Abstract
CD44 is involved in cancer-cell growth, invasion, proliferation and metastasis and is also a causal factor for acquisition of resistance to apoptosis. Therefore we evaluated different SNPs of CD44 gene viz. CD44rs187116 A/G, CD44rs4755392 A/T, CD44rs187115 C/T, CD44rs13347 C/T and CD44 rs353639 G/T for bladder cancer risk in North Indian population. 240 bladder cancer patients and 270 cancer free controls were recruited in this study. Genotyping was done by PCR–RFLP for CD44rs187116 A/G. However, CD44rs4755392 A/T, CD44rs187115 C/T, CD44rs13347 C/T and CD44 rs353639 G/T were genotyped by allelic discrimination Taqman® assay. Statistical analysis was done by SPSS. In-silico analysis was done using F-SNP. We found reduced risk in variant genotype, TT of rs4755392 (p = 0.011) as well as in variant allele, T (p = 0.045). No risk was seen in rs13347, heterozygous genotype, CT (p = 0.023) and variant allele, T (p = 0.007). The dominant model, CT + TT also revealed reduced risk (p = 0.009). A marginal risk was seen in dominant model, GT + TT of rs353639 (p = 0.044) and reduced risk in variant allele T (p = 0.040). A significant manifold risk was seen in smokers carrying variant genotype, TT of CD44rs353639 G/T (p = 0.038, OR 1.960). Haplotypic analysis revealed significant association in 4 sets viz. TCCGG p = 0.005, TTCGA p = 0.039, ACTGG p = 0.008 and TCTGA p = 0.006. In-silico analysis using F-SNP, showed altered transcriptional regulation for rs187115, rs13347 and rs353639. Our study suggests that rs353639 shows a marginal risk for bladder cancer susceptibility, whereas rs4755392 and rs13347 have reduced risk of bladder cancer and rs187115 and rs187116 had no effect on bladder cancer susceptibility in North Indians.
Keywords: BC, CD44, MIBC, NMIBC, BCG immunotherapy
Introduction
Cancer is a biggest threat globally and is the second most common disease in India responsible for maximum mortality with about 0.3 million deaths per year. Bladder cancer is the 9th most common cancer worldwide, with an estimated 74,000 new cases expected to occur in 2015 [1]. Bladder cancer incidence is about 4 times higher in men than in women. Bladder cancer incidence rates decreased from 2007 to 2011 by 1.6 % per year in men and by 1.1 % per year in women. An estimated 16,000 deaths will occur in 2015, 72 % of which will be in men [1]. In males, it is the fourth most common cancer (4 % of male total), whilst it is the 13th most common cancer in females (2 % of female total) [2]. As a general prevalence, in India, out of 1, 00,000 people 3 males and 1 female develop BC each year [3].
Cancer stem cells having the property to self renew, differentiate, proliferate and migrate and hence they play an important role in tumorigenesis and disease progression. CD44 is a cell surface transmembrane glycoprotein which is expressed in a variety of cells and tissues of hematopoietic, epithelial, endothelial, and mesodermal origins [4, 5]. Earlier studies predicted that CD44 is involved in a number of physiological processes including lymphocyte migration and extravasation, lymph node homing, and lymphocyte activation and apoptosis [6]. CD44 has a well documented role in tumor metastasis [7]. CD44 plays role in cellular signaling cascades by participating in signal transduction processes. For signal transduction it establish transmembrane complexes and also organize signaling cascades through association with actin cytoskeleton thereby promoting membrane motility and tumor cell migration. CD44 monitors change in ECM and hence influence cell growth, survival and differentiation [8]. CD44 signaling is crucial in cancer-cell growth, invasion, proliferation and metastasis. CD44 is also a causal factor for acquisition of resistance to apoptosis [9]. Considering the flourishing evidences entailing the role of CD44 in a wide variety of tumorigenic processes and the complexity of the CD44 gene it is credible that the functional genetic variants of CD44 gene may help sub-populations at high risk for early tumor recurrence in various cancers.
Thus, we hypothesized that CD44 polymorphisms may play an important role in bladder cancer development and may have potential significance as molecular prognostic markers, as CD44 polymorphisms have not been extensively studied in bladder cancer particularly in North Indian population.
Risk factors for the development of BC can be classified into different subsets like genetic and molecular abnormalities, chemical or environmental exposures, and chronic irritation [10]. Genetic susceptibility and gene environment interaction (e.g., gene-smoking and gene-occupational exposure interactions) in bladder cancer aetiology has been well documented in different populations [11]. So, in the present study, a case–control investigation was performed for 5 SNPs of CD44 gene out of which 2 SNPs i.e. CD44rs13347 C/T and CD44rs4755392 A/T are located in the 3′UTR region and the other 3 SNPs i.e. CD44rs187115 C/T, CD44 rs353639 G/T and CD44rs187116 A/G are situated on Intron-1 of CD44 gene, to analyze their contribution in risk prediction of BC and the associations between risk factors and bladder cancer clinicopathologic characteristics. Our investigations in the present paper are based on the hypothesis that CD44 gene polymorphism is associated with bladder cancer and may be effectively used in the risk assessment and genetic epidemiological analysis of bladder cancer.
Materials and Methods
Study Subjects
A total of 240 confirmed bladder cancer patients and 270 healthy controls were recruited in the present study. The patients were enrolled from outpatient department (OPD) of Urology. Those with a previous history of other cancer, cancer metastasized to other site of body from another origin and previous radiotherapy were excluded. 270 healthy controls (Mean age = 54.5 years, M:F = 249:21) were recruited from volunteers who came to the hospital for their routine checkups, unrelated to patients and were also age and ethnicity matched. The criteria for selecting controls included no evidence of any personal history of cancer or other malignant conditions or any other chronic diseases.
Among 240 patients ratio of male: female was 211:29 (mean age = 56.9 years). The disproportionate ratio between male and female bladder cancer in our population could be largely due to increased prevalence in males (3:1). The patients were subjected to detailed demographic, clinical and pathological investigations, which contained the details of age, stage, disease history, family history and other relevant details such as smoking history, occupation history and other lifestyle factors. At the end of the interview, a 5-ml blood sample was drawn into coded tubes. Informed and written consent was taken from all subjects when interviewing for the demographic details and blood sample collection. The Ethical Review Board of the Institute approved the study.
Epidemiology Data Collection
The demographic details were obtained by interviewing each individual. Individuals who smoked once a day for more than 5 years were defined as smokers. The individuals who had never smoked in their lifetime were regarded as non smokers. The demographic and clinical characteristics of the patients are demonstrated in Table 1.
Table 1.
Baseline demographic and clinical characteristics of bladder cancer patients and healthy controls
| Variables | Cases n = 240 [n (%)] |
Controls n = 270 [n (%)] |
Chi square#
p value |
|---|---|---|---|
| Sex | |||
| Female | 29 (12.1) | 21 (7.8) | 0.105 |
| Male | 211 (87.9) | 249 (92.2) | |
| Age (years) | |||
| Mean age- ±SD | 56.96 ± 13.86 | 54.50 ± 10.23 | 0.138$ |
| Smoking* | |||
| Non smokers | 48 (29.6) | 214 (79.3) | <0.001 |
| Smokers | 116 (70.4) | 56 (20.7) | |
| Tumor grade stage | |||
| TaG1 | 48 (20.0) | – | – |
| TaG2–3 + T1G1–3 | 128 (53.3) | – | |
| T2+ | 64 (26.7) | – | |
| Intravesical therapy | |||
| Non treated | 86 (47.7) | – | – |
| BCG induction (BCG i + m) | 94 (52.3) | – | |
| Event | |||
| Recurrence | 74 (43.9) | – | – |
| Non-recurrence | 95 (56.1) | – | |
$No significant age difference between controls and patients
#Student t test was used to determine the p value
* The sum could not add up to the total due to some missing values
BCG i + m, Bacillus Calmette-Guerin induction + maintenance
The statistically significant values are shown in bold
Clinical Data Collection
The clinical information about tumor stage and grade, intravesical therapy and dates of recurrence, radical cystectomy and pathological findings at cystectomy were provided by the urologists in our department. The classification tumor stages were as per the American Joint Committee on Cancer’s TNM staging system [12]. Of the 240 total patients enrolled in the study, 180 patients had non muscle invasive bladder cancer (NMIBC) while the rest 60 had muscle invasive bladder cancer (MIBC). Patients with NMIBC at high risk (high grade, multiple and large tumor) were treated with intravesical Bacillus Calmette-Guerin (BCG) (n = 94). The patients with NMI cancer of low risk (low grade and single small tumor) were kept on cystoscopic surveillance and considered as non-BCG patients. Subsequently, all the patients were examined by cystoscopy after every 3 months in first and second years and later at 6 monthly intervals as long as there was no tumor recurrence. BCG treatment consisted of 6 weekly instillation induction BCG (n = 94). Since the number of patients receiving maintenance BCG was too low, we did not categorize the patients according to BCG regime for statistical analysis. The end point of study included tumor recurrence, defined as a newly found bladder tumor following a previous negative follow-up cystoscopy, or end of study time (60 months). Patients with invasive BC (n = 60) were treated with radical cystectomy with or without adjuvant chemotherapy, which included cisplatin, gemcitabine followed by periodical cystoscopy.
SNP Selection
The potentially functional polymorphisms within the CD44 gene were selected by using the HapMap Project database (www.hapmap.org) based on the GIH population data of hapmap. We used certain criteria for the candidate gene polymorphisms viz., a minor allele frequency (MAF) >10 % in Caucasian population; situated in the 3′UTR, 5′UTR, intronic and exonic regions of the tested genes which shows some biological significance according to the location within the gene.
Tag SNPs were selected from the Haploview software 4.2 (Mark Daly’s lab of Broad Institute, Cambridge, MA, Britain) [13], based on the GIH population data of HapMap (HapMap Data Rel 27 PhaseII +III, Feb 09, on NCBI B36 assembly, dbSNP b126). TagSNPs that captured all the known common SNPs (with minor allele frequencies of .0.1) in the CD44 gene, with a pairwise correlation r2.0.8 were selected.
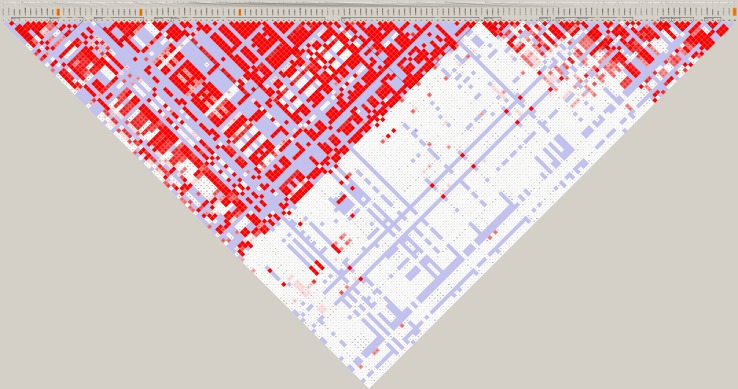
TaggerSNP rs353639 was found to represent the known SNPs in the haplotype blocks 3 and 4 in the CD44 gene of GIH population. Previously significantly reported SNP rs13347 in Chinese population also represents the haplotype block 9. rs187115 represent the known SNPs in the haplotype block 3. rs187116 represent the known SNPs in the haplotype block 2. In addition to these SNPs one more SNP, rs4755392 which is located on 3′UTR 98670 A/T is also included in this study. The LD Plot with SNPs is furnished in Fig. 1.
Fig. 1.
Linkage disequilibrium (LD) plot of CD44 gene in Hapmap- GIH population
Genotyping
Genomic DNA was extracted from venous blood by following standard salting out method [14]. Genotyping of CD44rs4755392 A/T, CD44rs187115 C/T, CD44rs13347 C/T and CD44 rs353639 G/T SNPs were carried out by using Taqman allelic discrimination assay. For the assay primers and probes were provided as predesigned assays by Applied Biosystems (Foster City, CA). Genotyping was performed with ABI 7500HT Fast Sequence Detection System (Applied Biosystems, Foster City, CA) using 96-well plates. Positive and negative controls were used in each genotyping assay plate, and 10 % of the samples were randomly selected and run in duplicates with 100 % concordance. The results were reproducible with no discrepancy in genotyping. The polymorphism in CD44 intron 1 +4883(CD44rs187116 A/G) was genotyped by PCR-based restriction fragment length polymorphism (PCR–RFLP) analysis. The primer sequence used for CD44rs187116 A/G was adopted from a previous study [15]. Genotyping was done on 10 % Poly-Acrylamide Gel and visualized after staining with ethidium bromide. Positive and negative controls were used in each genotyping assay, and 10 % of the samples were randomly selected and run in duplicates with 100 % concordance. The results were reproducible with no discrepancy in genotyping. About 5 % of the randomly selected samples were validated by sequencing.
Statistical Analysis
The power of the study was calculated using Quanto software, version 1.0 (available from: http://hydra.usc.edu/gxe). The present study achieved 80 % of the statistical power. The goodness-of-fit Chi square test was used to analyze any deviation from the Hardy–Weinberg equilibrium in controls. A binary logistic regression model was used to estimate the risk as the OR at the 95 % confidence interval. The statistical analysis was done using the Statistical Package for Social Sciences software, version 16.0 (SPSS, Chicago, IL), and p < 0.05 was considered statistically significant. Haplotypic analysis was done by using SNP analyzer version 1.2A.
Hardy–Weinberg equilibrium (HWE) test of SNP was performed using Michael H. Court’s (2005–2008) online calculator (http://www.tufts.edu/~mcourt01/Documents/Court%20lab%20-%20HW%20calculator.xls). Tests in bladder cancer patients and healthy unrelated controls did not show any significant deviation from HWE for any of the SNPs.
In Silico Analysis
The possible functional effects were determined in CD44 gene by online web server F-SNP (http://compbio.cs.queensu.ca/F-SNP/) [16].
Results
Characteristics of Study Subjects
Frequency distribution of the selected demographic characters of cases and controls are shown in Table 1. There was no significant difference between the patients and controls regarding age (p = 0.138), and sex (p = 0.105). However, there were more patients with a habit of smoking (70.4 %) among the cases than among the controls (20.7 %) (p < 0.001).
Genotypic and Allelic Frequency of CD44 gene Polymorphisms (CD44rs4755392 A/T, CD44rs187115 C/T, CD44rs13347 C/T, CD44 rs353639 G/T and CD44rs187116 A/G) in Bladder Cancer
The observed genotype frequencies of five SNPs studied in healthy controls were in accordance with Hardy–Weinberg Equilibrium. The genotypic and allelic frequencies of CD44rs4755392 A/T, CD44rs187115 C/T, CD44rs13347 C/T, CD44 rs353639 G/T and CD44rs187116 A/G gene polymorphisms in context with bladder cancer risk among patients and controls are depicted in Table 2.
Table 2.
Association of CD44 gene polymorphisms (CD44rs4755392 A/T, CD44rs187115 C/T, CD44rs13347 C/T, CD44 rs353639 G/T and CD44rs187116 A/G) in bladder cancer risk
| Genetic model | Genotypes | Controls n = 270 [n (%)] |
Patients n = 240 [n (%)] |
p value# | OR* (95 % CI) |
|---|---|---|---|---|---|
| CD44rs4755392 A/T | |||||
| Additive | AA | 72 (26.7) | 78 (32.5) | Ref | Ref |
| AT | 136 (50.3) | 123 (51.3) | 0.484 | 0.865 (0.576–1.299) | |
| TT | 62 (23.0) | 39 (16.3) | 0.011 | 0.578 (0.344–0.971) | |
| Dominant | AA | 72 (26.7) | 78 (32.5) | Ref | Ref |
| AT + TT | 198 (73.3) | 162 (67.5) | 0.149 | 0.755 (0.516–1.106) | |
| Multiple | A | 280 (51.9) | 279 (58.1) | Ref | Ref |
| T | 260 (48.1) | 201 (41.9) | 0.045 | 0.776 (0.606–0.994) | |
| CD44rs187115 C/T | |||||
| Additive | CC | 127 (47.0) | 101 (42.1) | Ref | Ref |
| CT | 101 (37.4) | 97 (40.4) | 0.199 | 1.290 (0.875–1.901) | |
| TT | 42 (15.6) | 42 (17.5) | 0.324 | 1.291 (0.777–2.143) | |
| Dominant | CC | 127 (47.0) | 101 (42.1) | Ref | Ref |
| CT + TT | 143 (53.0) | 139 (57.9) | 0.262 | 1.222 (0.861–1.735) | |
| Multiple | C | 355 (65.7) | 299 (62.3) | Ref | Ref |
| T | 185 (34.3) | 181 (37.7) | 0.252 | 1.162 (0.899–1.501) | |
| CD44rs13347 C/T | |||||
| Additive | CC | 140 (51.9) | 152 (63.3) | Ref | Ref |
| CT | 104 (38.5) | 73 (30.4) | 0.023 | 0.647 (0.443–0.943) | |
| TT | 26 (9.6) | 15 (6.3) | 0.067 | 0.531 (0.270–1.044) | |
| Dominant | CC | 140 (51.9) | 152 (63.3) | Ref | Ref |
| CT + TT | 130 (48.1) | 88 (36.7) | 0.009 | 0.623 (0.437–0.889) | |
| Multiple | C | 384 (71.1) | 377 (78.5) | Ref | Ref |
| T | 156 (28.9) | 103 (21.5) | 0.007 | 0.673 (0.505–0.896) | |
| CD44rs353639 G/T | |||||
| Additive | GG | 159 (58.9) | 120 (50.0) | Ref | Ref |
| GT | 92 (34.1) | 97 (40.4) | 0.077 | 1.397 (0.964–2.024) | |
| TT | 19 (7.0) | 23 (9.6) | 0.156 | 1.604 (0.835–3.079) | |
| Dominant | GG | 159 (58.9) | 120 (50.0) | Ref | Ref |
| GT + TT | 111 (41.1) | 120 (50.0) | 0.044 | 1.432 (1.009–2.034) | |
| Multiple | G | 410 (75.9) | 337 (70.2) | Ref | Ref |
| T | 130 (24.1) | 143 (29.8) | 0.040 | 1.338 (1.014–1.767) | |
| CD44rs187116 A/G | |||||
| Additive | AA | 78 (28.9) | 74 (30.8) | Ref | Ref |
| AG | 127 (47.0) | 120 (50.0) | 0.984 | 0.996 (0.665–1.492) | |
| GG | 65 (24.1) | 46 (19.2) | 0.245 | 0.746 (0.455–1.222) | |
| Dominant | AA | 78 (28.9) | 74 (30.8) | Ref | Ref |
| AG + GG | 192 (71.1) | 166 (69.2) | 0.632 | 0.911 (0.623–1.332) | |
| Multiple | A | 283 (52.4) | 268 (55.8) | Ref | Ref |
| G | 257 (47.6) | 212 (44.2) | 0.273 | 0.871 (0.680–1.115) | |
#Student’s t test was used to determine p value. Age-gender-smoking adjusted odds ratio
The statistically significant values are shown in bold
No significant differences were observed in the frequency distribution of rs187115 and rs187116 polymorphisms between bladder cancer patients and healthy controls, both at the genotypic and allelic levels. We found reduced risk in variant genotype, TT of additive model of rs4755392 (p = 0.011, Adjusted OR 0.578, 95 % CI 0.344–0.971) as well as in variant allele, T of allelic model (p = 0.045, OR 0.776, 95 % CI 0.606–0.994). Also, protective risk was seen in rs13347, heterozygous genotype, CT of additive model (p = 0.023, OR 0.647, 95 % CI 0.443–0.943) and the variant allele, T of allelic model (p = 0.007, OR 0.673, 95 % CI 0.505–0.896). The dominant model, CT + TT also revealed reduced risk (p = 0.009, OR 0.623, 95 % CI 0.437–0.889). A marginal risk was seen in dominant model, GT + TT of rs353639 (p = 0.044, OR 1.432, 95 % CI 1.009–2.034) and in allelic model, variant allele T (p = 0.040, OR 1.338, 95 % CI 1.014–1.767).
Association of CD44 gene Variants at Genotypic Level with Smoking
We correlated the genotypes of all five polymorphism of CD44 gene (CD44rs4755392 A/T, CD44rs187115 C/T, CD44rs13347 C/T, CD44 rs353639 G/T and CD44rs187116 A/G) with smoking habits among patients with the help of univariate analysis by Fischer’s exact test. For this analysis we stratified patients as smokers and non-smokers.
In case of CD44rs187115 C/T the heterozygous genotype, CT was associated with reduced risk of BC along with the smoking habits (p = 0.035, OR 0.438 95 % CI 0.203–0.942). A significant high risk was seen in those carrying variant genotype, TT of CD44rs353639 G/T (p = 0.038, OR 1.960, 95 % CI 1.128–3.190). No association was seen in the other variants of CD44 CD44rs4755392 A/T, CD44rs13347 C/T and CD44rs187116 A/G with respect to smoking (Table 3) .
Table 3.
Analysis of genotypes of CD44rs4755392 A/T, CD44rs187115 C/T, CD44rs13347 C/T, CD44 rs353639 G/T and CD44rs187116 A/G on the basis of smoking among bladder cancer patients
| Genotype | Patients non smokers n = 48 [n (%)] |
Patients smoker n = 116 [n (%)] |
p value | OR* (95 % CI) |
|---|---|---|---|---|
| CD44rs4755392 A/T | ||||
| AA | 19 (39.6) | 39 (33.6) | Ref | Ref |
| AT | 21 (43.8) | 60 (51.7) | 0.381 | 1.392 (0.664–2.917) |
| TT | 8 (16.7) | 17 (14.7) | 0.946 | 1.035 (0.380–2.824) |
| CD44rs187115 C/T | ||||
| CC | 14 (29.2) | 55 (47.4) | Ref | Ref |
| CT | 25 (52.1) | 43 (37.1) | 0.035 | 0.438 (0.203–0.942) |
| TT | 9 (18.8) | 18 (15.5) | 0.182 | 0.509 (0.189–1.373) |
| CD44rs13347 C/T | ||||
| CC | 30 (62.5) | 73 (62.9) | Ref | Ref |
| CT | 13 (27.1) | 37 (31.9) | 0.687 | 1.170 (0.546–2.505) |
| TT | 5 (10.4) | 6 (5.2) | 0.272 | 0.493 (0.140–1.740) |
| CD44rs353639 G/T | ||||
| GG | 28 (58.3) | 50 (43.1) | Ref | Ref |
| GT | 19 (39.6) | 50 (43.1) | 0.279 | 1.474 (0.730–2.974) |
| TT | 1 (2.1) | 16 (13.8) | 0.038 | 1.960 (1.128–3.190) |
| CD44rs187116 A/G | ||||
| AA | 16 (33.3) | 38 (32.8) | Ref | Ref |
| AG | 24 (50.0) | 57 (49.1) | 1.000 | 1.000 (0.470–2.126) |
| GG | 8 (16.7) | 21 (18.1) | 0.845 | 1.105 (0.406–3.011) |
OR odds ratio, CI confidence interval
The statistically significant values are shown in bold
Association of CD44rs4755392 A/T, CD44rs187115 C/T, CD44rs13347 C/T, CD44 rs353639 G/T and CD44rs187116 A/G Genotypes with Tumor Stage/Grade of Bladder Cancer Patients
To study the association of polymorphisms of CD44 gene with tumor stage/grade, the BC patients were stratified into three groups based on their tumor stage/grade [TaG1 (low risk NMIBC), TaG2–3 + T1G1–3 (High risk NMIBC) and T2+ (muscle invasive)]. TaG1 was taken as a reference. The patients with similar stage but with different grades respond to treatment differently. We did not find any association of CD44 gene variants CD44rs4755392 A/T, CD44rs187115 C/T, CD44rs13347 C/T, CD44 rs353639 G/T and CD44rs187116 A/G with any of the tumor stage/grade of BC patients (Data not shown).
Modulation of CD44 Genotype Variants and Outcome After BCG Immunotherapy
For analyzing the association of CD44rs4755392 A/T, CD44rs187115 C/T, CD44rs13347 C/T, CD44 rs353639 G/T and CD44rs187116 A/G gene variants and the risk of recurrence in NMIBC patients, the further analysis was throttled only to NMIBC patients (n = 180). We analyzed the association of genotypes and risk of recurrence after BCG immunotherapy. We grouped patients into BCG treated (n = 94) and non-treated (n = 86) as these were patients of low grade tumors and did not require BCG immunotherapy. None of the polymorphisms were associated with risk of recurrence (Data not shown).
Association of CD44 Haplotypes with Bladder Cancer Risk
Recent studies have demonstrated that haplotype analysis may be more manifesting in risk prediction and association of disease compared with an analysis of a single nucleotide polymorphism, as individual polymorphism is likely to confer modest effects to the risk of bladder cancer. Considering this we examined the effects of CD44 gene variants by constructing haplotype sets, taking combination ACCGA as a reference as these five alleles were wild alleles from the entire five candidate SNPs. We found significant association with high risk of bladder cancer in case of 4 sets of haplotype combinations (TCCGG p = 0.005, OR 2.304, 95 % CI 1.284–4.135; TTCGA p = 0.039, OR 2.281 95 % CI 1.041–4.999; ACTGG p = 0.008, OR 3.100 95 % CI 1.345–7.142 and TCTGA p = 0.006, OR 3.235 95 % CI 1.410–7.422), after applying Bonferroni correction (TCCGG pc = 0.001, TTCGA pc = 0.0078, ACTGG pc = 0.0016 and TCTGA pc = 0.0012) (Table 4).
Table 4.
Haplotypic analysis of CD44rs4755392 A/T, CD44rs187115 C/T, CD44rs13347 C/T, CD44 rs353639 G/T and CD44rs187116 A/G
| Haplotype combination | Controls [n (%)] |
Patients [n (%)] |
p value | OR (95 % CI) |
|---|---|---|---|---|
| ACCGA | 62 (12.0) | 46 (10.0) | Ref | Ref |
| TCCGG | 31 (6.0) | 53 (11.5) | 0.005 | 2.304 (1.284–4.135) |
| TTCGA | 13 (2.5) | 22 (4.8) | 0.039 | 2.281 (1.041–4.999) |
| ACTGG | 10 (1.9) | 23 (5.0) | 0.008 | 3.100 (1.345–7.142) |
| TCTGA | 10 (1.9) | 24 (5.2) | 0.006 | 3.235 (1.410–7.422) |
The statistically significant values are shown in bold
After applying Bonferroni corrections; TCCGG pc = 0.001, TTCGA pc = 0.0078, ACTGG pc = 0.0016 and TCTGA pc = 0.0012
In silico Analysis for the Functionality of CD44 gene Variants
SNPs rs4755392, rs187115, rs13347, rs353639 and rs187116 were selected for the present study. Their location in the CD44 gene is described in Table 5. In-silico analysis using F-SNP showed change in transcriptional regulation for three of the candidate SNPs (Table 5).
Table 5.
In silico analysis of CD44 gene polymorphisms by F–SNP
| SNPs of CD44 gene | Functional category | Prediction tool | Prediction result | FS score | Location |
|---|---|---|---|---|---|
| rs 4755392 | Transcriptional regulation | TF search | Not changed | 0 | 3′-UTR |
| rs 187115 | Transcriptional regulation | TF search | Changed | 0.176 | Intronic |
| rs 13347 | Transcriptional regulation | TF search | Changed | 0.176 | 3′-UTR |
| rs 353639 | Transcriptional regulation | TF search | Changed | 0.176 | Intronic |
| rs 187116 | Transcriptional regulation | TF search | Not changed | 0 | Intronic |
Association of High-Order Interactions with BC Risk by MDR Analysis
Multifactor Dimensionality Reduction (MDR) method is nonparametric, genetic model-free method for overcoming some of the limitations of logistic regression (i.e. sample size limitations) for the detection and characterization of gene–gene interactions. In MDR, multi-locus genotypes are pooled into high risk and low risk groups, effectively reducing the genotype predictors from n dimensions to one dimension (i.e. constructive induction). The MDR software (version 2.0 beta 8) was applied to identify high-order gene–gene interactions associated with BC risk. In our study, the best candidate interaction model was selected across all multi-locus models that maximized testing accuracy and the Cross-Validation Consistency (CVC).
The MDR permutation results were considered to be statistically significant at the 0.05 level. All the variables identified in the best model were combined and dichotomized according to the MDR software and their ORs and 95 % CI in relation to BC risk were calculated. Finally, combined effect of the variables in the best model by the number of risk genotypes was evaluated using logistic regression analysis (Table 6).
Table 6.
Association of high-order interactions with BC risk by MDR analysis
| S. No. | Gene combination | Odds ratio | p value | CVC | Testing accuracy |
|---|---|---|---|---|---|
| 1. | CD44 rs13347C/T | 1.400 (0.459–4.268) | 0.553 | 9/10 | 0.537 |
| 2. | CD44 rs13347C/T–CD44 rs353639 G/T | 0.925 (0.304–2.811) | 0.891 | 4/10 | 0.486 |
| 3. | CD44 rs13347C/T-CD44 rs353639G/T-CD44rs187116 A/G a | 1.678 (0.550–5.113) | 0.361 | 8/10 | 0.566 |
CVC cross validation consistency
aThe model with the maximum testing accuracy and maximum CVC cross was considered as the best model
Discussion
CD44 represents a family of class I trans-membrane glycoprotein and is involved in variety of cellular functions viz. proliferation, motility, invasion and survival [17]. Variety of function of CD44 depends upon binding of ligand-hyaluronic acid [18]. Any aberration in proper binding may lead to improper proliferation, motility, angiogenesis and metastasis in bladder cancer as well as other cancers. However CD44 polymorphism has not been studied extensively. Some studies are reported in breast cancer worldwide [4, 19, 20], gastric or colon cancer [21], head and neck cancer [6, 22] which states that CD44 is involved in cancer development and prognosis.
In this study we have studied five SNPs of CD44 (rs4755392, rs187115, rs13347, rs353639 and rs187116) among 240 confirmed bladder cancer patients and 270 healthy controls. We found protective risk of BC in rs4755392, rs13347, rs353639 and no association in the other two SNPs i.e. rs187115 and rs187116. No association was found after further stratification of patients on the basis of smoking habit, tumor stage/grade and BCG treatment.
In our study, CD44 rs4755392 revealed a significant reduced risk in BC as compared to the study of Gerger et al. [23] where they demonstrated colon cancer risk. Germline variant of CD44 rs4755392 revealed no significant statistical association in patients with localized gastric adenocarcinoma [15] and in patients with gastrointestinal (non-colorectal) cancer [24].
Our study suggested a protective risk to bladder cancer in heterozygous genotype of additive model (CT), dominant model (CT + TT) and variant allele (T) of rs13347 in contradictory to which a study by Jiang et al. [19] depicted 1.72-folds increased susceptibility in variant genotype (CT + TT) to breast cancer in Chinese population. Another study by Xiao. et al. [24] showed functional variation CD44 rs13347 CT and TT genotypes were associated with increased risk of nasopharyngeal cancer in Chinese population. rs13347C/T in 3′UTR of CD44 found to be a genetic modifier for developing acute myeloid leukemia in Chinese population [20]. This tagger SNP did also showed no effect in breast cancer susceptibility in North Indian females [25]. The variation in results may be due to varied ethnicity and different disease. CD44 rs13347 is also studied at translational and transcriptional level which also suggested high expression in various cancers like bladder, breast, colon [26] head and neck cancer [27]. Another tagger SNP rs353639 of CD44 gene was also evaluated to see the effect on bladder cancer. We found reduced risk to BC in dominant model (GT + TT) and variant allele (T) of rs353639. although, rs353639 did not show any effect on breast cancer susceptibility in North Indian females [25]. CD44 rs187116 and rs187115 in our study was not associated bladder cancer risk in our study.
Recently, Vazquez et al. [28] showed that the C/C genotype of intronic CD44 rs187115 germline variation was significantly associated with decreased cellular response to cytotoxic chemotherapeutics including doxorubicin, carboplatin, RNA/DNA and DNA antimetabolites in vitro strongly suggesting a functionally significant role for this SNP in tumor cells of soft tissue sarcoma. However, haplotype analysis of polymorphisms rs187115 and rs187116 provided evidence for association with tumor recurrence in soft tissue sarcoma. Winder et al. [15] found CD44rs 187116 to be significantly associated with tumor recurrence in gastric adenocarcinoma, Zhou et al. [20] found CD44 rs187116 to be involved in breast cancer progression. The CD44 rs187115 polymorphism has potential predictive significance in oral carcinogenesis in Taiwanian population [29].
The observations made so far in our study suggested that CD44 gene as well as its variants has a significant role in cancer development and prognosis. Nevertheless, this study provides the evidence for the first time that CD44 may represent one of the genes for the genetic risk of bladder cancer and CD44 polymorphisms may be effectively used in the risk assessment and genetic epidemiological analysis of bladder cancer among north Indians. This kind of hypothesis may be further used for individual specific cancer prognosis, if validated further at expression level.
Haplotype analysis among the SNPs is also done as these polymorphisms were in LD and can mask or change the genetic effects of those loci in the association analysis. We did the haplotypic analysis and got 26 combinations, out of which 4 combinations showed statistically significant association with BC risk. ACCGA is taken as reference and other four combinations viz. TCCGG (p = 0.005, OR 2.304), TTCGA (p = 0.039, OR 2.281), ACTGG (p = 0.008, OR 3.100) and TCTGA (p = 0.006, OR 3.235) showed significant high risk of BC. Our study was compatible with haplotype of CD44 variants significantly influenced risk of gastric cancer in Chinese patients [30]. Whereas, a study by Sharma et al. [31] showed reduced risk of gall bladder cancer in CCAT (p = 0.04, OR 0.47) and CAAT haplotype was marginally associated with low gall bladder cancer risk in patients from north India (p = 0.026, OR 0.53). Another study by Winder et al. [15] found T–A haplotype to be at low risk of tumor recurrence as compared with patients having T–G haplotype in gastric adenocarcinoma.
CD44 rs13347C/T SNP is supposed to be the strongest risk factor for BC among the polymorphisms examined in present study as an outcome of MDR analysis. Individuals carrying CD44 rs13347C/T-CD44 rs353639G/T-CD44rs187116A/G exhibited 1.6 folds risk i.e. the highest risk for BC. The model with the maximum testing accuracy and maximum CVC cross is considered as the best model in MDR analysis. The present study calculated, the best interaction model as the three-factor model including CD44rs13347C/T-CD44rs353639G/T-CD44rs187116A/G. The reported results represent the same results as in case of genotypic level for the entire gene with significant results. Thus, combining the single locus analysis, MDR we found that single genetic variants in CD44 gene may not be responsible in conferring high risk for disease but rather a higher order gene–gene interactions are likely to be involved in genetic susceptibility to BC.
In-silico analysis using bioinformatic tool F-SNP showed change in transcriptional regulation for CD44rs13347C/T, CD44rs353639G/T and CD44rs187116A/G supporting the results obtained at genotypic level of these SNPs taken for current study while some of them differed; the probable reason behind it may, as in silico analysis is based on logarithmic base module that is not present in natural statue as the study subjects exist.
Conclusion
Our study suggests that rs353639 shows a marginal risk for bladder cancer susceptibility, whereas, rs4755392 and rs13347 have reduced risk of bladder cancer. The other two candidate SNPs viz. rs187115 and rs187116 had no effect on bladder cancer susceptibility in North Indians. To the best of our knowledge, the present study is the first to report a group of five SNPs of CD44 gene polymorphisms with bladder cancer risk in North Indian population.
Compliance with Ethical Standards
Conflict of interest
Authors have no conflicts of interest in this work.
References
- 1.American Cancer Society . Cancer facts and figures 2015. Atlanta: American Cancer Society; 2015. [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012 v1.0, Cancer incidence and mortality worldwide: IARC CancerBase No. 11 [Internet]. Lyon: International Agency for Research on Cancer; 2013. http://globocan.iarc.fr. Accessed Dec 2013.
- 3.Murthy NS, Nandakumar BS, Pruthvish S, George PS, Mathew A. Disability adjusted life years for cancer patients in India. Asian Pac J Cancer Prev. 2010;11(3):633–640. [PubMed] [Google Scholar]
- 4.Götte M, Yip GW. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res. 2006;66:10233–10237. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- 5.Marhaba R, Zöller M. CD44 in cancer progression: adhesion, migration and growth regulation. J Mol Histol. 2004;35:211–231. doi: 10.1023/B:HIJO.0000032354.94213.69. [DOI] [PubMed] [Google Scholar]
- 6.Joshua B, Kaplan MJ, Doweck I, Pai R, Weissman IL, Prince ME, et al. Frequency of cells expressing CD44, a head and neck cancer stem cell marker: correlation with tumor aggressiveness. Head Neck. 2012;34:42–49. doi: 10.1002/hed.21699. [DOI] [PubMed] [Google Scholar]
- 7.Draffin JE, McFarlane S, Hill A, Johnston PG, Waugh DJ. CD44 potentiates the adherence of metastatic prostate and breast cancer cells to bone marrow endothelial cells. Cancer Res. 2004;64:5702–5711. doi: 10.1158/0008-5472.CAN-04-0389. [DOI] [PubMed] [Google Scholar]
- 8.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signaling regulators. Nat Rev Mol Cell Biol. 2003;4(1):33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 9.Park YS, Huh JW, Lee JH, Kim HR. shRNA against CD44 inhibits cell proliferation, invasion and migration, and promotes apoptosis of colon carcinoma cells. Oncol Rep. 2012;27(2):339–346. doi: 10.3892/or.2011.1532. [DOI] [PubMed] [Google Scholar]
- 10.Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009;374:239–249. doi: 10.1016/S0140-6736(09)60491-8. [DOI] [PubMed] [Google Scholar]
- 11.Walker CL, Ho S. Developmental reprogramming of cancer susceptibility. Nat Rev Cancer. 2012;12(7):479–486. doi: 10.1038/nrc3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colombel M, Soloway M, Akaza H. Epidemiology, staging, grading and risk stratification of bladder cancer. Eur Urol. 2008;7(Suppl.):618–626. doi: 10.1016/j.eursup.2008.08.002. [DOI] [Google Scholar]
- 13.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 14.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winder T, Ning Y, Yang D, Zhang W, Power DG, Bohanes P, et al. Germline polymorphisms in genes involved in the CD44 signaling pathway are associated with clinical outcome in localized gastric adenocarcinoma. Int J Cancer. 2011;129:1096–1104. doi: 10.1002/ijc.25787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee PH, Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2008;36:D820–D824. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tremmel M, Matzke A, Albrecht I, Laib AM, Olaku V, Ballmer-Hofer K, et al. A CD44v6 peptide reveals a role of CD44 in VEGFR-2 signaling and angiogenesis. Blood. 2009;114:5236–5244. doi: 10.1182/blood-2009-04-219204. [DOI] [PubMed] [Google Scholar]
- 18.Screaton GR, Bell MV, Jackson DG, Cornelis FB, Gerth U, Bell JI. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc Natl Acad Sci USA. 1992;89:12160–12164. doi: 10.1073/pnas.89.24.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang L, Deng J, Zhu X, Zheng J, You Y, Li N, et al. CD44 rs13347 C > T polymorphism predicts breast cancer risk and prognosis in Chinese populations. Breast Cancer Res. 2012;14:R105. doi: 10.1186/bcr3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J, Nagarkatti PS, Zhong Y, Creek K, Zhang J, Nagarkatti M. Unique SNP in CD44 intron 1 and its role in breast cancer development. Anticancer Res. 2010;30:1263–1272. [PMC free article] [PubMed] [Google Scholar]
- 21.Guo YJ, Liu G, Wang X, Jin D, Wu M, Ma J, et al. Potential use of soluble CD44 in serum as indicator of tumor burden and metastasis in patients with gastric or colon cancer. Cancer Res. 1994;54:422–426. [PubMed] [Google Scholar]
- 22.Van Hal NL, Van Dongen GA, Ten Brink CB, Heider KH, Rech-Weichselbraun I, Snow GB, et al. Evaluation of soluble CD44v6 as a potential serum marker for head and neck squamous cell carcinoma. Clin Cancer Res. 1999;5:3534–3541. [PubMed] [Google Scholar]
- 23.Gerger A, Zhang W, Yang D, Bohanes P, Ning Y, Winder T, et al. Common cancer stem cell gene variants predict colon cancer recurrence. Clin Cancer Res. 2011;17:6934–6943. doi: 10.1158/1078-0432.CCR-11-1180. [DOI] [PubMed] [Google Scholar]
- 24.Xiao M, Hu S, Zhang L, Huang J, Jiang H, Cai X. Polymorphisms of CD44 gene and nasopharyngeal carcinoma susceptibility in a Chinese population. Mutagenesis. 2013;28:577–582. doi: 10.1093/mutage/get035. [DOI] [PubMed] [Google Scholar]
- 25.Tulsyan S, Agarwal G, Lal P, Agrawal S, Mittal RD, Mittal B. CD44 gene polymorphisms in breast cancer risk and prognosis: a study in North Indian population. PLoS One. 2013;8:e71073. doi: 10.1371/journal.pone.0071073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodman AC, Sugiyama M, Yoshida K, Sugino T, Borgya A, Goodison S, et al. Analysis of anomalous CD44 gene expression in human breast, bladder, and colon cancer and correlation of observed mRNA and protein isoforms. Am J Pathol. 1996;149:1519–1530. [PMC free article] [PubMed] [Google Scholar]
- 27.Trapasso S, Allegra E. Role of CD44 as a marker of cancer stem cells in head and neck cancer. Biologics. 2012;6:379–383. doi: 10.2147/BTT.S37906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vazquez A, Grochola LF, Bond EE, Levine AJ, Taubert H, Müller TH, et al. Chemosensitivity profiles identify polymorphisms in the p53 network genes 14-3-3tau and CD44 that affect sarcoma incidence and survival. Cancer Res. 2010;70(1):172–180. doi: 10.1158/0008-5472.CAN-09-2218. [DOI] [PubMed] [Google Scholar]
- 29.Chou YE, Hsieh MJ, Hsin CH, Chiang WL, Lai YC, Lee YH, et al. CD44 gene polymorphisms and environmental factors on oral cancer susceptibility in Taiwan. PLoS One. 2014;9:e93692. doi: 10.1371/journal.pone.0093692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu Y, Hu Y, Zhang ZY, Ye L, Xu FH, Schneider ME, et al. Genetic association of osteopontin (OPN) and its receptor CD44 genes with susceptibility to Chinese gastric cancer patients. J Cancer Res Clin Oncol. 2014;140:2143–2156. doi: 10.1007/s00432-014-1761-9. [DOI] [PubMed] [Google Scholar]
- 31.Sharma KL, Yadav A, Gupta A, Tulsayan S, Kumar V, Misra S, et al. Association of genetic variants of cancer stem cell gene CD44 haplotypes with gallbladder cancer susceptibility in North Indian population. Tumour Biol. 2014;35:2583–2589. doi: 10.1007/s13277-013-1340-8. [DOI] [PubMed] [Google Scholar]