to perform further experiments.
Measurement of immune responses to α-GalCer
To test the function of thymic iNKT cells in vitro, CD8-negative thymocytes were selected using anti-CD8 conjugated to magnetic beads and MACS columns (Miltenyi Biotec), according to manufacturer’s protocol. Subsequently, cells were stimulated with 50 ng/ml of PMA and 5 µg/ml of ionomycin in culture medium (RPMI supplemented with 10% FBS) for 4 h. Brefeldin A was added for the last 2 h. To measure the in vivo responses to α-GalCer, mice were injected with 250 µg of brefeldin A in 200 µl of PBS, followed 30 min later by intraperitoneal injection of 2 µg of α-GalCer in 200 µl of PBS. Mice were sacrificed 2 h after α-GalCer injection, and livers were excised for preparation of liver mononuclear cell isolation and intracellular cytokine staining as described above.
Mixed bone marrow chimeric mice
Bone marrow cells were isolated from the femurs and tibias of wild-type B6.SJL (CD45.1+) or UTX-deficient (CD45.2+) mice. T cells were depleted using biotinylated anti-CD90, anti-biotin conjugated to magnetic beads, and MACS columns. Rag2−/− mice were irradiated with a cesium source (600 rad). Then, irradiated mice were injected intravenously with a 1:1 mixture of bone marrow cells (2 × 106 per mouse) from UTX-deficient or wild-type B6.SJL mice. Chimeras were analyzed 10–12 weeks after bone marrow cell injection.
Lentiviral transduction of bone marrow cells and UTX rescue experiment
Bone marrow cells from WT or UTX-KO mice were isolated and cultured with media containing IL-6, IL-3, IL-7, and SCF. After 24 h, cells were counted and spin-infected with lentivirus containing full-length UTX (pLVX-EF1a-IRES-mCherry-UTX-WT), or catalytically inactive UTX (pLVX-EF1a-IRES-mCherry-UTX-ED), and incubated for 3 h at 37 °C. After 24 h, cells were spin-infected again with the respective lentivirus and incubated for 3–4 h at 37 °C. Infected cells were collected and intravenously injected into irradiated (600 rad) Rag2−/− recipient mice.
qRT-PCR
Cells were sorted into TRI Reagent (Life Technologies), and total RNA was isolated according to the manufacturer's instructions with following modification: the aqueous phase containing total RNA was purified using the RNeasy plus kit (Qiagen). RNA was converted to cDNA with cDNA synthesis kit (Bio-Rad). qRT-PCR was performed with SYBR green master mix (Bio-Rad) on the Bio-Rad iCycler RT-PCR detection system. Actb was used as a housekeeping control. To calculate the relative fold change, the 2−ΔΔCT cycle threshold method was used. The following primer sequences were used:
Actb, forward, 5′-TCCAGCCTTCCTTCTTGGGTATGGA-3′,
Actb, reverse, 5′-CGCAGCTCAGTAACAGTCCGCC-3′,
Va14, forward, 5′-GTCCTCAGTCCCTGGTTGTC-3′,
Ja18, reverse, 5′-CAAAATGCAGCCTCCCTAAG-3′,
Tbx21, forward, 5′-AGCAAGGACGGCGAATGTT-3′,
Tbx21, reverse, 5′-GGGTGGACATATAAGCGGTTC-3′,
S100a6, forward, 5′-GCTCACCATTGGCTCCAAGC-3′,
S100a6, reverse, 5′-GGAAGGCGACATACTCCTGG-3′
Klrd1, forward, 5′-TCTAGGATCACTCGGTGGAGA-3′,
Klrd1, reverse, 5′-CACTTGTCCAGGCAAACACAG-3′,
Klrk1, forward, 5′-ACTCAGAGATGAGCAAATGCC-3′,
Klrk1, reverse, 5′-CAGGTTGACTGGTAGTTAGTGC-3′,
Cxcr3, forward, 5′-TACCTTGAGGTTAGTGAACGTCA-3′,
Cxcr3, reverse, 5′-CGCTCTCGTTTTCCCCATAATC-3′,
Il2rb, forward, 5′-TGGAGCCTGTCCCTCTACG-3′,
Il2rb, reverse, 5′-TCCACATGCAAGAGACATTGG-3′,
Junb, forward, 5′ -CGACTACAAACTCCTGAAAC-3′,
Junb, reverse, 5′-CTGTGTCTGATCCCTGAC-3′,
Utx-delq, forward, 5′-GGTCACTTCAACCTCTTATTGG-3′,
Utx-delq, reverse, 5′-CGACATAAAGCACCTCCTGA-3′,
Jmjd3-delq, forward, 5′-ATCCGAGGAACCAGACAGCACT-3′,
Jmjd3-delq, reverse, 5′-GAGCATGTTGCCTGTGGATG-3′
Il18r1, forward: 5′-ACTTTTGCTGTGGAGACGTTAC-3′
Il18r1, reverse: 5′- CCGGCTTTTCTCTATCAGTGAAT-3′
Il12rb1, forward: 5′- ATGGCTGCTGCGTTGAGAA-3′
Il12rb1, reverse: 5′- AGCACTCATAGTCTGTCTTGGA-3′
Eya2, forward: 5′- ACCGCTGGGCTCTATCAAG-3′
Eya2, reverse: 5′- GGTAGGACGGATAATCCTGGTG-3′
Gene expression microarray
Wild-type or UTX-KO thymic iNKT cells were sorted into TRI Reagent (Life Technologies), and total RNA was isolated as described above. RNA was reverse transcribed and amplified using Ovation Pico RNA Amplification System V2 (NuGEN), biotin-labeled (Encore Biotin Module, NuGEN), and hybridized to Affymetrix mouse 430A 2.0 arrays. Affymetrix CEL files were normalized using the Robust Multi-array Average (RMA) method52, and batch effects were removed using ComBat53. Differentially expressed genes were detected by lmFit function in the limma package54. Gene set enrichment analysis (http://www.broadinstitute.org/gsea/index.jsp) was conducted on a pre-ranked list of differentially expressed genes in UTX-KO compared to wild-type iNKT cells. Furthermore, Genomic Regions Enrichment of Annotations Tool (GREAT; http://bejerano.stanford.edu/great/public/html/splash.php) analysis was performed on differentially expressed genes in order to identify their functional annotations55.
Genome-wide ChIP-Seq analysis
ChIP-Seq for histone marks H3K4me3 and H3K27me3 was performed as described previously56. Briefly, 1 × 105 FACS-sorted thymic iNKT cells were fixed with 1% formaldehyde (EMS) for 5 min at 25 °C. Cells were lysed in 120 µl of ChIP buffer (0.5% SDS, 50 mM Tris, pH 8, 10 mM EDTA, 1× complete protease inhibitor cocktail, Roche) and were sonicated with an E210 Ultrasonicator (Covaris) in microtubes (Covaris) with the following sonication conditions: intensity, 5; duty cycle, 10%; 200 cycles per burst for 60 s and repeated 6 times. Sonicated chromatin was collected and diluted with four parts of ChIP dilution buffer (1.25% Triton X-100, 12.5 mM Tris, pH 8, 187.5 mM NaCl, 1× complete protease inhibitor cocktail). Input samples were collected and stored at −20 °C. Dynabeads Protein G (Life Technologies) coupled to specific antibodies were used for overnight immunoprecipitation using anti-H3K4me3 (ab8580, Abcam), or anti-H3K27me3 (07-449, Millipore). Subsequently, samples were washed and the ChIP immune complexes were eluted from the beads twice, using elution buffer (1% SDS and 0.1 M NaHCO3) under constant agitation for 30 min. Samples were pooled and incubated overnight at 65 °C for reversal of the formaldehyde crosslinks. The samples were then treated for 2 h at 37 °C with 200 µg RNase A (Qiagen) and 40 µg proteinase K (Life Technologies). ChIP DNA fragments were purified with a MinElute Reaction Cleanup kit (Qiagen). ChIP DNA was prepared for high-throughput Illumina sequencing with a NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina kit (New England BioLabs) and NEBNext Multiplex Oligos for Illumina (Index Primers 1–12) according to a modified manufacturer's protocol. For each ChIP, the ends of DNA fragments were repaired, dA-tailed and ligated to Illumina adaptors according to the kit instructions. DNA fragments 150–600 bp in length were then selected with Pippin Prep 2% Agarose Gel Cassettes and the Pippin Prep DNA Size Selection System (Sage Science). After each step, DNA was purified with a MinElute Reaction Cleanup Kit or a QIAquick PCR Purification Kit (Qiagen). The DNA obtained by ChIP was then amplified by 15 cycles of PCR with NEBNext Multiplex Oligos. Amplified DNA was purified with DNA to Agencourt AMPure XP beads (Beckman Coulter) in a ratio of 1:1. The 'multiplexed' DNA libraries were sequenced on the Illumina HiSeq2000. Chip-Seq raw reads were aligned using the Bowtie2 aligner57 on the mouse genome assembly mm9 (National Center for Biotechnology Information), and peaks were called using MACS258 with the default parameter. To compare the average profiles, the different tracks were down-sampled to the same number of reads, and duplicate reads were removed using the PICARD tool (http://broadinstitute.github.io/picard/). To assess the significance of differences between two average profiles in a given region, we computed empirically, using 100,000 permutations, the distribution of the differences between two profiles using random assignment.
Cluster analysis of H3K27me3 and H3K4me3
Cluster analysis was performed on promoter regions of different sets of genes. The promoter region was defined as transcription start site (TSS) −2 kb and TSS +2 kb. The number of clusters was determined using a Silhouette metric59, and K-means were performed thereafter using the Euclidean distance.
UTX and JunB ChIP-PCR
ChIP was performed as described with modifications56. Briefly, 2 × 106 FACS-sorted iNKT cells were fixed with 1% formaldehyde (EMS) for 5 min at 25 °C. UTX antibody (Bethyl, A302-374) and JunB antibody (CST, C37F9) were incubated with beads for 3–5 hours before incubating with sonicated chromatin overnight. ChIP DNA was purified and quantified by real-time PCR using the iQ SYBR Green Supermix (Bio-Rad). The following primer sequences were used:
S100a6 promoter, forward, 5′-GAAGGTTCAGCACACAAGCC-3′,
S100a6 promoter, reverse, 5′-CCCAAAGGAGACCAGTGCAA-3′,
Il2rb promoter, forward, 5′-TAAGATCTCCTCCTACGCCCA-3′,
Il2rb promoter, reverse, 5′-ATGTGTGAGATGTAGCGGGG-3′
Tbx21 promoter, forward, 5′-TGAAACTTCACTGGAGCGGG-3′,
Tbx21 promoter, reverse, 5′-TTCATAAAGCCACAGCAAAGGC-3′,
Klrd1 promoter, forward, 5′-TGCATCTGTGTCCCACCAAC-3′,
Klrd1 promoter, reverse, 5′-CCAAGGATGGTGCAGAGATGT-3′,
Actb promoter, forward, 5′-TGCCCCATTCAATGTCTCGG-3′,
Actb promoter, reverse, 5′-CCACACAAATAGGGTCCGGG-3′
Immunoprecipitation and Immunoblot
Cell lysates were incubated with 5 µg anti-UTX antibody (Bethyl), or anti-rabbit IgG control antibody (Santa Cruz) overnight at 4 °C followed by 2 h of incubation with Dynabeads Protein G for immunoprecipitation. Protein complexes bound to antibody and beads were washed 5 times and eluted with Laemmli sample buffer. Samples were resolved by SDS-PAGE. Protein interaction was analyzed by immunoblotting with anti-PLZF (R&D, AF2944), or anti-JunB primary antibodies, and goat anti-rabbit IgG-HRP secondary antibodies, followed by visualization using the Western Lightning Plus ECL detection kit (PerkinElmer).
Transcription factor (TF) target motif enrichment using Haystack
To identify potential TFs that mediate the observed gene expression changes, we scanned gene promoters for enriched TF motifs using the haystack_motifs utility from the Haystack Pipeline (https://github.com/lucapinello/Haystack)41. For analysis, we used the default parameters for the mouse genome: the mm9 genome assembly and the JASPAR motif database60. Subsequently, to remove potential false positives, we further integrated gene expression data in order to assess their specificity and concordance with gene expression changes. In particular, we filtered TF motifs based on visual exploration of the expression ratios of TFs and their targets between wild-type (WT) and UTX-KO (KO), using the haystack_tf_activity_plane utility.
Super-enhancer analysis
In order to call super-enhancers, we used ROSE, a software method previously developed by the Young lab (http://younglab.wi.mit.edu/super_enhancer_code.html)14. In particular, we first considered as enhancers peaks of H3K27ac obtained by the software MACS2, and then called ROSE with default parameters to define the super-enhancers. Using these settings, peaks closer than 12.5 kb are stitched together and then ranked based on the H3K27ac intensity. To assign super-enhancers to genes, we used again ROSE with default settings.
Genome-wide ATAC-Seq analysis
ATAC-Seq was performed as previously described42. Briefly, WT and UTX-KO thymic iNKT cells were sorted by FACS and lysed in lysis buffer containing 10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-360. The transposition reaction was carried out for 45 min using the Illumina Nextera DNA preparation kit (FC-121-1030). DNA was purified using the Qiagen MinElute PCR purification kit. Subsequently, library amplification was performed using Nextera primers and NEBNext high-fidelity PCR master mix. qPCR side reaction was done to determine optimum library amplification in order to reduce GC and size bias in the library. The amplified library was purified using the Qiagen PCR purification kit and sequenced on the Illumina Hi-Seq 2500. Paired reads were aligned to the reference genome using Bowtie2 in paired end mode and with the parameter -X2000 (fragments up to 2kb). ATAC-Seq peaks were called using MACS2 with the following parameters: macs2 callpeak --nomodel --shift -100 --extsize 200. Super-enhancers (SEs) more accessible in WT were defined as WT SE regions containing unique WT ATAC-Seq peaks, while SEs less accessible in WT were defined as WT SE regions containing unique KO ATAC-Seq peaks.
Integrative analysis of SE regions, ATAC-Seq, H3K27me3 levels, and gene expression
To study the potential connection between the level of H3K27me3 and chromatin accessibility profiled by ATAC-Seq, we used the defined SEs based on the unique ATAC-Seq peaks in WT or KO and profiled the average H3K27me3 level (RPM) in those regions. As a control set, we used random regions in the genome. To correlate chromatin accessibility in SEs with gene expression, we used the average gene expression level of the genes mapped with the ROSE pipeline in each group (closest genes and overlapping genes). As control sets, we used all the genes and the genes mapped to all the WT SEs in iNKT cells.
Statistical analysis
Data are presented as mean ± standard error or standard deviation. All samples represent independent experiments with biological replicates. Sample size was determined based on the results of preliminary experiments. No blinding was applied in this study. Two-tailed unpaired t test, Mann-Whitney U test, and one-way ANOVA with post multiple comparisons were applied as indicated, and the P-values are shown for each figure. If not otherwise indicated the P value was not statistically significant (P > 0.05). All statistical analyses for animal studies were calculated using Prism software (GraphPad).
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (S.H.O.), and by NIH P30DK0492 (S.H.O.) and RO1 AI083426 (F.W.). S.B. was supported by the Cancer Research Institute Predoctoral Emphasis Pathway in Tumor Immunology. J.H.K. was supported by the National Research Foundation of Korea (2012R1A6A3A03040248). L.P. was supported by NHGRI Career Development Award K99HG008399. We thank the NIH Tetramer Facility for CD1d-tetramers, the CCCB sequencing facility at DFCI for Illumina HiSeq2000 sequencing, Microarray core facility at the DFCI Molecular Biology Core Facilities for microarray analysis and the DFCI Jimmy Fund FACS core facility for cell sorting.
Footnotes
Author Contributions S.B. designed, performed and interpreted experiments involving gene expression analysis, ChIP-Seq, ATAC-Seq and lentiviral transduction. J.H.K. designed, performed and interpreted experiments involving iNKT cell analysis by flow cytometry with help from Y.H. L.P. designed, performed and interpreted bioinformatics analysis with help from J.H. S.B. utilized UTX-KO and JMJD3-KO mice with help from M.A.K. P.P.D., R.A.B. and R.D. assisted with ChIP-Seq analysis. M.E.X. performed immunoprecipitation, qRT-PCR and ChIP-PCR under the guidance of S.B. A.H. and E.P. provided JunB-KO mice. Ö.H.Y. and W.N.H. participated in the design and interpretation of experiments. G.C.Y. supervised bioinformatics analysis. S.H.O. and F.W. designed and supervised experiments. S.B., J.H.K., S.H.O. and F.W. wrote the manuscript with support from L.P.
Competing Financial Interests
The authors declare no competing financial interests.
Data availability
Data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) database under accession numbers GSE84238 and GSE84015.
Supplementary Information is available in the online version of the paper.
REFERENCES
- 1.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu. Rev. Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 2.Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d-restricted antigens by natural killer T cells. Nat. Rev. Immunol. 2012;12:845–857. doi: 10.1038/nri3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawano T, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 4.Cui J, et al. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 5.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J. Exp. Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazarevic V, et al. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat Immunol. 2009;10:306–313. doi: 10.1038/ni.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seiler MP, et al. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat Immunol. 2012;13:264–271. doi: 10.1038/ni.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuda JL, et al. Homeostasis of V alpha 14i NKT cells. Nat. Immunol. 2002;3:966–974. doi: 10.1038/ni837. [DOI] [PubMed] [Google Scholar]
- 11.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 12.Engel I, Kronenberg M. Transcriptional control of the development and function of Valpha14i NKT cells. Curr. Top. Microbiol. Immunol. 2014;381:51–81. doi: 10.1007/82_2014_375. [DOI] [PubMed] [Google Scholar]
- 13.Cohen NR, et al. Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nat Immunol. 2013;14:90–99. doi: 10.1038/ni.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell. Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 21.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 22.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 23.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 24.De Santa F, et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welstead GG, et al. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc. Natl. Acad. Sci. U S A. 2012;109:13004–13009. doi: 10.1073/pnas.1210787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansour AA, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488:409–413. doi: 10.1038/nature11272. [DOI] [PubMed] [Google Scholar]
- 28.Li Q, et al. Critical role of histone demethylase Jmjd3 in the regulation of CD4+ T-cell differentiation. Nat. Commun. 2014;5:5780. doi: 10.1038/ncomms6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S, Lee JW, Lee SK. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev. Cell. 2012;22:25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thieme S, et al. The histone demethylase UTX regulates stem cell migration and hematopoiesis. Blood. 2013;121:2462–2473. doi: 10.1182/blood-2012-08-452003. [DOI] [PubMed] [Google Scholar]
- 31.Ntziachristos P, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–517. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cook KD, et al. T Follicular Helper Cell-Dependent Clearance of a Persistent Virus Infection Requires T Cell Expression of the Histone Demethylase UTX. Immunity. 2015;43:703–714. doi: 10.1016/j.immuni.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manna S, et al. Histone H3 Lysine 27 demethylases Jmjd3 and Utx are required for T-cell differentiation. Nat. Commun. 2015;6:8152. doi: 10.1038/ncomms9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobenecker MW, et al. Coupling of T cell receptor specificity to natural killer T cell development by bivalent histone H3 methylation. J. Exp. Med. 2015;212:297–306. doi: 10.1084/jem.20141499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages of thymic NKT cell development. J. Exp. Med. 2005;202:485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14:1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat. Immunol. 2001;2:971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 39.Mao AP, et al. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc. Natl. Acad. Sci. U S A. 2016;113:7602–7607. doi: 10.1073/pnas.1601504113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinello L, Xu J, Orkin SH, Yuan GC. Analysis of chromatin-state plasticity identifies cell-type-specific regulators of H3K27me3 patterns. Proc. Natl. Acad. Sci. U S A. 2014;111:E344–E353. doi: 10.1073/pnas.1322570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanda M, et al. Transcriptional regulator Bhlhe40 works as a cofactor of T-bet in the regulation of IFN-gamma production in iNKT cells. Proceedings of the National Academy of Sciences of the United States of America. 2016 doi: 10.1073/pnas.1604178113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engel I, et al. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat. Immunol. 2016;17:728–739. doi: 10.1038/ni.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams KL, et al. BATF transgenic mice reveal a role for activator protein-1 in NKT cell development. J. Immunol. 2003;170:2417–2426. doi: 10.4049/jimmunol.170.5.2417. [DOI] [PubMed] [Google Scholar]
- 45.Jordan-Williams KL, Poston S, Taparowsky EJ. BATF regulates the development and function of IL-17 producing iNKT cells. BMC Immunol. 2013;14:16. doi: 10.1186/1471-2172-14-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawson VJ, Maurice D, Silk JD, Cerundolo V, Weston K. Aberrant selection and function of invariant NKT cells in the absence of AP-1 transcription factor Fra-2. J. Immunol. 2009;183:2575–2584. doi: 10.4049/jimmunol.0803577. [DOI] [PubMed] [Google Scholar]
- 47.Thomsen MK, et al. JUNB/AP-1 controls IFN-gamma during inflammatory liver disease. J. Clin. Invest. 2013;123:5258–5268. doi: 10.1172/JCI70405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pereira RM, et al. Jarid2 is induced by TCR signalling and controls iNKT cell maturation. Nat. Commun. 2014;5:4540. doi: 10.1038/ncomms5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vahedi G, et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520:558–562. doi: 10.1038/nature14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Methods
- 51.Passegue E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 2004;119:431–443. doi: 10.1016/j.cell.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 52.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 53.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 54.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 55.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kurachi M, et al. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat Immunol. 2014;15:373–383. doi: 10.1038/ni.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. Journal of Computational and Applied Mathematics. 1987;20:53–65. [Google Scholar]
- 60.Mathelier A, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.