Abstract
Accurate and timely detection of bacterial pathogens will improve the clinical management of infections. Herein, we demonstrate an electrochemical biosensor that directly detects bacteria in human blood samples, resulting in the rapid diagnosis of a bloodstream infection. The multiplex biosensor detects the species-specific sequences of the 16S ribosomal RNA of bacteria for pathogen identification in physiological samples without preamplification. The analytical performance characteristics of the biosensor, including the limit of detection and probe cross-reactivity, are evaluated systematically. The feasibility of the biosensor for a diagnosis of a bloodstream infection is demonstrated by identifying bacterial clinical isolates spiked in whole blood and blood culture samples that were tested positive for bacteria. The electrochemical biosensor correctly identifies all the species in the samples with 100% concordance to microbiological analysis.
Keywords: infection, biosensor, blood, diagnostics, RNA
Introduction
Sepsis is a severe bloodstream infection with a mortality rate over 30% and represents a major public health burden with over one million incidences per year in the United States.1,2 Bacteremia, the presence of bacteria in the bloodstream, can occur when bacteria enter into the bloodstream during surgery and dental procedures. Sepsis is most commonly caused by an infection in other parts of the body, such as the urinary tract, lung (e.g., pneumonia), and abdomen.3 For hospitalized patients, urinary catheters, bed sores, surgical incisions, and intravenous lines are common sites of infection leading to bloodstream infection. Patients with severe wounds, compromised immune systems, and diabetes are particularly susceptible to bloodstream infection. Bloodstream infection can lead to vigorous immune response, systemic inflammation, shock, and organ dysfunction.4 If left untreated, bloodstream infection can rapidly become life-threatening.
Rapid etiological diagnosis with sensitivities to determine appropriate antimicrobial treatment is essential for the proper clinical management of bloodstream infection. When bloodstream infection is suspected in a patient, blood cultures are used to detect the presence of bacteria. The Surviving Sepsis Campaign, sponsored by the Society for Critical Care Medicine, is a global initiative tasked with improving the management of patients with sepsis. One of the main recommendations is to encourage the administration of broad-spectrum antimicrobial therapy within the first hour of sepsis.5 After identification of the causative bacteria, treatment can be narrowed to a specific antimicrobial that is effective against the bacterium. Targeted antibiotics treatment has shown to improve the patient outcome and lower the mortality rate compared with a broad-spectrum therapy.6,7 Since bacterial bloodstream infections initially involve unknown species of bacteria at the time of clinical presentation, there is a need to develop a method for rapidly identifying the bacteria in the blood sample. The identification of bacterial pathogens recovered in positive blood culture often requires cumbersome procedures, such as gram staining, additional subculture, and overnight incubation to obtain isolated colonies for further testing. To address the issues, molecular techniques, such as fluorescence in situ hybridization, nucleic acid amplification, and high-resolution melt curve analysis, have been developed for rapid identification of the bacteria directly from blood cultures.8–10 The ability to rapidly identify bacteria in blood will accelerate the diagnosis process, allowing patients to receive timely appropriate antimicrobial treatment.
In this study, we report a multiplex electrochemical biosensor for rapid pathogen identification in blood samples. Electrochemical biosensors represent a sensitive, low-cost strategy for molecular diagnostics.11,12 The electrochemical sensor is robust to the matrix effects of physiological samples, which allows direct detection of bacteria in raw physiological samples.13–17 The electrochemical sensing platform is portable and therefore it is feasible for this testing to be done at the bedside, at the point of care. The assay does not require nucleic acid amplification, which minimizes false positives due to nonspecific amplification. This study seeks to evaluate the multiplex electrochemical biosensor for measuring and identifying bacterial pathogens, including Staphylococcus aureus (SA), Escherichia coli (EC), Pseudomonas aeruginosa (PA), and Proteus mirabilis (PM).18 The analytical performance characteristics of the multiplex electrochemical sensor for bacterial detection were studied systematically. The feasibility of the multiplex biosensor for bloodstream infection diagnosis was evaluated by spiking bacterial clinical isolates in whole blood and then testing blood culture bottles that grew bacteria on microbiologic culture.
Methods
Reagents and Samples
Six pairs of oligonucleotide probe pairs (defined in Table 1), including four specific probe pairs for EC, SA, PA, and PM; one Enterobacteriaceae (EB) probe pair for all EB; and a universal (UNI) probe pair for detecting the presence of all bacteria, were evaluated.19 The probes were synthesized by Integrated DNA Technologies (Coralville, IA). The capture and detector probes were modified with thiol and fluorescein at the 5′ end and 3′ end, respectively (Table 1). Four clinical isolates (EC, PA, PM, and SA) were obtained from the clinical microbiology laboratory at the Veterans Affairs Palo Alto Health Care System (VAPAHCS). The bacterial colonies were freshly incubated in Luria broth (LB) media and grown to logarithmic phase before the experiment. Human whole blood for spiking experiments was obtained from Innovative Research Inc. (Novi, MI). De-identified EC-positive blood culture samples were obtained from the clinical microbiology laboratory at the Penn State Hershey Medical Center.
Table 1.
Sequences of Capture and Detector Probe Pairs in This Study.
| Probea | Position (Length)b | Sequence (5′ to 3′) |
|---|---|---|
| Universal bacterial (UNI) | ||
| Capture | 798 (22) | TCGTTTACRGCGTGGACTACCA |
| Detector | 776 (22) | GGGTATCTAATCCTGTTTGCTC |
| Enterobacteriaceae (EB) | ||
| Capture | 1275 (23) | ACTTTATGAGGTCCGCTTGCTCT |
| Detector | 1252 (23) | CGCGAGGTCGCCTTCCTTTGTAT |
| Escherichia coli (EC) | ||
| Capture | 471 (24) | CTGCGGGTAACGTCAATGAGCAAA |
| Detector | 447 (24) | GGTATTAACTTTACTCCCTTCCTC |
| Proteus mirabilis (PM) | ||
| Capture | 1019 (22) | AGCGTTCCCGAAGGCACTCCTC |
| Detector | 997 (22) | TATCTCTAAAGGATTCGCTGGA |
| Pseudomonas aeruginosa (PA) | ||
| Capture | 594 (23) | CCCGGGGATTTCACATCCAACTT |
| Detector | 570 (23) | GCTGAACCACCTACGCGCGCTTT |
| Staphylococcus aureus (SA) | ||
| Capture | 91 (22) | CCCGTCCGCCGCTAACRTCAGA |
| Detector | 69 (22) | GRAGCAAGCTYCTCGTCYGTTC |
The capture and detector probes were modified with 5′ thiol and 3′ fluorescein, respectively.
Position of the 5′ nucleotide in alignment with the bacterial 16S ribosomal RNA molecule.
Electrochemical Sensors
Gold electrodes for electrochemical sensing were fabricated by sputtering with a shadow mask on plastic substrates. Each chip consisted of 16 sensors. The thiolated capture probes were immobilized on the gold sensor surface by self-assembly. Briefly, 6 μL of 0.05 μM thiolated capture probe and 300 μM hexanedithiol (HDT) in TE buffer (Tris-EDTA, 1X) were spotted on the working electrode and incubated overnight at 4 °C. After washing with water and drying with nitrogen gas, 6 μL of 1 mM 6-mercapto-1 hexanol (MCH) solution was spotted on the working electrode and incubated for 50 min at room temperature. A laser machining system (VLS2.30; Universal Laser System, Scottsdale, AZ) was used to create plastic wells for the electrochemical biosensor. Each sensor received 50 μL of substrate solution for amperometric sensing. The current signal was measured by a multichannel potentiostat. A negative control, LB medium or blood, was included on each sensor chip.
Detection of Bacterial 16S Ribosomal RNA
Bacterial lysis was performed using 50 μL of 100 mg/mL lysozyme (Sigma, St. Louis, MO) and 25 μL of 100 mg/mL lysostaphin (Sigma, St. Louis, MO) in 425 μL of lysis buffer. The lysis solution (20 μL) was added to the sample (50 μL) and incubated for 5 min at room temperature. Then, 13 μL of 1 M NaOH was added to the sample and incubated for 5 min at room temperature. After incubation, 83 μL of the detector probe solution with 2.5% bovine serum albumin (Sigma, St. Louis, MO) and 1 M phosphate buffer (pH 7.4) were added. After hybridization for 10 min at 37 °C, 6 μL of the sample was placed on each sensor. The sample was incubated on the sensor for 15 min at 37 °C. The sensor was washed with deionized water and dried with nitrogen. Then, 6 μL of 0.5 U/mL anti-fluorescein horseradish peroxidase (HRP) Fab fragments (Roche Diagnostics, Indianapolis, IN) in 1 M phosphate buffered saline containing 0.5% casein was added to each sensor. The chip was incubated at room temperature for 15 min. Plastic wells were then placed on the sensor chips, and 50 μL TMB-H2O2 solution (K-Blue Low Activity TMB Substrate; Neogen, Lexington, KY) was added in each well. For amperometric sensing, a bias voltage of 200 mV was applied to the potentiostat to measure the amperometric signals. The assay required less than 1 h.
Pathogen Identification in Spiked Blood
For detection in whole blood, four times the volumes of each reagent and sample were used. The larger volume was required to generate adequate solution for pipetting onto the sensor. To spike bacteria in blood, the bacterial samples were centrifuged to remove the media and resuspended in blood. The concentration of the spiked samples was ~108 colony-forming units (CFU)/mL. Each chip was immobilized with a capture probe. The samples with the detector probes were loaded to the sensor chips with the corresponding capture probes. To test the ability of the multiplex electrochemical sensor for bloodstream infection diagnosis, 40 blood samples were prepared by spiking different bacteria species into whole blood. The bacteria concentrations were on the order of 107 and 108 CFU/mL.
Results and Discussion
Amperometric Detection of 16S Ribosomal RNA
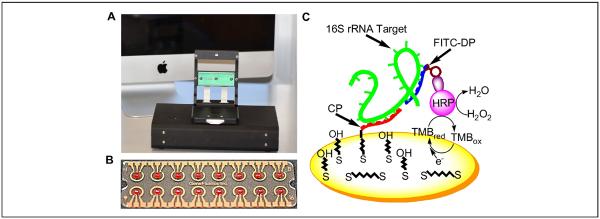
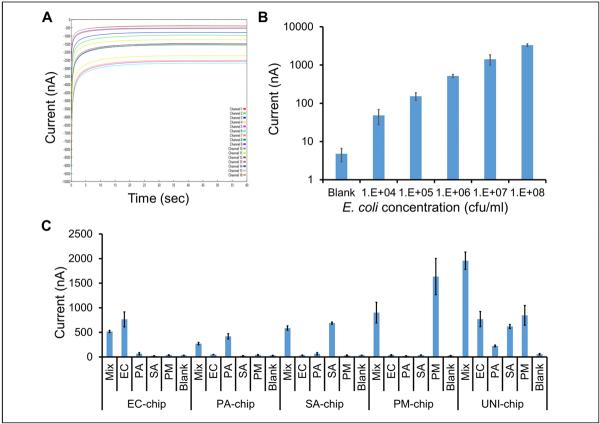
Electrochemical biosensing was performed using a portable, multichannel potentiostat (Fig. 1A). The multiplex electrochemical sensor chip consisted of 16 gold sensor electrodes with reference and auxiliary electrodes surrounding a circular working electrode (Fig. 1B). Each biosensor was coated with a self-assembled monolayer consisting of HDT, MCH, and a thiolated capture probe.20 The multiplex electrochemical sensor used a sandwich binding scheme with a capture probe and a detector probe for detecting the target 16S ribosomal RNA (rRNA) (Fig. 1C). The target 16S rRNA was immobilized by the capture probe and was detected by the detector probe bound with HRP. The HRP catalyzes the redox reaction to generate an amperometric signal with the addition of TMB and H2O2 substrates. An example of the current outputs of 16 electrochemical sensors simultaneously is shown in Figure 2A.
Figure 1.
A multiplex electrochemical sensor platform for detection of blood-borne pathogens. (A) A portable, multichannel potentiostat with a chip mount for amperometric sensing. (B) A disposable, 16-sensor chip (2.5 by 7.5 cm) fabricated by gold deposition on a plastic substrate. Each electrochemical sensor consists of a central working electrode, a circumferential reference electrode, and a short auxiliary electrode. (C) A schematic representation of the sandwich binding scheme on the central working electrode with the self-assembly monolayer. The target is captured on the sensor surface by the capture probe (CP) and detected using the detector probe (DP) amperometrically. HRP, horseradish peroxidase; rRNA, ribosomal RNA.
Figure 2.
Analytical performance of the multiplex electrochemical sensor. (A) A representative plot depicting the current outputs of 16 sensors over 60 s. (B) Analytical sensitivity of the electrochemical sensor for bacteria detection. The Escherichia coli sample was cultured to approximately ~108 colony-forming units (CFU)/mL and diluted to the appropriate concentrations with Luria broth (LB) media. (C) Analytical specificity of the multiplex electrochemical sensor platform. Escherichia coli (EC), Pseudomonas aeruginosa (PA), Proteus mirabilis (PM), Staphylococcus aureus (SA), and a mixture of all four bacteria (Mix) were spiked in whole blood. Blank represents blood only.
Analytical Performance of the Electrochemical Biosensor
The analytical performance of the multiplex electrochemical biosensor was first investigated and optimized in media. A serial dilution experiment was performed to determine the signal levels of EC from 104 to 108 CFU/mL. The sensor signal correlated with the concentration of EC for over four orders of magnitude (Fig. 2B). Fitting the data with the power law revealed a limit of detection (blank + 3× standard deviation) of 290 CFU/mL in culture media.
The ability of the electrochemical biosensor for pathogen identification in whole blood was then examined. EC, PA, PM, SA, and mixtures of all four bacteria (MIX) were spiked in blood. The samples were detected using sensor chips with specific probe pairs (EC-chip, PA-chip, SA-chip, and PM-chip). A universal probe pair targeting the conserved region of the bacterial 16S rRNA was also included in the experiment (UNI-chip). Figure 2C shows the results for detecting different bacteria with each chip. The multiplex biosensor detected the target pathogens with signal levels significantly higher than the blank (blood only) and nonspecific bacteria, suggesting a low cross-reactivity between the probes. The background level of the blank (whole blood) was approximately 35 nA for target-specific chips (EC, PM, PA, and SA). The signal level for nonspecific target increased to ~66 nA. In this study, a threshold value of 100 nA was defined to identify the existence of a pathogen in the sample.
The UNI-chip successfully detected the existence of bacteria in all samples (Fig. 2C). In agreement, the highest signal was observed when all four bacteria were mixed in the sample. In a clinical setting, the UNI probe is beneficial for first determining whether a bacterial infection is present, in contrast to false-positive or fungal infection. More important, the UNI probe is designed based on the conserved region of the bacterial 16S rRNA and can indicate the existence of bacteria when a specific probe is not available.
Optimization for Bloodstream Infection Diagnostics
Several aspects were considered in the design of the multiplex biosensor for bloodstream infection diagnostics. The multiplex electrochemical biosensor is designed to detect both gram-negative and gram-positive bacteria. For instance, SA is commonly observed in bloodstream infection.21 Without lysostaphin, a low signal was obtained due to the inefficiency in lysing the bacteria. Lysostaphin was, therefore, added in the lysis solution for cleaving the cell wall of staphylococci in all samples. Furthermore, a challenge of detecting bacteria in whole blood was coagulation and thickening of the sample after the addition of sodium hydroxide. Although an anticoagulant or a large volume of dilution buffer could be added to the sample, these procedures could reduce the signal-to-noise ratio of the biosensor. To facilitate pipetting of the sample, a larger volume was prepared in the protocol. At least four times the volumes of each reagent and sample were prepared to address the issue.
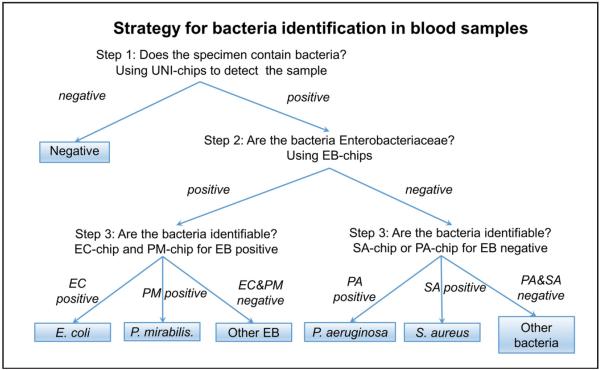
A test procedure was developed to demonstrate the ability of the multiplex electrochemical sensor for pathogen identification (Fig. 3). UNI, EB, EC, PM, SA, and PA probes were precoated on the sensor chips. In the first step, the samples were tested with the UNI-chip to detect the existence of bacterial pathogens in the samples. If the samples were positive, the samples were tested with the EB-chip to determine if the bacteria was Enterobacteriaceae or not. If the samples were EB positive, the samples were tested with the EC-chip and the PM-chip. If the samples were EB negative, the samples were tested with the PA-chip and SA-chip.
Figure 3.
Strategy to identify bacteria species in blood samples. In step 1, unknown samples are first detected with the universal probes (UNI-chip) to confirm the existence of bacteria pathogens. In step 2, UNI-positive samples are then detected with the Enterobacteriaceae probes (EB-chip). In step 3, EB-positive samples are further detected using the EC-chip and PM-chip. EB-negative samples are detected using the PA-chip and SA-chip. EC, Escherichia coli; PA, Pseudomonas aeruginosa; PM, Proteus mirabilis; SA, Staphylococcus aureus.
Pathogen Identification with Clinical Isolates Spiked in Whole Blood
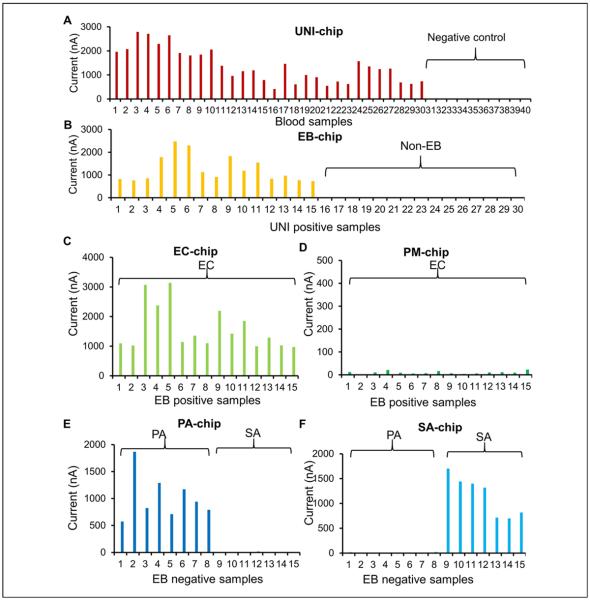
We designed a spiking study to test the ability of the electrochemical biosensor to detect bacteria in blood samples. Three common bacteria (EC, PA, and SA) were spiked into 30 blood specimens. Fifteen samples were spiked with EC, eight samples were spiked with PA, and seven samples were spiked with SA. In addition, 10 blood samples were included as a negative control. All 40 samples were first tested using the UNI-chip (Fig. 4A). The 30 positive and 10 negative samples were correctly identified using the threshold value. In the second step, the 30 UNI-positive specimens were detected using the EB-chip (Fig. 4B). The EB-chip had high signals for the 15 EC specimens and low signals for the eight PA and seven SA specimens. In the third step, the EC-chip and PM-chip were applied to detect the 15 EB-positive specimens, and the PA-chip and SA-chip were applied to detect the 15 EB-negative specimens (Fig. 4C–F). The chips correctly identified all EC, PA, and SA samples. For a non-specific target, the signal levels were below 25 nA for all sensors, compared with the threshold value of 100 nA for positive detection. These results support the selectivity of the species-specific probes for bloodstream infection diagnostics.
Figure 4.
A pathogen identification experiment by spiking EC (15, three different concentrations), PA (eight, one concentration), and SA (seven, two different concentrations) in whole blood. Ten blood samples were also included as a negative control. (A) Detection with the universal probes (UNI-chip). Sample numbers are sorted for clarity. Samples 1 to 30 were spiked with bacteria and samples 31 to 40 were blood only. (B) Detection with the Enterobacteriaceae probes (EB-chip). Samples 1 to 15 were EC, samples 16 to 23 were PA, and samples 24 to 30 were SA. (C, D) Detection of EB-positive samples with the EC-chip and PM-chip. All samples were EC. (E, F) Detection of EB-negative samples with the PA-chip and SA-chip. Samples 1 to 8 were PA and samples 9 to 15 were SA. EC, Escherichia coli; PA, Pseudomonas aeruginosa; PM, Proteus mirabilis; SA, Staphylococcus aureus.
Electrochemical Biosensors for Positive Blood Cultures
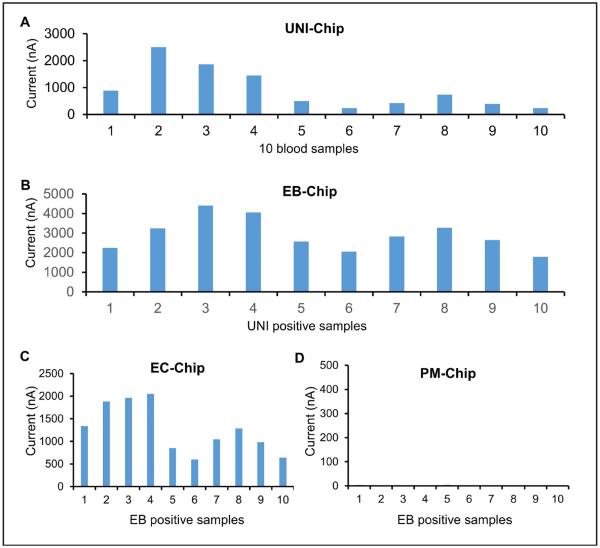
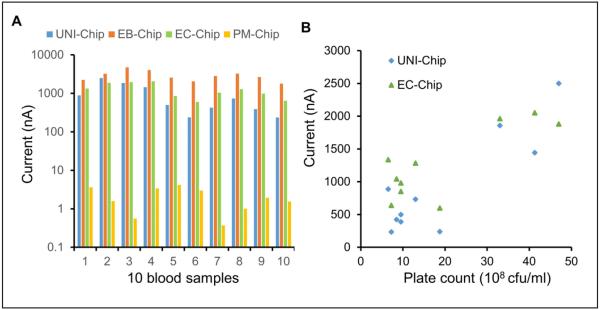
To evaluate the feasibility of the multiplex electrochemical sensor for bloodstream infection diagnosis, samples from positive blood culture bottles were tested. The samples were confirmed to be positive for EC in the clinical microbiology laboratory. All samples were first tested with the UNI-chip (Fig. 5A). The signals were over 100 nA in all the sensors, suggesting the existence of the bacterial pathogens in the samples. The EB-chip was then applied and resulted in high signals for all samples (Fig. 5B). Since all samples were EB positive, only the EC-chip and PM-chip were used in the third step (Fig. 5C). In agreement with the microbiological analysis, all samples were positive for EC and negative for PA. Overall, the multiplex electrochemical sensor correctly identified EC in all blood cultures (Fig. 6A). Since a large variation was observed among different samples, a plate count was performed to estimate the concentration of bacteria in the samples (Fig. 6B). A positive correlation was obtained between the plate count and current signals from the UNI-chip and EC-chip (UNI-chip, n = 10, R2 = 0.775, p = 0.0008; EC-chip, n = 10, R2 = 0.654, p = 0.00462).
Figure 5.
A clinical feasibility study using positive blood cultures. All samples were positive for E. coli. (A–D) Detection with the UNI-chip, EB-chip, EC-chip, and PM-chip. PA-chip and SA-chip were not tested since all samples were EB positive. EB, Enterobacteriaceae; EC, Escherichia coli; PA, Pseudomonas aeruginosa; PM, Proteus mirabilis; SA, Staphylococcus aureus; UNI, universal probes.
Figure 6.
Comparison of the sensor performance with blood cultures. (A) Current outputs of different sensor chips. (B) Correlation of the current signal with the plate count of the positive blood cultures. EB, Enterobacteriaceae; EC, Escherichia coli; PM, Proteus mirabilis; UNI, universal probes.
Discussion
In this study, we investigated the performance of a multiplex electrochemical biosensor for pathogen identification toward the rapid and point-of-care diagnosis of bacteremia. A small number of bacteria were tested as a proof of concept to evaluate the feasibility of the assay. In the future, several areas of improvement can be implemented to enhance the clinical significance of the biosensor. In particular, the panel of species-specific probe pairs should be expanded significantly to identify other bacteria that can result in human infections, such as Klebsiella, Enterobacter, Serratia, Citrobacter, and Enterococcus, for bloodstream and infections of other sites. Other bacterial pathogens should also be tested to evaluate the performance of the biosensor. Compared with other bacterial identification technology, the electrochemical biosensing system is portable and has a small footprint. Implementing the assay in an automated format, such as robotic and microfluidic systems, will facilitate the application of the biosensor in a clinical setting.22,23 Sample preparation and enrichment modules can also be integrated in the system to improve the limit of detection and enhance the clinical applicability of the biosensor.24 In the current procedure, the chips with different probe pairs were sequentially tested using different chips in a few hours. As the most rapid results are essential, different probes can be immobilized on the same chip to identify the pathogen within 1 h.
In conclusion, this study examines the feasibility of a multiplex electrochemical biosensor for pathogen identification toward rapid diagnosis of bloodstream infections. The sensor successfully identified bacterial species in spiked blood samples and positive blood cultures from the clinical microbiology laboratory. The system can accelerate the identification of bacterial pathogens in positive blood cultures and may improve the clinical management of bacteremia and sepsis by providing appropriate and timely antimicrobial treatment.
Acknowledgments
The authors thank Tinting Liu and Yi Lu for technical assistance.
Funding The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH R01AI117059, R44HD084033, and NSFC 21375068.
Footnotes
Declaration of Conflicting Interests The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Hall MJ, Williams SN, DeFrances CJ, et al. Inpatient Care for Septicemia or Sepsis: A Challenge for Patients and Hospitals. NCHS Data Brief. 2011;62:1–8. [PubMed] [Google Scholar]
- 2.Linde-Zwirble WT, Angus DC. Severe Sepsis Epidemiology: Sampling, Selection, and Society. Crit. Care. 2004;8:222–226. doi: 10.1186/cc2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stearns-Kurosawa DJ, Osuchowski MF, Valentine C, et al. The Pathogenesis of Sepsis. Annu. Rev. Pathol. 2011;6:19–48. doi: 10.1146/annurev-pathol-011110-130327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iskander KN, Osuchowski MF, Stearns-Kurosawa DJ, et al. Sepsis: Multiple Abnormalities, Heterogeneous Responses, and Evolving Understanding. Physiol. Rev. 2013;93:1247–1288. doi: 10.1152/physrev.00037.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dellinger RP, Levy MM, Rhodes A, et al. Surviving Sepsis Campaign Guidelines Committee Including the Pediatric Subgroup Surviving Sepsis Campaign: International Guidelines for Management of Severe Sepsis and Septic Shock: 2012. Crit. Care Med. 2013;41:580–637. doi: 10.1097/CCM.0b013e31827e83af. [DOI] [PubMed] [Google Scholar]
- 6.Kollef MH. Broad-Spectrum Antimicrobials and the Treatment of Serious Bacterial Infections: Getting It Right Up Front. Clin. Infect. Dis. 2008;47(Suppl 1):S3–13. doi: 10.1086/590061. [DOI] [PubMed] [Google Scholar]
- 7.Turenne CY, Witwicki E, Hoban DJ, et al. Rapid Identification of Bacteria from Positive Blood Cultures by Fluorescence-Based PCR-Single-Strand Conformation Polymorphism Analysis of the 16S rRNA Gene. J. Clin. Microbiol. 2000;38:513–520. doi: 10.1128/jcm.38.2.513-520.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Won H, Rothman R, Ramachandran P, et al. Rapid Identification of Bacterial Pathogens in Positive Blood Culture Bottles by Use of a Broad-Based PCR Assay Coupled with High-Resolution Melt Analysis. J. Clin. Microbiol. 2010;48:3410–3413. doi: 10.1128/JCM.00718-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogardt M, Trebesius K, Geiger AM, et al. Specific and Rapid Detection by Fluorescent In Situ Hybridization of Bacteria in Clinical Samples Obtained from Cystic Fibrosis Patients. J. Clin. Microbiol. 2000;38:818–825. doi: 10.1128/jcm.38.2.818-825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangold KA, Manson RU, Koay ES, et al. Real-Time PCR for Detection and Identification of Plasmodium spp. J. Clin. Microbiol. 2005;43:2435–2440. doi: 10.1128/JCM.43.5.2435-2440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sin ML, Mach KE, Wong PK, et al. Advances and Challenges in Biosensor-Based Diagnosis of Infectious Diseases. Expert Rev. Mol. Diagn. 2014;14:225–244. doi: 10.1586/14737159.2014.888313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mach KE, Wong PK, Liao JC. Biosensor Diagnosis of Urinary Tract Infections: A Path to Better Treatment? Trends Pharmacol. Sci. 2011;32:330–336. doi: 10.1016/j.tips.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu ML, Lawi W, Snyder ST, et al. Matrix Effect—A Challenge Toward Automation of Molecular Analysis. J. Assoc. Lab. Autom. 2010;15:233–242. [Google Scholar]
- 14.Altobellia E, Mohan R, Mach KE, et al. Integrated Biosensor Assay for Rapid Uropathogen Identification and Phenotypic Antimicrobial Susceptibility Testing. Eur. Urol. Focus. doi: 10.1016/j.euf.2015.12.010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu T, Lu Y, Gau V, et al. Rapid Antimicrobial Susceptibility Testing with Electrokinetics Enhanced Biosensors for Diagnosis of Acute Bacterial Infections. Ann. Biomed. Eng. 2014;42:2314–2321. doi: 10.1007/s10439-014-1040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mach KE, Mohan R, Baron EJ, et al. A Biosensor Platform for Rapid Antimicrobial Susceptibility Testing Directly from Clinical Samples. J. Urol. 2011;185:148–153. doi: 10.1016/j.juro.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu T, Sin ML, Pyne JD, et al. Electrokinetic Stringency Control in Self-Assembled Monolayer-Based Biosensors for Multiplex Urinary Tract Infection Diagnosis. Nanomedicine. 2014;10:159–166. doi: 10.1016/j.nano.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brock TK, Mecozzi DM, Sumner S, et al. Evidence-Based Point-of-Care Tests and Device Designs for Disaster Preparedness. Am. J. Disaster Med. 2010;5:285–294. [PMC free article] [PubMed] [Google Scholar]
- 19.Liao JC, Mastali M, Gau V, et al. Use of Electrochemical DNA Biosensors for Rapid Molecular Identification of Uropathogens in Clinical Urine Specimens. J. Clin. Microbiol. 2006;44:561–570. doi: 10.1128/JCM.44.2.561-570.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuralay F, Campuzano S, Haake DA, et al. Highly Sensitive Disposable Nucleic Acid Biosensors for Direct Bioelectronic Detection in Raw Biological Samples. Talanta. 2011;85:1330–1337. doi: 10.1016/j.talanta.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaasch AJ, Barlow G, Edgeworth JD, et al. Staphylococcus aureus Bloodstream Infection: A Pooled Analysis of Five Prospective, Observational Studies. J. Infect. 2014;68:242–251. doi: 10.1016/j.jinf.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sin ML, Gau V, Liao JC, et al. A Universal Electrode Approach for Automated Electrochemical Molecular Analyses. J. Microelectromech. Syst. 2013;22:1126–1132. doi: 10.1109/JMEMS.2013.2253545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouyang M, Mohan R, Lu Y, et al. An AC Electrokinetics Facilitated Biosensor Cassette for Rapid Pathogen Identification. Analyst. 2013;138:3660–3666. doi: 10.1039/c3an00259d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang TH, Wong PK. Transforming Microfluidics into Laboratory Automation. J. Assoc. Lab. Autom. 2010;15:A15–A16. [Google Scholar]