Abstract
The structural maintenance of chromosomes (SMC) family of proteins play key roles in the organization, packaging, and repair of chromosomes. Cohesin (Smc1+3) holds replicated sister chromatids together until mitosis, condensin (Smc2+4) acts in chromosome condensation, and Smc5+6 performs currently enigmatic roles in DNA repair and chromatin structure. The SMC heterodimers must associate with non-SMC subunits to perform their functions. Using both biochemical and genetic methods, we have isolated a novel subunit of the Smc5+6 complex, Nse3. Nse3 is an essential nuclear protein that is required for normal mitotic chromosome segregation and cellular resistance to a number of genotoxic agents. Epistasis with Rhp51 (Rad51) suggests that like Smc5+6, Nse3 functions in the homologous recombination based repair of DNA damage. We previously identified two non-SMC subunits of Smc5+6 called Nse1 and Nse2. Analysis of nse1-1, nse2-1, and nse3-1 mutants demonstrates that they are crucial for meiosis. The Nse1 mutant displays meiotic DNA segregation and homologous recombination defects. Spore viability is reduced by nse2-1 and nse3-1, without affecting interhomolog recombination. Finally, genetic interactions shared by the nse mutants suggest that the Smc5+6 complex is important for replication fork stability.
INTRODUCTION
Both endogenous and exogenous agents constantly threaten genomic integrity. The DNA double-strand break (DSB) is a potentially life-threatening lesion for a cell and can occur spontaneously during growth, as part of a programmed event such as meiosis or as the result of exposure to environmental genotoxic agents. Multiple mechanisms exist to repair DSBs, including nonhomologous end joining and homologous recombination (HR) (Paques and Haber, 1999; Symington, 2002). The HR pathway serves to maintain all original genetic information during DSB repair. Many components of that pathway have been identified and characterized (Paques and Haber, 1999; Symington, 2002). HR involves multiple steps. The DSB is first resected to generate a recombinogenic 3′ overhang that invades a homologous sequence (e.g., sister chromatid), forming a displacement or D-loop. The invasion step is catalyzed by a number of proteins including the Escherichia coli RecA homologue, Rad51. D-loop formation primes repair synthesis, which is followed by resolution of the recombined duplexes and ligation, producing intact duplex DNA molecules (Paques and Haber, 1999; Symington, 2002). The resolution of recombined DNA duplexes can yield products in which there has been reciprocal exchange of flanking markers (crossover) or not. During mitotic growth gene conversion is infrequently accompanied by crossover, whereas during meiosis, crossover recombination is much more prevalent (Paques and Haber, 1999).
Efficient DNA repair requires modulation of both local and higher order chromatin structure. Interestingly, the structural maintenance of chromosomes (SMC) proteins have recently been recognized as important players in DNA repair (Hirano, 2002; Jessberger, 2003). The SMC family of essential proteins includes cohesins (Smc1+3), required for sister chromatid cohesion; condensins (Smc2+4), involved in DNA compaction during mitosis; and the less well studied complex of Smc5+6, required for DNA repair (Hirano, 2002; Jessberger, 2003). SMC proteins contain Nand C-terminal nucleotide-binding motifs, called Walker A and B domains, separated by an extensive coiled-coil region that contains a central hinge (Hirano, 2002; Jessberger, 2003). The hinge allows SMC proteins to fold back on themselves, forming an intramolecular coiled-coil and creating an ATPase by juxtaposing the Walker A and B domains (Haering et al., 2002). SMC proteins form stable heterodimers, most likely through interactions mediated by their hinge regions (Haering et al., 2002).
Hypomorphic mutations of proteins in the SMC complexes render cells hypersensitive to genotoxic stress (Birkenbihl and Subramani, 1992; Lehmann et al., 1995; Sjogren and Nasmyth, 2001; Aono et al., 2002; Fujioka et al., 2002; Kim et al., 2002b; Yazdi et al., 2002; McDonald et al., 2003; Harvey et al., 2004). Cohesin may facilitate the homologous recombination repair of DSBs by holding sister-chromatids in proximity, promoting identification of an intact homologous duplex. However, cohesin is recruited to sites of laser-induced DSBs, supporting a more direct role in the repair process (Kim et al., 2002a). The role of condensin in DNA repair is unknown, but a direct involvement is suggested by the finding that a subunit of the complex interacts with DNA ligase IV (Przewloka et al., 2003).
Smc6 of the Smc5+6 heterodimer was initially identified by analysis of radiation-sensitive mutants in fission yeast and Smc5 was subsequently identified by its homology to Smc6 (Lehmann et al., 1995; Verkade et al., 1999; Fousteri and Lehmann, 2000). Like cohesin and condensin, the Smc5+6 complex is essential for viability and seems to control chromatin structure (Lehmann et al., 1995; Harvey et al., 2004). Studies on mutants of the complex in fission yeast have demonstrated that it has a role in the HR-based repair of DNA damage and that it may also influence checkpoint maintenance (Lehmann et al., 1995; Verkade et al., 1999; McDonald et al., 2003; Harvey et al., 2004).
SMC heterodimers need to associate with specific non-SMC family subunits to be functional. Non-SMC subunits of the cohesin, condensin and Smc5+6 complexes have been isolated and characterized (Fujioka et al., 2002; Hirano, 2002; Jessberger, 2003; McDonald et al., 2003; Harvey et al., 2004). Mutants of these non-SMC subunits display phenotypes very similar to those of the SMC mutants, consistent with their interdependent functions. Budding yeast cohesin subunit SCC1 controls the association of cohesin with chromosomes, perhaps by closing a loop-shaped complex via interactions with both head groups of SMC1+3, which can encircle chromosomes (Haering et al., 2002; Jessberger, 2003). At mitosis, separase (ESP1) cleaves SCC1, thus abrogating the cohesin loop structure and sister chromatid cohesion. Interestingly, the Smc5+6 non-SMC subunits Nse1 and Nse2 contain zinc finger domains related to the RING and Miztype domains, respectively (Fujioka et al., 2002; McDonald et al., 2003). This suggests that they are not only structural components of the complex, but likely act as E3-ligases to modify target proteins with ubiquitin or SUMO, thereby modulating their functions (Wu et al., 1997; Freemont, 2000; Joazeiro and Weissman, 2000; Hari et al., 2001; Takahashi et al., 2001).
We recently identified Nse1 and Nse2 as non-SMC subunits of the Smc5+6 complex (McDonald et al., 2003). However, previous biochemical analysis of Smc5+6 copurifying proteins indicated that there are more than two non-SMC subunits (Fousteri and Lehmann, 2000). Using both mass spectrometry and yeast two-hybrid methods we have identified a novel evolutionary conserved DNA repair protein, Nse3, as a third subunit of the Smc5+6 complex. Cells expressing hypomorphic Nse3 are hypersensitive to replication arrest, UV irradiation, and DSBs caused by gamma irradiation or camptothecin. Genetic studies support a role for Nse3 in DNA repair via homologous recombination. In addition, analysis of Smc5+6 subunit mutants nse1-1, nse2-1, and nse3-1, demonstrates a crucial role for the Smc5+6 complex in meiosis. Finally, nse mutants depend on Mus81-Eme1, Rqh1, and Brc1 for viability, suggesting that Smc5+6 may be important to reduce rates of replication fork stalling and collapse.
MATERIALS AND METHODS
General Techniques
Standard fission yeast methods and media were used in these studies (Moreno et al., 1991). UV and ionizing radiation sensitivity assays were performed as described previously (Boddy et al., 2000). For hydroxyurea (HU; Sigma-Aldrich, St. Louis, MO) and camptothecin (CPT; Sigma-Aldrich) sensitivity assays, plates were supplemented with the indicated concentrations of drug. Mitotic recombination assays were performed as described previously (Fortunato et al., 1996), with one modification. Instead of minimal (EMM) media to select ade+ recombinants, we used YEA supplemented with 200 μg/ml guanine.
Generation of Tagged, Deleted, and Mutated Genes
Nse3 was deleted by replacement of the entire open reading frame (start to stop codon) with the kanMx6 module as described in Bahler et al., 1998, producing heterozygous diploids (Bahler et al., 1998). Epitope-tagged Nse proteins and Smc5 were generated using a polymerase chain reaction (PCR)-based method to place a myc, green fluorescent protein (GFP), or TAP epitope at the C terminus of each protein and mark the allele with the kanMx6 gene (Bahler et al., 1998).
The nse mutant alleles were generated using PCR. Genomic DNA was isolated from yeast that contained the epitope tagged nse-myc:kanMx6 alleles. The entire genomic locus containing each allele was amplified by PCR by using standard conditions (from start codon to 100 bp downstream of KanMx6). The amplified loci were then reamplified in four parallel PCR reactions. The PCR reactions were pooled and transformed into Schizosaccharamyces pombe by using the transformation protocol described in Bahler et al., 1998, and transformants were selected by growth on YES media containing G418 (to select for kanMx6) at 25°C. Stable transformants were tested for replacement of the endogenous nse loci by the transformed nse-myc:kanMx6 alleles as described in Bahler et al., 1998. Stable transformants were then tested for temperature sensitivity and drug sensitivity by plating the strains on YES media at 36°C or on YES plates containing 5 mM hydroxyurea. Strains that displayed temperature and/or hydroxyurea sensitivity were transformed with an episomal plasmid containing the wild-type genomic nse genes to confirm that the strain defects were rescued by and therefore, allelic to the respective genes.
Immunoblotting and Microscopy Techniques
Immunoblotting was performed as described using extracts made from cells lysed in a bead beater (Boddy et al., 2000). Briefly, cells were lysed in buffer A (50 mM Tris pH 8, 150 mM NaCl, 2 mM EDTA, 10% glycerol, 0.2% Nonidet P-40, 5 μg/ml each of leupeptin, pepstatin, and aprotinin, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) and resolved in 10% SDS-PAGE. Proteins were transferred to Immobilon membrane, blocked in 5% milk in Tris-buffered saline and 0.3% Tween 20, and probed with antibodies to the epitope. Myc-tagged proteins were detected with anti-myc antibody (9E10 at 1:5000; Santa Cruz Biotechnology, Santa Cruz, CA), and the TAP tag was detected with peroxidase antiperoxidase reagent (at 1:2000 dilution; Sigma-Aldrich).
For TAP-tag immunoprecipitation experiments, cells were lysed in buffer A and IgG-Sepharose (Pfizer, New York, NY) was added to the lysates followed by incubation at 4°C for 1.5h with rotation. Complexes were collected by centrifugation and washed three times with buffer A before resuspension in SDS-PAGE loading buffer.
The entire Nse3 coding sequence was cloned into pHMTc, a derivative of the pMal-c2 × vector (NEB; Ryder et al., 2004) to express a maltose binding protein fusion of Nse3. MBP-Nse3 fusion protein was expressed in BL21 (DE3), purified on amylose resin (NEB) and used to inoculate rabbits. The resulting sera were affinity purified against the MBP-Nse3 protein and used at a 1:400 dilution for Western blotting.
Indirect immunofluorescence microscopy was performed using established methods (Lopez-Girona et al., 1999). GFP was visualized in live cells that were costained with 4,6-diamidino-2-phenylindole (DAPI) at 25 μg/ml. Cells were photographed with Nikon Eclipse E800 microscope equipped with a Photometrics Quantix charge-coupled device camera. Samples for visualization of meiotic figures were prepared as described previously (Boddy et al., 2001).
Identification of Smc5 Interacting Proteins
Proteins associating with Smc5-TAP were identified by multidimensional protein identification technology (MudPIT) by using established methods (Boddy et al., 2001; Washburn et al., 2001; McDonald and Yates, 2002). Briefly, cells (∼50 g wet weight) expressing Smc5-TAP at the genomic locus were frozen in liquid nitrogen and lysed using a motorized mortar and pestle (Retsch, Newtown, PA) in buffer A (50 mM Tris pH 8, 150 mM NaCl, 2 mM EDTA, 10% glycerol, 0.2% Nonidet P-40, 5 μg/ml each of leupeptin, pepstatin, and aprotinin, and 1 mM PMSF). Smc5-TAP was purified from clarified lysate as described previously (Rigaut et al., 1999). The final eluate was precipitated with trichloroacteic acid [25% (vol/vol)] for 1 h on ice. The precipitate was centrifuged (Eppendorf, Westbury, NY) at a relative centrifugal force of 16. The pellet was washed twice with acetone (-20°C) and air dried. The sample was reduced and alkylated using dithothreitol and iodoacetamide and then sequentially digested with endonuclease lyse-C (Roche Diagnostics, Indianapolis, IN) and soluble trypsin (Roche Diagnostics) (McCormack et al., 1997). The resulting peptide mixture was analyzed by MudPIT (Link et al., 1999; Washburn et al., 2001) with modifications described by McDonald et al. (2002) and MacCoss et al. (2002) (MacCoss et al., 2002; McDonald and Yates, 2002). Tandem mass spectra were searched against the latest version of the pompep database to which common contaminants such as keratin and trypsin were added (These sequence data were produced by the S. pombe Sequencing Group at the Sanger Centre and can be obtained from ftp://ftp.sanger.ac.uk/pub/yeast/Pombe/Protein_data/). Search results were filtered and grouped using the DTASelect program, and identifications were confirmed through manual evaluation of spectra. Common background proteins were also excluded by comparing the Smc5-TAP data set to the large number of other data sets obtained by purification of unrelated proteins in the laboratory.
Yeast Two-Hybrid Screen
The Nse1 cDNA was amplified using NdeI and BamH1 containing primers to allow cloning into pAS404 (Nakashima et al., 1999). pAS404-Nse1 was integrated at the TRP1 locus of Y190 yeast (Harper et al., 1993). This strain was used to screen an S. pombe cDNA library (BD Biosciences Clontech, Palo Alto, CA).
Strains
All strains are ura4-D18 leu1-32 unless otherwise stated. PR109, h-ura4-D18 leu1-32; NBY402, h+ smc5:myc:kanMx6; NBY460, h+ smc5:TAP:kanMx6; NBY468, h+ nse1:myc:kanMx6; NBY526, h+ nse1-1:myc:kanMx6; NBY527, h+ nse2-1:myc: kanMx6; NBY564, h-nse1-1:myc:kanMx6; NBY563, h-nse2-1:myc:kanMx6; NBY5111, h-nse3-1:myc:kanMx6; NBY511A, h+ nse3-1:myc:kanMx6; NBY531, nse2: TAP:kanMx ura4+; NBY770, h-nse4:TAP:kanMx6; NBY782, nse4:TAP:kanMx6 nse1:myc:kanMx6; NBY779, nse4:TAP:kanMx6 smc5:myc:kanMx6; NBY532, nse1: TAP:kanMx ura4+; NBY668, nse3:GFP:kanMx; NBY514, nse3::kanMx/nse3+ ade6-216/ade6-210; NBY648, nse3-1:myc:kanMx6 rhp51::ura4+; NBY557, nse1-1:myc: kanMx6 ade6-L469/pUC8/ura4+/ade6-M375; NBY558, nse2-1:myc:kanMx6 ade6-L469/pUC8/ura4+/ade6-M375; NBY645, nse3-1:myc:kanMx6 ade6-L469/pUC8/ura4+/ade6-M375; PS2345, rhp51::ura4+; PR2776, h-rec12::LEU2+ ade6-52; NBY282, h+ rec12::LEU2+; NBY573A, h+ nse1-1:myc:kanMx6 rec12::LEU2+; NBY573B, h-nse1-1:myc:kanMx6 rec12::LEU2+; NBY384, h+ ade7-152 ura4+ leu1+; NBY619, h-nse1-1:myc:kanMx6 ade7-152 ura4+ leu1+; NBY620, h-nse2-1: myc:kanMx6 ade7-152 ura4+ leu1+; NBY647, h-nse3-1:myc:kanMx6 ade7-152 leu1+; NBY651, h+ nse1-1:myc:kanMx6 ade6-L469; NBY650, h-nse1-1:myc:kanMx6 ade6-M26; NBY653, h-nse2-1:myc:kanMx6 ade6-M375; NBY654, h+ nse2-1:myc:kanMx6 ade6-L469; NBY645, h+ nse3-1:myc:kanMx6 ade6-L469; NBY646, h-nse3-1:myc: kanMx6 ade6-M375; PR2918, h+ ade6-M26 ura4+ leu1+; PR2919, h-ade6-L469 ura4+ leu1+; PR2914, h+ ade6-M375 ura4+ leu1+; NBY655, h+ eme1::ura4+; NBY125, h-mus81::kanMx; NBY231, h-brc1::kanMx6; NBY226, h-rad60-3; NBY202, h+ rqh1::ura4+.
RESULTS
Nse3, a Novel Non-SMC Component of the Smc5+6 Complex
We recently described the identification and characterization of two novel Smc5+6 non-SMC subunits, Nse1 and Nse2 (McDonald et al., 2003). Using both yeast two-hybrid and mass spectrometry-based methods, we have identified a third non-SMC component of the Smc5+6 complex (Figure 1A). Three independent clones encoding an uncharacterized protein, SPCC645.04, were obtained in a yeast two-hybrid screen by using Nse1 as bait. We also obtained single clones of the following genes in the Nse1 two-hybrid screen; SPBC685.06 (40s ribosome), SPBC14F5.04c (Pgk1), SPCC1827.05c (predicted RNA binding protein), and SPAC6G10.07 (largest component of nuclear cap binding complex). We did not identify any of the other known Smc5+6 complex components. It is possible that this reflects a direct interaction between Nse1 and SPCC645.04, because bridging of protein-protein interactions is less likely crossspecies. However, it is equally possible that the representation of the other Smc5+6 components is lower in the cDNA library.
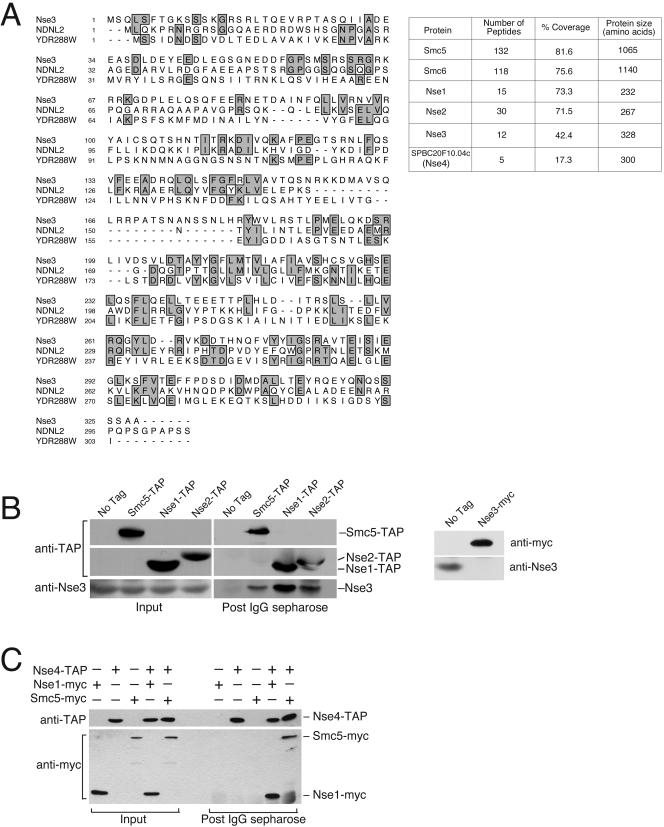
Figure 1.
Identification of Nse3, a novel Smc5+6 complex subunit. (A) Shows an alignment between fission yeast Nse3 (SPCC645.04), human NDNL2 (NP_619649.1) and budding yeast YDR288W. Regions of identity are boxed and shaded. The table on the right shows the number of Smc5, Smc6, Nse1, Nse2, Nse3, and SPBC20F10.04c (Nse4) peptides obtained in mass spectrometric analysis of Smc5-associated proteins. The primary sequence coverage and size of Smc5, Smc6, Nse1, Nse2, Nse3, and SPBC20F10.04c (Nse4) are also given. (B) Confirmation that Nse3 is part of the Smc5+6 complex. Smc5-TAP, Nse1-TAP, and Nse2-TAP proteins, expressed from their endogenous loci, were purified on IgG-Sepharose and the copurifying proteins were analyzed by Western blotting with a polyclonal anti-Nse3 antibody. The right-hand panel shows the specificity of the anti-Nse3 antibody. Nse3 is essential so to test the specificity of anti-Nse3 sera we probed total extracts from cells expressing endogenous Nse3 tagged, or not, with a 13myc epitope. A band of ∼35 kDa is detected with anti-Nse3 in cells without an epitope tag on Nse3. This band is absent in cells in which Nse3 is tagged with a 13myc epitope and is replaced by a lower mobility band corresponding to Nse3-myc that is detected by both anti-Nse3 and anti-myc antisera (anti-myc shown).
In addition to the two-hybrid analysis, we identified 12 peptides covering 42% of the SPCC645.04 primary sequence by mass spectrometric analysis of proteins associated with affinity-purified Smc5 (McDonald et al., 2003). We used MudPIT, which is described in our previous report on the Smc5+6 non-SMC subunits Nse1 and Nse2 (Washburn et al., 2001; McDonald et al., 2003). This system is highly sensitive and can identify individual proteins within a relatively complex mixture of peptides. This system therefore obviates the need for excision of protein bands from a gel for identification. Smc5 was purified in a single step, via an epitope tag that contains protein A repeats that bind to IgG immobilized on Sepharose beads. The protein and associated factors were eluted from the beads by cleavage of a specific protease site between the protein and epitope tag (Rigaut et al., 1999). Proteins in the eluate were precipitated and subjected to mass spectrometry analysis. We have used this system extensively for identification of protein complexes in S. pombe and have thus generated a comprehensive list of common background proteins (Boddy et al., 2001; Boddy et al., 2003; McDonald et al., 2003). Comparison of the Smc5-TAP data set with previous purifications allowed us to exclude common background such as ribosomal proteins, abundant metabolic enzymes, and actin. After removal of background, good peptide coverage was obtained for Smc5, Smc6, Nse1, Nse2, and Nse3 (Figure 1A, right). The identification of these known Smc5+6 components in addition to Nse3 validates the system. These independent identifications of SPCC645.04, which we call Nse3 (non-Smc element 3), strongly suggest that the interaction between Nse3 and components of the Smc5+6 complex is physiologically relevant.
To confirm that Nse3 interacts with the Smc5+6 complex in vivo, we affinity purified endogenous Smc5, Nse1, and Nse2 that were tagged at their C termini with the TAP epitope (Rigaut et al., 1999). Coprecipitating proteins were analyzed by Western blotting with an anti-Nse3 polyclonal antibody (Figure 1B). As anticipated, Nse3 specifically coprecipitated with all three components of the Smc5+6 complex.
Nse3 is conserved across evolution from yeast to man (Figure 1A). Interestingly, Nse3 shows homology to human MAGE-G1 (also called NDNL2), a protein that contains a melanoma antigen-encoding gene (MAGE) domain. Whereas the function of this family is currently unknown, NDNL2 suppresses cell growth when ectopically expressed and binds to the transcription factor E2F1 (Kuwako et al., 2004). Nse3 also shares homology with the uncharacterized essential Saccharomyces cerevisiae protein YDR288W.
In addition to Nse3, we identified peptides covering 17% of the primary sequence of an uncharacterized protein, SPBC20F10.04, which we propose to call Nse4 (Figure 1A, right). Nse4 is a 300-amino acid protein and was incorrectly annotated in the database as a 253-amino acid protein. We have confirmed that Nse4 interacts with the Smc5 and Nse1 components of the Smc5+6 complex (Figure 1C). Nse4 was TAP-tagged at its chromosomal locus in strains that also expressed Nse1-myc or Smc5-myc from their own promoters. Immunoprecipitation of Nse4-TAP resulted in the specific coprecipitation of both Smc5-myc and Nse1-myc (Figure 1C). Our preliminary studies show that Nse4 is an essential nuclear protein with terminal phenotypes that are consistent with its interaction with components of the Smc5+6 complex (our unpublished data). Nse4 is well conserved with homologues from yeast to human (human FJL20003, BLAST expect value 7e-19; Yeast YDL105w, BLAST expect value 7e-10). Notably, budding yeast YDL105w was recently identified in a high throughput mass spectrometry and two-hybrid screen as a component of the yeast SMC5+6 complex (Hazbun et al., 2003). YDL105w database annotations (SGD) show that it is an essential nuclear protein of unknown function. These observations suggest that Nse4 and YDL105w are functionally homologous components of the Smc5+6 complex. A detailed analysis of Nse4 will be presented elsewhere.
Nse3 Is an Essential Nuclear Protein
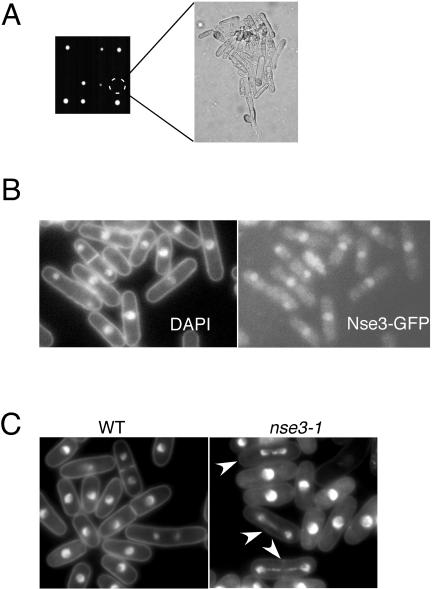
The Smc5+6 complex performs essential but currently unknown functions. If Nse3 has a central role in the Smc5+6 complex, then it should also be essential for growth. We generated a heterozygous diploid in which one allele of Nse3 was replaced by the kanMx6 gene. The diploid was sporulated and the asci produced were dissected for genetic analysis. Analysis of tetrads showed 2:0 segregation of viability and kanamycin resistance, demonstrating that Nse3 is essential (Figure 2A). Spores deleted for Nse3 germinated and produced microcolonies (∼15 cells) of highly elongated cells before growth stopped. This terminal phenotype matches that of deletions of any component of the Smc5+6 complex (Lehmann et al., 1995; Fousteri and Lehmann, 2000; McDonald et al., 2003). We also found that Nse3-GFP is a predominantly nuclear protein throughout the cell cycle (Figure 2B).
Figure 2.
Nse3 is an essential nuclear protein required for normal chromosome segregation. (A) A heterozygous nse3+/nse3::kanMx6 diploid was sporulated, and tetrads were dissected. Tetrad analysis demonstrates a 2:0 segregation of viability and the kanMx6 marker showing that Nse3 is essential for growth. The terminal phenotype of nse3::kanMx6 cells is shown in the right panel. (B) Nse3-GFP was localized in live cells and shows a predominantly nuclear localization. (C) Mutant nse3-1 cells grown at 36°C for 5 h show aberrant segregation of DAPI-stained chromosomes in ∼10% of cells (white arrowheads) compared with wild type. The DNA in nse3-1 mutants was often stretched along the axis of the mitotic spindle.
A hypomorphic mutant of Nse3, nse3-1, shows aberrant mitoses in ∼10% of cells at 36°C (Figure 2C). The majority of aberrations observed have DNA stretched out along the axis of the mitotic spindle. This phenotype matches well that recently described for the loss of function of two other components of the Smc5+6 complex, Nse1 and Smc6 (McDonald et al., 2003; Harvey et al., 2004). The nse3-1 phenotype could be consistent with a failure to fully condense chromosomes or, a physical impediment to their segregation such as sister chromatid catenation and unresolved recombination structures.
Nse3-1 Cells Are Hypersensitive to Genotoxic Stress
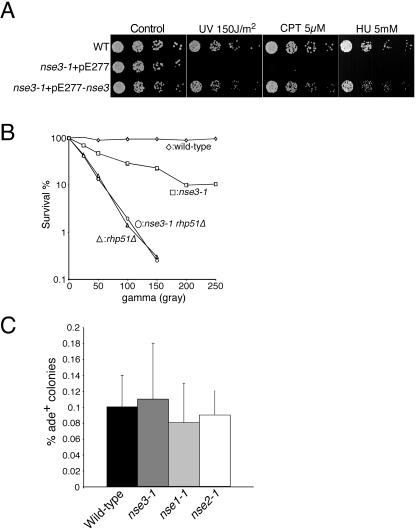
The Smc5+6 complex together with two recently described non-SMC subunits, Nse1 and Nse2, has been shown to mitigate the effects of various forms of DNA damage (Lehmann et al., 1995; Verkade et al., 1999; Fujioka et al., 2002; McDonald et al., 2003; Harvey et al., 2004). Consistently, we found that nse3-1 cells are sensitive to gamma irradiation, UV light, the topoisomerase I poison camptothecin, and replication inhibition by hydroxyurea (Figure 3, A and B). The sensitivity of nse3-1 cells to these agents is rescued by a plasmid carrying the wild-type nse3 gene, demonstrating that the phenotypes arise from nse3 dysfunction (Figure 3A). The sensitivity of nse3-1 cells to ionizing radiation is not additive with that of rhp51Δ cells, which are defective in the homologous recombination repair of DNA DSBs (Figure 3B). This result suggests that Nse3, like Smc5+6, facilitates the repair of DSBs via homologous recombination.
Figure 3.
Nse3 mutant cells are hypersensitive to genotoxic stress. (A) Wild-type (WT) and nse3-1 cells, covered by a control or nse3+ containing plasmid, were serially diluted and spotted at 2500, 500, 100, and 20 cells per spot. Plates contained the indicated concentrations of camptothecin (CPT) and HU or were irradiated with the indicated dose of UV, followed by growth at 25°C. (B) The indicated strains were counted and diluted to give a known number of cells per 100 μl at the start of the experiment. Cells were plated on YES plates after exposure to the indicated doses of gamma irradiation and grown at 25°C for 4-6 d, before counting and determination of survival. (C) Spontaneous mitotic recombination is unaffected in nse mutant cells. For each strain, multiple colonies were picked and suspended in water for counting. Cells were plated at ∼105 cells per YEA + guanine plate and a 400-fold dilution was plated on YES plates to determine actual cell titer. Conversion rates were determined by dividing the number of ade+ colonies on the YEA + guanine plates by the total number of cells plated.
Spontaneous Mitotic Recombination in Nse1, Nse2, and Nse3 Mutants
To gain insight into the function of the Smc5+6 holocomplex, we analyzed the recombination roles of the currently known non-SMC subunits Nse1, Nse2, and Nse3. Epistasis analysis between Nse1, Nse2, Nse3, and Rhp51 suggests that they play a role in the process of HR repair (Figure 3B; McDonald et al., 2003). Therefore, we wished to determine the effect of mutations in these genes on mitotic recombination events. A system has been established in fission yeast to measure recombination rates between heteroalleles of the ade6 gene, ade6-M26, and ade6-469 (Osman et al., 2000). We measured spontaneous recombination events that yield an ade+ phenotype in cells with wild-type and mutant alleles of Nse1, 2, and 3. In wild-type, ade+ cells were obtained at a rate of ∼0.1% (Figure 3C). Interestingly, the rates of spontaneous ade+ recombinants in nse1-1, nse2-1 and nse3-1 cells at 25°C were not notably different from wild type (Figure 3C). Furthermore, the system used allows broad classification of recombination modes that generate each ade+ recombinant (Osman et al., 2000). The ura4+ gene is inserted between the ade6 heteroalleles and so gene conversion events yield ade+ ura4+ cells, whereas other events such as unequal sister-chromatid exchange and intrachromosomal crossover yield ade+ ura4-- cells (Osman et al., 2000). We observed no significant differences in the proportion of gene conversion events between wild-type and the nse mutant backgrounds (our unpublished data). Therefore, although Nse1-3 work with Rhp51 in the repair of DSBs, they are not required for the modes of recombination that generate spontaneous ade+ recombinants in this assay.
Nse1, Nse2, and Nse3 Play a Critical Role in Meiosis
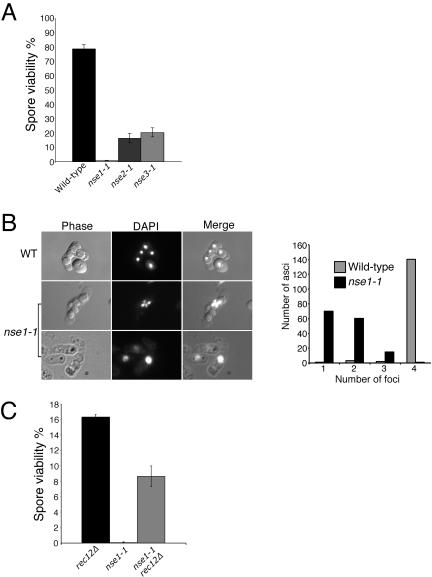
The formation and repair of DSBs is pivotal to the meiotic process, contributing to the correct segregation of chromosomes (Davis and Smith, 2001). Based on the mitotic DSB repair defects of nse cells, we hypothesized that these proteins may also have a role in the repair of meiotic DSBs. Initially, we mated h+ and h-nse mutant haploids at 25°C to determine the viability of spores obtained in these crosses. In comparison with wild-type crosses, nse mutant meioses exhibited low spore viability (Figure 4A). The nse2-1 and nse3-1 mutants yielded reduced (∼20%) spore viability, whereas the nse1-1 mutant gave very low (∼0.8%) spore viability.
Figure 4.
Nse1, Nse2, and Nse3 are critical for normal meiotic progression. (A) Wildtype, nse1-1, nse2-1, and nse3-1 mutant haploids (h+ and h-) were mated on SSALUAH plates at 25°C for 3 d. Spores produced after meiosis were counted and plated on YES plates at 25°C to determine spore viability. (B) Four wild-type spores each with a DAPI-stained genome are shown. Representative pictures of the aberrant nse1-1 meiotic phenotype are shown below. The right-hand panel shows quantification of the nse1-1 meiotic defect. For both wild-type and nse1-1 meiotic products, 146 asci were scored for the number of DAPI foci they contained. Even in nse1-1 asci that contained two or three foci, the DNA was unequally distributed between spores or actually excluded from them, undoubtedly contributing to the low viability of nse1-1 spores (C) The low spore viability of the nse1-1 mutant is largely rescued by abrogating the induction of meiotic recombination. Haploid crosses were performed as described above, but all strains lacked Rec12, which is necessary for double-strand break formation and subsequent recombination.
Wild-type fission yeast meiosis generates four haploid spores as a result of sequential nuclear divisions at meiosis I and II (Figure 4B). A discreet DAPI-stained focus of DNA is observed in each spore in wild-type meioses. We found that nse1-1 crosses produced highly aberrant asci, often containing one large spore, or multiple spores of different sizes (Figure 4B). Strikingly, the DNA was found in one large spore, or incompletely divided between two adjacent spores. The meiotic defects of nse1-1 compared with wild type were also quantified, underscoring the aberrant meioses in the nse1-1 mutant background (Figure 4B, right). The nse1-1 meiotic phenotype suggests that the normal meiotic nuclear divisions are blocked. This phenotype is reminiscent of that observed in the meiotic recombination-defective mutant mus81 (Boddy et al., 2001). In this mutant, DNA fails to segregate at meiosis I due to unresolved recombination structures. However, unlike the mus81 meiotic defect, overexpression of the Holliday junction resolvase RusA does not suppress the nse1-1 meiotic phenotype (our unpublished data).
To determine whether aberrant meiotic recombination was responsible for the nse1-1 phenotype, we used a mutant that is defective in the initiation of meiotic recombination. Before meiosis I, among other proteins an endonuclease called Rec12 (ScSpo11) produces DSBs that are repaired using the intact homologous chromosome as a template (Lin and Smith, 1994). In the absence of Rec12, meiotic recombination is almost totally abolished and chromosomes often segregate aberrantly. Despite aberrant segregation, rec12 mutants yield spores with ∼16% viability (Figure 4C). Interestingly, spores derived from the nse1-1 rec12Δ double mutant crosses display greatly improved spore viability (>50-fold) over the nse1-1 mutant alone (Figure 4C). This result indicates that the predominant role of Nse1 in meiosis is after the initiation of homologous recombination by Rec12. That the nse1-1 rec12Δ cross yields less viable spores than rec12Δ alone may indicate a minor recombination-independent role of Nse1 in meiosis, or reflect the inherently lower plating efficiency of nse1-1 strains. Epistasis with rec12Δ and the terminal meiotic phenotype of the nse1-1 mutant may support a late recombination role for Nse1 (see Discussion).
Meiotic Recombination in nse Mutants
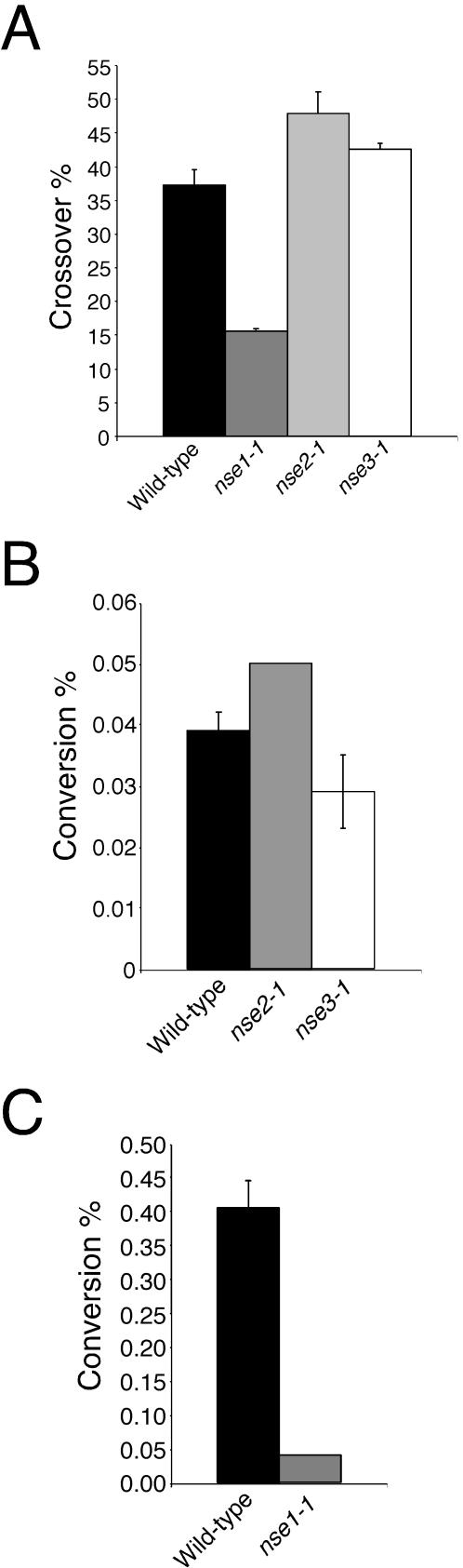
Based on the meiotic phenotypes of nse mutants and the vegetative roles of Nse proteins in DSB repair, we wished to determine their roles in meiotic recombination. We first measured the frequency of crossover in the ade7-leu1 interval on chromosome II. Wild-type, nse2-1, and nse3-1 gave similar rates of crossover in this interval (Figure 5A). However, compared with wild type, nse1-1 resulted in a significant (∼2.5-fold) reduction of crossover between the ade7 and leu1 alleles.
Figure 5.
Effect of nse mutants on the frequency of meiotic crossover in the ade7-leu1 interval and conversion at the ade6 locus. (A) Haploids of the indicated genotype that were additionally auxotrophic for ade7 or leu1 were mated on SSA-LUAH plates at 25°C for 3 d. Random spore analysis was performed by plating the spores first on YES plates and then replica plating colonies onto media selective for either ade7 or leu1. (B) Meiotic intragenic recombination between ade6-M375 and ade6-469 alleles. The effect of nse mutants on gene conversion rates were monitored as for crossover, except that spores (∼105) were plated directly to ade6+ selective YEA + guanine plates at 25°C. Dilutions of the spores were also plated to nonselective YES plates to determine the actual spore titer in each experiment. (C) The effect of nse1-1 on gene conversion was monitored using the recombination hotspot ade6-M26 instead of the ade6-M375 allele, but otherwise as described above.
We also measured gene conversion frequency between different alleles of ade6. Again, despite low spore viability, nse2-1 and nse3-1 crosses produced a comparable rate of ade+ progeny compared with wild-type (Figure 5B). We were unable to obtain sufficient numbers of viable spores to measure conversion rates in nse1-1 crosses by using the ade6-469 and ade6-M375 alleles. Therefore, we used the ade6-469 and ade6-M26 pair that gives an ∼10-fold increase in conversion frequency in wild-type crosses (Szankasi et al., 1988). Using these alleles, we found that nse1-1 strongly decreased gene conversion rates compared with wild type (Figure 5C).
It is important to note that the meiotic studies of the nse mutants were all performed using hypomorphic alleles of these essential genes at 25°C. Therefore, the lack of recombination defects of nse2-1 and nse3-1 may simply reflect a weaker defect in the function of these alleles. It is also possible that the Nse subunits play a role in recombination-dependent and -independent pathways of meiosis. In this case, Nse1 plays a role in both pathways whereas Nse2 and Nse3 play roles only in the latter.
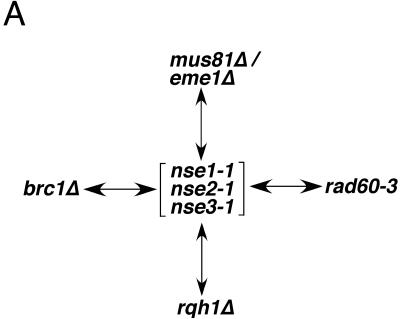
Genetic Interactions of nse Mutants
The physical interaction between the Nse proteins and the Smc5+6 complex, together with their mutant phenotypes, provides strong evidence that they are essential subunits of the Smc5+6 holocomplex. We also performed genetic tests to lend further support to this conclusion (Figure 6). We previously reported that smc6-X (rad18-X) is synthetically lethal in combination with a mutation in Rad60 (rad60-3), a protein that also physically interacts with the Smc5+6 complex (Boddy et al., 2003). In tetrad dissection analyses, we were unable to obtain double mutants between rad60-3 and any of the nse mutants, demonstrating that they are synthetically lethal (Figure 6). Therefore, the genetic interaction between rad60-3 and mutations in the Smc5+6 complex is not gene/allele specific. The Rad60-Smc5+6 physical interaction and the overlapping set of genetic interactions displayed by rad60 and nse mutants suggests that these proteins have codependent functions (Morishita et al., 2002; Boddy et al., 2003). Unlike the Nse proteins, however, Rad60 interacts weakly with components of the Smc5+6 complex (Morishita et al., 2002; Boddy et al., 2003; McDonald et al., 2003). That is, the Nse proteins coprecipitate robustly with the Smc5+6 complex components when expressed at their endogenous levels (Figure 1, B and C; McDonald et al., 2003). However, the Rad60-Smc5 interaction was not observed with standard immunoprecipitation. It was necessary to overexpress Rad60 from the potent nmt1 promoter to detect the relatively weak interaction with Smc5 and Smc6 (Boddy et al., 2003).
Figure 6.
Nse mutant cell viability depends on the activity of a number of genome and replication fork maintenance factors. Arrows indicate synthetic lethality between the mutants shown. Genetic interactions were determined by a combination of tetrad dissections and random spore analyses.
It was previously reported that the BRCT domain containing protein, Brc1, is also required for the viability of smc6-X (rad18-X) cells (Verkade et al., 1999). Consistent with codependent functions of Smc5+6 and the Nse proteins, we found that Brc1 is required for the viability of all nse mutants. That the nse mutants share these same genetic interactions as smc6-X (rad18-X) further supports the characterization of the Nse proteins as non-SMC components of the Smc5+6 complex.
We have observed that all nse mutants at restrictive temperature activate the DNA damage checkpoint, resulting in a Rad3and Chk1-dependent cell cycle arrest in late S phase/G2 (our unpublished data; Rhind and Russell, 2000). Such an arrest is often seen with “leaky” conditional mutants of proteins that are important for the normal completion of replication. In support of a replication-associated role, we identified the Smc5+6 proteins in association with Rad60, which is itself subject to regulation during S phase via its interaction with the replication checkpoint kinase Cds1 (Boddy et al., 2003). Finally, the hypomorphic nse alleles that we have isolated cause hypersensitivity to agents that perturb replication (Figure 3A; McDonald et al., 2003). To explore the nature of the DNA structure defects in nse mutants, we tested which genome maintenance factors are required for the viability of nse cells (Figure 6). In particular, we were interested to test genetic interactions between the nse alleles and mutants known to be defective in replication fork progression/stability.
Interestingly, all the nse mutants are lethal in combination with a loss of Mus81-Eme1 or Rqh1 function, proteins that are required for processing recombination structures that arise during replication (Figure 6; reviewed in Heyer, 2004). Notably, the Nse mutants are not dependent on the Mus81-Eme1-related endonuclease Rad16-Swi10 for viability, suggesting that the specific ability of Mus81-Eme1 to act on replication-associated structures is important in nse cells (our unpublished data).
DISCUSSION
We have identified a novel DNA repair protein that is evolutionarily conserved and essential for normal chromosome maintenance. Nse3 is a third non-SMC subunit of the fission yeast Smc5+6 complex. We and others previously reported the identification of two Smc5+6 subunits called Nse1 and Nse2 (Fujioka et al., 2002; McDonald et al., 2003; Harvey et al., 2004). These subunits contain RING and PIAS-like zinc fingers, respectively, implicating them in the modulation of target protein function via conjugation with ubiquitin and/or SUMO (McDonald et al., 2003). Notably, Nse3 contains a necdin-like or MAGE family domain, most closely related to that found in NDNL2 or MAGE-G1 (Barker and Salehi, 2002). Whereas the function of this domain is presently unknown, several studies in mammalian cells have implicated the family in cell cycle regulation, apoptosis, and neurological disorders. Whereas humans contain >25 MAGE genes, Drosophila, Aspergillus, and yeast seem to contain only one (our unpublished observation; Barker and Salehi, 2002).
In addition to Nse1-3, we have identified a fourth protein, Nse4, which associates with components of the Smc5+6 complex. Consistent with our proposal that Nse4 is a subunit of the Smc5+6 complex, it is an essential nuclear protein with terminal deletion phenotypes that are indistinguishable from those of the other Smc5+6 components (our unpublished observations). Several lines of evidence suggest that Nse1-4 are the core subunits of the Smc5+6 complex in fission yeast. A previous analysis of the ∼1.6-MDa complex found the Smc5+6 heterodimer in complex with four major silver-stained protein bands ranging in size from ∼35 to 45 kDa (Fousteri and Lehmann, 2000). Although the identity of these proteins was not reported, they closely match the size range of Nse1-4. We believe, therefore, that the best represented hits in our mass spectrometry analysis, Nse1-4, likely represent these unidentified bands. Furthermore, a recent high-throughput screen in budding yeast used mass spectrometry and yeast two-hybrid approaches to identify proteins that associate with essential uncharacterized yeast proteins (Hazbun et al., 2003). One of the proteins they purified was YDR288w, the yeast homologue of Nse3. The mass spectrometry analysis revealed interactions of YDR288w with SMC5, SMC6, MMS21 (SpNse2), NSE1 (SpNse1), and YDL105w (SpNse4). These observations strongly support our assignment of Nse1-4 as non-SMC subunits of the Smc5+6 complex and suggest that the subunits we have identified are functionally conserved across species.
Nse3 mutant cells are hypersensitive to a number of DNA damaging agents, as are mutants in other components of the Smc5+6 complex (Lehmann et al., 1995; McDonald et al., 2003; Harvey et al., 2004). The roles of the Smc5+6 holocomplex in the repair of UVand gamma irradiation-induced DNA damage are dependent on the RecA homologue Rhp51 (Rad51; Lehmann et al., 1995; McDonald et al., 2003; Harvey et al., 2004). However, we have found that the Smc5+6 subunits Nse1, Nse2, and Nse3 are not required for the normal modes and rates of spontaneous recombination at a nontandem ade6 heteroallele substrate. Previous studies have shown that Rhp51-deleted cells have an elevated (∼3-fold) ade6+ conversion frequency, but a complete loss of conversion-type recombinants (Osman et al., 2000). Recently, budding yeast SMC6 was also found to be dispensable for spontaneous recombination (Onoda et al., 2004). Interestingly, these authors also found that SMC6 mutants were specifically defective in damage-induced recombination. It is possible that the mechanisms generating spontaneous recombinants are not the same as those responsible for damage-induced recombination, with only Rhp51 required for the former and the latter requiring both Rhp51 and Smc5+6.
Excitingly, we have identified a pivotal role for Nse1, Nse2, and Nse3 in meiotic progression and recombination. Preliminary studies on the human Smc5+6 heterodimer showed that they localize to meiotic chromosomes, supporting a meiotic function of the complex (Taylor et al., 2001). The nse mutants all reduce the viability of spores, the products of fission yeast meiosis. The most severe defect is observed with nse1-1 cells, which yield <1% viable spores. The mitotic DSB repair phenotypes of nse mutants suggested that they may also be defective in the correct repair of programmed DSBs during meiosis. This is the case for nse1-1 because its meiotic defect is suppressed by deleting Rec12, a protein required for meiotic DSB formation and hence for recombination (Lin and Smith, 1994). Fission yeast, unlike budding yeast, does not have a robust meiotic recombination checkpoint. For example, budding yeast Rad54 mutants are DSB repair defective and arrest in meiotic prophase, whereas fission yeast lacking the Rad54 homologue Rhp54, continue through meiosis despite unrepaired DSBs (Shinohara et al., 1997; Catlett and Forsburg, 2003). In addition, Rhp51 mutants were shown to complete meiosis, despite persistent DSBs (Zenvirth and Simchen, 2000; Boddy et al., 2001). In both Rad54 and Rhp51 mutants DNA segregates remarkably well, with asci often containing four spores, each of which has DNA in it. Therefore, the terminal meiotic phenotype of the Nse1 mutant is not consistent with a simple failure to repair DSBs and suggests that there is a physical impediment to the segregation of chromosomes.
Based on the nse1-1 recombination-dependent defects in meiosis, we were interested to determine the effects of the nse mutants on meiotic crossover and intragenic recombination rates. Notably, we found that nse1-1 reduced the frequency of crossover approximately threefold in the ade7-leu1 interval of chromosome II, and intragenic recombination was also reduced between the ade6 heteroalleles. These results are very similar to those for rhp51 mutants in meiosis, which also display low spore viability (∼1%) and an apparent 2.5-fold reduction in crossover recombination (Muris et al., 1997). Considering the central role of rhp51 in DSB repair, this reduction in crossover seems small. It is possible, given the low spore viabilities, that both rhp51 and nse1-1 backgrounds select for those spores in which recombination has proceeded by a minor secondary mechanism. This would give an overestimate of the actual rates of recombination in the meiotic population as a whole.
The genetic interactions we have uncovered between the Nse mutants and genome maintenance factors supports a role for the complex in replication fork stability. In particular, the nse mutants are all synthetic lethal with mutations in the Mus81-Eme1 endonuclease or Rqh1 helicase. The Mus81-Eme1 endonuclease from humans and yeast has been shown to cleave replication fork and Holliday junction structures (reviewed in Heyer et al., 2003; Kai and Wang, 2003). Several lines of evidence suggest that Mus81-Eme1 processes replication-associated structures that form when replication forks stall. Fission yeast Mus81 was identified by virtue of its specific interaction with the replication checkpoint kinase Cds1 (Boddy et al., 2000). Mus81-Eme1 is required in situations that cause replication fork pausing or collapse (reviewed in Heyer et al., 2003; Kai and Wang, 2003). For example, Mus81 mutation severely lowers the restrictive temperature of thermosensitive alleles of DNA polymerases α and δ (Boddy et al., 2000). Furthermore, Mus81 mutants are hypersensitive to chemical agents known to arrest or collapse replication forks such as hydroxyurea, camptothecin (topoisomerase I poison), and methylmethane sulfonate (reviewed in Heyer et al., 2003; Kai and Wang, 2003). Therefore, the tight synthetic lethality between nse mutants and mus81-eme1 mutants strongly supports a role for the Smc5+6 complex in replication fork stability. Smc5+6 mutants may indirectly cause replication fork stalling due to repair or DNA structure defects that impede fork progression, or act directly at the fork to maintain its structure when it stalls at endogenous lesions.
The RecQ family helicases Rqh1 in fission yeast, BLM/WRN in humans, and SGS1 of budding yeast are important for genomic stability. As for Mus81-Eme1, the RecQ family of helicases are directly involved in replication fork maintenance (Constantinou et al., 2000; Doe et al., 2002; Fabre et al., 2002; Osborn et al., 2002) and reviewed in Kai and Wang (2003)). Therefore, the synthetic lethality we observed between nse mutants and rqh1 mutants again implicates Smc5+6 in replication fork maintenance.
We also found that, like smc6 mutants, nse mutants are synthetic lethal with a deletion of the BRCT domain protein Brc1 (Verkade et al., 1999). Little is known about the role of Brc1 in genome stability; however, the budding yeast Brc1 homologue Esc4 has been suggested to be involved in the restart of replication after genotoxic stress (Rouse, 2004).
Finally, we have found that the nse mutants all require Rad60 function for viability. We previously reported that Rad60 physically and genetically interacts with the Smc5+6 heterodimer (Boddy et al., 2003). In this case, partial inactivation of two proteins with codependent functions results in synthetic lethality, most likely by compromising the essential roles of the Smc5+6 holocomplex. Interestingly, we identified Rad60 as a physical interactor and target of the replication checkpoint kinase Cds1 (Boddy et al., 2003). Rad60 is hyperphosphorylated in a Cds1-dependent manner when replication is blocked by hydroxyurea. A mutant of Rad60, rad60-4, is largely refractory to Cds1-dependent regulation and renders cells hypersensitive to replication arrest (Boddy et al., 2003). Regulation of Rad60 during replication arrest supports an important replication fork associated role for the Smc5+6 complex.
In conclusion, we have identified a novel DNA repair protein that is conserved to humans and is a non-SMC subunit of the Smc5+6 complex. Furthermore, we have demonstrated a role for the Smc5+6 non-SMC subunits Nse1, Nse2, and Nse3 in meiosis. Finally, we have provided evidence that Nse1, Nse2, and Nse3 play an important role in mitigating the genome destabilizing effects of replication fork stalling. This role may be regulated by the replication checkpoint kinase Cds1, via Rad60, providing an important avenue for understanding how Cds1 maintains replication fork integrity.
Acknowledgments
We thank James Wohlschlegel for advice on mass spectrometry issues. We also thank Clare McGowan, Curt Wittenberg, and Rob deBruin for valuable comments on the manuscript and members of the Scripps Cell Cycle Groups for support and encouragement. W.H.M is supported by MERK-MGRI-241; J.R.Y. is supported by R01 EY1328801, MERK-MGRI-241, and CA81665 RR11823. This work was funded by National Institutes of Health grant GM-068608 awarded to M.N.B. S.P. is a fellow of the Swiss Cancer League-KLS-01525-02-2004.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-05-0436. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-05-0436.
References
- Aono, N., Sutani, T., Tomonaga, T., Mochida, S., and Yanagida, M. (2002). Cnd2 has dual roles in mitotic condensation and interphase. Nature 417, 197-202. [DOI] [PubMed] [Google Scholar]
- Bahler, J., Wu, J., Longtine, M.S., Shah, N.G., McKenzie, A., Steever, A.B., Wach, A., Phileppsen, P., and Pringle, J.R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951. [DOI] [PubMed] [Google Scholar]
- Barker, P.A., and Salehi, A. (2002). The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J. Neurosci. Res. 67, 705-712. [DOI] [PubMed] [Google Scholar]
- Birkenbihl, R.P., and Subramani, S. (1992). Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand break repair. Nucleic Acids Res. 20, 6605-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy, M.N., Gaillard, P.H., McDonald, W.H., Shanahan, P., Yates, J.R., 3rd, and Russell, P. (2001). Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107, 537-548. [DOI] [PubMed] [Google Scholar]
- Boddy, M.N., Lopez-Girona, A., Shanahan, P., Interthal, H., Heyer, W.D., and Russell, P. (2000). Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20, 8758-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy, M.N., Shanahan, P., McDonald, W.H., Lopez-Girona, A., Noguchi, E., Yates, I.J., and Russell, P. (2003). Replication checkpoint kinase Cds1 regulates recombinational repair protein Rad60. Mol. Cell. Biol. 23, 5939-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett, M.G., and Forsburg, S.L. (2003). Schizosaccharomyces pombe Rdh54 (TID1) acts with Rhp54 (RAD54) to repair meiotic double-strand breaks. Mol. Biol. Cell 14, 4707-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou, A., Tarsounas, M., Karow, J.K., Brosh, R.M., Bohr, V.A., Hickson, I.D., and West, S.C. (2000). Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1, 80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, L., and Smith, G.R. (2001). Meiotic recombination and chromosome segregation in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 98, 8395-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe, C.L., Ahn, J.S., Dixon, J., and Whitby, M.C. (2002). Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277, 32753-32759. [DOI] [PubMed] [Google Scholar]
- Fabre, F., Chan, A., Heyer, W.D., and Gangloff, S. (2002). Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 99, 16887-16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato, E.A., Osman, F., and Subramani, S. (1996). Analysis of spontaneous and double-strand break-induced recombination in rad mutants of S. pombe. Mutat. Res. 364, 14-60. [PubMed] [Google Scholar]
- Fousteri, M.I., and Lehmann, A.R. (2000). A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J. 19, 1691-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont, P.S. (2000). RING for destruction? Curr. Biol. 10, R84-R87. [DOI] [PubMed] [Google Scholar]
- Fujioka, Y., Kimata, Y., Nomaguchi, K., Watanabe, K., and Kohno, K. (2002). Identification of a novel non-structural maintenance of chromosomes (SMC) component of the SMC5-SMC6 complex involved in DNA repair. J. Biol. Chem. 277, 21585-21591. [DOI] [PubMed] [Google Scholar]
- Haering, C.H., Lowe, J., Hochwagen, A., and Nasmyth, K. (2002). Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9, 773-788. [DOI] [PubMed] [Google Scholar]
- Hari, K.L., Cook, K.R., and Karpen, G.H. (2001). The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 15, 1334-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J.W., Adami, G.R., Wei, N., Keyomarsi, K., and Elledge, S.J. (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805-816. [DOI] [PubMed] [Google Scholar]
- Harvey, S.H., Sheedy, D.M., Cuddihy, A.R., and O'Connell, M.J. (2004). Coordination of DNA damage responses via the Smc5/Smc6 complex. Mol. Cell. Biol. 24, 662-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazbun, T.R., et al. (2003). Assigning function to yeast proteins by integration of technologies. Mol. Cell 12, 1353-1365. [DOI] [PubMed] [Google Scholar]
- Heyer, W.D. (2004). A new deal for Holliday junctions. Nat. Struct. Mol. Biol. 11, 117-119. [DOI] [PubMed] [Google Scholar]
- Heyer, W.D., Ehmsen, K.T., and Solinger, J.A. (2003). Holliday junctions in the eukaryotic nucleus: resolution in sight? Trends Biochem. Sci. 28, 548-557. [DOI] [PubMed] [Google Scholar]
- Hirano, T. (2002). The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16, 399-414. [DOI] [PubMed] [Google Scholar]
- Jessberger, R. (2003). SMC proteins at the crossroads of diverse chromosomal processes. IUBMB Life 55, 643-652. [DOI] [PubMed] [Google Scholar]
- Joazeiro, C.A., and Weissman, A.M. (2000). RING finger proteins: mediators of ubiquitin ligase activity. Cell 102, 549-552. [DOI] [PubMed] [Google Scholar]
- Kai, M., and Wang, T.S. (2003). Checkpoint responses to replication stalling: inducing tolerance and preventing mutagenesis. Mutat. Res. 532, 59-73. [DOI] [PubMed] [Google Scholar]
- Kim, J.S., Krasieva, T.B., LaMorte, V., Taylor, A.M., and Yokomori, K. (2002a). Specific recruitment of human cohesin to laser-induced DNA damage. J. Biol. Chem. 277, 45149-45153. [DOI] [PubMed] [Google Scholar]
- Kim, S.T., Xu, B., and Kastan, M.B. (2002b). Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 16, 560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwako, K., Taniura, H., and Yoshikawa, K. (2004). Necdin-related MAGE proteins differentially interact with the E2F1 transcription factor and the p75 neurotrophin receptor. J. Biol. Chem. 279, 1703-1712. [DOI] [PubMed] [Google Scholar]
- Lehmann, A.R., Walicka, M., Griffiths, D.J., Murray, J.M., Watts, F.Z., McCready, S., and Carr, A.M. (1995). The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol. Cell. Biol. 15, 7067-7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y., and Smith, G.R. (1994). Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics 136, 769-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link, A.J., Eng, J., Schieltz, D.M., Carmack, E., Mize, G.J., Morris, D.R., Garvik, B.M., and Yates, J.R., 3rd. (1999). Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676-682. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona, A., Furnari, B., Mondesert, O., and Russell, P. (1999). Nuclear localization of Cdc25 regulated by DNA damage and 14-3-3 protein. Nature 397, 172-175. [DOI] [PubMed] [Google Scholar]
- MacCoss, M.J., et al. (2002). Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. USA 99, 7900-7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack, A.L., Schieltz, D.M., Goode, B., Yang, S., Barnes, G., Drubin, D., and Yates, J.R., 3rd. (1997). Direct analysis and identification of proteins in mixtures by LC/MS/MS and database searching at the low-femtomole level. Anal. Chem. 69, 767-776. [DOI] [PubMed] [Google Scholar]
- McDonald, W.H., Pavlova, Y., Yates, J.R., 3rd, and Boddy, M.N. (2003). Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J. Biol. Chem. 278, 45460-45467. [DOI] [PubMed] [Google Scholar]
- McDonald, W.H., and Yates, J.R., 3rd. (2002). Shotgun proteomics and biomarker discovery. Dis. Markers 18, 99-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. [DOI] [PubMed] [Google Scholar]
- Morishita, T., Tsutsui, Y., Iwasaki, H., and Shinagawa, H. (2002). The Schizosaccharomyces pombe rad60 gene is essential for repairing double-strand DNA breaks spontaneously occurring during replication and induced by DNA-damaging agents. Mol. Cell. Biol. 22, 3537-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muris, D.F.R., Vreeken, K., Schmidt, H., Ostermann, K., Clever, B., Lohman, P.H.M., and Pastink, A. (1997). Homologous recombination in the fission yeast Schizosaccharomyces pombe: different requirements for the rhp51+, rhp54+ and rad22+ genes. Curr. Genet. 31, 248-254. [DOI] [PubMed] [Google Scholar]
- Nakashima, N., Noguchi, E., and Nishimoto, T. (1999). Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics 152, 853-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoda, F., Takeda, M., Seki, M., Maeda, D., Tajima, J., Ui, A., Yagi, H., and Enomoto, T. (2004). SMC6 is required for MMS-induced interchromosomal and sister chromatid recombinations in Saccharomyces cerevisiae. DNA Repair 3, 429-439. [DOI] [PubMed] [Google Scholar]
- Osborn, A.J., Elledge, S.J., and Zou, L. (2002). Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 12, 509-516. [DOI] [PubMed] [Google Scholar]
- Osman, F., Adriance, M., and McCready, S. (2000). The genetic control of spontaneous and UV-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Curr. Genet. 38, 113-125. [DOI] [PubMed] [Google Scholar]
- Paques, F., and Haber, J.E. (1999). Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka, M.R., Pardington, P.E., Yannone, S.M., Chen, D.J., and Cary, R.B. (2003). In vitro and in vivo interactions of DNA ligase IV with a subunit of the condensin complex. Mol. Biol. Cell 14, 685-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind, N., and Russell, P. (2000). Chk1 and Cds 1, linchpins of the DNA damage and replication checkpoint pathways. J. Cell Sci. 113, 3889-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030-1032. [DOI] [PubMed] [Google Scholar]
- Rouse, J. (2004). Esc4p, a new target of Mec1p (ATR), promotes resumption of DNA synthesis after DNA damage. EMBO J. 23, 1188-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder, S.P., Frater, L.A., Abramovitz, D.L., Goodwin, E.B., and Williamson, J.R. (2004). RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1.Nat. Struct. Mol. Biol. 11, 20-28. [DOI] [PubMed] [Google Scholar]
- Shinohara, M., Shita-Yamaguchi, E., Buerstedde, J.M., Shinagawa, H., Ogawa, H., and Shinohara, A. (1997). Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics 147, 1545-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren, C., and Nasmyth, K. (2001). Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 11, 991-995. [DOI] [PubMed] [Google Scholar]
- Symington, L.S. (2002). Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66, 630-670, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szankasi, P., Heyer, W.-D., Pshuchert, P., and Kohli, J. (1988). DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. J. Mol. Biol. 204, 917-925. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., Toh-e, A., and Kikuchi, Y. (2001). A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene 275, 223-231. [DOI] [PubMed] [Google Scholar]
- Taylor, E.M., Moghraby, J.S., Lees, J.H., Smit, B., Moens, P.B., and Lehmann, A.R. (2001). Characterization of a novel human SMC heterodimer homologous to the Schizosaccharomyces pombe Rad18/Spr18 complex. Mol. Biol. Cell 12, 1583-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkade, H.M., Bugg, S.J., Lindsay, H.D., Carr, A.M., and O'Connell, M.J. (1999). Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol. Biol. Cell 10, 2905-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn, M.P., Wolters, D., and Yates, J.R., 3rd. (2001). Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242-247. [DOI] [PubMed] [Google Scholar]
- Wu, L., Wu, H., Ma, L., Sangiorgi, F., Wu, N., Bell, J.R., Lyons, G.E., and Maxson, R. (1997). Miz1, a novel zinc finger transcription factor that interacts with Msx2 and enhances its affinity for DNA. Mech. Dev. 65, 3-17. [DOI] [PubMed] [Google Scholar]
- Yazdi, P.T., Wang, Y., Zhao, S., Patel, N., Lee, E.Y., and Qin, J. (2002). SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 16, 571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenvirth, D., and Simchen, G. (2000). Meiotic double-strand breaks in Schizosaccharomyces pombe. Curr. Genet. 38, 33-38. [DOI] [PubMed] [Google Scholar]