Abstract
RNA granules are microscopically visible cellular structures that aggregate by protein–protein and protein-RNA interactions. Using stress granules as an example, we discuss the principles of RNA granule formation, which rely on the multivalency of RNA and multi-domain proteins as well as low-affinity interactions between proteins with prion-like/low-complexity domains (e.g. FUS and TDP-43). We then explore how dysregulation of RNA granule formation is linked to neurodegenerative diseases, such as amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD), and discuss possible strategies for therapeutic intervention.
Keywords: Stress Granules, Low-complexity region, Prion-like domain, RNA granules, Phase Separation
1 A Cellular World of RNA Granules
Cellular RNAs rarely act alone. Rather, their functions are mediated through RNA-binding proteins (RBPs) and other functional partners, resulting in the formation of ribonucleoprotein (RNP) complexes. MicroRNAs, for example, associate with their cognate binding partner Argonaute, which in turn serves as a platform to recruit other proteins to form the silencing complexes responsible for translation repression and/or decay of mRNA targets (Höck et al. 2007; La Rocca et al. 2015). Besides microRNA-induced silencing complexes, various types of other RNPs are formed to regulate gene expression over the life cycle of an mRNA. For example, the birth of a eukaryotic mRNA is mediated by a cascade of proteins that cap the 5' end, add a poly(A) tail at the 3' end, and carry out splicing to remove introns. The process generally terminates with the deposition of exon junction complexes following splicing, which signal for the mature transcript to be exported to the cytoplasm where it can be bound by ribosomes for translation or exosomes for degradation (Bono and Gehring 2011). Whether the mature mRNA is destined for translation or degradation is, in turn, determined by RBPs bound to the transcript, often located in the 3' untranslated regions (UTRs) (Schoenberg and Maquat 2012). Thus, from transcription in the nucleus to degradation in the cytoplasm, proteins of distinct function often associate as macrocomplexes of RNPs to fulfill essential regulatory functions. Strikingly, RNPs frequently undergo further aggregation to form microscopically visible structures commonly termed RNA granules (Anderson and Kedersha 2006; Anderson and Kedersha 2009; Buchan and Parker 2009). These structures can be nuclear or cytoplasmic, and their mechanisms of action are only just beginning to be understood. Classes of these structures include nucleoli, Cajal bodies, nuclear speckles and paraspeckles in the nucleus, as well as P-bodies and stress granules in the cytoplasm (Spector 2006).
A striking feature of RNA granules as a structural class is that they maintain defined protein and RNA compositions in the absence of an enveloping membrane. In this review, we will focus on the potential mechanism(s) for the formation and regulation of a specific class of cytoplasmic RNA granule known as the stress granule (SG). Unlike other RNA granules, these cytoplasmic structures are not constitutively present; instead, their formation is induced by cellular stress, such as heat shock or oxidative stress, and they disassemble once the perturbation subsides. Notably, morphologically similar cytoplasmic inclusions are observed in neurons of patients with amyotrophic lateral sclerosis (ALS), frontotemporal lobar degeneration (FTLD) and other neurodegenerative disease, which often exhibit compositional overlap with endogenous SGs. These morphological phenotypes suggest that the formation or disassembly process of these structures may be important for neurological pathogenesis. We will conclude this chapter by focusing on the pathogenic roles of two RBPs — FUS and TDP-43 — and how their mutations link dysregulation of stress granules to ALS/FTLD.
2 Stress Granules
Stress granules (SGs) were first discovered in mammalian cells (Collier and Schlesinger 1986; Collier et al. 1988; Arrigo et al. 1988; Kedersha et al. 1999), and similar structures were later identified in Drosphila and yeast cells (Farny et al. 2009; Hoyle et al. 2007; Buchan et al. 2008; Grousl et al. 2009). Electron microscopy revealed that mammalian SGs are spheroid or ellipsoid structures that are usually 1–2 μm, but can range from 0.4 to 5 μm (Spector 2006; Souquere et al. 2009). SGs can be induced upon a variety of stresses that inhibit translation initiation, including heat shock, oxidative stress, hypoxia, inhibition of mitochondrial function, glucose starvation, proteotoxic stress, and infection by certain viruses (Moeller et al. 2004; Kedersha and Anderson 2007; Anderson and Kedersha 2008; White and Lloyd 2012). The blocks to protein synthesis under these conditions is evident by the concomitant loss of conventional “polysomal rosettes” (Warner et al. 1962; Souquere et al. 2009). Although SGs are intimately tied to polysome disassembly, translation inhibition is not in itself sufficient to induce their aggregation since pharmacological inhibition of 60S subunit recruitment can halt translation without SG formation (Mokas et al. 2009). Rather, a defining feature for mammalian SGs has been the presence of stalled 48S pre-initiation complex subunits (Anderson and Kedersha 2006). In the following subsection, we proceed with an introduction to the molecular composition and proposed functions of SGs.
2.1 Molecular Composition
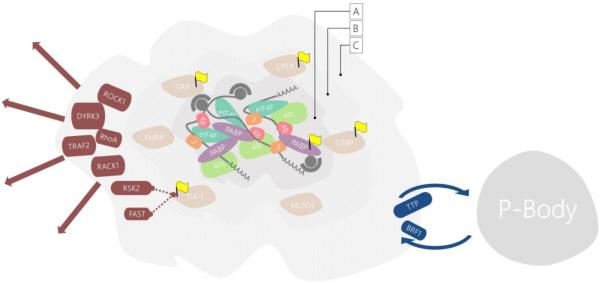
Inhibition of translation initiation is the trigger for SG formation, and subcomponents of the 48S complex — including eukaryotic initiation factors eIF3, eIF4F, eIF4B, the small ribosomal subunits, poly(A) binding protein PABP1, and stalled mRNA transcripts — likely initiate SG assembly through a series of concerted protein–protein and protein–RNA interactions. These macromolecules comprise the core class of SG components (Anderson and Kedersha 2008) (Figure 1).
Figure 1. Classes of SG components.
(A) The defining first class of SG components includes subcomponents of the 48S pre-initiation complex, along with stalled mRNA transcripts. (B) A second class of proteins that either bind mRNA directly or are partners of RBPs. Many of these proteins contain unstructured domains and can nucleate SG assembly. Some are commonly used as “markers” to detect SGs by immunofluorescence (yellow flags). (C) Tertiary “piggy back” proteins, many of which function in cell signaling pathways (red arrows).
Additional protein components are incorporated into SGs after the primary initating event, including a large number of other RBPs, many of which are known regulators of mRNA translation and stability (Anderson and Kedersha 2006). Intriguingly, proteins within this class, including TIA-1, G3BP1, CPEB, FXR1, FMRP, and MLN51 (Kedersha et al. 1999; Mazroui et al. 2002; Wilczynska et al. 2005; Kedersha et al. 2005; Baguet et al. 2007), can induce spontaneous formation of SGs when overexpressed even in the absence of stress. Many proteins in this class, such as TIA-1 and G3BP1, are often used as protein markers to identify SGs using fluorescence microscopy (Kedersha and Anderson 2007). A final class of SG components consists of tertiary “piggy-back” components, which might not function in RNA metabolism per se, but are recruited into SGs by protein–protein interactions (Anderson and Kedersha 2008). Recent exploration of these proteins, DYRK3 and RACK1 being notable examples, has suggested expanded functions for SGs in a variety of cell-signaling pathways including apoptosis and the mTOR pathway (Kedersha et al. 2013) (see section 2.2).
It is important to note that the composition of SGs is variable and depends on the type of stress used to induce them. For example, heat shock protein Hsp27 is found in SGs in heat-shocked mammalian cells, but not in cells subjected to arsenite treatment (Kedersha et al. 1999), while poliovirus-induced SGs contain a unique marker, Sam68 (Piotrowska et al. 2010). Similarly, in yeast, SGs induced by glucose deprivation contain the heterogeneous nuclear ribonucleoprotein (hnRNP) Hrp1, whereas those that form under inhibition of mitochondrial respiration do not (Buchan et al. 2011). Finally, while yeast SGs formed during robust heat shock at 46°C for 10 minutes contain the canonical components eIF3 and the 40S ribosomal units (Grousl et al. 2009), those formed under glucose-deprived conditions, surprisingly, do not (Hoyle et al. 2007; Buchan et al. 2008). These data suggest that, rather than a series of defined binding events proceeding orderly from a nucleating mRNP, SGs more likely exist along a continuum of compositional states dependent on the pathways activated by specific stress conditions (Buchan and Parker 2009). For example, TTP is removed from SGs when phosphorylated by MK2 (Stoecklin et al. 2004). Thus, SG composition depends on how these stressors cause the redistribution of various mRNPs between SGs and translation-competent polysomes, as well as between SGs and P-bodies (PBs) — another class of cytoplasmic RNA granule enriched for RNA decay factors. As will be emphasized in subsequent sections, SGs house mechanisms to selectively exclude certain RNA transcripts so as to prioritize translation of protein chaperones (Gilks et al. 2004; Tanaka et al. 2014). Besides these pathways, however, little is known about the identity of other RNA components, whether their recruitment is regulated in a stress-dependent manner, and how this might influence protein composition.
2.2 Proposed functions
Though a global proteomics approach has not yet been used to assess the full extent of proteins recruited into SGs, new components continue to be identified by co-localization studies using defined protein markers and fluorescence microscopy. Since these new components are not necessarily related to RNA metabolism, the proposed functions for SGs are rapidly expanding. Given the presence of translationally stalled mRNA complexes, SGs were originally hypothesized to function as storage sites for non-translating RNAs. However, fluorescence recovery after photobleaching (FRAP) analyses showed that both mRNA transcripts and nearly all protein components shuttle in and out of SGs with half-lives on the order of seconds to minutes (Kedersha et al. 2005; Mollet et al. 2008; Zhang et al. 2011). Thus, it is difficult to envisage that SGs act as static mRNA repositories. Instead, an emerging, working model for SG function posits that they are sites of dynamic mRNP remodeling and sorting, a process which has been coined mRNA triage (Kedersha and Anderson 2002). Under this framework, SGs function as transient structures induced by perturbations to translation initiation, during which they exchange components with polysomes and PBs to prioritize translation or degradation of some mRNA transcripts over others, thereby altering the proteome until stress subsides.
In support of this, strong evidence exists for a direct flow of mRNPs between polysomes and SGs. First, puromycin treatment, which releases ribosomes from translating mRNAs by inducing premature termination of translation, enhances SG formation (Kedersha et al. 2000). Emetine, which functions to the opposite effect by preventing mRNA-ribosome dissociation, both inhibits the formation of SGs in the presence of stress (e.g. sodium arsenite treatment) and even “dissolves” pre-formed SGs (Kedersha et al. 2000). Combined with the quick turnover rates as assessed by FRAP, these data suggest that SGs are not static structures, but are constantly “fed” by translationally stalled mRNAs originating from polysomes. Moreover, a specific pathway of mRNP trafficking between SGs and polysomes was recently characterized in yeast, facilitated by the ATP-dependent RNA helicase Ded1 (Hilliker et al. 2011). Ded1 binds eIF4G1 and interrupts translation, forming a translationally inactive pre-initiation complex with eIF4F and stalled mRNA which then accumulates in SGs (Hilliker et al. 2011). Upon ATP hydrolysis, Ded1 is thought to release the pre-initiation mRNP, regulating its return to translation (Hilliker et al. 2011). The mammalian orthologue DDX3 has been found to interact with eIF4E, a binding partner of eIF4G in the mRNA cap structure (Shih et al. 2008), though it remains unclear whether it carries specific roles in polysome/SG trafficking.
Another corner of this triage model is grounded in interactions between SGs and PBs. In a subset of cellular stress models, SGs and PBs have been observed to physically interact. Under sodium arsenite treatment, for example, PBs can make contact with SGs while remaining morphologically distinct (Souquere et al. 2009), or may otherwise exist in close proximity with them (Wilczynska et al. 2005; Kedersha et al. 2005). In a series of experiments utilizing time-lapse microscopy in conjunction with transient expression of tagged SG or PB protein markers, interactions between the two bodies were observed to vary — PBs appeared stably bound to SGs in some cases, whereas in others they were only intermittently attached (Kedersha et al. 2005). These interactions, which might only be observed in a fraction of imaged cells, can be significantly enhanced by overexpression of certain shared proteins. Most notably, transient expression of the mRNA destabilizing factors TTP and BRF1 were shown to induce quantitative SG/PB docking, extending the tethered interaction to 1 hour (Kedersha et al. 2005). Similarly, overexpression of CPEB1, a protein observed to localize to both types of RNA granules, resulted in PB clustering around spontaneously induced SGs (Wilczynska et al. 2005).
Beyond the mRNA triage model, an additional key functional property of SGs is the acute and localized enrichment of proteins responsible for RNA metabolism and various signaling pathways. Indeed, dozens of different proteins have been found to localize to SGs (Anderson and Kedersha 2008), and the local concentration of these components may either serve to increase the rate of biochemical reactions within SGs or reduce the cytosolic concentration of other proteins by sequestration, resulting in spillover effects that propagate through a variety of cell signaling pathways (Kedersha et al. 2013). For example, a number of emerging lines of evidence strongly suggest that one major function of SGs is to regulate apoptosis (Eisinger-Mathason et al. 2008; Arimoto et al. 2008; Gareau et al. 2011; Thedieck et al. 2013; Kedersha et al. 2013). First, SGs sequester pro-apoptotic factors such as RACK1 and TRAF2 from the cytosol to suppress the activation of cellular death pathways via MAPK or NF-kB signaling (Kim et al. 2005; Arimoto et al. 2008). Similarly, the sequestration of small GTPase RhoA and its downstream kinase ROCK1 by SGs prevents the activation of JNK which would otherwise trigger apoptosis (Tsai and Wei 2010). Second, SGs recruit anti-apoptotic factors, such as RSK2 and FAST, which bind and inactivate apoptosis promoting factor TIA-1 (Li et al. 2004; Eisinger-Mathason et al. 2008). Third, SG formation stabilizes mRNAs encoding anti-apoptotic protein p21WAF1/CIP1 (Gareau et al. 2011). Consistent with these findings, impairment of SG assembly often leads to a higher rate of cell death following stress exposure (Baguet et al. 2007; Kwon et al. 2007; Eisinger-Mathason et al. 2008). In addition, SG formation appears to regulate the mTOR pathway, which monitors nutrient and energy availability to promote either cell growth when conditions are favorable or catabolic processes during stress (Hofmann et al. 2012; Wippich et al. 2013; Fournier et al. 2013; Thedieck et al. 2013). In this case, the kinase DYRK3, which itself is recruited into SGs, phosphorylates the mTORC1 inhibitor PRAS40, allowing mTORC1 to exit SGs in an activated state to fulfill its signaling functions in cell growth and metabolism (Wippich et al. 2013).
In summary, SGs are implicated as regulatory hubs for mRNA sorting, stress signaling, and apoptosis. Besides various physiological stresses, SGs can also be induced by radiotherapy and chemotherapeutics, as is the case with the proteasome inhibitor Velcade in solid tumors (Moeller et al. 2004; Fournier et al. 2010). The persistent presence of cytoplasmic inclusions containing SG components in hypoxic tumor cores and ALS/FTLD patient neurons has been linked to chemotherapy resistance in cancers and neurodegeneration, respectively (Arimoto et al. 2008; Li et al. 2013). On the other hand, SGs seem to also serve in antiviral functions given that various viruses inhibit SG formation immediately upon infection (White and Lloyd 2012; Onomoto et al. 2014). Thus, it is critical to identify the molecular mechanisms that hold SGs together in physiological and pathophysiological contexts. In the next section, we will focus on a molecular model of how non-membranous RNA granules can be formed in a way that allows for select proteins to be retained while others move in and out readily.
3 RNA granule formation via multivalency and intrinsically disordered regions
An evolving framework of RNA granule formation, which works to explain assembly, regulation, and selectivity in recruitment, is grounded in the biophysical mechanisms facilitated by two properties: (1) multivalency mediated by proteins or RNA, and (2) low affinity protein–protein interactions involving intrinsically disordered regions of low complexity amino acid composition (i.e., a string of repetitive amino acids).
3.1 Multivalency
A number of recent experimental studies have highlighted how multivalent macromolecules, including both multi-domain proteins and RNA, are critical for the assembly of higher-order structures. Recently, the Rosen group systematically explored the importance of non-covalent interactions between multi-domain proteins in the formation of μm-sized cellular structures (Li et al. 2012; Banjade and Rosen 2014). Specifically, they studied the interactions between SRC homology 3 (SH3) domains and proline-rich motif (PRM) ligands, both of which are commonly found in cell signaling pathways. Chimeric proteins engineered with one to five SH3 domains were mixed incrementally with proteins containing up to 5 repeats of PRM ligand (Li et al. 2012). They found that a mixture of protein with four SH3 domains and four tandem PRM ligands resulted in an opalescent solution containing numerous droplets of 1–50 μm in diameter phase-separated from the bulk of the solution (Li et al. 2012). In contrast, no such structures could be formed in a solution of SH3 domains or PRM ligands alone. Proteins within the phase-separated droplet were concentrated by about 100-fold relative to the surrounding solution. This phenomenon, commonly referred to as liquid–liquid demixing, is reminiscent of non-membranous structures in vivo, which maintain high local protein concentrations while allowing for fluid component exchange with the surrounding bulk phase. As indicated by this study, the transition point for forming such microscopically visible structures is usually sharp and dependent on molecule valency (Li et al. 2012; Banjade and Rosen 2014). Consistently, studies have suggested that a critical threshold protein level is required to form RNA granules and that this amount is inversely correlated with the valency of the molecule (reviewed in Hyman and Simons 2012; Brangwynne 2013; see Box 1).
The assembly of these phase-separated higher-order structures is critical for the physiological functions of the proteins that are recruited to them. For example, SH3–PRM interactions are observed naturally in the nephrin–NCK–N-WASP system — a three-component interaction on the membranes of kidney podocytes that is involved in the formation of the glomerular filtration barrier, which participates in actin assembly. Applying the same methodology as described above, the Rosen group found that interactions between the three SH3 domains of NCK and the six PRMs in N-WASP were sufficient to induce the formation of phase-separated droplets and stimulate actin assembly in vitro. In addition to three SH3 domains, each NCK molecule has one SH2 domain that can bind to phosphorylated tyrosine. Addition of the phosphorylated nephrin tail, which contains three phosphorylated tyrosine sites, significantly reduced the critical amount of NCK and WASP proteins required to form phase-separated droplets (Li et al. 2012). Thus, in cells, protein kinases can regulate the level of nephrin phosphorylation, which in turn regulates a cascade of multivalent interactions to form higher-order structures.
In addition to multi-domain proteins, phase transitions can be triggered by other macromolecules capable of multivalent interactions, including RNA —the categorical component of RNA granules. A single RNA can serve as a scaffold of repeating units that allows for the binding of multiple RNA-binding proteins. Parallel to the multi-domain protein studies, the Rosen group has also demonstrated that phase-separated droplets can be formed in vitro by incubating a tetravalent RNA binding protein polypyrimidine-tract-binding protein (PTB) with an RNA oligonucleotide containing five repeats of the synthetic polypyrimidine tract, UCUCUAAAAA (Li et al. 2012). Analogous pathways of protein concentration via nucleating RNAs have been proposed for the higher-order assembly of FUS protein and nuclear bodies (Teixeira et al. 2005; Shevtsov and Dundr 2011; Schwartz et al. 2013). For example, Schwartz and colleagues (2013) have recently shown the importance of RNA for FUS to assemble and bind to the C-terminal domain of RNA polymerase II. The addition of less-than-stoichiometric amounts of RNA to a FUS protein solution induced liquid droplets much more readily than occurred in a solution of FUS alone (Schwartz et al. 2013). Combined with the finding that FUS binds RNA with high cooperativity (due to multiple RNA binding sites), the authors suggest that FUS oligomerization is nucleated by a tightly packed initiating FUS–RNA complex (Schwartz et al. 2013). Last but not least, it is worth noting that other cellular macromolecules besides proteins and RNA — such as poly-ubiquitin or poly(ADP-ribose) — are also found in SGs and could potentially serve as multivalent scaffolds by recruiting proteins to their repeating units (Kwon et al. 2007; Leung et al. 2011; Leung 2014) (see Section 3.3).
3.2 Intrinsically Disordered Regions
Intrinsic disorder has emerged as an essential mechanism in a diverse array of pathways, including the formation of RNA granules. Previously believed to be passive separators of functional domains, intrinsically disordered regions (IDRs) dynamically transition between structural states in order to serve multiple purposes in an efficient and highly regulated manner. A subclass of IDRs that is both essential to RNA granule physiology and holds important implications for neurological disease is the prion-like domain (PrLD). Prions are self-templating proteins originally discovered in yeast that serve in a variety endogenous functions and are capable of entering highly stable aggregates termed amyloids (Shorter and Lindquist 2005). These characteristics stem from a prion domain of low amino acid complexity, typically 60 residues in length and enriched in asparagine, glutamine, tyrosine, and/or glycine residues (King et al. 2012). Application of an experimentally validated bioinformatics algorithm (Alberti et al. 2009) to the human proteome revealed that 1% of proteins harbor predicted domains exhibiting a similar aggregation propensity to yeast prions (Couthouis et al. 2011). Within this 1%, however, there is a 12-fold enrichment for proteins that also contain at least one RNA recognition motif (RRM) (King et al. 2012). Significantly, many of these “prionogenic” RBPs are known components of SGs (Li et al. 2013) or RNA granules in general (Kato et al. 2012).
Studies from the McKnight group have elucidated possible roles for low complexity domains (LCDs) in the formation of RNA granules. McKnight and colleagues fortuitously discovered that a small molecule chemical, biotinylated isoxazole (b-isox), precipitated a class of proteins from cell-free lysate exhibiting remarkable overlap with known components of RNA granules (Kato et al. 2012). Despite an exceptionally high enrichment for RBPs within this precipitated class, the LCDs, not the RRMs, of these RBPs were found to be necessary and sufficient for b-isox mediated precipitation out of lysate (Kato et al. 2012). Notably, purified samples of the FUS or hnRNPA2 LCDs formed hydrogels at high concentrations via oligomerization-mediated phase transition (Kato et al. 2012). Besides this capacity for self-association (homotypic trapping), FUS and hnRNPA2 LCD-containing hydrogels were also able to retain the LCDs of other RBPs (Kato et al. 2012). As revealed by transmission electron microscopy and light microscopy, the hydrogels are composed of morphologically uniform amyloid-like fibers which incorporate other proteins via LCD interactions (Kato et al. 2012). However, unlike pathogenic fibers in amyloid plaques and prion diseases, the formation of these fibers is reversible. Conjoint to these studies, the McKnight group also analyzed the mRNAs pulled down by b-isox mediated precipitation using high-throughput sequencing (Han et al. 2012). They found that the precipitated mRNAs are also closely aligned with those known to partition into RNA granules. More intriguingly, the authors found that these mRNAs could also be retained by hydrogels made by the FUS LCD alone. This surprising selective mRNA retention suggests a model wherein RBPs associate with one another via their LCDs, pulling in select mRNAs via an RNA-binding domain distinct from the LCD-mediated oligomerization (Han et al. 2012).
The utility of PrLDs/LCDs is grounded in their ability to self-associate, and this propensity to rapidly oligomerize through low affinity protein–protein interactions might underpin the dynamic physical properties that characterize SGs and other RNA granules (Gilks et al. 2004; Li et al. 2013). Consistent with how readily SGs have been observed to exchange components and dissolve after stress release, an additional advantage for PrLD/LCD-mediated aggregation might therefore be high reversibility. The regulatory mechanisms that control these transient interactions are also being explored and will be discussed in subsequent sections. We note, finally, that PrLDs are typically also composed of low complexity amino acid sequences, frequently Q/N or QGSY-rich, and that both are subtypes of intrinsically disordered domains. The two terms PrLD and LCDs are often used interchangeably in the literature and are synonymous in this text as well.
3.3 SG assembly
As with their function, the precise mechanisms of SG assembly are not fully understood and likely vary alongside composition under different stress states or points of disruption to the translation initiation processes (Mokas et al. 2009; Buchan and Parker 2009). Since SGs form in response to translational arrest and almost always contain components of the translation initiation complex, the assembly process likely begins with a pool of repressed, translationally inactive mRNPs. Pathways of assembly that proceed from this starting pool of aborted complexes likely involve an mRNP-specific series of RNA–protein and protein–protein interactions mediated by PrLDs/LCDs. Although the initial stages of this process are not understood, there are some known examples of SG nucleation by oligomerization and multivalency-mediated cross linking. The SG component TIA-1, for example, contains a C-terminal PrLD enriched in glutamine residues, bearing high similarity to the aggregation-prone NM domain found in yeast prion Sup-35 (Perutz et al. 1994). In humans, both the prion-like and RNA binding domains of TIA-1 are necessary for SG assembly (Gilks et al. 2004). While the PrLD confers a propensity for aggregation by homotypic oligomerization, the RNA-binding domain is required to recruit RNAs to the complex (Gilks et al. 2004). Indeed, without the RRM, recombinant TIA-1 PrLD fragments form constitutive micro-aggregates when overexpressed, sequestering endogenous TIA-1 and inhibiting SG formation (Gilks et al. 2004). G3BP1, another common SG marker, might mediate assembly in a similar fashion. G3BP1 harbors both an NTF2-like domain prone to dimerization as well as an RNA-binding domain (Tourrière et al. 2003). Though not a PrLD, the NTF2-like domain in conjunction with RNA binding imbues G3BP1 with the ability to interact with multiple partners. As discussed in Sections 4 and 5, multiple RBPs which localize to SGs, such as TDP-43 and hnRNP A2, also contain LCDs. Pathological mutations in these regions might be responsible for impairing the reversibility of RNA granule formation in neurons, resulting in neurodegeneration. Thus, the preponderance of SG components with both LCDs and RBDs, the in vitro observation of liquid/liquid demixing facilitated by these properties, and the capacity for some of these components to trigger SG formation upon overexpression builds a strong case that reversible protein/RNA crosslinking plays a role in the assembly process. Importantly, pathways for how these mechanisms might be regulated are also beginning to be uncovered. Chaperone proteins and post translational modifications both play major roles in controlling the selective aggregation of heterogenous mRNPs into functional, higher order structures.
3.3.1 Regulation by chaperones
Prion-like interactions are a double-edged sword. They provide many of the key features of RNA granule physiology, but are prone to non-specific, potentially pathological aggregation as well. Recent data suggest that protein chaperones are required to continuously keep PrLDs in check, titrating the number of disordered domains available to interact in the cytoplasm, or altering their conformations to inhibit aggregation. In yeast, for example, highly stable amyloids can be returned to solubility by the heat shock protein HSP104, which works in conjunction with HSP40 and HSP70 to refold and reactivate denatured proteins locked within aggregates, thereby acting as a disaggregase (Glover and Lindquist, 1998). Indeed, the formation and propagation of TIA-1 aggregates in yeast is entirely dependent on the activity of HSP104, a known regulator of many Q/N-rich proteins (Li et al. 2014). Interestingly, HSP70 is expressed at higher levels after transfection with the PrLD of TIA-1 and can inhibit the latter's aggregation when cotransfected into COS7 cells (Gilks et al. 2004). It is also known that under sodium arsenite treatment, HSP70 mRNA is shielded from recruitment into SGs by the translation regulating protein YB-1, resulting in its preferential translation under stress (Tanaka et al. 2014). Besides YB-1, an additional mechanism of control has been elucidated by Boyault et al. (2007), based on the activation of heat-shock factor 1 (HSF1) by the cytoplasmic deacetylase HDAC6, which has known disaggregase function as well. Under basal conditions, HSF1 resides in a dormant complex along with HDAC6 and HSP90. Upon binding ubiquitin, HDAC6 releases HSF1 to transcriptionally activate a number of heat shock protein-encoding genes, including HSP70 (Boyault et al. 2007). SGs were found to be enriched for ubiquitin and HDAC6 was shown to be an indispensable component under a wide variety of stress conditions (Kwon et al. 2007). Fascinatingly, the HDAC6-mediated activation of HSF1 requires another disaggregase—the chaperone-like AAA ATPase P97/VCP which is itself a SG component—in order to “recycle” ubiquitinated HDAC6 back into the dormant HDAC6/HSF1/HSP90 complex (Boyault et al. 2007). As will be discussed later, mutations in the gene encoding VCP have been implicated in the neurodegenerative disease ALS. Thus, heat shock proteins may be a general protective mechanism in the regulation of unfolded proteins, either lowering the concentration of prion-like substrates or inducing conformational changes that make them less aggregation-prone (Rikhvanov et al. 2007). In both cases, it seems likely that proteins containing PrLDs/LCDs are continuously kept in check by heat shock proteins, and that a shift in chaperone concentration can induce aggregation (Gilks et al. 2004).
3.3.2 Regulation by post translational modifications
Finally, post-translational modifications have also been observed to enhance or inhibit assembly of SGs. In the case of G3BP1, oligomerization is blocked by phosphorylation on Ser149, and dephosphorylation is required for its ability to nucleate SGs (Tourrière et al. 2003). Similarly, hnRNP A1 is only recruited to SGs when phosphorylated by Mnk1/2 (Guil et al. 2006), whereas TTP and BRF1, two common SG components that bind to AU-rich element (ARE)-containing mRNAs, can be excluded by MK2 and PKB mediated phosphorylation respectively, with downstream effects on ARE transcript degradation (Stoecklin et al. 2004; Schmidlin et al. 2004). Phosphorylation of human antigen R (HuR) by Janus kinase 3 (JAK3) prevents its recruitment into SGs, accelerating the decay of SIRT1 and VHL mRNAs (Yoon et al. 2013). A recent study by the Pelkmans group has demonstrated a role for phosphorylation of LCDs in the regulation of SG assembly/disassembly. Dual specificity kinase DYRK3 cyclically partitions between SGs and the cytosol (Wippich et al. 2013) and such partition is proposed to be regulated by its auto-phosphorylation activities at the N-terminal LCDs. Consistent with this proposal, the expression of N-terminal LCDs alone or a kinase-dead mutant of DYRK3 induces SG formation in the absence of stress, while inhibition of this kinase by GSK-626616 prevents SG disassembly (Stoecklin et al. 2004; Wippich et al. 2013). Thus, phosphorylation functions as a protein-specific and reversible mechanism to either prevent or trigger recruitment of components to SGs.
Besides phosphorylation, poly(ADP-ribosyl)ation has been implicated in regulating SG assembly/disassembly (Leung et al. 2011). Poly(ADP-ribosyl)ation refers to the addition of two or more ADP-ribose units onto proteins, where the addition is regulated by a family of 17 ADP-ribosyltransferases, commonly referred to as PARPs (Hottiger et al. 2010). SGs contain five out of the 17 PARPs as well as two isoforms of the degradative poly(ADP-ribose) glycohydrolase PARG in humans (Leung et al. 2011). Three pieces of evidence suggest that poly(ADP-ribosyl)ation is intrinsically linked to SG structural integrity (Leung et al. 2011). First, overexpression of any of the SG-localized PARPs induces SG formation in the absence of stress. Second, overexpression of PARG inhibits SG formation in the presence of stress. Third, PARG knockdown delays SG disassembly. Several proteins, including G3BP1 and TIA-1, are poly(ADP-ribosyl)ated upon stress, but their roles of these modified substrates in SG assembly and functions remains to be determined. A recent informatics analysis indicate that RNA granule proteins enriched for LCDs are preferentially poly(ADP-ribosyl)ated, indicating poly(ADP-ribose) may potentially direct cellular organization (Leung 2014).
To summarize, multivalent protein–protein and RNA–protein interactions underlie the formation of RNA granules, which resemble liquid droplets phase-separated from the bulk of solution in vitro. The fluid-fluid demixing observed in the formation of these granules relies on a protein species' ability to oligomerize, forming a scaffold and incorporating other protein or RNA components as binding partners (Hyman and Simons 2012). Emerging research increasingly points towards the role of intrinsically disordered regions, such as PrLDs/LCDs, as necessary in the assembly of SGs and other RNA granules. These regions likely switch conformational states to facilitate/disengage interactions within RNA granules, which are regulated by a combination of post-translational modifications and chaperones (van der Lee et al. 2014). As a whole, the system relies on a delicate equilibrium between intrinsic disorder and the regulatory mechanisms that restrain misaggregation. However, the complexity of this balancing act translates to openings for disruption, and SGs specifically have been inextricably linked to the pathogenesis of two neurological diseases in particular – ALS and FTLD.
4 ALS and FTLD: when aggregation goes awry
At the present time, ALS and FTLD are among the best-characterized neurological diseases, and both increasingly appear to be at least partially linked to endogenous RNA granules. ALS is a clinical entity characterized by the progressive loss of motor neuron function, resulting in muscle wasting and eventually respiratory failure (Rowland and Shneider 2001). Approximately 10% of cases have familial origin (f.ALS), with the remaining majority occurring in individuals sporadically (s.ALS) (Rowland and Shneider 2001). In contrast, frontotemporal lobar degeneration (FTLD), the second most common form of dementia behind Alzheimer's, proceeds with neuronal atrophy in the frontal and temporal cortices, causing patients to undergo changes to personality, behavior, and language abilities (Rabinovici and Miller 2010). Both diseases are accompanied by a wide variety of possible genetic mutations resulting in clinical heterogeneity and notably, symptomatic overlap in a large subset of patients, suggesting that the two are linked by a spectrum of pathologies (Ling et al. 2013). This section will overview one facet of the many genetic and molecular factors that comprise the ALS/FTLD disease system: the pathological role of RBPs and RNA metabolism. Special emphasis will be paid to the mechanisms by which many of the same protein properties that allow for proper SG function can be co-opted or otherwise disrupted by genetic and environmental perturbations in these neurological diseases. Several hypothesized pathways of neurodegenerative toxicity are subsequently discussed.
4.1 SGs and cytoplasmic pathological inclusions
One of the few features that is common across the majority of ALS and FTLD variants is the presence of abnormal cytoplasmic inclusions in patient histology. The morphology, composition, and tissue distribution of these inclusions can vary but they nevertheless bear a resemblance to physiological non-membranous granules. It was the discovery that TAR DNA-binding Protein 43 (TDP-43) and Fused in Sarcoma (FUS) were components of the majority of these inclusions that both shifted attention to the role RBPs might play in neurodegeneration and suggested a connection to SGs in neurological pathogenesis (Robberecht and Philips 2013).
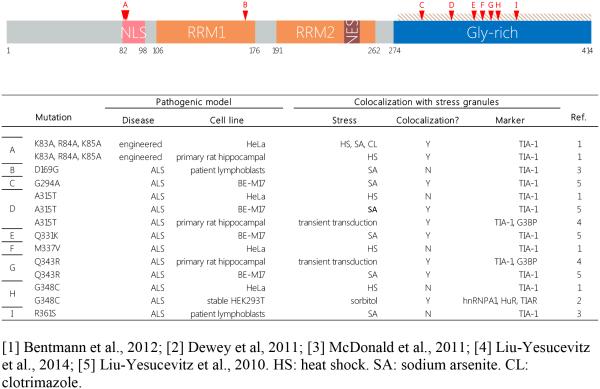
TDP-43 is a member of the hnRNP family and contains two putative RRMs biased towards GU-rich intronic targets (Tollervey et al. 2011) or exceptionally long introns (Lagier-Tourenne et al. 2012). In addition, it has a glycine-rich C-terminal PrLD and a bipartite nuclear localization signal (NLS) between the N-terminus and RRM1 (Figure 2). Following the revelation that TDP-43 was a component of pathological inclusions in ALS/FTLD models, the first concrete link to SGs came from observations that TDP-43 and some of its pathological mutants also co-localized with canonical SG markers in a diverse array of cell types, stress conditions, and mutation models. Indeed, though mutations to TARDBP, the gene encoding TDP-43, are implicated in only 4% of f.ALS cases and are rarer still in s.ALS (Chiò et al. 2012), TDP-43 protein is present in 45% of FTLD and a full 97% of ALS inclusions (Ling et al. 2013). It is now known that endogenous TDP-43 is a consistent component of SGs (Robberecht and Philips 2013). HeLa cells subjected to oxidative or heat stress, for example, exhibit up to 56% depletion of TDP-43 from the cytoplasm, corresponding with a quantitative increase in RIPA buffer-insoluble TDP-43 localized within putative SG aggregates (McDonald et al. 2011). Though TDP-43 knockdown does not in itself prevent SG assembly, lowered expression of TDP-43 has been shown to alter cytoplasmic levels of two other SG components, TIA-1 and G3BP1, causing a delay in SG formation, the formation of smaller SGs, lowered persistence after stress release, and diminished cell viability (McDonald et al. 2011). A follow-up report further confirmed that the modulation of G3BP1 mRNA levels by TDP-43 dictates SG size (Aulas et al. 2012). Most recently, the Vande-velde group showed that by indirectly controlling the G3BP1-dependent coalescence of smaller SGs into larger structures, TDP-43 also affects SG-PB docking, which partially determines mRNA fate after interruptions to translation (Aulas et al. 2015). siRNA against TDP-43 resulted in the progressive and step-wise loss of poly(A)mRNA after oxidative stress, suggesting a functional role in directing the flow of mRNA transcripts out of stalled polysomes. In the context of the RNA triage model, the fact that poly(A) levels are diminished despite the fact that SG/PB docking is also attenuated can only mean that transcripts are routed directly to PBs for degradation in the absence of G3BP1 (Aulas et al. 2015). Therefore, by regulating levels of TIA-1 and G3BP1, TDP43 contributes to both SG assembly and mRNA triage.
Figure 2. Schematic representation of the functional domains of TDP-43.
TDP-43 is a 414-amino acid RBP with functions in transcriptional repression, splicing, and translational regulation. The vast majority of ALS-associated mutations occur within the C-terminal glycine rich PrLD. Table only includes data on studied links between ALS mutations and SG association in cellular models. Mutations are marked by red arrows and letters corresponding to table entries. NLS = Nuclear localization signal; RRM = RNA recognition motif; NES = Nuclear export signal; Tan shading = PrLD.
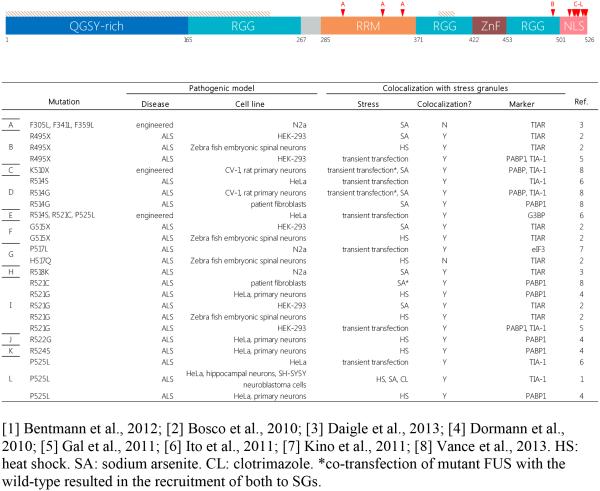
FUS carries a contiguous pair of PrLDs at the N-terminus, an RRM, two RGG domains flanking a zinc finger, and a NLS at the C-terminus (Iko et al. 2004) (Figure 3). FUS, though significantly less abundant in neurological inclusions (<1% in ALS, ~9% in FTLD), shares many functional similarities with TDP-43 – they both shuttle between the cytoplasm and nucleus, play roles in RNA metabolism, and exhibit prion-like behavior (Ling et al. 2013). Unlike TDP-43, SGs form and behave normally in FUS-depleted cells (Aulas et al. 2012). Furthermore, endogenous FUS is not a typical SG component, remaining predominantly nuclear during the stress response (Dormann et al. 2010; Gal et al. 2011; Aulas et al. 2012), though there have been observations of some WT FUS association with the SG markers TIA-1 and TIAR in a fraction of stressed cells (Andersson et al. 2008; Blechingberg et al. 2012), and more particularly in cells subjected to “hyperosmolar” stress (Aulas et al. 2012; Sama et al. 2013). Interestingly, however, siFUS-treated cells have been shown to undergo a 3-fold increase in the number of P-bodies labeled with Dcp1a, indicating a partial role in mRNA triage (Aulas et al. 2015).
Figure 3. Schematic representation of the functional domains of FUS.
FUS is a 526-amino acid RBP with functions in transcriptional repression, DNA damage repair, and splicing. The majority of ALS-linked FUS mutations occur within its NLS, linking its mislocalization to neurodegenerative pathogenesis. Table only includes data on studied links between ALS mutations and SG association in cellular models. Mutations are marked by red arrows and letters corresponding to table entries. NLS = Nuclear localization signal; RRM = RNA recognition motif; ZnF = Zinc finger domain; RGG = Arginine-Glycine-Glycine-rich domain; Tan shading = PrLD.
Strikingly, in ALS and FTLD disease models, recruitment of both proteins to SGs is frequently observed and can be dysregulated by the introduction of mutations. Multiple known SG components, such as eIF3, TIA-1, HuR, TIAR and G3BP1, have been shown to localize with cytoplasmic inclusions seen in ALS/FTLD patients (King et al. 2012). For example, overexpression of wild-type TDP-43 or transfection with truncated TDP-43 fragments commonly observed in patients exhibiting TDP-43 pathology yielded co-localization with SG markers in human neuroblastoma BE-M17 cells even in the absence of stress (Liu-Yesucevitz et al. 2010). In the same study, TDP-43+ inclusions were shown to overlap with eIF3 and TIA-1 in both ALS and FTLD donor brain samples (Liu-Yesucevitz et al. 2010). These parallels between cytoplasmic inclusions and SGs have spurred considerable interest in how endogenous mechanisms of RNA granule regulation can go awry, resulting in the altered composition, stability, and accumulation that categorically distinguish pathogenic inclusions from those observed in the physiological stress response.
4.2 FUS, TDP-43 and neurodegenerative pathogenesis
The multimerization-mediated phase transitions that imbue RNA granules with their dynamic, fluid-like properties rely on the tight regulation of protein concentration (see Box 1). A considerable body of evidence now indicates that disruptions to the equilibrium of protein concentration are causative factors in neurodegenerative pathogenesis. In both ALS and FTLD, a striking number of known mutations disrupt TDP-43 and FUS shuttling between the nucleus and cytoplasm or enhance their propensity to aggregate, with implications in the formation of insoluble cytoplasmic inclusions and ultimately cell toxicity.
4.2.1 Mislocalization of FUS from the nucleus to the cytoplasm
A number of C-terminal missense mutations linked to ALS have been shown to disrupt nuclear import of FUS, resulting in cytoplasmic accumulation (Dormann et al. 2010). Importantly, the degree to which nuclear import is disrupted seems to correlate with the severity of the disease. Mutations which result in higher levels of cytoplasmic FUS accumulation were identified in pedigrees exhibiting earlier age of onset (Dormann et al. 2010; Bosco et al. 2010). This trend may be partially explained by the crystal structure of the FUS NLS when bound to its nuclear receptor, Transportin. Unlike the localization signals of other hnRNPs which bind Transportin in a fully extended conformation, the FUS NLS forms an α-helix structure upon binding to Transportin (Zhang and Chook 2012). This α-helix is flanked by an N-terminal hydrophobic motif and a series of basic residues, both of which are conserved in the FUS-related proteins EWS and TAF15 (Zhang and Chook 2012). ALS-associated mutations are clustered within these two flanking epitopes, and a particularly severe mutation identified in early-onset juvenile ALS (P525L) — a residue that makes numerous contacts with Transportin — results in a 9-fold loss in binding affinity (Zhang and Chook 2012).
Dormann and colleagues (2012) subsequently found that the third RGG domain (RGG3) of FUS can also facilitate binding with Transportin, even if the NLS is disrupted by the P525L mutation. However, this FUS RGG3–Transportin interaction can be negated by methylation. This observation led to the surprising finding that cytoplasmic FUS/SG aggregates induced in cells by transient transfection of the P525L mutant were enriched for methylated FUS (Dormann et al. 2012). Post-mortem tissue samples from ALS-FUS patients exhibiting the same cytoplasmic aggregates also contained methylated FUS, whereas three FTLD-FUS subtypes did not (Dormann et al. 2012), possibly suggesting that the pathogenesis of these two closely related diseases may differ. Taken together, Dormann and colleagues posit that epitopes in the NLS anchor FUS to the Transportin binding pocket, allowing RGG3 to stabilize the interaction. Only when the NLS and RGG3 domains are both disrupted, by mutations and methylation, respectively, is nuclear import prevented (Dormann et al. 2012).
4.2.2 Prionogenicity of FUS and TDP-43
Whereas endogenous SGs are reversible and dynamic by nature, stability and persistence are a token trait of pathological inclusions in patient histology (Ito et al. 2011). One particularly compelling explanation for this physical misbehavior has implicated the PrLDs/LCDs present in both FUS and TDP-43 as susceptible to mutation-induced fibrilization (King et al. 2012). Indeed, the endogenous forms of both proteins are known to be aggregation-prone, coalescing into phase-separated droplets (Johnson et al. 2009), or a polymer hydrogel (Kato et al. 2012) when purified in vitro. In bioinformatics assays, based on the predictive algorithm developed by Alberti and colleagues (2009), both FUS and TDP-43 exhibit an exceptionally high prion propensity, ranking 15th and 69th, respectively, out of 27,879 analyzed proteins (Alberti et al. 2009; Cushman et al. 2010). The predicted FUS PrLD encompasses amino acids 1-239, spanning the entirety of its QGSY-rich N-terminus, and most of the adjacent glycine-rich domain. The LCD used by Kato and colleagues to generate a hydrogel in vitro covers residues 2–214 (See Section 3.2) (Kato et al. 2012). In the case of TDP-43, a PrLD spans 138 residues at the C-terminus (Cushman et al. 2010). Emergent data now suggests that mislocalization allows these physical properties to come to bear in the cytoplasm, resulting in aggregation and association with SGs. ALS-linked mutations exacerbate this behavior by altering the PrLD itself in the case of TDP-43, or by elevating cytoplasmic FUS concentration.
4.2.3 Aggregation-prone mutations in FUS and TDP-43
Current models indicate that FUS aggregation is driven by its mislocalization, facilitated by mutations to its NLS and exacerbated by stress-induced recruitment into SGs (Dormann et al. 2010; Bosco et al. 2010). For example, HEK293 cells stably expressing FUS truncation mutants R495X or G515X at near endogenous levels exhibited stable FUS aggregates, corresponding with a cytoplasmic:nuclear FUS expression ratio 30–50 times greater than in cells expressing the wild-type (Bosco et al. 2010). Unlike TDP-43, the first RGG domain of FUS is required in addition to the PrLD in order to aggregate (Fushimi et al. 2011; Sun et al. 2011). Interestingly, this RGG domain houses a short second sequence of prion-like residues which narrowly misses the cutoff of Alberti's prediction algorithm (Sun et al. 2011). In addition, aggregation of FUS with SGs is also RNA-dependent as demonstrated by the fact that FUS association with canonical SG markers can be abolished by RNAse treatment (Vance et al. 2013). Finally, although FUS localization to the cytoplasm is a prerequisite for aggregation with SGs, mutant FUS has been shown to trap endogenous (presumably nuclear) FUS into SG aggregates, most likely by protein–protein interactions facilitated by the FUS PrLD (Vance et al. 2013).
TDP-43 appears to form inclusions by a different sequence of events, perhaps due to its endogenous cytoplasmic function during the stress response. Specifically, the vast majority of TDP-43 mutations are clustered in its C-terminal PrLD, and an emerging model is that these mutations prevent its return to the cytoplasm post stress release by strengthening the protein–protein interactions that drive its incorporation into SGs (Barmada et al. 2010). As an example, ALS-associated mutations have been found to promote TDP-43 aggregate stability (Ling et al. 2010). Specifically, radioactive pulse labeling showed that TDP-43 mutants are degraded as much as 4 times slower than wild-type (Ling et al. 2010). Furthermore, two PrLD mutations in TDP-43 — Q331K and M337V — were found to significantly increase association between TDP-43 and FUS in the nucleus, an interaction that is not normally observed and not present in patient histology (Ling et al. 2010). These same two mutations consistently form more cytoplasmic puncta in yeast models and aggregate more quickly in vitro, demonstrating that point mutations can enhance PrLD-mediated interactions (Johnson et al. 2009). Though this study did not examine co-localization with SG markers, mutations in the TDP-43 PrLD have also been found to alter the dynamics of SG assembly under sorbitol treatment, an oxidative and osmotic stressor capable of targeting endogenous TDP-43 to cytoplasmic structures enriched for the canonical SG markers TIAR and HuR (Dewey et al. 2011). Under prolonged sorbitol exposure, a G348C mutation was found to increase the fraction of primary glial cells containing TDP-43+ SGs, accompanied by the formation of fewer, larger SGs compared to cells expressing the wild-type (Dewey et al. 2011).
4.2.4 Mislocalization without aggregation
An important caveat is that in many cellular models of FUS or TDP-43 pathogenesis, cytoplasmic relocalization is not sufficient to induce aggregation. Absent cellular stress, FUS bearing ALS-associated NLS mutations, for example, was not observed to aggregate or associate with SGs despite extensive cytoplasmic localization (Dormann et al. 2010). Indeed, in most cellular models, even if aggregates do ensue from the mislocalization of FUS alone, they are typically only detected in a minority of cells (Bosco et al. 2010) and exposure to stress increases both the number of cells with cytoplasmic aggregates and the average number of inclusions per cell (Bosco et al. 2010). Dorman and colleagues propose a “2-hit” model for pathogenicity, whereby mutations to the C-terminal NLS first causes FUS to mislocalize to the cytoplasm (step 1), where it then becomes susceptible to stress-induced aggregation and recruitment into SGs (step 2). Similarly, the TDP-43 mutants observed to alter SG assembly dynamics remained nuclear until prompted to relocalize by an external stressor (Dewey et al. 2011). In a separate study, three ALS-associated TDP-43 mutations, all occurring in the C-terminal PrLD, were confined to the nucleus when transiently transfected into HeLa cells and therefore did not recruit to SGs upon heat shock (Bentmann et al. 2012). Consistent with the working “2-hit” model of TDP-43/FUS proteinopathy, all three of these mutations, when introduced in conjunction with a disrupted NLS, resulted in cytoplasmic localization, aggregation, and SG recruitment (Bentmann et al. 2012).
4.3 Aggregation, toxicity, and a role for SGs
Explicit clarification is warranted here to elucidate the relation of cytoplasmic aggregation to cellular toxicity. At the present time, the data for whether aggregation is necessary to convey cellular toxicity is mixed, and how pathogenesis proceeds from cytoplasmic accumulation of TDP-43 or FUS is unknown. Observations from cellular models that toxicity is accompanied by cytoplasmic aggregation are legion, but many reports question whether inclusion formation plays a causative role in pathogenesis or is instead an endogenous cellular response to other disease factors. Gitler and colleagues evaluated molecular determinants for both TDP-43 and FUS mediated toxicity in a yeast model recapitulating the cytoplasmic aggregation observed in ALS patient histology (Johnson et al. 2008; Sun et al. 2011). While the C-terminal PrLD of TDP-43 was found to be necessary and sufficient for both cytoplasmic localization and aggregation, toxicity required an intact RRM2 domain as well (Johnson et al. 2008). Interestingly, constructs bearing only the two RRMs or the C-terminal PrLD were observed to localize and aggregate in the cytoplasm, despite the fact that neither was associated with abnormal cell growth (Johnson et al. 2008). FUS was found to depend on a more complex array of molecular determinants, requiring its N-terminal PrLD, RRM, and first RGG domain to aggregate in the cytoplasm and promote cell death (Sun et al. 2011). Notably, as with TDP-43, this toxicity has been reported to depend on RNA binding as deletion of the RRM rescues the deleterious effects associated with NLS mutants (Daigle et al. 2013). In a final example, Bosco et al. (2010) observed that cells expressing R495X or G515X FUS mutants, both of which robustly localize to the cytoplasm and form aggregates, remained perfectly viable, and no signs of toxicity were identified. As will be discussed, these conflicting data on TDP-43/FUS toxicity might be partially explained by the intrinsic features that are unique to neurons, making the selection of proper cellular systems especially critical for studies examining pathways of toxicity.
4.3.1 Neuron-specific toxicity
Some have argued that cytoplasmic localization, not the formation of inclusion bodies, provides a superior correlate for cell death (Barmada et al. 2010). In support of this, preventing TDP-43 shuttling out of the nucleus by a mutation in the nuclear export signal rescued the toxicity of A315T mutants (Barmada et al. 2010). Studies arguing either that mutations in the PrLD do not cause cytoplasmic mislocalization until stress induction, or that mislocalization is not toxic — typically do not use a primary neuronal cell line, neglecting possible “neuron-specific mechanisms that govern metabolism and distribution” (Barmada et al. 2010). Indeed, the TDP-43 mutations A315T, G348C, and A382T, were only found to aggregate in the cytoplasm and confer toxicity when expressed in motor neurons derived from primary mouse spinal cord cultures, as opposed to tissue culture cell lines COS1 and N2A in which they remained confined to the nucleus and did not influence cell viability (Kabashi et al. 2010). In Drosophila, ectopic localization of FUS was found to be crucial to development of a neurodegenerative phenotype and a simple deletion of the FUS nuclear export signal rescued toxicity (Lanson et al. 2011). One possible explanation is that nervous tissue is enriched for transcripts carrying longer introns compared to other tissues, a characteristic which might carry heightened susceptibility to FUS abnormalities (Tollervey et al. 2011; Polymenidou and Cleveland 2011).
4.3.2 Stress and inclusion solubility
Another notable difference between in vitro studies and patient pathology is that the majority of cell studies aiming to model attributes of ALS and FTLD pathology do not recapitulate the irreversibility of TDP-43 and FUS inclusions in patient histology, with FUS or TDP-43 positive aggregates disassembling readily after removal of stress (Bentmann et al. 2012). In a unique study utilizing mild but prolonged conditions of oxidative stress induced by paraquat in the neuron-like cell line SH-SY5Y, TDP-43 co-localized with the SG markers HuR and TIAR in cytoplasmic inclusions (Meyerowitz et al. 2011). Intriguingly however, these TDP-43 aggregates persisted after stress release or cycloheximide treatment despite the fact that HuR and TIAR had both returned to solubility (Parker et al. 2012). These observations align with the developing notion that the PrLD-mediated assembly of RNA granules might “seed” the progression of aberrant protein aggregates by raising the local concentration of proteins bearing prion-like properties. Alternatively, a combination of post-translational modifications, chaperone-facilitated protein remodeling, and other mechanisms normally involved in RNA granule regulation may be disrupted under conditions of abnormal cellular stress. Indeed the paraquat-induced TDP-43 inclusions were heavily enriched for ubiquitin (Parker et al. 2012). These phenomena might be unique to both cell and stress type, potentially explaining the conflicting findings of many in vitro studies of ALS/FTLD pathogenesis.
4.3.2 Disruptions to endogenous TDP-43/FUS function
If toxicity does not require aggregation, what might be the driving mechanism for TDP-43/FUS-mediated neurodegeneration? To date, TDP-43 and FUS functions have not been fully characterized but both proteins are known to play prolific roles in diverse pathways. Based on what is known, besides a common PrLD-mediated predisposition towards aggregation, FUS and TDP-43 do share some functional overlap. For example, the two proteins have been shown to complex in order to co-regulate the mRNA of HDAC6, the SG component with disaggregase properties and a primary role in activating HSF1 as discussed in section 3.3 (Kim et al. 2010). Moreover, TDP-43 mediates the alternative splicing of FUS mRNAs (Polymenidou et al. 2011). However, there is also considerable evidence for the divergence of TDP-43 and FUS-mediated pathogenesis. In addition to differences in domain requirements for toxicity, yeast screens have shown that there is very little overlap between genetic modifiers of TDP-43 and FUS toxicity (Sun et al. 2011). Depletion of FUS alters the splicing of approximately 1000 mRNAs, most of which differ from those altered upon TDP-43 depletion (Lagier-Tourenne et al. 2012). Furthermore, TDP-43 and FUS have only very rarely been observed to co-localize in immunostaining of pathological inclusions in patient histology (Sun et al. 2011) and their binding partners share little overlap (Lagier-Tourenne et al. 2012). Thus, the root cause of pathogenesis might be the loss of TDP-43/FUS function, which can certainly be facilitated by cytoplasmic aggregation, but not necessarily so. In this model, aggregation induced by mutations, exogenous stressors, or some other as-of-yet unidentified pathway would be an easily visible symptom of disrupted TDP-43/FUS function. The direct cause of pathogenesis, however, would be the resulting dysregulation of RNA homeostasis, which would differ between TDP-43 and FUS pathologies. What is increasingly certain is that the expression of both TDP-43 and FUS is tightly regulated, and a variety of cellular and animal models have demonstrated the deleterious outcomes of altering expression levels of either.
Studies in C. elegans have recapitulated neurological impairment due to expression of human or mutant TDP-43. A transgenic line expressing human WT TDP-43 or a truncation mutant, for example, exhibited severe locomotor deficits, impaired synaptic transmission, and growth defects despite an absence of detectable neuronal loss (Zhang et al., 2011). In this case, though nuclear aggregates were detected in WT-expressing worms and cytoplasmic aggregates were observed in those expressing the truncation mutant, both sets of inclusions were assessed to be relatively immobile by FRAP (Zhang et al., 2011). Importantly, RNAi of HSF1 was also found to dramatically worsen the neurological phenotype (Zhang et al., 2011). In mammals, an important clue for how TDP-43 loss of function might proceed comes from transgenic mouse models in which mislocalization and aggregation are either rarely observed or not observed at all in post-mortem brain samples (Igaz et al. 2011; Arnold et al. 2013). In one study, expression of human TDP-43 was found to correspond with a loss of endogenous mouse TDP-43 in the nucleus – not to the cytoplasm, but through TDP-43's ability to autoregulate its own mRNA by non-sense mediated decay (Igaz et al. 2011). The fact that regions of the mouse brain that did not exhibit signs of neurodegeneration also did not show altered levels of endogenous mouse TDP-43 strongly indicates that the progression of the neurodegenerative phenotype is caused by TDP-43 depletion (Igaz et al. 2011). Genome-wide splicing arrays have also been used to show that depletion of endogenous mouse TDP-43 causes an enhancement of some splicing activities but an abrogation of others, while mice expressing a Q331K TDP-43 mutant exhibited mutation-unique splicing alterations (Arnold et al. 2013). Taken together, these studies demonstrate a strong dependence of TDP-43 function on levels of TDP-43 expression, which is not dependent on inclusion formation, but which does not exclude a role for cytoplasmic aggregation either.
In vivo models for FUS pathologies also yield insight into the deleterious outcomes of altering its expression levels. Transgenic mice homozygous for an inactive FUS allele, for example, fail to suckle and die soon after birth (Hicks et al. 2000). Knock down of the FUS homologue Caz in Drosophila causes degeneration of motor neurons and abnormal locomotive behavior (Sasayama et al. 2012) whereas expression of wild-type or pathogenic mutations of human FUS in Drosophila induces a similarly severe neural impairment (Chen et al. 2011). Therefore, the expression level of FUS protein needs to be critically maintained for normal neuronal function. As in the case for TDP-43, although cytoplasmic aggregates seem unnecessary for pathogenesis, the formation of such higher-order structures likely disrupts the delicate equilibrium of protein concentration required for the endogenous functions of these RBPs. In support of this, two studies have successfully generated transgenic mice expressing the ALS-associated FUS mutation, R521C, a point substitution in the middle of the FUS NLS. In the first of these, expression of mutant human, but not WT human FUS, caused the degeneration of motor axons, the cortex, and the hippocampus, resulting in progressive paralysis (Huang et al. 2011). Interestingly, these ALS/FTLD phenotypes were also accompanied by ubiquitinated aggregates, though neither FUS nor TDP-43 were present within them. A study conducted 3 years later using the same R521C mutation found that an overly stable mutant FUS complex with brain-derived neurotrophic factor RNA was linked to neurological damage to dendrites and synapses (Qiu et al. 2014). Both studies managed to recapitulate a neurodegenerative phenotype exhibiting clear neuronal toxicity without any detectable cytoplasmic aggregation of TDP-43 or FUS, once more implicating RNA dysregulation as the true root of pathogenesis. Interestingly, an in vitro study utilizing patient fibroblasts expressing the exact same R521C mutation caused extensive cytoplasmic FUS localization but also did not contain FUS inclusions under basal conditions (Vance et al. 2013). Upon sodium arsenite treatment, however, the R521C fibroblasts exhibited FUS aggregation and colocalization with SGs whereas the WT did not (Vance et al. 2013). Taken together, these data support the case that FUS mutations are capable of inducing neuronal toxicity without aggregation but that inclusions can still develop under external oxidative stress. This susceptibility will be discussed in more detail in section 4.4.
One final note on FUS proteinopathies is that despite colocalization with common SG markers, pathological FUS granules may have unique properties. Yasuda and colleagues (2013) recently reported that FUS facilitates preferential translation of RNA transcripts within granules that localize to cellular protrusions during cell migration. These RNA granules stained positive for both the SG marker TIA-1 and the tumor suppressor protein adenomatous polyposis coli (APC), but were shown to persist under cycloheximide treatment. Strikingly, tracking nascent protein synthesis with azidohomoalanine labeling revealed that APC-containing FUS granules were translationally active (Yasuda et al. 2013). These data suggest that FUS granules may be distinct from endogenous SGs, which do not possess translation machinery and are sensitive to cycloheximide treatment. Notably, APC RNA granules could be induced by ALS associated FUS mutants, and tissue samples taken from FTLD-FUS patients immunostained positive for APC (Yasuda et al. 2013). Taken together, these studies point towards a more complex picture for neurodegeneration wherein pathological inclusions may vary significantly in composition and behavior between disease subtypes. This variation warrants further characterization and poses a significant challenge to designing therapeutic interventions.
4.4 TDP-43, FUS, and oxidative stress
Besides disruptions to RNA homeostasis, a separate angle from which to consider the pathogenicity of TDP-43 and FUS (along with the role that SGs might play) stems from the unique susceptibility of neuronal cells to oxidative stress. First, the larger energy requirements of neurons might strain mitochondria, resulting in the production of more reactive oxygen species (ROS) and lowered mitochondrial efficiency, outcomes that are only exacerbated with age (Beal 2002; Barber and Shaw 2010). Additionally, biomarkers for oxidative stress have long been identified in motor neurons and spinal cords of ALS patients (Carrì et al. 2015). These include a 2-fold increase in protein carbonyl levels (Shaw et al. 1995), elevated levels of DNA 8-hydroxy-2'-deoxyguanosine (OH8dG) (Ferrante et al. 1997; Bogdanov et al. 2000; Ihara et al. 2005) and high immunoreactivity for lipid peroxidation and protein glycoxidation (Shibata et al. 2001). Mutations in other proteins implicated in pathogenesis might further predispose neurons to high levels of oxidative stress. For example, mutations in the chaperone protein VCP increase neuronal susceptibility to ROS build-up upon depletion of glutathione (GSH), a free radical scavenger which is also diminished in the motor cortices of ALS patients (Hirano et al. 2014, Weiduschat et al. 2014, Carrì et al. 2015). Interestingly, GSH depletion in NSC34 cells has been shown to result in the formation of insoluble inclusions containing the hyperphosphorylated, truncated TDP-43 fragments commonly found in ALS histology (Iguchi et al. 2012). Sensitivity to oxidative stress might also explain the age dependency of ALS, FTLD, and related neurological diseases. Given that SGs are a natural defense mechanism against sources of oxidative stress, the unresolved issue has been whether pathological inclusions constitute a root cause of pathogenesis, a symptom of RBP dysregulation, or a beneficial cellular response to exogenous stressors.
5 Concluding remarks
Significant progress over the past decade has revealed that PrLDs/LCDs are critical for SG formation and that disruptions to these regions in ALS/FTLD patients are related to the development of pathological inclusions. To fulfill their functions as transient hubs of mRNP sorting and cellular signaling, SGs rely on dynamic, low affinity protein–protein and protein–RNA interactions. It is increasingly certain that these interactions are facilitated by PrLD-mediated oligomerization in conjunction with multivalent proteins, RNA, or other scaffolds mediated by post-translational modifications. SGs were first linked to neurological disease with the discovery that FUS and TDP-43, both RBPs with roles in mRNA metabolism, were frequent components of pathological inclusions found in ALS/FTLD patient histology. TDP-43 is now understood to be an endogenous SG component with a potential role in regulating mRNA triage and evidence that FUS also participates in cytoplasmic mRNP remodeling is growing. More importantly, both proteins robustly co-localize with SG markers in a wide range of cellular disease models, either due to cytoplasmic mislocalization, prolonged stress to which neurons are especially susceptible, or the mutation-induced enhancement of aggregation propensity. Whether this phenomenon plays a core role in neurodegenerative pathogenesis remains uncertain, largely due to the heterogeneous and highly variable nature of the disease class.
Interestingly, another SG RBP bearing prion-like properties has emerged as a candidate in neurodegenerative pathogenesis. Kim and colleagues (2013) showed that three distinct mutations in familial ALS pedigrees all resulted in a valine substitution in the center of the putative PrLD of the protein hnRNPA2B1, resulting in the similar pathological phenotype of mislocalization to the cytoplasm, aggregation into irreversible inclusions, and co-localization with SG markers. Interestingly, the valine mutations were computationally predicted to increase the prion-like propensity of the protein by enhancing homotypic oligomerization. Constructs of the hnRNPA2B1 PrLD formed amyloid fibers in vitro, and astoundingly, full length mutants fibrillized even quicker and were capable of hetero-oligomerization with other proteins (Kato et al. 2012; Kim et al. 2013b). FUS-related proteins TAF15 and EWSR1 have also been recently linked to both ALS and FTLD (Couthouis et al. 2011; Mackenzie and Neumann 2012; Couthouis et al. 2012). Of note, these PrLD-mediated RNA aggregates are not restricted to ALS/FLTD pathologies, but have also been described in tauopathies, Alzheimer and Parkinson diseases (Banks et al. 2008; Vanderweyde et al. 2012; Ash et al. 2014).
Given the intrinsic link between pathological inclusions and SGs, one potential approach to therapeutic intervention might be the formulation of drugs to disrupt SG formation (Kim et al. 2013a; Leung 2014; Sidrauski et al. 2015). Because eIF2 alpha phosphorylation is known to induce SG assembly, a small molecule chemical which inhibits this pathway (GSK2606414) has been evaluated and shown to rescue TDP-43 toxicity in fly models and mammalian neurons (Kim et al. 2013a). Recent data suggest that aggregation due to oligomerization of PrLDs/LCDs is regulated by post-translational modifications within these protein domains (Kwon et al. 2013; Kwon et al. 2014; Gambetta and Müller 2014). By first identifying the enzymes responsible for these modifications, tissue-specific (in)activation of these proteins could provide new therapeutic avenues.
In addition, a disaggregase exhibiting remarkable effectiveness against misfolded protein aggregates, including those comprised of FUS, TDP-43, and TAF15, was engineered from the conserved yeast protein HSP104 (Jackrel et al. 2014). Through screening of a random mutagenesis library of HSP104 variants and systematic optimization of the resulting potentiated variants, Shorter and colleagues abolished a putative auto-inhibitory structural motif in the protein, reprogramming HSP104 with the capacity to solubilize TDP-43 and FUS aggregates, normalize proteotoxic mislocalization, and even rescue a neurological phenotype in nematode models (Jackrel et al. 2014). An alternative strategy would center on how cytoplasmic aggregates are actively cleared. Genetic screens in yeast have revealed that different components in the autophagy pathway, such as VCP, are involved in such processes, consistent with recent genomic and biochemical data from ALS patients (Buchan et al. 2013; Meyer and Weihl 2014; Cirulli et al. 2015). Indeed, VCP is known to play a role in returning TDP-43 back to solubility in the cytoplasm after stress release (Barmada et al. 2014), and TDP-43 has even been shown to be preferentially cleared by autophagy (Wang et al. 2010; Wang et al. 2012; Wang et al. 2013) Taken together, these data suggest a potential synergistic therapy by preventing prionogenic aggregation while enhancing autophagy. The ALS/FTLD disease system may display confounding heterogeneity, but where there are numerous openings to dysregulation there must also be diverse opportunities for intervention. Targeting prion-like interactions may constitute just one of many future complimentary approaches.
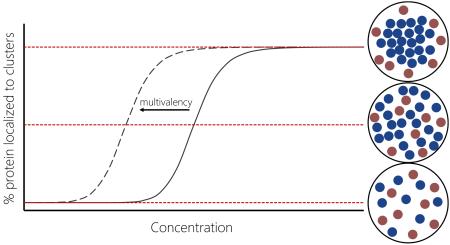
Box 1 | Phase Separation in Biology.
A phase is a spatial region with uniform physical properties. In cells, discrete phases can form via a process of liquid-liquid demixing, resulting in droplet-like structures that remain distinct from the cytosol. This physical phenomenon has recently emerged in connection to non-membranous structures as a paradigm to understand cellular compartmentalization. Just as oil and water separate due to hydrophobic effects, proteins and nucleic acids can demix into condensed droplets above a certain concentration threshold. Beyond this transition point, multivalent and low affinity interactions mediated by PrLDs/LCDs create stronger intermolecular interactions within the droplet than in the surrounding cytoplasm. The components that localize to these structures therefore exist along the boundary between solubility and solid-like aggregation because their components bind transiently without entering a more ordered state. In other words, phase separation occurs when low-affinity interactions bring molecules together without arresting their dynamics (Weber and Brangwynne 2012).
A graphical representation of the dependence of phase separation on protein concentration
Multiple studies have indicated that the transition from solubility to a phase-separated state is sharp (Kato et al. 2012; Schwartz et al. 2013; Banjade and Rosen 2014) and multivalent interactions have been shown to lower this transition threshold (see main text).
The criteria for liquid-liquid demixing in biological systems are poorly understood. Though “nucleators” of some RNA granules have been identified — stalled initiation factors in SG assembly, for example — how these proteins initiate demixing while other complexes do not remains uncertain. Nevertheless, conceptualizing RNA granules as μm-sized cellular structures with unique biophysical properties allows us to gain insight into functions that might not be as evident from single-protein approaches, especially given the staggering complexity of RNA granule composition.
What might be the biological advantage of such non-membranous structures? First, since demixing itself is strongly concentration-dependent, a point of localized protein enrichment could regulate the rate of biochemical reactions, accelerating some by acting as “micro reactors” while sequestering away components to slow others (Weber and Brangwynne 2012). Incorporating only the proteins with an affinity for LCD-mediated interactions might provide RNA granules with a passive selectivity mechanism, aided by multivalent proteins that contribute a “branching” scaffold to the assembly process. Furthermore, by virtue of their reversibility, which is mediated in large part by protein modifications, phase-separated structures can assemble only when needed, allowing for their composition to vary based on cellular requirements. For a more comprehensive discussion of liquid-liquid phase separations in biology, interested readers are directed to the recent review by Hyman, Weber and Jülicher (Hyman et al. 2014).
Acknowledgements
We would like to thank Drs. Phillip Sharp, Nancy Kedersha, Anaïs Aulas, and Voula Mili for critical reading of the manuscript, Drs. Steve McKnight and Masato Kato on sharing their insights on the role of low-complexity domains in RNA granule formation. This work was partly supported by Johns Hopkins Catalyst Award and NIH R01-GM104135 to A.K.L.L.
References
- Alberti S, Halfmann R, King O, et al. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha NL. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha NL. RNA granules. The Journal of Cell Biology. 2006;172:803–808. doi: 10.1083/jcb.200512082. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha NL. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Andersson MK, Ståhlberg A, Arvidsson Y, et al. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 2008;9:37. doi: 10.1186/1471-2121-9-37. doi: 10.1186/1471-2121-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto K, Fukuda H, Imajoh-Ohmi S, et al. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- Armakola M, Higgins MJ, Figley MD, et al. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat Genet. 2012;44:1302–1309. doi: 10.1038/ng.2434. doi: 10.1038/ng.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ES, Ling S-C, Huelga SC, et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc Natl Acad Sci USA. 2013;110:E736–45. doi: 10.1073/pnas.1222809110. doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo AP, Suhan JP, Welch WJ. Dynamic changes in the structure and intracellular locale of the mammalian low-molecular-weight heat shock protein. Mol Cell Biol. 1988;8:5059–5071. doi: 10.1128/mcb.8.12.5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PEA, Vanderweyde TE, Youmans KL, et al. Pathological stress granules in Alzheimer's disease. Brain Res. 2014;1584:52–58. doi: 10.1016/j.brainres.2014.05.052. doi: 10.1016/j.brainres.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A, Caron G, Gkogkas CG, et al. G3BP1 promotes stress-induced RNA granule interactions to preserve polyadenylated mRNA. The Journal of Cell Biology. 2015;209:73–84. doi: 10.1083/jcb.201408092. doi: 10.1083/jcb.201408092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A, Stabile S, Vande Velde C. Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Mol Neurodegener. 2012;7:54. doi: 10.1186/1750-1326-7-54. doi: 10.1186/1750-1326-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguet A, Degot S, Cougot N, et al. The exon-junction-complex-component metastatic lymph node 51 functions in stress-granule assembly. J Cell Sci. 2007;120:2774–2784. doi: 10.1242/jcs.009225. doi: 10.1242/jcs.009225. [DOI] [PubMed] [Google Scholar]
- Banjade S, Rosen MK. Phase transitions of multivalent proteins can promote clustering of membrane receptors. Elife. 2014 doi: 10.7554/eLife.04123. doi: 10.7554/eLife.04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks GT, Kuta A, Isaacs AM, Fisher EMC. TDP-43 is a culprit in human neurodegeneration, and not just an innocent bystander. Mamm Genome. 2008;19:299–305. doi: 10.1007/s00335-008-9117-x. doi: 10.1007/s00335-008-9117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber SC, Shaw PJ. Oxidative stress in ALS: key role in motor neuron injury and therapeutic target. Free Radic Biol Med. 2010;48:629–641. doi: 10.1016/j.freeradbiomed.2009.11.018. doi: 10.1016/j.freeradbiomed.2009.11.018. [DOI] [PubMed] [Google Scholar]
- Barmada SJ, Serio A, Arjun A, et al. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014;10:677–685. doi: 10.1038/nchembio.1563. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barmada SJ, Skibinski G, Korb E, et al. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF. Oxidatively modified proteins in aging and disease. Free Radic Biol Med. 2002;32:797–803. doi: 10.1016/s0891-5849(02)00780-3. [DOI] [PubMed] [Google Scholar]
- Bentmann E, Neumann M, Tahirovic S, et al. Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43) J Biol Chem. 2012;287:23079–23094. doi: 10.1074/jbc.M111.328757. doi: 10.1074/jbc.M111.328757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechingberg J, Luo Y, Bolund L, et al. Gene expression responses to FUS, EWS, and TAF15 reduction and stress granule sequestration analyses identifies FET-protein non-redundant functions. PLoS ONE. 2012;7:e46251. doi: 10.1371/journal.pone.0046251. doi: 10.1371/journal.pone.0046251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Brown RH, Matson W, et al. Increased oxidative damage to DNA in ALS patients. Free Radic Biol Med. 2000;29:652–658. doi: 10.1016/s0891-5849(00)00349-x. [DOI] [PubMed] [Google Scholar]
- Bono F, Gehring NH. Assembly, disassembly and recycling: the dynamics of exon junction complexes. RNA Biol. 2011;8:24–29. doi: 10.4161/rna.8.1.13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco DA, Lemay N, Ko HK, et al. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet. 2010;19:4160–4175. doi: 10.1093/hmg/ddq335. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyault C, Zhang Y, Fritah S, et al. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007;21:2172–2181. doi: 10.1101/gad.436407. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP. Phase transitions and size scaling of membrane-less organelles. The Journal of Cell Biology. 2013;203:875–881. doi: 10.1083/jcb.201308087. doi: 10.1083/jcb.201308087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis R-M, Taylor JP, Parker R. Eukaryotic Stress Granules Are Cleared by Autophagy and Cdc48/VCP Function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. The Journal of Cell Biology. 2008;183:441–455. doi: 10.1083/jcb.200807043. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic Stress Granules: The Ins and Outs of Translation. Molecular Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Yoon J-H, Parker R. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci. 2011;124:228–239. doi: 10.1242/jcs.078444. doi: 10.1242/jcs.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrì MT, Valle C, Bozzo F, Cozzolino M. Oxidative stress and mitochondrial damage: importance in non-SOD1 ALS. Front Cell Neurosci. 2015;9:41. doi: 10.3389/fncel.2015.00041. doi: 10.3389/fncel.2015.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yang M, Deng J, et al. Expression of human FUS protein in Drosophila leads to progressive neurodegeneration. Protein Cell. 2011;2:477–486. doi: 10.1007/s13238-011-1065-7. doi: 10.1007/s13238-011-1065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Calvo A, Mazzini L, et al. Extensive genetics of ALS: a population-based study in Italy. Neurology. 2012;79:1983–1989. doi: 10.1212/WNL.0b013e3182735d36. doi: 10.1212/WNL.0b013e3182735d36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Lasseigne BN, Petrovski S, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015 doi: 10.1126/science.aaa3650. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier NC, Heuser J, Levy MA, Schlesinger MJ. Ultrastructural and biochemical analysis of the stress granule in chicken embryo fibroblasts. J Cell Biol. 1988;106:1131–1139. doi: 10.1083/jcb.106.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier NC, Schlesinger MJ. The dynamic state of heat shock proteins in chicken embryo fibroblasts. J Cell Biol. 1986;103:1495–1507. doi: 10.1083/jcb.103.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Erion R, et al. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21:2899–2911. doi: 10.1093/hmg/dds116. doi: 10.1093/hmg/dds116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Shorter J, et al. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci USA. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman M, Johnson BS, King OD, et al. Prion-like disorders: blurring the divide between transmissibility and infectivity. J Cell Sci. 2010;123:1191–1201. doi: 10.1242/jcs.051672. doi: 10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle JG, Lanson NA, Smith RB, et al. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum Mol Genet. 2013;22:1193–1205. doi: 10.1093/hmg/dds526. doi: 10.1093/hmg/dds526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey CM, Cenik B, Sephton CF, et al. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol Cell Biol. 2011;31:1098–1108. doi: 10.1128/MCB.01279-10. doi: 10.1128/MCB.01279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Madl T, Valori CF, et al. Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J. 2012;31:4258–4275. doi: 10.1038/emboj.2012.261. doi: 10.1038/emboj.2012.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Rodde R, Edbauer D, et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger-Mathason TSK, Andrade J, Groehler AL, et al. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Molecular Cell. 2008;31:722–736. doi: 10.1016/j.molcel.2008.06.025. doi: 10.1016/j.molcel.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farny NG, Kedersha NL, Silver PA. Metazoan stress granule assembly is mediated by P-eIF2alpha-dependent and -independent mechanisms. RNA. 2009;15:1814–1821. doi: 10.1261/rna.1684009. doi: 10.1261/rna.1684009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante RJ, Browne SE, Shinobu LA, et al. Evidence of increased oxidative damage in both sporadic and familial amyotrophic lateral sclerosis. J Neurochem. 1997;69:2064–2074. doi: 10.1046/j.1471-4159.1997.69052064.x. [DOI] [PubMed] [Google Scholar]
- Fournier M-J, Coudert L, Mellaoui S, et al. Inactivation of the mTORC1-eukaryotic translation initiation factor 4E pathway alters stress granule formation. Mol Cell Biol. 2013;33:2285–2301. doi: 10.1128/MCB.01517-12. doi: 10.1128/MCB.01517-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier M-J, Gareau C, Mazroui R. The chemotherapeutic agent bortezomib induces the formation of stress granules. Cancer Cell Int. 2010;10:12. doi: 10.1186/1475-2867-10-12. doi: 10.1186/1475-2867-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushimi K, Long C, Jayaram N, et al. Expression of human FUS/TLS in yeast leads to protein aggregation and cytotoxicity, recapitulating key features of FUS proteinopathy. Protein Cell. 2011;2:141–149. doi: 10.1007/s13238-011-1014-5. doi: 10.1007/s13238-011-1014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal J, Zhang J, Kwinter DM, et al. Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol Aging. 2011;32:2323.e27–40. doi: 10.1016/j.neurobiolaging.2010.06.010. doi: 10.1016/j.neurobiolaging.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta MC, Müller J. O-GlcNAcylation Prevents Aggregation of the Polycomb Group Repressor Polyhomeotic. Developmental Cell. 2014 doi: 10.1016/j.devcel.2014.10.020. doi: 10.1016/j.devcel.2014.10.020. [DOI] [PubMed] [Google Scholar]
- Gareau C, Fournier M-J, Filion C, et al. p21(WAF1/CIP1) upregulation through the stress granule-associated protein CUGBP1 confers resistance to bortezomib-mediated apoptosis. PLoS ONE. 2011;6:e20254. doi: 10.1371/journal.pone.0020254. doi: 10.1371/journal.pone.0020254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha NL, Ayodele M, et al. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Grousl T, Ivanov P, Frydlova I, et al. Robust heat shock induces eIF2 -phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J Cell Sci. 2009;122:2078–2088. doi: 10.1242/jcs.045104. doi: 10.1242/jcs.045104. [DOI] [PubMed] [Google Scholar]
- Guil S, Long JC, Cáceres JF. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol Cell Biol. 2006;26:5744–5758. doi: 10.1128/MCB.00224-06. doi: 10.1128/MCB.00224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Hicks GG, Singh N, Nashabi A, et al. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat Genet. 2000;24:175–179. doi: 10.1038/72842. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
- Hilliker A, Gao Z, Jankowsky E, Parker R. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Molecular Cell. 2011;43:962–972. doi: 10.1016/j.molcel.2011.08.008. doi: 10.1016/j.molcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, et al. VCP gene analyses in Japanese patients with sporadic amyotrophic lateral sclerosis identify a new mutation. Neurobiol. Aging. 2015;36:1604.e1–6. doi: 10.1016/j.neurobiolaging.2014.10.012. [DOI] [PubMed] [Google Scholar]
- Hofmann S, Cherkasova V, Bankhead P, et al. Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol Biol Cell. 2012;23:3786–3800. doi: 10.1091/mbc.E12-04-0296. doi: 10.1091/mbc.E12-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Lüscher B, et al. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010 doi: 10.1016/j.tibs.2009.12.003. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Hoyle NP, Castelli LM, Campbell SG, et al. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höck J, Weinmann L, Ender C, et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Zhou H, Tong J, et al. FUS transgenic rats develop the phenotypes of amyotrophic lateral sclerosis and frontotemporal lobar degeneration. PLoS Genet. 2011;7:e1002011. doi: 10.1371/journal.pgen.1002011. doi: 10.1371/journal.pgen.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Simons K. Cell biology. Beyond oil and water--phase transitions in cells. Science. 2012;337:1047–1049. doi: 10.1126/science.1223728. doi: 10.1126/science.1223728. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Jülicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Igaz LM, Kwong LK, Lee EB, et al. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J Clin Invest. 2011;121:726–738. doi: 10.1172/JCI44867. doi: 10.1172/JCI44867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi Y, Katsuno M, Takagi S, et al. Oxidative stress induced by glutathione depletion reproduces pathological modifications of TDP-43 linked to TDP-43 proteinopathies. Neurobiol Dis. 2012;45:862–870. doi: 10.1016/j.nbd.2011.12.002. doi: 10.1016/j.nbd.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Ihara Y, Nobukuni K, Takata H, Hayabara T. Oxidative stress and metal content in blood and cerebrospinal fluid of amyotrophic lateral sclerosis patients with and without a Cu, Zn-superoxide dismutase mutation. Neurol Res. 2005;27:105–108. doi: 10.1179/016164105X18430. doi: 10.1179/016164105X18430. [DOI] [PubMed] [Google Scholar]
- Iko Y, Kodama TS, Kasai N, et al. Domain architectures and characterization of an RNA-binding protein, TLS. J Biol Chem. 2004;279:44834–44840. doi: 10.1074/jbc.M408552200. doi: 10.1074/jbc.M408552200. [DOI] [PubMed] [Google Scholar]
- Ito D, Seki M, Tsunoda Y, et al. Nuclear transport impairment of amyotrophic lateral sclerosis-linked mutations in FUS/TLS. Ann Neurol. 2011;69:152–162. doi: 10.1002/ana.22246. doi: 10.1002/ana.22246. [DOI] [PubMed] [Google Scholar]
- Jackrel ME, DeSantis ME, Martinez BA, et al. Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell. 2014;156:170–182. doi: 10.1016/j.cell.2013.11.047. doi: 10.1016/j.cell.2013.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci USA. 2008;105:6439–6444. doi: 10.1073/pnas.0802082105. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, Snead D, Lee JJ, et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Lin L, Tradewell ML, et al. Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum Mol Genet. 2010;19:671–683. doi: 10.1093/hmg/ddp534. doi: 10.1093/hmg/ddp534. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Anderson P. Mammalian stress granules and processing bodies. Methods in Enzymology. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–969. doi: 10.1042/bst0300963. doi: 10.1042/ [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Cho MR, Li W, et al. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, et al. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. The Journal of Cell Biology. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013 doi: 10.1016/j.tibs.2013.07.004. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Stoecklin G, Ayodele M, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. The Journal of Cell Biology. 2005;169:871–884. doi: 10.1083/jcb.200502088. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-J, Raphael AR, LaDow ES, et al. Therapeutic modulation of eIF2α phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat Genet. 2013a doi: 10.1038/ng.2853. doi: 10.1038/ng.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang Y-D, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013b;495:467–473. doi: 10.1038/nature11922. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Shanware NP, Bowler MJ, Tibbetts RS. Amyotrophic lateral sclerosis-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to co-regulate HDAC6 mRNA. J Biol Chem. 2010;285:34097–34105. doi: 10.1074/jbc.M110.154831. doi: 10.1074/jbc.M110.154831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WJ, Back SH, Kim V, et al. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol Cell Biol. 2005;25:2450–2462. doi: 10.1128/MCB.25.6.2450-2462.2005. doi: 10.1128/MCB.25.6.2450-2462.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino Y, Washizu C, Aquilanti E, et al. Intracellular localization and splicing regulation of FUS/TLS are variably affected by amyotrophic lateral sclerosis-linked mutations. Nucleic Acids Res. 2011;39:2781–2798. doi: 10.1093/nar/gkq1162. doi: 10.1093/nar/gkq1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppers M, van Blitterswijk MM, Vlam L, et al. VCP mutations in familial and sporadic amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:837.e7–13. doi: 10.1016/j.neurobiolaging.2011.10.006. doi: 10.1016/j.neurobiolaging.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, et al. Phosphorylation-Regulated Binding of RNA Polymerase II to Fibrous Polymers of Low-Complexity Domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, et al. Poly-dipeptides encoded by the C9ORF72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014 doi: 10.1126/science.1254917. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 2007;21:3381–3394. doi: 10.1101/gad.461107. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rocca G, Olejniczak SH, González AJ, et al. In vivo, Argonaute-bound microRNAs exist predominantly in a reservoir of low molecular weight complexes not associated with mRNA. Proc Natl Acad Sci USA. 2015 doi: 10.1073/pnas.1424217112. doi: 10.1073/pnas.1424217112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Hutt KR, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–1497. doi: 10.1038/nn.3230. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanson NA, Maltare A, King H, et al. A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum Mol Genet. 2011;20:2510–2523. doi: 10.1093/hmg/ddr150. doi: 10.1093/hmg/ddr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AKL. Poly(ADP-ribose): An organizer of cellular architecture. The Journal of Cell Biology. 2014;205:613–619. doi: 10.1083/jcb.201402114. doi: 10.1083/jcb.201402114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AKL, Vyas S, Rood JE, et al. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Molecular Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng H-C, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Simarro M, Kedersha NL, Anderson P. FAST is a survival protein that senses mitochondrial stress and modulates TIA-1-regulated changes in protein expression. Mol Cell Biol. 2004;24:10718–10732. doi: 10.1128/MCB.24.24.10718-10732.2004. doi: 10.1128/MCB.24.24.10718-10732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Rayman JB, Kandel ER, Derkatch IL. Functional Role of Tia1/Pub1 and Sup35 Prion Domains: Directing Protein Synthesis Machinery to the Tubulin Cytoskeleton. Molecular Cell. 2014 doi: 10.1016/j.molcel.2014.05.027. doi: 10.1016/j.molcel.2014.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. The Journal of Cell Biology. 2013;201:361–372. doi: 10.1083/jcb.201302044. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S-C, Albuquerque CP, Han JS, et al. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci USA. 2010;107:13318–13323. doi: 10.1073/pnas.1008227107. doi: 10.1073/pnas.1008227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S-C, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bilgutay A, Zhang Y-J, et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS ONE. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Lin AY, Ebata A, et al. ALS-linked mutations enlarge TDP-43-enriched neuronal RNA granules in the dendritic arbor. J Neurosci. 2014;34:4167–4174. doi: 10.1523/JNEUROSCI.2350-13.2014. doi: 10.1523/JNEUROSCI.2350-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IRA, Neumann M. FET proteins in frontotemporal dementia and amyotrophic lateral sclerosis. Brain Res. 2012;1462:40–43. doi: 10.1016/j.brainres.2011.12.010. doi: 10.1016/j.brainres.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Mamiya N, Worman HJ. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem. 1999;274:15751–15756. doi: 10.1074/jbc.274.22.15751. [DOI] [PubMed] [Google Scholar]
- Mazroui R, Huot M-E, Tremblay S, et al. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum Mol Genet. 2002;11:3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- McDonald KK, Aulas A, Destroismaisons L, et al. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum Mol Genet. 2011;20:1400–1410. doi: 10.1093/hmg/ddr021. doi: 10.1093/hmg/ddr021. [DOI] [PubMed] [Google Scholar]
- Meyer H, Weihl CC. The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J Cell Sci. 2014;127:3877–3883. doi: 10.1242/jcs.093831. doi: 10.1242/jcs.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz J, Parker SJ, Vella LJ, et al. C-Jun N-terminal kinase controls TDP-43 accumulation in stress granules induced by oxidative stress. Mol Neurodegener. 2011;6:57. doi: 10.1186/1750-1326-6-57. doi: 10.1186/1750-1326-6-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller BJ, Cao Y, Li CY, Dewhirst MW. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5:429–441. doi: 10.1016/s1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- Mokas S, Mills JR, Garreau C, et al. Uncoupling stress granule assembly and translation initiation inhibition. Mol Biol Cell. 2009;20:2673–2683. doi: 10.1091/mbc.E08-10-1061. doi: 10.1091/mbc.E08-10-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet S, Cougot N, Wilczynska A, et al. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol Biol Cell. 2008;19:4469–4479. doi: 10.1091/mbc.E08-05-0499. doi: 10.1091/mbc.E08-05-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onomoto K, Yoneyama M, Fung G, et al. Antiviral innate immunity and stress granule responses. Trends Immunol. 2014 doi: 10.1016/j.it.2014.07.006. doi: 10.1016/j.it.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SJ, Meyerowitz J, James JL, et al. Endogenous TDP-43 localized to stress granules can subsequently form protein aggregates. Neurochem Int. 2012;60:415–424. doi: 10.1016/j.neuint.2012.01.019. doi: 10.1016/j.neuint.2012.01.019. [DOI] [PubMed] [Google Scholar]
- Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowska J, Hansen SJ, Park N, et al. Stable formation of compositionally unique stress granules in virus-infected cells. J Virol. 2010;84:3654–3665. doi: 10.1128/JVI.01320-09. doi: 10.1128/JVI.01320-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Cleveland DW. The seeds of neurodegeneration: prion-like spreading in ALS. Cell. 2011;147:498–508. doi: 10.1016/j.cell.2011.10.011. doi: 10.1016/j.cell.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Lee S, Shang Y, et al. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest. 2014;124:981–999. doi: 10.1172/JCI72723. doi: 10.1172/JCI72723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovici GD, Miller BL. Frontotemporal lobar degeneration: epidemiology, pathophysiology, diagnosis and management. CNS Drugs. 2010;24:375–398. doi: 10.2165/11533100-000000000-00000. doi: 10.2165/11533100-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikhvanov EG, Romanova NV, Chernoff YO. Chaperone effects on prion and nonprion aggregates. Prion. 2007;1:217–222. doi: 10.4161/pri.1.4.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robberecht W, Philips T. The changing scene of amyotrophic lateral sclerosis. Nat Rev Neurosci. 2013;14:248–264. doi: 10.1038/nrn3430. doi: 10.1038/nrn3430. [DOI] [PubMed] [Google Scholar]
- Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. N Engl J Med. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- Sama RRK, Ward CL, Kaushansky LJ, et al. FUS/TLS assembles into stress granules and is a prosurvival factor during hyperosmolar stress. J Cell Physiol. 2013;228:2222–2231. doi: 10.1002/jcp.24395. doi: 10.1002/jcp.24395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasayama H, Shimamura M, Tokuda T, et al. Knockdown of the Drosophila fused in sarcoma (FUS) homologue causes deficient locomotive behavior and shortening of motoneuron terminal branches. PLoS ONE. 2012;7:e39483. doi: 10.1371/journal.pone.0039483. doi: 10.1371/journal.pone.0039483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidlin M, Lu M, Leuenberger SA, et al. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J. 2004;23:4760–4769. doi: 10.1038/sj.emboj.7600477. doi: 10.1038/sj.emboj.7600477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–259. doi: 10.1038/nrg3160. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Wang X, Podell ER, Cech TR. RNA Seeds Higher-Order Assembly of FUS Protein. Cell Rep. 2013 doi: 10.1016/j.celrep.2013.11.017. doi: 10.1016/j.celrep.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Ince PG, Falkous G, Mantle D. Oxidative damage to protein in sporadic motor neuron disease spinal cord. Ann Neurol. 1995;38:691–695. doi: 10.1002/ana.410380424. doi: 10.1002/ana.410380424. [DOI] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- Shibata N, Nagai R, Uchida K, et al. Morphological evidence for lipid peroxidation and protein glycoxidation in spinal cords from sporadic amyotrophic lateral sclerosis patients. Brain Res. 2001;917:97–104. doi: 10.1016/s0006-8993(01)02926-2. [DOI] [PubMed] [Google Scholar]
- Shih J-W, Tsai T-Y, Chao C-H, Wu Lee Y-H. Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene. 2008;27:700–714. doi: 10.1038/sj.onc.1210687. doi: 10.1038/sj.onc.1210687. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, McGeachy AM, Ingolia NT, Walter P. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. Elife. 2015 doi: 10.7554/eLife.05033. doi: 10.7554/eLife.05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souquere S, Mollet S, Kress M, et al. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J Cell Sci. 2009;122:3619–3626. doi: 10.1242/jcs.054437. doi: 10.1242/jcs.054437. [DOI] [PubMed] [Google Scholar]
- Spector DL. SnapShot: Cellular bodies. Cell. 2006;127:1071. doi: 10.1016/j.cell.2006.11.026. doi: 10.1016/j.cell.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Stubbs T, Kedersha NL, et al. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Diaz Z, Fang X, et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Ohashi S, Kobayashi S. Roles of YB-1 under arsenite-induced stress: translational activation of HSP70 mRNA and control of the number of stress granules. Biochim Biophys Acta. 2014;1840:985–992. doi: 10.1016/j.bbagen.2013.11.002. doi: 10.1016/j.bbagen.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, et al. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thedieck K, Holzwarth B, Prentzell MT, et al. Inhibition of mTORC1 by Astrin and Stress Granules Prevents Apoptosis in Cancer Cells. Cell. 2013;154:859–874. doi: 10.1016/j.cell.2013.07.031. doi: 10.1016/j.cell.2013.07.031. [DOI] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H, Chebli K, Zekri L, et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. The Journal of Cell Biology. 2003;160:823–831. doi: 10.1083/jcb.200212128. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tsai N-P, Wei L-N. RhoA/ROCK1 signaling regulates stress granule formation and apoptosis. Cell Signal. 2010;22:668–675. doi: 10.1016/j.cellsig.2009.12.001. doi: 10.1016/j.cellsig.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R, Buljan M, Lang B, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Scotter EL, Nishimura AL, et al. ALS mutant FUS disrupts nuclear localization and sequesters wild-type FUS within cytoplasmic stress granules. Hum Mol Genet. 2013;22:2676–2688. doi: 10.1093/hmg/ddt117. doi: 10.1093/hmg/ddt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderweyde T, Yu H, Varnum M, et al. Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J Neurosci. 2012;32:8270–8283. doi: 10.1523/JNEUROSCI.1592-12.2012. doi: 10.1523/JNEUROSCI.1592-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I-F, Guo B-S, Liu Y-C, et al. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc Natl Acad Sci USA. 2012;109:15024–15029. doi: 10.1073/pnas.1206362109. doi: 10.1073/pnas.1206362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I-F, Tsai K-J, Shen C-KJ. Autophagy activation ameliorates neuronal pathogenesis of FTLD-U mice: a new light for treatment of TARDBP/TDP-43 proteinopathies. Autophagy. 2013;9:239–240. doi: 10.4161/auto.22526. doi: 10.4161/auto.22526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Fan H, Ying Z, et al. Degradation of TDP-43 and its pathogenic form by autophagy and the ubiquitin-proteasome system. Neurosci Lett. 2010;469:112–116. doi: 10.1016/j.neulet.2009.11.055. doi: 10.1016/j.neulet.2009.11.055. [DOI] [PubMed] [Google Scholar]
- Warner JR, Rich A, Hall CE. Electron Microscope Studies of Ribosomal Clusters Synthesizing Hemoglobin. Science. 1962;138:1399–1403. doi: 10.1126/science.138.3548.1399. doi: 10.1126/science.138.3548.1399. [DOI] [PubMed] [Google Scholar]
- Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Weiduschat N, et al. Motor cortex glutathione deficit in ALS measured in vivo with the J-editing technique. Neurosci Lett. 2014;570:102–107. doi: 10.1016/j.neulet.2014.04.020. [DOI] [PubMed] [Google Scholar]
- White JP, Lloyd RE. Regulation of stress granules in virus systems. Trends Microbiol. 2012;20:175–183. doi: 10.1016/j.tim.2012.02.001. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska A, Aigueperse C, Kress M, et al. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- Wippich F, Bodenmiller B, Trajkovska MG, et al. Dual Specificity Kinase DYRK3 Couples Stress Granule Condensation/Dissolution to mTORC1 Signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- Yasuda K, Zhang H, Loiselle D, et al. The RNA-binding protein Fus directs translation of localized mRNAs in APC-RNP granules. The Journal of Cell Biology. 2013;203:737–746. doi: 10.1083/jcb.201306058. doi: 10.1083/jcb.201306058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J-H, Abdelmohsen K, Srikantan S, et al. Tyrosine phosphorylation of HuR by JAK3 triggers dissociation and degradation of HuR target mRNAs. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt903. doi: 10.1093/nar/gkt903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Okabe K, Tani T, Funatsu T. Dynamic association-dissociation and harboring of endogenous mRNAs in stress granules. J Cell Sci. 2011;124:4087–4095. doi: 10.1242/jcs.090951. doi: 10.1242/jcs.090951. [DOI] [PubMed] [Google Scholar]
- Zhang T, Mullane PC, Periz G, Wang J. TDP-43 neurotoxicity and protein aggregation modulated by heat shock factor and insulin/IGF-1 signaling. Hum Mol Genet. 2011;20:1952–1965. doi: 10.1093/hmg/ddr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZC, Chook YM. Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS) Proc Natl Acad Sci USA. 2012;109:12017–12021. doi: 10.1073/pnas.1207247109. doi: 10.1073/pnas.1207247109. [DOI] [PMC free article] [PubMed] [Google Scholar]