ABSTRACT
Cellular responses to the graded Sonic Hedgehog (Shh) morphogenic signal are orchestrated by three Gli genes that give rise to both transcription activators and repressors. An essential downstream regulator of the pathway, encoded by the tumor suppressor gene Suppressor of fused (Sufu), plays critical roles in the production, trafficking, and function of Gli proteins, but the mechanism remains controversial. Here, we show that Sufu is upregulated in active Shh responding tissues and accompanies Gli activators translocating into and Gli repressors out of the nucleus. Trafficking of Sufu to the primary cilium, potentiated by Gli activators but not repressors, was found to be coupled to its nuclear import. We have identified a nuclear export signal (NES) motif of Sufu in juxtaposition to the protein kinase A (PKA) and glycogen synthase kinase 3 (GSK3) dual phosphorylation sites and show that Sufu binds the chromatin with both Gli1 and Gli3. Close comparison of neural tube development among individual Ptch1−/−, Sufu−/−, and Ptch1−/−; Sufu−/− double mutant embryos indicates that Sufu is critical for the maximal activation of Shh signaling essential to the specification of the most-ventral neurons. These data define Sufu as a novel class of molecular chaperone required for every aspect of Gli regulation and function.
KEYWORDS: Sonic Hedgehog, Suppressor of Fused, Gli, nucleocytoplasmic trafficking, molecular chaperone
INTRODUCTION
The morphogenic factor Sonic Hedgehog (Shh) specifies spatial pattern and developmental cell fate in a wide array of embryonic tissues during development and organizes stem cell differentiation in the adults (1–3). In the developing neural tube, for example, Shh emanates from the notochord and floor plate (FP) provides positional cues to five classes of ventral neuronal progenitors, which adopt their respective future fate according to the level of Shh signal received (4, 5). Shh is also a strong mitogenic factor for sustaining the proliferation of granule neuron precursors in the neonatal cerebellar anlagen (6, 7). As myriad developmental anomalies and cancer syndromes are associated with genetic lesions in genes that transduce Shh signal, a thorough depiction of the Shh signaling mechanism not only is crucial to the understanding of how tissues and organs are formed but also may pave ways to the development of novel classes of cancer treatment (8–11).
A key feature of Shh signaling is the ability to discern incremental changes in the ligand gradient and mete out appropriate transcriptional output accordingly (12, 13). At the ground state, the pathway is kept inactive by a resistance, nodulation and division domain-containing receptor, Patched1 (Ptch1) (14–16), which occupies the primary cilium, a microtubule-based protrusion present in interface cells (17), and acting through a still poorly characterized mechanism prevents an intrinsically active G-protein coupled receptor, Smoothened (Smo), from being transported into the primary cilium (18–20). When Shh signal appears, binding to the ligand alleviates the inhibition of Ptch1 on Smo, allowing the latter to move into the primary cilium and turn on the downstream pathway (21). Ligand engagement also promotes endocytic turnover of Ptch1 in lysosomes by mobilizing the homology to E6AP carboxyl terminus (HECT) domain-containing E3 ligases Smurf1 and Smurf2, which catalyze its ubiquitination in lipid rafts (22). This system allows Shh to control the degree of Ptch1 inhibition on the pathway activity, of which Ptch1 itself is a target (23).
Downstream from the Ptch1-Smo receptor system, the signaling responses are orchestrated by three Krüppel family transcription factors, Gli1, Gli2, and Gli3, each of which uses a highly conserved zinc finger DNA-binding domain to bind a common cis-acting DNA element in target gene promoters (24–27). Early data from Drosophila showed that the fly Gli-homologous protein encoded by cubitus interruptus (Ci) undergoes a partial proteolysis that converts the full-length 150-kDa protein into a carboxyl-terminally truncated 75-kDa transcriptional repressor, Ci75D, in unstimulated cells (28). Mammalian Gli2 and Gli3 can be processed in a similar fashion into Gli2R and Gli3R repressors, respectively, whereas Gli1 functions purely as an auxiliary activator (29–33). Production of these Gli repressors is governed by a conserved phosphorylation cascade involving protein kinase A (PKA), glycogen synthase kinase 3 (GSK3), and casein kinase I (CK1), which render the nascent Gli2 and Gli3 to be recognized by ubiquitin E3 ligase Slimb/βTRCP and send them for limited degradation in proteasomes (31, 34–37). Activation by Smo blocks the phosphorylation and processing, causing these Gli proteins to be stabilized into the full-length activators. In essence, the expression level of any given Shh target gene is determined by a unique combination of these Gli activators and repressors, dubbed a “Gli code” (38).
Suppressor of Fused (Sufu) is a pan-Gli-binding protein and plays an indispensable role during embryonic development (39). Mouse embryos lacking Sufu closely resemble the Ptch1 null mutants; both die around embryonic day 9.5 (e9.5) with ventralized open neural tubes (40, 41). In the absence of Sufu, Gli1 is upregulated as the result of pathway activation, but Gli2 and Gli3 become unstable and cannot support the generation of truncated repressors (42, 43). In humans, Sufu is encoded by a tumor suppressor gene, mutations of which have been found in Gorlin syndromic cancers, namely, medulloblastoma and basal cell carcinoma (44). A large body of literature has described many aspects of Sufu function in negatively regulating Shh signaling, but its mechanism of action remains controversial. Early studies with cultured mammalian cells and Drosophila indicated that Sufu has a capacity to restrain Ci/Gli in the cytoplasm (45–47). In line with these observations, Sufu was recently reported to dissociate from Gli3 after Gli3 is processed into truncated repressors or stabilized into the full-length activator upon Shh signaling; in either case, the unrestricted Gli3R or Gli3A was proposed to enter the nucleus to regulate target gene expression without the company of Sufu (42, 48). However, in Drosophila salivary glands and wing imaginal discs, ectopically expressed Sufu was shown to enter the nucleus with Ci (49), and mammalian Sufu was also shown to be capable of recruiting the transcriptional corepressor complex through an interaction with SAP18 (50). Sufu is known to form two contact points with Gli proteins (51, 52); recent data indicate that Sufu impedes the nuclear trafficking of Ci by masking a proline-tyrosine nuclear localization signal (PY-NLS) in the N terminus and blocks the recruitment of transcriptional coactivator CBP to the C-terminal binding site (51, 53). Moreover, Sufu was reported to interact with two nuclear proteins, p66β and MycBP (54), strongly suggesting that it possesses crucial nuclear functions.
Using immunohistochemistry (IHC) staining, we determined in this study that the Sufu level is surprisingly elevated in active Shh receiving tissues, and Sufu accompanies both Gli activators and repressors trafficking into the nucleus, where it interacts with the chromatin at Gli-binding sites. We also report that Sufu is essential to the maximal activation of Shh signaling required for specification of the most-ventral neuronal progenitors in the neural tube. These diverse roles indicate that Sufu is required for every aspect of Gli functions, an attribute consistent with a molecular chaperone.
RESULTS
Sufu is required for the active expression and nuclear localization of Gli1.
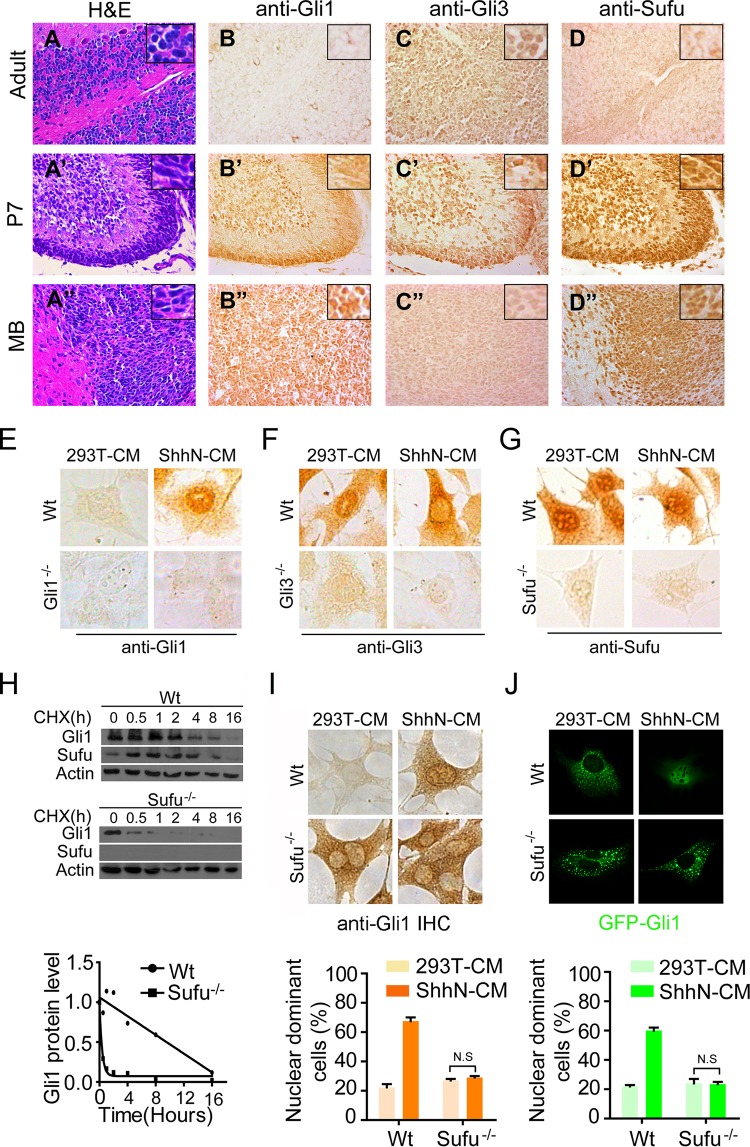
The prevailing view on Sufu in the field regards it as a negative regulator of Shh signaling, acting as a cytoplasmic constraint for the nuclear translocation of Gli transcription factors (42, 48, 55). In the external germinal layer (EGL) of the developing P7 cerebellum (Fig. 1A to 1D′), where active Shh signaling sustains the proliferation of granule neuron precursor cells (GNPCs) (6), we detected a high expression of Gli1 but a low expression of Gli3 by IHC staining (Fig. 1B′ and C′), as expected (7). This differential expression pattern of Gli1 and Gli3 in the EGL is consistent with their roles as a transcriptional activator and a repressor, respectively (27). However, we were surprised to find a very high level of Sufu in P7 EGL (Fig. 1D′), which is counterintuitive to the current consensus view of Sufu as a negative regulator of Shh signaling. We further detected a high level of Sufu expression in spontaneous medulloblastomas (MB) derived from Ptch−/+ mice, where the Shh pathway is reactivated or remains active (Fig. 1A″ to D″). Moreover, the Sufu staining in both normal cerebellar and MB tissues appeared to be enriched in the nucleus (Fig. 1D′ and D″), but to precisely measure the subcellular distribution of Sufu, we performed IHC staining in wild-type mouse embryonic fibroblasts (MEFs). Here, treatment with ShhN-conditioned medium (ShhN-CM) in 67.5% of the cells induced the nuclear enrichment of Gli1, while in 97% of the cells, Gli3 faded into the cytoplasm (Fig. 1E and F); regardless of the treatment, however, Sufu was clearly enriched in the nucleus in 70% of the cells (Fig. 1G). Although the anti-Gli3 used in this study recognizes both full-length and truncated Gli3, the IHC signal should reflect the movement of the truncated repressor, Gli3R, since it is much more prominent than the full-length activator (see Fig. S1A in the supplemental material). The specificity of these IHC staining patterns was verified using mutant MEFs that lack Gli1, Gli3, or Sufu (Fig. 1E to G, lower images). Although no evidence indicates that Shh induces Sufu gene expression, our results imply that Sufu may in fact be positively required for supporting Gli1 functions in addition to its negative role in Shh signaling.
FIG 1.
Sufu is essential for the upregulation and nuclear localization of Gli1. Hematoxylin and eosin (H&E) staining (A to A″) and immunohistochemistry staining (B to B″, C to C″, and D to D″) were carried out on adult (6 months old) and developing (P7) cerebellar and medulloblastoma sections. (E to G) IHC staining in wild-type and mutant MEFs. The proportions of cells showing Shh-induced nuclear enrichment are approximately 67.5% (Gli1 [E]), 3% (Gli3 [F]), and 70% (Sufu [G]), respectively. (H) Western analyses showing the turnover rate of Gli1 in wild-type and Sufu−/− MEFs. (I) IHC staining of Gli1 in wild-type and Sufu−/− MEFs. The cells were treated with control (293T cells) or ShhN-conditioned medium (ShhN-CM) for 24 h before they were fixed in 4% PFA. The graphs were calculated based on two independent experiments (n = 100). (J) Immunofluorescence visualization of GFP-Gli1 in transiently transfected MEFs. The cells were treated and the graphs were calculated as for panel I (n = 50).
Previously, Sufu was shown to be an essential stabilizing factor for full-length Gli2 and Gli3, but both Gli1mRNA transcript and protein were found in abundance even in unstimulated Sufu−/− MEFs (Fig. S2A and B) (42, 43, 56). Despite the high expression, blocking protein synthesis with cycloheximide showed that Gli1 is actually rather unstable without Sufu (Fig. 1H; see also Fig. S1C), and in the absence of Sufu, the Shh-induced nuclear localization of either the endogenous protein assessed by IHC staining (Fig. 1I) or the exogenously expressed green fluorescent protein (GFP)-Gli1 (Fig. 1J) was completely blocked. Thus, these data unified the role of Sufu in stabilizing all Gli activators; the high level of Gli1 in Sufu−/− MEFs can be attributed to a high rate of synthesis due to the absence of the transcriptional repression by the truncated Gli3 or Gli2 repressors. Taken together, the results described thus far suggest that the high level of Sufu in active Shh-receiving tissues may be necessary to stabilize the high level of Gli1 induced by Shh and allow for its nuclear accumulation and function.
Sufu accompanies Gli1 translocating into and Gli3R translocating out of the nucleus upon Shh induction.
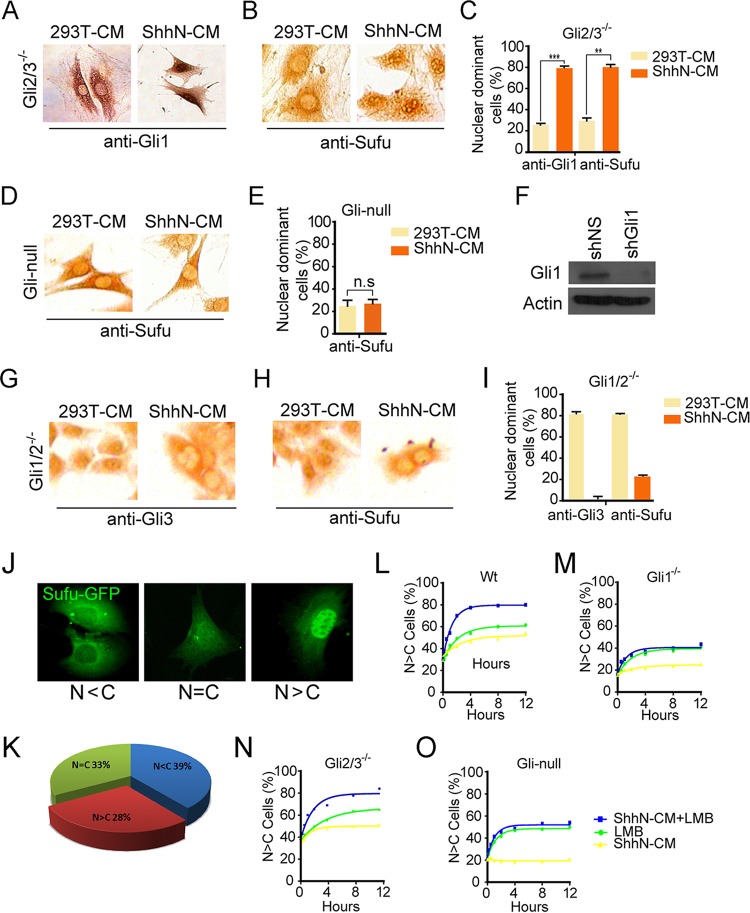
Sufu is known to bind both Gli activators and Gli repressors. So the fact that we detected Shh-induced opposing subcellular movement of the activating Gli1 and repressing Gli3R proteins raised an intriguing possibility that the stationary appearance of Sufu in the nucleus is a “zero sum” effect of Sufu accompanying Gli activators, including Gli1, moving into and truncated repressors out of the nucleus. To test this hypothesis, we examined the nucleocytoplasmic distribution of Sufu in a panel of mutant MEFs lacking each individual Gli or in binary combinations (57). These cells faithfully recapitulated a spectrum of Shh signaling responses, including transcriptional activation of Gli1 and proteolytic processing of Gli3 (see Fig. S1). In support of our view, IHC staining of Gli2−/−; Gli3−/− MEFs clearly showed that ShhN-CM treatment not only induced the nuclear localization of Gli1 (Fig. 2A and C) but also changed the percentage of cells with nuclear Sufu from about 29.7% to 80.5% (Fig. 2B and C). Since Gli2−/−; Gli3−/− MEFs express only Gli1, it is likely that Shh induces Sufu to accompany Gli1 translocating into the nucleus. Indeed, when Gli1 was removed in Gli-null cells derived from Gli2−/−; Gli3−/− MEFs carrying genomically integrated shGli1, ShhN-CM treatment was completely unable to alter the cytoplasmic localization of Sufu (Fig. 2D to F). Knocking down Gli1 expression by a second shGli1 in transiently transfected Gli2−/−; Gli3−/− MEFs also curtailed the ability of ShhN-CM to induce the nuclear translocation of Sufu (see Fig. S3A to C), precluding an off-target effect of Gli1-specific short hairpin RNAs (shRNAs). IHC staining of Gli1−/−; Gli2−/− MEFs, which express only Gli3, also lent weight to the bidirectional movement of Sufu. A total of 79.8% of untreated cells showed Sufu in the nucleus, as they did with Gli3, but when the cells were treated with ShhN-CM, only 21.7% of them retained Sufu in the nucleus, whereas close to none had nuclear Gli3 (Fig. 2G to I). Finally, experiments using chromatin immunoprecipitation (ChIP) to detect formation of Gli1-Sufu and Gli3-Sufu complexes on target gene promoters described below provided further support to the bidirectional movement of Sufu (see Fig. 5B and C). Thus, our data indicate that Sufu chaperones the nucleocytoplasmic trafficking of all forms of Gli proteins.
FIG 2.
Shh induces Sufu-Gli1 moving into and Sufu-Gli3 out of the nucleus. (A and B) Gli2−/−; Gli3−/− double mutant MEFs were treated with control 293T-CM or ShhN-CM and analyzed by IHC for anti-Gli1 (A) or anti-Sufu (B). Quantification for panels A and B was done in duplicate samples and is shown in panel C (n ≥ 35). (D) Gli2−/−; Gli3−/− MEFs carrying a genomically integrated shGli1-expressing unit (Gli-null) were treated with control 293T-CM or ShhN-CM and analyzed by IHC for anti-Sufu. Quantification was done in duplicates and is shown in panel E (n ≥ 38). (E) Quantification of results in panel D. (F) Western analysis of Gli1 expression in Gli1−/−; Gli2−/− (shNS) and Gli-null (shGli1) MEFs showing the knockdown effect of shGli1. (G and H) Anti-Gli3 (G) and anti-Sufu (H) IHC staining in Gli1−/−; Gli2−/− double mutant MEFs. The cells were treated with control 293T-CM or ShhN-CM for 24 h before the analysis. (I) Quantification of results in panels G and H (n ≥ 50). (J and K) Representative immunofluorescence images (J) and subcellular distribution (K) of Sufu-GFP transiently expressed in normal MEFs. (L to O) Kinetics of nuclear accumulation of Sufu-GFP in wild-type (L), Gli1−/− (M), Gli2−/−; Gli3−/− (N), and Gli-null (O) MEFs.
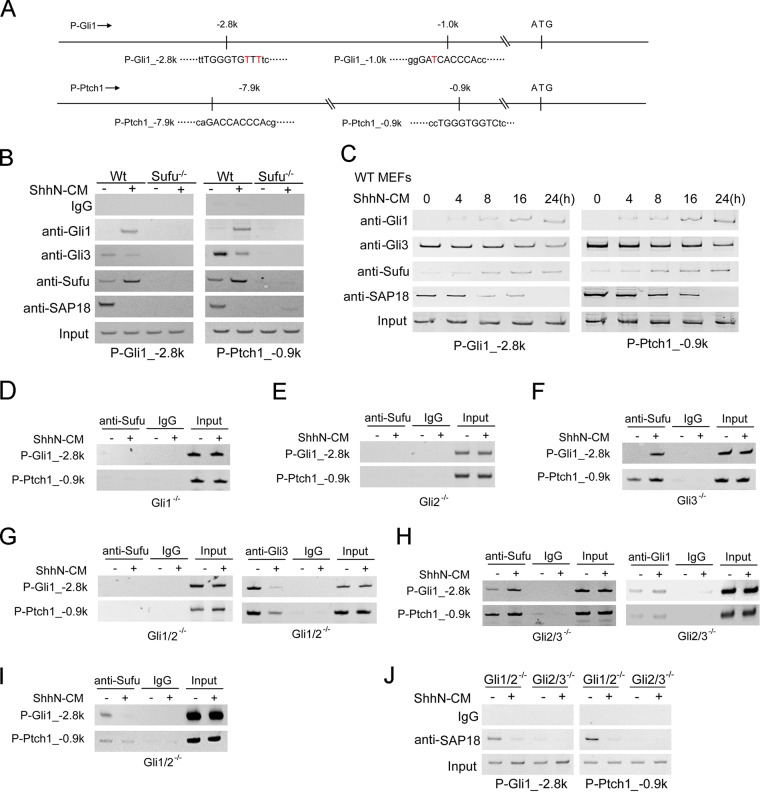
FIG 5.
Shh induces Sufu-Gli1 binding to and Sufu-Gli3R dissociation from the chromatin at target gene promoters. (A) Schematic representation of Gli binding sites on mouse Gli1 and Ptch1 promoters. The coordinates are referenced to the translational start codon. (B) ChIP analyses at a representative site each in the mouse Gli1 and Ptch1 promoter. Normal MEFs were treated with ShhN-CM or the control medium for 24 h before the cells were harvested. (C) Time course of Shh-induced association of Gli1 and Sufu with and dissociation of Gli3 and SAP18 from the Gli1 (P-Gli1_-2.8k) and Ptch1 (P-Ptch1_-0.9k) promoters in normal MEFs. ShhN-CM treatment was for 24 h. (D to H) Anti-Sufu ChIP analyses on the mouse Gli1 and Ptch1 promoters in Gli1−/− (D), Gli2−/− (E), Gli3−/− (F), Gli1−/−; Gli2−/− (G), and Gli2−/−; Gli3−/− (H) MEFs were carried out as described for panel B. Anti-Gli3 and anti-Gli1 ChIP assays in Gli1−/−; Gli2−/− and Gli2−/−; Gli3−/− MEFs are also shown in panels G and H, respectively, for comparison. (I) Anti-Sufu ChIP assay of panel H was rerun with a higher number of PCR amplification cycles to show Gli3-mediated Sufu binding of the promoters with the increased sensitivity of detection. (J) Anti-SAP18 ChIP assays in Gli1−/−; Gli2−/− and Gli2−/−; Gli3−/− MEFs.
Kinetics of Shh-induced nuclear translocation of Sufu.
To kinetically quantify the dependence of Sufu nuclear translocation on various Gli proteins, we generated a Sufu-GFP fusion protein and measured its movement in the Gli mutant MEFs. When expressed in normal MEFs, Sufu-GFP exhibited a mixed pattern of mostly nuclear (N>C), mostly cytoplasmic (N<C), or evenly distributed (N=C) (Fig. 2J and K). About 29% of transfected MEFs had Sufu-GFP enriched in the nucleus in the absence of Shh, but within 8 h of ShhN-CM treatment, the fraction of MEFs with mostly nuclear Sufu-GFP reached to 54% and ShhN-CM showed an additive effect with leptomycin B (LMB), an inhibitor of Crm1-mediated nuclear export (58) (Fig. 2L), lending further weight to the notion that Shh promotes the nuclear translocation of Sufu. In various Gli mutant MEFs, whereas the kinetics of Sufu-GFP translocating autonomously into the nucleus, as shown by LMB induction, remained the same, the percentage of MEFs with mostly nuclear Sufu-GFP induced by Shh appeared to be a function of the level of Gli1 (Fig. 2L to O). Thus, in cells that lack Gli1 (Gli1−/−, Gli1−/−; Gli2−/−, and Gli-null MEFs), little Sufu-GFP was induced into the nucleus by ShhN-CM treatment (Fig. 2M and O; see also Fig. S2D to F); however, in cells that lack Gli3 (Gli3−/− and Gli2−/−; Gli3−/− MEFs), the basal level of MEFs with mostly nuclear Sufu-GFP was already close to 40%, and ShhN-CM treatment induced that percentage even higher (Fig. 2N; see also Fig. S3D to F). These data demonstrate that while Sufu is capable of shuttling into the nucleus on its own, the Shh-induced nuclear localization is dependent on Gli proteins, particularly the most abundant Gli1.
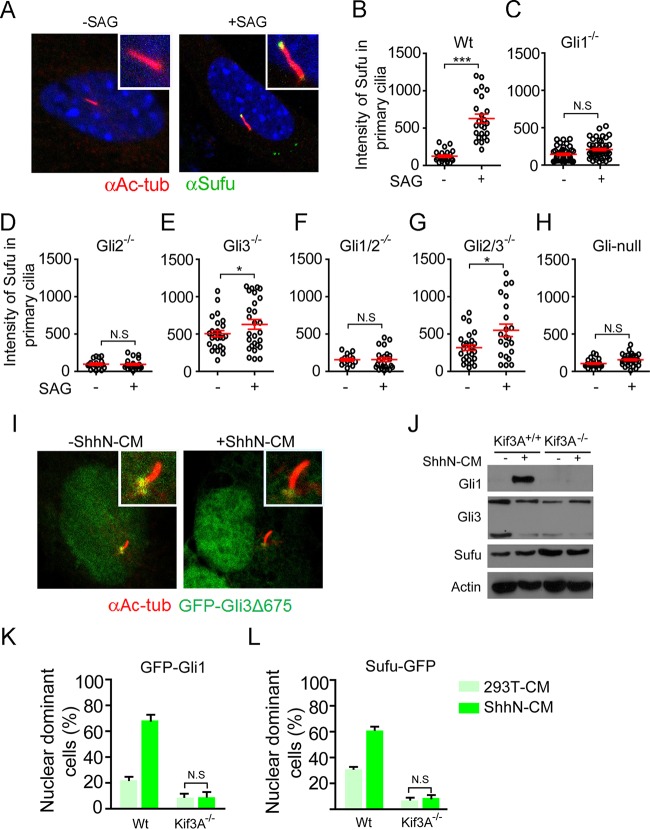
Sufu accompanies Gli activators but not repressors to the ciliary tip upon Shh signaling.
We and others previously showed that Shh induces Sufu-Gli complexes trafficking to the tip of the primary cilium (48, 59), but it was not clear if Sufu accompanies the nascent full-length form, the activator, the truncated repressor, or all of these forms of Gli proteins in this movement. Therefore, we reexamined this issue with the panel of Gli mutant MEFs using Smo agonist SAG to activate the pathway. As with nuclear import, localization of Sufu to the ciliary tip was completely blocked in Gli1-deficient Gli1−/−, Gli2−/−, Gli1−/−; Gli2−/−, and Gli-null MEFs (Fig. 3A to D, F, and H). Since full-length Gli2 activator is the major mediator of Shh-induced transcription, Gli1 was not induced without Gli2 (see Fig. S1A). Conversely, truncated Gli3R is the major repressor of Gli1, so in Gli3-deficient Gli3−/− and Gli2−/−; Gli3−/− MEFs, Gli1 was constitutively expressed at high levels (Fig. S1A) and Sufu was constitutively localized at the ciliary tip (Fig. 3E and G). A parsimonious interpretation of these data would be that Sufu accompanies the activator forms of Gli proteins, particularly the abundant Gli1, to the ciliary tip upon Shh signaling, whereas the repressor forms of Gli proteins, namely, Gli2R and Gli3R, would never move to the ciliary tip with Sufu. To corroborate the latter notion, we expressed a fluorescence-tagged Gli3 repressor, GFP-Gli3R, in normal MEFs, and we found that it was present exclusively at basal bodies without or with SAG treatment (Fig. 3I). In light of our finding that Sufu is required for transporting Gli1 into the nucleus, we speculated that the ciliary trafficking of Gli1, and perhaps other activators as well, would be coupled to their nuclear import. Indeed, in Kif3A−/− MEFs that lack the primary cilium (60) and are incapable of responding to Shh to induce Gli1 expression (Fig. 3J), we found that Shh was unable to induce exogenously expressed GFP-Gli1 into the nucleus as it did in the control MEFs (Fig. 3K), and consequently the nuclear content of Sufu-GFP was also very low in the mutant MEFs (Fig. 3L).
FIG 3.
The nuclear accumulation of Sufu-Gli1 is coupled to its localization at the ciliary tip. (A) Representative fluorescence image of SAG-induced localization of Sufu-GFP to the ciliary tip. (B to H) Quantification of SAG-induced ciliary localization of Sufu-GFP in wild-type (B), Gli1−/− (C), Gli2−/− (D), Gli3−/− (E), Gli1−/−; Gli2−/− (F), Gli2−/−; Gli3−/− (G), and Gli-null (H) MEFs (n ≥ 42). (I) Representative fluorescence images showing that GFP-Gli3Δ673 is localized exclusively at the base of the primary cilium without or with ShhN-CM treatment. (J) Western analyses of Gli1, Gli3, and Sufu in Kif3A−/− and control MEFs. (K and L) Quantification of IHC analysis of ShhN-induced nuclear accumulation of Gli1 (K) and Sufu (L) in Kif3A−/− and control MEFs. N.S, not significant.
Identification of a Crm1-mediated nuclear export signal of Sufu.
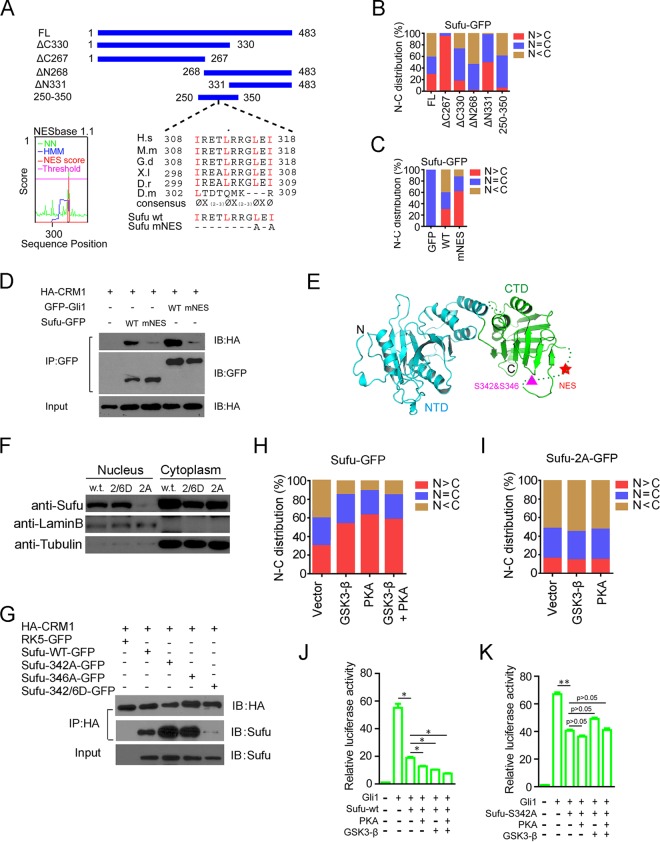
Although Sufu exhibited a strong dependence on Gli proteins for its Shh-induced nuclear movement, there still could be signals present in its primary sequence that regulate the nuclear localization of Sufu because of its ability to shuttle in unbound form (Fig. 2O). We thus conducted deletion mapping to identify such signals by scoring subcellular distribution of GFP-labeled Sufu deletion fragments in normal MEFs. Consistent with a previous report (52), SufuΔC267-GFP was found exclusively in the nucleus, whereas its C-terminal complement, SufuΔN268-GFP, was mostly cytoplasmic (Fig. 4A and B), implying the presence of a C-terminal nuclear export signal (NES). Further deletion analysis indicated that this signal is likely confined to the region between residues 250 and 350 (Fig. 4A and B). Using the online search tool NESbase (http://www.cbs.dtu.dk/databases/NESbase/) (61), we identified a sequence motif in this region that closely matches the NES consensus Φx(2-3)Φx(2-3) ΦxΦ, which is highly conserved in all vertebrate Sufu proteins examined but is not present in Drosophila Sufu (Fig. 4A). No nuclear localization signal (NLS) was found in Sufu using any available online tools. The functional relevance of the NES motif that we identified was confirmed by the nuclear enrichment of the mutant Sufu-mNES, in which L316 and I318 were replaced with alanine (Fig. 4C). As the leucine-rich NES is known to be recognized by Crm1 (62), we carried out coimmunoprecipitation (co-IP) experiments and found that HA-Crm1 could be readily brought down by GFP-Sufu and GFP-Gli1 but not either of the NES mutants Sufu-mNES and Gli1-mNES (Fig. 4D), indicating that Crm1 specifically binds Sufu via the NES sequence motif. These data indicate that Sufu has an intrinsic property to augment the nuclear export of Gli1, rather than strictly blocking the nuclear access of Ci/Gli from the cytoplasm (42, 48, 53).
FIG 4.
Identification of a Crm1-mediated nuclear export signal of Sufu and its regulation at a juxtaposing dual phosphorylation site. (A) Schematic representation of amino acid sequences of full-length and deletion mutant Sufu. Alignments of sequences between residues 308 and 318 (mouse) are shown. The mutant mouse Sufu-mNES contains replacements of L316 and I318 by alanine. NESbase 1.1 search scores are shown on the left. (B and C) The nuclear and cytoplasmic distribution (percentage) of various GFP-Sufu deletion constructs (B) and the NES mutant (C) was determined by transient expression in normal MEFs (n ≥ 57). (D) Co-IP analyses of Sufu-GFP and HA-Crm1 interaction in HEK293 cells. GFP-Gli1 was used as a positive control. (E) Three-dimensional structural model of full-length Sufu showing the position of NES and the dual phosphorylation site in the amorphous loop in the C terminus (61). (F) Western analyses of the nuclear and cytoplasmic fractionated samples from transiently transfected cells. Lamin B and tubulin were used as the nuclear and cytoplasmic markers, respectively. (G) Co-IP analyses of interaction between HA-Crm1 and various phosphorylation site mutants of Sufu-GFP. (H and I) Effects of PKA and GSK3β on nucleocytoplasmic distribution of wild-type (H) and the nonphosphorylatable mutant Sufu342A (I) were determined in normal MEFs (n ≥ 65). (J and K) Shh-responsive reporter luciferase assays for the inhibition of Gli1-mediated transcription by wild-type Sufu (J) and the nonphosphorylatable mutant Sufu342A (K).
PKA- and GSK3-mediated dual phosphorylation of Sufu promotes its nuclear localization.
The recently resolved crystal structure of the full-length Sufu protein revealed two globular domains straddling a β strand of Gli1 (63). The C-terminal domain is interrupted by an amorphous loop covering residues 268 to 345 (Fig. 4E). Since the newly identified NES falls in this loop and is in juxtaposition to S342 and S346, which are the dual phosphorylation sites recognized by GSK3 and PKA, respectively (59), we speculated that the Crm1-mediated nuclear export of Sufu might be subject to phosphorylation control. In support of this possibility, a biochemical fractionation experiment showed that a mutant Sufu with S342 replaced by alanine (S342A; the double mutant S342A/S346A was unstable) was primarily cytoplasmic, whereas another mutant, with S342 and S346 replaced by aspartic acid (S342D/S346D), mimicking phosphorylation, was distributed in both compartments (Fig. 4F). Moreover, the nonphosphorylatable S342A and S346A mutants also bound Crm1 more strongly than wild-type Sufu, and the phosphomimicking S342D/S346D mutant failed to bind Crm1 (Fig. 4G). Coexpressing Sufu-GFP with GSK3β or PKA, or the two enzymes together, increased its nuclear content, but these kinases have no such effect on the nucleocytoplasmic distribution of the S342A mutant fusion protein (Fig. 4H and I). Coexpressing GSK3β or PKA also enhanced the repression of wild-type Sufu on Gli1-mediated transcription but showed no such effect on the S342A mutant (Fig. 4J and K). Thus, these dual phosphorylation sites and their cognate kinases may represent a cross talk mechanism allowing Shh signaling output to be fine-tuned by other pathways (64).
Shh regulates the formation of Sufu-Gli chromatin complexes on Gli-binding sites.
The nuclear presence of Sufu implies that it may directly participate in the Gli-mediated transcriptional control, a possibility raised previously from results showing that Sufu binds DNA as a complex with Gli1 (46, 50). To determine if this occurs in vivo, we performed ChIP experiments to demonstrate that Sufu-Gli complexes are formed on Gli-binding sites in Shh target gene promoters. As shown on several representative sites in Ptch1 and Gli1 promoters (Fig. 5A), activation by Shh induced Gli1binding to the chromatin in normal MEFs, while it concomitantly reduced Gli3 binding (Fig. 5B; see also Fig. S4A and B in the supplemental material). Strikingly, Shh also induced the binding of Sufu to these Gli-binding sites on the chromatin but simultaneously reduced that of SAP18 (Fig. 5B), a Sufu binding protein and a component of the transcriptional core-repressor complex (50). Binding of Sufu and SAP18 to the chromatin was observed at other Gli-binding sites of Gli1 and Ptch1 promoters as well (Fig. S4A). The specificity of the anti-Sufu ChIP analysis was demonstrated in Sufu−/− cells, in which no binding was detected; however, in the absence of Sufu, Gli1 was also not detected on the chromatin (Fig. 5B), despite its abundance (Fig. S1). This is likely because in Sufu−/− cells the unstable Gli1 is not able to traffic into the nucleus (Fig. 1J) or bind the chromatin. Consistent with the requirement of Sufu for its recruitment to Gli proteins (50), SAP18 was also not detected on the chromatin in Sufu−/− cells (Fig. 5B). Kinetic measurements showed that association of the Gli1-Sufu complex with and dissociation of the Gli3-Sufu-SAP18 complex from the chromatin occur concurrently and begin immediately following ShhN treatment (Fig. 5C; see also Fig. S4C), implying a dynamic mode of transcriptional regulation of target genes by Shh signaling. To rigorously demonstrate that Shh induces dissociation of the Gli3-Sufu-SAP18 and association of the Gli1-Sufu complexes on the chromatin, we conducted the ChIP assay in Gli3-only (Gli1−/−; Gli2−/−) and Gli1-only (Gli2−/−; Gli3−/−) MEFs, respectively, and treated the cells with LMB to block nuclear export. The results indicated that following ShhN treatment, the levels of both Gli1 and Sufu on the chromatin increased, and they were further increased by blocking nuclear export (Fig. S5). In contrast, the levels of Gli3 and SAP18 showed an opposite outcome in responding to Shh signaling, in that they both decreased, but LMB treatment reversed that decrease (Fig. S5). In light of the Shh-regulated nuclear translocation, these results suggested that in resting cells a Gli3R-Sufu-SAP18 complex occupies the chromatin surrounding target gene promoters for actively repressing transcription, whereas in activated cells that complex is replaced by Gli1-Sufu for activation. To address which Gli proteins support Sufu binding to the chromatin, we repeated the ChIP experiment in the panel of Gli mutant MEFs, and the results showed that in Gli1−/− and Gli1−/−; Gli2−/− cells that lack Gli1, Shh was unable to induce Sufu binding to the chromatin (Fig. 5D and G), indicating that Sufu-Gli1 is the major transcriptional activation complex. Since full-length Gli2 is the major activator required for Shh induction of Gli1, we also did not detect Sufu on the chromatin in Gli2−/− cells (Fig. 5E), whereas chromatin-bound Sufu was detected even prior to Shh induction in Gli3−/− and Gli2−/−; Gli3−/− cells that constitutively express high level of Gli1 (Fig. 5F and H; see also Fig. S1A). In contrast, in Gli1−/−; Gli2−/− cells that express only Gli3, ShhN-CM treatment induced the dissociation of Gli3R repressor from the chromatin (Fig. 5G), and with an increased cycle number of the PCR, we detected constitutive presence of Sufu on the chromatin, which was abolished by the ShhN-CM treatment (Fig. 5H). Once again, SAP18 was found constitutively on the chromatin in the Gli3-only Gli1−/−; Gli2−/− cells but not the Gli1-only Gli2−/−; Gli3−/− cells, and Shh induced its dissociation (Fig. 5J). These data indicate that Sufu accompanies both activator Gli1 and repressor Gli3R to bind the chromatin, but SAP18 is present only in the repressor complex with Sufu-Gli3R.
Sufu is required for the maximal Shh signaling output that specifies FP cells in the neural tube.
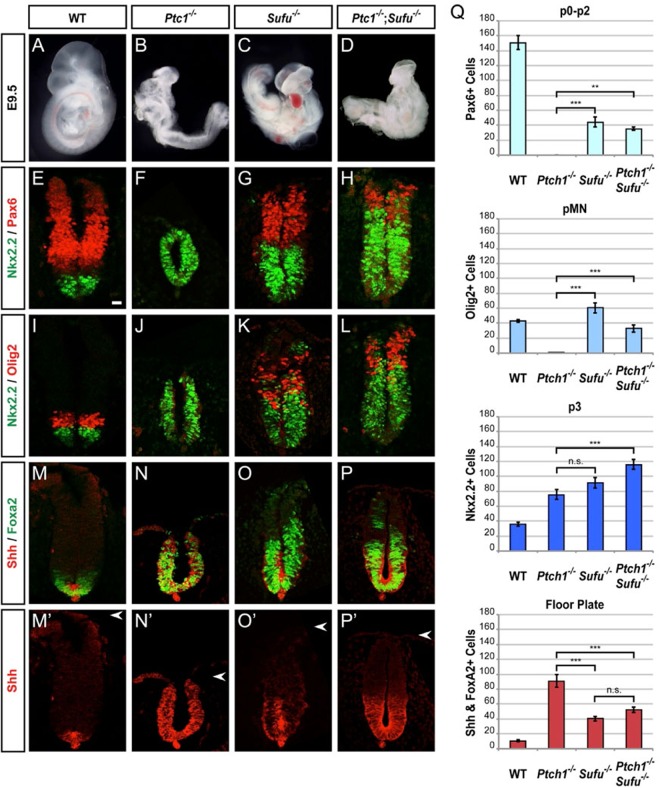
To validate the positive role of Sufu in Shh signaling and to elucidate its physiological significance, we examined the neural tube development in Ptch1−/− and Sufu−/− individual mutant or Ptch1−/−; Sufu−/− double mutant embryos. A strong piece of implicating evidence for Sufu as a negative regulator of Shh signaling is the close resemblance of Sufu−/− embryos to Ptch1−/− embryos, which at e9.5 showed complete ventral expansion as the result of maximal Shh pathway activation (40, 41). In comparison with wild-type embryos, the highly ventralized spinal cords of Ptch1 mutants exhibited a 2-fold increase in Nkx2.2+ cells and a complete loss of Pax6+ or Olig2+ cells (Fig. 6E and F, I and J, and Q). In contrast, Pax6+ and Olig2+ cells remained specified in Sufu−/− mutants, despite severe ventralization of cell fates, including a 2.5-fold increase in Nkx2.2+ cells, a 70% reduction of Pax6+ cells, and the dorsal shift of pMN and other ventral domains (Fig. 6G, K, and Q). More revealingly, Ptch1−/−; Sufu−/− double mutants showed gross morphology comparable to that of Sufu−/− single mutants (Fig. 6C and D) and exhibited a severe but incomplete ventralization phenotype with a 3-fold increase in the number of Nkx2.2+ cells and a dorsal shift of the Pax6+ and Olig2+ domains (Fig. 6H, L, and Q). Since efficient Gli repressor formation is similarly abrogated in Ptch1−/−, Sufu−/−, and Ptch1−/−; Sufu−/− mutants, partial ventralization and thus submaximal activation of the Shh signaling pathway in Sufu−/− and Ptch1−/−; Sufu−/− mutants but not Ptch1−/− mutants are consistent with an essential requirement for Sufu to elicit robust Gli activator function. To address the role of Sufu in Gli activator function directly, we examined floor plate (FP) development, which is a Gli activator-dependent process that requires prompt initiation and sustained robust pathway activation (65). Ptch1−/− mutants exhibited an 8-fold increase in the induction of Shh+ Foxa2+ FP cells, and their specification extended to the dorsal limit of the spinal cord (Fig. 6M, N, M′, N′, and Q). In contrast, Sufu−/− mutants showed a less substantial (4-fold) increase in FP specification (Fig. 6O, O′, and Q). Importantly, Ptch1−/−; Sufu−/− mutants exhibited levels of FP induction similar to those of Sufu−/− mutants (Fig. 6P, P′, and Q). Thus, despite the primary role of Sufu−/− as a negative regulator that prevents ectopic expression of FP cells (66), the lack of complete ventralization in Ptch1−/−; Sufu−/− mutants confirms the positive role for Sufu in Shh signaling. In light of the requirement for stabilizing all forms of full-length Gli proteins (Fig. 1H; Fig. S3D and E) (42, 43, 56) and the Shh-induced nuclear localization of Gli1 (Fig. 1I and J), our results indicate that Sufu is not merely a less potent negative regulator in comparison to Ptch1, but rather, it has a unifying role in regulating all three Gli activators. Our analyses also indicate that both the negative repression and the optimal Gli activation function of Sufu are indispensable for attaining proper floor plate development.
FIG 6.
Sufu is essential to promote maximal activation of the Hh pathway for complete spinal cord ventralization in Ptch1 mutants. (A to D) e9.5 embryos contrast the severe morphological defects of Ptch1−/− mutants with Sufu−/− mutants, which are similar to Ptch1−/−; Sufu−/− double mutants. (E to H) Nkx2.2 and Pax6 immunofluorescence labeling shows reduced spinal cord ventralization in Sufu−/− and Ptch1−/−; Sufu−/− mutants compared with Ptch1−/− mutants. (I to L) Nkx2.2 and Olig2 expression shows that the ventralization of Ptch1−/−; Sufu−/− mutants is similar to that of Sufu−/− mutants, which is reduced in comparison with that of Ptch1−/− mutants. (M to P′) Shh and Foxa2 expression shows complete dorsal expansion of ectopic FP induction in Ptch1−/− mutants in contrast to moderate ectopic FP induction in Sufu−/− mutants. Ptch1−/−; Sufu−/− mutants demonstrate a Sufu requirement for complete ventralization. Scale bar, 25 μm. (Q) Graphs indicate the number of Pax6HIGH (p0 to p2 and some pDI), Olig2+ pMN, Nkx2.2+ p3, and Shh+ Foxa2+ FP cells, represented as the means ± SEMs (n ≥ 4). **, P < 0.01; ***, P < 0.001. n.s., not significant.
DISCUSSION
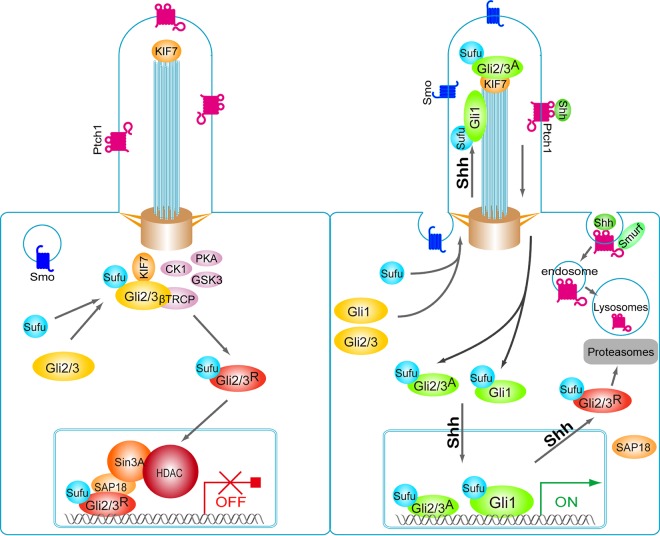
A major downstream regulator of Shh signaling, Sufu plays pivotal roles in the production of Gli repressors and restraining the activities of Gli activators, but current literature differs on the precise mechanism of how these crucial functions are executed. Using IHC staining and by taking advantages of a panel of various Gli mutant cells, we showed that Sufu accompanies Gli activators and repressors trafficking in opposite directions between the nucleus and the cytoplasm in response to Shh, and it binds the chromatin with both Gli activators and repressors. In light of the previously demonstrated stabilizing effect on nascent Gli2 and Gli3 (42, 43, 56), we think our data well support a model in which Sufu functions as a novel molecular chaperone that accompanies every step of Gli regulation and function (Fig. 7). This classification is also compatible with the positive roles of Sufu in stabilizing Gli1 and accompanying its nuclear translocation for the maximal pathway activation as revealed in the Ptch1−/−; Sufu−/− double mutant neural tube.
FIG 7.
Model depicting roles of Sufu in chaperoning Gli functions during Shh signaling. In the absence of Shh little Gli1 is made, whereas Sufu associates, stabilizes, and accompanies nascent Gli2 and Gli3 to the basal body of the primary cilium, where the latter two are phosphorylated by PKA/GSK3/CK1 kinases and processed into the truncated repressors. The resultant Sufu-GliR complexes traverse unrestrictedly to the nucleus, forming repressive transcriptional complexes on the chromatin. Here, Sufu also actively repress transcription via its interaction with SAP18 and the corepressor complex. In the presence of Shh, formation of truncated Gli2 and Gli3 repressors is blocked, and Sufu-GliR complexes fall off the chromatin and are exported out of the nucleus either as dissociated components or together for turnover, resulting in the induction of Gli1 and other pathway target genes. Sufu accompanies nascent Gli1 and full-length Gli2 and Gli3 traversing through the primary cilium, where they are modified by Smo-dependent events and become competent to be imported into the nucleus, where Sufu-GliA complexes bind the chromatin and further activate target gene expression. Note that in this model, Shh promotes the movement of Sufu-GliA into and Sufu-GliR complexes out of the nucleus concurrently, but the nuclear import of Sufu-GliA complexes is coupled to its trafficking through the primary cilium.
Molecular chaperones are proteins that interact with and aid in the folding or assembly of another protein without being part of its final structure (67). Classic molecular chaperones, such as heat shock and oxidative stress proteins, are induced under stress conditions to aid protein folding, aggregate dissociation, protein transport, and degradation (67). However, certain inherently unstable proteins require a lifetime company of a molecular chaperone to maintain their proper structure (68). Since three-dimensional (3D) structural data for full-length Gli2 or Gli3 are not available, we can only speculate if these two large proteins require constant structural maintenance, but since Sufu interacts with Gli proteins at two contact points (51, 52), it is conceivable that binding to Sufu would lock Gli in a conformation optimized to their functions. The newly synthesized Gli2 and Gli3 and their Drosophila counterpart, Ci, are recognized, respectively, by the related speckle-type POZ protein SPOP and the MATH and BTB domain-containing protein HIB, which target them to degradation in the ubiquitin-proteasome system (43, 69). The Sufu binding sites on Ci/Gli2/Gli3 were found to overlap those of HIB/SPOP, such that the binding could competitively protect these Gli proteins from proteasomal degradation (43). Sufu also likely competes with Numb for the binding of Gli1, whose stability is controlled by the Numb-mediated proteasomal degradation (70). Since these protective roles are conferred to the nascent Gli proteins, the function of Sufu in a way is akin to that of those orthodoxy chaperones guiding the folding of newly synthesized proteins as they emerge from the endoplasmic reticulum (ER) (67). The chaperone model stipulates that Sufu assumes a “licensing” role for Gli functions such that Sufu must be supplied in sufficient quantity in active Shh-responding tissues to accommodate the elevated level of Gli1. This notion is strongly supported by our observation that both Gli1 and Sufu are upregulated in EGL of the developing cerebellum and in medulloblastomas (Fig. 1A to D″). Furthermore, we also observed that removal of both Sufu alleles prevented MB formation in a Ptch-null or Smo-M2-overexpressing background that are known to develop MB spontaneously (unpublished results). Two types of mice were generated in these experiments: (i) GFAP-cre; Ptch1fl/fl; Sufufl/fl and (ii) Rosa26-SmoM2; Sufufl/fl; GFAP-cre. In both cases, deletion of Sufu suppressed MB tumorigenesis. These observations strongly support our view that Sufu has a positive role in supporting Gli1 function.
When ectopically expressed in cultured cells, Sufu exhibits a widely reported activity to restrain Ci/Gli from moving into the nucleus; however, paradoxically both Drosophila and mammalian Sufu were also identified in the nucleus (45, 46, 49). This has been a key contentious point in interpreting how Sufu exactly exerts its control over Gli activities, which is exacerbated by the scarcity of data on the subcellular distribution of endogenous Sufu. By reducing the complexity of multiple Gli proteins imposed on Sufu localization, we demonstrated that rather than serving as a link to an unknown molecular anchor in the cytoplasm, Sufu accompanies Gli activators moving into and Gli repressors moving out of the nucleus concomitantly in response to Shh signaling. Thus, the apparent inertia in Sufu movement to ligand induction is well accounted for by the underlying movements in opposite directions across the nuclear envelope. Although Sufu is capable of shuttling on its own, we only identified a NES in the unstructured loop in the C-terminal globular domain, and the Shh-induced movement appeared to be mediated solely by its association with Gli proteins. This observation is consistent with a recent finding that Sufu blocks the nuclear import of Gli2 and Gli3 by masking a transportin/Kapβ2-mediated PY-NLS signal that overlaps with the Sufu binding site in the N-terminal region (53). It is conceivable that signaling by Shh may cause certain conformation changes in Gli which would expose this NLS and set forth the motion into the nucleus. In considering this model, however, a caution should be noted in that Gli proteins contain another bipartite NLS in their C-terminal domain as well, and Sufu also interacts with Gli through a C-terminal sequence (51, 52). This configuration of dual binding and dual NLS regulation may play a critical role in setting up Shh control over the nuclear movement of Gli proteins.
Results from our ChIP and nuclear transport experiments indicated that Sufu is induced by Shh to accompany Gli1 to Gli-binding sites on the chromatin but is exported out of the nucleus with or dissociated from Gli3R, which preoccupies the chromatin sites at the ground signaling state. At the physiological level, the capacity to stabilize Gli activators and guide their nuclear import, particularly Gli1, underpins the positive role of Sufu in guaranteeing the maximal activation of the Shh pathway. However, the literature is laden with reports showing Sufu as a negative component of the pathway both in vitro and in vivo. There are at least three documented mechanisms that could account for the negative activity of Sufu; these include abilities to facilitate Gli repressor production (42, 43), drive the nuclear export of Gli1 (46, 47, 51, 53), and recruit transcription corepressors via SAP18 or other negative regulators, such as MycBP (50, 54). Under overexpressed culture conditions, the capacity to drive nuclear export of Gli1 becomes dominant, thus revealing Sufu as a potent transcription repressor; however, in the absence of Sufu, Gli1 is unable to drive the specification of the most-ventral neurons, which require the highest Shh signaling input, thus revealing the positive role of Sufu. These myriad roles argue that Sufu should not be simply classified either as a negative or a positive regulator in the Shh pathway; rather, a molecular chaperone would be much more befitting its close association with every aspect of Gli function.
MATERIALS AND METHODS
Animals and phenotypic analysis.
All experimental procedures were performed with approval from the Hospital for Sick Children Animal Care Committee and the ethics committee for animal use of Nanjing Medical University. Ptch+/− and Sufu+/− mutant mice were used to generate Ptch; Sufu double mutant embryos, which are genotyped as described previously (66). Embryos were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) at 4°C overnight, and paraffin sections were used for immunofluorescence staining with mouse anti-Nkx2.2 and anti-Foxa2 (Developmental Studies Hybridoma Bank), rabbit anti-Olig2 (Chemicon), rabbit anti-Pax6 (Covance), and rabbit anti-Shh (Santa Cruz) antibodies. Immunofluorescence images were acquired with a Zeiss LSM510 META laser scanning confocal microscope and subjected to quantification and statistical analysis. Spontaneous medulloblastomas were dissected from Ptch+/− mice housed in the animal facility of Nanjing Medical University, fixed in 4% paraformaldehyde in PBS at 4°C overnight, and paraffin embedded for sectioning and IHC staining.
Plasmids, cell lines, and antibodies.
GFP- or epitope-tagged versions and deletion mutants of Crm1, Gli1, and Sufu were generated by PCR and subcloned into the pRK5 vector. The primers for PCR amplification were as follows: Sufu-1-Forward, 5′-CCATCGATATGGCGGAGCTGCGGCCTA; Sufu-267-Reverse, 5′-TGCGGTCGACGTACATGCTAACATAGTCCA; Sufu-330-Reverse, 5′-GACGTCGACAGGGTTGATTGGTGGAAGGA; Sufu-268-Forward, 5′-CCATCGATATGGGTGTCAGTGCCAAGTGTG; Sufu-331-Forward, 5′-CCATCGATATGCAGCGGCAGAATGGCCTCGC; Sufu-250-Forward, 5′-CCCATCGATATGCACCTGCAAGAGAGAGTTG; Sufu-350-Reverse, 5′-GCGTCGACGTCACTTTCCAGGCTGTCT; and Sufu-484-Reverse, 5′-TGCGGTCGACGTGTAGCGGACTGTCGAACA. The NES mutants of Sufu and Gli1 were generated using the QuikChange site-directed mutagenesis kit (Stratagene), and their sequences have been verified. The Gli-null stable cell line was generated by transfecting shGli1 into Gli2−/−; Gli/3−/− mutant MEFs using Lipofectamine Plus reagent (Invitrogen) and selection under 2 μg/ml of puromycin for 2 weeks. Resistant cells were pooled for subsequent experiments. The sequence for mouse Gli1 shRNA is 5′-CCACAAGTCAATAGCTATA. Mutant Gli1−/−, Gli2−/−, Gli3−/−, Gli1−/−; Gli2−/−, and Gli2−/−; Gli3−/− MEFs were generous gifts from the Wade Bushman laboratory. Various antibodies were purchased and used according to the vendors' suggestions: anti-Gli1 (Cell Signaling Technology), anti-Gli3 (R&D), and anti-Sufu (Santa Cruz Biotechnology) were used for Western and ChIP analyses, and anti-Sufu (Epitomics) was used for IHC staining.
Immunoprecipitation.
Transfected HEK293T cells were lysed in IP buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 1× Roche complete protease inhibitor cocktail), and the lysates were clarified by centrifugation for 15 min at 14,000 × g. The protein concentration of each cell lysate sample was determined by bicinchoninic acid (BCA) assay. Immunoprecipitation was carried out with anti-GFP (Sigma) coupled to protein G-agarose beads (Millipore, USA), and the isolated proteins were subjected to 8% SDS-PAGE followed by Western analyses.
Protein turnover assay.
To measure protein turnover of endogenous Gli1, normal and Sufu−/− MEFs were starved in low-serum medium (0.5% fetal bovine serum [FBS]) and induced with ShhN-CM to increase Gli1 expression. After 24 h, the cells were treated with cycloheximide (20 μm; Sigma) to block protein synthesis. At each time point, the cells were lysed in RIPA buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1× Roche complete protease inhibitor cocktail) for Western analysis.
Immunofluorescence staining and determining subcellular localization of Sufu and Gli1 in MEFs.
Approximately 5 × 104 cells per well were seeded in three wells of a Lab-Tek chambered slide for each treatment condition described. The cells were starved in Dulbecco modified Eagle medium (DMEM) containing 0.5% FBS for 24 h before addition of conditioned medium or compounds as indicated. At each time point, MEFs were fixed with 4% paraformaldehyde (PFA) at 4°C for 10 min. Fluorescence-labeled proteins were visualized with a Leica DMI 3000b microscope. A scoring matrix with N>C, N=C, and N<C was established as shown in Fig. 2J. The percentage of cells with nuclear enriched (N>C) Gli1 or Sufu was calculated by randomly counting over 50 cells in each of the three triplicated chambers and plotted against time.
Confocal microscopy.
Confocal images of the primary cilium were acquired on a Carl Zeiss LSM710 microscope. Quantification of the fluorescence intensity of Sufu in primary cilia was carried out using Image-Pro as described previously (59). The primary antibodies used were mouse anti-acetylated tubulin (1:2,000; Sigma) and goat anti-Sufu (1:200; Santa Cruz Biotechnology). The secondary antibodies used were donkey anti-mouse antibody–Alexa Fluor 594 (1:200) and donkey anti-goat antibody–Alexa Fluor 488 (1:200), both of which were purchased from Invitrogen.
ChIP assay.
Chromatin was cross-linked with 1% formaldehyde. Cells were incubated in chromatin immunoprecipitation (ChIP) lysis buffer (150 mM NaCl, 25 mM Tris [pH 7.5], 1% Triton X-100, 0.1% SDS, 0.5% deoxycholate, 1× Roche complete protease inhibitor cocktail). The reaction was stopped by the addition of 125 mM glycine. DNA was fragmented into ∼200-bp to ∼500-bp pieces using a Sonics VCX130 sonicator. Aliquots of lysates containing 400 μg of protein were used for each immunoprecipitation reaction with anti-Gli1 (CST), anti-Gli3 (R&D), anti-Sufu, and anti-Sap18 (Santa Cruz Biotechnology). Precipitated genomic DNA was amplified by PCR with primers as follows: for P-Gli1_-2.8k, 5′-TATGGGGTTGGGAGAGTTTG (forward) and 5′-AAAGAGACCTGGGACAGACAC (reverse); for P-Ptch1_-0.9k, 5′-GGGTTGCCTACCTGGGTGGTCT (forward) and 5′-AACGCGATTGGCTCTTGGAG (reverse).
Immunohistochemistry.
e9.5 developing mouse cerebellum, adult cerebellum, and spontaneous medulloblastoma from Ptch+/− mice were embedded in paraffin and cut into 5-μm-thick sections. After mounting on glass slides, the sections were dewaxed, hydrated, and subsequently incubated for 10 min in 3% hydrogen peroxide to block endogenous peroxidase. Primary antibodies, rabbit anti-Gli1 antibody (Cell Signaling Technology), goat anti-Gli3 (R&D), and rabbit anti-Sufu (Epitomics) were incubated at 4°C for overnight, and secondary antibodies were incubated for 30 min. Streptavidin-peroxidase conjugate (3′-3′ diaminobenzidine) was used in the detection procedure. For determining nucleocytoplasmic distribution of Suf and Gli proteins, various MEFs were seeded on glass coverslips at 5 × 105 per well in 6-well plates. The cells were starved in DMEM containing 0.5% FBS for 24 h before the treatment. The cells were then fixed with 4% PFA for IHC staining. The percentage of cells with nuclear enriched Sufu or Gli proteins was calculated based on over 35 cells in three nonoverlapping random fields. The statistical analyses were performed using Student's t test.
RNA isolation, RT, and real-time PCR.
Total RNAs were isolated from cultured cells using the RNAiso reagent (TaKaRa, Shiga, Japan), and reverse transcription (RT) was carried out using the PrimeScript RT reagent kit (TaKaRa). Standard RT-PCR primers used were mouse Ptch1 (5′-GAAGCCACAGAAAACCCTGTC and 5′-GCCGCAAGCCTTCTCTAGG), mouse Gli1 (5′-TTCGTGTGCCATTGGGGAGG and 5′-CTTGGGCTCCACTGTGGAGA), mSufu (5′-CTCCAGGTTACCGCTATCGTC and 5′-CACTTGGTCCGTCTGTTCCTG), and Hprt (5′-TATGGACAGGACTGAAAGAC and 5′-TAATCCAGCAGGTCAGCAAA). Real-time PCR was carried out using the FastStart SYBR green master mix (Roche) on a 7500 real-time PCR system (Applied Biosystems) with primers for mouse Gli1 (5′-GCTTGGATGAAGGACCTTGTG and 5′-GCTGATCCAGCCTAAGGTTCTC), mouse Ptch1 (5′-GAAGCCACAGAAAACCCTGTC and 5′-GCCGCAAGCCTTCTCTAGG), and mouse Hprt (5′-TATGGACAGGACTGAAAGAC and 5′-TAATCCAGCAGGTCAGCAAA). Experiments were repeated at least three times, and samples were analyzed in triplicates.
Supplementary Material
ACKNOWLEDGMENTS
We thank Wade Bushman for the generous gift of Gli mutant cells.
This work was supported by grants from the Chinese National Science foundation (81272238 and 81261120386) and the National Basic Research Program of China (973 Program, 2012CB945003, and 2009CB918403) to S.Y.C., NSFC grant (81572720) to S.Y., and the Canadian Cancer Society Research Institute (700320) to C.-C.H.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00421-16.
REFERENCES
- 1.Jiang J, Hui CC. 2008. Hedgehog signaling in development and cancer. Dev Cell 15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrova R, Joyner AL. 2014. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development 141:3445–3457. doi: 10.1242/dev.083691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan KE, Chiang C. 2012. Hedgehog secretion and signal transduction in vertebrates. J Biol Chem 287:17905–17913. doi: 10.1074/jbc.R112.356006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choudhry Z, Rikani AA, Choudhry AM, Tariq S, Zakaria F, Asghar MW, Sarfraz MK, Haider K, Shafiq AA, Mobassarah NJ. 2014. Sonic hedgehog signalling pathway: a complex network. Ann Neurosci 21:28–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ribes V, Briscoe J. 2009. Establishing and interpreting graded Sonic Hedgehog signaling during vertebrate neural tube patterning: the role of negative feedback. Cold Spring Harb Perspect Biol 1:a002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wechsler-Reya RJ, Scott MP. 1999. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron 22:103–114. doi: 10.1016/S0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 7.Dahmane N, Ruiz i Altaba A. 1999. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 126:3089–3100. [DOI] [PubMed] [Google Scholar]
- 8.Amakye D, Jagani Z, Dorsch M. 2013. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat Med 19:1410–1422. doi: 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- 9.Scales SJ, de Sauvage FJ. 2009. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci 30:303–312. doi: 10.1016/j.tips.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Yao E, Chuang PT. 2015. Hedgehog signaling: from basic research to clinical applications. J Formos Med Assoc 114:569–576. doi: 10.1016/j.jfma.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Aberger F, Ruiz IAA. 2014. Context-dependent signal integration by the GLI code: the oncogenic load, pathways, modifiers and implications for cancer therapy. Semin Cell Dev Biol 33:93–104. doi: 10.1016/j.semcdb.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balaskas N, Ribeiro A, Panovska J, Dessaud E, Sasai N, Page KM, Briscoe J, Ribes V. 2012. Gene regulatory logic for reading the Sonic Hedgehog signaling gradient in the vertebrate neural tube. Cell 148:273–284. doi: 10.1016/j.cell.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stecca B, Ruiz IAA. 2010. Context-dependent regulation of the GLI code in cancer by HEDGEHOG and non-HEDGEHOG signals. J Mol Cell Biol 2:84–95. doi: 10.1093/jmcb/mjp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein EH Jr, Scott MP. 1996. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 15.Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, Noll M, Hooper JE, de Sauvage F, Rosenthal A. 1996. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 16.Taipale J, Cooper MK, Maiti T, Beachy PA. 2002. Patched acts catalytically to suppress the activity of Smoothened. Nature 418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- 17.Goetz SC, Anderson KV. 2010. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet 11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. 2003. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 19.Rohatgi R, Scott MP. 2007b. Patching the gaps in Hedgehog signalling. Nat Cell Biol 9:1005–1009. [DOI] [PubMed] [Google Scholar]
- 20.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. 2005. Vertebrate Smoothened functions at the primary cilium. Nature 437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 21.Rohatgi R, Milenkovic L, Scott MP. 2007a. Patched1 regulates hedgehog signaling at the primary cilium. Science 317:372–376. [DOI] [PubMed] [Google Scholar]
- 22.Yue S, Tang LY, Tang Y, Shen QH, Ding J, Chen Y, Zhang Z, Yu TT, Zhang YE, Cheng SY. 2014. Requirement of Smurf-mediated endocytosis of Patched1 in sonic hedgehog signal reception. eLife 3:e02555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP. 1996. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev 10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- 24.Vokes SA, Ji H, Wong WH, McMahon AP. 2008. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev 22:2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vokes SA, Ji H, McCuine S, Tenzen T, Giles S, Zhong S, Longabaugh WJ, Davidson EH, Wong WH, McMahon AP. 2007. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development 134:1977–1989. doi: 10.1242/dev.001966. [DOI] [PubMed] [Google Scholar]
- 26.Kinzler KW, Vogelstein B. 1990. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol Cell Biol 10:634–642. doi: 10.1128/MCB.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falkenstein KN, Vokes SA. 2014. Transcriptional regulation of graded Hedgehog signaling. Semin Cell Dev Biol 33:73–80. doi: 10.1016/j.semcdb.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aza-Blanc P, Ramirez-Weber FA, Laget MP, Schwartz C, Kornberg TB. 1997. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89:1043–1053. doi: 10.1016/S0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 29.Ding Q, Motoyama J, Gasca S, Mo R, Sasaki H, Rossant J, Hui CC. 1998. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development 125:2533–2543. [DOI] [PubMed] [Google Scholar]
- 30.Hui CC, Joyner AL. 1993. A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat Genet 3:241–246. doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]
- 31.Pan Y, Wang C, Wang B. 2009. Phosphorylation of Gli2 by protein kinase A is required for Gli2 processing and degradation and the Sonic Hedgehog-regulated mouse development. Dev Biol 326:177–189. doi: 10.1016/j.ydbio.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, Joyner AL. 2000. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127:1593–1605. [DOI] [PubMed] [Google Scholar]
- 33.Wang B, Fallon JF, Beachy PA. 2000. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100:423–434. doi: 10.1016/S0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 34.Jiang J, Struhl G. 1998. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature 391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- 35.Price MA, Kalderon D. 2002. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by glycogen synthase kinase 3 and casein kinase 1. Cell 108:823–835. doi: 10.1016/S0092-8674(02)00664-5. [DOI] [PubMed] [Google Scholar]
- 36.Tempé D, Casas M, Karaz S, Blanchet-Tournier MF, Concordet JP. 2006. Multisite protein kinase A and glycogen synthase kinase 3beta phosphorylation leads to Gli3 ubiquitination by SCFbetaTrCP. Mol Cell Biol 26:4316–4326. doi: 10.1128/MCB.02183-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang B, Li Y. 2006. Evidence for the direct involvement of βTrCP in Gli3 protein processing. Proc Natl Acad Sci U S A 103:33–38. doi: 10.1073/pnas.0509927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruiz i Altaba A, Nguyen V, Palma V. 2003. The emergent design of the neural tube: prepattern, SHH morphogen and GLI code. Curr Opin Genet Dev 13:513–521. doi: 10.1016/j.gde.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 39.Cheng SY, Yue S. 2008. Role and regulation of human tumor suppressor SUFU in Hedgehog signaling. Adv Cancer Res 101:29–43. doi: 10.1016/S0065-230X(08)00402-8. [DOI] [PubMed] [Google Scholar]
- 40.Svärd J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergstrom A, Ericson J, Toftgard R, Teglund S. 2006. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell 10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 41.Cooper AF, Yu KP, Brueckner M, Brailey LL, Johnson L, McGrath JM, Bale AE. 2005. Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development 132:4407–4417. doi: 10.1242/dev.02021. [DOI] [PubMed] [Google Scholar]
- 42.Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. 2010. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev 24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen MH, Wilson CW, Li YJ, Law KK, Lu CS, Gacayan R, Zhang X, Hui CC, Chuang PT. 2009. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev 23:1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor MD, Liu L, Raffel C, Hui CC, Mainprize TG, Zhang X, Agatep R, Chiappa S, Gao L, Lowrance A, Hao A, Goldstein AM, Stavrou T, Scherer SW, Dura WT, Wainwright B, Squire JA, Rutka JT, Hogg D. 2002. Mutations in SUFU predispose to medulloblastoma. Nat Genet 31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 45.Ding Q, Fukami S, Meng X, Nishizaki Y, Zhang X, Sasaki H, Dlugosz A, Nakafuku M, Hui C. 1999. Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr Biol 9:1119–1122. doi: 10.1016/S0960-9822(99)80482-5. [DOI] [PubMed] [Google Scholar]
- 46.Kogerman P, Grimm T, Kogerman L, Krause D, Unden AB, Sandstedt B, Toftgard R, Zaphiropoulos PG. 1999. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat Cell Biol 1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- 47.Méthot N, Basler K. 2000. Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development 127:4001–4010. [DOI] [PubMed] [Google Scholar]
- 48.Tukachinsky H, Lopez LV, Salic A. 2010. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol 191:415–428. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sisson BE, Ziegenhorn SL, Holmgren RA. 2006. Regulation of Ci and Su(fu) nuclear import in Drosophila. Dev Biol 294:258–270. doi: 10.1016/j.ydbio.2006.02.050. [DOI] [PubMed] [Google Scholar]
- 50.Cheng SY, Bishop JM. 2002. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc Natl Acad Sci U S A 99:5442–5447. doi: 10.1073/pnas.082096999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han Y, Shi Q, Jiang J. 2015. Multisite interaction with Sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc Natl Acad Sci U S A 112:6383–6388. doi: 10.1073/pnas.1421628112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merchant M, Vajdos FF, Ultsch M, Maun HR, Wendt U, Cannon J, Desmarais W, Lazarus RA, de Vos AM, de Sauvage FJ. 2004. Suppressor of fused regulates Gli activity through a dual binding mechanism. Mol Cell Biol 24:8627–8641. doi: 10.1128/MCB.24.19.8627-8641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi Q, Han Y, Jiang J. 2014. Suppressor of fused impedes Ci/Gli nuclear import by opposing Trn/Kapbeta2 in Hedgehog signaling. J Cell Sci 127:1092–1103. doi: 10.1242/jcs.142828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin C, Yao E, Wang K, Nozawa Y, Shimizu H, Johnson JR, Chen JN, Krogan NJ, Chuang PT. 2014. Regulation of Sufu activity by p66beta and Mycbp provides new insight into vertebrate Hedgehog signaling. Genes Dev 28:2547–2563. doi: 10.1101/gad.249425.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nozawa YI, Lin C, Chuang PT. 2013. Hedgehog signaling from the primary cilium to the nucleus: an emerging picture of ciliary localization, trafficking and transduction. Curr Opin Genet Dev 23:429–437. doi: 10.1016/j.gde.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang C, Pan Y, Wang B. 2010. Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development 137:2001–2009. doi: 10.1242/dev.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lipinski RJ, Bijlsma MF, Gipp JJ, Podhaizer DJ, Bushman W. 2008. Establishment and characterization of immortalized Gli-null mouse embryonic fibroblast cell lines. BMC Cell Biol 9:49. doi: 10.1186/1471-2121-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res 242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 59.Chen Y, Yue S, Xie L, Pu XH, Jin T, Cheng SY. 2011. Dual phosphorylation of suppressor of fused (Sufu) by PKA and GSK3beta regulates its stability and localization in the primary cilium. J Biol Chem 286:13502–13511. doi: 10.1074/jbc.M110.217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin F, Hiesberger T, Cordes K, Sinclair AM, Goldstein LS, Somlo S, Igarashi P. 2003. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc Natl Acad Sci U S A 100:5286–5291. doi: 10.1073/pnas.0836980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S. 2003. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res 31:393–396. doi: 10.1093/nar/gkg101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fornerod M, Ohno M, Yoshida M, Mattaj IW. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051–1060. doi: 10.1016/S0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Fu L, Qi X, Zhang Z, Xia Y, Jia J, Jiang J, Zhao Y, Wu G. 2013. Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci. Nat Commun 4:2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hsu SH, Zhang X, Cheng S, Wunder JS, Hui CC, Alman BA. 2012. Suppressor of fused (Sufu) mediates the effect of parathyroid hormone-like hormone (Pthlh) on chondrocyte differentiation in the growth plate. J Biol Chem 287:36222–36228. doi: 10.1074/jbc.M112.382275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ribes V, Balaskas N, Sasai N, Cruz C, Dessaud E, Cayuso J, Tozer S, Yang LL, Novitch B, Marti E, Briscoe J. 2010. Distinct Sonic Hedgehog signaling dynamics specify floor plate and ventral neuronal progenitors in the vertebrate neural tube. Genes Dev 24:1186–1200. doi: 10.1101/gad.559910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Law KK, Makino S, Mo R, Zhang X, Puviindran V, Hui CC. 2012. Antagonistic and cooperative actions of Kif7 and Sufu define graded intracellular Gli activities in Hedgehog signaling. PLoS One 7:e50193. doi: 10.1371/journal.pone.0050193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. 2013. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 68.Dunker AK, Silman I, Uversky VN, Sussman JL. 2008. Function and structure of inherently disordered proteins. Curr Opin Struct Biol 18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. 2006. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell 10:719–729. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 70.Di Marcotullio L, Ferretti E, Greco A, De Smaele E, Po A, Sico MA, Alimandi M, Giannini G, Maroder M, Screpanti I, Gulino A. 2006. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol 8:1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.