Abstract
Cullin-dependent ubiquitin ligases regulate a variety of cellular and developmental processes by recruiting specific proteins for ubiquitin-mediated degradation. Cullin proteins form a scaffold for two functional modules: a catalytic module comprised of a small RING domain protein Roc1/Rbx1 and a ubiquitin-conjugating enzyme (E2), and a substrate recruitment module containing one or more proteins that bind to and bring the substrate in proximity to the catalytic module. Here, we present evidence that the three Drosophila Roc proteins are not functionally equivalent. Mutation of Roc1a causes lethality that cannot be rescued by expression of Roc1b or Roc2 by using the Roc1a promoter. Roc1a mutant cells hyperaccumulate Cubitus interruptus, a transcription factor that mediates Hedgehog signaling. This phenotype is not rescued by expression of Roc2 and only partially by expression of Roc1b. Targeted disruption of Roc1b causes male sterility that is partially rescued by expression of Roc1a by using the Roc1b promoter, but not by similar expression of Roc2. These data indicate that Roc proteins play nonredundant roles during development. Coimmunoprecipitation followed by Western or mass spectrometric analysis indicate that the three Roc proteins preferentially bind certain Cullins, providing a possible explanation for the distinct biological activities of each Drosophila Roc/Rbx.
INTRODUCTION
Protein modification by ubiquitin is widely used by eukaryotic organisms to regulate many important cellular and developmental processes (Ciechanover et al., 2000; Ben-Neriah, 2002; Conaway et al., 2002; Wojcik, 2002). Three enzymatic activities, ubiquitin activation (E1), conjugation (E2), and ligation (E3), lead to the covalent attachment of ubiquitin, a 76-amino acid protein, to specific target proteins (Hershko and Ciechanover, 1998). Monoubiquitylation can influence the activity or subcellular localization of the target protein (Pickart, 2001; Raiborg et al., 2003). Repeated rounds of the E2-E3 reaction result in the formation of polyubiquitin chains that generally serve as a signal for destruction by the 26S proteosome, but they can also have nonproteolytic effects on protein function (Hershko and Ciechanover, 1998; Kaiser et al., 2000; Pickart, 2000, 2001). The E3 plays a crucial role in this pathway not only because it mediates the transfer of ubiquitin to the target protein but also because it provides substrate specificity (Jackson et al., 2000).
One well characterized E3 is the multisubunit SCF complex (Deshaies, 1999; Jackson and Eldridge, 2002). Consisting of four members (Skp1, CUL-1/Cdc53, an F-box-containing protein, and Roc1/Rbx1/Hrt1), the SCF regulates many developmental processes such as the cell cycle, signaling pathways, circadian rhythms, and apoptosis (Koepp et al., 1999; Maniatis, 1999; DeSalle and Pagano, 2001; Grima et al., 2002; Ko et al., 2002; Nateri et al., 2004). The specificity of SCF complexes is conferred by the F-box subunit, which binds to the target protein through a protein-protein interaction motif such as WD40 or leucine-rich repeats (Skowyra et al., 1997; Craig and Tyers, 1999; Kipreos and Pagano, 2000). Skp1 serves as an adapter by binding to the F-box domain of the F-box protein and the N-terminal portion of CUL-1 (Bai et al., 1996; Feldman et al., 1997; Zheng et al., 2002). CUL-1 is a scaffold that brings together the F-box/substrate complex and the E2 enzyme, which is recruited by the Roc subunit bound to the C terminus of CUL-1 (Kipreos et al., 1996; Patton et al., 1998; Kamura et al., 1999; Ohta et al., 1999; Seol et al., 1999; Furukawa et al., 2000, 2002).
The SCF complex is just one member of a family of Cullin/Roc-based E3 ubiquitin ligases. The VBC complex, which regulates the stability of transcription factors involved in the response to hypoxia and has been associated with hypervascularization of tumors and cancer progression (Kim and Kaelin, 2003), contains CUL-2, Elongins B and C, and the VHL tumor suppressor protein (Iwai et al., 1999; Kamura et al., 2001). Elongins B and C also interact with CUL-5 to form a distinct E3 ligase that functions in HIV-1 replication (Kamura et al., 2001; Yu et al., 2003). CUL-3, which is required for normal development in mice, Drosophila, and Caenorhabditis elegans, uses BTB proteins as adaptors to recruit substrates (Ou et al., 2002; Furukawa et al., 2003; Geyer et al., 2003; Pintard et al., 2003; Xu et al., 2003). It is thought that the DNA-damage binding protein DDB1 is one adaptor for human CUL-4A ubiquitin ligase complexes (Nag et al., 2001; Groisman et al., 2003; Wertz et al., 2004), and in C. elegans and Drosophila S2 cells CUL-4 has been shown to target the DNA replication licensing factor CDT-1 for degradation (Higa et al., 2003; Zhong et al., 2003). Like the Cullin proteins, there are multiple Roc proteins in higher eukaryotes that fall within two subfamilies, Roc1 and Roc2 (Ohta et al., 1999). With a few exceptions, it has not been demonstrated which of the Roc family members participates in a given Cullin complex.
With regard to the SCF complex, current data indicate that the F-box subunit is the major factor in determining functional specificity (Craig and Tyers, 1999; Kipreos and Pagano, 2000). Many different F-box proteins exist, and of those that have been analyzed each seems to recruit different sets of target proteins. In yeast, the Cdc4 F-box protein regulates the stability of Sic1, Far1, and Cdc6, whereas Grr1 targets CLN1 and CLN2 (Tyers and Jorgensen, 2000). In Drosophila, targets of the F-box protein Slimb include Cubitus interruptus (Ci) and Armadillo (Arm), transcription factors in the Hedgehog and Wingless signaling cascades, respectively (Jiang and Struhl, 1998), the Dorsal/(nuclear factor κB) inhibitor Cactus (inhibitor of κB) (Spencer et al., 1999), as well as the circadian rhythm-regulating proteins Period (Per) and Timeless (Tim) (Grima et al., 2002; Ko et al., 2002). Cyclin E and Myc degradation are controlled by a different F-box protein, Archipelago (Moberg et al., 2001, 2004). A common theme emerging from analyses of these F-box proteins and their targets is that phosphorylation of the target seems to be a prerequisite for recognition.
Intriguingly, the Drosophila genome contains three members of the Roc gene family (Roc1a, Roc1b, and Roc2, which share 57-70% amino acid similarity), whereas the genomes of other higher eukaryotes such as worms, mice, and humans contain two (Roc1 and Roc2). Previously, we have shown that clones of cells mutant for Roc1a fail to proliferate, implicating a role for Roc1a in cell cycle progression (Noureddine et al., 2002). These mutant clones also hyperaccumulate the full-length form of Ci (Noureddine et al., 2002), suggesting that Roc1a is part of an SCFSlimb complex that targets this transcription factor for proteolytic processing (Jiang and Struhl, 1998). In this study, we sought to determine if Roc1b or Roc2 could substitute for Roc1a in the context of both the proliferation defect and the SCFSlimb complex that targets Ci. Surprisingly, expression of Roc1b or Roc2 was insufficient to fully compensate for Roc1a loss, although overexpression of Roc1b could rescue the Ci hyperaccumulation phenotype. Homologous recombination-mediated gene targeting (Rong and Golic, 2000) was used to generate mutations in Roc1b, and we show that these mutations cause male sterility. Using similar in vivo complementation assays, we demonstrate that Roc1a, but not Roc2, is able to partially compensate for Roc1b loss. Finally, we provide evidence that each Roc protein preferentially associates with different members of the Cullin family, which may likely explain the inability of the three Roc proteins to fully compensate for each other.
MATERIALS AND METHODS
Fly Stocks and Crosses
To test for rescue of Roc1a lethality, Roc1aG1, FRT/FM7, Act-GFP females were mated with males from stocks expressing a given Roc transgene under control of the Roc1a promoter and the lethal phase of green fluorescent protein (GFP)-negative males was analyzed. To examine the ability of each Roc to rescue Ci hyperaccumulation, clones of Roc1a mutant cells were generated using the MARCM system as described previously (Noureddine et al., 2002), except that the flies also carried the appropriate Roc transgene. HomozygousRoc1b mutant males carrying the appropriate Roc transgene were generated to analyze the rescue of the male sterility.
Rescue Constructs
The Roc1a genomic rescue fragment was described previously (Noureddine et al., 2002) and contains 980 bp of sequence upstream of the ATG initiation codon and 620 bp downstream of the translational Stop, except that a FLAG tag was inserted at the N-terminus. To create the rescue constructs in which the Roc1a coding region was replaced with that of Roc1b or Roc2, an NruI site was introduced into the above-mentioned construct immediately downstream of the sequences coding for the FLAG epitope, and an AatII site was introduced immediately after the Stop codon. The Roc1b and Roc2 coding regions were amplified from the Origene RapidScan library with primers containing the appropriate linkers and subcloned as NruI/AatII fragments into the modified Roc1a rescue construct. To obtain the Roc1b genomic rescue transgene, the Roc1b genomic locus, including 840 bp upstream of the ATG codon and 330 bp downstream of the translational Stop site, was amplified from BACR13F06. To generate the constructs in which the Roc1a and Roc2 open reading frames (ORFs) replaced the Roc1b ORF, Roc1a and Roc2 were amplified from the Origene RapidScan library with a forward primer containing an NcoI linker and a reverse primer with an EagI linker and these polymerase chain reaction (PCR) products were subcloned into the Roc1b genomic rescue construct. To generate Roc1bgrf::FLAG-Roc1b, the 840-bp Roc1b promoter and 330 bp downstream region were amplified in separate PCR reactions and joined in the appropriate orientation through an AgeI linker introduced during PCR. Using Roc1agrf::FLAG-Roc1b as a template, the FLAG-Roc1b ORF was amplified with primers also containing AgeI linkers and inserted in the correct orientation between the promoter and downstream region. Each of these genomic rescue constructs was cloned into pCaSpeR-4. To generate the UAS-Roc transgenes, each Roc ORF was cloned into pUAST (Brand and Perrimon, 1993). Microinjection of Drosophila embryos was done using standard methods, and multiple independent lines for each construct were analyzed.
Immunofluorescence
For detection of the FLAG-Roc proteins in situ, larvae from each of the Roc1agrf::FLAG-Roc stocks were mixed with UAS-FLAG-dribble; en-Gal4 larvae, dissected, fixed in 4% paraformaldehyde, and stained with mouse anti-FLAG M2 (diluted 1:250) and Cy3-conjugated goat anti-mouse (diluted 1:1000; Jackson Immunoresearch Laboratories, West Grove, PA). Discs were mounted in Fluoromount-G and photographed with a Nikon Eclipse E800 equipped with a charge-coupled device camera. For detection of Ci protein accumulation, discs were stained with rat anti-Ci 2A1 (gift of R. Holmgren, Northwestern University, Evanston, IL) and processed as described previously (Noureddine et al., 2002).
Western Blotting and Immunoprecipitation
To verify expression of the FLAG-Roc transgenes, 0- to 8-h embryos from each of the stocks were homogenized in sample buffer (Harlow and Lane, 1999) and boiled for 5 min. The extracts were then run on a 15% acrylamide gel and transferred to nitrocellulose in methanol-free transfer buffer (38 mM glycine, 5 mM Tris base). The blots were rinsed several times with phosphate-buffered saline plus 0.1% Tween-20 (PBT), blocked with 5% bovine serum albumin (BSA), and incubated for 1 h at room temperature with a mouse anti-FLAG M2 antibody (diluted 1:100 in PBT plus BSA; F3165, Sigma-Aldrich, St. Louis, MO). For detection, horseradish peroxidase (HRP)-conjugated goat anti-mouse secondary antibody (diluted 1:15,000; Amersham Biosciences, Piscataway, NJ) and ECL-Plus (Amersham Biosciences) were used. For coimmunoprecipitation experiments, 4- to 8-h (for Western) or overnight (for mass spectrometry) collections of embryos were dechorionated in 50% bleach for 5 min and then lysed in NET buffer (50 mM Tris pH 7.0, 400 mM NaCl, 5 mM EDTA, 1% NP-40, 50 μg/ml phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1.4 μg/ml pepstatin). Samples were incubated with mouse IgG-agarose beads (A0919; Sigma-Aldrich) for 1 h at 4°C to reduce nonspecific binding, and the supernatants were then incubated with anti-FLAG-M2-conjugated agarose gel (A2220; Sigma-Aldrich) for 2 h at 4°C. After three washes with NET buffer, immunocomplexes were eluted with 66 μg/ml 3×-FLAG peptide for 30 min at 37°C and analyzed by SDS-PAGE. For Western analysis, the gel was cut in half so that the larger migrating CUL-1 bands could be blotted with traditional Western transfer buffer, whereas the smaller, FLAG-Roc bands could be blotted with methanol-free buffer. Detection of CUL-1 was performed with a rabbit anti-CUL-1 antibody (diluted 1:250; Zymed Laboratories, South San Francisco, CA) and goat anti-rabbit-HRP secondary (diluted 1:10,000; Chemicon International, Temecula, CA). Detection of the FLAG-Rocs was as described above. For mass spectrometry, the gel was fixed in 25% acetic acid/10% isopropanol for 20 min, stained overnight in 0.01% G-250 Coomassie Blue solution, and destained in 10% acetic acid. Specific bands were excised, digested with trypsin, and analyzed by matrix-assisted laser desorption ionization/time of flight mass spectrometry by the University of North Carolina Proteomics Core Facility.
Construction of Roc1b Targeting Vectors
Approximately 7 kb of DNA homologous to the Roc1b locus was used for targeting. Recombinant PCR was used to obtain the region of homology distal to Roc1b (with respect to the centromere), which contains the last exon of CG1228, the gene CG1231, and the 5′ part of Roc1b, as well as to introduce an I-SceI cut site ∼500 base pairs upstream of the Roc1b translation start site. The outside primers for these reactions were 5′-CCTCAGCGGCCGCCCTATTTCAGATGACTGCAC-3′ (which has a 5′ NotI linker) and 5′-CTCAACCTCTAGATCTCCTCG-3′ (which introduces two bases [underlined] into the Roc1b coding region, creating and XbaI site). The inside primers were 5′-TATTACCCTGTTATCCCTACATTATTATTAAGGAAGCTTTAC-3′ and 5′-GTAGGGATAACAGGGTAATACATTCGAGTTTGGGAAACAG (the 18 bases corresponding to the I-SceI site are underlined). The recombinant PCR product was cloned into pCR-BluntII (Invitrogen, Carlsbad, CA) and shuttled to pBluescript KS+ (pBS) as a NotI/XbaI fragment. To obtain the region of homology proximal to Roc1b, which includes most of Roc1b and the gene CG6905, and simultaneously generate the desired mutations, two separate PCR reactions were performed, one for each allele. The forward primers (both of which create an XbaI site, underlined) used to generate the frameshift and deletion alleles were 5′-CGAGGAGATCTAGAGGTTGAGG-3′ and 5′-AATCATCTAGACAACAAGGAGTGGGTCTAC-3′, respectively. The reverse primer for both reactions was 5′-GGAGTAGGTACCACACTGTCGCGTTATGTTATG-3′ (which has a 5′ Asp718 linker). Each PCR reaction was cloned into pCR-BluntII and subsequently inserted into pBS containing the distal region as an XbaI/Asp718 fragment. All PCR reactions used the BAC clone BACR13F06 as a template and were sequenced. Each targeting construct was then cloned into pTargetB (a gift of Sarah Radford and Jeff Sekelsky, University of North Carolina, Chapel Hill, NC) for injection into embryos.
Genetics of Targeting
Thirty-two potential donor constructs (18 frameshift, 14 deletion) were first crossed to flies constitutively expressing the FLP recombinase (w; P{70FLP}10; a gift of Kent Golic, University of Utah, Salt Lake City, UT) and the number of white-eyed progeny was divided by the total number of progeny (including those with mosaic eyes) to obtain a mobilization percentage (% Mob; Table 1) indicative of the ability of each donor line to excise. Fifteen lines with a high mobilization percentage were chosen to be sent through the targeting scheme. Virgin females carrying the donor construct were crossed to yw; P{hsp70-FLP}, P{hsp70-I-SceI}, Sco/Cyo, S (a gift of Kent Golic, University of Utah) males, and the progeny were heat shocked in a 37°C water bath for 1 h 3 d after egg laying. Next, w+, Sco virgin female progeny were mated to w; P{70FLP}10 males (20 females, 10 males per bottle) to screen for the presence of a w+ element that does not mobilize in the presence of constitutive FLP expression, indicative of a potential homologous recombination (HR) event. The number of female germlines screened for each donor construct is given in Table 1. w+ progeny were then subjected to a second round of screening by backcrossing to w; P{70FLP}10; this step eliminated more than half of w+ flies obtained from the first round of screening. Flies that still contained the w gene after two rounds of screening were then analyzed for HR events in two ways. First, we verified that w+ segregated with the third chromosome and then we performed inverse PCR to specifically identify class II (tandem duplication) events (Rong and Golic, 2000). Inverse PCR was performed as described on the Berkeley Drosophila Genome Project Web site (http://www.fruitfly.org/about/methods/inverse.pcr.html) by using SacI, except that 1 fly equivalent of ligated genomic DNA was used as a template. The primers (designed to amplify a fragment only when a class II HR event occurred) were 5′-CTCTCTTGCTGTACCATG-3′ (which anneals to part of the w gene) and 5′-GTCAGCACACGATCATCG-3′ (which anneals to genomic sequence just outside the tandem duplication). Fragments obtained by inverse PCR were cloned into pCR-II (Invitrogen) and sequenced.
Table 1.
Summary of the scheme used to target Roc1b
| Donor line | % Mob (n)a | No. female germlines | No. potential HR events | No. non-HR events | No. verified | % reduction |
|---|---|---|---|---|---|---|
| F9 | 100 (86) | 720 | 1 | 1 | 1 | ND |
| F13 | 100 (93) | 880 | 0 | 1 | NA | NA |
| F14 | 100 (76) | 400 | 0 | 0 | NA | NA |
| F15 | 98.8 (83) | 260 | 1 | 0 | ND | NA |
| F16 | 98.9 (95) | 360 | 0 | 0 | NA | NA |
| F17 | 98.7 (77) | 400 | 2 | 0 | ND | NA |
| F30 | 95.9 (98) | 360 | 2 | 0 | ND | NA |
| F32 | 98.9 (88) | 280 | 2 | 0 | 1 | 60.5b |
| D5 | 91.7 (97) | 200 | 0 | 0 | NA | NA |
| D6 | 100 (81) | 460 | 1 | 1 | ND | NA |
| D10 | 88.4 (95) | 160 | 0 | 1 | NA | NA |
| D13 | 73.3 (90) | 360 | 0 | 0 | NA | NA |
| D19 | 94.0 (67) | 460 | 0 | 0 | NA | NA |
| D24 | 98.3 (115) | 460 | 3 | 1 | 1 | 16.0c |
| D27 | 98.0 (49) | 120 | 0 | 0 | NA | NA |
NA, not applicable; ND, not done.
Number of flies scored for eye color
Reduction attempted in both males and females
Reduction attempted in males only
We positively identified three independent HR events. Potential HR events (Table 1) met three criteria: the w+ is stable when crossed to P{70FLP}10, maps to the third chromosome, and is similar in color to the three HR events verified by inverse PCR and sequencing. We also obtained at least five nonhomologous recombination events, each from a different donor line. Two of these (D6 and D10) occurred on a chromosome that was not chromosome 3. Of the three others that were on chromosome 3, one (from F9) did not give an eye color similar to the verified targeting events, one (from D24) was unlinked from a verified targeting event (tested by meiotic recombination), and one (from F13) was sequenced and found to target a location at 89E. Males from two verified HR events (F32 and D24) were then crossed to w1118;P{ν+, hsp70-I-CreI}, Sb/TM6 (a gift of Kent Golic, University of Utah) females to reduce the tandem duplication to a single copy. Progeny were heat shocked in a 36°C water bath for 30 min 3 d after egg laying. White-eyed, Sb males were collected and mated individually to w; TM3/TM6 females. Stocks were then created from Sb+/TM6B progeny and analyzed by PCR for the presence of the desired Roc1b mutations by using the primers 5′-CGCCGTTGTTATTTCGTAG-3′ and 5′-CTCTGTCTCACTCTCGAC-3′. The products were then digested with XbaI to confirm the presence of the mutation and subsequently sequenced. For reasons unknown, we observed a difference in the frequencies of reduction to generate the two mutant alleles. Reduction of the tandem duplication generated the frameshift allele at a higher frequency (60.5%) than that for the deletion allele (16%).
Characterization of Male Sterility
A single male 1-2 d posteclosion was mated to three w1118 virgin females and transferred daily. The number of eggs was counted immediately after transfer, and 7-10 d later the number of pupae was scored (Kusano et al., 2001). At least 30 males of each genotype were tested. Two aspects of male sterility were measured; the percentage of viable offspring produced (no. pupae/no. eggs) and the number of males that were completely sterile. Pairwise comparisons of the average hatching rates of each genotype were performed using a Student's t test.
Reverse Transcription (RT)-PCR
RNA was isolated from tissue samples (20 testes or ∼50 embryos) with TRIzol (Sigma-Aldrich) according to manufacturer's instructions. For the RT reaction, 1 μg of total RNA and 3.5 μM anchored oligo-dT23 were mixed in 10 μl of total volume and heated to 70°C for 10 min. Next, 2 μl of 10× RT buffer (750 mM KCl, 500 mM Tris-HCl, 30 mM MgCl2, 100 mM dithiothreitol, pH 8.3), 1 μl of dNTP mix (10 mM each), 1 μl of RNasin (Promega, Madison, WI), 1 μl (200 U) of M-MuLV-RT (New England Biolabs, Beverly, MA), and 5 μl of distilled H2O were added, and the reaction was incubated at 25°C for 15 min and then at 42°C for 1 h. Three microliters of cDNA was amplified with 35 cycles by using 66°C as the annealing temperature for the Roc1b and Rp49 primer sets and 58°C for the Roc1a and Roc2 primer sets by using Taq polymerase (Roche Diagnostics, Indianapolis, IN) and analyzed on a 1% agarose gel.
RESULTS
Mutations in Roc1a Cause Lethality That Cannot Be Rescued by Roc1b or Roc2
Previously, we reported that Roc1a mutant animals die as late first/early second instars (Noureddine et al., 2002), demonstrating a unique function for Roc1a that cannot be compensated by Roc1b or Roc2. This could be the result of functional differences between the Roc proteins, or because the Roc1b/Roc2 proteins are functionally equivalent but not expressed in the same spatial or temporal pattern or at the same level as Roc1a. To directly test these possibilities, we placed FLAG-tagged versions of each Roc open reading frame under control of the Roc1a promoter and 3′ untranslated sequence and asked whether this was sufficient to rescue the lethality caused by the Roc1a mutation. Like a native Roc1a genomic rescue fragment (grf) (Noureddine et al., 2002), Roc1agrf::FLAG-Roc1a was able to rescue Roc1a mutant flies to adulthood, indicating that the presence of the FLAG epitope does not significantly affect the function of the Roc1a protein. In contrast, Roc1a mutant flies expressing either Roc1agrf::FLAG-Roc1b or Roc1agrf::FLAG-Roc2 did not survive beyond the early second instar, precisely the same time at which Roc1a mutants die. Western blotting and immunostaining of wing discs revealed that each of the FLAG-Roc proteins was expressed (Figure 1). Moreover, the level of Roc1b was greater than that of Roc1a, which is sufficient for rescue (Figure 1D). These data suggest that there is at least one essential protein targeted by a Cullin-dependent E3 ligase complex containing Roc1a that is unable to be efficiently targeted by complexes containing either Roc1b or Roc2.
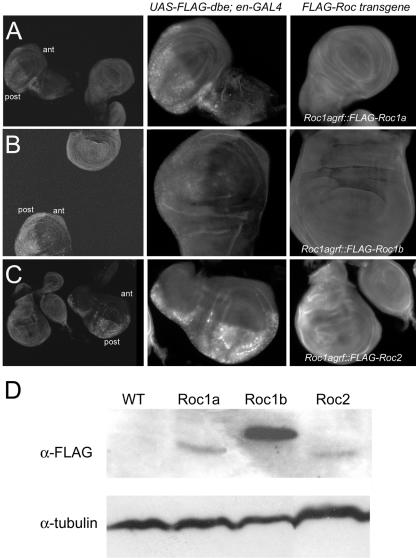
Figure 1.
Expression of Roc1agrf::FLAG-Roc transgenes. (A-C) Expression of FLAG-Roc proteins in the wing disc. Larvae from stocks expressing either FLAG-Roc1a (A), FLAG-Roc1b (B), or FLAG-Roc2 (C) from the Roc1a promoter were combined with UAS-FLAG-dribble; en-GAL4 larvae, dissected, fixed, and stained with an anti-FLAG antibody. Left, discs from both UAS-FLAG-dribble; en-GAL4 and Roc1agrf::FLAG-Roc genotypes in the same frame. Middle and right, close-ups of the same control (center) and experimental (right) discs, taken with same camera settings. Because it was difficult to discern background staining from the actual signal of the low, ubiquitous expression from the Roc1a promoter, UAS-FLAG-dribble; en-GAL4 discs served as both a positive and negative control for FLAG staining, because FLAG-Dribble is expressed only in the posterior compartment (post). Notice that the level of expression of each Roc protein is greater than that of the anterior compartments (ant) of the control discs where FLAG-Dribble is not expressed. (D) Western blot of embryo extracts showing expression of FLAG-Roc proteins. This level of FLAG-Roc1a expression is sufficient to rescue Roc1a mutant animals to adulthood. FLAG-Roc1b expression is significantly higher, but it is still unable to rescue the Roc1a mutation. Likewise, FLAG-Roc2 is expressed but also unable to rescue Roc1a mutants. Twice as much extract was loaded to the Roc2 lane. Similar results were observed from wing disc extracts (our unpublished data).
Roc1b, but Not Roc2, Can Partially Rescue Phenotypes of Roc1a Mutant Cells
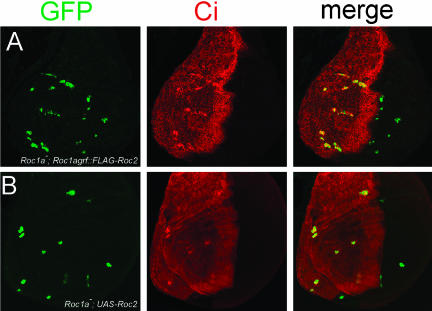
Given that the lethality of the Roc1a mutation may likely be the result of the hyperaccumulation of many different SCF targets, we asked whether Roc1b or Roc2 could substitute in the absence of Roc1a in regulating the stability of one known SCF target. We previously showed that Roc1a mutant clones of cells in the wing disc do not grow very large and also fail to process the Hh effector Ci from a 175-kDa transcriptional activator form to the 75-kDa repressor (Figure 2A; Noureddine et al., 2002). This suggests that neither Roc1b nor Roc2 normally play a major role in targeting Ci for processing, either because they are not expressed appropriately or because they are unable to assemble with the SCF complex responsible for targeting Ci. We used the MARCM system (Lee and Luo, 1999) to generate GFP-positive, Roc1a mutant clones in wing imaginal discs of flies carrying a specific Roc transgene and analyzed whether that transgene was able to supply a sufficient amount of Roc function to rescue either of the two Roc1a mutant phenotypes, namely, small clones and Ci hyperaccumulation.
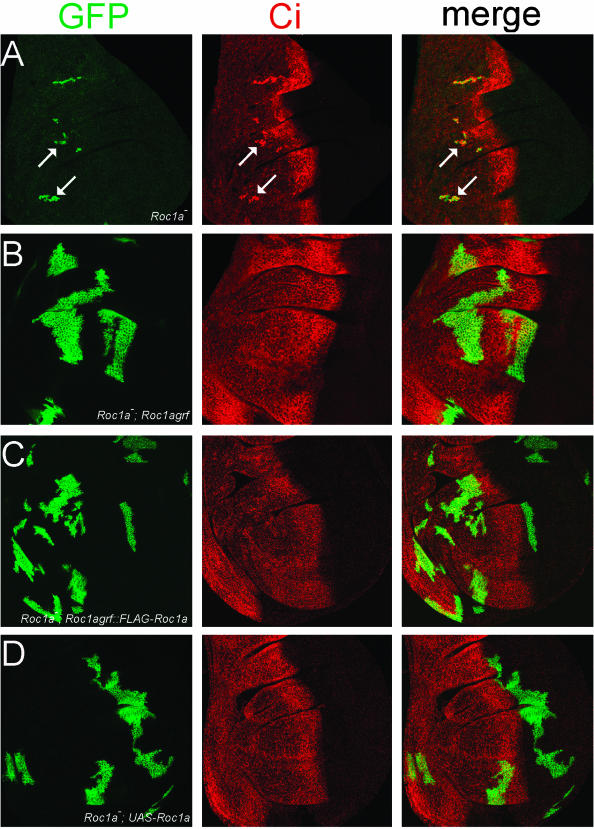
Figure 2.
Expression of Roc1a can fully rescue Roc1a mutant phenotypes. (A) Roc1a mutant clones (positively marked with GFP, arrows) have a proliferation defect and accumulate full-length Ci. (B-D) Expression of native Roc1a (B) or FLAG-Roc1a (C) with the Roc1a promoter or UAS-Roc1a (D) can rescue both the proliferation and Ci hyperaccumulation defects of the Roc1a mutation.
As expected, a Roc1a genomic rescue fragment expressing either a native or FLAG-tagged version of Roc1a was able to rescue both phenotypes; the Roc1a mutant clones grew to a large size and did not hyperaccumulate Ci (Figure 2, B and C). When Roc1agrf::FLAG-Roc1b was expressed in the Roc1a mutant cells, we observed a partial rescue of both phenotypes. In approximately half of the discs, the clones were similar in size to Roc1a mutant clones, and these clones always displayed elevated levels of full-length Ci protein (Figure 3A). However, in the other half of the discs, the mutant clones grew larger than clones not expressing a transgene, but not quite as large as clones expressing Roc1agrf::FLAG-Roc1a. In addition, Ci hyperaccumulation was not observed in these clones (Figure 3, C-E). This partial phenotypic rescue was not caused by a reduction in Roc1b function due to inclusion of the FLAG epitope, because FLAG-Roc1b rescues the null Roc1b phenotype as well as untagged Roc1b, which is described below (Figure 5C and Table 2). In contrast to the effects we observed with both Roc1a and Roc1b, Roc2 was completely unable to rescue the Roc1a mutant phenotypes. Roc1a mutant clones expressing Roc1agrf::FLAG-Roc2 were always small and always hyperaccumulated Ci (Figure 4A).
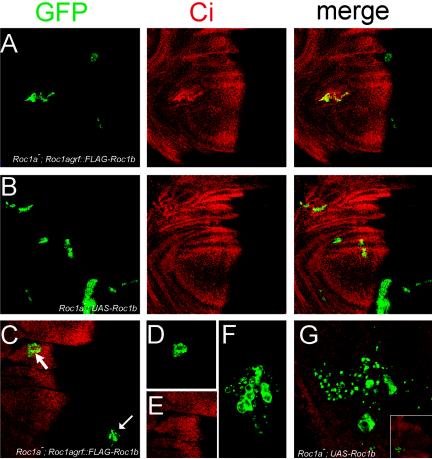
Figure 3.
Roc1b can partially substitute for Roc1a. (A) Roc1a mutant clones expressing Roc1agrf::FLAG-Roc1b. The clones are small and hyperaccumulate unprocessed Ci in about half of the wing discs. (B) Roc1a mutant clones expressing UAS-Roc1b. A partial rescue of the proliferation and Ci hyperaccumulation defects is observed. (C-F) A wing disc containing Roc1a mutant clones, depicting an example where Roc1agrf::FLAG-Roc1b expression rescues the Roc1a phenotypes (i.e., a normal level of Ci is observed). Single channels of the clone in the merged image (C, arrowhead) are shown in D and E. (F) Enlarged image of Roc1a mutant clone (C, arrow) showing the cell death phenotype. (G) Cell death phenotype of Roc1a mutant clones expressing UASRoc1b. Notice the area of the clone is larger than that in (F), consistent with the fact that UAS expression of Roc1b provides a higher degree of rescue than expression with the Roc1a promoter. Inset shows a lower magnification view of the wing pouch, indicating the position of the clone.
Figure 5.
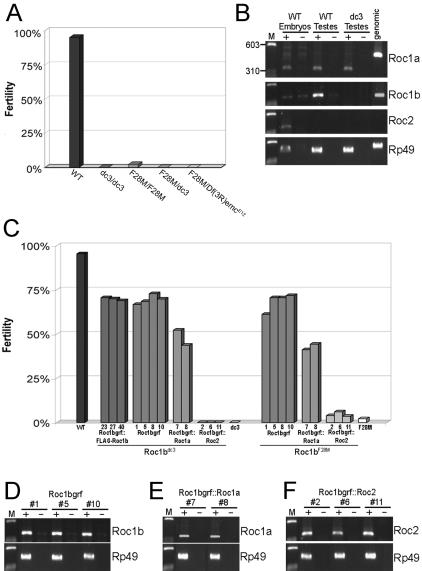
The Roc1b mutation causes male sterility. (A) Graph showing fertility of wild-type and various Roc1b mutant males. “Fertility” is the percentage of viable progeny produced by 30 males mated individually to three virgin females. (B) RT-PCR performed on indicated tissues using primer sets designated on the right. Genomic DNA was used as a control for each primer set. + and - indicate reactions with or without reverse transcriptase. The Roc1a and Rp49 primer sets span small introns and thus produce slightly larger PCR fragments. The Roc2 primer set spans a 26-kb intron. WT embryos were also used as a control to test primer sets that do not amplify messages in the testes (i.e., Roc2). This also reaffirms our previous observation that each Roc is expressed in the embryo (Noureddine et al., 2002). Roc1b does not contain an intron and the primers amplify a band in the no-RT control, indicating the presence of some contaminating DNA. Both Roc1a and Roc1b are expressed in the testis, but Roc2 is not. Note the absence of Roc1b message in the Roc1bdc3 mutant. M indicates Marker and is the same in other panels. (C) Expression of FLAG-Roc1b, Roc1b, and Roc1a, but not Roc2, can rescue the male sterile phenotype. Roc transgenes expressed with the Roc1b promoter were crossed into the Roc1bdc3 (null, left) or Roc1bF28M (hypomorph, right) backgrounds and the percentage of viable progeny from individual males was determined. Three transgenic lines expressing FLAG-Roc1b (#23, #27, and #40) and four lines expressing native Roc1b (#1, #5, #8, and #10) were found to rescue the mutations. Two transgenic lines expressing Roc1a (#7 and #8) also rescue the mutations, but to a significantly lower degree (see Table 2). None of three Roc2 transgenic lines (#2, #6, and #11) showed any significant rescue. (D-F) RT-PCR of testes from Roc1bdc3 mutant males expressing a Roc1bgrf transgene. Three of the four individual Roc1b lines (D), both Roc1a lines (E), and all three Roc2 lines (F) used in the rescue assay were tested, and each indicates a similar level of expression.
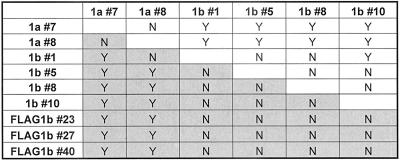
Table 2.
Roc1a does not fully rescue the male sterility of the Roc1b mutations
Pairwise comparisons of the ability of each Roc1bgrf (transgenic lines #1,5,8, and 10), Roc1bgrf::FLAG-Roc1b (transgenic lines #23, 27, and 40) and Roc1bgrf::Roc1a (transgenic lines #7 and 8) transgene to rescue the Roc1b– male sterile phenotype. Y indicates a significant difference (p < 0.02) in the ability of the indicated transgenes to rescue the Roc1bF28M hypomorphic (unshaded) or Roc1bdc3 null (shaded) mutations (see Materials and Methods). N indicates no significant difference. Note that for the most part, 1) the transgenic lines of a given construct are not significantly different from each other, and 2) the Roc1a transgenic lines are significantly different from the Roc1b lines.
Figure 4.
Roc2 cannot substitute for Roc1a. tk;2Expression Roc1agrf::FLAG-Roc2 (A) or UAS-Roc2 (B) in Roc1a mutant clones does not rescue either the proliferation or Ci hyperaccumulation defects caused by the Roc1a mutation.
Although the Roc1agrf::FLAG-Roc1a transgene we used was able to rescue the phenotypes of the mutant clones as well as viability of Roc1a mutant animals, we suspected that using the endogenous Roc1a promoter to express the Roc transgenes may provide at best only moderate levels of gene expression. This could potentially explain the inability of either Roc1b of Roc2 to fully rescue the Roc1a mutant clones, if these proteins shared some redundancy. Therefore, we tested whether higher levels of Roc1b or Roc2 expression could provide additional rescue of Roc1a mutant clones. For this we used the UAS-Gal4 system (Brand and Perrimon, 1993) to express each of the Roc genes, which when used in the MARCM system expresses at high levels and only in the cells that become mutant for Roc1a. Just as with Roc1agrf, expression of UAS-Roc1a was able to fully rescue both the small clone size and Ci hyperaccumulation phenotypes associated with the Roc1a mutation (Figure 2D). Expression of UAS-Roc1b was also able to rescue the Ci hyperaccumulation phenotype similarly to UAS-Roc1a (Figure 3B). However, even though the UAS-Roc1b-rescued clones were larger than Roc1a mutant clones, they were not quite as large as those obtained by rescue with either a Roc1a genomic fragment or UAS-Roc1a (Figure 3B). In addition, unlike the rescue observed with Roc1agrf::FLAG-Roc1b, the effect of UASRoc1b expression was fully penetrant; every disc analyzed showed some degree of rescue. In sharp contrast, expression of UAS-Roc2 was unable to rescue either of the Roc1a mutant phenotypes; we always observed small clones that always hyperaccumulated Ci (Figure 4B).
We observed one additional phenotype in Roc1a mutant clones expressing Roc1agrf::FLAG-Roc1b and (to a lesser extent) UAS-Roc1b. Occasionally, these clones were associated with small punctate regions of GFP expression (Figure 3, F and G). We attribute this to apoptosis of the Roc1a mutant cells, which as they died left behind fragments of cell membranes still expressing the CD8-tagged, membrane-anchored GFP. This was not observed in any other Roc1a mutant clones, perhaps because the clones were either rescued (as those expressing exogenous Roc1a) or were never able to get large enough before dying to notice the fragments of membranes (as those expressing Roc2 or no transgene).
Mutations in Roc1b Cause Male Sterility
To further our analysis of potential redundancies between Roc proteins, we generated mutations in Roc1b by using homologous recombination-mediated gene targeting (Rong and Golic, 2000). We obtained two mutant alleles of Roc1b. One allele is a deletion of the Roc1b coding region (Roc1bdc3) and is both a protein and genetic null. In the other allele, a frameshift/premature stop codon was introduced after the fifth codon of the Roc1b open reading frame (Roc1bF28M; see Materials and Methods).
Flies homozygous for either of the Roc1b mutant alleles are viable, indicating that Roc1b is not essential for Drosophila development. However, these mutations caused male-specific sterility. To examine this male sterility in detail, we calculated the percentage of viable progeny produced from 30 individual males and compared those values among wild-type and various mutant genotypes (see Materials and Methods; Figure 5A). Homozygous null Roc1bdc3 males were completely sterile; none of the eggs laid by the females of the cross hatched. In contrast, ∼2% of the eggs laid by females mated with homozygous Roc1bF28M males hatched. Additionally, about half of these males were completely sterile, whereas the fertility of the other half ranged from 0.3 to 12%. Although the frameshift allele would be expected to be a protein null, this partial sterility suggests that this allele is actually a hypomorph, perhaps due to reinitiation of translation at a downstream methionine. All males transheterozygous for both alleles (Roc1bdc3/Roc1bF28M), or for the frameshift allele over a deficiency that uncovers Roc1b (Roc1bF28M/Df(3L)emcE12), were also completely sterile, thus confirming the hypomorphic nature of the Roc1bF28M allele. The sterility induced by both Roc1b mutant alleles was rescued by a transgene containing the Roc1b genomic locus (Figure 5C), although the fertility of the rescued males was still somewhat lower than wild type (∼70% compared with 95%).
Squashed preparations of live testes from Roc1bdc3 homozygous males revealed the presence of sperm bundles that complete the individualization process normally (our unpublished data). However, these sperm cells were completely immotile. This is the most common of the male sterile phenotypes and suggests that the mitotic and meiotic cell divisions are normal and that there is a defect in some aspect of sperm differentiation or maturation. Consistent with the hypomorphic nature of the Roc1bF28M allele, homozygotes produce some live, motile sperm, but in quantities much less than heterozygous or wild-type males.
Roc1a but Not Roc2 Can Partially Rescue the Male Sterile Phenotype of Roc1b Mutations
The male sterile phenotype of Roc1b mutants indicates a unique role for Roc1b in spermatogenesis. The other two Roc proteins may not be able to compensate for loss of Roc1b because they may not be expressed in the correct time or place in the testes. To test this, we first performed RT-PCR of testes from wild-type and Roc1bdc3 mutant males. Both Roc1a- and Roc1b-specific primers were able to amplify bands of the correct size from wild-type testes, and as expected no Roc1b mRNA was detected in the Roc1bdc3 null mutant line (Figure 5B). Roc2 mRNA is detected in embryos, but not in the testes (Figure 5B). This was somewhat unexpected because Roc2 is expressed in all stages of Drosophila development and in many mammalian tissues, including the testis (Duan et al., 1999; Noureddine et al., 2002). This suggests that all necessary Cullin-mediated ubiquitylation reactions in the testes can be carried out using either Roc1a or Roc1b.
Next, we placed Roc1a and Roc2 ORFs under the control of the Roc1b promoter and 3′ untranslated region, and asked whether these transgenes could rescue the male sterility of the Roc1b mutants. Expression of Roc1a was able to rescue the male sterile phenotype of the Roc1b mutation to some degree (Figure 5C). For example, Roc1bF28M homozygous males expressing Roc1bgrf::Roc1a had hatching rates ranging from 2 to 85%, and none were completely sterile (compared with approximately half of Roc1bF28M homozygous males). However, the rescue of both Roc1b alleles with Roc1bgrf::Roc1a differed significantly (p < 0.02) from that observed with Roc1bgrf, indicating that Roc1a is not fully able to compensate for loss of Roc1b (Table 2).
In contrast, Roc2 was completely unable to rescue the Roc1b male sterile phenotype. Roc1bdc3 males expressing Roc1bgrf::Roc2 were completely sterile, and the hatching rates from crosses of Roc1bF28M males with or without the transgene were not significantly different, even at p < 0.001 (Figure 5C). Using RT-PCR, we verified that mRNA from each transgenic line was expressed (Figure 5, D-F), suggesting that the lack of complete rescue is due to biological differences of the Roc proteins.
The Drosophila Roc Proteins Preferentially Bind Different Members of the Cullin Family
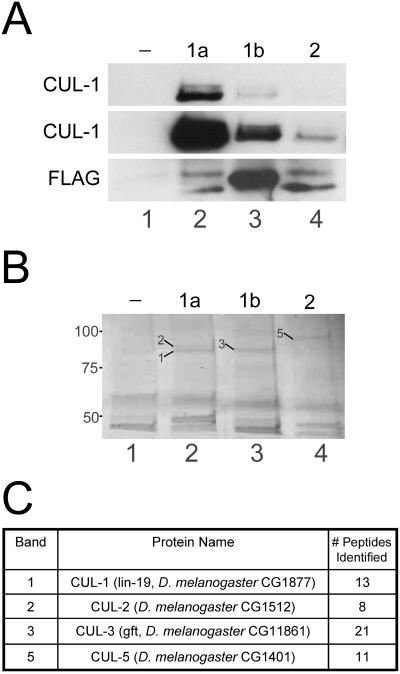
One possible explanation for the inability of a given Roc protein to rescue the phenotype of a different Roc mutant is that each Roc protein may form a unique set of E3 ubiquitin ligase complexes by preferentially interacting with different Cullin family members. To test this, we performed coimmunoprecipitation experiments with our Roc1agrf::FLAG-Roc transgenes. Lysates from control, nontransgenic (w1118) embryos or embryos expressing each of the FLAG-Roc transgenes were incubated with anti-FLAG-agarose and immunocomplexes were analyzed by Western blotting or mass spectrometry. Western analysis with a CUL-1 antibody showed that CUL-1 efficiently coprecipitates with FLAG-Roc1a (Figure 6A). Relatively little, but still above-background, amounts of CUL-1 was present in immunocomplexes from FLAG-Roc1b or FLAG-Roc2 lysates (Figure 6A). This result shows that whereas Roc1a, Roc1b, and Roc2 are each able to bind to CUL-1 when expressed from the Roc1a promoter, Roc1a does so much more efficiently. We also analyzed immunocomplexes from each of the FLAG-Roc transgenic lysates by mass spectrometry. Proteins from a Coomassie-stained polyacrylamide gel that migrated with the predicted molecular weight of the Cullins and that were present in one or more of the transgenic lines but absent from wild-type, nontransgenic lysate (Figure 6B) were excised and identified by tandem mass spectrometry. Using this approach, we identified CUL-1 and CUL-2 in Roc1a immunocomplexes, CUL-3 in Roc1b immunocomplexes, and CUL-5 in Roc2 immunocomplexes (Figure 6C). Because weaker Cullin-Roc interactions may not permit the precipitation of enough Cullin protein to be visible on a Coomassie-stained gel, this technique does not rule out any particular Cullin-Roc interactions. However, the data do suggest that there is a preference for the formation of certain Cullin-Roc complexes.
Figure 6.
The three Drosophila Roc proteins preferentially bind different Cullins. (A) Lysates from embryos expressing no transgene (-; lane 1) or one of the Roc1agrf::FLAG-Roc proteins (1a, 1b, or 2; lanes 2-4) were incubated with anti-FLAG agarose and immunocomplexes were analyzed by Western blot with anti-CUL-1. Top, short exposure showing that Roc1a efficiently immunoprecipitates both neddylated and unmodified CUL-1. Middle, longer exposure of the same blot showing that Roc1b and Roc2 can also immunopurify CUL-1, although much less efficiently than Roc1a. Bottom, blot probed with anti-FLAG to demonstrate immunoprecipitation of the FLAG-Roc proteins. The slower migrating bands in lanes 2 and 4 correspond to unidentified proteins, possibly read-through translation products. (B and C) Immunocomplexes from the embryo lysates of the same genotypes as in A were analyzed by mass spectrometry. (B) Coomassie-stained gel showing that each Roc protein immunoprecipitates a unique set of proteins in the size range expected for the Cullins. Bands indicated with arrows were excised and analyzed by mass spectrometry (the corresponding Cullin protein is identified by the number). (C) Protein identification of the bands excised from the gel in B. CUL-1 and CUL-2 were each identified FLAG-Roc1a immunocomplexes, CUL-3 was identified in FLAG-Roc1b immunocomplexes, and CUL-5 was identified in FLAG-Roc2 immunocomplexes. The number of peptides matching a theoretical digest of the corresponding protein is indicated.
DISCUSSION
Drosophila Roc Proteins Have Distinct Functions
Our results indicate that there are significant differences in the biological roles of the three Drosophila Roc proteins and that these differences are not simply the result of distinct expression patterns during development. In all of our experimental paradigms, Roc1a and Roc1b could partially, but not completely, substitute for one another, whereas Roc2 showed no ability to substitute for either Roc1 paralogue. Results of coimmunoprecipitation experiments suggest that these differences are due to preferential interactions between Roc and Cullin family members. For example, CUL-1 seems to interact most strongly with Roc1a, suggesting that a majority of SCF (i.e., CUL-1) targets require Roc1a. However, we cannot rule out that Roc1b or Roc2 function within the context of an SCF complex, as both showed weak interactions with CUL-1. Indeed, Roc1b seems to be capable of participating in SCF-mediated ubiquitylation, because it was able to rescue the aberrant accumulation of Ci, a bona fide SCF target, when overexpressed.
Because the Drosophila Roc proteins share between 40 and 60% overall sequence identity, it is somewhat surprising that we did not observe a higher degree of complementation in our rescue assays. Most of the conservation is within the C-terminal 67 residues, which contains the catalytic RING domain. Roc1a and Roc1b share 76% identity and 88% similarity in this domain, whereas Roc1a and Roc2 are 45% identical and 59% similar. In the N-terminal regions, the sequence identity/similarity is lower (38%/50% between Roc1a and Roc1b; 41%/57% between Roc1a and Roc2). It was found previously that deletion of the Rbx1 (Roc1) N-terminus prevents interaction with CUL-1 in 293T cells (Furukawa et al., 2002). The crystal structure of the SCF complex (Zheng et al., 2002) shows that the association between Rbx1 and the C-terminal portion of the CUL-1 protein (termed the Cullin homology domain or CHD) consists of two parts. First, the RING domain of Rbx1 packs into a V-shaped groove formed by the α/B and WH-B domains of CUL-1. Second, the Rbx1 N terminus threads into CUL-1 and makes a five-stranded intermolecular β-sheet (four strands provided by CUL-1 and one by Rbx1). This intermolecular β-sheet seems to provide the primary mechanism of Rbx1 recruitment. Together, these data implicate the N terminus of the Drosophila Roc proteins as the region responsible for mediating the differential binding to Cullins.
Additional factors may also contribute to the differences in the biological roles of Roc proteins. Specific interactions between E2s and RING domains have been observed, suggesting that the identity of the Roc protein can influence which E2 gets recruited to the complex. Roc1-containing immunoprecipitates from Cullin-transfected cells possess ubiquitin ligase activity with both UbcH5 and Ubc3/Cdc34; however, similar complexes containing Roc2 are only active with UbcH5 (Furukawa et al., 2002). The RING proteins RAD5 and RAD18 form a macromolecular complex with the E2s RAD6 and Ubc13-MMS2 that affects postreplicative DNA damage. RAD5 is able to directly interact with Ubc13-MMS2, but not with RAD6, and the converse is true for RAD18 (Ulrich and Jentsch, 2000). Protein mapping studies have identified specific residues important for the interaction between Ubc13-MMS2 and the RING domain of RAD5 (Ulrich, 2003). Finally, although the RING domain protein BARD1 interacts with UbcH5, the BARD1 RING domain is not required for this and the interaction is instead mediated by the RING domain protein BRCA1 (Brzovic et al., 2003). Thus, one possible model is that Roc1a, Roc1b, and Roc2 recruit a unique (set of) E2(s) that each act on a different set of targets, thereby providing an additional level of functional specificity in vivo. Other issues such as protein half-life or subcellular localization may also be important contributing factors to the biological differences among the three Drosophila Roc proteins.
The Roc1b Mutant Phenotype
We were able to use the two step method of homologous recombination mediated gene targeting to obtain two mutant alleles of Roc1b. Our results confirm the validity of the technique for use in generating specific mutations in a gene of interest. Furthermore, our results are consistent with previous work (Rong et al., 2002) stating that a minimum distance of 400 base pairs between the I-SceI cut site and the mutation to be introduced is sufficient to prevent the mutation from being lost due to gap enlargement. Our mutation was ∼520 base pairs away from the I-SceI site and all three targeting events that we analyzed molecularly retained the mutation. Our results also demonstrate that a simple deletion of the start codon or the introduction of a premature Stop codon early in the reading frame may not be sufficient to generate a null mutation due to the possibility of translation initiation at downstream AUG codons. Although we did not definitively determine what causes the Roc1bF28M mutation to be hypomorphic, we suspect that this is the result of a low frequency of translation reinitiation that leads to a reduction in the total Roc1b protein levels and/or the production of an N-terminally truncated protein with reduced activity.
We do not know the precise cause of the sterility of Roc1b mutant males. Given the role that SCF complexes play in regulation of the cell cycle, it is attractive to speculate that the Roc1b mutation may disrupt some aspect of the mitotic or meiotic divisions of the germ cells. However, this seems unlikely given the fact that the null mutation is completely sterile. Aberrant mitotic or meiotic divisions generally only reduce fertility and result in other cytological defects not seen in the Roc1b mutant. Furthermore, the only discernable phenotype of the Roc1b mutation is the production of elongated individualized, but nonmotile sperm, suggesting that the mitotic and meiotic divisions are normal.
The fact that the Roc1b mutation results in a male sterile phenotype is somewhat surprising given the observation that Roc1a is also expressed in adult (Figure 5B) and larval (Noureddine and Duronio, unpublished data) testes. However, Roc1a and Roc1b may be expressed in different cell types in the testes (e.g., soma vs. germline). This idea is supported by the observation that Roc1a partially rescues the male sterile phenotype when expressed with the Roc1b promoter. However, the difference in expression pattern may not be the only difference between the two proteins, because the phenotypic rescue observed with Roc1a was less than that with Roc1b.
Ubiquitylation is believed to play a variety of roles in spermatogenesis. In addition to the proposed role in the regulation of the meiotic cell cycle, ubiquitylation may affect aspects such as synaptonemal complex formation or the chromatin remodeling and reorganization that occurs during nuclear condensation and elongation (Roest et al., 1996; Grootegoed et al., 1998; Baarends et al., 2003). Furthermore, many components of the ubiquitin system show high and/or unique expression patterns in the testes. For example, the UbcD1 gene of Drosophila, which is involved in maintaining telomere structure, encodes three transcripts, one of which is expressed solely in the male germline (Cenci et al., 1997). Mutations in this gene disrupt male meiosis, leading to infertility (Cenci et al., 1997). Another Drosophila E2, Ubc7, also encodes a male specific transcript and plays a role in spermatogenesis, although mutations are pleiotropic and affect other processes such as courtship behavior and neural development/function (Orgad et al., 2000).
At the molecular level, the Roc1b mutant male sterile phenotype is likely the result of a specific target protein of a Cullin-Roc1b E3 ligase that fails to be ubiquitylated. Presumably, this ubiquitylation occurs in the context of an SCF-like complex for several reasons. First, Roc1a has been shown to associate with SCF components (Bocca et al., 2001) and is able to partially rescue the Roc1b mutation. Second, we observed enhancement of the Roc1bF28M induced sterility by halving the gene dose of either CUL-1 or CUL-3 (Donaldson and Duronio, unpublished results), suggesting that both of these Cullins have specific targets within the testes. This is consistent with the observation from the coimmunoprecipitation experiments that Roc1b and CUL-3 interact strongly and that Roc1b can interact with CUL-1 in vivo. Interestingly, mutations in the Slimb homologue β-TRCP1 result in male infertility in the mouse (Guardavaccaro et al., 2003). However, unlike the Roc1b mutation, the lack of β-TRCP1 disrupts the meiotic divisions, and mutant testes contain spermatocytes that arrest in metaphase I, reducing the number of postmeiotic spermatids (Guardavaccaro et al., 2003). This, however, does not exclude the possibility that Roc1b and Slimb might be part of the same complex, because Slimb may also have additional functions later in spermatogenesis. Along these lines, mutations in both Slimb and SkpA (the Drosophila Skp1 homologue) have been associated with centrosome overduplication in other tissues (Wojcik et al., 2000; Murphy, 2003) and mutations in centrosomin result in defects in cytokinesis, karyokinesis, and growth of the axoneme during spermatogenesis (Megraw et al., 1999). Thus, it is possible that an SCFSlimb complex with Roc1a has a function early in sperm development, and an SCFSlimb complex with Roc1b regulates later stages.
The Roc Subunit of Cullin-dependent E3 Ligases
In this study, we have used the powerful genetic techniques of the fruit fly to assess how the RING domain subunit contributes to the function of Cullin-dependent ubiquitin ligases. We have found that the Drosophila Roc proteins have nonredundant roles during development and that these differences may be mediated by the formation of specific Cullin-Roc ligase complexes. Our results are consistent with studies of mammalian Roc proteins showing that although both Rbx1 and mammalian Roc2 can associate with all Cullin proteins, these interactions, as well as the associated ligase activities of the different complexes, seem to show certain preferences (Furukawa et al., 2002). Because each Cullin family member may use a distinct mechanism to target nonoverlapping sets of proteins for ubiquitylation (Kamura et al., 2001; Nag et al., 2001; Groisman et al., 2003; Yu et al., 2003; van den Heuvel, 2004; Wertz et al., 2004), preferential Cullin binding provides a sufficient, if not the only, explanation for the functional differences among the three Drosophila Roc proteins. Further experiments are needed identify which complexes exist in vivo and to determine exactly what mediates these specific Cullin-Roc interactions.
Acknowledgments
We thank Kent Golic, Jin Jiang, Jeff Sekelsky, Bob Holmgren, and the Bloomington Stock Center for fly stocks and other reagents; John Tomkiel for help analyzing testis phenotypes; Mark Hall, Carol Parker, Christoph Borchers, and the Michael Hooker Proteomics Core Facility at University of North Carolina-Chapel Hill for the mass spectrometric analysis; and Melissa Adams and members of the Duronio laboratory for helpful discussion. This work was supported by a grant from the von Hipple-Lindau Family Alliance and a National Institutes of Health postdoctoral training grant (CA-09156) to T.D.D. and National Institutes of Health grant GM-57859 and the Cancer Research Fund of the Damon-Runyon-Walter Winchell Foundation to R.J.D.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-03-0180. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-03-0180.
References
- Baarends, W.M., Wassenaar, E., Hoogerbrugge, J.W., van Cappellen, G., Roest, H.P., Vreeburg, J., Ooms, M., Hoeijmakers, J.H., and Grootegoed, J.A. (2003). Loss of HR6B ubiquitin-conjugating activity results in damaged synaptonemal complex structure and increased crossing-over frequency during the male meiotic prophase. Mol. Cell. Biol. 23, 1151-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J.W., and Elledge, S.J. (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263-274. [DOI] [PubMed] [Google Scholar]
- Ben-Neriah, Y. (2002). Regulatory functions of ubiquitination in the immune system. Nat. Immunol. 3, 20-26. [DOI] [PubMed] [Google Scholar]
- Bocca, S.N., Muzzopappa, M., Silberstein, S., and Wappner, P. (2001). Occurrence of a putative SCF ubiquitin ligase complex in Drosophila. Biochem. Biophys. Res. Commun. 286, 357-364. [DOI] [PubMed] [Google Scholar]
- Brand, A.H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415. [DOI] [PubMed] [Google Scholar]
- Brzovic, P.S., Keeffe, J.R., Nishikawa, H., Miyamoto, K., Fox, D., 3rd, Fukuda, M., Ohta, T., and Klevit, R. (2003). Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl. Acad. Sci. USA 100, 5646-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci, G., Rawson, R.B., Belloni, G., Castrillon, D.H., Tudor, M., Petrucci, R., Goldberg, M.L., Wasserman, S.A., and Gatti, M. (1997). UbcD1, a Drosophila ubiquitin-conjugating enzyme required for proper telomere behavior. Genes Dev. 11, 863-875. [DOI] [PubMed] [Google Scholar]
- Ciechanover, A., Orian, A., and Schwartz, A.L. (2000). The ubiquitin-mediated proteolytic pathway: Mode of action and clinical implications. J. Cell. Biochem. 77, 40-51. [DOI] [PubMed] [Google Scholar]
- Conaway, R.C., Brower, C.S., and Conaway, J.W. (2002). Emerging roles of ubiquitin in transcription regulation. Science 296, 1254-1258. [DOI] [PubMed] [Google Scholar]
- Craig, K.L., and Tyers, M. (1999). The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog. Biophys. Mol. Biol. 72, 299-328. [DOI] [PubMed] [Google Scholar]
- DeSalle, L.M., and Pagano, M. (2001). Regulation of the G1 to S transition by the ubiquitin pathway. FEBS Lett. 490, 179-189. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435-467. [DOI] [PubMed] [Google Scholar]
- Duan, H., Wang, Y., Aviram, M., Swaroop, M., Loo, J.A., Bian, J., Tian, Y., Mueller, T., Bisgaier, C.L., and Sun, Y. (1999). SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol. Cell. Biol. 19, 3145-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, R.M., Correll, C.C., Kaplan, K.B., and Deshaies, R.J. (1997). A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91, 221-230. [DOI] [PubMed] [Google Scholar]
- Furukawa, M., He, Y.J., Borchers, C., and Xiong, Y. (2003). Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 5, 1001-1007. [DOI] [PubMed] [Google Scholar]
- Furukawa, M., Ohta, T., and Xiong, Y. (2002). Activation of UBC5 ubiquitinconjugating enzyme by the RING finger of ROC1 and assembly of active ubiquitin ligases by all cullins. J. Biol. Chem. 277, 15758-15765. [DOI] [PubMed] [Google Scholar]
- Furukawa, M., Zhang, Y., McCarville, J., Ohta, T., and Xiong, Y. (2000). The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 20, 8185-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer, R., Wee, S., Anderson, S., Yates, J., and Wolf, D.A. (2003). BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 12, 783-790. [DOI] [PubMed] [Google Scholar]
- Grima, B., Lamouroux, A., Chelot, E., Papin, C., Limbourg-Bouchon, B., and Rouyer, F. (2002). The F-box protein slimb controls the levels of clock proteins period and timeless. Nature 420, 178-182. [DOI] [PubMed] [Google Scholar]
- Groisman, R., Polanowska, J., Kuraoka, I., Sawada, J., Saijo, M., Drapkin, R., Kisselev, A.F., Tanaka, K., and Nakatani, Y. (2003). The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113, 357-367. [DOI] [PubMed] [Google Scholar]
- Grootegoed, J.A., Baarends, W.M., Roest, H.P., and Hoeijmakers, J.H. (1998). Knockout mouse model and gametogenic failure. Mol. Cell. Endocrinol. 145, 161-166. [DOI] [PubMed] [Google Scholar]
- Guardavaccaro, D., Kudo, Y., Boulaire, J., Barchi, M., Busino, L., Donzelli, M., Margottin-Goguet, F., Jackson, P.K., Yamasaki, L., and Pagano, M. (2003). Control of meiotic and mitotic progression by the F box protein beta-Trcp1 in vivo. Dev. Cell 4, 799-812. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1999). Using Antibodies: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425-479. [DOI] [PubMed] [Google Scholar]
- Higa, L.A., Mihaylov, I.S., Banks, D.P., Zheng, J., and Zhang, H. (2003). Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5, 1008-1015. [DOI] [PubMed] [Google Scholar]
- Iwai, K., Yamanaka, K., Kamura, T., Minato, N., Conaway, R.C., Conaway, J.W., Klausner, R.D., and Pause, A. (1999). Identification of the von Hippellindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. USA 96, 12436-12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, P.K., and Eldridge, A.G. (2002). The SCF ubiquitin ligase: an extended look. Mol. Cell 9, 923-925. [DOI] [PubMed] [Google Scholar]
- Jackson, P.K., Eldridge, A.G., Freed, E., Furstenthal, L., Hsu, J.Y., Kaiser, B.K., and Reimann, J.D. (2000). The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10, 429-439. [DOI] [PubMed] [Google Scholar]
- Jiang, J., and Struhl, G. (1998). Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature 391, 493-496. [DOI] [PubMed] [Google Scholar]
- Kaiser, P., Flick, K., Wittenberg, C., and Reed, S.I. (2000). Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell 102, 303-314. [DOI] [PubMed] [Google Scholar]
- Kamura, T., Burian, D., Yan, Q., Schmidt, S.L., Lane, W.S., Querido, E., Branton, P.E., Shilatifard, A., Conaway, R.C., and Conaway, J.W. (2001). Muf1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J. Biol. Chem. 276, 29748-29753. [DOI] [PubMed] [Google Scholar]
- Kamura, T., et al. (1999). Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284, 657-661. [DOI] [PubMed] [Google Scholar]
- Kim, W., and Kaelin, W.G., Jr. (2003). The von Hippel-Lindau tumor suppressor protein: new insights into oxygen sensing and cancer. Curr. Opin. Genet. Dev. 13, 55-60. [DOI] [PubMed] [Google Scholar]
- Kipreos, E.T., Lander, L.E., Wing, J.P., He, W.W., and Hedgecock, E.M. (1996). cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 85, 829-839. [DOI] [PubMed] [Google Scholar]
- Kipreos, E.T., and Pagano, M. (2000). The F-box protein family. Genome Biol. 1, REVIEWS3002. [DOI] [PMC free article] [PubMed]
- Ko, H.W., Jiang, J., and Edery, I. (2002). Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 420, 673-678. [DOI] [PubMed] [Google Scholar]
- Koepp, D.M., Harper, J.W., and Elledge, S.J. (1999). How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell 97, 431-434. [DOI] [PubMed] [Google Scholar]
- Kusano, K., Johnson-Schlitz, D.M., and Engels, W.R. (2001). Sterility of Drosophila with mutations in the Bloom syndrome gene-complementation by Ku70. Science 291, 2600-2602. [DOI] [PubMed] [Google Scholar]
- Lee, T., and Luo, L. (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451-461. [DOI] [PubMed] [Google Scholar]
- Maniatis, T. (1999). A ubiquitin ligase complex essential for the NF-kappaB, Wnt/Wingless, and Hedgehog signaling pathways. Genes Dev. 13, 505-510. [DOI] [PubMed] [Google Scholar]
- Megraw, T.L., Li, K., Kao, L.R., and Kaufman, T.C. (1999). The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126, 2829-2839. [DOI] [PubMed] [Google Scholar]
- Moberg, K.H., Bell, D.W., Wahrer, D.C., Haber, D.A., and Hariharan, I.K. (2001). Archipelago regulates cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature 413, 311-316. [DOI] [PubMed] [Google Scholar]
- Moberg, K.H., Mukherjee, A., Veraksa, A., Artavanis-Tsakonas, S., and Hariharan, I.K. (2004). The Drosophila F box protein archipelago regulates dMyc protein levels in vivo. Curr. Biol. 14, 965-974. [DOI] [PubMed] [Google Scholar]
- Murphy, T.D. (2003). Drosophila skpA, a component of SCF ubiquitin ligases, regulates centrosome duplication independently of cyclin E accumulation. J. Cell Sci. 116, 2321-2332. [DOI] [PubMed] [Google Scholar]
- Nag, A., Bondar, T., Shiv, S., and Raychaudhuri, P. (2001). The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol. Cell. Biol. 21, 6738-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nateri, A.S., Riera-Sans, L., Da Costa, C., and Behrens, A. (2004). The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303, 1374-1378. [DOI] [PubMed] [Google Scholar]
- Noureddine, M.A., Donaldson, T.D., Thacker, S.A., and Duronio, R.J. (2002). Drosophila Roc1a encodes a RING-H2 protein with a unique function in processing the Hh signal transducer Ci by the SCF E3 ubiquitin ligase. Dev. Cell 2, 757-770. [DOI] [PubMed] [Google Scholar]
- Ohta, T., Michel, J.J., Schottelius, A.J., and Xiong, Y. (1999). ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3, 535-541. [DOI] [PubMed] [Google Scholar]
- Orgad, S., Rosenfeld, G., Greenspan, R.J., and Segal, D. (2000). courtless, the Drosophila UBC7 homolog, is involved in male courtship behavior and spermatogenesis. Genetics 155, 1267-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, C.Y., Lin, Y.F., Chen, Y.J., and Chien, C.T. (2002). Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 16, 2403-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, E.E., Willems, A.R., Sa, D., Kuras, L., Thomas, D., Craig, K.L., and Tyers, M. (1998). Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box proteincomplexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 12, 692-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart, C.M. (2000). Ubiquitin in chains. Trends Biochem. Sci. 25, 544-548. [DOI] [PubMed] [Google Scholar]
- Pickart, C.M. (2001). Ubiquitin enters the new millennium. Mol. Cell 8, 499-504. [DOI] [PubMed] [Google Scholar]
- Pintard, L., et al. (2003). The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425, 311-316. [DOI] [PubMed] [Google Scholar]
- Raiborg, C., Rusten, T.E., and Stenmark, H. (2003). Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15, 446-455. [DOI] [PubMed] [Google Scholar]
- Roest, H.P., et al. (1996). Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell 86, 799-810. [DOI] [PubMed] [Google Scholar]
- Rong, Y.S., and Golic, K.G. (2000). Gene targeting by homologous recombination in Drosophila. Science 288, 2013-2018. [DOI] [PubMed] [Google Scholar]
- Rong, Y.S., Titen, S.W., Xie, H.B., Golic, M.M., Bastiani, M., Bandyopadhyay, P., Olivera, B.M., Brodsky, M., Rubin, G.M., and Golic, K.G. (2002). Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16, 1568-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol, J.H., et al. (1999). Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 13, 1614-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra, D., Craig, K.L., Tyers, M., Elledge, S.J., and Harper, J.W. (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209-219. [DOI] [PubMed] [Google Scholar]
- Spencer, E., Jiang, J., and Chen, Z.J. (1999). Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes Dev. 13, 284-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers, M., and Jorgensen, P. (2000). Proteolysis and the cell cycle: with this RING I do thee destroy. Curr. Opin. Genet. Dev. 10, 54-64. [DOI] [PubMed] [Google Scholar]
- Ulrich, H.D. (2003). Protein-protein interactions within an E2-RING finger complex. Implications for ubiquitin-dependent DNA damage repair. J. Biol. Chem. 278, 7051-7058. [DOI] [PubMed] [Google Scholar]
- Ulrich, H.D., and Jentsch, S. (2000). Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19, 3388-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel, S. (2004). Protein degradation: CUL-3 and BTB - partners in proteolysis. Curr. Biol. 14, R59-R61. [PubMed] [Google Scholar]
- Wertz, I.E., ÓRourke, K.M., Zhang, Z., Dornan, D., Arnott, D., Deshaies, R.J., and Dixit, V.M. (2004). Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303, 1371-1374. [DOI] [PubMed] [Google Scholar]
- Wojcik, C. (2002). Regulation of apoptosis by the ubiquitin and proteasome pathway. J. Cell Mol. Med. 6, 25-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik, E.J., Glover, D.M., and Hays, T.S. (2000). The SCF ubiquitin ligase protein slimb regulates centrosome duplication in Drosophila. Curr. Biol. 10, 1131-1134. [DOI] [PubMed] [Google Scholar]
- Xu, L., Wei, Y., Reboul, J., Vaglio, P., Shin, T.H., Vidal, M., Elledge, S.J., and Harper, J.W. (2003). BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425, 316-321. [DOI] [PubMed] [Google Scholar]
- Yu, X., Yu, Y., Liu, B., Luo, K., Kong, W., Mao, P., and Yu, X.F. (2003). Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302, 1056-1060. [DOI] [PubMed] [Google Scholar]
- Zheng, N., et al. (2002). Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703-709. [DOI] [PubMed] [Google Scholar]
- Zhong, W., Feng, H., Santiago, F.E., and Kipreos, E.T. (2003). CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423, 885-889. [DOI] [PubMed] [Google Scholar]