Abstract
Pro bone morphogenetic protein-4 (BMP-4) is initially cleaved at a consensus furin motif adjacent to the mature ligand domain (the S1 site), and this allows for subsequent cleavage at an upstream motif (the S2 site). Previous studies have shown that S2 cleavage regulates the activity and signaling range of mature BMP-4, but the mechanism by which this occurs is unknown. Here, we show that the pro- and mature domains of BMP-4 remain noncovalently associated after S1 cleavage, generating a complex that is targeted for rapid degradation. Degradation requires lysosomal and proteosomal function and is enhanced by interaction with heparin sulfate proteoglycans. Subsequent cleavage at the S2 site liberates mature BMP-4 from the prodomain, thereby stabilizing the protein. We also show that cleavage at the S2, but not the S1 site, is enhanced at reduced pH, consistent with the possibility that the two cleavages occur in distinct subcellular compartments. Based on these results, we propose a model for how cleavage at the upstream site regulates the activity and signaling range of mature BMP-4 after it has been released from the prodomain.
INTRODUCTION
Bone morphogenetic protein-4 (BMP-4) is a signaling molecule that acts as a morphogen to influence cell fate in a concentration-dependent manner. BMP-4 was originally identified as a protein that is capable of inducing ectopic bone formation, but more recent studies have shown that it plays many different roles during embryonic development and in adults (Hogan, 1996).
BMP-4 function is essential for normal embryogenesis as illustrated by the fact that mice homozygous for a null allele of BMP-4 form little or no mesoderm and die near the time of gastrulation (Winnier et al., 1995). BMP-4 heterozygous mutant mice are viable but display a variety of birth defects, including reduced numbers of primordial germ cells, polydactyly, and kidney, eye, and craniofacial abnormalities (Dunn et al., 1997; Lawson et al., 1999; Miyazaki et al., 2000; Chang et al., 2001). These data indicate that control of BMP-4 gene dosage is essential for normal embryonic patterning.
Excess BMP-4 activity also leads to birth defects. Mice mutant for the BMP antagonists gremlin, noggin, and/or chordin show early lethality and/or defects in the spinal cord, forebrain, somites, skeleton, and kidney (Brunet et al., 1998; McMahon et al., 1998; Gong et al., 1999; Khokha et al., 2003). In humans, mutations in the noggin gene are responsible for multiple synostoses syndrome, a genetic disease characterized by fusion of the joints (Gong et al., 1999), and abnormally high levels of BMP-4 protein are a key feature of fibrodysplasia ossificans progressiva, a crippling hereditary disorder in which ectopic bone forms throughout the body (Kaplan and Shore, 1998).
The requirement for strict regulation of BMP-4 dosage is met by controlling BMP-4 activity at multiple levels. At the extracellular level, BMP-4 is regulated by binding proteins, such as chordin and noggin, that block activation of cell surface receptors, and by the protease Tolloid, which cleaves chordin to liberate active BMP-4 (Nakayama et al., 2000). BMP-4 also binds to cell surface heparin sulfate proteoglycans (HSPGs), and these interactions can promote or restrict activity (Selleck, 2001; Ohkawara et al., 2002). At the intracellular level, BMP signaling is negatively regulated in responding cells by Smad6 and Smad7, which block transmission of signals from the membrane to the nucleus, and by the Smurf family of ubiquitin-protein ligases that target BMP receptors and other components of the intracellular signal transduction cascade for proteosomal or lysosomal degradation (Shi and Massague, 2003).
The bioactivity of BMP-4 also is regulated at the level of proteolytic activation. BMP-4 is synthesized as an inactive precursor that is cleaved after the multibasic motif RSKR to yield the active, carboxyl-terminal mature protein dimer (Aono et al., 1995). Specific members of the proprotein convertase (PC) family of endoproteases cleave proBMP-4 (Cui et al., 1998; Constam and Robertson, 1999). In mammals, seven members of this family have been characterized, and these exhibit overlapping but distinct substrate specificities. The well characterized PC furin activates proproteins at the carboxy-terminal side of the preferred consensus sequence -RXR/KR-, but it also can cleave after the minimal sequence -RXXR- (Thomas, 2002). Two other family members, PACE4 and PC6, recognize these same consensus motifs (Rockwell et al., 2002).
We have previously shown that proBMP-4 is sequentially cleaved by furin at two sites and that this ordered proteolysis regulates the activity and signaling range of mature BMP-4 (Cui et al., 2001). The initial cleavage event takes place at an optimal furin motif (-RSKR-) at a site (S1) adjacent to the mature ligand, and this is followed by cleavage at a minimal furin motif (-RISR-) at an upstream site (S2) within the prodomain (Figure 1A). Our previous studies in Xenopus embryos have shown that the first cleavage releases mature BMP-4, whereas the second cleavage serves a regulatory function. Specifically, ectopically expressed proBMP-4 carrying a point mutation that renders the S2 site noncleavable generates a ligand that shows less activity, signals over a shorter range, and accumulates at lower levels than does BMP-4 cleaved from native precursor (Cui et al., 2001). Analysis of mice harboring this same point mutation demonstrates that cleavage at the S2 site is essential for normal embryonic development (Goldman, Hackenmiller, and Christian, unpublished data). Given that S1 cleavage of native and mutant precursors liberates an identical ligand from the prodomain, it is unclear how subsequent cleavage of the prodomain can alter the activity of this ligand.
Figure 1.
Failure to cleave proBMP-4 at the S2 site targets mature BMP-4 for degradation. (A) Schematic illustration of myc- and HA-epitope–tagged wild-type and cleavage mutant forms of proBMP-4. (B–D) RNA encoding wild-type or cleavage mutant proBMP-4 was injected into oocytes together with [35S]Met/Cys, and cleavage products were analyzed by SDS-PAGE. The position of precursor-, S1-, or S2-cleaved prodomain and mature BMP-4 is illustrated schematically to the right of each gel. (B) Oocyte lysates were immunoprecipitated with a myc-specific antibody, and duplicate samples were treated with or without Endo H or PNGase F before SDS-PAGE under reducing or nonreducing conditions as indicated. Bands corresponding to Endo H-sensitive (asterisks) and Endo H-resistant (arrowheads) mature BMP-4 or Endo H-resistant, PNGase F-sensitive proBMP-4 dimers (arrows) are indicated. (C and D) Oocytes were pulse labeled for 3 h and then transferred to media containing cold methionine and cysteine. At increasing time intervals, lysates and conditioned media were subjected to immunoprecipitation by using antibodies specific for the HA- (C) or myc (D)-epitope, and cleavage products were analyzed by SDS-PAGE under reducing (C) or nonreducing (D) conditions.
In the present study, we examine the molecular mechanism by which cleavages within the prodomain regulate the activity of mature BMP-4. Our results show that differential use of the S2 site regulates BMP-4 activity by directing intracellular trafficking of the cleaved ligand to either degradatory or secretory/recycling pathways. In addition, our results suggest possible mechanisms by which tissue-specific cleavage at the S2 site might be regulated.
MATERIALS AND METHODS
cDNA Constructs
The FLAG epitope tag in the prodomain of wild-type and mutant forms of proBMP-4FLAG-Myc (Cui et al., 2001) was replaced with a hemagglutinin (HA) epitope tag by using polymerase chain reaction (PCR). Mutation of the P6 histidine at the S2 site, and deletion of the three-amino acid HSPG binding motif in the mature domain of BMP-4 was accomplished using the PCR-based splicing by overlap extension technique (Horton et al., 1990). Regions of cDNAs generated by PCR were sequenced.
Oocyte Isolation and Analysis of Proteins
Ovaries were surgically removed from mature female frogs, and oocytes were dissociated with 0.2 U/ml liberase blendzyme 3 (Roche Diagnostics, Indianapolis, IN) in OR-2. Stage VI oocytes were cultured overnight at 18°C in ND-96 (96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 5 mM HEPES, pH 7.5) with 1.5% horse serum and 50 μg/ml gentamicin. Oocytes were injected with in vitro synthesized RNAs (Moon and Christian, 1989) together with 700 nCi of [35S]Met/Cys. For pulse-chase analysis, 5 mM Met/Cys was added to the culture media to initiate the chase. After further culture, 6–10 oocytes were harvested and proteins immunoprecipitated from clarified lysates (Xiong et al., 1997) or culture media by incubation overnight at 4°C with antibodies specific for HA (12CA5) or myc (9E10) epitope tags and protein A-Sepharose 4B beads. Beads were washed, boiled in 0.5% SDS with or without 1% β-mercaptoethanol, and immunoprecipitated proteins were treated with or without endoglycosidase H (Endo H) or peptide N-glycosidase F (PNGase F). Radiolabeled proteins were separated by electrophoresis on 11.5% SDS-PAGE, fixed in 7.5% acetic acid/15% methanol, enhanced using Amplify (Amersham Biosciences, Piscataway, NJ), and visualized by autoradiography.
Inhibition of Lysosomal or Proteosomal Function
Ooyctes were coinjected with 5 ng of RNA and 700 nCi of [35S]Met/Cys, cultured for 16 h, washed, and incubated for 3 h in the absence or presence of chloroquine. For proteosomal inhibition, oocytes were coinjected with 2 ng of RNA and 700 nCi of [35S]Met/Cys, labeled for 3 h, transferred to media containing 5 mM cold methionine/cysteine for 12 h, and then incubated in the absence or presence of 10 μM epoximicin or10 μM lactacystin for 4 h. Mature BMP-4 was immunoprecipitated from the culture media by using myc-specific antibodies as described above.
Coimmunoprecipitation and Western Blot Analysis
Oocytes were injected with 5 ng of synthetic RNA and cultured overnight before resuspending in 30 μl of homogenization buffer (0.25 mM sucrose, 50 mM Tris, pH 7.5, 50 mMkAc, 5 mMMgU2, ImMDTT) (Xiong et al., 1997) with 10 mM phenylmethylsulfonyl fluoride and lysing in 600 μl of NETT (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.2% Triton X-100) for 1 h on ice. Proteins were precipitated from one-third of the culture media by using 10% tricholoracetic acid (TCA), washed once with acetone, once with ethanol, and resuspended in NETT. BMP-4 was immunoprecipitated from clarified lysates or TCA-precipitated media by overnight incubation with myc-conjugated beads (Eimon and Harland, 2002). Conjugates were washed three times in NETT and twice in phosphate-buffered saline (PBS). Proteins were deglycosylated with PNGase F, separated by SDS-PAGE, and transferred onto polyvinylidene difluoride membrane, which was probed with HA- or myc-specific antibodies in 5% milk/Tris-buffered saline-Tween 20 (10 mM Tris, pH 8.0, 150 mM NaCl, 0.1% Tween 20). Immunoreactive proteins were detected using chemiluminescence.
In Vitro Digestion with Furin
Radiolabeled precursor proteins were generated by coinjecting oocytes with 5 ng of RNA encoding wild-type or mutant proBMP-4, 700 nCi of [35S]Met/Cys, and 0.5 ng of RNA encoding the furin inhibitor α1-PDX (Anderson et al., 1993). Precursor proteins were immunoprecipitated from oocyte lysates 16 h postinjection by using myc-specific antibodies, and 5000 cpm of each precursor was resuspended in furin digestion buffer (Anderson et al., 1997) adjusted to the appropriate pH with HEPES. Then, 120 nM recombinant furin (Jean et al., 1998) was added to start the reaction. Aliquots were removed at various time intervals and added directly to denaturation buffer (0.5% SDS, 1% β-mercaptoethanol), boiled, and deglycosylated with PNGase F before electrophoresis.
To determine relative rates of cleavage, [35S]Met/Cys-labeled proBMP-4 was synthesized in rabbit reticulocyte lysates and immunoprecipitated using myc-specific antibodies. Protein concentration was determined from incorporated cpm by using an input of 0.91 μM labeled methionine and assuming 5 μM methionine in the reticulocyte lysates. ProBMP-4 (0.8–20 nM) was incubated with 2.2 nM recombinant furin at pH 7.0, 6.5, and 6.0, and aliquots removed at various intervals and analyzed by SDS-PAGE and autoradiography. Cleavage products were visualized using an Amersham Biosciences PhosphorImager, and band intensities were quantified using the IP lab gel program (LifeScience Software, Longlake, MN). Cleavage is ordered and therefore the reaction proceeds as follows:
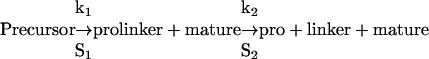 |
where the linker is the 35-amino acid fragment between the S1 and S2 sites and pro is the N-terminal prodomain fragment lacking the linker. The following assumptions are made for burst rate kinetics: 1) each step is irreversible; 2) k1 > k2; 3) initially, [prolinker] = [mature] = [i - precursor], where i is the initial concentration at t0; 4) at later times [mature] = [prolinker] + [pro]; and 5) [pro] = [linker]. The concentration of S1 and S2 cleaved prodomain was calculated using the following equation:
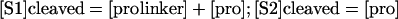 |
[S1] cleaved = [prolinker] + [pro]; [S2] cleaved = [pro]
V0(S1) and V0(S2) were obtained from Michaelis-Menten plots of [S1]/min and [S2]/min, respectively. Data was plotted and analyzed using Kaleidograph (Synergy Software, Reading, PA) to obtain relative rates.
RESULTS
Inability to Cleave proBMP-4 at the S2 Site Targets Mature Ligand for Degradation
Previous studies have shown that failure to cleave proBMP-4 at the S2 site leads to lower steady-state levels of mature BMP-4 in Xenopus embryos (Cui et al., 2001). The reduced accumulation of mature BMP-4 could result from misfolding of the precursor in the endoplasmic reticulum (ER) and subsequent degradation before cleavage, inefficient cleavage of the precursor after it exits the ER, or targeted degradation of mature BMP-4 after cleavage at the S1 site. To distinguish between these possibilities, we compared synthesis, processing, and degradation of epitope-tagged wild-type proBMP-4 with that of a precursor in which the S2 site cannot be cleaved [BMP-4(mS2G)] (Figure 1A) in Xenopus oocytes.
To determine whether mutation of the S2 site prevents proper folding and thus promotes degradation of proBMP-4(mS2G) before exiting the ER, we asked whether this precursor is dimerized and present in post-ER compartments at levels comparable with wild-type proBMP-4. RNAs (5 ng) encoding wild-type or S2 cleavage mutant proBMP-4 were injected into Xenopus oocytes together with [35S]Met/Cys, and oocytes were cultured for 20 h to label newly synthesized proteins. Precursor and mature BMP-4 were immunoprecipitated from lysates by using antibodies specific for the myc-tag and were treated with or without deglycosylating agents. Carbohydrates that are transferred onto proteins in the ER are sensitive to Endo H digestion. When further modified in the Golgi, these moieties become Endo H resistant but remain sensitive to PNGase F. Thus, Endo H resistance/PNGase F sensitivity is a hallmark of proteins that are properly folded and able to traffic from the ER. As shown in Figure 1B, Endo H-sensitive (asterisks) and Endo H-resistant/PNGase F-sensitive (arrowheads) forms of mature BMP-4 cleaved from wild-type and cleavage mutant precursors were detected under reducing and nonreducing conditions. This indicates that high mannose, Endo H-sensitive carbohydrates are retained at one or more glycosylation site(s) on mature BMP-4, even after it has trafficked through the Golgi. A similar glycosylation pattern is observed for the closely related protein, BMP-2 (Israel et al., 1992), suggesting that a subset of carbohydrates present in the mature domains of BMP-2 and -4 become inaccessible to modifying enzymes in the Golgi once these proteins adopt their fully folded conformation. Significantly less mature BMP-4 accumulated in oocytes expressing proBMP-4(mS2G) than in those expressing wild-type precursor, consistent with our previous studies showing that failure to cleave at the S2 site leads to reduced levels of mature BMP-4 in embryos. The majority of wild-type and S2 mutant proBMP-4 was present in an Endo H-sensitive, monomeric form, but a small amount of dimerized precursor was detected under nonreducing conditions. Relatively equivalent levels of dimerized, Endo H-resistant, PNGase-sensitive-proBMP-4 and proBMP-4(mS2G) were detected (arrows), indicating that the amino acid substitution at the S2 site does not cause misfolding and degradation of the precursor before cleavage.
To determine whether the reduced level of mature BMP-4 generated from proBMP-4(mS2G) is due to inefficient cleavage of the precursor at the S1 site, we examined the time course of proBMP processing by pulse-chase analysis. Oocytes were injected with RNA (5 ng) encoding BMP-4 precursors together with [35S]Met/Cys, and after 3 h, label was chased by incubation in media containing unlabeled methionine and cysteine. BMP-4 precursor and cleaved prodomain were immunoprecipitated from cell or media fractions at increasing time intervals by using antibodies specific for the HA tag and separated on reducing gels. A band corresponding to prodomain cleaved at both the S1 and S2 sites was observed in lysates and media from oocytes made to express wild-type proBMP-4, whereas a lower Mr band corresponding to prodomain cleaved only at the S1 site accumulated with equal kinetics in lysates and media from oocytes made to express proBMP-4(mS2G) (Figure 1C). These data demonstrate that the S1 site of proBMP-4(mS2G) is efficiently cleaved.
To directly examine accumulation of mature BMP-4 generated from each precursor, we repeated the pulse-chase experiment, but injected less RNA (0.45 ng) to avoid saturating the system. BMP-4 precursor and mature protein were immunoprecipitated from cell or media fractions at increasing time intervals by using antibodies specific for the myc-tag and were separated on nonreducing gels. As shown in Figure 1D, wild-type and cleavage mutant precursors dimerized and both disappeared from lysates within the time course of the experiment, although the wild-type precursor disappeared slightly more quickly. Mature BMP-4 was readily detected in lysates, and less so in media, from oocytes made to express proBMP-4 but was barely or non-detectable in lysates or media from oocytes made to express proBMP-4(mS2G). Under these same experimental conditions, prodomain cleaved from proBMP-4(mS2G) is barely detectable relative to that cleaved from native precursor (our unpublished data). Together, these data demonstrate that failure to cleave proBMP-4 at the S2 site has no effect on folding of the precursor and does not prevent cleavage at the S1 site, but it leads to rapid degradation of the cleaved prodomain and ligand.
Degradation of Mature BMP-4 Requires Lysosomal and Proteosomal Function
To test whether degradation of mature BMP-4 requires lysosomal function, we asked whether treatment with the deacidifying agent chloroquine rescues accumulation of ligand cleaved from proBMP-4(mS2G). Oocytes were injected with RNA encoding wild-type or mutant precursor together with [35S]Met/Cys, cultured for 16 h to label newly synthesized proteins, and then cultured for an additional 3 h in the presence or absence of chloroquine. Mature BMP-4 was immunoprecipitated from culture media by using antibodies specific for the myc-tag. Consistent with our previous data, very little mature BMP-4 was detected in the media of untreated oocytes made to express proBMP-4(mS2G) relative to those expressing proBMP-4 (Figure 2, top). Treatment with chloroquine increased the accumulation of mature BMP-4 generated from proBMP-4(mS2G) by 4- to 15-fold in five independent experiments. Chloroquine also increased accumulation of BMP-4 cleaved from wild-type precursor in other experiments (our unpublished data), although to a lesser extent (1.8- to 6-fold).
Figure 2.
Degradation of BMP-4 requires lysosomal and proteosomal function. RNA encoding wild-type or cleavage mutant proBMP-4 was injected into oocytes together with [35S]Met/Cys. Oocytes were cultured in the presence or absence of a lysosomal (chloroquine [chloroq]) or proteosomal (epoximicin or lactacystin) inhibitor for varying amounts of time. Mature BMP-4 was immunoprecipitated from the media by using myc-specific antibodies and analyzed by SDS-PAGE and autoradiography.
To test whether proteosomal function also is required for degradation of mature BMP-4, the above-mentioned experiment was repeated, but oocytes were cultured in the presence or absence of 10 μM epoximicin or lactacystin. Levels of mature BMP-4 cleaved from proBMP-4(mS2G) were significantly increased (1.5- to 5.4-fold in 4 independent experiments) by inhibition of proteosomal function (Figure 2, bottom). Mature BMP-4 cleaved from wild-type precursor showed a more modest increase (1.3- to 2-fold in four independent experiments) under the same conditions. Together, our results show that failure to cleave the S2 site leads to rapid degradation of mature BMP-4 and this requires both proteosomal and lysosomal function.
Cleavage at the S2 Site Disrupts Noncovalent Association of the Prodomain with mature BMP-4
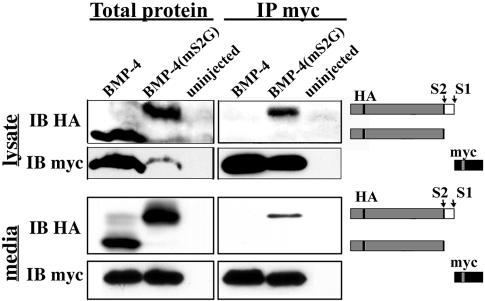
Our observation that cleavage at the S2 site can regulate the activity of mature BMP-4 after it has been excised from the prodomain raises the possibility that these two fragments remain noncovalently associated after cleavage. Coimmunoprecipitation experiments were performed to test this possibility. RNA encoding proBMP-4 or proBMP-4(mS2G) was injected into oocytes, and cleavage products were analyzed 24 h later. Mature BMP-4 was immunoprecipitated from oocyte lysates or media by using an anti-myc–conjugated antibody. Western blots of immunoprecipitates or total proteins were probed with antibodies specific for HA or myc epitopes. As shown in Figure 3, the intact HA-tagged prodomain generated by cleavage at the S1 site coimmunoprecipitated with mature BMP-4, both inside (top) and outside (bottom) of the oocyte, whereas the N-terminal prodomain fragment generated by cleavage at the S2 site did not. These data show that the intact prodomain of BMP-4 remains bound to the mature domain after cleavage at the S1 site and that cleavage at the S2 site disrupts this association.
Figure 3.
Cleavage at the S2 site disrupts noncovalent association between the S1-cleaved prodomain and mature BMP-4. Epitope tagged wild-type or S2 cleavage mutant proBMP-4 was expressed in Xenopus oocytes. Cleavage products present in lysates or media were analyzed by probing immunoblots (IB) of total protein or α-myc immunoprecipitates (IP myc) with antibodies specific for HA- or myc-epitopes as indicated. The position of S1- or S2-cleaved prodomain and mature BMP-4 is illustrated schematically to the right.
Cleavage at the S2 Site, but Not the S1 Site, Is Enhanced under Acidic Conditions
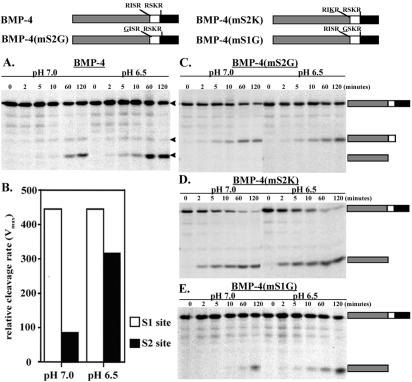
To begin to ask whether cleavage of proBMP-4 at the S1 and S2 sites may occur in distinct subcellular compartments, we analyzed the pH dependence of the relative rate of cleavage at each site. [35S]proBMP-4 synthesized in oocytes was incubated with recombinant furin in vitro under neutral (pH 7) or acidic (pH 6.5) conditions, and cleavage products were analyzed by SDS-PAGE and autoradiography. Under neutral conditions, a band corresponding to the prodomain cleaved at the S1 site alone was apparent within 5 min of furin addition, and subsequent cleavage at the S2 site converted this band into the faster migrating species that was barely detectable after 5 min but was readily apparent at 60 min (Figure 4A). By contrast, under acidic conditions proBMP-4 was cleaved with equal efficiency at both the S1 and S2 sites within 2–5 min, as evidenced by the appearance of bands corresponding to the intact (S1 cleaved) and N-terminal (S1 and S2 cleaved) prodomain fragments. The band intensities of the S1- and S2-cleaved products over a range of substrate concentration (0.8–20.0 nM) and pH (6.5–7.0) were quantified and used to calculate the relative rate of cleavage under neutral and acidic conditions. As shown in Figure 4B, the S1 site was cleaved with equal efficiency at neutral and acidic pH, demonstrating that the acidic environment does not enhance furin activity in general. By contrast, the S2 site was cleaved more efficiently under acidic conditions. Lowering the pH to 6.0 did not lead to further enhancement of cleavage at the S2 site nor did it affect the rate of cleavage at the S1 site (our unpublished data). Consistent with the above-mentioned data showing that cleavage at the S1 site is insensitive to pH, no difference was seen in the rate of cleavage of the S1 site of BMP-4(mS2G) under acidic or neutral conditions (Figure 4C).
Figure 4.
Cleavage at the S2, but not the S1, site is enhanced at acidic pH. Radiolabeled wild-type (A) or cleavage mutant (C–E) forms of proBMP-4 (illustrated schematically above figure) were incubated with recombinant furin at neutral or acidic pH. Aliquots were removed and analyzed by SDS-PAGE at the indicated times. The position of precursor-, S1-, and S2-cleaved prodomain is illustrated schematically to the right of gels in C–E and is marked by arrowheads in A. (B) ProBMP-4 was synthesized in vitro and cleaved by recombinant furin over a range of concentrations and pHs to generate relative rates of cleavage (arbitrary units) for the S1 (white bars) and S2 (black bars) sites.
To ask whether enhanced cleavage of the S2 site at acidic pH is dependent upon the presence of a minimal furin cleavage motif, we analyzed cleavage of proBMP-4(mS2K) in which the upstream cleavage site is converted to an optimal furin motif (RISR→RIKR). We had previously shown that both sites of this precursor are cleaved rapidly and with equal efficiency, rather than sequentially (Cui et al., 2001). As shown in Figure 4D, no difference was seen in the rate of cleavage of the optimal furin motifs under neutral and acidic conditions.
To test whether enhanced cleavage of the S2 site at acidic pH requires prior cleavage at the S1 site, we analyzed cleavage of a precursor in which the furin motif at the S1 site had been disrupted [RSKR→GSKR; BMP-4(mS1G)]. As shown in Figure 4E, a band corresponding to the prodomain cleaved at the S2 site alone was first detected at 10 min after furin addition under neutral conditions but was apparent within 5 min under acidic conditions. Thus, enhanced cleavage of the S2 site at acidic pH can occur independently of cleavage at the S1 site.
Together, our results demonstrate that sequential cleavage of proBMP-4 is driven by the presence of optimal and minimal furin motifs at the S1 and S2 sites, respectively, and that cleavage of the minimal furin motif at the S2 is enhanced at acidic pH. Given that organelles of the secretory pathway become increasingly acidic from the ER to the trans-Golgi network (TGN) to secretory granules, our results are consistent with the possibility that cleavage at the S1 site occurs in a proximal, less acidic subcellular compartment (most likely the TGN, where furin is first active) and that the prodomain/ligand complex then traffics to a more acidic, post-TGN compartment in which the S2 site becomes accessible for cleavage.
A Conserved P6 Histidine at the S2 Site Inhibits Cleavage at Neutral pH
ProBMP-4 contains an evolutionarily conserved P6 histidine residue at the S2 site (underlined in Figure 5A), raising the possibility that this amino acid functions as a pH sensor. Specifically, at neutral pH the deprotonated histidine may mask the S2 site, whereas at acidic pH the additional charge might induce a conformational change that enhances accessibility to the S2 site. To test this hypothesis, we analyzed pH dependence of cleavage of precursors in which the P6 histidine was substituted with a basic residue [HVRISR→RVRISR; BMP-4(mH-R)], to mimic the protonated state, or with a neutral residue [HVRISR→TVRISR; (BMP-4(mH-T)], to mimic the uncharged, deprotonated state. If our hypothesis is correct, substitution with a basic residue should generate a precursor that is efficiently cleaved at the S2 site under both acidic and neutral conditions, whereas substitution with a neutral residue should generate a precursor that is inefficiently cleaved at both neutral and acidic pH. Precursor proteins were synthesized in oocytes and subjected to in vitro cleavage with recombinant furin at neutral and acidic pH as described above. Consistent with our prediction, cleavage of the S2 site was enhanced at both acidic and neutral pH by substitution of the P6 histidine with arginine [BMP-4(mH-R)] (Figure 5B). Contrary to our prediction, however, substitution of the P6 histidine with threonine [BMP-4(mH-T)] enhanced, rather than inhibited, cleavage at both pH values (Figure 5C). These results are consistent with a simpler model in which the deprotonated P6 histidine adopts a conformation that negatively regulates cleavage of the S2 site at neutral pH and protonation of this histidine at acidic pH, or substitution with other amino acids that disrupt the inhibitory conformation, enhance S2 cleavage.
Figure 5.
A conserved P6 histidine at the S2 site inhibits cleavage at neutral pH. Radiolabeled wild-type (A) or P6 histidine mutant (B and C) forms of proBMP-4 (illustrated schematically above each panel) were incubated with recombinant furin at neutral or acidic pH. Aliquots were removed and analyzed by SDS-PAGE at the indicated times. The position of precursor-, S1-, and S2-cleaved prodomain is illustrated schematically to the right of each gel.
Binding to HSPGs Facilitates Degradation of Mature BMP-4
Binding to HSPGs has been shown to restrict BMP-4 diffusion (Ohkawara et al., 2002) and to target transforming growth factor (TGF)-β family ligands for internalization and degradation (Hashimoto et al., 1997). To test whether binding to HSPGs facilitates targeted degradation of mature BMP-4 generated from proBMP-4(mS2G), we deleted three basic residues in the mature domain (illustrated above Figure 6) that have been shown to be necessary and sufficient for association with HSPGs (Ohkawara et al., 2002) and asked whether this affected accumulation of mature BMP-4 in a pulse-chase assay. Mature BMP-4 cleaved from proBMP-4(mS2G) was undetectable in oocyte lysates and culture media (Figure 6A), whereas deletion of the heparin-binding motif [BMP-4(mS2G)ΔRKK] resulted in significant accumulation of mature BMP-4 in both lysate and media fractions (Figure 6B). Deletion of the heparin-binding motif from the wild-type precursor (BMP-4ΔRKK) also enhanced accumulation of mature BMP-4 in lysates and culture media (Figure 6, C and D). These results are consistent with the possibility that binding to HSPGs facilitates internalization and/or lysosomal targeting of mature BMP-4.
Figure 6.
Deletion of a heparin-binding motif in mature BMP-4 inhibits degradation. RNA encoding BMP-4 precursors containing (A and C) or lacking (B and D) the RKK motif that mediates binding to HSPGs (underlined in schematic diagram above figure) was injected into oocytes together with [35S]Met/Cys. Oocytes were pulse labeled for 3 h and then transferred to media containing cold methionine and cysteine. BMP-4 was immunoprecipitated from lysates and media at the indicated times postchase by using anti-myc antibodies. The position of mature BMP-4 dimers and proBMP-4 monomers or dimers is indicated to the right.
DISCUSSION
Model for Regulation of BMP-4 Activity by Sequential Cleavage
Our data support the following model for how cleavage at the S2 site within the prodomain regulates the activity of mature BMP-4. As illustrated in Figure 7, proBMP-4 dimerizes and folds before exiting the ER. Within the TGN, the S1 site of proBMP-4 is cleaved but the excised prodomain remains noncovalently associated with the mature ligand. This complex then traffics to a more acidic post-TGN compartment where the S2 site is cleaved, promoting dissociation of the prodomain fragments from mature BMP-4. If cleavage of the S2 site does not occur, the prodomain remains associated with mature BMP-4, and the complex is targeted for degradation. At present, it is unknown whether the small peptide that is liberated from the prodomain by cleavage at the S2 site (white box in Figure 7) remains associated with mature BMP-4 or the prodomain fragment or whether it plays a role in regulating ligand stability. This peptide is, however, required to generate bioactive mature BMP-4 because deletion mutant forms of proBMP-4 that lack this 35-amino acid fragment do not exit the ER and thus are not cleaved when expressed in Xenopus oocytes (Degnin and Christian, unpublished data).
Figure 7.
Model for regulation of BMP-4 activity by sequential cleavage. See text for details.
Our results suggest that a conserved histidine residue at the P6 position of the S2 site of proBMP-4 mediates pH-dependent cleavage. Specifically, we propose that at neutral pH the deprotonated histidine adopts a conformation that masks the S2 site and that protonation at acidic pH disrupts this conformation to allow cleavage to occur. A somewhat analogous situation exists in the case of α4 integrin where a P6 histidine compensates for the lack of a basic residue at the P4 position and renders cleavage sensitive to cellular pH (Bergeron et al., 2003). The P6 position is often not included in the consensus motif for cleavage by PCs, but others have shown that the S6 subsite of furin contributes to substrate binding and prefers basic residues (Rockwell et al., 2002). Consistent with this preference, analysis of the crystal structure of furin has revealed an overall negative surface potential for the S6 subsite of the enzyme (Henrich et al., 2003). Furthermore, this binding site resides in a relatively solvent-exposed region (Siezen et al., 1994) and specificity for P6 basic residues can be modulated by pH (Krysan et al., 1999).
Sequential Cleavage of proBMP-4 Provides a Mechanism for Tissue-specific Regulation of BMP Activity and Signaling Range
Furin undergoes a two-step proteolytic activation, analogous to that of proBMP-4, which serves to ensure that the zymogen is not activated prematurely (Thomas, 2002). Specifically, the propeptide of furin is autocatalytically excised within the ER at a consensus furin motif, but it remains noncovalently bound within the active site to keep the enzyme inactive. This complex is trafficked to the TGN where reduced pH, coupled with increased calcium, enables furin to cleave the bound propeptide at a nonconsensus motif, releasing it from the catalytic cleft so that furin can then cleave its substrates in trans (Anderson et al., 1997, 2002).
By contrast with furin, sequential cleavage of proBMP-4 is not required to prevent premature activation but may instead provide a mechanism for tissue-specific regulation of activity and signaling range. BMP-4, and its Drosophila ortholog Decapentaplegic (Dpp) function as either short- or long-range signaling molecules, depending on the tissue in which they are expressed (Neumann and Cohen, 1997), but the mechanisms that confer this tissue specificity are unknown. Constitutive cleavage of the S1 site in all tissues, coupled with selective cleavage of the S2 site in the subset of tissues where BMP-4 signals at long distance, could provide a mechanism for tissue-specific regulation of BMP signaling range. Consistent with this possibility, studies in Xenopus embryos (Cui et al., 2001), together with the current data, demonstrate that cleavage at the S2 site generates a stable ligand that possesses long-range signaling properties, whereas cleavage at the S1 site alone generates an identical ligand that is targeted for rapid degradation and thus can signal only to adjacent cells. Our analysis of mice carrying a targeted mutation that prevents cleavage of the proBMP-4 S2 site provide further support for the hypothesis that tissue-specific use of the S2 site regulates the activity of endogenous BMP-4 (Goldman, Hackenmiller, and Christian, unpublished data).
Potential Mechanisms for Tissue-specific Regulation of S2 Cleavage
The current results suggest several mechanisms through which tissue-specific cleavage of the S2 site might be achieved. First, our data showing that cleavage at the S2 site is enhanced at low pH raise the possibility that cleavage of the S2 site only occurs in tissues in which BMP-4 is sorted from the TGN into an acidic, endosomal transport pathway. Proteins destined for secretion can be targeted to the endosomal system for delivery to the cell surface or can be shunted directly from the TGN to the plasma membrane (Bonifacino and Traub, 2003). S1 cleaved BMP-4 could potentially be sorted to endosomes before secretion in a subset of cells, where the acidic environment would allow for cleavage at the S2 site. In other cell types, S1 cleaved BMP-4 might be delivered directly to the plasma membrane from the TGN, thereby bypassing the acidic compartment where S2 cleavage occurs.
Selective cleavage of the S2 site of proBMP-4 also could be mediated by tissue-specific expression of a convertase that resides in a post-TGN compartment and cleaves the S2, but not the S1 site. Furin is broadly expressed and resides predominantly in the TGN (Thomas, 2002), whereas PC6B is expressed in only a few tissues (Nakagawa et al., 1993; Seidah et al., 1994) and is localized to a distinct, post-TGN compartment (Xiang et al., 2000). Thus, it is possible that the S1 site is constitutively cleaved by furin, whereas the S2 site is cleaved only in tissues that coexpress PC6B.
The idea that different PCs might cleave a single substrate is not unprecedented. Activation of proinsulin, for example, requires sequential cleavage by PC1 and then PC2 (Zhou et al., 1999). Furthermore, PC1- and/or PC2-mediated cleavage of proopiomelanocortin or proglucagon generates distinct end products that are expressed in tissue-specific patterns, reflecting the differential distribution of PC1 and PC2. Identification of endogenous proBMP-4 convertase(s) will be required to test the hypothesis that the S1 and S2 sites are cleaved by distinct PCs.
Cleavages within the Prodomain Direct Intracellular Trafficking and Degradation of BMP-4
We have shown that failure to cleave proBMP-4 at the S2 site promotes rapid degradation of the mature ligand, and this requires both lysosomal and proteosomal function. Proteosome inhibitors have been shown to impair endosomal sorting and degradation of a number of cell surface receptors and their bound ligands. These receptors are not direct targets of the proteosome, however, but they are degraded by lysosomal hydrolysis (Rocca et al., 2001; van Kerkhof et al., 2001; Longva et al., 2002; Melman et al., 2002). In these cases, the proteosome seems to play a broader function in regulating the trafficking of receptors into the degradation pathway (Hicke and Dunn, 2003). By analogy, proteosomal involvement in degrading mature BMP-4 may reflect its role in lysosomal sorting rather than degradation.
Our studies do not address whether mature BMP-4 is targeted for degradation within the biosynthetic or endocytic pathway. Newly synthesized transmembrane proteins can be delivered directly from the TGN to the lysosome for degradation without ever reaching the cell surface. In yeast, for example, GAP1 is delivered directly to the lysosome under certain environmental conditions (Soetens et al., 2001), and the Drosophila axon guidance molecule Robo undergoes regulated transport from the secretory pathway to the lysosome as a mechanism to decrease cell surface activity (Keleman et al., 2002). It is possible that motifs within the BMP-4 prodomain bind to a sorting receptor in the secretory pathway and this preferentially targets the prodomain/ligand complex for degradation. Dissociation of the prodomain after cleavage at the S2 site would thus enable free mature BMP-4 to bypass the lysosome and be secreted. More commonly, ligands are secreted, bind their cognate transmembrane receptors, and are sorted to either recycling or lysosomal compartments after endocytosis (Seto et al., 2002). Both pathways require ubiquitination of the cytoplasmic tail of transmembrane proteins as well as protein sorting complexes that specifically recognize the ubiquitinated cargo (Babst, 2004).
Targeted degradation of BMP-4 may be mediated by binding to its receptor because BMPs are known to undergo receptor-mediated endocytosis and degradation (Jortikka et al., 1997). Furthermore, the Smurf family of ubiquitin-protein ligases has been shown to be recruited to the cytoplasmic tail of activated BMP and TGF-β receptors, thereby targeting them for degradation in a proteosome- and lysosome-dependent manner (Kavsak et al., 2000; Podos et al., 2001; Murakami et al., 2003). Our data are consistent with the possibility that the prodomain of BMP-4 enhances the affinity of the ligand for its receptor such that both are targeted for lysosomal degradation. By contrast, mature BMP-4 that is released from the prodomain after cleavage at the S2 site may more readily dissociate from the receptor, enabling it to bypass the lysosome. An analogous situation exists in the case of epidermal growth factor (EGF) and TGFα, which differ in their ability to target a common receptor for degradation. EGF remains bound to the receptor after internalization and is proteolytically degraded in the lysosome, whereas TGFα rapidly dissociates, resulting in recycling of the receptor to the cell surface (Sorkin and Waters, 1993).
Recent studies have revealed a role for receptor-mediated endocytosis and lysosomal degradation in shaping the BMP/Dpp concentration gradient in flies (Entchev et al., 2000; Gonzalez-Gaitan, 2003). These studies suggest that Dpp is transported across the embryonic wing disk by consecutive rounds of endocytosis, active intracellular transport, and exocytosis. When clones of cells are generated that are mutant for either the Dpp receptor or for components of the endocytic machinery, Dpp is not internalized and cannot spread across the patch of endocytosis-defective cells. Overexpression of Rab5, which promotes early endocytic trafficking, broadens the Dpp gradient, whereas overexpression of a constitutively active form of Rab7, which enhances trafficking from endosomes to the lysosome, reduces the spread of Dpp. A requirement for endocytosis in ligand transport is highly controversial, however, and conflicting evidence suggests that the extracellular Dpp gradient forms instead by free diffusion (Lander et al., 2002). Regardless of whether Dpp moves through the intracellular or extracellular space, the role of lysosomal degradation in shaping the gradient of Dpp and other morphogens is well established (Seto et al., 2002). Our results are consistent with a model in which cleavage at the S2 site occurs in selected tissues, such as the wing disk, preferentially directing the ligand away from the lysosome and enabling a long-range concentration gradient to form. In other tissues, where cleavage at the S2 site does not occur, the ligand would be sorted to the lysosome and signal only at short range.
Role of HSPGs in Targeting BMP-4 for Degradation
Our data showing that deletion of the heparin-binding motif in BMP-4 stabilizes the mature ligand raise the possibility that transmembrane HSPGs contribute to internalization and/or lysosomal targeting of mature BMP-4. Consistent with this possibility, binding to HSPGs has been shown to accelerate endocytosis and lysosomal degradation of Activin A (Hashimoto et al., 1997). Furthermore, addition of excess free heparin to the outside of cultured cells inhibits association of exogenous BMP-2 with its signaling receptor and suppresses intracellular accumulation of this ligand (Takada et al., 2003). These data suggest that HSPGs promote receptor binding and internalization of BMPs. In Drosophila, mutations in HSPG core proteins or in the enzymes responsible for HSPG biosynthesis lead to a reduction in Dpp signaling and loss of cell surface Dpp protein, suggesting that HSPGs sequester and concentrate Dpp at the cell surface (Tabata and Takei, 2004). Cells mutant for HSPG core proteins, however, also show an inability to down-regulate Dpp signaling, consistent with an additional role in clearing Dpp from the system (Fujise et al., 2003). Future studies will be required to determine whether HSPGs play a general role in promoting BMP-4 degradation or whether they contribute to preferential targeting of S1 cleaved BMP-4 to the lysosome.
Acknowledgments
We thank Drs. W. Skach and T. O'Hare and L. Shamieh and members of the Christian laboratory for helpful comments. This work was supported in part by grants from the National Institutes of Health to J.L.C. (HD37976 and HD042598) and G.T. (DK37274, AI49793, and AI48585). C.D. was supported by a predoctoral training grant from the National Institutes of Health (T32 HL007781). F.J. is a Canadian Institutes of Health Research/Health Canada Research Initiative on Hepatitis C Virus Scholar.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–08–0673. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–08–0673.
References
- Anderson, E.D., Molloy, S.S., Jean, F., Fei, H., Shimamura, S., and Thomas, G. (2002). The ordered and compartment-specfific autoproteolytic removal of the furin intramolecular chaperone is required for enzyme activation. J. Biol. Chem. 277, 12879-12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, E.D., Thomas, L., Hayflick, J.S., and Thomas, G. (1993). Inhibition of HIV-1 gp160-dependent membrane fusion by a furin-directed alpha 1-antitrypsin variant. J. Biol. Chem. 268, 24887-24891. [PubMed] [Google Scholar]
- Anderson, E.D., VanSlyke, J.K., Thulin, C.D., Jean, F., and Thomas, G. (1997). Activation of the furin endoprotease is a multiple-step process: requirements for acidification and internal propeptide cleavage. EMBO J. 16, 1508-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono, A., Hazama, M., Notoya, K., Taketomi, S., Yamasaki, H., Tsukuda, R., Sasaki, S., and Fujisawa, Y. (1995). Potent ectopic bone-inducing activity of bone morphogenetic protein-4/7 heterodimer. Biochem. Biophys. Res. Commun. 210, 670-677. [DOI] [PubMed] [Google Scholar]
- Babst, M. (2004). GGAing ubiquitin to the endosome. Nat. Cell Biol. 6, 175-177. [DOI] [PubMed] [Google Scholar]
- Bergeron, E., Basak, A., Decroly, E., and Seidah, N.G. (2003). Processing of alpha4 integrin by the proprotein convertases: histidine at position P6 regulates cleavage. Biochem. J. 373, 475-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino, J.S., and Traub, L.M. (2003). Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395-447. [DOI] [PubMed] [Google Scholar]
- Brunet, L.J., McMahon, J.A., McMahon, A.P., and Harland, R.M. (1998). Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science 280, 1455-1457. [DOI] [PubMed] [Google Scholar]
- Chang, B., et al. (2001). Haploinsufficient Bmp4 ocular phenotypes include anterior segment dysgenesis with elevated intraocular pressure. BMC Genet. 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constam, D.B., and Robertson, E.J. (1999). Regulation of bone morphogenetic protein activity by pro domains and proprotein convertases. J. Cell Biol. 144, 139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Y., Hackenmiller, R., Berg, L., Jean, F., Nakayama, T., Thomas, G., and Christian, J.L. (2001). The activity and signaling range of mature BMP-4 is regulated by sequential cleavage at two sites within the prodomain of the precursor. Genes Dev. 15, 2797-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Y., Jean, F., Thomas, G., and Christian, J.L. (1998). BMP-4 is proteolytically activated by furin and/or PC6 during vertebrate embryonic development [In Process Citation]. EMBO J. 17, 4735-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, N.R., Winnier, G.E., Hargett, L.K., Schrick, J.J., Fogo, A.B., and Hogan, B.L. (1997). Haploinsufficient phenotypes in Bmp4 heterozygous null mice and modification by mutations in Gli3 and Alx4. Dev. Biol. 188, 235-247. [DOI] [PubMed] [Google Scholar]
- Eimon, P.M., and Harland, R.M. (2002). Effects of heterodimerization and proteolytic processing on Derriere and Nodal activity: implications for mesoderm induction in Xenopus. Development 129, 3089-3103. [DOI] [PubMed] [Google Scholar]
- Entchev, E.V., Schwabedissen, A., and Gonzalez-Gaitan, M. (2000). Gradient formation of the TGF-beta homolog Dpp. Cell 103, 981-991. [DOI] [PubMed] [Google Scholar]
- Fujise, M., Takeo, S., Kamimura, K., Matsuo, T., Aigaki, T., Izumi, S., and Nakato, H. (2003). Dally regulates Dpp morphogen gradient formation in the Drosophila wing. Development 130, 1515-1522. [DOI] [PubMed] [Google Scholar]
- Gong, Y., et al. (1999). Heterozygous mutations in the gene encoding noggin affect human joint morphogenesis. Nat. Genet. 21, 302-304. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gaitan, M. (2003). Signal dispersal and transduction through the endocytic pathway. Nat. Rev. Mol. Cell. Biol. 4, 213-224. [DOI] [PubMed] [Google Scholar]
- Hashimoto, O., Nakamura, T., Shoji, H., Shimasaki, S., Hayashi, Y., and Sugino, H. (1997). A novel role of follistatin, an activin-binding protein, in the inhibition of activin action in rat pituitary cells. Endocytotic degradation of activin and its acceleration by follistatin associated with cell-surface heparan sulfate. J. Biol. Chem. 272, 13835-13842. [DOI] [PubMed] [Google Scholar]
- Henrich, S., Cameron, A., Bourenkov, G.P., Kiefersauer, R., Huber, R., Lindberg, I., Bode, W., and Than, M.E. (2003). The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat. Struct. Biol. 10, 520-526. [DOI] [PubMed] [Google Scholar]
- Hicke, L., and Dunn, R. (2003). Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell. Dev. Biol. 19, 141-172. [DOI] [PubMed] [Google Scholar]
- Hogan, B.L. (1996). Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 10, 1580-1594. [DOI] [PubMed] [Google Scholar]
- Horton, R.M., Cai, Z., Ho, Z.N., and Pease, L.R. (1990). Gene splicing by overlap extension: tailor made genes using the polymerase chain reaction. Biotechniques 8, 528-536. [PubMed] [Google Scholar]
- Israel, D.I., Nove, J., Kerns, K.M., Moutsatsos, I.K., and Kaufman, R.J. (1992). Expression and characterization of bone morphogenetic protein-2 in Chinese hamster ovary cells. Growth Factors 7, 139-150. [DOI] [PubMed] [Google Scholar]
- Jean, F., Stella, K., Thomas, L., Liu, G., Xiang, Y., Reason, A.J., and Thomas, G. (1998). alpha1-Antitrypsin Portland, a bioengineered serpin highly selective for furin: application as an antipathogenic agent. Proc. Natl. Acad. Sci. USA 95, 7293-7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jortikka, L., Laitinen, M., Lindholm, T.S., and Marttinen, A. (1997). Internalization and intracellular processing of bone morphogenetic protein (BMP) in rat skeletal muscle myoblasts (L6). Cell Signal. 9, 47-51. [DOI] [PubMed] [Google Scholar]
- Kaplan, F.S., and Shore, E.M. (1998). Encrypted morphogens of skeletogenesis: biological errors and pharmacologic potentials. Biochem. Pharmacol. 55, 373-382. [DOI] [PubMed] [Google Scholar]
- Kavsak, P., Rasmussen, R.K., Causing, C.G., Bonni, S., Zhu, H., Thomsen, G.H., and Wrana, J.L. (2000). Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol. Cell 6, 1365-1375. [DOI] [PubMed] [Google Scholar]
- Keleman, K., Rajagopalan, S., Cleppien, D., Teis, D., Paiha, K., Huber, L.A., Technau, G.M., and Dickson, B.J. (2002). Comm sorts robo to control axon guidance at the Drosophila midline. Cell 110, 415-427. [DOI] [PubMed] [Google Scholar]
- Khokha, M.K., Hsu, D., Brunet, L.J., Dionne, M.S., and Harland, R.M. (2003). Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nat. Genet. 34, 303-307. [DOI] [PubMed] [Google Scholar]
- Krysan, D.J., Rockwell, N.C., and Fuller, R.S. (1999). Quantitative characterization of furin specificity. Energetics of substrate discrimination using an internally consistent set of hexapeptidyl methylcoumarinamides. J. Biol. Chem. 274, 23229-23234. [DOI] [PubMed] [Google Scholar]
- Lander, A.D., Nie, Q., and Wan, F.Y. (2002). Do morphogen gradients arise by diffusion? Dev. Cell 2, 785-796. [DOI] [PubMed] [Google Scholar]
- Lawson, K.A., Dunn, N.R., Roelen, B.A., Zeinstra, L.M., Davis, A.M., Wright, C.V., Korving, J.P., and Hogan, B.L. (1999). Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 13, 424-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longva, K.E., Blystad, F.D., Stang, E., Larsen, A.M., Johannessen, L.E., and Madshus, I.H. (2002). Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J. Cell Biol. 156, 843-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon, J.A., Takada, S., Zimmerman, L.B., Fan, C.M., Harland, R.M., and McMahon, A.P. (1998). Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev. 12, 1438-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melman, L., Geuze, H.J., Li, Y., McCormick, L.M., van Kerkhof, P., Strous, G.J., Schwartz, A.L., and Bu, G. (2002). Proteasome regulates the delivery of LDL receptor-related protein into the degradation pathway. Mol. Biol. Cell 13, 3325-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki, Y., Oshima, K., Fogo, A., Hogan, B.L., and Ichikawa, I. (2000). Bone morphogenetic protein 4 regulates the budding site and elongation of the mouse ureter. J. Clin. Investig. 105, 863-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, R.T., and Christian, J.L. (1989). Microinjection and expression of synthetic mRNAs in Xenopus embryos. Technique 1, 76-89. [Google Scholar]
- Murakami, G., Watabe, T., Takaoka, K., Miyazono, K., and Imamura, T. (2003). Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol. Biol. Cell 14, 2809-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T., Murakami, K., and Nakayama, K. (1993). Identification of an isoform with an extremely large Cys-rich region of PC6, a Kex2-like processing endoprotease. FEBS Lett. 327, 165-171. [DOI] [PubMed] [Google Scholar]
- Nakayama, T., Cui, Y., and Christian, J.L. (2000). Regulation of BMP/Dpp signaling during embryonic development. Cell. Mol. Life Sci. 57, 943-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, C., and Cohen, S. (1997). Morphogens and pattern formation. Bioessays 19, 721-729. [DOI] [PubMed] [Google Scholar]
- Ohkawara, B., Iemura, S., ten Dijke, P., and Ueno, N. (2002). Action range of BMP is defined by its N-terminal basic amino acid core. Curr. Biol. 12, 205-209. [DOI] [PubMed] [Google Scholar]
- Podos, S.D., Hanson, K.K., Wang, Y.C., and Ferguson, E.L. (2001). The DSmurf ubiquitin-protein ligase restricts BMP signaling spatially and temporally during Drosophila embryogenesis. Dev. Cell 1, 567-578. [DOI] [PubMed] [Google Scholar]
- Rocca, A., Lamaze, C., Subtil, A., and Dautry-Varsat, A. (2001). Involvement of the ubiquitin/proteasome system in sorting of the interleukin 2 receptor beta chain to late endocytic compartments. Mol. Biol. Cell 12, 1293-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell, N.C., Krysan, D.J., Komiyama, T., and Fuller, R.S. (2002). Precursor processing by kex2/furin proteases. Chem. Rev. 102, 4525-4548. [DOI] [PubMed] [Google Scholar]
- Seidah, N.G., Chretien, M., and Day, R. (1994). The family of subtilisin/kexin like pro-protein and pro-hormone convertases: divergent or shared functions. Biochimie 76, 197-209. [DOI] [PubMed] [Google Scholar]
- Selleck, S.B. (2001). Genetic dissection of proteoglycan function in Drosophila and C. elegans. Semin. Cell Dev. Biol. 12, 127-134. [DOI] [PubMed] [Google Scholar]
- Seto, E.S., Bellen, H.J., and Lloyd, T.E. (2002). When cell biology meets development: endocytic regulation of signaling pathways. Genes Dev. 16, 1314-1336. [DOI] [PubMed] [Google Scholar]
- Shi, Y., and Massague, J. (2003). Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113, 685-700. [DOI] [PubMed] [Google Scholar]
- Siezen, R.J., Creemers, J.W., and Van de Ven, W.J. (1994). Homology modelling of the catalytic domain of human furin. A model for the eukaryotic subtilisin-like proprotein convertases. Eur. J. Biochem. 222, 255-266. [DOI] [PubMed] [Google Scholar]
- Soetens, O., De Craene, J.O., and Andre, B. (2001). Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J. Biol. Chem. 276, 43949-43957. [DOI] [PubMed] [Google Scholar]
- Sorkin, A., and Waters, C.M. (1993). Endocytosis of growth factor receptors. Bioessays 15, 375-382. [DOI] [PubMed] [Google Scholar]
- Tabata, T., and Takei, Y. (2004). Morphogens, their identification and regulation. Development 131, 703-712. [DOI] [PubMed] [Google Scholar]
- Takada, T., Katagiri, T., Ifuku, M., Morimura, N., Kobayashi, M., Hasegawa, K., Ogamo, A., and Kamijo, R. (2003). Sulfated polysaccharides enhance the biological activities of bone morphogenetic proteins. J. Biol. Chem. 278, 43229-43235. [DOI] [PubMed] [Google Scholar]
- Thomas, G. (2002). Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell. Biol. 3, 753-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kerkhof, P., Alves dos Santos, C.M., Sachse, M., Klumperman, J., Bu, G., and Strous, G.J. (2001). Proteasome inhibitors block a late step in lysosomal transport of selected membrane but not soluble proteins. Mol. Biol. Cell 12, 2556-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnier, G., Blessing, M., Labosky, P.A., and Hogan, B.L. (1995). Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 9, 2105-2116. [DOI] [PubMed] [Google Scholar]
- Xiang, Y., Molloy, S.S., Thomas, L., and Thomas, G. (2000). The PC6B cytoplasmic domain contains two acidic clusters that direct sorting to distinct trans-Golgi network/endosomal compartments. Mol. Biol. Cell 11, 1257-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, X., Bragin, A., Widdicombe, J.H., Cohn, J., and Skach, W.R. (1997). Structural cues involved in endoplasmic reticulum degradation of G85E and G91R mutant cystic fibrosis transmembrane conductance regulator. J. Clin. Investig. 100, 1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, A., Webb, G., Zhu, X., and Steiner, D.F. (1999). Proteolytic processing in the secretory pathway. J. Biol. Chem. 274, 20745-20748. [DOI] [PubMed] [Google Scholar]