Abstract
Background & Aims
The transcription factor atonal homolog 1 (ATOH1) controls the fate of intestinal progenitors downstream of the Notch signaling pathway. Intestinal progenitors that escape Notch activation express high levels of ATOH1 and commit to a secretory lineage fate, implicating ATOH1 as a gatekeeper for differentiation of intestinal epithelial cells. Although some transcription factors downstream of ATOH1, such as SPDEF, have been identified to specify differentiation and maturation of specific cell types, the bona fide transcriptional targets of ATOH1 still largely are unknown. Here, we aimed to identify ATOH1 targets and to identify transcription factors that are likely to co-regulate gene expression with ATOH1.
Methods
We used a combination of chromatin immunoprecipitation and messenger RNA–based high-throughput sequencing (ChIP-seq and RNA-seq), together with cell sorting and transgenic mice, to identify direct targets of ATOH1, and establish the epistatic relationship between ATOH1 and SPDEF.
Results
By using unbiased genome-wide approaches, we identified more than 700 genes as ATOH1 transcriptional targets in adult small intestine and colon. Ontology analysis indicated that ATOH1 directly regulates genes involved in specification and function of secretory cells. De novo motif analysis of ATOH1 targets identified SPDEF as a putative transcriptional co-regulator of ATOH1. Functional epistasis experiments in transgenic mice show that SPDEF amplifies ATOH1-dependent transcription but cannot independently initiate transcription of ATOH1 target genes.
Conclusions
This study unveils the direct targets of ATOH1 in the adult intestines and illuminates the transcriptional events that initiate the specification and function of intestinal secretory lineages.
Keywords: ATOH1, SPDEF, Transcription, Intestinal Epithelium, Villin-creER, TRE-Spdef, Atoh1GFP, Atoh1Flag
Abbreviations used in this paper: ATOH1, atonal homolog 1; ChIP, chromatin immunoprecipitation; ChIP-seq, chromatin immunoprecipitation sequencing; CRC, colorectal cancer; DBZ, dibenzazepine; FACS, fluorescence-activated cell sorting; FDR, false-discovery rate; Gfi1, growth factor independent 1; GFP, green fluorescent protein; GO, gene ontology; ISC, intestinal stem cell; mRNA, messenger RNA; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; QES, Q-enrichment-score; RT-qPCR, reverse-transcription quantitative polymerase chain reaction; Spdef, SAM pointed domain containing ETS transcription factor; TSS, transcription start site
See editorial on page 2.
Summary.
The transcription factor ATOH1 is critical for Notch-mediated differentiation and maturation of intestinal secretory cells. Here we identify direct targets of ATOH1 in mouse small intestine and colon.
The adult intestinal epithelium proliferates rapidly with average cellular lifespans of approximately 5–7 days. To maintain epithelial integrity and perform its major function of nutrient digestion and absorption, intestinal stem cells (ISCs) located at the base of crypts of Lieberkühn must self-renew and produce transit-amplifying cells, which subsequently differentiate into 1 of 2 cell classes: absorptive lineage cells, including enterocytes and colonocytes; and secretory lineage cells, including mucus-secreting goblet cells, hormone-secreting enteroendocrine cells, and antimicrobial peptide-secreting Paneth cells.1, 2, 3 Under physiological conditions, signaling pathways, such as Notch and Wnt, modulate homeostasis and differentiation of the intestinal epithelium, directing ISCs/progenitors toward either the absorptive or secretory fate by controlling the expression of a downstream transcriptional network.4, 5 Dysregulated ISC proliferation or aberrant differentiation may cause gastrointestinal diseases, such as inflammatory bowel disease and intestinal cancer.5, 6
Canonical Notch signaling relies on direct cell–cell contact and plays an important role in modulating homeostasis and differentiation of the intestinal epithelium. In the intestines, Notch signaling controls the fate of ISCs/progenitors by regulating the expression of the basic helix-loop-helix transcription factor atonal homolog 1 (ATOH1).5 Previous studies have suggested that ATOH1 is required for the differentiation of all secretory cells.7 Germ-line Atoh1 deletion causes mice to die shortly after birth and fail to form any secretory cells without affecting enterocytes.7 Consistent with these observations, conditional deletion of Atoh1 in the adult intestinal epithelium results in the loss of all secretory cells.8 In contrast, overexpression of ATOH1 directs progenitor cells to the secretory cell fate in the embryonic intestine.9 Previous studies have indicated that pharmacologic inhibition of Notch signaling using γ-secretase inhibitors or specific antibodies blocking the Notch receptors results in loss of proliferative progenitor cells and secretory cell hyperplasia.10, 11, 12 However, Atoh1-deficient intestines fail to respond to Notch inhibition, indicating that the primary role of Notch is to regulate the expression of Atoh1, and in doing so control secretory vs absorptive cells fate.13, 14, 15 Consistent with the concept, a recent study suggested that ATOH1 controls Notch-medicated lateral inhibition in the adult intestinal epithelium.16 These results indicate that ATOH1 is a critical gatekeeper for the program of Notch-mediated differentiation and cell fate determination of intestinal epithelial cells. Although previous studies have suggested that some transcription factors, such as SAM pointed domain containing ETS transcription factor (Spdef) and growth factor independent 1 (Gfi1), are downstream of ATOH1 and are important for differentiation of specific secretory cell types,17, 18, 19 the bona fide targets of endogenous ATOH1 at the genome-wide level in the adult intestine still largely are unknown.
To better understand the molecular functions of ATOH1 in vivo, we used a combination of chromatin immunoprecipitation (ChIP) and RNA-based, high-throughput sequencing techniques to identify direct transcriptional targets of ATOH1 in ileal and colonic crypts. In addition, our data unveiled a novel molecular mechanism whereby SPDEF functions as a transcriptional co-regulator of ATOH1, amplifying ATOH1-dependent transcription of a subset of secretory genes. This study provides novel insight toward understanding cell fate decisions within the intestines.
Materials and Methods
Animals
VilCreERT2; Fabp1Cre; Atoh1fl/fl; Rosa26 LSL-rtta-ires-EGFP; TRE-Spdef; Spdef null; Atoh1GFP/GFP; and Atoh1Flag/Flag mice have been described previously.8, 18, 20, 21, 22, 23, 24 To achieve deletion of Atoh1 from intestinal epithelium, Atoh1fl/fl; VilCreERT2 mice and littermate controls were given an intraperitoneal injection of 1 mg/mouse tamoxifen (Sigma, St. Louis, MO) dissolved in corn oil for 3 consecutive days. Animals were killed 5 days after the first injection. To achieve SPDEF induction, Fabp1Cre; Atoh1fl/fl; Rosa26 LSL-rtta-ires-EGFP; TRE-Spdef mice, and littermate controls were given 2 mg/mL tetracycline in water for 5 consecutive days. To achieve Notch inhibition, mice were treated either with vehicle or GSI-20 (also called dibenzazepine [DBZ]; EMD–Calbiochem, Darmstadt, Germany) at 15 μmol/L/kg once a day for 5 days. All mouse studies were approved by the Institutional Animal Care and Use Committee.
Crypt Isolation
Intestinal crypts were prepared as previously described.25 Entire colons and 6–7 cm distal small intestine were dissected out and flushed with ice-cold phosphate-buffered saline (PBS) with 5 mmol/L phenylmethylsulfonyl fluoride. Intestines were opened lengthwise and cut into 1-cm pieces. Tissues were incubated with shaking buffer (25 mmol/L EDTA, protease inhibitor cocktail; Calbiochem) at 4°C for 30 minutes by gentle shaking. Shaking buffer was replaced by ice-cold Ca2+/Mg2+-free Dulbecco’s PBS followed by vigorous shaking for approximately 8–10 minutes to generate disassociated crypts. For the colon, it takes 15 minutes to disassociate crypts. Intestinal crypts were isolated by filtering through a 70-μm cell strainer (BD Falcon) for small intestinal crypts and a 100-μm cell strainer (BD Falcon, Tewksbury, MA) for colonic crypts, and then spun down at 150g for 10 minutes.
Cell Culture
Human colorectal cancer cell line HCT was grown in RPMI1640 (10-040-CV; Corning, New York, NY) supplemented with 10% fetal bovine serum (S1200-500; BioExpress, Kaysville, UT), penicillin, and streptomycin (17-602E; Lonza, Basel, Switzerland).
Plasmids and DNA Transfection
Expression plasmid of ATOH1-GFP was a gift from Dr Tiemo Klisch (Baylor College of Medicine).20 HCT116 green fluorescent protein (GFP) cells were transfected by using Lipofectamine 2000 (11668-019; Invitrogen, Waltham, MA) following the manufacturer’s instructions.
Chromatin Immunoprecipitation
Crypts and transfected cells were used in chromatin immunoprecipitation (ChIP) experiments with antibodies against GFP (NB600-303; Novus, Littleton, CO), Flag M2 (F1804; Sigma), H3k27Ac (ab4729; Abcam, Cambridge, MA), or H3K27me3 (ab6002; Abcam). For each ChIP sample, 2–3 μg of antibodies were used to bind to 10 μL Protein G Dynabeads (100-03D; Invitrogen) following the manufacturer’s instructions. Samples from either crypts or 5–10 × 106 HCT116 cells transfected with ATOH1-GFP were cross-linked in 1% formaldehyde (15710; Electron Microscopy Sciences, Hatfield, PA) in cross-linking buffer (50 mmol/L HEPES pH 8.0, 1 mmol/L EDTA pH 8.0, 1 mmol/L ethylene glycol-bis[β-aminoethyl ether]-N,N,N′,N′-tetraacetic acid pH 8.0, 100 mmol/L NaCl, RPMI1640) at room temperature for 30 minutes and then quenched by adding glycine to a final concentration of 135 mmol/L on ice for 5 minutes. Cross-linked cells were washed twice with ice-cold PBS and stored at -80°C before sonication. Chromatin was sheared to 300- to 1000-bp fragments in 1 mL ice-cold sonication buffer (10 mmol/L Tris-HCl pH 8.0, 1 mmol/L EDTA pH 8.0, 1 mmol/L ethylene glycol-bis[β-aminoethyl ether]-N,N,N′,N′-tetraacetic acid pH 8.0, supplemented with a protease inhibitor cocktail; 539134; Calbiochem), using a 250D Sonifier Ultrasonic Processor Cell Disruptor (Branson, Danbury, CT) with a one-eighth inch microtip (50% power output, interval 1-second on/1-second off, for a total of 24 minutes). Sarkosyl was added to a final concentration of 0.5% and the sheared chromatin was incubated at room temperature for 10 minutes and then spun down to remove debris. For immunoprecipitation, 500 μL sheared chromatin was mixed with 150 μL binding buffer (440 mmol/L NaCl, 0.44% sodium deoxycholate, 4.4% Triton X-100) and incubated with 10 μL antibody-bound protein G Dynabeads at 4°C overnight. ChIP samples were washed in washing buffer (1% Nonidet P-40, 1% sodium deoxycholate, 1 mmol/L EDTA pH 8.0, 50 mmol/L HEPES pH 8.0, 500 mmol/L LiCl) 5 times and then eluted in elution buffer (50 mmol/L Tris-HCl pH 8.0, 10 mmol/L EDTA pH 8.0, 1% sodium dodecyl sulfate) at 65°C for 15 minutes. Both ChIP and input samples were incubated at 65°C overnight to reverse formaldehyde cross-linking. DNA was purified by phenol-chloroform extraction. Precipitated DNA fragments were used for ChIP sequencing (ChIP-seq) or polymerase chain reaction (PCR). The ChIP-seq library was made following the instructions of the NEBNext ChIP-Seq Library Prep Master Mix Set (E6240; New England Biolabs, Ipswich, MA). The primers used for ChIP-PCR are listed in Supplementary Table 1.
RNA Preparation
Sorted ATOH1-GFP–positive cells and purified crypts from either Atoh1 deletion or littermate control mice were collected immediately in TRIzol reagent (Invitrogen). RNA was isolated following the manufacture’s instructions and subsequently purified with the RNeasy kit (Qiagen), using on-column DNAse digestion (Qiagen, Hilden, Germany). RNA quality controls were performed by the Gene Expression Core at Cincinnati Children’s Hospital Medical Center using an Agilent (Santa Clara, CA) Bioanalyzer nanochip. The RNA integrity number of the RNA samples for RNA-seq was at least 8.8.
Reverse-Transcription and Real-Time PCR
A total of 1 μg RNA was used to synthesize complementary DNA using Superscript III First Strand Synthesis System (Invitrogen) following the manufacturer’s instructions. Quantitative PCR was performed with Brilliant III Ultra Fast SYBR Green Master Mix (Agilent Technologies) using the primers listed in Supplementary Table 2.
Tissue Staining
Intestinal tissues were fixed in 4% paraformaldehyde in PBS at 4°C overnight, transferred to 70% ethanol, paraffin-embedded, and sectioned at 5-μm thickness. Paraffin-embedded sections were deparaffinized and rehydrated before staining. For immunofluorescence, antigen retrieval was achieved in sodium citrate buffer (10 mmol/L sodium citrate pH 6.0). The sections were blocked in 4% normal donkey serum in PBS at room temperature for 1 hour. Primary antibodies against GFP (1:1000; Abcam) and chromagranin A (1:5000; Immuostar, Hudson, WI), mucin 2 (1:1000; Santa Cruz, Dallas, TX), or lysozyme (1:5000; Zymed Laboratories, San Francisco, CA) were co-incubated on the sections in blocking buffer (4% normal donkey serum in PBS) at 4°C overnight. After washing 3 times by PBS, donkey anti-goat–Alexa 488 and donkey anti-rabbit–Alexa 594 secondary antibodies (1:200; Invitrogen) were incubated on the sections at room temperature for 1 hour. All sections were washed 3 times by PBS and mounted with Vectashield medium with 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA).
In Situ Hybridization
Fresh intestinal tissues were harvested from mice and lightly fixed by 4% paraformaldehyde on ice for 15 minutes. Fixed tissues were cryoprotected with 30% sucrose in PBS and then embedded in optimal cutting temperature compound (OCT). In situ hybridization staining was performed as previously described.26 In situ hybridization staining was performed by the RNA In Situ Hybridization Core at Baylor College of Medicine.
Fluorescence-Activated Cell Sorting
Isolated crypts were dissociated as previously described.25 Briefly, crypts were dissociated with TrypLE express (Invitrogen) supplemented with 10 μmol/L Y-27632 and 1 mmol/L N-acetylcysteine (Sigma-Aldrich) for 5 minutes at 37°C. Cell clumps were removed using a 35-μm cell strainer (Fisher Scientific, Waltham, MA) and the flow-through was pelleted at 500 × g at 4°C for 5 minutes. Cell pellets were resuspended in 5% bovine serum albumin, 1 mmol/L EDTA, and 10 μmol/L Y27632 (Sigma-Aldrich) in PBS at 2–5 × 106 cells/mL. 7-AAD was added 20 minutes before fluorescence-activated cell sorting (FACS) to evaluate cell viability. A FACSAria II equipped with a 100-μm nozzle was used (BD Biosciences, San Jose, CA). GFP-positive and 7-AAD–negative single cells were sorted into 500 μL TRIzol reagent (Invitrogen) for RNA sequencing.
RNA-Seq Data Preprocessing and Analysis
Total RNA from 14 samples from mouse colon and ileum (2 biological replicates of wild-type and Atoh1-mutant crypts, 3 biological replicates of GFP+ cells) were prepared for RNA sequencing using the Illumina HiSeq 2000 with single-end, 50-bp reads (Illumina, San Diego, CA). For each sample, 14–23 million of 50-bp, single-end reads were generated. The raw reads were aligned to the Mus musculus genome (Ensembl mm9) using TopHat v1.4.1 (http://tophat.cbcb.umd.edu/) with default parameters.27 The mappability for each sample was greater than 80%. To measure the expression level from aligned sequence reads for differential gene analysis, we used the free Python program HTSeq.28 The htseq-count function of HTSeq (http://www-huber.embl.de/users/anders/HTSeq/) allowed us to quantify the number of aligned reads that align with the exons of the gene (union of all the exons of the gene). The read counts obtained were analogous to the expression level of the gene. By using the raw counts, differential gene analysis was performed using the DESeq package in the R environment. DESeq includes functions to test for gene expression changes between samples in different conditions by the use of the negative binomial distribution and a shrinkage estimator for the distribution’s variance.29 The nbinomTest function of DESeq was used to test if each gene was expressed differentially. The significance of the observed changes are indicated by the P value, and the false-discovery rate (FDR) reported in this study is the P values adjusted for multiple testing with the Benjamini–Hochberg procedure implemented within DESeq. Heatmaps of gene expressions were plotted using the heatmap.2 function implemented in the gplots package in R.
ChIP-Seq Data Preprocessing and Analysis
ChIP samples from mouse colon and ileum were sequenced using an Illumina HiSeq 2000. Fourteen samples were prepared: H3k27me3, H3k27Ac, 2 replicates of ATOH1-GFP, and 3 replicates of input for each tissue. Each sample was sequenced at a depth of 9–18 million, 50-bp, single-end reads. Reads were trimmed from both ends before mapping to the reference genome. The trimmed reads were first mapped to the Mus musculus genome (Ensembl mm9) using Bowtie2 with the preset of the very-sensitive setting (specific parameters are as follows: -D 20 -R 3 -N 0 -L 20 -i S,1,0.50).30 The mappability for each sample was greater than 75% except for the GFP samples. By using the mapping files, regions with enriched ATOH1 binding were identified using well-established, peak-calling software, Model-based Analysis of ChIP-Seq (MACS).31 The input samples were used as the control for calling peaks from the ATOH1, histone methylation, or histone acetylation data sets. Peaks (ATOH1 and histone-bound regions) then were annotated using the mouse mm9 gene model.32 In the binding site comparison analysis, we used deeptools to generate peak-based correlation heatmaps and scatterplots.33 ATOH1 ChIP-Seq data from the cerebellum were obtained from GEO DataSets (GSE22111). First, all aligned ChIP-seq data in bam format were ratio-normalized to their respective inputs and converted to bigwig format using the bamCompare module from deeptools. Next, the whole genome was binned into 10-kb windows and respective coverage was computed across the 3 different tissues (ileum, colon, and cerebellum), using the computeMatrix module of deeptools. PlotCorrelation was used to compute genome level correlations and to generate scatterplots. To directly compare ileal vs colonic ATOH1 binding sites (Figure 3C), log2-normalized enrichment of binding regions were plotted on both axes, which was defined using the following formula. This is analogous to the average peak height (RPM) analyzed as described previously.34
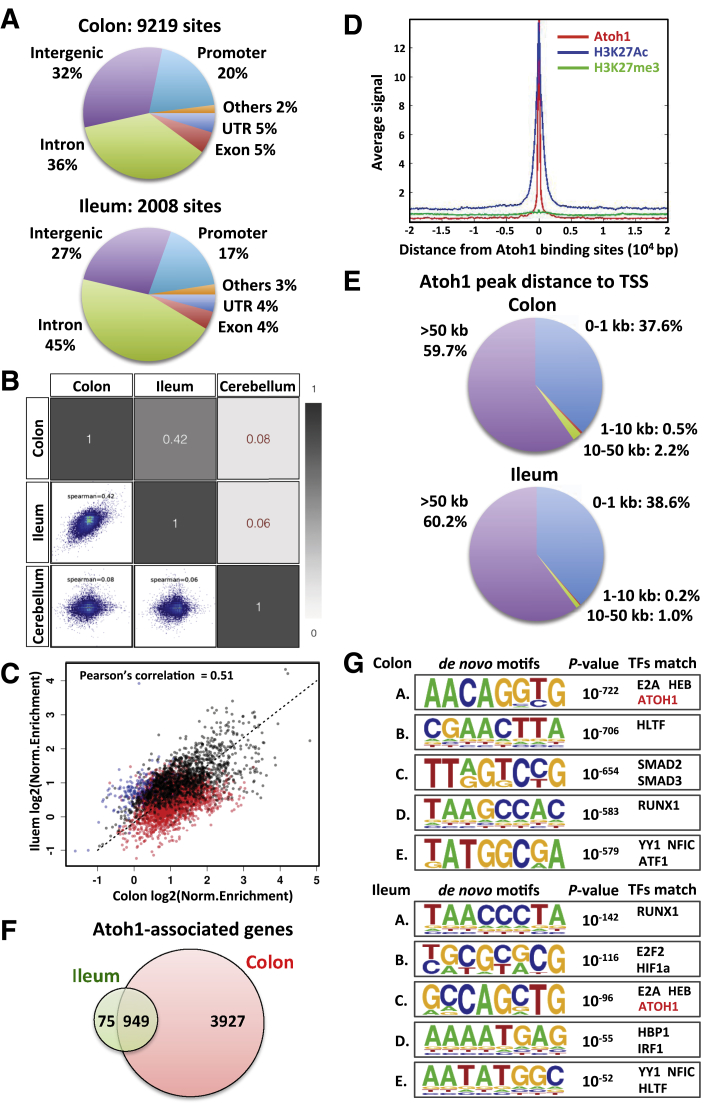
Figure 3.
ATOH1 genomic binding regions in ileum and colon. (A) Genome distribution of ATOH1 ChIP-seq peaks. (B) Comparison of colonic ATOH1, H3K27Ac, and H3K27me3 signals generated from ChIP-seq fragment counts in the 40 kb surrounding ATOH1 peaks. (C) Genome-wide ATOH1 binding sites were compared between ileum, colon, and cerebellum. Scatterplots show the distribution of enrichment scores for the entire genome separated into 10-kb segments. Numbers in the figure indicate Spearman correlation coefficients. (D) Comparison of enrichment scores in the regions with significant enrichment in ATOH1-bound chromatin from either ileum or colon. Black points indicate regions with significant peaks from both the ileum and colon, red points are significant peaks from the colon but not ileum, and blue points are significant peaks from the ileum only. (E) Distribution of ATOH1 ChIP-seq peaks according to the distance from TSS. (F) Genes that have ATOH1 binding sites within 20 kb of the TSS are defined as ATOH1-associated genes. Venn diagram indicates overlap of ATOH1-associated genes in the ileum and colon. (G) Logos for the top motifs enriched in ATOH1-binding sites are identified by HOMER de novo motif analysis. P values are for motif enrichment. Transcription factors matched to each motif were listed. TFs, transcription factors.
ATOH1 De Novo Motif Analysis
The hypergeometric optimization of motif enrichment (HOMER) software suite was used to identify DNA motifs enriched in the ChIP-seq data sets. First, sites bound by ATOH1 were subjected to de novo motif identification using findMotifsGenome.pl within HOMER. Second, de novo motif identification was performed on ATOH1 binding sites within the colon ATOH1 targetome. Significantly enriched motifs were matched to the most similar transcription factor–binding motifs from the JASPAR 2014 database. FIMO was used to retrieve genes with SPDEF binding motifs from the ATOH1 targetome.
Gene Ontology Analysis
Gene ontology (GO) analysis was performed with Database for Annotation, Visualization, and Integrated Discovery (DAVID; available: http://david.abcc.ncifcrf.gov/) using the ATOH1 targetome genes lists (Figure 5) to identify the biological processes and molecular functions in which the input gene lists are enriched. The -log10(FDR) of the enriched functions were plotted to indicate the significance of the enrichment of each function.
Figure 5.
Direct ATOH1 transcriptional targets in adult intestines. (A) Overlap of ATOH1-associated genes from ChIP-seq and genes significantly enriched in ATOH1-positive cells from RNA-seq identifies the ATOH1 targetome, a list of putative direct transcriptional ATOH1 targets in the ileum and colon. (B) GO analysis using DAVID identified ontology terms associated significantly with ATOH1 targetome. (C) Venn diagram indicates overlap of ATOH1-bound genes from ChIP-seq and genes decreased significantly in ATOH1-positive cells from RNA-seq. (D) GO analysis using DAVID identified ontology terms. BMP, bone morphogenetic proteins; GTPase, guanosine triphosphatase.
Results
ATOH1 Transcriptional Profile in the Adult Distal Small Intestinal and Colon Crypts
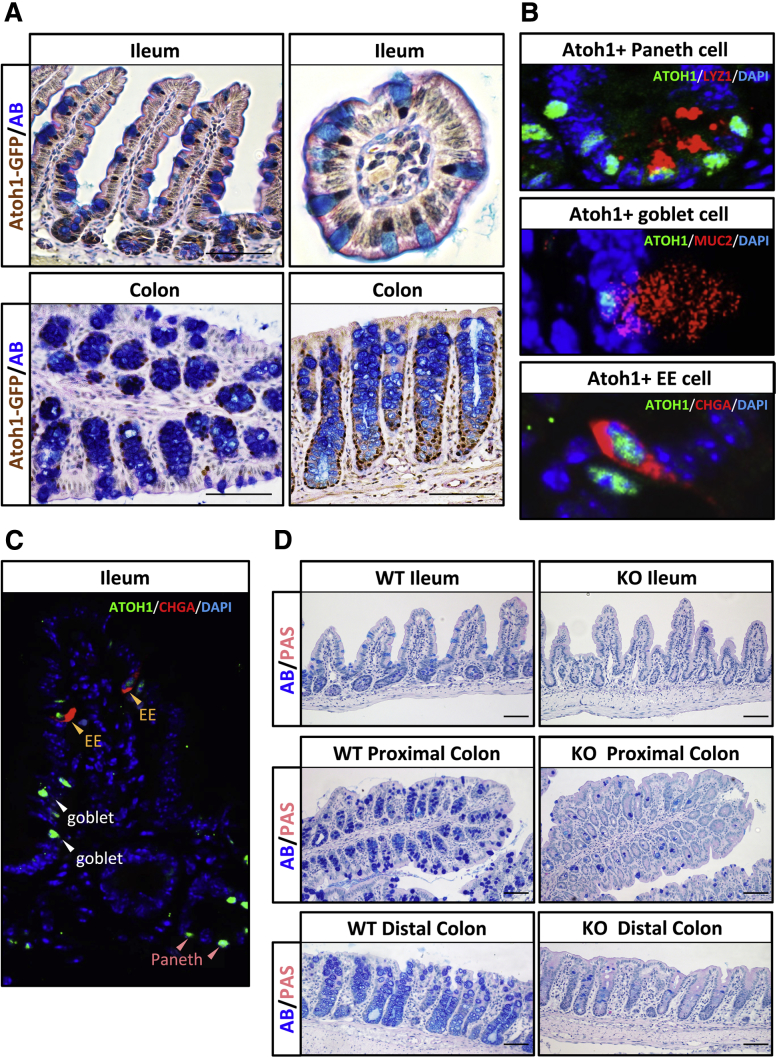
Previous studies have shown that ATOH1 is required for the differentiation of secretory cell lineages in the intestines.7 Conditional deletion of Atoh1 in the adult intestine confirmed that ATOH1 is expressed in and essential for the formation of all secretory cells.8 Consistent with these observations, Alcian blue and periodic acid–Schiff staining indicated that mice with an ATOH1–GFP fusion protein inserted into the Atoh1 locus (Atoh1GFP/GFP) express ATOH1–GFP in goblet and Paneth cells (Figure 1A). Specifically, ATOH1–GFP expression is co-localized with all secretory cells, including mucin 2–positive goblet cells, lysozyme 1–positive Paneth cells, as well as chromogranin A–positive enteroendocrine cells (Figure 1B and C).35
Figure 1.
ATOH1 is required for all secretory lineages in ileum and colon. (A) Immunohistochemistry combined with Alcian blue (AB) and periodic acid–Schiff (PAS) staining of the ileum and colon from transgenic mice Atoh1GFP/GFP indicates that endogenous ATOH1 is expressed in goblet and Paneth cells. Scale bars: 100 μm. (B) Immunofluorescent analysis of ileum and colon from Atoh1GFP/GFP mice indicates that endogenous ATOH1 is expressed in all secretory lineages. Goblet cells were labeled by mucin 2 (MUC2). Paneth cells were labeled by lysozyme 1 (LYZ1). Enteroendocrine cells were labeled by chromogranin A (CHGA). (C) Immunofluorescence analysis indicates the expression level of ATOH1 is lower in enteroendocrine (EE) cells compared with goblet and Paneth cells in Atoh1GFP/GFP mice. (D) Conditional deletion of ATOH1 in the intestinal epithelium was achieved by using Atoh1lox/lox;VilCreERT2 mice. After tamoxifen injection for 3 consecutive days, Alcian blue staining showed that the secretory lineages were nearly absent in both the ileum and colon compared with wild-type control Atoh1lox/WT;VilCreERT2 mice. Scale bars: 100 μm. DAPI, 4′,6-diamidino-2-phenylindole; KO, knockout; WT, wild type.
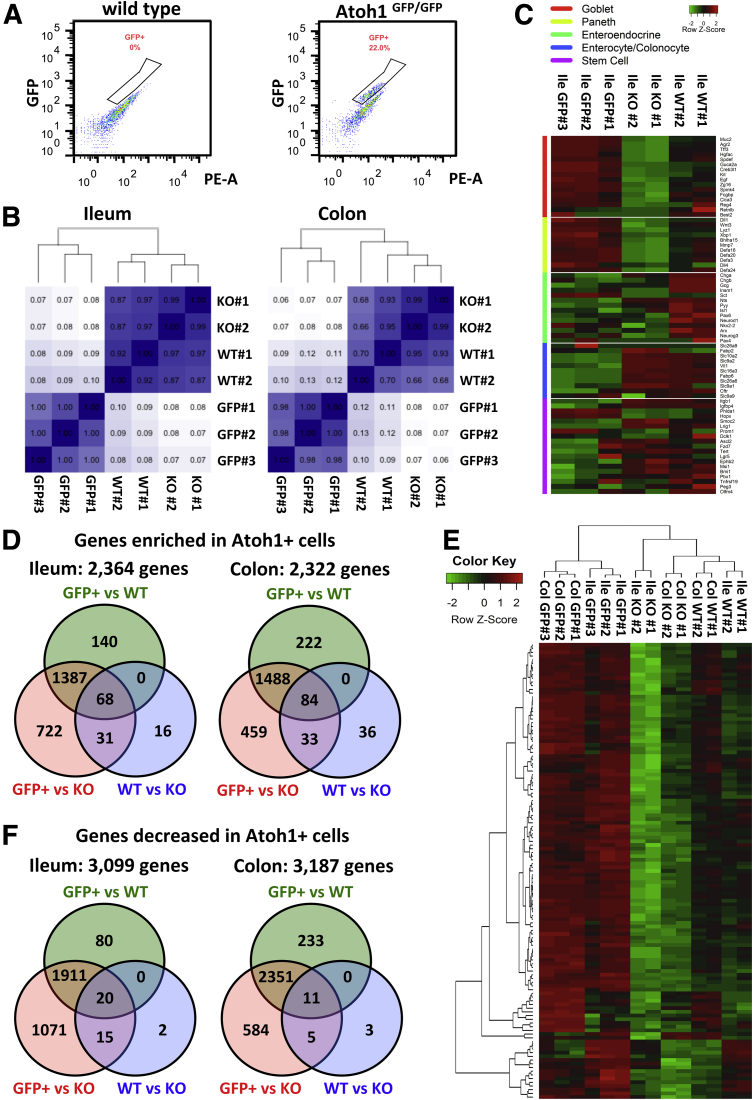
To define ATOH1-associated transcripts in adult intestines, we first generated 3 messenger RNA (mRNA) expression profiles by RNA-seq of the following: (1) wild-type crypts, (2) Atoh1 deletion crypts, and (3) purified ATOH1-positive cells. We isolated Atoh1 deletion and littermate wild-type crypts from 6- to 8-week old Atoh1lox/lox;VilCreERT2 and Atoh1lox/WT;VilCreERT2 mice, respectively. After tamoxifen injection for 3 consecutive days, secretory lineages were nearly absent throughout the entire intestinal epithelium (Figure 1D). ATOH1-positive cells were isolated by flow cytometry of 7AAD-negative (live), GFP-positive cells from either ileal or colonic crypts of Atoh1GFP/GFP mice (Figure 2A). RNA sequencing was performed on the Illumina Hi-Seq 2000 with single-end, 50-bp reads. Three purified ATOH1–GFP–positive, 2 Atoh1 wild-type, and 2 Atoh1 deletion samples were collected from the ileum and colon of independent animals with corresponding genotypes (total, 14 samples). By using hierarchal clustering analysis, we observed that samples generated from independent experiments for each group clustered together, indicating that the RNA-seq data were highly reproducible and reliable (Figure 2B). To evaluate whether these RNA-seq data represent a secretory cell–associated gene signature, we assessed the expression of genes characteristic of individual cell types in the intestine. We selected 71 genes representing 5 different intestinal cell types, and created a heat map of gene expression from our RNA-seq data sets (Figure 2C). As expected, compared with wild-type crypts, the expression of goblet and Paneth cell genes was enriched in purified ATOH1-positive cells, but decreased in Atoh1 deletion crypts (Figure 2C). Of note, compared with wild-type crypts, we did not observe significant enrichment of enteroendocrine genes in isolated ATOH1-positive cells. However, the expression of enteroendocrine genes was decreased in Atoh1 deletion crypts, indicating that although enteroendocrine cells require ATOH1 for their formation, they were not efficiently purified during FACS of ATOH1–GFP cells, likely owing to their low level of ATOH1–GFP (Figure 1C). As expected, ATOH1-positive cells expressed lower absorptive enterocyte/colonocyte and intestinal stem cell genes (Figure 2C). Finally, to identify genes that are regulated by ATOH1, we compared these 3 expression groups with each other and identified genes with at least a 1.5-fold difference in expression level with an adjusted P value less than .05. We identified 2322 genes in the colon and 2364 genes in the ileum that were enriched in ATOH1-positive cells (Figure 2D, Supplementary Table 3). Hierarchal clustering analysis for the intersection area (68 genes in the ileum and 84 genes in the colon enriched in ATOH1-expressing cells) verified the sample-to-sample reproducibility of the transcripts we identified (Figure 2E, Supplementary Table 4). On the other hand, we identified 3187 genes in the colon and 3099 genes in the ileum that were expressed at a lower level in ATOH1–GFP–positive cells (Figure 2F, Supplementary Table 5). Taken together, we generated ATOH1-associated transcripts in adult small and large intestines under homeostatic conditions.
Figure 2.
Transcriptional profile of ATOH1-positive cells. (A) Live ATOH1-positive cells were sorted by flow cytometry from either ileal or colonic crypts of Atoh1GFP/GFP mice for RNA-seq. (B) Hierarchal clustering analysis of independent RNA-seq samples generated from ATOH1-positive cell sorting, wild-type crypts, and Atoh1 deletion crypts. Numbers in the figure indicate Pearson correlation coefficients. (C) Heat map of gene expression of individual cell type markers in the intestine. (D) Venn diagram indicates overlap of genes that are enriched significantly at least 1.5-fold in each group. (E) Heat map of mRNA expression of genes we identified shows a significant enrichment in ATOH1-positive cells in the ileum and colon. (F) Venn diagram indicates overlap of genes that are decreased significantly at least 1.5-fold in each group. KO, knockout; WT, wild type
ATOH1 Genomic Binding Sites in Adult Ileal and Colonic Crypts
To identify targets of ATOH1 binding in the adult intestinal epithelium, we performed chromatin immunoprecipitation-sequencing (ChIP-seq). Ileal or colonic crypts were isolated from 10- to 12-week-old adult Atoh1GFP/GFP mice (which express a functional ATOH1-GFP fusion protein and are phenotypically normal35) followed by ATOH1, H3K27Ac, and H3K27me3 ChIP-Seq. We prepared duplicate ATOH1 ChIP-seq libraries from 2 independent experiments for both tissues. Analysis of pooled data identified 2008 ATOH1 binding sites in the ileum and 9219 ATOH1 binding sites in the colon (FDR < 1e-10) across the entire genome (Figure 3A, Supplementary Tables 6 and 7). Next, we performed Q enrichment score (QES) analysis to verify the quality of these ATOH1 binding sites.36 Compared with the reference values for the quality metrics generated from 392 data sets from ENCODE (https://www.encodeproject.org/), the QES from our ATOH1 ChIP-seq (QES, 0.24 in colon; QES, 0.16 in ileum) was ranked at a level between moderate high to very high (http://charite.github.io/Q/tutorial.html#output_of_q), suggesting a high quality of these ATOH1 peaks (Supplementary Table 8). To study the ATOH1 binding patterns between tissues, we first assessed the co-occurrence of all ATOH1 binding sites in the ileum and colon; for comparison, we included ATOH1 ChIP-seq results from the developing cerebellum20 in this analysis (Figure 3B). Our results showed that ATOH1 binding sites were similar between the ileum and colon (Spearman correlation coefficient, 0.42) as compared with cerebellum (correlation coefficient, 0.06–0.08). Next, we restricted our comparison with sites that were enriched significantly in either ileum or colon (shown in Figure 3A), which showed stronger correlation of co-occurring ATOH1 binding sites (Figure 3C). Although a fair portion of the ATOH1-bound peaks called from colon were not considered significant peaks in ileum (Figure 3C, red points), the enrichments were correlated highly (ie aligned with the x = y line, shown as a dotted line), indicating strong enrichment for ATOH1 at similar sites in the small intestine and colon. For each tissue, the distribution of peaks across functional domains in the genome was analyzed (Figure 3A). ATOH1 peaks were enriched strongly in gene-associated functional domains, such as promoter (by default defined from -1 kb to +100 bp of transcription start site [TSS]), untranslated region, intron, and exon, where they usually mapped within 1 kb of the TSS, indicating that the peaks generated from our ATOH1 ChIP-seq were not located randomly on the genome but instead were associated with core promoters (Figure 3E). Consistent with its predicted activity as a transcription activator, ATOH1 binding sites were highly co-localized with active enhancer marker H3K27Ac, but not inactive chromatin-associated H3K27me3 (Figure 3D). Because we aimed to identify ATOH1 direct transcriptional targets, we defined genes that have ATOH1 binding sites within 20 kb of the TSS as ATOH1-associated genes. We identified 1024 and 4876 ATOH1-associated genes in ileum and colon, respectively (Figure 3F). Based on initial overlap analysis, 92.7% (949 of 1024 genes) of ATOH1-associated genes in the ileum also were bound by ATOH1 in the colon (Figure 3F). Taken together, using ChIP-seq, we identified bona fide ATOH1 binding sites in intestinal tissues under homeostatic conditions.
Motif Analysis of ATOH1 Binding Regions
Previous studies in the developing cerebellum have indicated that ATOH1 binds to a 10-nucleotide motif (AtEAM) containing a consensus E-box (5’-CANNTG-3’) binding motif of basic helix-loop-helix transcription factors.20 We performed de novo motif analysis for our ATOH1 ChIP-seq data using HOMER.32 As expected ATOH1-bound chromatin was enriched significantly in consensus E-box motifs in both colon and ileum (P = 1e-96 in ileum and 1e-722 in colon) (Figure 3G). This indicated that direct binding sites of ATOH1 were enriched in our ChIP-seq data. In addition to E-box, additional DNA binding motifs for several other transcription factor classes were enriched significantly within ATOH1-bound chromatin (Figure 3G). According to our RNA-seq data, we identified several transcription factors that were highly expressed within ATOH1–GFP–purified cells, and whose consensus DNA binding motif matched to these binding sequences derived de novo from ATOH1 ChIP-seq analysis (Figure 3G). Included in this list of transcription factors are E2A, HEB, RUNX1, YY1, NFIC, and HLTF, suggesting that these factors may bind cooperatively with ATOH1 to regulate secretory cell transcription. In fact, E2A and HEB are class I basic helix-loop-helix proteins known to interact with ATOH1,37 suggesting that these are its relevant partners within the intestine. Taken together, these results show that our ATOH1 ChIP-seq comprehensively identified ATOH1 targets in small and large intestines.
Validation of ATOH1 Binding Sites
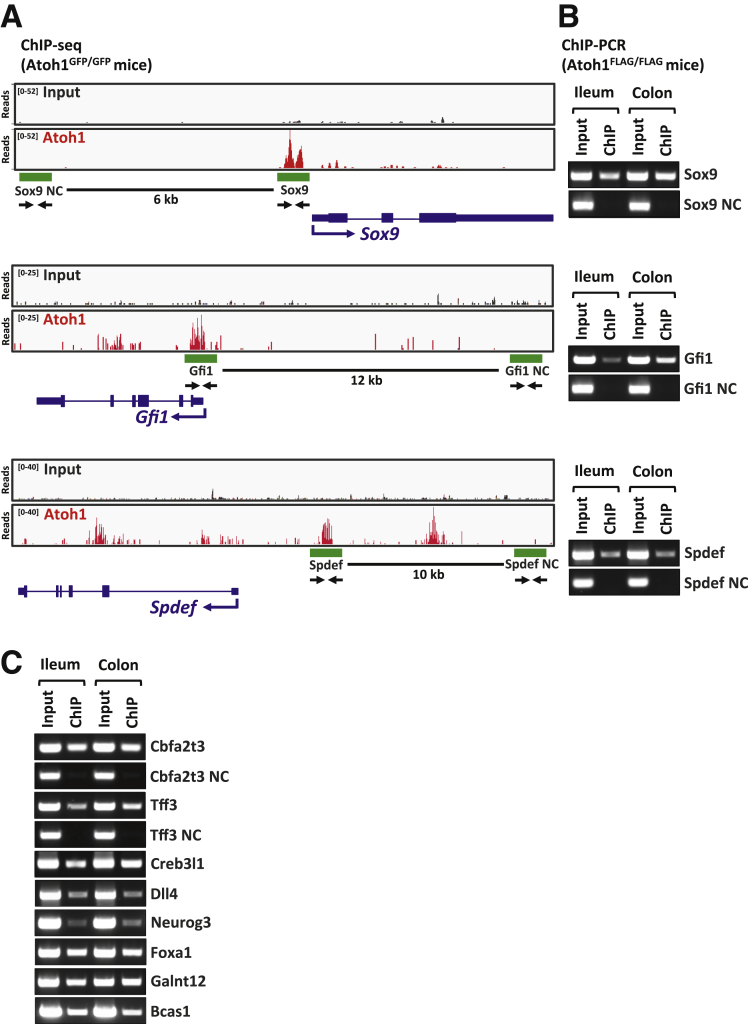
To validate our ATOH1 ChIP-seq data, we performed ChIP PCR in a different transgenic mouse model, which has an ATOH1–Flag fusion protein inserted into the Atoh1 locus (Atoh1Flag/Flag).38 We first focused on the ATOH1-associated genes Sox9, Gfi1, and Spdef, which have been implicated in secretory lineage differentiation. The high mobility group-box transcription factor SOX9 is expressed in the epithelial cells of the intestinal crypts and is required for goblet and Paneth cell differentiation.39, 40 Previous studies have suggested that both the zinc-finger transcription factor Gfi1 and the Ets-transcription factor SPDEF are downstream targets of ATOH1.17, 19 GFI1 directs secretory progenitors toward a goblet or Paneth cell fate, in part by repression of the pro-endocrine transcription factor NEUROG3.41 SPDEF plays an important role in goblet and Paneth cell terminal differentiation in the intestines.17, 18 Our ATOH1 ChIP-seq data indicated that ATOH1 binds to the core promoter regions of Sox9, Gfi1, and Spdef (Figure 4A). Consistent with our ChIP-seq results, we confirmed by ChIP PCR that ATOH1–Flag was enriched at the promoters of these target genes, but not upstream negative control regions (Figure 4B). Next, to confirm our ATOH1 ChIP-seq data further, we selected another 8 ATOH1-associated genes for validation. All 8 ATOH1 binding regions were validated by ChIP PCR, including Neurog3, Dll4, Tff3, Creb3l1, Galnt12, Bcas1, Foxa1, and Cbfa2t3 (Figure 4B). Taken together, these results confirm that ATOH1 binding sites identified by our ChIP-seq analysis were robust and highly reliable.
Figure 4.
Validation of ATOH1 binding sites. (A) ATOH ChIP-seq data generated from Atoh1GFP/GFP mice indicate ATOH1 binds to the promoter regions of Sox9, Gfi1, and Spdef. The peak density plots show fragment counts across the indicated genomic interval. Sox9, Gfi1, and Spdef genes are labeled in blue with exons as thick rectangles (coding sequence is slightly thicker) and introns as lines connecting rectangles. The arrows show primers designed for ChIP PCR. The green rectangles indicate regions selected for ATOH1-Flag ChIP PCR. (B, C) Ileal and colonic crypts isolated from Atoh1Flag/Flag mice were used to validate ATOH1 ChIP-seq peaks by ChIP PCR. ATOH1 was enriched in all ATOH1 binding regions predicted by our ATOH1 ChIP-seq, but not the negative control (NC) regions.
Identifying ATOH1 Transcriptional Targets
To identify direct ATOH1 transcriptional targets in the intestines, we compared ATOH1-associated genes identified by our ATOH1 ChIP-seq analysis (Figure 3F) with up-regulated genes in ATOH1-positive cells identified by our RNA-seq analysis (Figure 2D). We defined the ATOH1 targetome as the 658 genes in the colon and 193 genes in the ileum with significantly enriched expression in ATOH1-positive cells that also were bound by ATOH1 (Figure 5A, Supplementary Table 9). Consistent with the concept that ATOH1 functions as a key transcription factor for differentiation of the intestinal epithelium, several ATOH1 target genes were known to be involved in intestinal secretory lineage differentiation and function, such as Notch ligands Dll1 and Dll4; transcription factors Spdef,17, 18 Sox9,39, 40 Gfi1,19, 41 and Creb3l142; transcription co-repressors Cbfa2t2 and Cbfa2t343, 44, 45, 46; and secretory lineage-specific genes such as Best2, Spink4, Muc2, Sct, EphB3, Xbp1, and Clca3.17, 47, 48, 49 To gain broader insight into ATOH1 target genes in the intestines, we performed GO analysis using DAVID. ATOH1 target genes were associated with ontology terms including positive regulation of transcription machinery (adjusted P < .03), suggesting that ATOH1 is a regulator of other transcriptional regulators, consistent with a function as a master regulator of intestinal differentiation (Figure 5B). In addition, ATOH1 target genes are members of ontology groups such as intracellular transport, guanosine triphosphatase regulator activity, Rab guanosine triphosphatase binding, acetylgalactosaminyltransferase activity, and positive regulation of metabolic process, and so forth, indicating roles for ATOH1 in directing the program of modification and secretion of proteins from intestinal secretory cells (Figure 5B, Supplementary Table 10). We also found that ATOH1 targets were enriched in BMP signaling pathway constituents, suggesting a previously undefined role for ATOH1 in intestinal BMP signaling (Figure 5B). To gain more insight into the function of ATOH1 in the intestines, we next asked whether genes significantly de-enriched in ATOH1-positive cells (Figure 2F) also were bound by ATOH1 (Figure 3F). Surprisingly, a large number of genes, 1085 genes in the colon and 194 genes in the ileum, were identified (Figure 5C, Supplementary Table 11). Of note, among these genes, 2 important Notch pathway genes, Notch receptor Notch1 and transcription factor Hes1, were identified. Interestingly, we also found several genes that previously have been described to be important for Microfold cells and enterocytes, such as Spib, Elf3, and Ppargc1b.50, 51, 52 In this scenario, one possibility is that ATOH1 functions as a transcriptional activator of these genes in a subset of ATOH1-positive cells, but other factors repress their expression in the majority of cells, or drive stronger expression in ATOH1-negative cells, resulting in stronger relative expression in ATOH1-negative cells. However, we cannot exclude the possibility that ATOH1 functions as a negative regulator of transcription of these genes. GO analysis indicated that these de-enriched ATOH1 targets were associated significantly with several biological processes including nucleotide binding, positive regulation of cellular biosynthetic process, positive regulation of transcription, actin cytoskeleton organization, guanosine triphosphatase regulator activity, negative regulation of transcription, and epithelium development (Figure 5D, Supplementary Table 12). Taken together, these results indicated that ATOH1 functions as a master transcription factor, directly regulating the program of differentiation and function within secretory cells in the intestines.
In Situ Validation of ATOH1 Targetome Identifies Novel Secretory Cell Markers
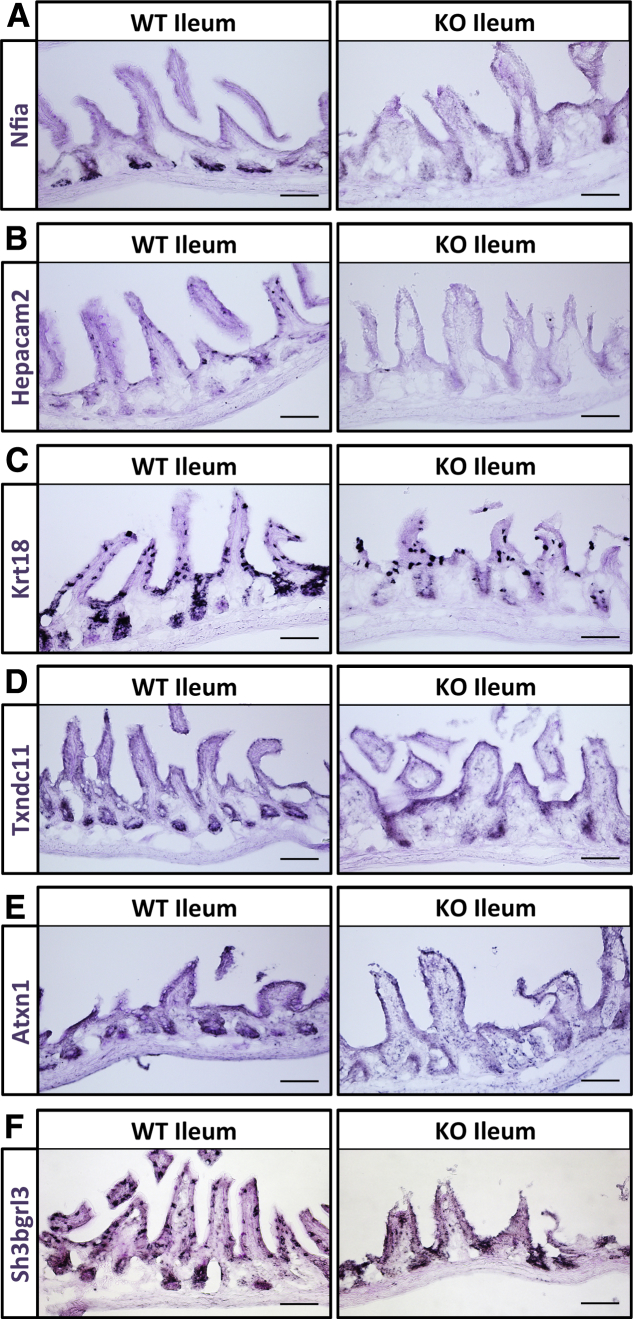
To validate the mRNA expression of the genes identified in the ATOH1 targetome, we performed in situ hybridizations on the ileum of transgenic mice where ATOH1 is deleted (Fabp1Cre; Atoh1lox/lox) and in littermate controls (Fabp1Cre; Atoh1+/+).8 Six genes that have not been fully studied in the intestinal secretory cells were selected randomly from the list of ATOH1 targets (Supplementary Table 9). These included the transcription factor nuclear factor I/A (Nfia), HEPACAM family member 2 (Hepacam2), keratin 18 (Krt18), thioredoxin domain-containing 11 (Txndc11), ataxin 1 (Atxn1), and SH3 domain binding glutamate-rich protein–like 3 (Sh3bgrl3). We found that the mRNA expression of Nfia is restricted in Paneth cells and completely depleted in Atoh1 deletion tissues (Figure 6A). In addition, we identified Hepacam2 as an ATOH1-dependent goblet cell gene in the ileum (Figure 6B). Krt18 expression is scattered in what appear to be progenitor cells in the crypts and a minority of cells in the villus. In Atoh1 mutant tissues, Krt18-positive cells in villus, but not in crypts, retain the expression of Krt18, suggesting these Krt18-positive cells in villus are not derived from ATOH1-positive secretory lineage (Figure 6C). Finally, we found that Txndc11, Atxn1, and Sh3bgrl3 are expressed not only in goblet cells, Paneth cells, and transit amplifying cells, but also in the other epithelial cell types (Figure 6D–F). Although the mRNA level of these 3 genes are decreased in Atoh1 mutant tissues, it is clear that they also are expressed in some remaining cells through ATOH1-independent transcription. Taken together, these results indicated that the ATOH1 targetome we generated in this study is a valuable resource for identifying novel secretory cell genes.
Figure 6.
In situ validation of ATOH1 targetome. Fresh-frozen ileal tissues were generated from Atoh1 deletion (Fabp1Cre; Atoh1lox/lox; knockout [KO]) or littermate control (Fabp1Cre; Atoh1+/+; wild type [WT]) mice. The mRNA expression of ATOH1 target genes, including (A) Nfia, (B) Hepacam2, (C) Krt18, (D) Txndc11, (E) Atxn1, and (F) Sh3bgrl3, were shown by in situ hybridizations. Light periodic acid–Schiff staining was performed after in situ hybridization to provide contrast for imaging. Scale bars: 100 μm.
ATOH1 Transcriptional Targets in Human Colorectal Cancer Cells
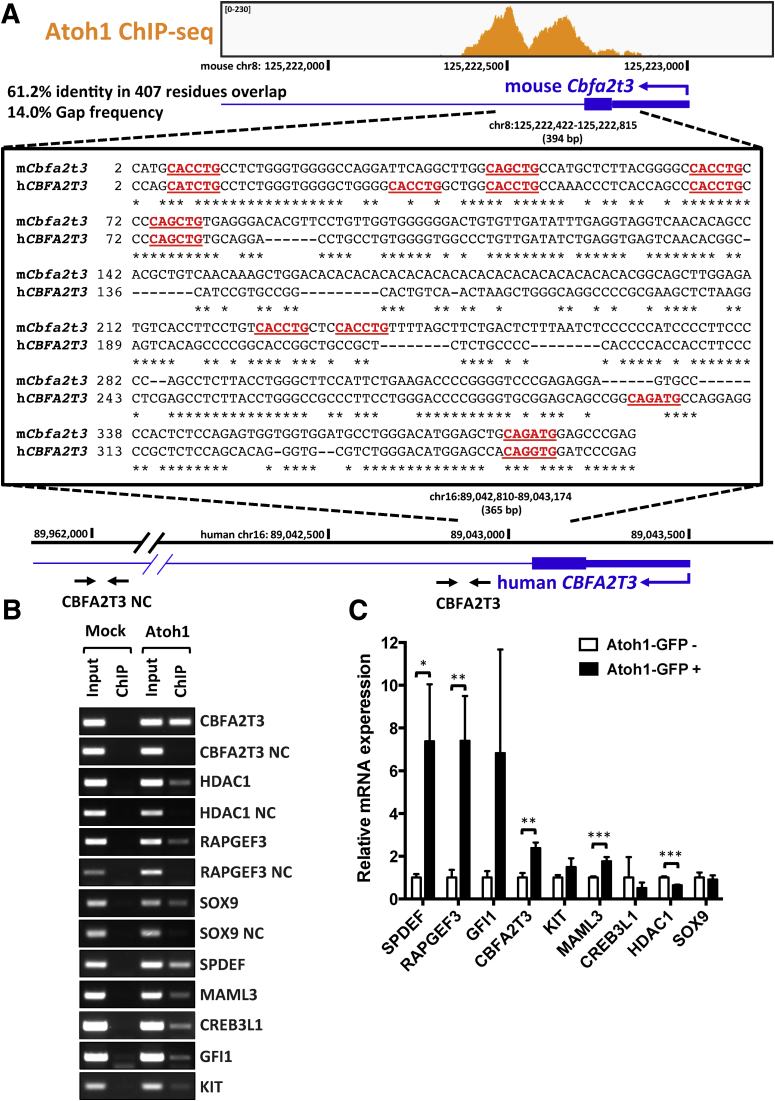
ATOH1 is highly conserved between species.53 In colorectal cancers (CRCs), ATOH1 functions as a tumor suppressor.54 Re-expression of ATOH1 in colon cancer cells not only inhibits proliferation but also promotes apoptosis, suggesting a potential window for new CRC therapeutics. Therefore, identification of ATOH1 targets in human CRCs will provide novel insights into CRC therapeutics. We asked whether ATOH1 shares similar transcriptional targets between normal intestines and human CRC cells. First, we focused on Cbfa2t3, a direct ATOH1 target that we identified in mouse colon. CBFA2T3 (also referred to as MTG16 or ETO2) is one of the MTG family of transcriptional corepressors that contributes to intestinal crypt proliferation and regeneration after injury.45, 46 Our ATOH1 ChIP-seq data indicated that ATOH1 strongly binds to the first exon/intron of Cbfa2t3 (Figure 7A). By using the University of California Santa Cruz genome browser (genome.ucsc.edu), we identified a corresponding region within the human CBFA2T3 promoter that contained several putative ATOH1 binding motifs (Figure 7A). To determine whether ATOH1 binds to the CBFA2T3 promoter in human CRC cells, we performed ChIP-PCR for transiently expressed ATOH1-GFP in human colon cancer cell line HCT116. Compared with mock-transfected cells, ATOH1 was enriched in the promoter region of CBFA2T3, but not in the downstream negative control region (Figure 7B). We extended our analysis of potentially conserved ATOH1 targets by examining another 8 ATOH1 colonic target genes, including HDAC1, RAPGEF3, SOX9, GFI1, SPDEF, MAML3, KIT, and CREB3L1. By using a similar approach as described for CBFA2T3 earlier, we identified orthologous human sequences with predicted ATOH1 binding sites for all 8 genes. ChIP PCR confirmed that ATOH1 bound to all 8 predicted ATOH1 binding regions, but not in the negative control regions, indicating strong conservation of ATOH1 binding sites across species (Figure 7B). To further determine whether ATOH1 could functionally regulate the expression of these genes in human CRCs, we isolated ATOH1-positive cells by flow cytometry followed by reverse-transcription quantitative PCR (RT-qPCR). Compared with ATOH1-negative cells, the expression of CBFA2T3, SPDEF, RAPGEF3, and MAML3 were up-regulated significantly in ATOH1-positive cells (Figure 7C). In addition, the expression of GFI1 and KIT was increased in ATOH1-positive cells. In contrast, ATOH1 induced a small but significant decrease in HDAC1 expression (Figure 7C). Taken together, these results suggested that ATOH1 functionally regulates the majority of these genes not only in mouse colon, but also in human CRCs.
Figure 7.
ATOH1 transcriptional targets in human colorectal cancer cells. (A) Highly conserved promoter sequence of Cbfa2t3 bound by ATOH1 between human and mouse genome. Several putative ATOH1 binding motifs 5’-CANNTG-3’ within the ATOH1 peak were highlighted (red). (B) Human colon cancer cell line HCT116 was transfected transiently with ATOH1-GFP and used for ChIP PCR. Anti-GFP antibodies were used for ChIP. (C) Real-time PCR analysis of complementary DNAs synthesized from mRNA isolated from FACS purified ATOH1-GFP–positive or ATOH1-GFP–negative HCT116 cells 48 hours after transient transfection. Relative fold change is presented as means ± SEM of 3 independent experiments (*P < .05, **P < .01, and ***P < .001).
SPDEF Cooperates With ATOH1 to Amplify Target Gene Expression
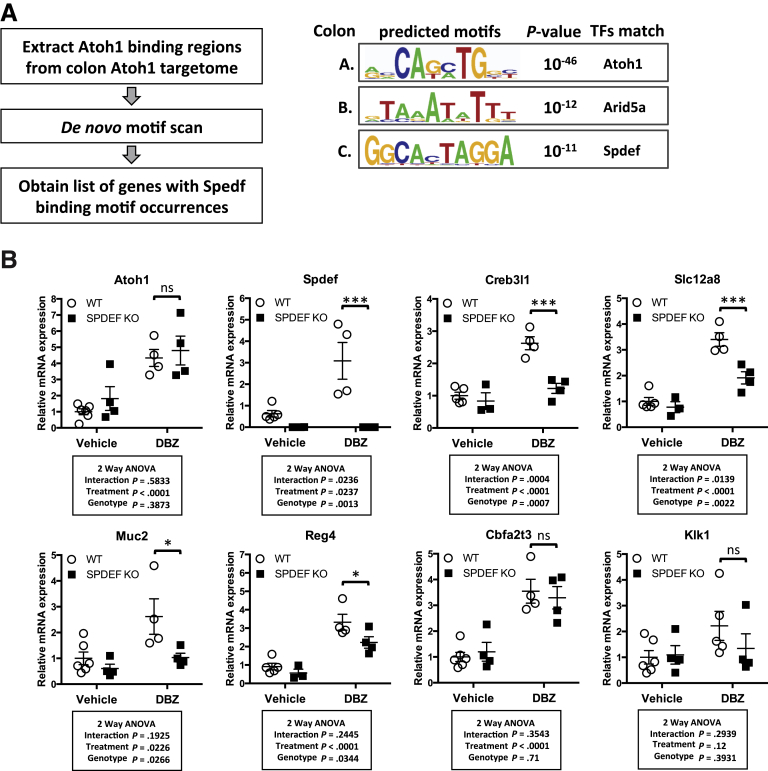
We next sought to identify transcription factors that are likely to co-regulate gene expression with ATOH1. Our unbiased de novo motif analysis (Figure 3E) identified many potential co-regulators, but most of these sites included intergenic regions of unknown significance. Therefore, we performed a motif scan analysis of the colon-specific ATOH1 targetome. Specifically, HOMER was used to scan for 10-mer motifs that were enriched significantly in the ATOH1 targetome while optimized for 50 motifs during the search (findMotifsGenome.pl –len 10 –S 50). This analysis showed SPDEF binding motifs enriched within the ATOH1 targetome (Figure 8A), with SPDEF motifs associated with 75 of 658 (11%) of ATOH1 target genes in the colon (Supplementary Table 13). Of note, among these genes we found several goblet cell–associated genes, including Atoh1, Spdef, Muc2, Reg4, Klk1, Creb3l1, and Slc12a8. Previous studies have suggested that SPDEF plays a critical role in controlling goblet cell terminal differentiation.17, 18 To determine the interdependence between ATOH1 and SPDEF to control expression of these putative co-regulated genes, we assessed the effect of overexpression of ATOH1 or SPDEF in the absence of the other protein. We enhanced ATOH1 expression using the γ-secretase inhibitor, DBZ, in wild-type and Spdef null mice, and assessed target gene expression in colonic crypts by RT-qPCR. As expected, DBZ treatment increased the expression of Atoh1 and all downstream target genes in wild-type mice (Figure 8B, open circles). Deletion of Spdef significantly blunted the effects of DBZ-ATOH1–mediated transcription in a subset of ATOH1 target genes, including Creb3l1, Slc12a8, Muc2, and Reg4, but not others, such as Cbfa2t3 and Klk1 (Figure 8B, closed squares). These results suggested that ATOH1 is sufficient to drive target gene expression and that as a direct target of ATOH1, SPDEF provides positive feedback to amplify ATOH1-dependent transcription of a subset of secretory cell–associated genes, especially goblet cell genes.
Figure 8.
SPDEF functions as a transcriptional co-regulator of ATOH1. (A) Flow chart for identifying SPDEF binding motifs within ATOH1 binding regions in the colon ATOH1 targetome. P values are for motif enrichment. (B) Real-time PCR analysis of complementary DNAs synthesized from mRNA isolated from mouse colonic crypts. Spdef null or littermate wild-type control mice were treated with either vehicle or dibenzazepine (DBZ) for 5 consecutive days. Relative fold change is presented as means ± SEM (*P < .05 and ***P < .001). ANOVA, analysis of variance; TFs, transcription factors.
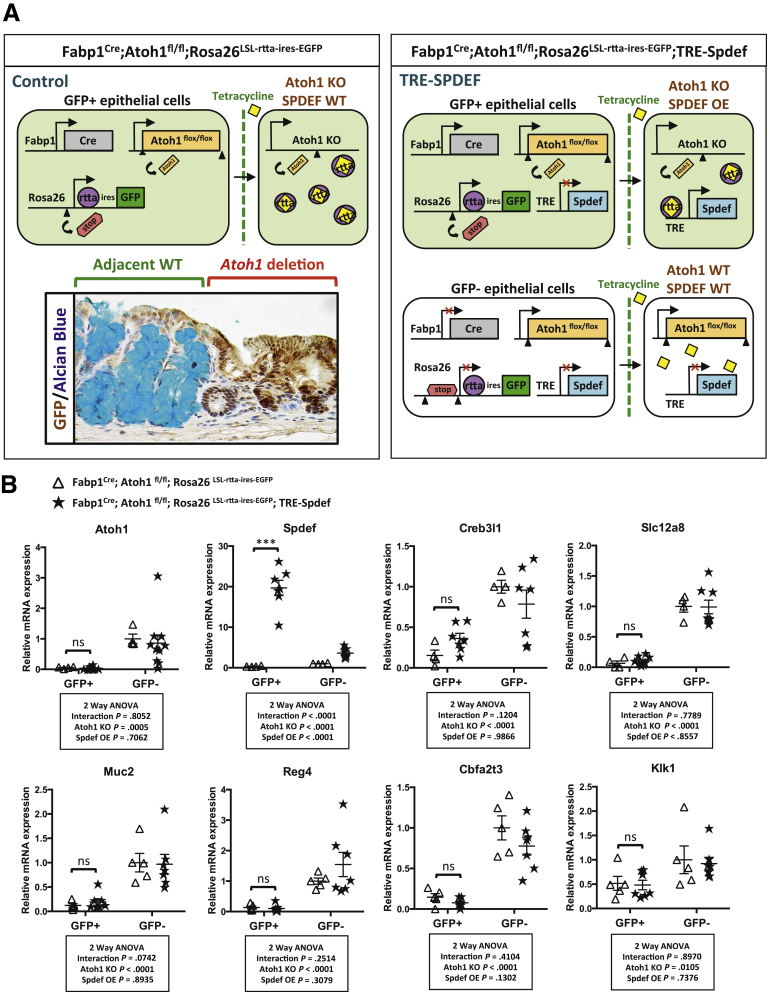
To further establish the relationship between ATOH1 and SPDEF, we asked whether SPDEF could activate expression of secretory cell genes in the absence of ATOH1. To test this hypothesis, transgenic mice in which ATOH1 is deleted in the intestinal epithelium (Fabp1Cre; Atoh1lox/lox) were bred with tetracycline-inducible SPDEF transgenic mice (Rosa26LSL-rtta-ires-EGFP; TRE-Spdef). In this mouse model, Fabp1-Cre is expressed in a patchy pattern in the ileum and colon (Figure 9A).8 With the Rosa LSL-rtta-ires-EGFP reporter, we were able to sort GFP-positive Atoh1 deletion cells from control (Fabp1Cre; Atoh1lox/lox; Rosa26 LSL-rtta-ires-EGFP) and littermate (Fabp1Cre; Atoh1lox/lox; Rosa26 LSL-rtta-ires-EGFP; TRE-Spdef) mice; we induced SPDEF expression in Atoh1-mutant cells by treating these mice with tetracycline in water for 5 consecutive days (Figure 9A). Thus, after isolating 7AAD-negative (live) cells by flow cytometry from control or SPDEF-induced colonic crypts, we were able to analyze the mRNA expression by RT-qPCR of the following: (1) wild-type (GFP-negative cells from either control or TRE-SPDEF mice), (2) Atoh1 deletion (GFP-positive cells from control mice), and (3) Atoh1 deletion and Spdef overexpression (GFP-positive cells from TRE–SPDEF mice) cells (Figure 9A). As expected, in Atoh1 deletion (GFP-positive) cells, the mRNAs of ATOH1 targets were decreased significantly compared with wild-type (GFP-negative) cells (Figure 9B, open triangles). In contrast, despite robust transgene induction (∼20-fold), SPDEF was not sufficient to activate transcription of Creb3l1, Slc12a8, Muc2, Reg4, Cbfa2t3, and Klk1 in Atoh1 deletion cells (Figure 9B). Taken together, these results indicated that SPDEF amplifies ATOH1-mediated transcription of secretory cell genes, but is insufficient to drive secretory cell gene expression in the absence of ATOH1 (Figure 10).
Figure 9.
SPDEF functions as a transcriptional co-regulator of ATOH1. (A) Experimental strategy using the inducible mouse model (Fabp1Cre; Atoh1lox/lox; Rosa26 LSL-rtta-ires-EGFP; TRE-Spdef). Arrows indicate the direction of transcription. Arrowheads indicate loxP sites. SPDEF expression was induced by feeding mice with water containing tetracycline (2 mg/mL). Fabp1-Cre is expressed in a patchy pattern in the ileum and colon. Immunohistochemistry staining of GFP (Atoh1 deletion region) combined with Alcian blue staining (for goblet cells) staining indicates the Atoh1 deletion and the adjacent wild-type colonic epithelium. (B) GFP-positive cells were FACS-purified from colonic crypts followed by real-time PCR analysis. Relative fold change is presented as means ± SEM (***P < .001). ANOVA, analysis of variance; KO, knockout; OE, overexpression; WT, wild type.
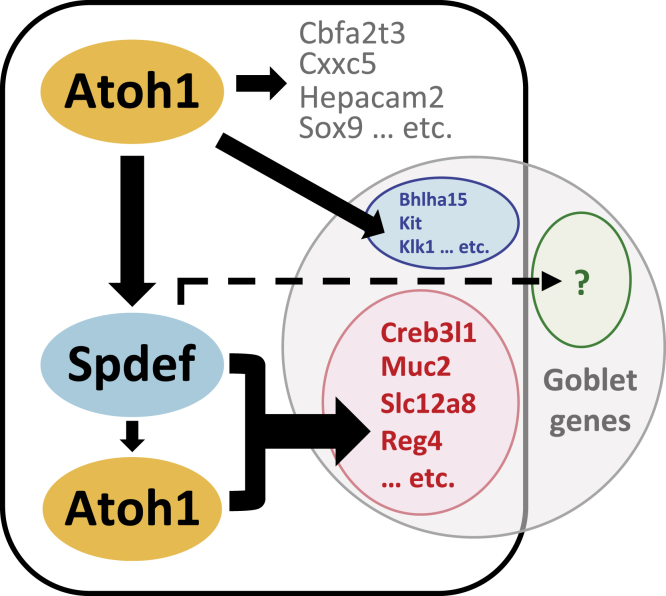
Figure 10.
Proposed model of transcriptional co-regulation by ATOH1 and SPDEF.
Discussion
In this study, we used a combination of RNA-seq and ChIP-seq techniques together with cell sorting and state-of-the-art transgenic mice to identify more than 700 direct transcriptional targets of ATOH1 in the small and large intestines. Of note, these unbiased genome-wide approaches were performed in primary ileal and colonic crypts under homeostatic conditions, thereby increasing the relevance and credibility of identified target genes. Our data showed that ATOH1 strongly binds to core promoter and enhancer regions, which were marked by the active chromatin histone modification H3K27Ac, suggesting that ATOH1 likely functions as a transcriptional activator. Although the physiological function of ileum and colon are very different, the ATOH1-associated genes were highly similar in these 2 tissues. The ontology analysis indicated that ATOH1 directly regulates several important biological processes and controls the transcription machinery of secretory lineage differentiation, suggesting that ATOH1 is required for specifying and maintaining secretory cells throughout the intestinal epithelium.
The Notch signaling pathway is critical for gastrointestinal cell fate determination.5, 55, 56 In the adult intestines, activation of Notch signaling induces the expression of HES1, which directly represses Atoh1, and thus directs progenitors to differentiate along the absorptive lineage. On the other hand, adjacent progenitors that escape Notch activation express ATOH1, which commits these cells to the secretory lineage. Considerable genetic evidence suggests that ATOH1 is a key transcription factor that controls Notch-mediated lateral inhibition.16 However, the details underlying this mechanism are characterized incompletely. Previous studies have suggested that delta-like protein (DLL) 1 and DLL4 are key Notch ligands required for maintaining ISC homeostasis and differentiation.57 Simultaneous deletion of Dll1 and Dll4 phenocopies the loss of Notch activity and causes the complete conversion of proliferating progenitors into postmitotic secretory cells, resulting in loss of the active ISC population.57 In this study, we identified Dll1 and Dll4 as direct targets of ATOH1, confirming a central role for ATOH1 in control of lateral inhibition from ATOH1-positive secretory progenitors to adjacent absorptive progenitors/stem cells. In addition to Dll1 and Dll4, several other Notch signaling pathway components were identified as ATOH1-associated genes, such as CBF1, Suppressor of hairless, Lag-1 (CSL) transcriptional co-activator Maml3 and Crebbp, CSL transcriptional co-repressor Hdac1, Ncor2, Ctbp1 and Ctbp2, Notch ligand Jag1, Notch receptor Notch1, and Notch antagonist Numb.58, 59, 60, 61, 62, 63 Taken together, our data suggest that ATOH1 functions as a master transcription factor for Notch-mediated lateral inhibition by directly activating Notch ligands to reinforce secretory cell fate commitment. Expression of ATOH1 is likely to be the key event in commitment of differentiating cells to the secretory lineage.
ATOH1 is required for the differentiation of all intestinal secretory cells.7 Consistent with these observations, the expression of goblet cell–, Paneth cell–, and enteroendocrine cell–specific genes were decreased after conditional deletion of ATOH1 throughout the intestinal epithelium. Interestingly, our immunofluorescence staining suggested that ATOH1 is expressed at much lower levels in enteroendocrine cells than in goblet and Paneth cells. This observation can explain why we did not find enteroendocrine-specific genes in ATOH1-positive cells purified from Atoh1GFP/GFP mice. The lower expression level of ATOH1 in enteroendocrine cells may be caused by post-translational modification or by the other negative transcriptional feedback. We speculate that different levels of ATOH1 specify different subtypes of secretory cells, which may contribute to secretory cell allocation.64
Several transcription factors downstream of ATOH1, such as SPDEF and GFI1, were shown to regulate secretory cell differentiation.17, 18, 19 However, little is known about how these transcription factors modulate secretory gene expression. Our data indicated that SPDEF amplifies ATOH1-dependent transcription of a subset of goblet cell genes (Figure 8B). Although we cannot determine whether the amplification of ATOH1-dependent transcription is contributed directly by SPDEF binding to the chromatin or caused indirectly by the other critical components lost in Spdef null mice, de novo motif analysis indicated a significant enrichment of the SPDEF binding motif within the ATOH1 targetome, suggesting the possibility that SPDEF coordinates with ATOH1 on the promoter or enhancer regions of these genes (Figure 8A). We further found that SPDEF itself is not sufficient to activate ATOH1 targets, suggesting a hierarchy of transcription factor–mediated gene expression during intestinal cell differentiation (Figure 9B). One caveat of this experiment was that Atoh1 deletion tissues lack specified secretory cells, therefore the majority of these cells are enterocytes. Thus, SPDEF might not be able to regulate secretory gene transcription in the enterocyte context owing to limited chromatin accessibility. However, our unpublished data suggested that SPDEF is able to drive mucus-like production in Atoh1 deletion tissues, indicating SPDEF retains at least part of its biological function in enterocytes (data not shown). Moreover, a previous study indicated that secretory and absorptive progenitors show similar distributions of histone marks and DNase hypersensitivity, suggesting intestinal lineage determination is not dependent on chromatin priming.16 Based on our findings in this and previous studies, as a master transcription factor, it is most likely that ATOH1 is expressed at the earliest step of secretory progenitor differentiation, and it must be continuously expressing in all secretory lineages for their maintenance. Within secretory progenitor cells, an unknown mechanism results in NEUROG3 or GFI1 expression; those cells that express GFI1 commit to the Paneth/goblet cell fate; we suggest that ATOH1 expression levels may mediate this decision. Subsequently, when SPDEF is activated in the progenitors, it strengthens the expression of ATOH1-dependent goblet genes, resulting in goblet cell terminal differentiation. We suggest that in addition to ATOH1-dependent targets, SPDEF also may regulate transcription of ATOH1-independent goblet gene expression (Figure 10). Future studies to determine how transcription networks select alternate secretory cell fates will expand our current knowledge of stem cell biology and chromatin biology of the intestinal cells.
Previous studies have suggested that ATOH1 functions as a tumor suppressor in human CRCs.54 To gain more insight into this activity, we examined whether ATOH1 shares similar transcriptional targets in mouse colonic crypts and human colon cancer cell line HCT116. Interestingly, even though ATOH1 binds to all of the human ATOH1 targets predicted by our murine ChIP-seq analysis, only 4 of 9 of these ATOH1 targets were regulated in a similar manner at the transcriptional level. Because canonical Wnt/β-catenin signaling is hyperactivated in HCT116 owing to a gain-of-function β-catenin mutation, it is possible that this interferes with ATOH1 target gene expression in colon cancer cells. It also is possible that Wnt/β-catenin target genes, such as SOX9, are expressed at maximal levels in CRC cells and further transcriptional activation by ATOH1 is not possible.65 Alternatively, the transcriptional machinery of ATOH1 might rely on different cofactors that are not available in these cancer cells. These data highlight the difficulty of using cancer cell lines to extrapolate information about transcriptional targets in normal tissues.
We previously identified SPDEF as a tumor suppressor in both murine and cell culture CRC models.66 Consistent with these observations, in this study, we show that ATOH1 binds to SPDEF and directly regulates its expression in both mouse intestines and human colon cancer cells. Given previous findings that ATOH1 is a colorectal tumor suppressor,13, 54, 67 our study suggests that SPDEF may be a key mediator of ATOH1's tumor-suppressive activity. Further studies of direct transcriptional targets of ATOH1, such as SPDEF, in human CRCs will provide insight into therapeutic strategies for targeting human CRCs through the Notch–ATOH1 axis.
Next-generation sequencing provides unbiased genome-wide approaches to studying transcriptional machinery. However, there are some caveats to this study. First, although we performed the ATOH1 ChIP-seq in purified intestinal crypts, these data derive from a mixed cell population. Thus, we cannot distinguish whether ATOH1 binding sites were present in all ATOH1-positive cells or are found only in a subpopulation. Second, ChIP-seq cannot identify binding sites in relatively rare subpopulations of cells (eg, enteroendocrine cells), and therefore these may be missed in this study. We noted that the ATOH1 ChIP-seq from colonic crypts identified more binding sites than from ileal crypts. This is possibly owing to the gradient of endogenous ATOH1 expression in the adult intestine—much higher in the colon than ileum. Advanced ChIP-seq and RNA-seq techniques for small amounts of sorted cells will be helpful to address these caveats in the future. Further integration of the ATOH1 transcriptional network with other pathways regulating intestinal differentiation and homeostasis is an important future direction for this project.68, 69, 70
In summary, this study unveiled the direct targets of ATOH1 in the adult intestine, providing novel insight toward understanding the cell differentiation and biological function of intestinal secretory lineages. We further showed interaction between ATOH1 and SPDEF to regulate the expression of a subset of target genes, suggesting that basal expression of secretory cell genes may require amplification factors to achieve full expression. Thus, our results identify novel interactions between secretory lineage-specific transcription factors that control cellular differentiation and maturation in the adult intestines.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This project was supported in part by the Texas Medical Center Digestive Disease Center with funding from the National Institutes of Health (P30DK56338), the Integrated Microscopy Core at Baylor College of Medicine with funding from the National Institutes of Health (DK56338 and CA125123), the Dan L. Duncan Cancer Center, the John S. Dunn Gulf Coast Consortium for Chemical Genomics, the RNA In Situ Hybridization Core facility at Baylor College of Medicine, which is supported by a Shared Instrumentation grant from the National Institutes of Health (1S10OD016167) and a Public Health Service grant (DK56338), and by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the National Institutes of Health (P30 AI036211, P30 CA125123, and S10 RR024574). The authors also received assistance from the Research Flow Cytometry Core in the Division of Rheumatology at Cincinnati Children’s Hospital Medical Center, supported in part by the National Institutes of Health (AR-47363, DK78392, and DK90971).
Contributor Information
Joo-Seop Park, Email: joo-seop.park@cchmc.org.
Noah F. Shroyer, Email: noah.shroyer@bcm.edu.
Supplementary Material
References
- 1.van der Flier L.G., Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 2.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 3.Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Clevers H., Loh K.M., Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 5.Noah T.K., Shroyer N.F. Notch in the intestine: regulation of homeostasis and pathogenesis. Annu Rev Physiol. 2013;75:263–288. doi: 10.1146/annurev-physiol-030212-183741. [DOI] [PubMed] [Google Scholar]
- 6.Clevers H., Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 7.Yang Q., Bermingham N.A., Finegold M.J. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 8.Shroyer N.F., Helmrath M.A., Wang V.Y. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132:2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 9.VanDussen K.L., Samuelson L.C. Mouse atonal homolog 1 directs intestinal progenitors to secretory cell rather than absorptive cell fate. Dev Biol. 2010;346:215–223. doi: 10.1016/j.ydbio.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y., Cain-Hom C., Choy L. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 11.Wong G.T., Manfra D., Poulet F.M. Chronic treatment with the gamma-secretase inhibitor LY-411,575 inhibits beta-amyloid peptide production and alters lymphopoiesis and intestinal cell differentiation. J Biol Chem. 2004;279:12876–12882. doi: 10.1074/jbc.M311652200. [DOI] [PubMed] [Google Scholar]
- 12.Milano J., McKay J., Dagenais C. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol Sci. 2004;82:341–358. doi: 10.1093/toxsci/kfh254. [DOI] [PubMed] [Google Scholar]
- 13.Kazanjian A., Noah T., Brown D. Atonal homolog 1 is required for growth and differentiation effects of notch/gamma-secretase inhibitors on normal and cancerous intestinal epithelial cells. Gastroenterology. 2010;139:918–928. doi: 10.1053/j.gastro.2010.05.081. 928 e1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim T.H., Shivdasani R.A. Genetic evidence that intestinal Notch functions vary regionally and operate through a common mechanism of Math1 repression. J Biol Chem. 2011;286:11427–11433. doi: 10.1074/jbc.M110.188797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Es J.H., de Geest N., van de Born M. Intestinal stem cells lacking the Math1 tumour suppressor are refractory to Notch inhibitors. Nat Commun. 2010;1:18. doi: 10.1038/ncomms1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim T.H., Li F., Ferreiro-Neira I. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature. 2014;506:511–515. doi: 10.1038/nature12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noah T.K., Kazanjian A., Whitsett J. SAM pointed domain ETS factor (SPDEF) regulates terminal differentiation and maturation of intestinal goblet cells. Exp Cell Res. 2010;316:452–465. doi: 10.1016/j.yexcr.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregorieff A., Stange D.E., Kujala P. The ets-domain transcription factor Spdef promotes maturation of goblet and paneth cells in the intestinal epithelium. Gastroenterology. 2009;137:1333–1345. doi: 10.1053/j.gastro.2009.06.044. e1-3. [DOI] [PubMed] [Google Scholar]
- 19.Shroyer N.F., Wallis D., Venken K.J. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005;19:2412–2417. doi: 10.1101/gad.1353905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klisch T.J., Xi Y., Flora A. In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc Natl Acad Sci U S A. 2011;108:3288–3293. doi: 10.1073/pnas.1100230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park K.S., Korfhagen T.R., Bruno M.D. SPDEF regulates goblet cell hyperplasia in the airway epithelium. J Clin Invest. 2007;117:978–988. doi: 10.1172/JCI29176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saam J.R., Gordon J.I. Inducible gene knockouts in the small intestinal and colonic epithelium. J Biol Chem. 1999;274:38071–38082. doi: 10.1074/jbc.274.53.38071. [DOI] [PubMed] [Google Scholar]
- 23.Belteki G., Haigh J., Kabacs N. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 2005;33:e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.el Marjou F., Janssen K.P., Chang B.H. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 25.Mahe M.M., Aihara E., Schumacher M.A. Establishment of gastrointestinal epithelial organoids. Curr Protoc Mouse Biol. 2013;3:217–240. doi: 10.1002/9780470942390.mo130179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yaylaoglu M.B., Titmus A., Visel A. Comprehensive expression atlas of fibroblast growth factors and their receptors generated by a novel robotic in situ hybridization platform. Dev Dyn. 2005;234:371–386. doi: 10.1002/dvdy.20441. [DOI] [PubMed] [Google Scholar]
- 27.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anders S., Pyl P.T., Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Liu T., Meyer C.A. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinz S., Benner C., Spann N. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramirez F., Dundar F., Diehl S. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Everett L.J., Le Lay J., Lukovac S. Integrative genomic analysis of CREB defines a critical role for transcription factor networks in mediating the fed/fasted switch in liver. BMC Genomics. 2013;14:337. doi: 10.1186/1471-2164-14-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose M.F., Ren J., Ahmad K.A. Math1 is essential for the development of hindbrain neurons critical for perinatal breathing. Neuron. 2009;64:341–354. doi: 10.1016/j.neuron.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen P., Hecht J., Ibrahim D.M. Saturation analysis of ChIP-seq data for reproducible identification of binding peaks. Genome Res. 2015;25:1391–1400. doi: 10.1101/gr.189894.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akazawa C., Ishibashi M., Shimizu C. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J Biol Chem. 1995;270:8730–8738. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- 38.Flora A., Klisch T.J., Schuster G. Deletion of Atoh1 disrupts Sonic Hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science. 2009;326:1424–1427. doi: 10.1126/science.1181453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bastide P., Darido C., Pannequin J. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol. 2007;178:635–648. doi: 10.1083/jcb.200704152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mori-Akiyama Y., van den Born M., van Es J.H. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology. 2007;133:539–546. doi: 10.1053/j.gastro.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 41.Bjerknes M., Cheng H. Cell Lineage metastability in Gfi1-deficient mouse intestinal epithelium. Dev Biol. 2010;345:49–63. doi: 10.1016/j.ydbio.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 42.Asada R., Saito A., Kawasaki N. The endoplasmic reticulum stress transducer OASIS is involved in the terminal differentiation of goblet cells in the large intestine. J Biol Chem. 2012;287:8144–8153. doi: 10.1074/jbc.M111.332593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parang B., Rosenblatt D., Williams A.D. The transcriptional corepressor MTGR1 regulates intestinal secretory lineage allocation. FASEB J. 2015;29:786–795. doi: 10.1096/fj.14-254284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amann J.M., Chyla B.J., Ellis T.C. Mtgr1 is a transcriptional corepressor that is required for maintenance of the secretory cell lineage in the small intestine. Mol Cell Biol. 2005;25:9576–9585. doi: 10.1128/MCB.25.21.9576-9585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams C.S., Bradley A.M., Chaturvedi R. MTG16 contributes to colonic epithelial integrity in experimental colitis. Gut. 2013;62:1446–1455. doi: 10.1136/gutjnl-2011-301439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poindexter S.V., Reddy V.K., Mittal M.K. Transcriptional corepressor MTG16 regulates small intestinal crypt proliferation and crypt regeneration after radiation-induced injury. Am J Physiol Gastrointest Liver Physiol. 2015;308:G562–G571. doi: 10.1152/ajpgi.00253.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Batlle E., Henderson J.T., Beghtel H. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 48.Adolph T.E., Tomczak M.F., Niederreiter L. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leverkoehne I., Gruber A.D. The murine mCLCA3 (alias gob-5) protein is located in the mucin granule membranes of intestinal, respiratory, and uterine goblet cells. J Histochem Cytochem. 2002;50:829–838. doi: 10.1177/002215540205000609. [DOI] [PubMed] [Google Scholar]
- 50.Kanaya T., Hase K., Takahashi D. The Ets transcription factor Spi-B is essential for the differentiation of intestinal microfold cells. Nat Immunol. 2012;13:729–736. doi: 10.1038/ni.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng A.Y., Waring P., Ristevski S. Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology. 2002;122:1455–1466. doi: 10.1053/gast.2002.32990. [DOI] [PubMed] [Google Scholar]
- 52.Bellafante E., Morgano A., Salvatore L. PGC-1beta promotes enterocyte lifespan and tumorigenesis in the intestine. Proc Natl Acad Sci U S A. 2014;111:E4523–E4531. doi: 10.1073/pnas.1415279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mulvaney J., Dabdoub A. Atoh1, an essential transcription factor in neurogenesis and intestinal and inner ear development: function, regulation, and context dependency. J Assoc Res Otolaryngol. 2012;13:281–293. doi: 10.1007/s10162-012-0317-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bossuyt W., Kazanjian A., De Geest N. Atonal homolog 1 is a tumor suppressor gene. PLoS Biol. 2009;7:e39. doi: 10.1371/journal.pbio.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koch U., Lehal R., Radtke F. Stem cells living with a Notch. Development. 2013;140:689–704. doi: 10.1242/dev.080614. [DOI] [PubMed] [Google Scholar]
- 56.Willet S.G., Mills J.C. Stomach organ and cell lineage differentiation: from embryogenesis to adult homeostasis. Cell Mol Gastroenterol Hepatol. 2016;2:546–559. doi: 10.1016/j.jcmgh.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pellegrinet L., Rodilla V., Liu Z. Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology. 2011;140:1230–1240. doi: 10.1053/j.gastro.2011.01.005. e1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu L., Sun T., Kobayashi K. Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors. Mol Cell Biol. 2002;22:7688–7700. doi: 10.1128/MCB.22.21.7688-7700.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oswald F., Tauber B., Dobner T. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol Cell Biol. 2001;21:7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kao H.Y., Ordentlich P., Koyano-Nakagawa N. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oswald F., Winkler M., Cao Y. RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol Cell Biol. 2005;25:10379–10390. doi: 10.1128/MCB.25.23.10379-10390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nickoloff B.J., Qin J.Z., Chaturvedi V. Jagged-1 mediated activation of notch signaling induces complete maturation of human keratinocytes through NF-kappaB and PPARgamma. Cell Death Differ. 2002;9:842–855. doi: 10.1038/sj.cdd.4401036. [DOI] [PubMed] [Google Scholar]
- 63.Frise E., Knoblich J.A., Younger-Shepherd S. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc Natl Acad Sci U S A. 1996;93:11925–11932. doi: 10.1073/pnas.93.21.11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stamataki D., Holder M., Hodgetts C. Delta1 expression, cell cycle exit, and commitment to a specific secretory fate coincide within a few hours in the mouse intestinal stem cell system. PLoS One. 2011;6:e24484. doi: 10.1371/journal.pone.0024484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blache P., van de Wetering M., Duluc I. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J Cell Biol. 2004;166:37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Noah T.K., Lo Y.H., Price A. SPDEF functions as a colorectal tumor suppressor by inhibiting beta-catenin activity. Gastroenterology. 2013;144:1012–1023 e6. doi: 10.1053/j.gastro.2013.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leow C.C., Romero M.S., Ross S. Hath1, down-regulated in colon adenocarcinomas, inhibits proliferation and tumorigenesis of colon cancer cells. Cancer Res. 2004;64:6050–6057. doi: 10.1158/0008-5472.CAN-04-0290. [DOI] [PubMed] [Google Scholar]
- 68.Heuberger J., Kosel F., Qi J. Shp2/MAPK signaling controls goblet/paneth cell fate decisions in the intestine. Proc Natl Acad Sci U S A. 2014;111:3472–3477. doi: 10.1073/pnas.1309342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McElroy S.J., Castle S.L., Bernard J.K. The ErbB4 ligand neuregulin-4 protects against experimental necrotizing enterocolitis. Am J Pathol. 2014;184:2768–2778. doi: 10.1016/j.ajpath.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Watanabe N., Mashima H., Miura K. Requirement of Gαq/Gα11 signaling in the preservation of mouse intestinal epithelial homeostasis. Cell Mol Gastroenterol Hepatol. 2016;2:767–782. doi: 10.1016/j.jcmgh.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.