Abstract
Developmental plasticity may accelerate the evolution of phenotypic novelty through genetic accommodation, but studies of genetic accommodation often lack knowledge of the ancestral state to place selected traits in an evolutionary context. A promising approach for assessing genetic accommodation involves using a comparative framework to ask whether ancestral plasticity is related to the evolution of a particular trait. Bees are an excellent group for such comparisons because caste-based societies (eusociality) have evolved multiple times independently and extant species exhibit different modes of eusociality. We measured brain and abdominal gene expression in a facultatively eusocial bee, Megalopta genalis, and assessed whether plasticity in this species is functionally linked to eusocial traits in other bee lineages. Caste-biased abdominal genes in M. genalis overlapped significantly with caste-biased genes in obligately eusocial bees. Moreover, caste-biased genes in M. genalis overlapped significantly with genes shown to be rapidly evolving in multiple studies of 10 bee species, particularly for genes in the glycolysis pathway and other genes involved in metabolism. These results provide support for the idea that eusociality can evolve via genetic accommodation, with plasticity in facultatively eusocial species like M. genalis providing a substrate for selection during the evolution of caste in obligately eusocial lineages.
Keywords: social evolution, genetic accommodation, eusociality, gene expression, selection
1. Introduction
Phenotypic plasticity may accelerate the evolution of phenotypic novelty through genetic accommodation ([1–4], cf. [5]), but studies of genetic accommodation often lack knowledge of the ancestral state to place selected traits in an evolutionary context. Most empirical support for genetic accommodation employs artificial selection on experimentally induced phenotypes [6–8]. Other empirical studies demonstrate the necessary phylogenetic relationships between environmentally sensitive phenotypes in ancestral lineages and more fixed phenotypes in derived lineages required for genetic accommodation [9,10] but do not show evidence of selection. For condition-sensitive traits, the ancestral condition cannot always be inferred accurately from traits of extant forms [11].
As genomic tools are deployed across a wider array of species, a promising approach to investigate genetic accommodation is to look for both plasticity in ancestral lineages and evidence of selection [3,12,13]. Groups with repeated evolution of traits are ideal for this approach, as comparative genomics can be used to identify genes under selection for a trait of interest. Social insects, and bees in particular, are especially promising for addressing the role of genetic accommodation in phenotypic innovation because eusocial behaviour has evolved independently multiple times [14–17].
A growing volume of published data has identified genes undergoing selection in social lineages of bees [18–20], providing a framework to test the role of genetic accommodation in the evolution of eusociality. Still missing, however, is knowledge of the ancestral phenotypic plasticity of different bee lineages, and whether this ancestral plasticity is related to genes under selection in eusociality.
Gene expression data have been used as a measure of plasticity in many different contexts, including those related to genetic accommodation [3]. In particular, genes that are differentially expressed are thought to experience reduced genetic constraint and evolve more quickly, which has been confirmed in empirical studies [21,22]. Brain gene expression differences have been described across caste and many behavioural contexts in bumblebees [23] and honeybees [24,25]. Queen (Q) and worker (W) honeybees are morphologically distinct and adapted to reproductive and non-reproductive functions, respectively; these differences are reflected in a transcriptomic study which reported over 2000 differentially expressed genes (DEGs) in the brain [26]. These genes, along with caste-biased genes in other obligately eusocial insects, have been found to evolve more rapidly than genes unrelated to caste [19,21,27]. However, whether these genes also showed facultative expression along with plasticity in organismal-level phenotypic traits prior to the evolution of obligate eusociality is unknown.
Robust phylogenetic studies point to a solitary ancestral lifestyle for bees [28,29]. Some species of bees display facultative eusociality, with both solitary and social nests existing either across geographical gradients ([30–34], reviewed in [35]) or even within the same population [36,37]. It is thus likely that at least some mechanisms underpinning phenotypic differentiation in these facultatively eusocial bees play roles in evolutionary transitions from solitary to social life histories. If so, then testing the predictions of genetic accommodation in eusocial evolution can use facultatively eusocial species as proxies for the ancestral state. We did this by using the facultatively eusocial bee, Megalopta genalis, to measure environmentally induced plasticity in gene expression and then compared differentially expressed genes from this species with genes previously shown to be under selection in obligately eusocial taxa.
Megalopta genalis (Halictidae) is a Neotropical sweat bee that displays facultative eusociality [37–40]. Many facultatively eusocial species exhibit social plasticity across geographical gradients [31–33], but in M. genalis both social and solitary nests exist within a single population. This strongly suggests that M. genalis eusociality is at least partially environmentally determined. The plasticity arises through variation in reproductive behaviour of nest-founding females [39–42]. Solitary nests form when females produce only males in their first broods, with subsequent female production resulting in dispersal rather than retention of female workers [42]. By contrast, social nests form when one or more females produced in the first brood remain as non-reproductive workers. Expression of alternative reproductive phenotypes in Megalopta is related to social competition linked to body size and nutrition [39,42,43], as well as ovary size [40] and hormonal differences [44]. Workers in social nests remain sensitive to environmental and social conditions and have the ability to mate and reproduce after queen loss or supersedure, becoming replacement queens with reproductive outputs equivalent to those of solitary females [41].
We used the naturally occurring phenotypic variation of M. genalis and comparative genomics to explore the mechanisms of genetic accommodation. We compared gene expression for four female phenotypes (solitary, queen, worker and replacement queen) in both brain and abdominal tissues. We then assessed the degree to which differences in expression associated with phenotypic variation in M. genalis are common across other species of bees to ask whether similar molecular mechanisms are implicated in caste determination. Finally, we tested for commonality of caste-biased genes in M. genalis with genes previously identified as undergoing selection in eusocial bee lineages to address whether ancestral plasticity is consistent with genetic accommodation for social traits in the evolution of eusociality in bees.
2. Material and methods
See the electronic supplementary material for detailed methods.
(a). Sample collection
Megalopta genalis females were collected on Barro Colorado Island in the Republic of Panama where they are abundant during the dry season [45]. Frozen tissues were exported with permission from the National Authority for the Environment of the Government of Panama (permit Nos. SEX/A-53-13 and SEX/AH-4-15). Observation nests were created with newly emerged females and monitored daily until offspring emergence enabled classification of nests as solitary or social [37–39,46]. Solitary females (S) and queens (Q) were collected following an egg-laying event in solitary and social nests, respectively. In half of the social nests, workers (W) were removed on the same day as queens, and in the other half workers were allowed to transition into replacement queens (R) before collection (4–18 days following queen removal). All females were collected during inactive periods so that gene expression differences were not likely due to acute differences in activity or short-term behaviours, but rather stable gene expression differences between groups. Detailed sample information is provided in the electronic supplementary material, S1.
(b). RNA preparation
Whole brains (n = 30; 7 S, 7 Q, 9 W, 7 R) and abdomens (n = 25; 7 S, 7 Q, 6 W, 5 R; gut tissue and Dufour's glands removed) were dissected as in [47], and ovaries were imaged to confirm reproductive state. Abdominal tissues extracted were primarily fat body and ovarian tissue, but also include the sting sac, muscle and nervous tissues. To facilitate communication, results will refer to ‘abdominal tissues’ which denotes these tissues collectively. Total RNA was extracted using QIAGEN RNeasy columns following the manufacturer's protocol.
(c). Library preparation and RNA-sequencing
Poly-A RNA was enriched from total RNA and strand-specific cDNA libraries were prepared using the Bioo Scientific NEXTflex Directional RNA-Sequencing Kit (dUTP Based) for Illumina. Paired-end sequencing was performed on an Illumina HiSeq 2500 at the W. M. Keck Center (University of Illinois). Quality was assessed using FASTQC and read trimming was performed with Trimmomatic prior to alignment with Bowtie to a previously assembled transcriptome for M. genalis [47].
(d). Differential expression analyses
Estimated read counts at the putative gene level were obtained using RSEM following alignment with scripts packaged in Trinity r20140413 [48]. Gene counts were filtered to include only genes with at least 1 count per million in the minimum number of samples per group per tissue type. A surrogate variable analysis [49,50] was performed on each tissue dataset to identify potential batch effects due to collection year, library preparation batch, sequencing lane or other unidentified technical differences. Dispersion estimates and pairwise tests of differential expression were conducted in edgeR [51] with FDR correction. DEG lists can be found in the electronic supplementary material, S2. Annotation of differentially expressed genes for GO enrichment was conducted using PANTHER for pairwise lists of DEGs, and a statistical overrepresentation test with Bonferroni correction for multiple testing was used to identify PANTHER pathways and GO-Slim Biological Processes overrepresented in pairwise lists as presented in the electronic supplementary material, S3 [52].
(e). Overlap with other studies
Putative orthologues between species were identified using BLAST reciprocal best hits (RBH) between predicted peptides. For M. genalis and Bombus terrestris transcriptomes [47,53], predicted peptides were obtained using TransDecoder. Conversion lists between microarray probes and annotation versions of the Apis mellifera genome, along with RBH results, are found in the electronic supplementary material, S4. For Representation Factor (RF) gene overlap tests, only genes tested in the study with a putative orthologue in both species were compared between any two given studies. Gene lists and complete RF results are given in the electronic supplementary material, S5 (gene expression studies) and S6 (selection studies).
3. Results
(a). Caste differences in gene expression
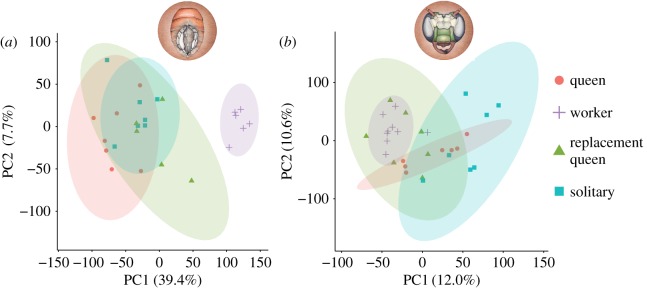
Female castes of M. genalis differed in gene expression in both brain and abdominal tissues (figure 1 and table 1). In abdominal tissues, variance in gene expression was largely explained by reproductive activity. The first principal component (figure 1a) explained nearly 40% of the variance in abdominal gene expression and separated workers (who are non-reproductive) from all reproductive groups. All reproductive females showed similar patterns of abdominal gene expression relative to workers, regardless of sociality (figure 2). Nearly 95% (3618/3827) of the genes that were more highly expressed in solitary females compared to workers were also more highly expressed in queens compared with workers, a highly significant overlap (representation factor: 3.34, p < 0.0001). These 3618 genes were strongly enriched for GO-Slim Biological Processes related to DNA metabolism and repair, chromatin organization and cell cycle (all GO enrichments listed in the electronic supplementary material, S3).
Figure 1.
Plot of first two principal components for (a) abdominal and (b) brain gene expression patterns across four female behavioural groups. Points represent individual samples and shaded ellipses show 95% CIs. Percentage of variance explained by each principal component is shown on axes. Drawings of M. genalis brain and abdominal tissues by Julie Himes.
Table 1.
Number of differentially expressed genes (DEGs, FDR < 0.05) for each pairwise comparison of female groups in both brain and abdominal tissues. Numbers in parentheses indicate number of genes more highly expressed in first group of pair (e.g. Q versus W comparison has 157 total DEGs, 49 of which are more highly expressed in Q compared to W). Q, queen; W, worker; S, solitary reproductive; R, replacement queen.
| comparison | abdomen DEGs | brain DEGs |
|---|---|---|
| Q versus W | 8127 (4044) | 157 (49) |
| S versus W | 6708 (3827) | 542 (219) |
| R versus W | 4206 (3048) | 0 |
| Q versus R | 510 (88) | 18 (8) |
| S versus R | 38 (9) | 133 (30) |
| S versus Q | 37 (35) | 16 (11) |
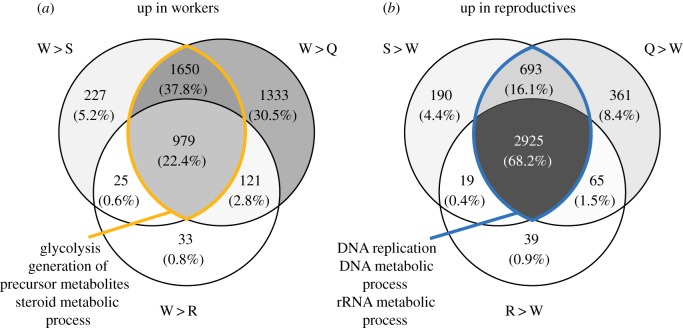
Figure 2.
Venn diagrams show genes (a) upregulated in workers relative to reproductive individuals and (b) upregulated in reproductive individuals relative to workers. Numbers in each subregion show the number of DEGs with an FDR < 0.05, and percentage of genes in that subregion relative to the universe of genes. Shading is relative to percentage of genes in each subregion, with darker regions corresponding to regions with higher percentages of genes. Yellow and blue highlighted regions show overlap between solitary and queen versus worker DEGs, respectively, and text refers to the top 3 significant enriched GO terms for genes in those regions. Q, queen; W, worker; S, solitary reproductive; R, replacement queen. (Online version in colour.)
Among abdominal worker-biased genes (figure 2a), 2629 genes were more highly expressed in both W > Q and W > S comparisons. The most enriched GO-Slim Biological Process for these genes was glycolysis (GO:0006096), with a nearly fivefold enrichment. Also enriched in worker-biased abdominal genes compared with queens and solitary females were steroid metabolic process (GO:0008202), respiratory electron transport chain (GO:0022904) and monosaccharide metabolic process (GO:0005996).
Brain differences in expression were much less pronounced than abdominal differences (table 1), but variance in brain gene expression explained by the first two principal components (22.6% of total variance) roughly mapped to the variance in behaviour seen across castes. Queens and workers were least variable, while both solitary and replacement queen females showed large variation in brain gene expression. The latter are the two groups that, at the time of collection, performed all activities in the nest (cell building, foraging, provisioning and reproduction). Many of the DEG lists in brain tissue did not have significant functional enrichment. However, heterotrimeric G-protein signalling pathways (P00026 and P00027) and both muscarinic (P00042 and P00043) and nicotinic (P00044) acetylcholine receptor signalling pathways were upregulated in both workers and replacement queens relative to solitary females. In solitary females, extracellular transport (GO:0006858) was enriched relative to workers.
(b). Replacement queens shift gene expression to reproductive-like phenotypes
The shift to reproductive activity in replacement queens following queen removal was associated with a near-complete shift in abdominal gene expression from worker-like to queen- and solitary-like (figures 1a and 2). Of the 3618 genes in common between Q > W and S > W abdominal DEGs, 81% were also more highly expressed in replacement queens compared with workers.
Replacement queens displayed high variation in brain gene expression compared with workers and queens, possibly reflecting the increased behavioural variation in this group. Three individuals (all collected in 2014) more closely resembled the reproductive phenotype along the first two principal components shown in figure 1b, while four individuals (two collected in 2014 and two in 2015) appeared worker-like in brain gene expression.
(c). Cross-species expression comparisons
Abdominal caste-specific differences in gene expression in M. genalis overlapped significantly with caste-specific DEGs in B. terrestris and five worker-related DEG lists in A. mellifera. In addition, brain caste-specific differences in gene expression in M. genalis overlapped significantly with differences in gene expression in two out of four comparisons with A. mellifera DEGs.
Caste-specific abdominal expression in M. genalis was compared with previous studies of B. terrestris caste differences [23] and A. mellifera workers [54,55]. Significant overlap was observed when comparing queen-biased and worker-biased genes in M. genalis abdomen and B. terrestris whole-body extractions (figure 3; RF: 1.5 for Q > W and RF: 1.6 for W > Q, both p < 0.0001). Of the genes more highly expressed in B. terrestris queens, 49% were also more highly expressed in the abdomens of M. genalis queens compared with workers. These genes were enriched for DNA metabolic process (GO:0006259), cell cycle (GO:0007049) and nucleobase-containing compound metabolic process (GO:0006139).
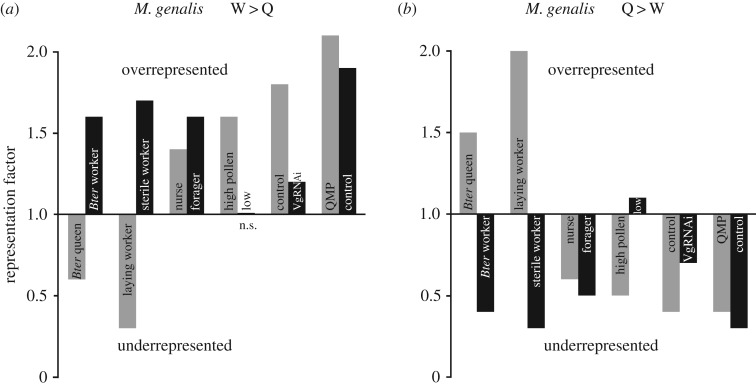
Figure 3.
Representation factors for overlap of M. genalis abdominal (a) worker > queen DEGs and (b) queen > worker DEGs with previous studies in Bombus terrestris (whole body [23]) and Apis mellifera (laying versus sterile worker, abdomen [54]; all other comparisons, fat body [55]). For each comparison, DEGs are split into two bars and labelled with the group more highly expressed (e.g. the black bar labelled ‘Bter worker’ in (a) shows the representation factor for W > Q genes in M. genalis and W > Q genes in B. terrestris). A representation factor (RF) of 1 indicates a level of overlap expected by chance, while RF > 1 indicates more overlap than expected and RF < 1 indicates less overlap than expected; n.s. not significantly different from RF = 1 (hypergeometric p > 0.05), all other bars are significantly different from RF = 1 (hypergeometric p < 0.05) in the direction shown.
Very strong overlap was observed when comparing queen-biased and worker-biased genes in M. genalis abdomens with DEGs in the abdomens of laying worker (LW) and sterile worker (S) honeybees (figure 3; RF: 2.0 for Q > W,LW > S and 1.7 for W > Q,S > LW; both p < 0.0001; [54]). Among the reproductive-related (Q > W and LW > S) overlap, enriched GO-Slim terms included chromatin organization (GO:0006325), DNA replication (GO:0006260) and mitosis (GO:0007067). No significant GO enrichment was observed for the overlapping genes between M. genalis W > Q and A. mellifera S > LW.
Significant overlap was also observed when comparing the M. genalis Q versus W abdomen DEG list and genes differentially expressed in the fat body of worker honeybees fed high versus low pollen diets [55]: Q > W genes had marginal overlap with Low > High pollen (RF: 1.1, p < 0.049) and W > Q genes had strong overlap with High > Low pollen-responsive genes (RF: 1.6, p < 0.0001; figure 3). For the remaining comparisons, M. genalis workers showed gene expression consistent with both worker phenotypes in honeybees. Genes that were more highly expressed in the abdomens of workers than in queens of M. genalis overlapped significantly with nurse (N) and forager (F) biased genes, genes both upregulated and downregulated by exposure to QMP, and vitellogenin (Vg)-responsive genes in the honeybee fat body ([55], figure 3).
No significant overlap was observed when comparing brain Q versus W DEGs in M. genalis and A. mellifera [26]. The majority of A. mellifera brain gene expression studies have compared behaviourally distinct subcastes of workers. Honeybee workers as a whole resemble M. genalis castes in terms of their behavioural flexibility (e.g. performance of both nurse-like and forager-like tasks). We therefore tested for overlap of our M. genalis Q versus W brain DEGs and DEGs from two A. mellifera N versus F experiments [56,57], as well as from an RNAi experiment looking at the effects of peripheral Vg knockdown on worker brain gene expression [58].
Significant overlap (RF: 2.0, p < 0.021) was found when comparing M. genalis Q versus W DEGs and DEGs from one N versus F study [56] but not the other (p < 0.379, [57]). Among the 11 overlapping DEGs between the M. genalis Q versus W and A. mellifera N versus F lists are serine/threonine-protein kinase ICK-like, apyrase precursor and two transporter proteins. A significant degree of overlap was found when comparing worker-biased genes in M. genalis (W > Q) and genes more highly expressed in Vg knockdown bees relative to control bees [58] (RF: 2.1, p < 0.017). Four of the 10 VgRNAi-overlapping genes were also within the 11 genes that overlap between M. genalis Q versus W and A. mellifera N versus F lists, including serine/threonine-protein kinase ICK-like, apyrase precursor, and an excitatory amino acid transporter. Lists of overlapping genes and tests for significance are found in the electronic supplementary material, S5.
(d). Insights from comparative molecular evolution studies
Worker-biased genes expressed in the abdomen of M. genalis were enriched for many GO terms related to metabolism. This was noteworthy because previous molecular evolution studies identified metabolic genes as one of the more prominent categories of genes undergoing selection in social bee lineages (figure 4; [18,20]). To more formally test the association between worker-biased genes in M. genalis and genes under selection in other bee species, we compared our abdomen W > Q DEG list with genes identified in three studies of molecular evolution [18–20]. In all three comparisons, the genes that were more highly expressed in the abdomens of workers relative to queens in M. genalis were also overrepresented among genes undergoing positive selection in social lineages of bees (table 2; electronic supplementary material, S6).
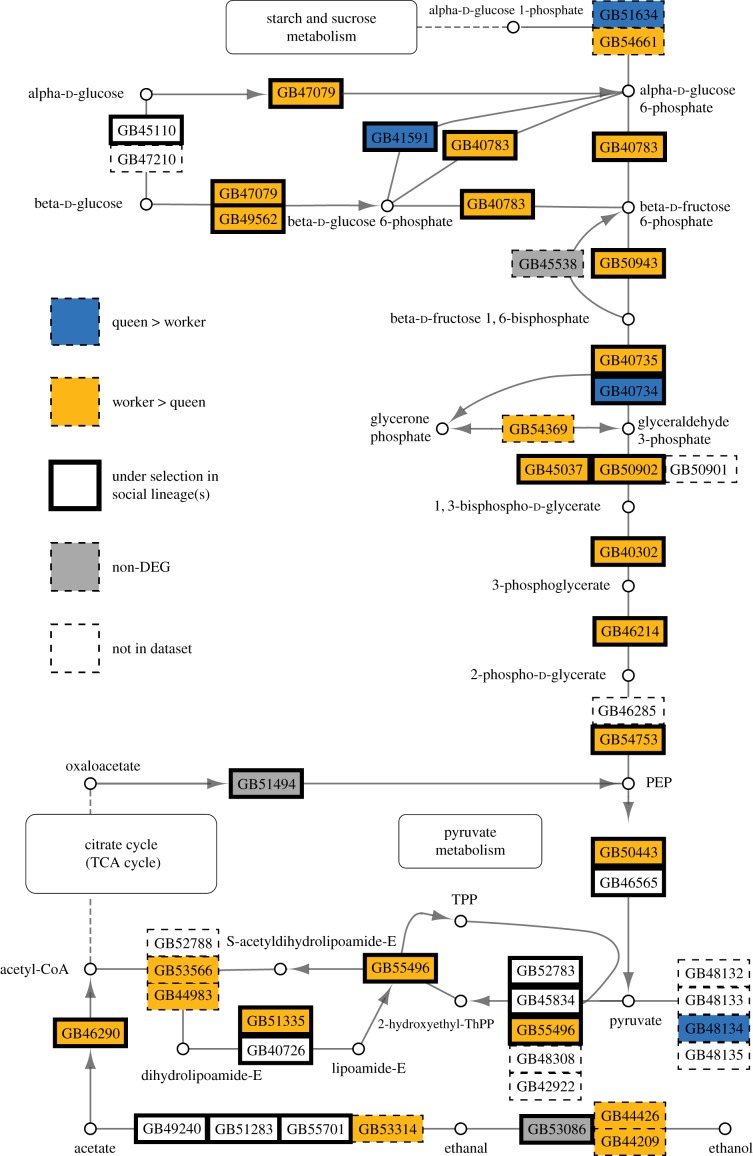
Figure 4.
Differentially expressed genes (FDR < 0.05) between queen and worker M. genalis abdominal tissues in the glycolysis/gluconeogenesis pathway. DEGs were mapped onto putative honeybee orthologues modified from KEGG pathway ame00010 (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/) and [18]. Genes labelled as under selection were identified as undergoing positive selection in at least one of three studies of selection in bees [18–20]. Genes not in dataset are honeybee genes without a BLAST reciprocal best hit to the M. genalis transcriptome or genes that are not expressed in the abdomen of M. genalis.
Table 2.
Overlap of genes more highly expressed in the abdomens of M. genalis workers than queens and three independent studies of selection across bees. Woodard et al. [18] genes are those identified as rapidly evolving in highly eusocial lineages of bees. Harpur et al. [19] genes are those with signatures of positive selection in A. mellifera. Kapheim et al. [20] genes are those undergoing positive selection across two independent origins of eusociality. Gene lists were restricted to those that had a putative orthologue in M. genalis (based on BLAST reciprocal best hit) and were expressed in the abdomen of M. genalis females. RF: Representation Factor, p-value is from hypergeometric test of overlap. GO terms listed are from PANTHER overrepresentation tests (Bonferroni corrected p < 0.05).
| source | genes under selection | no. W > Q | RF | p-value | GO enrichment |
|---|---|---|---|---|---|
| Woodard et al. | 130 | 50 | 1.3 | 0.015 | cellular amino acid metabolic process |
| Harpur et al. | 639 | 254 | 1.4 | <0.0001 | respiratory electron transport chain, generation of precursor metabolites and energy |
| Kapheim et al. | 70 | 28 | 1.5 | 0.014 | none |
Among the Harpur et al. [19] genes showing positive selection in A. mellifera that have putative M. genalis orthologues, nearly 40% (254/639) were worker-biased in expression (RF: 1.4, p < 0.0001; table 2). These genes were enriched for GO processes relating to respiratory electron transport chain (GO:0022904) and the generation of precursor metabolites and energy (GO:0006091). In contrast with worker-biased genes, queen-biased genes were underrepresented among the Harpur et al. [19] genes showing positive selection (RF: 0.7, p < 0.0001).
Genes identified under positive selection in Woodard et al. [18] included 50 that were also worker-biased in expression in M. genalis, eight of which are in the glycolysis pathway (figure 4; RF: 1.3, p = 0.015). These 50 genes were enriched for cellular amino acid metabolic process (GO:0006520), and seven were also among the Harpur et al. [19] genes, including aldehyde oxidase-like, aspartate aminotransferase, prostaglandin reductase 1-like, bifunctional ATP-dependent dihydroxyacetone kinase and 6-phosphofructokinase.
Genes identified as undergoing positive selection (103, 70 of which have M. genalis putative orthologues) across social lineages in Kapheim et al. [20] also overlapped significantly with abdominal worker-biased genes in M. genalis (28 genes; RF: 1.5, p = 0.014). These genes were not enriched for GO terms, but four of the 28 genes are in the glycolysis pathway (figure 4), including beta-lactamase-like protein 2 homologue, glyceraldehyde-3-phosphate dehydrogenase 2 isoform 1, phosphoglycerate mutase 2-like and enolase-like.
4. Discussion
The fact that eusociality has evolved independently multiple times in bees has provided rich material for comparative analyses. Recent work has demonstrated that different origins of eusociality have involved both common and unique pathways, often with changes in gene regulatory networks [18,20,59]. We found that abdominal caste-specific differences in gene expression in a facultatively eusocial bee, M. genalis, overlapped significantly with caste-specific differences in gene expression in other eusocial bee species. Our findings suggest common mechanisms involved in caste regulation across lineages with different types of eusociality. Moreover, we found that worker-biased abdominal genes in M. genalis overlapped significantly with genes shown to be rapidly evolving in eusocial lineages of bees from three independent studies of selection. With M. genalis serving as a proxy for ancestral variation, these results provide support for genetic accommodation in the evolution of eusociality in bees. According to this hypothesis, phenotypic plasticity exhibited by facultatively eusocial species may have enabled selection for permanent reproductive and non-reproductive castes.
Dramatic differences in gene expression were observed in abdominal tissues when comparing workers with reproductively active females. While DNA-related processes dominated the reproductive signal in abdominal tissues, workers showed a bias for expression of many metabolic pathways, including glycolysis (figure 4). In contrast with the abdominal results, brain gene expression differences were more subtle. The number of DEGs between queens and workers was more similar to that seen in the primitively eusocial wasp Polistes dominula [60] than in the highly derived eusocial honeybee [26].The striking differences in DEG numbers between the brain and abdomen suggest that signalling between the reproductive system and the brain plays a role in mediating behaviour of M. genalis females. This is similar to hypotheses from the honeybee literature [61–63], where it has been reported that surgical implantation of supernumerary ovaries in workers leads to a shift in the age at onset of foraging [63]. An alternative explanation for the differences in DEG numbers between brain and abdominal comparisons is that collection of individuals during inactive periods led to a reduction in the genes that were differentially expressed in the brain.
Theory predicts that social traits may be subject to relaxed selective constraint and higher levels of polyphenism [64,65]. Our results are consistent with predicted relaxed constraints on worker traits, which may predispose worker-biased genes to accumulate mutations to be screened by selection [12,66]. Consistent with theory, the genes identified by Harpur et al. [19] with signatures of adaptive evolution in honeybees were also worker-biased, and worker-biased genes have been reported to be more derived in multiple social insect species [19,67,68]. Overlap of M. genalis worker-biased genes and those under selection in eusocial lineages include many involved in glycolysis (figure 4), which has been implicated in caste determination not only in bees but also in other social insects [59].
Phenotypes that are originally environmentally induced can be selected upon and shaped, such that inherited variants can express the trait absent the environmental induction [4]. Genetic accommodation predicts selection on genes that are environmentally sensitive if there is a selective advantage to the environmentally induced phenotype. Eusocial insects have evolved a worker caste that is reproductively inactive and often specialized for non-reproductive behaviours relative to the ancestral solitary state. In multiple social insect lineages, this specialization was enabled by selection on genes that are now worker-biased in their expression [19,67,68]. Our study implicates some of the same genes in the flexible worker phenotype of M. genalis, which suggests a role for genetic accommodation in the evolution of specialized worker castes. If genetic accommodation facilitated worker specialization, we would expect ancestral plasticity in gene expression associated with worker-related genes. Consistent with this prediction, we found that worker-biased genes in M. genalis share significant identity with genes identified as undergoing selection in three independent tests of positive selection across eusocial lineages of bees [18–20]. Plasticity in these genes in the ancestors of obligately eusocial species may have facilitated the evolution of derived worker castes through genetic accommodation, leading to a more fixed caste determination system as seen in many obligately eusocial groups.
We provide support for the idea that genetic accommodation may have played a role in the evolution of caste, with ancestral environmentally induced plasticity leading to selection on worker traits in the evolution of eusociality in bees. Future research investigating allele frequency change within M. genalis itself would strengthen this support if caste-biased genes were found to be under selection in this species. As molecular data from other incipiently social lineages are integrated with knowledge from complex eusocial species [35,69], it will be possible to explore the role of genetic accommodation in eusociality more rigorously.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Betzi Perez, Gabriel Trujillo, Isis Lopez, Esther Velasquez and Ernesto Gomez-Perez for their assistance with collections in the field. We also thank Amy Cash Ahmed for assistance with molecular work, and Claudia Lutz and members of the Robinson lab for helpful discussion and comments on the manuscript.
Data accessibility
Raw sequencing data have been deposited in the short read archive (NCBI) under the accession SRP079663. The transcriptome used for alignment is available as a TSA submission under the accession PRJNA282469. Electronic supplementary material associated with this article is available online.
Authors' contributions
B.M.J., C.J.K., W.T.W. and G.E.R. conceived and designed the experiments; B.M.J. and C.J.K. performed fieldwork; B.M.J. performed molecular and bioinformatic experiments; B.M.J., C.J.K., W.T.W. and G.E.R. wrote the paper.
Competing interests
We declare we have no competing interests.
Funding
Research was funded through the University of Illinois NSF IGERT grant (A. Suarez, PI; DGE 1069157) and fellowship support to B.M.J. and C.J.K. from the Smithsonian Tropical Research Institute (STRI). Additional support was provided by STRI research funds to W.T.W. and the NIH Director's Pioneer Award (DP1 OD006414) to G.E.R. Much of the computing work for this research was supported by the National Science Foundation funded MRI-R2 project #DBI-0959894 (Data Intensive Academic Grid).
References
- 1.Yampolsky LY, Glazko GV, Fry JD. 2012. Evolution of gene expression and expression plasticity in long-term experimental populations of Drosophila melanogaster maintained under constant and variable ethanol stress. Mol. Ecol. 21, 4287–4299. ( 10.1111/j.1365-294X.2012.05697.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pichancourt J-B, van Klinken RD. 2012. Phenotypic plasticity influences the size, shape and dynamics of the geographic distribution of an invasive plant. PLoS ONE 7, e32323 ( 10.1371/journal.pone.0032323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leichty AR, Pfennig DW, Jones CD, Pfennig KS. 2012. Relaxed genetic constraint is ancestral to the evolution of phenotypic plasticity. Integr. Comp. Biol. 52, 16–30. ( 10.1093/icb/ics049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West-Eberhard MJ. 2003. Developmental plasticity and evolution. Oxford, UK: Oxford University Press. [Google Scholar]
- 5.Ghalambor CK, Hoke KL, Ruell EW, Fischer EK, Reznick DN, Hughes KA. 2015. Non-adaptive plasticity potentiates rapid adaptive evolution of gene expression in nature. Nature 525, 372–375. ( 10.1038/nature15256) [DOI] [PubMed] [Google Scholar]
- 6.Waddington CH. 1942. Canalization of development and the inheritance of acquired characters. Nature 150, 563–656. ( 10.1038/150563a0) [DOI] [PubMed] [Google Scholar]
- 7.Waddington CH. 1953. Genetic assimilation of an acquired character. Evolution 7, 118–126. ( 10.2307/2405747) [DOI] [Google Scholar]
- 8.Suzuki Y, Nijhout F. 2006. Evolution of a polyphenism by genetic accommodation. Science 311, 650–652. ( 10.1126/science.1118888) [DOI] [PubMed] [Google Scholar]
- 9.Heil M, Greiner S, Meimberg H, Kruger R, Noyer JL, Heubl G, Linsenmair KE, Boland W. 2004. Evolutionary change from induced to constitutive expression of an indirect plant resistance. Nature 430, 205–208. ( 10.1038/nature02703) [DOI] [PubMed] [Google Scholar]
- 10.Santana SE, Dumont ER. 2009. Connecting behaviour and performance: the evolution of biting behaviour and bite performance in bats. J. Evol. Biol. 22, 2131–2145. ( 10.1111/j.1420-9101.2009.01827.x) [DOI] [PubMed] [Google Scholar]
- 11.Piperno DR, Holst I, Winter K, McMillan O. 2015. Teosinte before domestication: experimental study of growth and phenotypic variability in Late Pleistocene and early Holocene environments. Quat. Int. 363, 65–77. ( 10.1016/j.quaint.2013.12.049) [DOI] [Google Scholar]
- 12.Moczek AP, Sultan S, Foster S, Ledon-Rettig C, Dworkin I, Nijhout HF, Abouheif E, Pfennig DW. 2011. The role of developmental plasticity in evolutionary innovation. Proc. R. Soc. B 278, 2705–2713. ( 10.1098/rspb.2011.0971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ledon-Rettig CC, Pfennig DW, Nascone-Yoder N. 2008. Ancestral variation and the potential for genetic accommodation in larval amphibians: implications for the evolution of novel feeding strategies. Evol. Dev. 10, 316–325. ( 10.1111/j.1525-142X.2008.00240.x) [DOI] [PubMed] [Google Scholar]
- 14.Cameron SA, Mardulyn P. 2001. Multiple molecular data sets suggest independent origins of highly eusocial behavior in bees (Hymenoptera: Apinae). Syst. Biol. 50, 194–214. ( 10.1080/10635150120230) [DOI] [PubMed] [Google Scholar]
- 15.Schwarz MP, Richards MH, Danforth BN. 2007. Changing paradigms in insect social evolution: insights from halictine and allodapine bees. Annu. Rev. Entomol. 52, 127–150. ( 10.1146/annurev.ento.51.110104.150950) [DOI] [PubMed] [Google Scholar]
- 16.Cardinal S, Straka J, Danforth BN. 2010. Comprehensive phylogeny of apid bees reveals the evolutionary origins and antiquity of cleptoparasitism. Proc. Natl Acad. Sci. USA 107, 16 207–16 211. ( 10.1073/pnas.1006299107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danforth BN, Sipes S, Fang J, Brady SG. 2006. Recent and simultaneous origins of eusociality in halictid bees. Proc. Natl Acad. Sci. USA 103, 15 118–15 123. ( 10.1098/rspb.2006.3496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodard SH, Fischman BJ, Venkat A, Hudson ME, Varala K, Cameron SA, Clark AG, Robinson GE. 2011. Genes involved in convergent evolution of eusociality in bees. Proc. Natl Acad. Sci. USA 108, 7472–7477. ( 10.1073/pnas.1103457108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harpur BA, Kent CF, Molodtsova D, Lebon JMD, Alqarni AS, Owayss AA, Zayed A. 2014. Population genomics of the honey bee reveals strong signatures of positive selection on worker traits. Proc. Natl Acad. Sci. USA 111, 2614–2619. ( 10.1073/pnas.1315506111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapheim KM, et al. 2015. Genomic signatures of evolutionary transitions from solitary to group living. Science 348, 1139–1143. ( 10.1126/science.aaa4788) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunt BG, Wyder S, Elango N, Werren JH, Zdobnov EM, Yi SV, Goodisman MAD. 2010. Sociality is linked to rates of protein evolution in a highly social insect. Mol. Biol. Evol. 27, 497–500. ( 10.1093/molbev/msp225) [DOI] [PubMed] [Google Scholar]
- 22.Snell-Rood EC, Cash A, Han MV, Kijimoto T, Andrews J, Moczek AP. 2011. Developmental decoupling of alternative phenotypes: insights from the transcriptomes of horn-polyphenic beetles. Evolution 65, 231–245. ( 10.1111/j.1558-5646.2010.01106.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison MC, Hammond RL, Mallon EB. 2015. Reproductive workers show queenlike gene expression in an intermediately eusocial insect, the buff-tailed bumble bee Bombus terrestris. Mol. Ecol. 24, 3043–3063. ( 10.1111/mec.13215) [DOI] [PubMed] [Google Scholar]
- 24.Chandrasekaran S, Ament SA, Eddy JA, Rodriguez-Zas SL, Schatz BR, Price ND, Robinson GE. 2011. Behavior-specific changes in transcriptional modules leads to distinct and predictable neurogenomic states. Proc. Natl Acad. Sci. USA 108, 18 020–18 025. ( 10.1073/pnas.1114093108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ament SA, et al. 2012. New meta-analysis tools reveal common transcriptional regulatory basis for multiple determinants of behavior. Proc. Natl Acad. Sci. USA 109, E1801–E1810. ( 10.1073/pnas.1205283109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grozinger CM, Fan Y, Hoover SER, Winston ML. 2007. Genome-wide analysis reveals differences in brain gene expression patterns associated with caste and reproductive status in honey bees (Apis mellifera). Mol. Ecol. 16, 4837–4848. ( 10.1111/j.1365-294X.2007.03545.x) [DOI] [PubMed] [Google Scholar]
- 27.Hunt BG, Ometto L, Wurm Y, Shoemaker D, Yi SV, Keller L, Goodisman MAD. 2011. Relaxed selection is a precursor to the evolution of phenotypic plasticity. Proc. Natl Acad. Sci. USA 108, 15 936–15 941. ( 10.1073/pnas.1104825108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson EO. 1971. The insect societies. Cambridge, MA: Harvard University Press. [Google Scholar]
- 29.Wilson EO, Hölldobler B. 2005. Eusociality: origin and consequences. Proc. Natl Acad. Sci. USA 205, 13 367–13 371. ( 10.1073/pnas.0505858102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Packer L. 1990. Solitary and eusocial nests in a population of Augochlorella striata (Provancher) (Hymenoptera, Halictidae) at the northern edge of its range. Behav. Ecol. Sociobiol. 27, 339–344. ( 10.1007/BF00164004) [DOI] [Google Scholar]
- 31.Eickwort GC, Eickwort JM, Gordon J, Eickwort MA. 1996. Solitary behavior in a high-altitude population of the social sweat bee Halictus rubicundus (Hymenoptera: Halictidae). Behav. Ecol. Sociobiol. 38, 227–233. ( 10.1007/s002650050236) [DOI] [Google Scholar]
- 32.Plateaux-Quenu C, Plateaux L, Packer L. 2000. Population-typical behaviours are retained when eusocial and non-eusocial forms of Evylaeus albipes (F.) (Hymenoptera, Halictidae) are reared simultaneously in the laboratory. Insectes Soc. 47, 263–270. ( 10.1007/PL00001713) [DOI] [Google Scholar]
- 33.Cronin AL, Hirata M. 2003. Social polymorphism in the sweat bee Lasioglossum (Evylaeus) baleicum (Cockerell) (Hymenoptera, Halictidae) in Hokkaido, northern Japan. Insectes Soc. 50, 379–386. () [DOI] [Google Scholar]
- 34.Field J, Paxton RJ, Soro A, Bridge C. 2010. Cryptic plasticity underlies a major evolutionary transition. Curr. Biol. 20, 2028–2031. ( 10.1016/j.cub.2010.10.020) [DOI] [PubMed] [Google Scholar]
- 35.Kocher SD, Paxton RJ. 2014. Comparative methods offer powerful insights into social evolution in bees. Apidologie 45, 289–305. ( 10.1007/s13592-014-0268-3) [DOI] [Google Scholar]
- 36.Yanega D. 1988. Social plasticity and early-diapausing females in a primitively social bee. Proc. Natl Acad. Sci. USA 85, 4374–4377. ( 10.1073/pnas.85.12.4374) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wcislo WT, Arneson L, Roesch K, Gonzalez V, Smith A, Fernandez H. 2004. The evolution of nocturnal behaviour in sweat bees, Megalopta genalis and M. ecuadoria (Hymenoptera: Halictidae): an escape from competitors and enemies? Biol. J. Linn. Soc. 83, 377–387. ( 10.1111/j.1095-8312.2004.00399.x) [DOI] [Google Scholar]
- 38.Wcislo WT, Gonzalez VH. 2006. Social and ecological contexts of trophallaxis in facultatively social sweat bees, Megalopta genalis and M. ecuadoria (Hymenoptera, Halictidae). Insectes Soc. 53, 220–225. ( 10.1007/s00040-005-0861-6) [DOI] [Google Scholar]
- 39.Kapheim KM, Bernal SP, Smith AR, Nonacs P, Wcislo WT. 2011. Support for maternal manipulation of developmental nutrition in a facultatively eusocial bee, Megalopta genalis (Halictidae). Behav. Ecol. Sociobiol. 65, 1179–1190. ( 10.1007/s00265-010-1131-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapheim KM, Smith AR, Ihle KE, Amdam GV, Nonacs P, Wcislo WT. 2012. Physiological variation as a mechanism for developmental caste-biasing in a facultatively eusocial sweat bee. Proc. R. Soc. B 279, 1437–1446. ( 10.1098/rspb.2011.1652) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith AR, Kapheim KM, O'Donnell S, Wcislo WT. 2009. Social competition but not subfertility leads to a division of labour in the facultatively social sweat bee Megalopta genalis (Hymenoptera: Halictidae). Anim. Behav. 78, 1043–1050. ( 10.1016/j.anbehav.2009.06.032) [DOI] [Google Scholar]
- 42.Kapheim KM, Smith AR, Nonacs WT, Wayne RK. 2013. Foundress polyphenism and the origins of eusociality in a facultatively eusocial sweat bee, Megalopta genalis (Halictidae). Behav. Ecol. Sociobiol. 67, 331–340. ( 10.1007/s00265-012-1453-x) [DOI] [Google Scholar]
- 43.Smith AR, Wcislo WT, O'Donnell S. 2008. Body size shapes caste expression, and cleptoparasitism reduces body size in the facultatively eusocial bees Megalopta (Hymenoptera: Halictidae). J. Insect Behav. 21, 394–406. ( 10.1007/s10905-008-9136-1) [DOI] [Google Scholar]
- 44.Smith AR, Kapheim KM, Pérez-Ortega B, Brent CS, Wcislo WT. 2012. Juvenile hormone levels reflect social opportunities in the facultatively eusocial sweat bee Megalopta genalis (Hymenoptera: Halictidae). Horm. Behav. 63, 1–4. ( 10.1016/j.yhbeh.2012.08.012) [DOI] [PubMed] [Google Scholar]
- 45.Wolda H, Roubik DW. 1986. Nocturnal bee abundance and seasonal bee activity in a Panamanian forest. Ecology 67, 426–433. ( 10.2307/1938586) [DOI] [Google Scholar]
- 46.Smith AR, Wcislo WT, O'Donnell S. 2007. Survival and productivity benefits to social nesting in the sweat bee Megalopta genalis (Hymenoptera: Halictidae). Behav. Ecol. Sociobiol. 61, 1111–1120. ( 10.1007/s00265-006-0344-4) [DOI] [Google Scholar]
- 47.Jones BM, Wcislo WT, Robinson GE. 2015. Developmental transcriptome for a facultatively eusocial bee, Megalopta genalis. G3 5, 2127–2135. ( 10.1534/g3.115.021261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 15, 644–652. ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmean B, Johnson WE, Geman D, Baggerly K, Irizarry RA. 2010. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 11, 733–739. ( 10.1038/nrg2825) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883. ( 10.1093/bioinformatics/bts034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. ( 10.1093/nar/gks042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mi H, Muruganujan A, Thomas PD. 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386. ( 10.1093/nar/gks1118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colgan TJ, Carolan JC, Bridgett SJ, Sumner S, Blaxter ML, Brown MJF. 2011. Polyphenism in social insects: insights from a transcriptome-wide analysis of gene expression in the life stages of the key pollinator, Bombus terrestris. BMC Genomics 12, 623 ( 10.1186/1471-2164-12-623) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galbraith DA, Kocher SA, Glenn T, Albert I, Hunt GJ, Strassmann JE, Queller DC, Grozinger CM. 2015. Testing the kinship theory of intragenomic conflict in honey bees (Apis mellifera). Proc. Natl Acad. Sci. USA 113, 1020–1025. ( 10.1073/pnas.1516636113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ament SA, Chan QW, Wheeler MM, Nixon SE, Johnson SP, Rodriguez-Zas SL, Foster LJ, Robinson GE. 2011. Mechanisms of stable lipid loss in a social insect. J. Exp. Biol. 214, 3808–3821. ( 10.1242/jeb.060244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alaux C, Le Conte Y, Adams HA, Rodriguez-Zas S, Grozinger CM, Sinha S, Robinson GE. 2009. Regulation of brain gene expression in honey bees by brood pheromone. Genes Brain Behav. 8, 309–319. ( 10.1111/j.1601-183X.2009.00480.x) [DOI] [PubMed] [Google Scholar]
- 57.Whitfield CW, Cziko AM, Robinson GE. 2003. Gene expression profiles in the brain predict behavior in individual honey bees. Science 302, 296–299. ( 10.1126/science.1086807) [DOI] [PubMed] [Google Scholar]
- 58.Wheeler MM, Ament SA, Rodriguez-Zas SL, Robinson GE. 2013. Brain gene expression changes elicited by peripheral vitellogenin knockdown in the honey bee. Insect Mol. Biol. 22, 562–573. ( 10.1111/imb.12043) [DOI] [PubMed] [Google Scholar]
- 59.Berens AJ, Hunt JH, Toth AL. 2015. Comparative transcriptomics of convergent evolution: different genes but conserved pathways underlie caste phenotypes across lineages of eusocial insects. Mol. Biol. Evol. 32, 690–703. ( 10.1093/molbev/msu330) [DOI] [PubMed] [Google Scholar]
- 60.Standage DS, Berens AJ, Glastad KM, Severin AJ, Brendel VP, Toth AL. 2016. Genome, transcriptome and methylome sequencing of a primitively eusocial wasp reveal a greatly reduced DNA methylation system in a social insect. Mol. Ecol. 25, 1769–1784. ( 10.1111/mec.13578) [DOI] [PubMed] [Google Scholar]
- 61.Amdam GV, Csondes A, Fondrk MK, Page RE Jr. 2006. Complex social behaviour derived from maternal reproductive traits. Nature 439, 76–78. ( 10.1038/nature04340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, Amdam GV, Rueppell O, Wallrichs MA, Fondrk MK, Kaftanoglu O, Page RE Jr. 2009. PDK1 and HR46 gene homologs tie social behavior to ovary signals. PLoS ONE 4, e4899 ( 10.1371/journal.pone.0004899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Kaftanoglua O, Siegela AJ, Page RE Jr, Amdam GV. 2010. Surgically increased ovarian mass in the honey bee confirms link between reproductive physiology and worker behavior. J. Insect Physiol. 56, 1216–1224. ( 10.1016/j.jinsphys.2010.07.013) [DOI] [PubMed] [Google Scholar]
- 64.Gadagkar R. 1997. The evolution of caste polymorphism in social insects: genetic release followed by diversifying evolution. J. Genetics 76, 167–179. ( 10.1007/bf02932215) [DOI] [Google Scholar]
- 65.Linksvayer TA, Wade MJ. 2009. Genes with social effects are expected to harbor more sequence variation within and between species. Evolution 63, 1685–1696. ( 10.1111/j.1558-5646.2009.00670.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Dyken J, Wade MJ. 2010. The genetic signature of conditional expression. Genetics 184, 557–570. ( 10.1534/genetics.109.110163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feldmeyer B, Elsner D, Foitzik S. 2014. Gene expression patterns associated with caste and reproductive status in ants: worker-specific genes are more derived than queen-specific ones. Mol. Ecol. 23, 151–161. ( 10.1111/mec.12490) [DOI] [PubMed] [Google Scholar]
- 68.Ferreira PG, Patalano S, Chauhan R, Ffrench-Constant R, Gabaldon T, Guigó R, Sumner S. 2013. Transcriptome analyses of primitively eusocial wasps reveal novel insights into the evolution of sociality and the origin of alternative phenotypes. Genome Biol. 14, R20 ( 10.1186/gb-2013-14-2-r20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rehan SM, Toth AL. 2015. Climbing the social ladder: the molecular evolution of sociality. Trends Ecol. Evol. 30, 426–433. ( 10.1016/j.tree.2015.05.004) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data have been deposited in the short read archive (NCBI) under the accession SRP079663. The transcriptome used for alignment is available as a TSA submission under the accession PRJNA282469. Electronic supplementary material associated with this article is available online.