Abstract
Ancient DNA of extinct species from the Pleistocene and Holocene has provided valuable evolutionary insights. However, these are largely restricted to mammals and high latitudes because DNA preservation in warm climates is typically poor. In the tropics and subtropics, non-avian reptiles constitute a significant part of the fauna and little is known about the genetics of the many extinct reptiles from tropical islands. We have reconstructed the near-complete mitochondrial genome of an extinct giant tortoise from the Bahamas (Chelonoidis alburyorum) using an approximately 1 000-year-old humerus from a water-filled sinkhole (blue hole) on Great Abaco Island. Phylogenetic and molecular clock analyses place this extinct species as closely related to Galápagos (C. niger complex) and Chaco tortoises (C. chilensis), and provide evidence for repeated overseas dispersal in this tortoise group. The ancestors of extant Chelonoidis species arrived in South America from Africa only after the opening of the Atlantic Ocean and dispersed from there to the Caribbean and the Galápagos Islands. Our results also suggest that the anoxic, thermally buffered environment of blue holes may enhance DNA preservation, and thus are opening a window for better understanding evolution and population history of extinct tropical species, which would likely still exist without human impact.
Keywords: Bahamas, biogeography, extinction, palaeontology, phylogeny
1. Introduction
Post-mortem degradation of DNA is climate dependent, being greatly accelerated in warm tropical and subtropical regions [1,2]. As a result, extinct Late Pleistocene megafauna from cold climates has been widely studied using ancient DNA (aDNA) approaches [3], providing valuable insights in ecology, evolution, and biogeography, and causes of extinction of vanished species. By contrast, aDNA from tropical and subtropical environments remains largely unexplored, apart from some notable exceptions [4–8]. A further consequence is that aDNA studies are biased toward taxa that are abundant at higher latitudes, in particular, mammals. Other groups, such as non-avian reptiles, which are highly diverse in warm climates, remain little studied [9–13]. Subtropical and tropical islands are systems that would benefit greatly from information from aDNA because they have experienced substantial losses of both megafauna and small-bodied species after the Holocene arrival of humans [14–16]. The Bahamas are one such example, with much of the original vertebrate fauna (reptiles, birds, and mammals) having disappeared within a few centuries after the arrival of human settlers about 1 000 years before present (BP) [17].
Among the extinct Bahamian species is an endemic giant tortoise, Chelonoidis alburyorum, which is believed to have gone extinct around 780 BP [18]. Complete tortoise fossils, with a shell length of up to 47 cm, have been discovered in Sawmill Sink, a deep inland blue hole and cave system, on Great Abaco Island in the northern Bahamas [19]. The fossils were retrieved from anoxic saltwater, and were found to contain substantial collagen [20], suggesting at least the potential for DNA preservation. Although samples from temperate saltwater deposits have yielded endogenous DNA [21,22], the validity of a recent report on aDNA from a Mexican underwater cave [23] has been questioned [24]. Thus, the preservation potential of DNA in tropical aquatic or water-logged environments is poorly understood. In this study, we have analysed aDNA from an almost 1 000-year-old subfossil humerus of C. alburyorum from Sawmill Sink. Although DNA preservation in the sample is poor, we have recovered an almost complete mitochondrial genome sequence from the sample, which provides new insights into the origin and relationships of this enigmatic giant tortoise and contributes to a better understanding of the biogeography of the Bahamas.
2. Material and methods
(a). Studied specimens
The following specimens from the collections of the National Museum of The Bahamas, Marsh Harbour, Bahamas (NMB) and the Museum of Zoology, Senckenberg Dresden, Germany (MTD) were studied: C. alburyorum, subfossil humerus of specimen NMB.AB50.0008 (Sawmill Sink, Abaco Island, Bahamas); C. carbonarius, fresh tissue sample MTD-T 5138 (Iracoubo, French Guiana); C. chilensis, fresh tissue sample MTD-T 5754 (−40.787778, −65.316389, Río Negro Province, Argentina); C. denticulatus, fresh tissue sample MTD-T 7255 (from pet trade); C. vicina (‘Poldi’ kept at Reptile Zoo Happ, Klagenfurt, Austria), blood sample MTD-T 14174; Geochelone sulcata, fresh tissue sample MTD-T 872 (captive bred).
(b). Processing of the ancient sample
All stages of sample processing prior to polymerase chain reaction (PCR) amplification were carried out in dedicated aDNA facilities at the University of Potsdam, following established guidelines [25]. Negative controls (water blanks) were included during DNA extraction and library preparation and screened for evidence of contamination. Two 50 mg bone powder samples were obtained from the C. alburyorum humerus. DNA was extracted from each sample using a published protocol optimized for the recovery of short aDNA fragments [26]. DNA extracts were treated with uracil-DNA glycosylase (UDG) to remove uracil residues probably resulting from DNA damage and then converted into Illumina sequencing libraries using a protocol based on single-stranded DNA [27]. An initial assessment of DNA preservation and contamination was made by low-level shotgun sequencing of the libraries on an Illumina NextSeq 500 sequencing platform generating 75 bp paired-end reads. Owing to low abundance of endogenous DNA fragments in the sequencing libraries, we performed two-rounds of in-solution hybridization capture to enrich for mitochondrial DNA fragments [28,29], using DNA baits generated from long-range PCR products of the congeneric species C. chilensis (see below). Sequencing of enriched libraries was as described above.
(c). Processing of modern samples
DNA of extant relatives of C. alburyorum was extracted using commercial kits (Analytik Jena AG, Jena, Germany), and served as template for amplicon sequencing (C. chilensis, C. vicina, and G. sulcata), or in-solution hybridization capture enrichment (C. carbonarius and C. denticulatus), depending on DNA quality. Amplicon sequencing involved PCR amplification of mitogenomes using standard methods (for primer sequences and PCR conditions see electronic supplementary material, Amplicon sequencing, and table S1). Amplification products were sheared and converted into Illumina sequencing libraries using a published protocol based on double-stranded DNA [30] with modifications [31]. Hybridization capture enrichment of degraded samples followed the procedures described previously for the ancient sample. All modern sample libraries were sequenced on an Illumina NextSeq 500 sequencing platform generating 150 bp paired-end reads.
(d). Assessment of endogenous and contaminant DNA content
Prior to analysis, adapter sequences were trimmed from the 3′ read ends, overlapping paired-end reads were merged, and any merged reads less than 20 bp discarded, using the program SeqPrep [32]. Analysis of shotgun data from the ancient C. alburyorum sample involved estimation of endogenous DNA content by calculating the proportion of sequence reads that could be mapped to the reference nuclear genome assembly of the painted turtle (Chrysemys picta bellii) [33] using bwa [34] with a mismatch value of 0.001. Reads with low mapping quality (less than 30) and likely PCR duplicates were removed from the alignment using SAMtools [35]. Cow, dog, cat, human, and mouse were then investigated as potential sources of contamination using fastqscreen [36]. To assess the authenticity of the ancient reads obtained, the shotgun data as well as the assembled reads from the enriched libraries were re-mapped to the reference nuclear genome assembly of Ch. picta and the newly generated mitogenome of C. alburyorum, respectively, in order to generate nucleotide misincorporation plots using mapDamage 2.0 [37]. Finally, we estimated the preservation of DNA in a bone sample deposited in the terrestrial environment of the Bahamas at 25.04829° latitude and −77.432848° longitude and buried under a 20 m layer of silt-loam soil using the online resource http://thermal-age.eu (Job 1337), for comparison to the empirical data obtained from the C. alburyorum sample.
(e). Assembly of mitogenome sequences
Assembly of mitogenome sequences from the enriched and amplicon libraries involved a two-step baiting and iterative mapping approach in MITObim [38]. Prior to assembly, duplicate read pairs were removed from each dataset using FastUniq [39] and the order of the remaining unique reads randomized using fastq-sort [36]. Only reads more than 31 bp were used for assembly, which corresponded to the k-mer size used for baiting. Various levels of coverage and mapping stringency were tested and optimal values selected based on visual assessment of the final alignments in Tablet v. 1.15.09.1 [40]. After assembly, PCR priming sites were removed from amplicon assemblies. Mitogenome annotation was performed using MITOS [41].
(f). Phylogenetic analyses and molecular dating
Novel sequences were aligned with all Testudinidae mitochondrial genomes available on GenBank, plus representatives of the turtle genera Mauremys and Emys as outgroups (electronic supplementary material, Mitochondrial genomes from GenBank used for phylogenetic analyses), using the ClustalW algorithm [42] with default settings, resulting in 22 485 aligned positions. Alternative data partitioning schemes were compared using the software PartitionFinder [43] using the Bayesian Information Criterion (BIC).
Phylogenetic analysis using Bayesian Inference was conducted with MrBayes 3.2.1 [44] and optimal models selected by PartitionFinder (electronic supplementary material, table S2), with two parallel runs (each with four chains) and default parameters. Parameter convergence, sampling adequacy, and appropriate burn-in was determined using the software Tracer 1.6 [45]. A 50% majority rule consensus tree was then generated from the posterior sample of trees. Phylogenetic analysis was additionally conducted under Maximum Likelihood using RAxML 7.2.8 [46] and the GTR + G substitution model. Clade support was assessed by bootstrap analysis, involving multiple independent runs using both fast and thorough bootstrap algorithms.
Molecular dating was conducted with BEAST 1.8.2 [47]. Two calibration points were specified using normally distributed priors. Based on the fossil species C. hesternus from the middle Miocene La Venta Fauna of Colombia, thought to be close to the last common ancestor of C. carbonarius and C. denticulatus [48], the split between these two species was identified with La Ventan age, 13.5–11.8 million years ago (mya) [49,50]. Accordingly, the node age was set to a mean of 12.55 mya with a standard deviation of 0.6. The Geoemydidae (Mauremys) + Testudinidae node was dated to 50.3–66.99 mya, based on the fossil tortoise species Hadrianus majusculus [51], using a mean of 58.65 mya and a standard deviation of 5.08. Analyses involved the HKY substitution model, estimated base frequencies, an uncorrelated lognormal relaxed molecular clock, and the Yule tree prior. MCMC chains were inspected as described above, and the maximum clade credibility tree was extracted using TreeAnnotator and viewed in FigTree 1.4.2 [52].
3. Results
(a). DNA preservation of the Chelonoidis alburyorum sample
Analysis of the C. alburyorum shotgun data indicated high levels of degradation and contamination (electronic supplementary material, figure S1). Only 1.4% of reads could be mapped to the Ch. picta reference genome, although this is almost certainly an underestimate of endogenous DNA content due to the considerable evolutionary divergence of Ch. picta from C. alburyorum (approx. 86 mya) [51]. To corroborate the presence of ancient endogenous DNA molecules, misincorporation plots were generated for the 24 362 reads resulting from shotgun sequencing that mapped against the full genome of Ch. picta (electronic supplementary material, figure S1a), as well as for 25 913 captured reads of C. alburyorum that re-mapped to the assembled mitochondrial genome (electronic supplementary material, figure S1b). The observed C to T substitutions increase towards the ends of the fragments, which is consistent with the expectation for aDNA fragments [53,54]. The relatively low misincorporation rates for the re-mapped mitochondrial reads can be attributed to the use of UDG during library preparation, which removes the majority of deaminated cytosines. Overall, this result validates the ancient origin of the C. alburyorum mitogenome.
Contamination analysis using fastqscreen revealed multiple potential sources of contamination, in particular, human (electronic supplementary material, figure S1c). Yet, more reads could be uniquely assigned to the Chrysemys genome than to any of the alternative genomes tested. Predicted DNA preservation for a bone sample deposited in a terrestrial environment of the Bahamas indicated a mean fragment length of just 24 bp, and a probability of 0.012 for the survival of an intact 100 bp fragment (electronic supplementary material, figure S1d). However, DNA preservation appears to be substantially better in the C. alburyorum sample; the mean length of recovered mitochondrial fragments is 65 bp and 7.88% of recovered fragments are at least 100 bp long (electronic supplementary material, figure S1e).
(b). Mitochondrial phylogeny of Chelonoidis
The assembly of the C. alburyorum mitogenome comprised 19 929 reads, resulting in an average 85-fold read-depth and included the nearly complete mtDNA gene and tRNA complement, covering 15 328 bp and ranging from 12S to cyt b, but lacking the control region. Assemblies of modern relatives were all of a similar standard (table 1). Read information of sequenced voucher specimens, including European Nucleotide Archive (ENA) accession numbers and sequenced blanks, can be found in the electronic supplementary material, tables S3 and S4.
Table 1.
Assembly of mitochondrial genomes using MITObim: settings and results. k-mer size (= length of bait) was 31 (default). Allowed mismatches per read was set to 8. Cropped contig length is the assembled contig cropped to 5′ and 3′ long-range PCR primers.
| taxon | size of readpool | # reads assembled | # MITObim iterations | average consensus quality (max. = 90) | allowed mismatches | assembled contig length | cropped contig length | average total coverage |
|---|---|---|---|---|---|---|---|---|
| Chelonoidis alburyorum | 100 000 | 19 929 (19.9%) | 203 | 90 | 8 | 15 328 bp | 15 150 bp | 85 |
| Chelonoidis carbonarius | 40 000 | 23 929 (59.8%) | 99 | 90 | 8 | 15 475 bp | 15 163 bp | 146 |
| Chelonoidis chilensis | 5 000 | 4 732 (94.6%) | 73 | 90 | 8 | 15 177 bp | 15 136 bp | 48 |
| Chelonoidis denticulatus | 15 000 | 9 027 (60.2%) | 79 | 90 | 8 | 15 649 bp | 15 146 bp | 78 |
| Chelonoidis vicina | 10 000 | 9 713 (97.1%) | 166 | 90 | 8 | 15 204 bp | 15 151 bp | 59 |
| Geochelone sulcata | 5 000 | 4 643 (92.9%) | 74 | 90 | 8 | 14 244 bp | 14 192 bp | 51 |
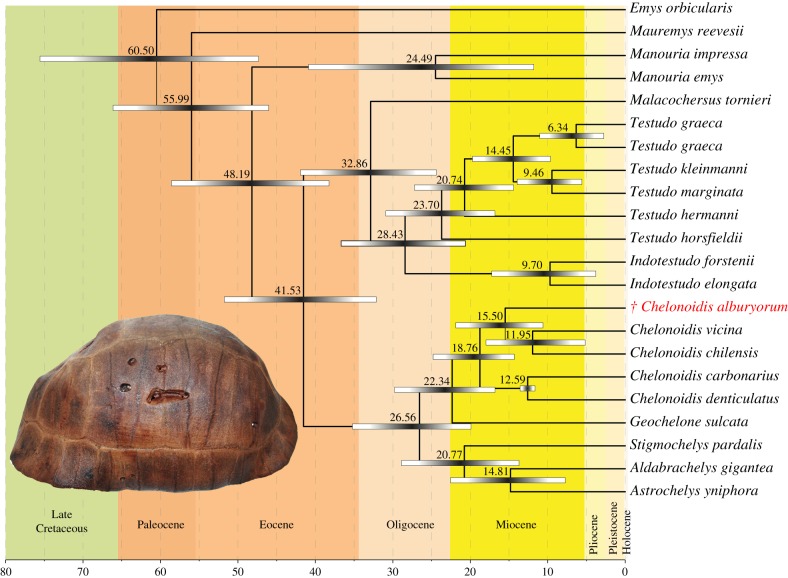
Phylogenetic analyses unambiguously placed C. alburyorum in a clade together with C. chilensis and C. vicina (figure 1; electronic supplementary material, figure S2), with the latter two suggested as weakly supported sister taxa. C. carbonarius and C. denticulatus together constituted the sister clade to the previous three taxa. The relationships of the remaining testudinid species corresponded to expectations from previous papers based on less sequence data [55,56]. According to our molecular clock calculations, C. alburyorum diverged from the last common ancestor of C. chilensis and the Galápagos tortoises (represented by C. vicina) about 15.5 mya, whereas C. chilensis and C. vicina diverged approximately 12 mya, similar to C. carbonarius and C. denticulatus (figure 1).
Figure 1.
Phylogenetic position of Chelonoidis alburyorum and divergence times of land tortoises based on complete or nearly complete mitochondrial genomes. Shown are mean divergence dates and corresponding upper and lower bounds of 95% highest posterior density intervals. Inset: shell of C. alburyorum from holotype specimen.
4. Discussion
(a). Biogeography of Chelonoidis
Despite advanced DNA degradation and high levels of contamination, we successfully recovered a high-quality mitogenome from the extinct tropical tortoise C. alburyorum. Our results both shed new light on the biogeography of Chelonoidis and have wider implications for aDNA research on tropical taxa.
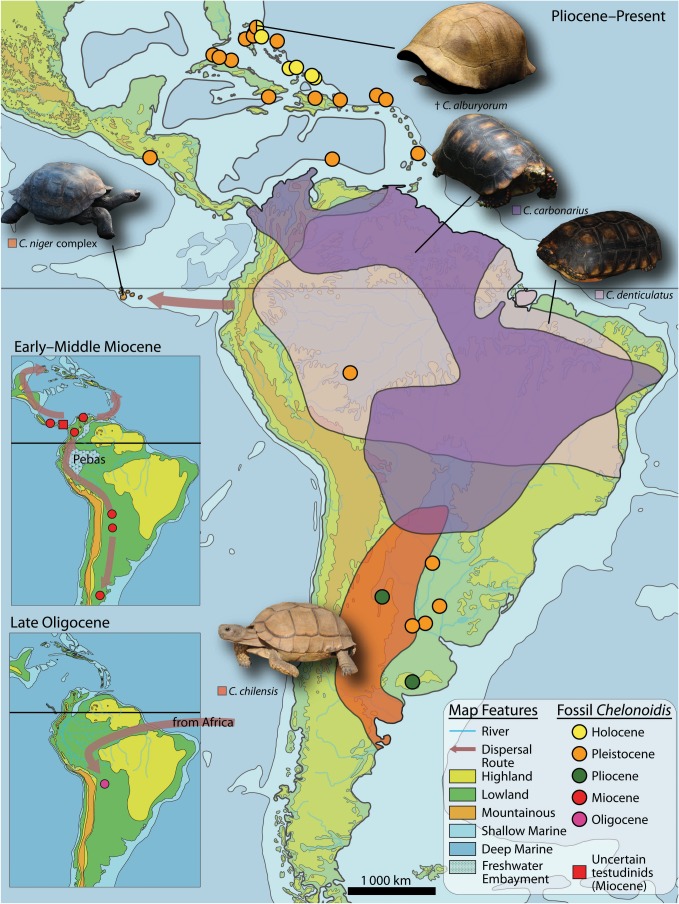
Chelonoidis represents a South American radiation, including the Galápagos and the Caribbean Islands (figure 2; electronic supplementary material, table S5). All Caribbean species are extinct. Chelonoidis is most closely related to African tortoises; fossils of related tortoises are unknown from North America. Thus, overseas dispersal from Africa has been postulated to explain its occurrence in South America [55], as in New World monkeys [57] and rodents [58]. According to our molecular clock calculations, and in agreement with the oldest record of a fossil tortoise in South America, the divergence of Chelonoidis from the African Geochelone sulcata and subsequent dispersal to South America would have occurred distinctly later than in the two other groups (Eocene), around the Oligocene–Miocene transition (figure 1). For the colonization of the Caribbean islands, two transoceanic routes have to be considered: directly from South America or via southern Central America. The originally wide Caribbean distribution of Chelonoidis is indicated by records of extinct species from 10 Bahamian islands as well as from Cuba, Hispaniola, Mona, Navassa, Barbados, Curacao, Grand Turk, Caicos, Anguilla, and Bermuda [19]. The extent to which Caribbean terrestrial ecosystems have been altered by the loss of these ‘ecosystem engineers' is fertile ground for new research in palaeoecology and restoration ecology [59,60].
Figure 2.
Extant and fossil occurrences of Chelonoidis and dispersals through time. For details, see electronic supplementary material, table S1.
With a proposed divergence date of approximately 15.5 mya, this Caribbean island radiation postdates the divergence of South American Chelonoidis from African Geochelone by only approximately 7 mya and predates the divergence of Galápagos and Chaco tortoises by approximately 3.5 mya. Owing to human activities during the mid- to late Holocene, the entire Caribbean tortoise radiation was lost, as was the case for the sloths that once occupied the Greater Antilles [61]. This loss of the Caribbean tortoises is another example of the massive impoverishment of evolutionary diversity that accompanied human colonization of oceanic islands worldwide [15,16]. The extent of this depletion only increases as the insular fossil record continues to grow. Because these eliminated species and lineages would still exist if not for human interference, we should endeavour to incorporate them into studies of ‘modern’ biodiversity, including their genetic diversity. Until aDNA analyses are done on other Caribbean forms of Chelonoidis, we cannot evaluate, for example, how many dispersal events from South America were required to account for the Caribbean radiation of tortoises, or how much of the Holocene diversity of Chelonoidis was lost due to human activity.
(b). Implications for the study of tropical ancient DNA
The recovery of genetic information from tropical and subtropical fossils remains a challenge. A unique property of the C. alburyorum fossil analysed here is its deposition environment: the Sawmill Sink blue hole. It is well known that certain microenvironments can provide conditions that enhance DNA preservation, e.g. cave environments greatly improve the probability of DNA survival relative to the external landscape [2]. Marine environments in general are also known to provide promising potential for DNA preservation, as evidenced by studies of Late Pleistocene remains retrieved from temperate oceans [22,62]. Although the estimated endogenous DNA content and preservation of the C. alburyorum sample is poor, it is nevertheless sufficient for mitogenome sequencing using methods optimized for the retrieval of aDNA. Moreover, preservation in this sample is substantially better than that predicted for a bone sample deposited for the same time in a terrestrial environment of the Bahamas. Although any conclusions based on this single sample are tentative, we propose that the anoxic, thermally buffered marine environment of blue holes and similar preservation contexts may provide conditions that enhance DNA preservation—even in tropical regions, where DNA recovery from ancient samples is often considered to be unachievable. These findings indicate a future direction with high potential for aDNA research in the tropics.
Supplementary Material
Acknowledgements
We would like to thank Christoph Hahn (Kingston-upon-Hull) and Stefanie Hartmann (Potsdam) for bioinformatic support, Brian Kakuk for assistance with collecting the specimen in Sawmill Sink, Hayley Singleton and Oona Takano (University of Florida) for laboratory assistance, Keith Tinker and The National Museum of The Bahamas for continued support of collections and studies, and the Friends of the Environment in Marsh Harbour for local support. Helga Happ (Reptilienzoo Happ, Klagenfurth) kindly provided samples of her Galápagos tortoise ‘Poldi’.
Data accessibility
All DNA sequences have been deposited in the European Nucleotide Archive ENA under accession numbers LT599482-LT599492. DNA sequence assemblies are available through Dryad and can be accessed at http://dx.doi.org/10.5061/dryad.728hn [63].
Authors' contributions
U.F., M.H., A.K.H., and D.W.S. conceived the project. C.K., A.B., and J.L.A.P. designed and carried out the laboratory experiments and coordinated NGS data analysis and mitogenome assembly. C.K. conducted NGS data analysis and mitogenome assembly. C.K. and M.V. conducted sequence alignment, and M.V. calculated phylogenetic and molecular clock analyses. N.A.A. collected samples and facilitated government approvals in The Bahamas. D.W.S. evaluated samples for organic content. R.F. and A.K.H. contributed manuscript parts on tortoise fossils and geological context. U.F., A.B., M.H., and D.W.S. coordinated writing of the manuscript, with all authors participating.
Competing interests
We have no competing interests.
Funding
Field and museum research of D.W.S. was funded by U.S. National Science Foundation grants BCS-1118369 and GSS-1461496.
References
- 1.Smith CI, Chamberlain AT, Riley MS, Stringer C, Collins MJ. 2003. The thermal history of human fossils and the likelihood of successful DNA amplification. J. Hum. Evol. 45, 203–217. ( 10.1016/S0047-2484(03)00106-4) [DOI] [PubMed] [Google Scholar]
- 2.Hofreiter M, Paijmans JLA, Goodchild H, Speller CF, Barlow A, Fortes GG, Thomas JA, Ludwig A, Collins MJ. 2014. The future of ancient DNA: technical advances and conceptual shifts. BioEssays 37, 284–293. ( 10.1002/bies.201400160) [DOI] [PubMed] [Google Scholar]
- 3.Lorenzen ED, et al. 2011. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature 479, 359–364. ( 10.1038/nature10574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutiérrez-García TA, Vázquez-Domínguez E, Arroyo-Cabrales J, Kuch M, Enk J, King C, Poinar HN. 2014. Ancient DNA and the tropics: a rodent's tale. Biol. Lett. 10, 20140224 ( 10.1098/rsbl.2014.0224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Llorente MG, et al. 2015. Ancient Ethiopian genome reveals extensive Eurasian admixture in Eastern Africa. Science 350, 820–822. ( 10.1126/science.aad2879) [DOI] [PubMed] [Google Scholar]
- 6.Brace S, Turvey ST, Weksler M, Hoogland MLP, Barnes I. 2015. Unexpected evolutionary diversity in a recently extinct Caribbean mammal radiation. Proc. R. Soc. B 282, 20142371 ( 10.1098/rspb.2014.2371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brace S, Thomas JA, Dalén L, Burger J, MacPhee RDE, Barnes I, Turvey ST. 2016. Evolutionary history of the Nesophontidae, the last unplaced recent mammal family. Mol. Evol. Biol. 12, 3033–3041. ( 10.1093/molbev/msw186) [DOI] [PubMed] [Google Scholar]
- 8.Mohandesan E, Speller CF, Peters J, Uerpmann H-P, Uerpmann M, De Cupere B, Hofreiter M, Burger PA. 2016. Combined hybridization capture and shotgun sequencing for ancient DNA analysis of extinct wild and domestic dromedary camel. Mol. Ecol. Resour. ( 10.1111/1755-0998.12551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Austin JJ, Arnold EN. 2001. Ancient mitochondrial DNA and morphology elucidate an extinct island radiation of Indian Ocean giant tortoises (Cylindraspis). Proc. R. Soc. Lond. B 268, 2515–2523. ( 10.1098/rspb.2001.1825) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Austin JJ, Arnold EN. 2006. Using ancient and recent DNA to explore relationships of extinct and endangered Leiolopisma skinks (Reptilia: Scincidae) in the Mascarene islands. Mol. Phylogenet. Evol. 39, 503–511. ( 10.1016/j.ympev.2005.12.011) [DOI] [PubMed] [Google Scholar]
- 11.Austin JJ, Arnold EN, Bour R. 2003. Was there a second adaptive radiation of giant tortoises in the Indian Ocean? Using mitochondrial DNA to investigate speciation and biogeography of Aldabrachelys (Reptilia, Testudinidae). Mol. Ecol. 12, 1415–1424. ( 10.1046/j.1365-294X.2003.01842.x) [DOI] [PubMed] [Google Scholar]
- 12.Austin JJ, Arnold EN, Jones CG. 2004. Reconstructing an island radiation using ancient and recent DNA: the extinct and living day geckos (Phelsuma) of the Mascarene islands. Mol. Phylogenet. Evol. 31, 109–122. ( 10.1016/j.ympev.2003.07.011) [DOI] [PubMed] [Google Scholar]
- 13.Sommer RS, Lindqvist C, Persson A, Bringsøe H, Rhodin AGJ, Schneeweiss N, Široký P, Bachmann L, Fritz U. 2009. Unexpected early extinction of the European pond turtle (Emys orbicularis) in Sweden and climatic impact on its Holocene range. Mol. Ecol. 18, 1252–1262. ( 10.1111/j.1365-294X.2009.04096.x) [DOI] [PubMed] [Google Scholar]
- 14.Case TJ, Bolger DT, Richman AD. 1992. Reptilian extinctions: the last ten thousand years. In Conservation biology (eds Fiedler PL, Jain SK), pp. 91–125. New York, NY: Chapman and Hall. [Google Scholar]
- 15.Steadman DW. 2006. Extinction and biogeography of tropical Pacific birds. Chicago, IL: University of Chicago Press. [Google Scholar]
- 16.Rhodin AGJ, et al. 2015. Turtles and tortoises of the world during the rise and global spread of humanity: first checklist and review of extinct Pleistocene and Holocene chelonians. In Conservation biology of freshwater turtles and tortoises: a compilation project of the IUCN/SSC tortoise and freshwater turtle specialist group (eds Rhodin AGJ, Pritchard PCH, Dijk PP, Saumure RA, Buhlmann KA, Iverson JB, Mittermeier RA), pp. 000.e1–000.e66. Lunenburg, MA: Chelonian Research Foundation; (Chelonian Research Monographs No. 5). [Google Scholar]
- 17.Steadman DW, Albury NA, Maillis P, Mead JI, Slapcinsky J, Krysko KL, Singleton HM, Franklin J. 2014. Late-Holocene faunal and landscape change in the Bahamas. Holocene 24, 220–230. ( 10.1177/0959683613516819) [DOI] [Google Scholar]
- 18.Hastings AK, Krigbaum J, Steadman DW, Albury NA. 2014. Domination by reptiles in a terrestrial food web of the Bahamas prior to human occupation. J. Herpetol. 48, 380–388. ( 10.1670/13-091R1) [DOI] [Google Scholar]
- 19.Franz R, Franz SE. 2009. A new fossil land tortoise in the genus Chelonoidis (Testudines: Testudinidae) from the northern Bahamas, with an osteological assessment of other neotropical tortoises. Bull. Florida Mus. Nat. Hist. 49, 1–44. [Google Scholar]
- 20.Steadman DW, et al. 2007. Exceptionally well preserved late Quaternary plant and vertebrate fossils from a blue hole on Abaco, The Bahamas. Proc. Natl Acad. Sci. USA 104, 19 897–19 902. ( 10.1073/pnas.0709572104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohland N, Pollack JL, Nagel D, Beauval C, Airvaux J, Pääbo S, Hofreiter M. 2005. The population history of extant and extinct hyenas. Mol. Biol. Evol. 22, 2435–2443. ( 10.1093/molbev/msi244) [DOI] [PubMed] [Google Scholar]
- 22.Alter SE, et al. 2015. Climate impacts on transocean dispersal and habitat in gray whales from the Pleistocene to 2100. Mol. Ecol. 24, 1510–1522. ( 10.1111/mec.13121) [DOI] [PubMed] [Google Scholar]
- 23.Chatters JC, et al. 2014. Late Pleistocene human skeleton and mtDNA link Paleoamericans and modern Native Americans. Science 344, 750–754. ( 10.1126/science.1252619) [DOI] [PubMed] [Google Scholar]
- 24.Prüfer K, Meyer M. 2015. Comment on ‘Late Pleistocene human skeleton and mtDNA link Paleoamericans and modern Native Americans’. Science 347, 835 ( 10.1126/science.1260617) [DOI] [PubMed] [Google Scholar]
- 25.Fulton TL. 2012. Setting up an ancient DNA laboratory. In Ancient DNA: methods and protocols. Methods in molecular biology, vol. 840 (eds Shapiro B, Hofreiter M), pp. 1–11. Berlin, Germany: Springer. [DOI] [PubMed] [Google Scholar]
- 26.Dabney J, et al. 2013. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl Acad. Sci. USA 110, 15 758–15 763. ( 10.1073/pnas.1314445110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gansauge M-T, Meyer M. 2013. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protoc. 8, 737–748. ( 10.1038/nprot.2013.038) [DOI] [PubMed] [Google Scholar]
- 28.Maricic T, Whitten M, Pääbo S. 2010. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS ONE 5, e14004 ( 10.1371/journal.pone.0014004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horn S. 2012. Target enrichment via DNA hybridization capture. In Ancient DNA: methods and protocols. Methods in molecular biology, vol. 840 (eds Shapiro B, Hofreiter M), pp. 177–188. Berlin, Germany: Springer. [DOI] [PubMed] [Google Scholar]
- 30.Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, pdb.prot5448. ( 10.1101/pdb.prot5448) [DOI] [PubMed] [Google Scholar]
- 31.Fortes GG, Paijmans JLA.2015. Analysis of whole mitogenomes from ancient samples. ( https://arxiv.org/abs/1503.05074)
- 32.St John J. 2013. SeqPrep v1.1. See https://github.com/jstjohn/SeqPrep (accessed 3 October 2016).
- 33.Shaffer HB, et al. 2013. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 14, R28 ( 10.1186/gb-2013-14-3-r28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. ( 10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones D. 2012. fastq-tools. See http://homes.cs.washington.edu/~dcjones/fastq-tools (accessed 3 October 2016).
- 37.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. 2013. mapDamage 2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684. ( 10.1093/bioinformatics/btt193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41, 1–9. ( 10.1093/nar/gkt371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu H, Luo X, Qian J, Pang X, Song J, Qian G, Chen J, Chen S. 2012. FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS ONE 7, e52249 ( 10.1371/journal.pone.0052249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief. Bioinform. 14, 193–202. ( 10.1093/bib/bbs012) [DOI] [PubMed] [Google Scholar]
- 41.Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69, 313–319. ( 10.1016/j.ympev.2012.08.023) [DOI] [PubMed] [Google Scholar]
- 42.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. ( 10.1093/nar/22.22.4673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. ( 10.1093/molbev/mss020) [DOI] [PubMed] [Google Scholar]
- 44.Ronquist F et al. . 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v1.6. See http://beast.bio.ed.ac.uk/tracer 2014 (accessed 30 June 2015).
- 46.Stamatakis A. 2006. RAxML-VI-HPC: Maximum Likelihood based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. ( 10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 47.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. ( 10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Auffenberg W. 1971. A new fossil tortoise, with remarks on the origin of South American Testudinines. Copeia 1971, 106–117. ( 10.2307/1441604) [DOI] [Google Scholar]
- 49.Madden RH, Guerrero J, Kay RF, Flynn JJ, Swisher CC, Walton AH. 1997. The Laventan stage and age. In Vertebrate paleontology in the neotropics. The Miocene Fauna of La Venta, Colombia (eds Kay RF, Madden RH, Cifelli RL, Flynn JJ), pp. 499–519. Washington, D.C: Smithsonian Institution Press. [Google Scholar]
- 50.Suarez C, Forasiepi AM, Goin FJ, Jaramillo C. 2016. Insights into the neotropics prior to the Great American Biotic Interchange: new evidence of mammalian predators from the Miocene of Northern Colombia. J. Vert. Paleontol. 36, e1029581 ( 10.1080/02724634.2015.1029581) [DOI] [Google Scholar]
- 51.Joyce WG, Parham JF, Lyson TR, Warnock RCM, Donoghue PCJ. 2013. A divergence dating analysis of turtles using fossil calibrations: an example of best practices. J. Paleontol. 87, 612–634. ( 10.1666/12-149) [DOI] [Google Scholar]
- 52.Rambaut A. 2014. FigTree 1.4.2. See http://tree.bio.ed.ac.uk/software/figtree (accessed 3 October 2016).
- 53.Briggs AW, et al. 2007. Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl Acad. Sci. USA 104, 14 616–14 621. ( 10.1073/pnas.0704665104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brotherton P, Endicott P, Sanchez JJ, Beaumont M, Barnett R, Austin J, Cooper A. 2007. Novel high-resolution characterization of ancient DNA reveals C > U-type base modification events as the sole cause of post mortem miscoding lesions. Nucleic Acids Res. 35, 5717–5728. ( 10.1093/nar/gkm588) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le M, Raxworthy CJ, McCord WP, Mertz L. 2006. A molecular phylogeny of tortoises (Testudines: Testudinidae) based on mitochondrial and nuclear genes. Mol. Phylogenet. Evol. 40, 517–531. ( 10.1016/j.ympev.2006.03.003) [DOI] [PubMed] [Google Scholar]
- 56.Fritz U, Bininda-Emonds ORP. 2007. When genes meet nomenclature: tortoise phylogeny and the shifting generic concepts of Testudo and Geochelone. Zoology, 110, 298–307. ( 10.1016/j.zool.2007.02.003) [DOI] [PubMed] [Google Scholar]
- 57.Bond M, Tejedor MF, Campbell KE, Chornogubsky L, Novo N, Goin F. 2015. Eocene primates of South America and the African origins of New World monkeys. Nature 520, 538–541. (doi:0.1038/nature14120) [DOI] [PubMed] [Google Scholar]
- 58.Antoine P-O, et al. 2011. Middle Eocene rodents from Peruvian Amazonia reveal the pattern and timing of caviomorph origins and biogeography. Proc. R. Soc. B 279, 1319–1326. ( 10.1098/rspb.2011.1732) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Griffiths CJ, Jones CG, Hansen DM, Puttoo M, Tatayah RV, Müller CB, Harris S. 2010. The use of extant non-indigenous tortoises as a restoration tool to replace extinct ecosystem engineers. Restoration Ecol. 18, 1–7. ( 10.1111/j.1526-100X.2009.00612.x) [DOI] [Google Scholar]
- 60.Hansen DM, Donlan CJ, Griffiths CJ, Campbell KJ. 2010. Ecological history and latent conservation potential: large and giant tortoises as a model for taxon substitutions. Ecography 33, 272–284. ( 10.1111/j.1600-0587.2010.06305.x) [DOI] [Google Scholar]
- 61.Steadman DW, Martin PS, MacPhee RDE, Jull AJT, McDonald HG, Woods CA, Iturralde-Vinent M, Hodgins GWL. 2005. Asynchronous extinction of late Quaternary sloths on continents and islands. Proc. Natl Acad. Sci. USA 102, 11 763–11 768. ( 10.1073/pnas.0502777102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Foote AD, et al. 2013. Ancient DNA reveals that bowhead whale lineages survived Late Pleistocene climate change and habitat shifts. Nat. Commun. 4, 1677 ( 10.1038/ncomms2714) [DOI] [PubMed] [Google Scholar]
- 63.Kehlmaier C, Barlow A, Hastings AK, Vamberger M, Paijmans JLA, Steadman DW, Albury NA, Franz R, Hofreiter M, Fritz U. 2017. Data from: Tropical ancient DNA reveals relationships of the extinct Bahamian giant tortoise Chelonoidis alburyorum. Dryad Digital Repository. ( 10.5061/dryad.728hn) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All DNA sequences have been deposited in the European Nucleotide Archive ENA under accession numbers LT599482-LT599492. DNA sequence assemblies are available through Dryad and can be accessed at http://dx.doi.org/10.5061/dryad.728hn [63].