Abstract
Satellite cells are stem cells with an essential role in skeletal muscle repair. Precise regulation of gene expression is critical for proper satellite cell quiescence, proliferation, differentiation and self -renewal. Nuclear proteins required for gene expression are dependent on the nucleocytoplasmic transport machinery to access to nucleus, however little is known about regulation of nuclear transport in satellite cells. The best characterized nuclear import pathway is classical nuclear import which depends on a classical nuclear localization signal (cNLS) in a cargo protein and the heterodimeric import receptors, karyopherin alpha (KPNA) and beta (KPNB). Multiple KPNA1 paralogs exist and can differ in importing specific cNLS proteins required for cell differentiation and function. We show that transcripts for six Kpna paralogs underwent distinct changes in mouse satellite cells during muscle regeneration accompanied by changes in cNLS proteins in nuclei. Depletion of KPNA1, the most dramatically altered KPNA, caused satellite cells in uninjured muscle to prematurely activate, proliferate and undergo apoptosis leading to satellite cell exhaustion with age. Increased proliferation of satellite cells led to enhanced muscle regeneration at early stages of regeneration. In addition, we observed impaired nuclear localization of two key KPNA1 cargo proteins: p27, a cyclin-dependent kinase inhibitor associated with cell cycle control and lymphoid enhancer factor 1, a critical co-transcription factor for β-catenin. These results indicate that regulated nuclear import of proteins by KPNA1 is critical for satellite cell proliferation and survival and establish classical nuclear import as a novel regulatory mechanism for controlling satellite cell fate.
Keywords: nuclear import, importin alpha 5, satellite cells, cNLS proteins, karyopherin alpha 1, KPNA1
Introduction
Proper repair of muscle after injury is dependent on activation and repression of numerous genes in satellite cells, which are tissue-specific stem cells. Under basal conditions, satellite cells are mitotically quiescent [1], but in response to injury, satellite cells become activated to proliferate and eventually differentiate and fuse with one another or with myofibers to restore normal tissue architecture [2], [3]. Large changes in gene expression occur when satellite cells transition from quiescence to activation with over 5,000 genes being differentially expressed [4]. More than half of these genes, such as cell cycle inhibitory or RNA modifying genes, are decreased with satellite cell activation, whereas 44% of these genes including cell cycle progression genes, are increased during satellite cell activation. Gene expression is dependent upon proteins such as transcription factors and chromatin remodeling factors that are synthesized in the cytoplasm but act in the nucleus. The subcellular localization of such nuclear proteins must be tightly controlled because altered nuclear import or export could result in aberrant gene expression compromising satellite cell function. Little is known about how key nuclear regulatory proteins gain access to nuclei in satellite cells.
In all cells, transport between the nucleus and the cytoplasm is mediated by nuclear pore complexes (NPC) embedded in the nuclear envelope. NPC are multiprotein superstructures (~ 120 MDa) which provide channels for the nucleocytoplasmic exchange of ions and macromolecules [5, 6]. While smaller ions and molecules can diffuse through the NPC, molecules larger than ~40 kDa re quire nuclear transport receptors that recognize cargo proteins and mediate transport through these nuclear pores [7-9]. Classical nuclear import is the best characterized of the nuclear import pathways. In mouse, 35-55% of nuclear proteins may depend on classical nuclear import for nuclear targeting as determined using a bioinformatics approach [10]. Classical nuclear import depends on a classical nuclear localization signal (cNLS) in a cargo protein that is bound by a heterodimeric receptor consisting of a classical nuclear import receptor karyopherin alpha (KPNA) and a pore targeting subunit karyopherin beta1 (KPNB1) [11]. Once in the nucleus, the cNLS protein is released and both KPNA and KPNB1 are recycled separately back to the cytoplasm for another round of import [12, 13]. In humans, a single KPNB1 can function with any of seven KPNA paralogs: KPNA1, KPNA2, KPNA3, KPNA4, KPNA5, KPNA6 and KPNA7 [14-17]. Six KPNA paralogs exist in mouse with which the corresponding human homologues share 80-90% amino acid identity. Each KPNA paralog likely transports a large num ber of cargo proteins [18, 19]. Furthermore, each KPNA paralog has differential affinity for a subset of cargo proteins [15] with the binding affinity and concentration of the specific KPNA being determinants of the rate of protein transport [20-22]. While all KPNA paralogs play a role in classical nuclear import, their roles can differ in importing specific cNLS proteins that are required for cell differentiation and function [23-25].
The purpose of our study was to gain insights into the classic nuclear import pathway in satellite cells and how perturbations in this system affect satellite cell biology. Here, we show distinct expression of cNLS proteins and Kpna paralogs during satellite cell activation in response to muscle injury in mice. Depletion of KPNA1 (also known as importin alpha 5 in human) led not only to increased satellite cell proliferation and muscle regeneration but also to satellite cell apoptosis in both uninjured and injured muscles. Impaired nuclear localization of key proteins associated with cell cycle control and Wnt signaling was noted in KPNA1 knock out (KO) satellite cells. Taken together, our data suggest that the cNLS proteins imported by KPNA1 into the nucleus are critical for maintaining satellite cells quiescence and promoting their survival.
Material and methods
Mice
C57BL/6 mice were purchased from Charles River Laboratories. WT and KPNA1 KO mice were produced by Kpna1+/− breeding pairs [26]. Mice were genotyped using the following primers: for W T allele: F: 5'–TAGCTTGTTTTGGGCTGGTG-3', R: 5'–GGGTAAAGTTCTAGGCTAGC-3' and for Kpna1 KO allele; F: 5'–CTCGAGGCTAGAACTAGTGG-3', R: 5'–AGGCGATTAAGTTGGGTAACG-3'. Male and female mice from 12 to 16 weeks old were used for all experiments unless described otherwise. Experiments involving animals were performed in accordance with approved guidelines and ethical approval from Emory University's Institutional Animal Care and Use Committee.
Muscle injury
Mice were anesthetized with intraperitoneal injection of 80 mg/kg ketamine/5 mg/kg xylazine. Injury was induced in gastrocnemius (GA) or tibialis anterior (TA) muscles of anesthetized mice by injection of 1.2% BaCl2 using either 40 μl for GA or 25 μl for TA. For analgesia, mice were injected subcutaneously with 0.1mg/kg buprenorphine before and after muscle injury. Muscles were collected 2-90 days after injury depending on the experiment.
Satellite cell isolation and Fluorescence Activated Cell Sorting (FACS)
Satellite cells were isolated as previously described [27] from hindlimb muscles. Cells were labeled using the following antibodies: 1:400 CD31-PE (clone 390; eBiosciences), 1:400 CD45-PE (clone 30-F11; BD Pharmingen), 1:4000 Sca-1-PE-Cy7 (clone D7; BD Pharmingen), 1:500 α7-integrin-APC (Clone R2F2; AbLab). Dead cells were excluded by propidium iodide (PI) staining. Cell sorting was performed using a BD FACSAria II cell sorter (Becton-Dickinson). Sorted cells were immediately reanalyzed to ensure purity. Analyses of flow cytometry data were performed using FACSDiva (BD version 8.0.1).
Nuclear enrichment
Nuclei were enriched from satellite cells by fractionation over a sucrose cushion [28]. The cell pellet was homogenized with a dounce homogenizer in lysis buffer (0.32 M sucrose, 5 mM CaCl2, 3 mM MgCl2, 0.1 mM EDTA, 10 mM Tris-HCl, pH 8.0, 0.1% Triton X-100, 1 mM DTT, protease inhibitor cocktail (Roche)). This homogenate was layered over a 1.8 M sucrose cushion (1.8 M sucrose, 3 mM MgCl2, 10 mM Tris-HCl, pH 8.0) and centrifuged at 222,000×G for 120 min at 4°C in a Beckman SWTi 41 rotor. The pelleted nuclear fraction was resuspended in phosphate buffered saline (PBS) containing protease inhibitor cocktail (Roche) and pelleted at 1000×G for 2 minutes.
Proteomics
The prepared protein samples were resuspended in 8M Urea, 10mM Tris, 100mM Na2PO4 and protein content was determined by BCA assay. Equal protein from each sample was then subjected to in-solution trypsin digest [29] and high pressure liquid chromatography resolution by a NanoAcquity UPLC system (Waters Corporation) [30] according to established protocols. Peptides were ionized with 2.0 kV electrospray ionization voltage from a nano-ESI source (Thermo Scientific) on a hybrid LTQ XL Orbitrap mass spectrometer (Thermo Scientific). MS spectra and MS/MS spectra were obtained in the LTQ following collision induced dissociation (collision energy 3 5%, activation Q 0.25, activation time 30 ms) for the top 10 precursor ions with charge z ≥ 2. The MS/MS spectra were matched to the complete semi-tryptic mouse proteome (refseq version 62) using the SageN Sorcerer SEQUEST 3.5 algorithm [31, 32] with a 20 ppm mass accuracy threshold. The normalized score and search dependent score for b and y ions were dynamically increased to remove false positive hits [33].
Peptide/protein quantification was performed based on the extracted ion current (XIC) measurements of identified peptides. Peptide-specific ion current intensities were extracted and compared across cases using DQuan [34, 35].
Cell proliferation and apoptosis assays by flow cytometry
To analyze satellite cell proliferation, 5-Bromo-2’-deoxyuridine (BrdU) (100mg/g of body weight; Sigma) was injected intraperitoneally twice a day for 2 days. Muscles were dissected and digested as described above. Isolated mononucleated cells were labeled with CD31-PE, CD45-PE, Sca-1-PE-Cy7 and α7-integrin-APC. Subsequently, cells were immunostained for BrdU using an FITC-BrdU flow kit in accordance with the manufacturer's instructions (BD Pharmingen).
To analyze hematopoietic stem cell proliferation, bone marrow cells were collected as previously described [36] and labeled with 1:400 FITC-c-kit (clone 2B8; Biolegend), 1:200 PE-lineage (Lin) antibodies (CD3/Gr-1/CD11b/CD45R/Ter-119; Biolegend), 1:200 Sca-1-PE-Cy7, and 1:200 AF700-CD34 (BD Pharmigen). Subsequently, cells were immunostained for BrdU as described for satellite cells. Hematopoietic stem cells were defined with Lin−/Sca-1+/c-kit+/CD34− gating.
To analyze satellite cell apoptosis, isolated mononucleated cells from hindlimb muscle were labeled with CD31-PE, CD45-PE, Sca-1-PE-Cy7 and α7-integrin-APC. Subsequently, cells were labeled with PI and 1:40 annexin V-FITC (Biolegend) in 10mM HEPES, 140mM NaCl, 2.5mM CaCl2 (pH 7.4). Apoptotic cells were defined as Annexin V+/PI− from the satellite cell population.
Culture of satellite cells after magnetic-activated cell sorting (MACS)
Mononucleated cells were isolated from the hindlimb muscles as described previously [37] and satellite cells were purified using a satellite cell isolation kit in accordance with the manufacturer's instructions (Miltenyi Biotech). Isolated satellite cells were cultured in Ham's F-10, 20% fetal bovine serum, 5 ng/ml basic fibroblast growth factor, 100 U/ml penicillin and 100 μg/ml streptomycin on collagen-coated plates for 3 or 5 days.
Single-myofiber isolation and floating culture
Single myofibers were isolated, as previously described [38] from gastrocnemius muscles. Myofibers were cultured for 72 hours in DMEM 20% fetal bovine serum, 10% horse serum, 1% chicken embryo extract and 100 U/ml penicillin and 100 μg/ml streptomycin. After 72 hours, myofibers were fixed with 3.7% paraformaldehyde for 10 minutes.
Immunohistochemistry/Immunofluorescence
Muscle tissues were frozen in Tissue Freezing Medium (Triangle Biomedical Sciences) and stored at −80°C. Tissue cross sections of 14 μm thickness were collected every 200 μm using a Leica CM1850 cryostat. To quantitate myofiber cross-sectional areas, muscle sections were stained with hematoxylin and eosin and analyzed using ImageJ 1.43u.
All antibody incubations were performed at room temperature unless otherwise noted. All biotinylated or fluorophore-conjugated secondary antibodies were purchased from Jackson ImmunoResearch.
For immunostaining Pax7 and laminin, tissue sections were fixed in freshly prepared 4% paraformaldehyde (Electron microscopy sciences). Tissue sections were heated in citrate buffer (10mM sodium citrate, 0.05% Tween-20, pH 6.0) for antigen retrieval with a pressure cooker (Cuisinart model CPC-600) for 6 minutes. Subsequently, the M.O.M. Kit (Vector Laboratories Inc.) was used to block endogenous Fc receptor binding sites followed by a 1 hour incubation with 5% goat serum, 5% donkey serum, 0.5% BSA, 0.25% Triton-X 100 in PBS (blocking buffer). Sections were then labeled with blocking buffer containing anti-Pax7 (3.8 μg/ml, Developmental Studies Hybridoma Bank) and anti-laminin (2 μg/ml, Sigma) antibodies or isotype controls overnight at 4°C. After 1 hour incubation with biotinylated goat-anti-mouse F(ab’)2 IgG fragments (2.5 μg/ml), a TSA (Tyramide Signal Amplification; Perkin Elmer) Green kit was used for Pax7 imaging. Sections were further incubated with AF594-conjugated donkey anti-rabbit F(ab’)2 IgG fragments (3 μg/ml) for 40 minutes for laminin imaging. For immunostaining active caspase 3, Pax7 and laminin, methods were identical as described above, except active caspase -3 immnostaining (Cell signaling technology, clone D3E9, 1:100 dilution and overnight incubation at 4°C) before antigen retrieval step. For immunostaining active caspase 3 and MyoD, methods were identical as described above, except using MyoD (Santa cruz Biotechnology, 1 μg/ml) instead of Pax7 antibodies. Sections were labeled with 4',6-diamidino-2-phenylindole (DAPI) and mounted with Vectashield (Vector lab).
To analyze satellite cell fusion in vivo, BrdU (Sigma) at 100mg/g of body weight was injected intraperitoneally twice a day for 7 days and BrdU water (0.8mg/ml) was provided for the following 7 days. Tissue sections were incubated with the M.O.M kit and followed by blocking buffer as described above. Sections were labeled with anti-dystrophin (13.5 μg/ml, clone MANDYS8, Sigma) antibody or mouse isotype control overnight at 4°C. Sections were fixed in freshly prepared 4% paraformaldehyde (Electron microscopy sciences) and heated in citrate buffer from the BrdU in situ kit (BD Pharmigen) for antigen retrieval with a pressure cooker for 10 minutes. Sections were then labeled with biotinylated anti-BrdU in diluent buffer (1:10; BrdU in situ kit) for 1 hr. A TSA Green kit was used for subsequent steps as described above. Sections were further incubated with Texas Red-conjugated goat anti-mouse F(ab’)2 IgG fragments (2.5 μg/ml) for 40 minutes for dystrophin imaging. Sections were labeled with DAPI and mounted with Vectashield.
For immunostaining of isolated myofibers, myofibers were incubated with the M.O.M kit and followed by blocking buffer as described above. Myofibers were then labeled with anti-Pax7 and anti-MyoD antibodies or isotype controls overnight at 4°C. After 1 hr incubation with biotinylated donkey anti-mouse F(ab’)2 IgG fragments (2.5 μg/ml), the TSA Green kit was used for subsequent steps as described above. Myofibers were incubated with AF594-conjugated donkey anti-rabbit F(ab’)2 IgG fragments (3 μg/ml) for 1 hr for MyoD imaging. Myofibers were labeled with DAPI and mounted with Vectashield.
For immunostaining of cultured satellite cells, cells were fixed with 2% paraformaldehyde for 15 minutes and incubated with blocking buffer as described above. Cells were labeled with anti-p27 (2 μg/ml, Abcam) antibody or anti-LEF1(1:50, Cell Signaling Technology) or isotype control overnight at 4°C followed by a 1 hour incubation with biotinylated donkey anti-rabbit F(ab’)2 IgG fragments (2.5 μg/ml). A TSA Green kit was used for subsequent steps as described above. For dual immunolabeling studies, cells were heated in citrate buffer with a pressure cooker for 6 minutes and then incubated with anti-Pax7 antibody or isotype control overnight at 4°C. After 1 hr incubation with biotinylated goat anti-mouse F(ab’)2 IgG fragments (2.5 μg/ml), the TSA Red kit was used for subsequent steps as described above. Cells were labeled with DAPI and mounted with Vectashield.
Confocal images for cultured satellite cells were acquired using an Olympus FV1000 inverted microscope with 40x objective with FV10 ASW ver. 4.2 software. All other images were acquired using an Axioplan microscope with a 0.5 NA 20x Plan-Neofluar objective (Carl Zeiss MicroImaging, Inc.) and charge-coupled device camera (Carl Zeiss MicroImaging, Inc.) with Scion Image 1.63 (Scion Corp.). All images were globally processed for size, brightness and contrast using Photoshop CS3 (Adobe).
Real time-PCR analyses
Total RNA was isolated from FACS-sorted satellite cells using a Picopure RNA isolation kit (Applied Biosystems) in accordance with the manufacturer's instructions. The reverse transcriptase reaction was performed using 250 ng of total RNA/sample using random hexamers and M -MLV reverse transcriptase (Invitrogen). cDNA was amplified using the SYBR select master mix (Applied Biosystems) and 2.5 μM of each primer. All RNA samples were tested for DNA contamination by PCR. Amplified cDNA signals were detected and analyzed by StepOne software v2.2.2 (Applied Biosystems) using GAPDH as an internal control. Fold change of gene expression was determined using the ΔΔCt method [39]. Three to four independent experiments were performed. All Kpna1, Pax7, myogenin, and Gapdh primers were purchased from Qiagen's RT2 qPCR Primer Assay library. All primers for cell cycle related proteins were from PrimerBank (http://pga.mgh.harvard.edu/primerbank/). Primer sequences were: p27 (F: 5′-TCAAACGTGAGAGTGTCTAACG-3′ and R: 5′-CCGGGCCGAAGAGATTTCTG-3′); Cyclin d1 (F: 5′-GCGTACCCTGACACCAATCTC-3′ and R: 5′-CTCCTCTTCGCACTTCTGCTC-3′); Fos-l1 (F: 5′-ATGTACCGAGACTACGGGGAA-3′ and R: 5′-CTGCTGCTGTCGATGCTTG-3′); Lef1 (F:5’-TGTTTATCCCATCACGGGTGG-3’ and R: 5’-CATGGAAGTGTCGCCTGACAG-3’), and Survivin (F:5’-GAGGCTGGCTTCATCCACTG-3’ and R: 5’-CTTTTTGCTTGTTGTTGGTCTCC-3’).
Statistical analyses
To determine the statistical significance between two groups, comparisons were made using an unpaired Student's t-test. To determine the statistical significance of frequency distributions between two groups, comparisons were made using a non-parametric one way ANOVA with Kruskal-Wallace post-test correction. The significance of the results obtained from multiple groups was evaluated by one -way ANOVA with Bonferroni's post-test correction. The significance of the results obtained from multiple time points between genotypes was evaluated by two-way ANOVA with Bonferroni's post-test correction. Statistical analyses were performed using GraphPad Prism v.4 software (GraphPad Software). A p value of less than 0.05 was considered significant.
Results
Changes in the expression of Kpna paralogs and cNLS proteins in satellite cells during muscle regeneration
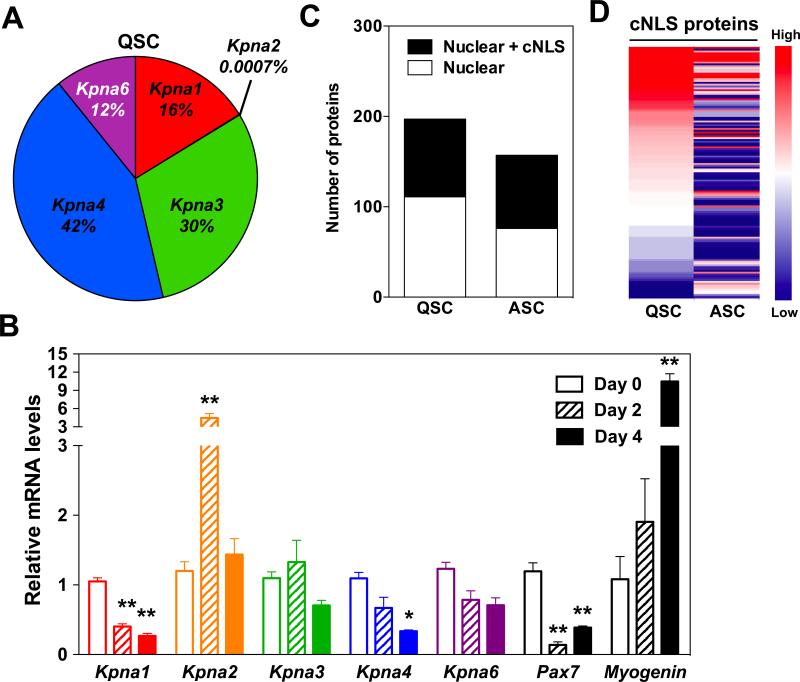
To begin to study the classical nuclear import pathway in satellite cells, we first isolated satellite cells from mouse hindlimb muscles using flow cytometry with established cell surface markers (Sca1−/CD31−/CD45−/α7-integrin+) (Supplementary Figure 1) [40] and confirmed expression of markers of quiescence (Pax7) and activation (myogenin) using quantitative real-time PCR (qRT-PCR). As expected, Pax7 mRNA levels were high in quiescent satellite cells (QSC) in uninjured muscles (Day 0), and myogenin mRNA levels were increased in activated satellite cells (ASC) 4 days post-injury (Figure 1B). Subsequently, we analyzed the steady-state mRNA levels of Kpna paralogs in QSC and ASC using qRT-PCR. In the mouse, six Kpna paralogs exist designated Kpna1, Kpna2, Kpna3, Kpna4, Kpna6, and Kpna7 [16]. In QSC, Kpna1, Kpna2, Kpna3, Kpna4, Kpna6 mRNAs were present but not Kpna7. Kpna3 and Kpna4 mRNAs were the most abundant, whereas Kpna2 mRNA was the least abundant (Figure 1A). In response to muscle injury, we observed differential Kpna expression patterns in ASC. No significant changes in Kpna3 or Kpna6 mRNA levels occurred in satellite cells after injury. In contrast, Kpna2 mRNA levels increased in ASC 2 days after injury while Kpna4 mRNA levels decreased 4 days after injury. Kpna7 mRNA was not detected in ASC. Interestingly, only Kpna1 mRNA levels decreased in ASC at both 2 and 4 days post-injury (Figure 1B). The distinct changes in Kpna mRNAs that occur in satellite cells in the transition from quiescence to activation suggest that various KPNA paralogs may transport different subsets of cNLS proteins that are critical for regulating satellite cell behavior.
Figure 1. Changes in the expression of Kpna paralogs and cNLS-containing proteins in satellite cells during muscle regeneration.
(A, B) qRT-PCR analyses of FACS sorted satellite cells from uninjured or injured gastrocnemius muscles for Kpna paralogs. (A) Relative mRNA levels of Kpna paralogs in quiescent satellite cells (QSC). (B) mRNA levels of Kpna paralogs, Pax7, and myogenin at different days after muscle injury. All data were normalized to uninjured values (Day 0) for each Kpna paralog. Data represent the mean ± SEM. n=3, **p<0.01 and *p<0.05. (C, D) Proteomic analysis of nuclei isolated from FACS sorted QSC and activated satellite cells (ASC) 3 days after injury. (C) The number of nuclear proteins and classical nuclear localization signal (cNLS) nuclear proteins identified by mass spectrometry from QSC and ASC. (D) Heat map showing abundance of cNLS-containing nuclear proteins in QSC and ASC.
We next analyzed whether changes in cNLS proteins, which are cargo proteins of KPNA paralogs, occur in satellite cells during muscle regeneration. We isolated QSC from uninjured hindlimb muscles or ASC from gastrocnemius muscles 3 days after BaCl2-induced injury using the flow cytometry approach in Supplementary Figure 1. Subsequently, nuclei were enriched from the sorted satellite cells and mass spectrometry was performed. Approximately 200 nuclear proteins were identified in QSC, whereas 157 nuclear proteins were identified in ASC. These nuclear proteins were then additionally analyzed using NLStradamus [41] to identify predicted cNLS proteins. Fifty-three percent of the nuclear proteins in QSC and 56% of the nuclear proteins in ASC contained a predicted cNLS (Figure 1C), in agreement with a previous bioinformatics study that estimated that 30-55% of nuclear proteins in mice contain a predicted cNLS [10]. Among the predicted 136 cNLS proteins in QSC and ASC, 68% of the cNLS proteins were decreased while 15% of the cNLS proteins were increased during satellite cell activation as defined by a 2-fold change. Only 17% of the cNLS proteins were present at similar levels in the nuclei of QSC and ASC (Figure 1D). In the cNLS proteome of QSC, mRNA processing, translation, and anatomical structure development-related proteins were enriched by gene ontology (GO) molecular function analysis (Supplementary Table 1). Particularly, more than half of the mRNA processing proteins were related to the spliceosome. These results are consistent with a previous microarray study that showed RNA processing and splicing-related genes were highly expressed in QSC [4]. Due to the limited number of predicted cNLS proteins in ASC, only one protein group related to biosynthesis was enriched by GO analysis (Supplementary Table 1), in agreement with the increased mitochondrial activity and cell size noted with satellite activation [42]. Taken together, our results show that Kpna transport receptors and cNLS proteins in the nucleus undergo dynamic changes between QSC and ASC. These results suggest that the classical nuclear import pathway plays an important role in satellite cells.
Increased satellite cell proliferation under basal conditions in KPNA1 KO mice
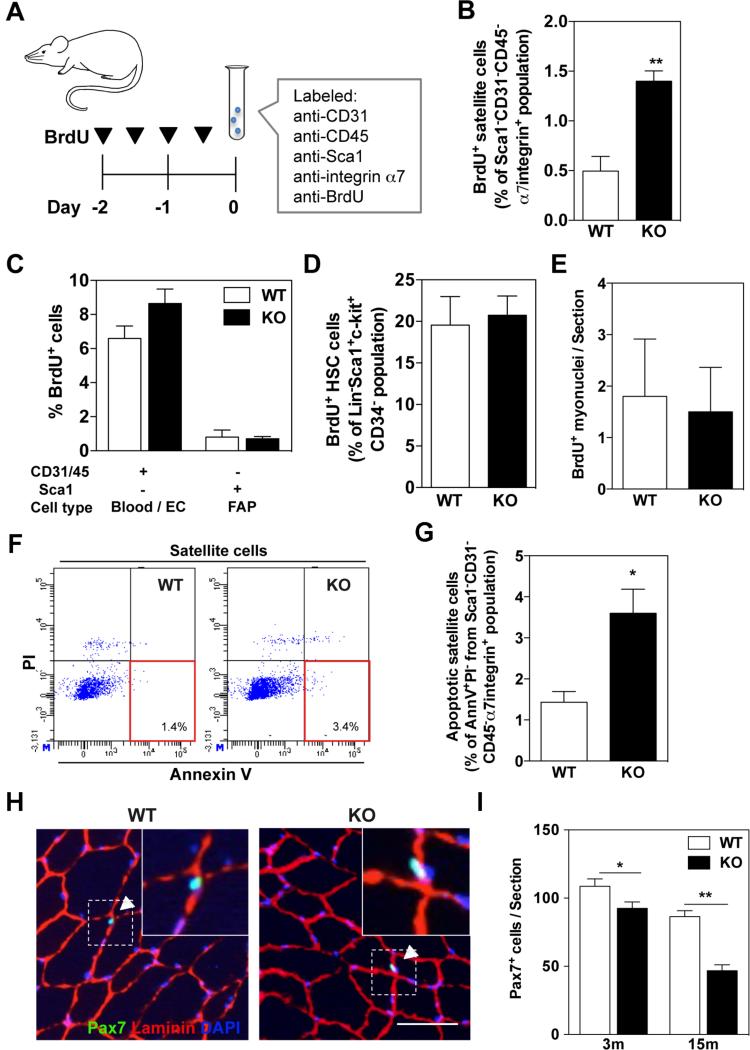
Given that Kpna1 mRNA levels negatively correlated with the proliferation state of satellite cells (Figure 1B), we reasoned that KPNA1 (importin alpha 5) may transport nuclear proteins that function to inhibit proliferation of QSC. To address the role of KPNA1 in QSC, we utilized KPNA1 knockout (KO) mice. These mice were born in Mendelian ratios and displayed 10% reduced body weight. Myofiber cross-sectional area was also reduced 10% compared to wild type (W T) mice (Supplementary Figure 2A). Furthermore, tissue development is reported to be normal except in female reproductive organs [26, 43]. We performed in vivo BrdU proliferation assays using 3 months old KPNA1 KO mice. Mice were injected with BrdU for 2 days, and then satellite cells were isolated from hindlimb muscles and labeled with antibodies against CD31, CD45, Sca1, α7-integrin and BrdU to identify proliferating satellite cells by flow cytometry (Figure 2A). The percentage of proliferating KPNA1 KO satellite cells was 2.8-fold higher than WT satellite cells (Figure 2B). Of note, no signs of degeneration or regeneration were observed in KPNA1 KO muscle sections indicating that satellite cell proliferation did not result from muscle injury in these mice (Supplementary Figure 2B). To determine whether increased proliferation was specific to satellite cells, we also analyzed the proliferation of other cell types in BrdU-injected KPNA1 W T and KO mice. However, no significant differences were noted in the percentage of proliferating blood and endothelial cells (EC) or fibro-adipogenic progenitor cells (FAP) [44], in the hindlimb muscles of KPNA1 W T and KO mice (Figure 2C). Similarly, no proliferation differences were observed in hematopoietic stem cells of the bone marrow (Figure 2D).
Figure 2. Increased satellite cell proliferation and apoptosis in uninjured muscle s of KPNA1 null mice.
(A) Schematic of BrdU injection and immunolabeling of satellite cells for FACS analysis. (B) The percentage of BrdU+ satellite cells in KPNA1 null (KO) hindlimb muscles was increased approximately 3-fold relative to wild type (WT). (C) The percentage of BrdU+ blood/endothelial cells (EC) and fibro/adipocyte progenitors (FAP) did not significantly differ between WT and KO hindlimb muscles. (D) By flow cytometry, the percentage of BrdU+ hematopoietic stem cells (HSC) did not significantly differ between W T and KO mice. Lin = CD3/Gr-1/CD11b/CD45R/Ter-119 (E) The number of BrdU+ myonuclei in sections of tibialis anterior (TA) muscles did not differ between uninjured WT and KO mice. (F) Representative flow plots depicting apoptotic satellite cells positive for Annexin V and negative for propidium iodide (PI). (G) The percentage of apoptotic satellite cells was increased approximately 2.5-fold in KO hindlimb muscles relative to WT. AnnV = Annexin V. (H) Representative images of TA muscles immunostained for Pax7 (green), laminin (red), and DAPI (blue). Bar = 50 μm. White arrowheads indicate Pax7+ satellite cells co-stained with DAPI and located inside of laminin outlines. (I) Pax7+ satellite cells were significantly decreased in TA muscles of KO mice relative to W T at both 3 and 15 months of age. Data represent the mean ± SEM. n=3 for all experiments. **p<0.01 and *p<0.05.
We next investigated the fate of KPNA1 KO satellite cells. To analyze differentiation of satellite cells in KPNA1 KO muscle, we immunostained TA muscle sections with BrdU antibodies after 2 weeks BrdU treatment. Since myonuclei are post-mitotic, any BrdU+ nuclei within muscle fibers must have arisen from the fusion of BrdU+ satellite cells with the myofiber. We quantified the number of BrdU+ myonuclei in KPNA1 WT and KO muscles but observed no significant difference (Figure 2E). These results indicate that ASC in KPNA1 KO muscles under basal conditions do not undergo enhanced myogenic differentiation. To further investigate the fate of KPNA1 KO satellite cells, apoptosis was measured by flow cytometry [45]. Mononucleated cells were isolated from hindlimb muscles and labeled with antibodies against CD31, CD45, Sca1, and α7-integrin to detect satellite cells and Annexin V and PI to detect apoptotic cells (Figure 2F). The percentage of early apoptotic (Annexin V+/PI−) KPNA1 KO satellite cells was 2.5-fold higher than WT (Figure 2G). Similar to the flow cytometry results, sections of uninjured KPNA1 KO TA muscles contained 3 times higher active caspase -3+ cells (a marker of apoptosis) among satellite cells (Pax7+ cells) than W T (Supplementary Figure 3). To determine the effect of apoptosis on the numbers of satellite cells, muscle sections were immunolabeled for Pax7 and laminin (Figure 2H) and the number of Pax7+ satellite cells was quantified (Figure 2I). The number of Pax7+ cells was decreased by 15% in KPNA1 KO muscles at 3 months of age compared to W T with further age-related declines in satellite cell number (40%) in 15 months old KPNA1 KO muscles. Together, these results indicate that KPNA1 KO satellite cells proliferate but do not differentiate under basal conditions and undergo apoptosis leading to an age-related exhaustion of the satellite cell pool.
Increased satellite cell proliferation in injured muscle of KPNA1 KO mice
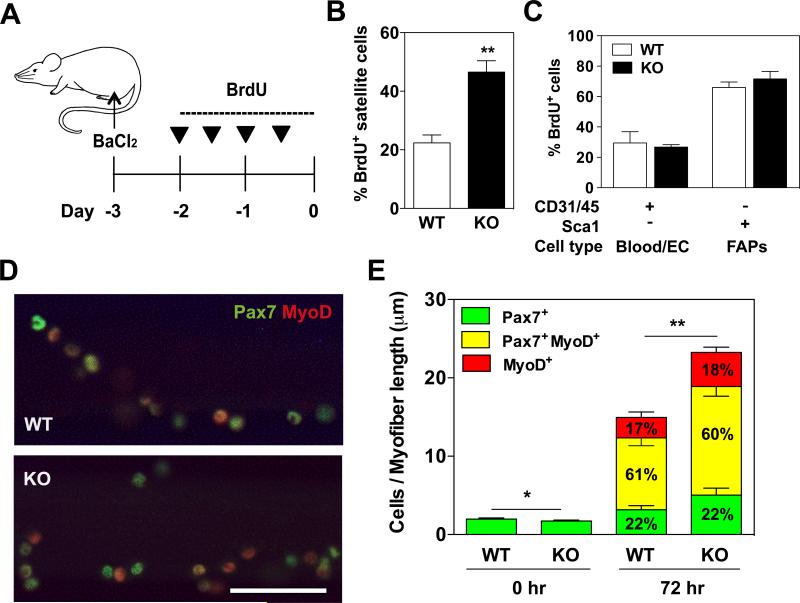
We next investigated the role of KPNA1 in ASC during BaCl2-induced muscle regeneration in gastrocnemius muscles. Mice were injected with BrdU after injury and the muscles collected 3 days after injury (Figure 3A). Proliferating satellite cells in KPNA1 W T and KO mice were analyzed by flow cytometry. The number of proliferating satellite cells in regenerating muscles was about 2-fold higher in KPNA1 KO mice compared to WT, consistent with the increased satellite cell proliferation observed in uninjured muscles (Figure 3B). The increased proliferation was specific for satellite cells as blood and endothelial cells as well as FAP cells in the same muscles did not show any proliferative difference between KPNA1 WT and KO (Figure 3C). To analyze the myogenic lineage progression of proliferating satellite cells, suspension cultures of isolated myofibers, an ex vivo model of muscle injury, were performed using KPNA1 WT and KO myofibers. At 0 and 72 hours in culture, satellite cells were immunostained for Pax7 and MyoD, two myogenic transcription factors whose expression levels change during myogenic differentiation, and the number of satellite cells expressing these markers was quantified (Figure 3D). After 72 hours of culture, the number of satellite cells associated with KPNA1 KO myofibers was 30% greater than with WT myofibers suggesting that the early stages of satellite cell expansion are accelerated by the loss of KPNA1. However, no significant differences between genotypes were noted in the percentage of satellite cells at each stage of myogenic differentiation (Figure 3E). Taken together, these results indicate that the loss of KPNA1 alters satellite cell proliferation but not differentiation during muscle injury.
Figure 3. Increased satellite cell proliferation in regenerating muscles of KPNA1 null mice.
(A) Schematic of muscle injury induced by BaCl2 and BrdU injection protocol. (B) The percentage of BrdU+ satellite cells in regenerating KPNA1 null (KO) gastrocnemius muscles was increased about 2-fold relative to wild type (WT). (C) The percentage of BrdU+ blood/endothelial cells (EC) and fibro/adipocyte progenitors (FAPs) did not significantly differ between regenerating gastrocnemius muscles of WT and KO mice. (D) Representative images of WT and KO myofibers immunostained for Pax7 (green) and myoD (red) after 72 hours of culture. Bar = 50 μm. (E) Pax7+, Pax7+MyoD+ and MyoD+ satellite cells were quantified on myofibers at 0 and 72 hours of culture in order to analyze lineage progression. The total number of KO satellite cells was increased by approximately 30% after 72 hours of culture. The percentage of satellite cells at each stage of lineage progression did not significantly differ between WT and KO myofibers. WT myofibers=15; KO myofibers=21. Data represent the mean ± SEM. n=3-5 for all experiments. **p<0.01 and *p<0.05.
Enhanced muscle regeneration and normal satellite cell self-renewal in injured KPNA1 KO muscles
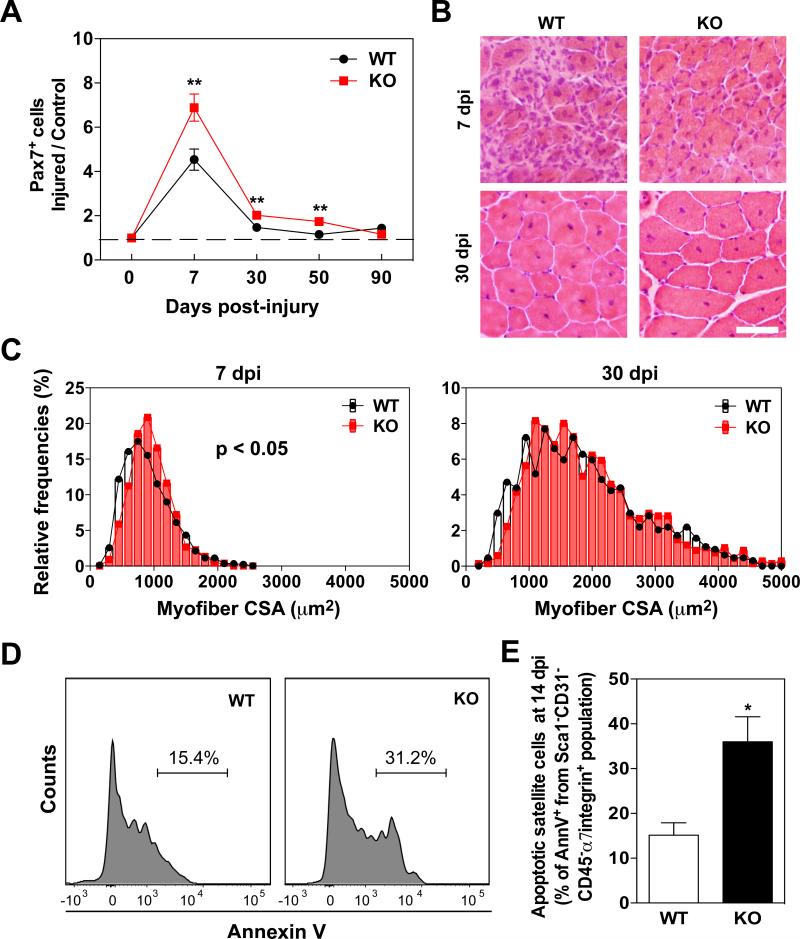
Subsequently, we analyzed later stages of muscle regeneration in order to determine if loss of KPNA1 affects satellite cell self-renewal or the restoration of normal tissue architecture after muscle degeneration. TA muscles of KPNA1 WT and KO mice were injured with BaCl2 and collected 7, 30, 50, and 90 days post-injury. Muscle sections were immunostained for Pax7 and the number of Pax7+ satellite cells was quantified at each time point (Figure 4A). Consistent with previous data, more Pax7+ cells were observed in KPNA1 KO muscles at 7 days post-injury; however, similar numbers of Pax7+ cells were present in both genotypes after 30 days post-injury. Since satellite cell renewal is complete in WT muscles by 50 days after injury [46] these results suggest that self-renewal is normal in KPNA1 KO satellite cells under these conditions. The increased number of satellite cells present in KPNA1 KO muscles contributed to accelerated muscle regeneration at early stages as myofiber cross-sectional area in KPNA1 KO muscle was increased relative to W T at 7 days post-injury. However, this hypertrophic effect in KPNA1 KO muscle was not retained after 30 days post-injury (Figure 4B, C). To investigate whether KPNA1 KO satellite cells undergo apoptosis in regenerating muscles, we quantified Annexin V+ satellite cells by flow cytometry as previously described for uninjured muscles. The percentage of apoptotic KPNA1 KO satellite cells was ~2-fold higher than WT in gastrocnemius muscles 14 days after injury (Figures 4D, E). Similar to the flow cytometry results, the percentage of active caspase-3+MyoD+ satellite cells was 2-fold higher in KPNA1 KO TA muscle sections 7 days after injury compared to WT (Supplementary Figure 4). These results indicate that the enhanced number of KPNA KO satellite cells is beneficial for muscle regeneration in the early phases after injury, but this effect is not maintained at later phases likely due to the high level of apoptotic satellite cells.
Figure 4. Increased numbers of satellite cells and enhanced regeneration but normal self-renewal in KPNA1 null muscles.
(A) The number of Pax7+ cells was increased at 7 days post-injury in TA muscles of KPNA1 null (KO) mice relative to WT demonstrating the higher proliferative capacity of KO satellite cells. (B) Representative hematoxylin and eosin stained TA muscle sections 7 and 30 days post-injury (dpi) in W T and KO mice. Bar = 50 μm (C) TA myofiber cross-sectional area (CSA) of KO 7 days post-injury (dpi) was significantly increased relative to WT indicating enhanced muscle regeneration at early phases. However, no significant differences were noted 30 dpi. Total 903 W T myofibers (n=5) and 748 KO myofibers (n=3) for 7 dpi and total 637 WT myofibers (n=4) and 674 KO myofibers (n=4) for 30 dpi were analyzed, respectively. (D) Representative histogram plots depicting apoptotic satellite cells positive for Annexin V. (E) The percentage of apoptotic satellite cells was increased approximately 2.4-fold in KO gastrocnemius muscles relative to W T. AnnV = Annexin V. Data represent the mean ± SEM. n=3 for all experiments. **p<0.01 and *p<0.05.
Reduced nuclear translocation of p27 and increased Cyclin d1 mRNA in KPNA1 KO satellite cells
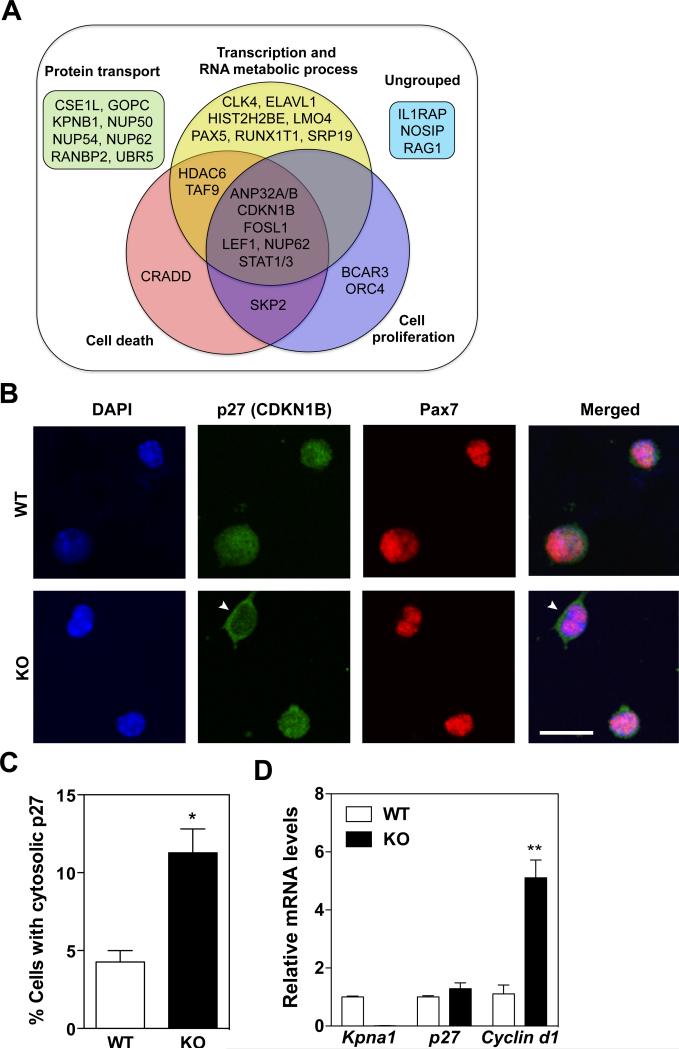
In order to gain mechanistic insights into the proliferative phenotype of KPNA1 KO satellite cells, we searched for potential cargo proteins of KPNA1 from a protein-protein interaction database (Wiki-Pi) [47]. In humans, 32 proteins are known to interact with KPNA1 and these proteins were grouped into 4 categories by Gene Ontology analysis (Figure 5A). More than half of the known KPNA1-interacting proteins fall into the category of transcription and RNA metabolism, which implies a critical role for KPNA1 in regulating gene expression. Moreover, 44% of cargo proteins were categorized as being involved in cell death or cell proliferation, which are likely the most relevant cargo candidates for KPNA1 given the phenotype of KPNA1 KO satellite cells.
Figure 5. Reduced nuclear translocation of the cell cycle inhibitor p27/CDKN1B in KPNA1 null satellite cells.
(A) Venn diagram of 32 KPNA1-interacting proteins identified by Wiki-Pi and grouped into 4 categories by Gene Ontology (GO) analysis. (B) Representative images of p27/CDKN1B (green) and Pax7 (red) immunostaining and DAPI (blue) of wild type (WT) and KPNA1 null (KO) satellite cells isolated by magnetic activated cell sorting and cultured for 3 days. White arrowheads indicate satellite cells with cytosolic p27. Bar = 20 μm. (C) The number of satellite cells with cytosolic p27/CDKN1B immunostaining was increased 3-fold in KO relative to WT indicating impaired nuclear import of p27/CDKN1B. Approximately 400 cells were analyzed for each genotype. (D) Relative mRNA levels of Kpna1, p27, and Cyclin d1 in WT and KO isolated satellite cells. No significant difference was noted in p27 mRNA levels. The increased levels of Cyclin d1 in KO satellite cells are indicative of the increased cell proliferation observed in KO satellite cells relative to WT. Data represent the mean ± SEM. n=3 for all experiments. **p<0.01 and *p<0.05.
Among the cargo candidates shown in Figure 5A, the cell cycle inhibitory protein, Cyclin-Dependent Kinase Inhibitor 1B (CDKN1B), known as p27, prevents activation of the cyclin D1-CDK4 complex to stall cell cycle entry into G1 [48] and keep cells quiescent [49]. Previous work indicated that p27 binds KPNA1 in order to be transported into the nucleus [50, 51]. Therefore, we hypothesized that p27 may be mislocalized in KPNA1 KO satellite cells thereby inducing cyclin D1 activation and leading to the observed proliferative phenotype in these cells. To test this hypothesis, we isolated satellite cells from uninjured W T and KPNA1 KO muscles and immunostained for p27 after 3 days in culture. We observed two-fold more KPNA1 KO satellite cells with cytosolic localization of p27 compared to WT cells (Figures 5B, C), while levels of p27 mRNA did not differ in satellite cells from W T and KPNA1 KO (Figure 5D). We also noted a 5-fold increase in Cyclin D1 mRNA in KPNA1 KO satellite cells consistent with the increased proliferation of these cells (Figure 5D). Taken together, inefficient nuclear localization of the cell cycle inhibitor p27 likely contributes to the proliferative phenotype observed in KPNA1 KO satellite cells.
Down-regulation of Wnt signaling in KPNA1 KO satellite cells
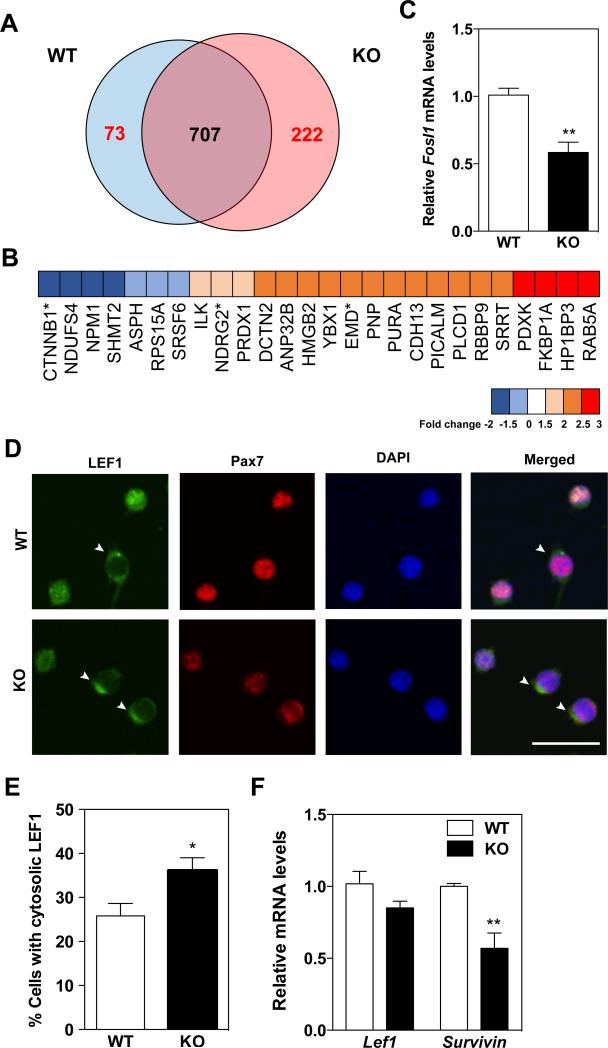
To examine alterations in the proteome of satellite cells in the absence of KPNA1, QSC were isolated from hindlimb muscles of W T and KPNA1 KO mice and mass spectrometry was performed. A total of 1,002 proteins were identified with 73 proteins enriched in WT satellite cells and 222 proteins enriched in KPNA1 KO satellite cells as defined by a 1.5-fold change (Figure 6A). Using Gene Ontology analysis, in the cell proliferation category we noted a proteomic signature indicating down-regulation of canonical Wnt signaling in KPNA1 KO satellite cells (Figure 6B, Supplementary Table 2). For example, the levels of β-catenin (CTNNB1, a critical transcriptional co-activator in Wnt signaling [52]) were lower in KPNA1 KO satellite cells. In contrast, levels of N-myc downstream regulated gene 2 (NDRG2) [53] and emerin (EMD) [54], which all inhibit β-catenin-mediated Wnt signaling, were higher in KPNA1 KO satellite cells. Reduced Wnt signaling can affect satellite cell proliferation [55], survival [56] and lineage conversion [57]. We used qRT-PCR to quantitate levels of Fosl1, a target gene of Wnt signaling [58] as well as a KPNA1-interacting protein [59], in KPNA1 W T and KO satellite cells and observed decreased Fosl1 mRNA in KPNA1 KO satellite cells (Figure 6C). Interestingly, Lymphoid enhancer factor 1 (LEF1), a critical co-transcription factor for Wnt signaling through interaction with β-catenin in the nucleus [60], is a known KPNA1-interacting protein (Figure 5A, [61]). To examine whether loss of KPNA1 alters LEF1 nuclear import in satellite cells, we immunostained for LEF1 in WT and KPNA1 KO satellite cells after 5 days in culture. Although levels of Lef1 mRNA did not differ (Figure 6F), approximately 1.4-fold more KPNA1 KO satellite cells showed cytosolic localization of LEF1 compared to WT cells (Figures 6D, E), implying down-regulation of LEF1-dependent transcription of Wnt target genes. Since β-catenin-mediated Wnt signaling regulates cell survival-related genes [62], reduced Wnt signaling may increase the susceptibility of KPNA1 KO satellite cells to apoptosis. To investigate this possibility, we used qRT-PCR to analyze Survivin mRNA, an anti-apoptotic gene regulated by Wnt signaling, in 5 day-cultured satellite cells and observed 45% lower levels in KPNA1 KO satellite cells compared to WT (Figure 6F). Taken together, these results suggest that the nuclear proteins transported by KPNA1 in satellite cells modulate apoptosis through regulation of Wnt signaling.
Figure 6. Down-regulation of Wnt signaling in KPNA1 null satellite cells.
(A) Venn diagram of proteins identified by mass spectrometry in KPNA1 W T and KO satellite cells purified by flow cytometry from hindlimb muscles. A total of 893 proteins were identified with 70 proteins enriched in W T and 211 proteins enriched in KO as defined by a 1.5-fold change. (B) Heat map of down-regulated (< −1.5 fold) and up-regulated (> 1.5 fold) cell proliferation categorized proteins by Gene Ontology (GO) analysis in KO QSC. *proteins indicate down-regulation of Wnt signaling in KO satellite cells. (C) Reduced levels of Fosl1 mRNA in KO QSC indicates decreased Wnt signaling in these cells. (D) Representative images of Lymphoid enhancer factor 1 (LEF1, (green)) and Pax7 (red) immunostaining and DAPI (blue) of satellite cells isolated by magnetic activated cell sorting and cultured for 5 days. White arrowheads indicate satellite cells with cytosolic LEF1. Bar = 30 μm. (E) The number of satellite cells with cytosolic LEF1 immunostaining was increased about 40% in KO satellite cells relative to WT indicating impaired nuclear transport of LEF1. Approximately 800-1000 cells were analyzed for each genotype. (F) Relative mRNA levels of Lef1 and Survivin in W T and KO 5 day-cultured satellite cells. No significant difference was noted in Lef1 mRNA levels. The decreased levels of Survivin mRNA in KO satellite cells is indicative of the higher apoptosis observed in these cells relative to WT. Data represent the mean ± SEM. n=3 for all experiments. **p<0.01 and *p<0.05.
Discussion
These studies establish that the nuclear import receptor KPNA1 plays a role in regulating the balance between quiescence and activation of muscle satellite cells. In the absence of KPNA1, satellite cells prematurely activate and proliferate in uninjured muscle but they undergo apoptosis leading to satellite cell exhaustion with age. Increased proliferation of satellite cells was also observed in injured muscle leading to enhanced regeneration at the early but not late phases of regeneration likely due to the high level of apoptosis in KPNA1 KO satellite cells. These results indicate that regulated nuclear import of proteins by the classical nuclear localization pathway is critical for satellite cell proliferation and survival.
During the transition between satellite cell quiescence and activation, we observed differential changes in the transcript levels of six Kpna paralogs. For example, Kpna1 mRNA decreased from day 0 to day 2 after injury but Kpna2 mRNA increased during the same time period. Plasticity in KPNA paralog expression also occurs during differentiation of other cell types both in vivo and in vitro although cell types differ in the particular types of KPNA paralogs expressed [23, 63-67]. Interestingly, distinct cellular roles for KPNA paralogs have been identified in several cell types, including the current study. In mouse ES cells, KPNA2 plays a critical role in inhibiting cell differentiation, whereas KPNA1 induces neuronal differentiation [63, 68]. During mouse spermatogenesis, KPNA4 acts to protect these cells against oxidative stress [69]. In lower organisms such as Planaria, knockdown of the KPNA homolog, Smed-ima-1, leads to defects in differentiation and tissue patterning [70]. Furthermore, we previously reported distinct roles for KPNA1, KPNA2, and KPNA4 in mouse myoblast proliferation and myotube growth during in vitro myogenesis [24]. Although KPNA paralogs play critical roles in cell differentiation of multiple cell types, little is known about the molecular mechanisms which link signaling pathways to changes in their expression. The observed plasticity in KPNA paralog expression during satellite cell activation would be predicted to alter the nuclear proteome since individual KPNA paralogs can preferentially bind to specific cNLS-containing proteins [71]. Indeed, during satellite cell activation the levels of more than 80% of cNLS proteins identified in satellite cell nuclei changed. Taken together, our data suggest that changes in KPNA paralog expression during satellite cell activation are a regulatory mechanism by which the nuclear entry of unique sets of proteins is controlled in a precise temporal manner to regulate specific molecular programs required for satellite cell function. Nuclear proteins such as transcription factors required at specific stages of a satellite cell's life cycle could lie dormant in the cytoplasm, but then quickly gain access to the genome as a group once a particular KPNA is expressed. Such a system would bypass the need to wait for the cell to transcribe and/or translate multiple genes in response to external signals.
We focused on studying the role of KPNA1 in satellite cells because this was the only KPNA paralog that decreased during satellite cell activation suggesting that nuclear import of KPNA1-dependent proteins must be down-regulated during the early phases of muscle regeneration. Depletion of KPNA1 resulted in increased satellite cell proliferation and early exhaustion of the satellite cell pool in uninjured mouse muscle. The finding of increased satellite cell proliferation in uninjured muscle is a rare phenotype. The only other report of such re-entry into the cell cycle by quiescent satellite cells occurred with knockout of retinoblastoma tumor suppressor protein (Rb1), a cell cycle regulatory factor [72]. However, unlike the expanded satellite cell pool found in Rb1 null muscle, the satellite cell pool of KPNA1-depleted muscle was reduced with aging due to apoptosis of the satellite cells.
We also observed increased proliferation and apoptosis of satellite cells in regenerating KPNA1 KO muscle as in uninjured muscle. Increased proliferation of satellite cells in regenerating muscles has been previously noted with the knockout of Notch signaling-related proteins [73, 74], Hexamethylene Bis-Acetamide Inducible 1 [75] and Mitogen-Activated Protein kinase phosphatase 5 [76] resulting in enhanced hypertrophy of regenerated myofibers. We noted enhanced myofiber size in the early phases of regeneration in KPNA1 KO muscles but this was not maintained at late phases likely due to apoptotic satellite cells. However, we cannot rule out that the loss of KPNA1 in other cell types, such as nerves or myofibers, may have also contributed to the absence of myofiber hypertrophy at later phases.
Several lines of evidence suggest that the hyperproliferation observed in KPNA1 KO satellite cells both in uninjured and injured muscle is likely due to a cell intrinsic role for KPNA1 rather than an extrinsic effect by KPNA1 loss in other cell types. We have previously demonstrated that siRNA-mediated knockdown of KPNA1 in pure cultures of primary mouse myoblasts results in enhanced cell proliferation [24]. In addition, in the current study, 30% higher numbers of KPNA1 KO satellite cells were observed after 72 hrs in single myofiber cultures without other mononucleated cell types. Furthermore, increased levels of transcripts for cell cycle regulators were observed in sorted KPNA1 satellite cells. We attempted to analyze the cell intrinsic role of KPNA1 in satellite cells by generating Pax7creER-Kpna1 floxed mice but despite multiple modifications of the tamoxifen treatment protocol, we could only obtain a 50% decrease in Kpna1 levels, which were not sufficient to alter satellite cell proliferation (data not shown).
To study the molecular mechanisms underlying the proliferative phenotype of KPNA1 KO satellite cells, we mined a protein-protein interaction database (Wiki-Pi) [47] based on verified data, and identified 32 KPNA1-interacting proteins. We demonstrated that one of these KPNA1 cargo proteins, p27 (CDKN1B, a cell cycle inhibitor protein), was localized to the cytoplasm in a greater proportion of KPNA1 KO satellite cells than WT although both genotypes showed similar transcript levels. Loss of p27 nuclear localization in KPNA1 KO satellite cells would be predicted to stimulate cell proliferation [77], consistent with the hyperproliferation observed in KPNA1 KO cells in both uninjured and injured muscle. Not all KPNA1 satellite cells showed the defect in nuclear localization of p27, perhaps because p27 also binds KPNA4 [51], which may compensate for the absence of KPNA1-mediated nuclear transport of p27 in a subset of satellite cells. Such heterogeneity could arise from differences in the levels of KPNA1 among individual satellite cells. Studies of KPNA1 at the single cell level await the generation of new antibodies that are highly specific to KPNA1 in mouse satellite cells. In addition to redundant nuclear transport receptors for p27, p27 also has functional redundancy with p57, which maintains stem cell quiescence [49] and is not known to interact with KPNA1.
To gain further insights into the molecular mechanisms underlying the phenotype of KPNA1 KO satellite cells, we analyzed QSC from WT and KPNA1 KO muscles by mass spectrometry. Reduced Wnt signaling was suggested by the proteomic data, which was confirmed by decreased mRNA levels of Fosl1, a downstream gene target of Wnt signaling [58]. Although Wnt signaling is critical for cellular proliferation and muscle differentiation [55, 78], KPNA1 KO satellite cells were characterized by greater proliferation, normal myogenic lineage progression and enhanced regeneration. Likely other nuclear proteins which positively regulate proliferation and differentiation could overcome the effects of decreased Wnt signaling on these cellular parameters. Interestingly, LEF1, a known KPNA1 interacting protein and a critical transcription factor together with β-catenin for activating Wnt target genes, was present in the cytosol of a higher proportion of KPNA1 KO satellite cells than WT. LEF1 also binds KPNA2 [61], which may compensate for the absence of KPNA1-mediated nuclear transport of LEF1 in a subset of satellite cells. Loss of LEF1 nuclear localization in a limited subset of KPNA1 KO satellite cells would lead to less activation of Wnt signaling but would not be sufficient to appreciably change myogenic lineage commitment in activated satellite cells. Among Wnt target genes, mRNA of Survivin [62], an inhibitor of apoptosis, was reduced in KPNA1 KO satellite cells, which correlates to the high levels of apoptosis observed in KPNA1 KO satellite cells. Although we identified several nuclear proteins (p27 and LEF1) that are mislocalized in KPNA1 KO satellite cells whose known functions correlate with the hyperproliferation and apoptosis phenotypes, likely the observed phenotype results from a net effect of multiple mislocalized nuclear proteins.
Conclusion
This work establishes cNLS nuclear import as a novel regulatory mechanism for controlling satellite cell proliferation and survival. Changes in the levels of individual KPNA paralogs in satellite cells during muscle regeneration could provide an additional level of cellular control by acting as a gate keeper for access of specific subsets of cargo proteins to the nucleus in addition to transcriptional and translational regulation of individual cargo proteins. Loss of KPNA1 led specifically to enhancement of cell proliferation and apoptosis without any effects on differentiation. Additional studies are require d to assess the role of other KPNA paralogs in satellite cells. Our results suggest that changes in the nuclear transport machinery with oxidative stress [79], aging [80], or disease [81] could lead to loss of satellite cells or changes in myogenic ability with adverse outcomes on skeletal muscle.
Supplementary Material
Acknowledgements
We thank Dr. M. Hall for initial experiments for the project; Dr. J.P. Canner for critical reading of the manuscript; C. Lam for technical assistance; A. Kotlar for technical assistance with bioinformatics analysis; Emory University School of Medicine Core Facility for Flow Cytometry and the Emory Integrated Proteomics Core for experimental assistance. This work was supported by National Institute of Health grants AR062483 (GP) and AR067645 (AC).
Biography
Hyo-Jung Choo: Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript
Alicia Cutler: Conception and design, financial support, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript
Grace Pavlath: Conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript
Footnotes
Disclosure of Potential Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- 1.Grounds MD, Garrett KL, Lai MC, et al. Identification of skeletal muscle precursor cells in vivo by use of MyoD1 and myogenin probes. Cell and tissue research. 1992;267:99–104. doi: 10.1007/BF00318695. [DOI] [PubMed] [Google Scholar]
- 2.Bischoff R. Proliferation of muscle satellite cells on intact myofibers in culture. Developmental biology. 1986;115:129–139. doi: 10.1016/0012-1606(86)90234-4. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff R. A satellite cell mitogen from crushed adult muscle. Developmental biology. 1986;115:140–147. doi: 10.1016/0012-1606(86)90235-6. [DOI] [PubMed] [Google Scholar]
- 4.Farina NH, Hausburg M, Betta ND, et al. A role for RNA post-transcriptional regulation in satellite cell activation. Skeletal muscle. 2012;2:21. doi: 10.1186/2044-5040-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alber F, Dokudovskaya S, Veenhoff LM, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 6.Reichelt R, Holzenburg A, Buhle EL, Jr., et al. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. The Journal of cell biology. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabut G, Lenart P, Ellenberg J. Dynamics of nuclear pore complex organization through the cell cycle. Current opinion in cell biology. 2004;16:314–321. doi: 10.1016/j.ceb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 9.Freitas N, Cunha C. Mechanisms and signals for the nuclear import of proteins. Current genomics. 2009;10:550–557. doi: 10.2174/138920209789503941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marfori M, Mynott A, Ellis JJ, et al. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochimica et biophysica acta. 2011;1813:1562–1577. doi: 10.1016/j.bbamcr.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 11.Lange A, Mills RE, Lange CJ, et al. Classical nuclear localization signals: definition, function, and interaction with importin alpha. The Journal of biological chemistry. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hood JK, Silver PA. Cse1p is required for export of Srp1p/importin-alpha from the nucleus in Saccharomyces cerevisiae. The Journal of biological chemistry. 1998;273:35142–35146. doi: 10.1074/jbc.273.52.35142. [DOI] [PubMed] [Google Scholar]
- 13.Kutay U, Bischoff FR, Kostka S, et al. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 14.Kohler M, Ansieau S, Prehn S, et al. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS letters. 1997;417:104–108. doi: 10.1016/s0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- 15.Kohler M, Speck C, Christiansen M, et al. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Molecular and cellular biology. 1999;19:7782–7791. doi: 10.1128/mcb.19.11.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsuji L, Takumi T, Imamoto N, et al. Identification of novel homologues of mouse importin alpha, the alpha subunit of the nuclear pore-targeting complex, and their tissue-specific expression. FEBS letters. 1997;416:30–34. doi: 10.1016/s0014-5793(97)01092-2. [DOI] [PubMed] [Google Scholar]
- 17.Kelley JB, Talley AM, Spencer A, et al. Karyopherin alpha7 (KPNA7), a divergent member of the importin alpha family of nuclear import receptors. BMC cell biology. 2010;11:63. doi: 10.1186/1471-2121-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blazek E, Meisterernst M. A functional proteomics approach for the detection of nuclear proteins based on derepressed importin alpha. Proteomics. 2006;6:2070–2078. doi: 10.1002/pmic.200500449. [DOI] [PubMed] [Google Scholar]
- 19.Fukumoto M, Sekimoto T, Yoneda Y. Proteomic Analysis of Importin alpha-interacting Proteins in Adult Mouse Brain. Cell Struct Funct. 2011;36:57–67. doi: 10.1247/csf.10026. [DOI] [PubMed] [Google Scholar]
- 20.Hu W, Jans DA. Efficiency of importin alpha/beta-mediated nuclear localization sequence recognition and nuclear import. Differential role of NTF2. The Journal of biological chemistry. 1999;274:15820–15827. doi: 10.1074/jbc.274.22.15820. [DOI] [PubMed] [Google Scholar]
- 21.Hodel AE, Harreman MT, Pulliam KF, et al. Nuclear localization signal receptor affinity correlates with in vivo localization in Saccharomyces cerevisiae. The Journal of biological chemistry. 2006;281:23545–23556. doi: 10.1074/jbc.M601718200. [DOI] [PubMed] [Google Scholar]
- 22.Riddick G, Macara IG. A systems analysis of importin-{alpha}-{beta} mediated nuclear protein import. The Journal of cell biology. 2005;168:1027–1038. doi: 10.1083/jcb.200409024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohler M, Fiebeler A, Hartwig M, et al. Differential expression of classical nuclear transport factors during cellular proliferation and differentiation. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2002;12:335–344. doi: 10.1159/000067903. [DOI] [PubMed] [Google Scholar]
- 24.Hall MN, Griffin CA, Simionescu A, et al. Distinct roles for classical nuclear import receptors in the growth of multinucleated muscle cells. Developmental biology. 2011;357:248–258. doi: 10.1016/j.ydbio.2011.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogarth CA, Calanni S, Jans DA, et al. Importin alpha mRNAs have distinct expression profiles during spermatogenesis. Developmental dynamics : an official publication of the American Association of Anatomists. 2006;235:253–262. doi: 10.1002/dvdy.20569. [DOI] [PubMed] [Google Scholar]
- 26.Shmidt T, Hampich F, Ridders M, et al. Normal brain development in importin -alpha5 deficient-mice. Nature cell biology. 2007;9:1337–1338. doi: 10.1038/ncb1207-1337. author reply 1339. [DOI] [PubMed] [Google Scholar]
- 27.Cheung TH, Quach NL, Charville GW, et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–528. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dammer EB, Duong DM, Diner I, et al. Neuron enriched nuclear proteome isolated from human brain. Journal of proteome research. 2013;12:3193–3206. doi: 10.1021/pr400246t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herskowitz JH, Seyfried NT, Duong DM, et al. Phosphoproteomic analysis reveals site-specific changes in GFAP and NDRG2 phosphorylation in frontotemporal lobar degeneration. Journal of proteome research. 2010;9:6368–6379. doi: 10.1021/pr100666c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dammer EB, Duong DM, Diner I, et al. Neuron Enriched Nuclear Proteome Isolated from Human Brain. Journal of Proteome Research. 2013;12:3193–3206. doi: 10.1021/pr400246t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 32.Xu P, Duong DM, Peng J. Systematical optimization of reverse-phase chromatography for shotgun proteomics. J Proteome Res. 2009;8:3944–3950. doi: 10.1021/pr900251d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seyfried NT, Gozal YM, Donovan LE, et al. Quantitative analysis of the detergent-insoluble brain proteome in frontotemporal lobar degeneration using SILAC internal standards. J Proteome Res. 2012;11:2721–2738. doi: 10.1021/pr2010814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donovan LE, Higginbotham L, Dammer EB, et al. Analysis of a membrane -enriched proteome from postmortem human brain tissue in Alzheimer's disease. Proteomics Clinical applications. 2012;6:201–211. doi: 10.1002/prca.201100068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gozal YM, Duong DM, Gearing M, et al. Proteomics analysis reveals novel components in the detergent-insoluble subproteome in Alzheimer's disease. J Proteome Res. 2009;8:5069–5079. doi: 10.1021/pr900474t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossi L, Challen GA, Sirin O, et al. Hematopoietic stem cell characterization and isolation. Methods in molecular biology. 2011;750:47–59. doi: 10.1007/978-1-61779-145-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jansen KM, Pavlath GK. Mannose receptor regulates myoblast motility and muscle growth. The Journal of cell biology. 2006;174:403–413. doi: 10.1083/jcb.200601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kafadar KA, Yi L, Ahmad Y, et al. Sca-1 expression is required for efficient remodeling of the extracellular matrix during skeletal muscle regeneration. Developmental biology. 2009;326:47–59. doi: 10.1016/j.ydbio.2008.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Pasut A, Oleynik P, Rudnicki MA. Isolation of muscle stem cells by fluorescence activated cell sorting cytometry. Methods in molecular biology. 2012;798:53–64. doi: 10.1007/978-1-61779-343-1_3. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen Ba AN, Pogoutse A, Provart N, et al. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC bioinformatics. 2009;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodgers JT, King KY, Brett JO, et al. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature. 2014;510:393–396. doi: 10.1038/nature13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moriyama T, Nagai M, Oka M, et al. Targeted disruption of one of the importin alpha family members leads to female functional incompetence in delivery. FEBS J. 2011;278:1561–1572. doi: 10.1111/j.1742-4658.2011.08079.x. [DOI] [PubMed] [Google Scholar]
- 44.Joe AW, Yi L, Natarajan A, et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nature cell biology. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fulle S, Sancilio S, Mancinelli R, et al. Dual role of the caspase enzymes in satellite cells from aged and young subjects. Cell death & disease. 2013;4:e955. doi: 10.1038/cddis.2013.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abou-Khalil R, Brack AS. Muscle stem cells and reversible quiescence: the role of sprouty. Cell cycle. 2010;9:2575–2580. doi: 10.4161/cc.9.13.12149. [DOI] [PubMed] [Google Scholar]
- 47.Orii N, Ganapathiraju MK. Wiki-pi: a web-server of annotated human protein-protein interactions to aid in discovery of protein function. PloS one. 2012;7:e49029. doi: 10.1371/journal.pone.0049029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larrea MD, Liang J, Da Silva T, et al. Phosphorylation of p27Kip1 regulates assembly and activation of cyclin D1-Cdk4. Molecular and cellular biology. 2008;28:6462–6472. doi: 10.1128/MCB.02300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zou P, Yoshihara H, Hosokawa K, et al. p57(Kip2) and p27(Kip1) cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell stem cell. 2011;9:247–261. doi: 10.1016/j.stem.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 50.Shin I, Rotty J, Wu FY, et al. Phosphorylation of p27Kip1 at Thr-157 interferes with its association with importin alpha during G1 and prevents nuclear re-entry. The Journal of biological chemistry. 2005;280:6055–6063. doi: 10.1074/jbc.M412367200. [DOI] [PubMed] [Google Scholar]
- 51.Sekimoto T, Fukumoto M, Yoneda Y. 14-3-3 suppresses the nuclear localization of threonine 157-phosphorylated p27(Kip1). The EMBO journal. 2004;23:1934–1942. doi: 10.1038/sj.emboj.7600198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Behrens J, Jerchow BA, Wurtele M, et al. Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 53.Kim YJ, Yoon SY, Kim JT, et al. NDRG2 expression decreases with tumor stages and regulates TCF/beta-catenin signaling in human colon carcinoma. Carcinogenesis. 2009;30:598–605. doi: 10.1093/carcin/bgp047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Markiewicz E, Tilgner K, Barker N, et al. The inner nuclear membrane protein emerin regulates beta-catenin activity by restricting its accumulation in the nucleus. The EMBO journal. 2006;25:3275–3285. doi: 10.1038/sj.emboj.7601230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Otto A, Schmidt C, Luke G, et al. Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. Journal of cell science. 2008;121:2939–2950. doi: 10.1242/jcs.026534. [DOI] [PubMed] [Google Scholar]
- 56.Block GJ, Narayanan D, Amell AM, et al. Wnt/beta-catenin signaling suppresses DUX4 expression and prevents apoptosis of FSHD muscle cells. Human molecular genetics. 2013;22:4661–4672. doi: 10.1093/hmg/ddt314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brack AS, Conboy MJ, Roy S, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 58.Abe K, Takeichi M. NMDA -receptor activation induces calpain-mediated beta-catenin cleavages for triggering gene expression. Neuron. 2007;53:387–397. doi: 10.1016/j.neuron.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 59.Wang J, Huo K, Ma L, et al. Toward an understanding of the protein interaction network of the human liver. Mol Syst Biol. 2011;7:536. doi: 10.1038/msb.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eastman Q, Grosschedl R. Regulation of LEF -1/TCF transcription factors by Wnt and other signals. Current opinion in cell biology. 1999;11:233–240. doi: 10.1016/s0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 61.Prieve MG, Guttridge KL, Munguia JE, et al. The nuclear localization signal of lymphoid enhancer factor-1 is recognized by two differentially expressed Srp1-nuclear localization sequence receptor proteins. The Journal of biological chemistry. 1996;271:7654–7658. doi: 10.1074/jbc.271.13.7654. [DOI] [PubMed] [Google Scholar]
- 62.Ma H, Nguyen C, Lee KS, et al. Differential roles for the coactivators CBP and p300 on TCF/beta-catenin-mediated survivin gene expression. Oncogene. 2005;24:3619–3631. doi: 10.1038/sj.onc.1208433. [DOI] [PubMed] [Google Scholar]
- 63.Yasuhara N, Shibazaki N, Tanaka S, et al. Triggering neural differentiation of ES cells by subtype switching of importin-alpha. Nature cell biology. 2007;9:72–79. doi: 10.1038/ncb1521. [DOI] [PubMed] [Google Scholar]
- 64.Giarre M, Torok I, Schmitt R, et al. Patterns of importin -alpha expression during Drosophila spermatogenesis. Journal of structural biology. 2002;140:279–290. doi: 10.1016/s1047-8477(02)00543-9. [DOI] [PubMed] [Google Scholar]
- 65.Young JC, Major AT, Miyamoto Y, et al. Distinct effects of importin alpha2 and alpha4 on Oct3/4 localization and expression in mouse embryonic stem cells. FASEB J. 2011;25:3958–3965. doi: 10.1096/fj.10-176941. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki T, Ishigami Y, Okada N, et al. Differentiation-associated alteration in gene expression of importins and exportins in human leukemia HL-60 cells. Biomedical research. 2008;29:141–145. doi: 10.2220/biomedres.29.141. [DOI] [PubMed] [Google Scholar]
- 67.Umegaki N, Tamai K, Nakano H, et al. Differential regulation of karyopherin alpha 2 express ion by TGF-beta1 and IFN-gamma in normal human epidermal keratinocytes: evident contribution of KPNA2 for nuclear translocation of IRF-1. The Journal of investigative dermatology. 2007;127:1456–1464. doi: 10.1038/sj.jid.5700716. [DOI] [PubMed] [Google Scholar]
- 68.Yasuhara N, Yamagishi R, Arai Y, et al. Importin alpha subtypes determine differential transcription factor localization in embryonic stem cells maintenance. Developmental cell. 2013;26:123–135. doi: 10.1016/j.devcel.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 69.Young JC, Ly-Huynh JD, Lescesen H, et al. The nuclear import factor importin alpha4 can protect against oxidative stress. Biochimica et biophysica acta. 2013;1833:2348–2356. doi: 10.1016/j.bbamcr.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 70.Hubert A, Henderson JM, Cowles MW, et al. A functional genomics screen identifies an Importin-alpha homolog as a regulator of stem cell function and tissue patterning during planarian regeneration. BMC genomics. 2015;16:769. doi: 10.1186/s12864-015-1979-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pumroy RA, Cingolani G. Diversification of importin-alpha isoforms in cellular trafficking and disease states. The Biochemical journal. 2015;466:13–28. doi: 10.1042/BJ20141186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hosoyama T, Nishijo K, Prajapati SI, et al. Rb1 gene inactivation expands satellite cell and postnatal myoblast pools. The Journal of biological chemistry. 2011;286:19556–19564. doi: 10.1074/jbc.M111.229542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hindi SM, Paul PK, Dahiya S, et al. Reciprocal interaction between TRAF6 and notch signaling regulates adult myofiber regeneration upon injury. Molecular and cellular biology. 2012;32:4833–4845. doi: 10.1128/MCB.00717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kitamoto T, Hanaoka K. Notch3 null mutation in mice causes muscle hyperplasia by repetitive muscle regeneration. Stem cells. 2010;28:2205–2216. doi: 10.1002/stem.547. [DOI] [PubMed] [Google Scholar]
- 75.Hong P, Chen K, Huang B, et al. HEXIM1 controls satellite cell expansion after injury to regulate skeletal muscle regeneration. The Journal of clinical investigation. 2012;122:3873–3887. doi: 10.1172/JCI62818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi H, Verma M, Zhang L, et al. Improved regenerative myogenesis and muscular dystrophy in mice lacking Mkp5. The Journal of clinical investigation. 2013;123:2064–2077. doi: 10.1172/JCI64375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin J, Della-Fera MA, Li C, et al. P27 knockout mice: reduced myostatin in muscle and altered adipogenesis. Biochemical and biophysical research communications. 2003;300:938–942. doi: 10.1016/s0006-291x(02)02949-2. [DOI] [PubMed] [Google Scholar]
- 78.Jones AE, Price FD, Le Grand F, et al. Wnt/beta-catenin controls follistatin signalling to regulate satellite cell myogenic potential. Skeletal muscle. 2015;5:14. doi: 10.1186/s13395-015-0038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stochaj U, Rassadi R, Chiu J. Stress-mediated inhibition of the classical nuclear protein import pathway and nuclear accumulation of the small GTPase Gsp1p. FASEB J. 2000;14:2130–2132. doi: 10.1096/fj.99-0751fje. [DOI] [PubMed] [Google Scholar]
- 80.Pujol G, Soderqvist H, Radu A. Age-associated reduction of nuclear protein import in human fibroblasts. Biochemical and biophysical research communications. 2002;294:354–358. doi: 10.1016/S0006-291X(02)00492-8. [DOI] [PubMed] [Google Scholar]
- 81.Lee HG, Ueda M, Miyamoto Y, et al. Aberrant localization of importin alpha1 in hippocampal neurons in Alzheimer disease. Brain research. 2006;1124:1–4. doi: 10.1016/j.brainres.2006.09.084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.