Abstract
To ensure successful parasitism, parasitoid wasps inject venom along with their eggs into their hosts. The venom serves to suppress host immune responses, including melanization. Venom from Pteromalus puparum, a pupal endoparasitoid, inhibits melanization of host hemolymph in vitro in a dose-dependent manner. Using assay-guided fractionation, a serpin splicing isoform with phenoloxidase inhibitory activity was identified as P. puparum serpin-1, venom isoform (PpS1V). This serpin gene has 16 predicted splicing isoforms that differ only in the C-terminal region. RT-PCR results show that the specific serpin isoform is differentially expressed in the venom gland. Recombinant PpS1V (rPpS1V) suppresses host prophenoloxidase (PPO) activation rather than inhibiting the phenoloxidase directly. Pulldown assays show that PpS1V forms complexes with two host hemolymph proteins, here named Pieris rapae hemolymph proteinase 8 (PrHP8) and P. rapae prophenoloxidase-activating proteinase 1 (PrPAP1), based on gene sequence blasting and phylogenetic analysis. The role of rPrPAP1 in the PPO activation cascade and its interaction with rPpS1V were confirmed. The stoichiometry of inhibition of PrPAP1 by PpS1V is 2.3. PpS1V also inhibits PPO activation in a non-natural host, Ostrinia furnacalis, through forming a complex with O. furnacalis serine protease 13 (OfSP13), an ortholog to PrPAP1. Our results identify a venom-enriched serpin isoform in P. puparum that inhibits host PPO activation, probably by forming a complex with host hemolymph proteinase PrPAP1.
Keywords: alternative splicing, insect immunity, parasitology, proteinase, serpin, melanization, parasitoid wasp, venom
Introduction
Insect immune systems consist of cellular and humoral immunity (1, 2). Melanization is a conserved part of humoral immunity in insects (3). The melanization pathway is initiated in response to infection and wounding (4) and is followed by a sequential activation of serine proteinases (5). The final step of this serine proteinase cascade is activation of prophenoloxidase (PPO)2-activating proteinases (PAPs), which further catalyze PPO into phenoloxidase (PO) (2). Active PO oxidizes tyrosine and o-diphenols to quinones, which further polymerize to form melanin. Melanin is then deposited on the surface of invaders and seals them from spreading within the host or, in the case of parasitoid eggs, from hatching and completing development (3, 6).
Diverse cytotoxic molecules, such as reactive oxygen and nitrogen species, are produced in this process, and they act in killing invaders via chemical processes (7). These toxic compounds can also be harmful to hosts, which helps explain why melanization is tightly regulated by endogenous protease inhibitors like serine protease inhibitors (serpins) (8–11). The serpins are a superfamily of proteins that have similar structures (12), generally 40–60 kDa with three β-sheets, eight or nine α-helices, and a reactive center loop (RCL) near the C terminus. Most serpins are serine protease inhibitors, which act through a “suicide mechanism,” forming permanent covalent complexes with target serine proteases (13, 14). Through their inhibitory activities, endogenous serpins in insects are primary regulators of the PPO cascade and play a role in Toll pathway regulation (15).
Insect pathogens and parasites have evolved diverse strategies to evade host immune functions (16–18). Parasitoid wasps often inject virulence factors, such as polydnavirus (PDV) or venom proteins, that inhibit host immunity. Inhibition of melanization is particularly important as this is a common mechanism for encapsulating and killing parasitoid eggs (19–21). Possibly because of the very small sizes of parasitoids, a limited number of components from their virulence factors have been identified and characterized (22–26).
Pteromalus puparum is a generalist endoparasitoid wasp that parasitizes the pupal stage of several butterfly species, including the small cabbage white butterfly, Pieris rapae, an agricultural pest (27, 28). Other virulence factors found in some parasitoids, such as PDVs, virus-like particles, or teratocytes, have not been recorded in P. puparum (29). As the major maternal virulence factor in P. puparum, P. puparum venom regulates host development and metabolism (30) and suppresses cellular (27, 28, 31) and humoral immunity of the host (32–34). Here we report a serpin gene with 16 predicted splicing isoforms in P. puparum and show that one of these isoforms is a venom protein, which inhibits host PPO activation by forming complexes with host hemolymph proteinases.
Results
Venom Suppresses Host Melanization
Parasitoid wasps often inject venoms that suppress host immunity. In particular, they inhibit melanization of the parasitoid eggs by the host hemolymph (23–25, 35, 36). P. puparum venom inhibits melanization by hemolymph from P. rapae pupae and larvae (Fig. 1A; Student's t test: for hemolymph from pupae, t = 5.68, df = 4, p < 0.01; for hemolymph from larvae, t = 37.86, df = 4, p < 0.001). Although P. rapae pupae are the natural hosts for P. puparum, collecting hemolymph from larvae is far more efficient compared with pupae. Thus, the following assays were conducted using larval hemolymph.
FIGURE 1.
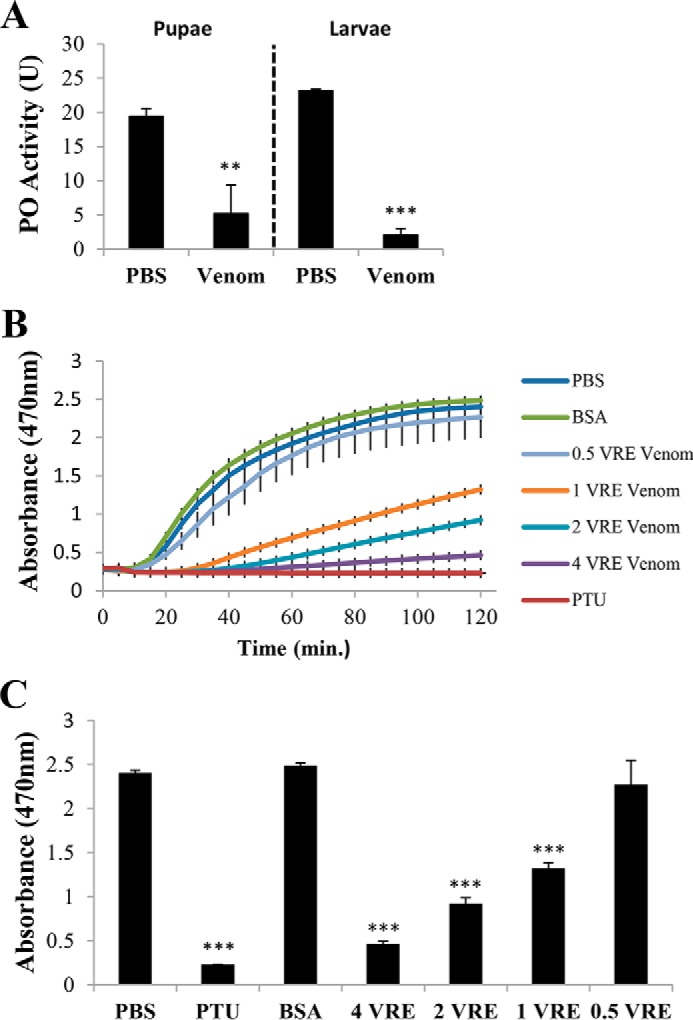
Venom from P. puparum inhibits P. rapae hemolymph PO activity. A, effect of P. puparum venom on PO activity of P. rapae hemolymph from larvae and pupae. B, dose-dependent suppression of host hemolymph PO activity by P. puparum venom. Dopa chrome or dopamine chrome (melanization intermediates) was monitored at A470 every 5 min for 2 h. C, summary of absorbance A470 at 120 min from B. All values are presented as the mean (n = 3). Error bars represent S.E. Asterisks indicate significant differences from PBS as the control. **, p < 0.01; ***, p < 0.001.
To increase the throughput of our PO activity assay, the protocol was modified as follows. The substrate (l-dopa), elicitor (Micrococcus luteus), and inhibitors (phenylthiourea, venom, and other proteins) were mixed in wells of a 384-well plate. The diluted hemolymph samples were loaded on another 384-well plate. After placing one plate on top of the other and sealing them with tape, reaction samples were mixed simultaneously by centrifugation. For samples without inhibitor, spectrophotometric monitoring at 470 nm documented a ∼5-min lag phase before absorbance rapidly rose to a plateau (Fig. 1B). Samples with venom had longer lag phases, ∼20 min for 1 venom reservoir equivalent (VRE), ∼35 min for 2 VREs, and ∼40 min for 4 VREs. Absorbances rose more slowly and to lower final absorbances in venom-exposed hemolymph samples. Absorbance at 120 min was chosen for further statistical analysis (Fig. 1C; analysis of variance (ANOVA): df = 6, F = 47.245, p < 0.001). Results show that P. puparum venom inhibits host hemolymph melanization in vitro in a dose-dependent manner.
Venom Serpin Isoform Fraction and Identification
In a previous study, 70 venom proteins were identified in P. puparum (37). Protease inhibitors, serine proteases, a serine proteinase homolog, a β-1,3-glucan-binding protein, and several venom proteins with no similarities to other known proteins were present and might have a role in the host's melanization inhibition. To determine which of these many proteins are responsible for the melanization inhibitory activity, an assay-guided venom fractionation program was conducted.
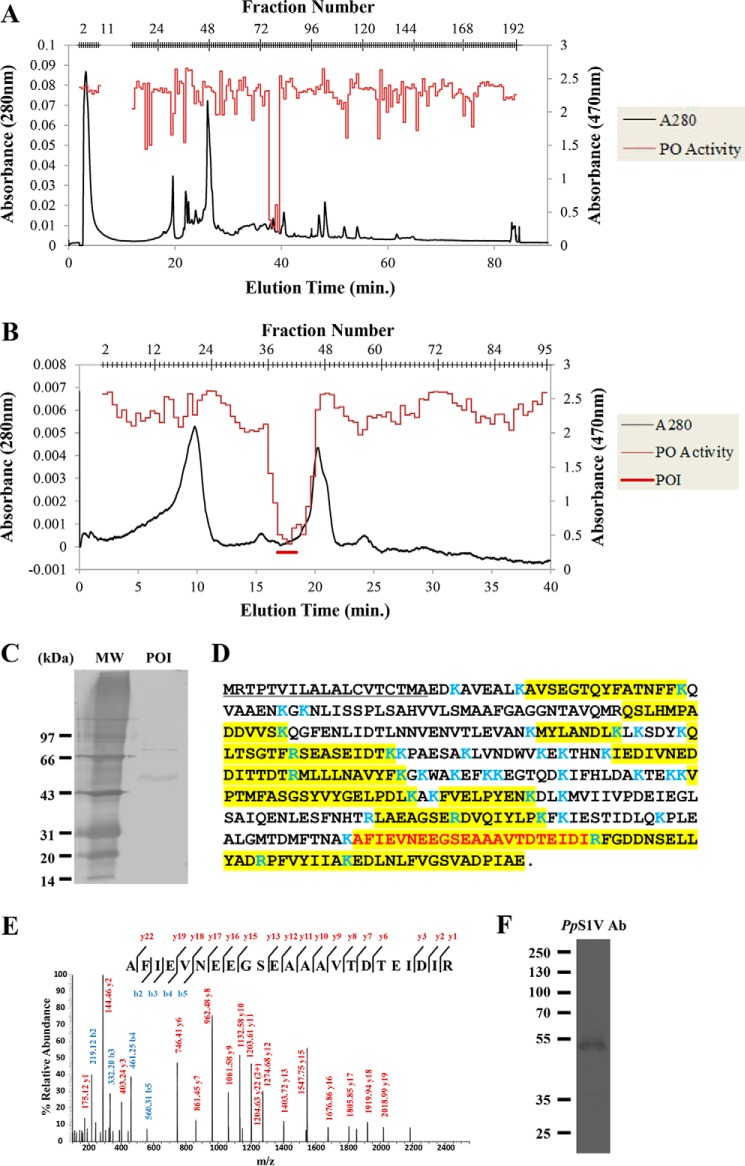
Venom proteins from ∼1000 venom reservoirs were separated on an anion exchange column (Fig. 2A). In total, 182 fractions were collected. Of these, fractions 76–81 showed strong inhibitory activity. The fractions with PO inhibitory activity were pooled and separated on the same column using a slower gradient (e.g. 20–25% buffer B in 40 min; Fig. 2B), leading to a collection of 95 fractions. The peak of inhibitory activity did not correspond to the peak of absorbance, indicating that a low abundance venom component is responsible for inhibiting melanization.
FIGURE 2.
Partial separation and identification of PpS1V. A, chromatograph of total P. puparum venom by Bio SAX column. Venom protein (∼1000 RVEs) was eluted with a gradient of buffer B from 0 to 100% at a flow rate of 0.5 ml/min. B, chromatograph of pooled fractions 76–81 from A on the same column with a shallow gradient of buffer B from 20 to 25% in 40 min. Fractions 36–45 were denoted as phenoloxidase inhibitor (POI) and are indicated by the red bar. These fractions were pooled together for further SDS-PAGE and LC-MS/MS analysis. C, SDS-PAGE analysis of pooled fraction POI followed by silver staining. D, amino acid sequence of PpS1V with peptides identified by MS (highlighted residues). Trypsin cleavage sites are demarcated in blue. The first isoform-specific tryptic peptide of PpS1V is colored in red. E, MS/MS map marked with b ions (blue) and y ions (red) for the first isoform-specific tryptic peptide of PpS1V. F, SDS-PAGE analysis of pooled fraction POI followed by immunoblotting using antibody (Ab) against PpS1V.
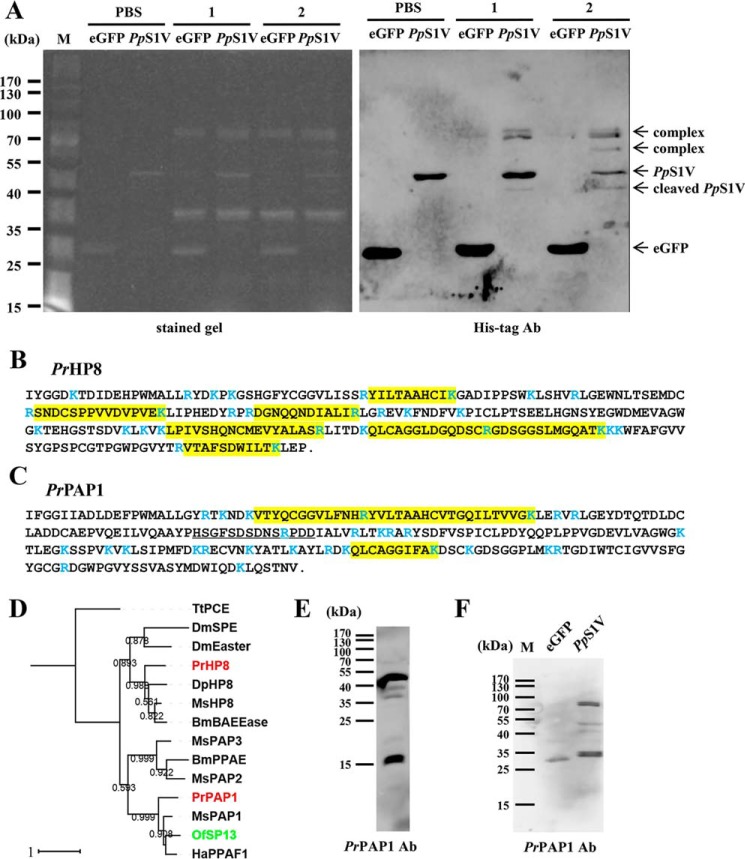
Fractions 36–45 were pooled, and proteins were separated by SDS-PAGE followed by silver staining (Fig. 2C). Two major bands (at 55 and 77 kDa) and several minor bands were present. Simultaneously, the pooled fraction was digested by trypsin and analyzed by LC-MS/MS. After searching against the P. puparum transcriptomic database, a splicing isoform of serpin (comp44322_c1_seq8; from UniGene GECT01032828.1) (37) was identified with the highest score (Fig. 2, D and E). As this isoform is identified in partially isolated venom, it was named P. puparum serpin-1, venom isoform (PpS1V). In total, 14 trypsin-digested peptides matched PpS1V, and three of these 14 were unique to this isoform. No isoform-specific trypsin-digested peptides matched other splicing isoforms. The best blastp match for PpS1V in the NCBI Protein Database is Nasonia vitripennis ovalbumin-related protein X isoform X12 (XP_001606111), which is a splicing isoform of gene LOC100122505. Ovalbumin is the main protein of egg white and belongs to the serpin superfamily (38). N. vitripennis gene LOC100122505 has 18 splicing isoforms in the NCBI Protein Database. All of these 18 isoforms have a serpin domain (cd00172). Other proteins in the P. puparum pooled fraction included venom allergen 3-like isoform 1 (comp45101_c0_seq1), α-amylase 1-like (comp22216_c0_seq1), hypothetical protein LOC100117405 (comp44498_c8_seq1), and endonuclease-like venom protein precursor (comp42418_c0_seq1). The presence of PpS1V in the pooled fraction was confirmed by Western blotting (Fig. 2F).
Sequence Analysis
The PpS1V transcript (comp44322_c1_seq8) is 1806 bp long with a 1197-bp open reading frame (ORF), a 220-bp 5′-non-coding region, and a 389-bp 3′-non-coding region containing a poly(A) tail. The ORF encodes a protein of 399 amino acids with a predicted signal peptide consisting of the first 19 residues. The calculated molecular mass of the mature protein without the signal peptide is 42.1 kDa, and the calculated isoelectric point is 4.8. To determine whether this isoform was caused by arbitrary assembly, the sequence was confirmed by PCR using cDNA from female adults and specific primers spanning the whole ORF (forward primer, 5′-GCGTTAGCGTCTGGAACTCA-3′; reverse primer, 5′-AAACAGATTGAGTTTGCGGA-3′). The results confirmed that this isoform is a true transcript in wasp female adults.
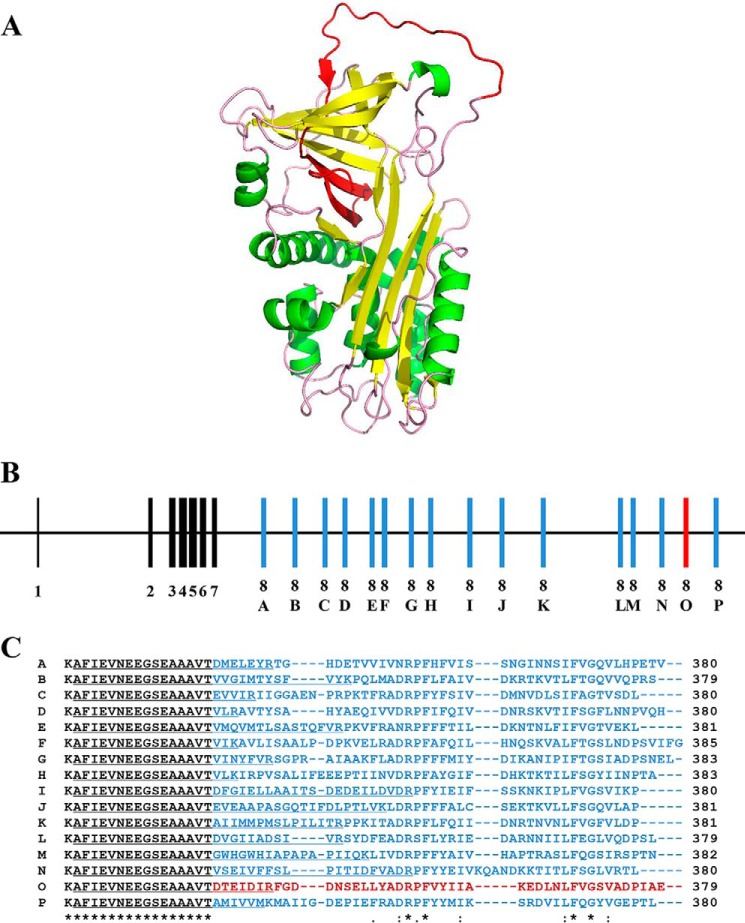
To investigate the gene structure of PpS1V, its genomic sequence was retrieved from the assembly of P. puparum genome,3 and the gene was named P. puparum serpin-1 (PpSerpin-1, GenBankTM accession number KX268468). Using available RNA sequencing data of P. puparum and 18 isoforms from N. vitripennis gene LOC100122505, 16 putative splicing isoforms were predicted for PpSerpin-1 and alphabetically named isoforms A–P; isoform O was the identified PpS1V in the isolated venom fractions (Fig. 3, A and B). These 16 isoforms share the first seven exons and differ in exon 8. This variable exon 8 includes the RCL, which determines the inhibitor selectivity of the serpin (Fig. 3, A and B). Although these 16 exon variants are different from each other in exon 8, the amino acid alignment showed that four positions were fully identical, and six more were identified as conservations in the variable region (Fig. 3C). These conserved positions may be important to the inhibitory function of serpin.
FIGURE 3.
Alternative splicing of the serpin gene for PpS1V with different versions of exon 8 producing different reactive center loops. A, predicted tertiary protein structure of PpS1V. The alternative splicing region in the PpS1V is colored in red. Yellow and green indicate α-helices and β-sheets, respectively. B, structure of the serpin gene for PpS1V. The constant coding regions from exons 1–7 are colored in black. Coding regions of variable exon 8 are colored in red for PpS1V and blue for other isoforms. C, amino acid alignment of the C termini of the 16 predicted isoforms starting with Thr335. The isoform-specific sequences are colored in red for PpS1V and blue for other isoforms. The first isoform-specific tryptic peptides are underlined. An “*” (asterisk) indicates positions that have a single, fully conserved residue. A “:” (colon) indicates conservation between groups of strongly similar properties. A “.” (period) indicates conservation between groups of weakly similar properties.
To reveal the possible evolutionary history of gene PpSerpin-1, phylogenetic analysis was performed using the amino acid sequence of PpS1V excluding the variable region coded by exon 8 (Fig. 4). If there were multiple splicing isoforms in serpins from other insects, the first isoforms were chosen for analysis. Our analysis shows that PpS1V is closely related to N. vitripennis antichymotrypsin-2 isoform X1 (XP_008201829.1), which is the first isoform of N. vitripennis gene LOC100122505. Alternative splicing was found in serpins from other parasitoid wasps, such as Copidosoma floridanum, Trichogramma pretiosum, and Orussus abietinus, and non-parasitoid hymenopterans, such as Apis mellifera, Athalia rosae, and Ceratosolen solmsi marchali. Splicing isoforms were also identified in serpins from several non-venom dipterans, coleopterans, lepidopterans, and hemipterans (Fig. 4).
FIGURE 4.
Phylogenetic tree of PpS1V. The amino acid sequence of PpS1V excluding the region coded by exon 8 was used for maximum likelihood tree construction. For serpins with multiple splicing isoforms in other insects, the first isoforms were chosen.
Isoform-specific RT-PCR
To investigate the distribution of PpS1V expression, specific RT-PCR was performed using isolated heads, thoraces, ovaries, venom glands, and carcasses (abdomen without ovary and venom apparatus) from female adults (Fig. 5). Result showed that PpS1V is highly differentially expressed in venom gland, although weak bands were also present in thorax and ovary. This indicates that PpS1V is more likely to be a real venom isoform rather than leakage during sample processing. However, because of retention of the intron between exons 8N and 8O in transcript of isoform N, it is not possible to design specific primers for real time quantitative PCR of isoform O (e.g. PpS1V) (Fig. 5).
FIGURE 5.
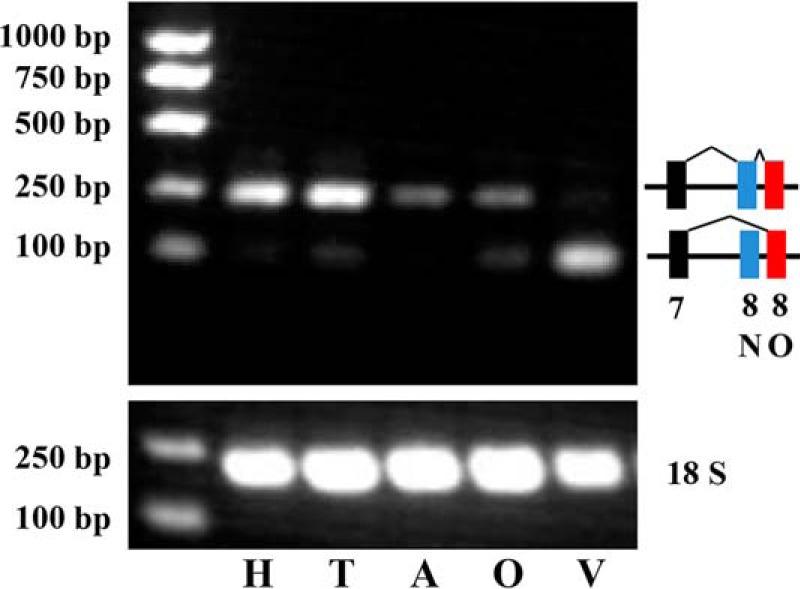
Expression pattern of PpS1V in wasp female adult. Isoform-specific PCR was conducted using cDNAs of different tissues dissected from wasp female adults. H, head; T, thorax; A, abdomen without ovary and venom apparatus; O, ovary; V, venom gland.
Effect of rPpS1V on Host Melanization
As serpins are primary regulators in PPO activation and PpS1V was identified with the highest score in the partially isolated venom fractions, we hypothesized that PpS1V could be responsible for the host's PPO inhibition in P. puparum venom. To test the function of PpS1V, we produced recombinant proteins and examined their effects on the host's PPO activation pathway.
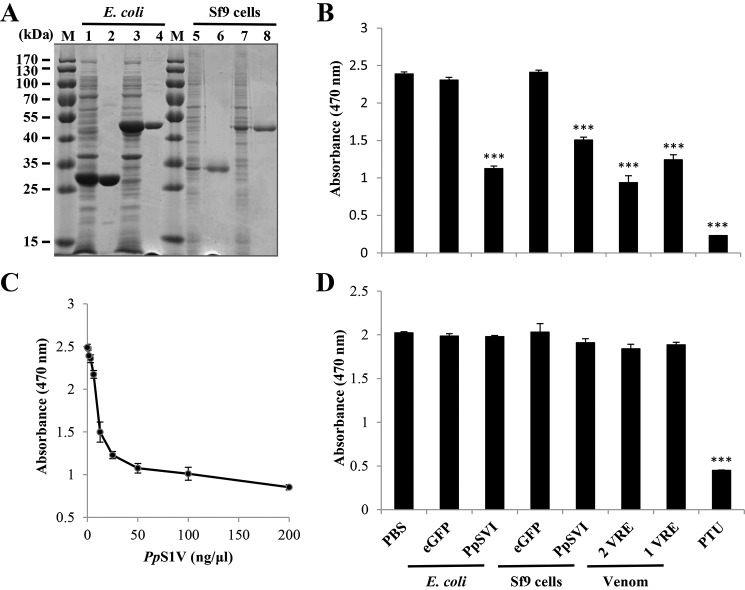
The coding sequence of mature PpS1V was cloned into pFast-HTB and pET-28a vectors. rPpS1V was successfully expressed in both Escherichia coli and the baculovirus system (Fig. 6A). The protein expressed in E. coli showed an observed mass of 48 kDa, which was close to the predicted size of 45.7 kDa. The protein expressed in Sf9 cells showed an observed mass of 45 kDa, which was close to the predicted size of 44.3 kDa. The predicted size of rPpS1V expressed in E. coli was bigger than that in the baculovirus system. This was caused by a longer introduced linker to the His tag at the N terminus in pET-28a vector.
FIGURE 6.
Inhibition of host PPO activation by rPpS1V. A, expression and purification of rPpS1V. Lane M, marker; lane 1, cell lysate of E. coli transformed with pET-28a/eGFP; lane 2, purified recombinant eGFP from E. coli; lane 3, cell lysate of E. coli transformed with pET-28a/PpS1V; lane 4, purified rPpS1V from E. coli; lane 5, cell lysate of Sf9 cells infected by baculovirus-eGFP; lane 6, purified recombinant eGFP from Sf9 cells; lane 7, cell lysate of Sf9 cells infected by baculovirus-PpS1V; lane 8, purified rPpS1V from Sf9 cells. B, effect of rPpS1V on the host's PO activity. Recombinant proteins (0.5 μg) were incubated with naïve P. rapae hemolymph for 5 min before adding M. luteus. C, dose-dependent suppression of host PO activity by rPpS1V purified from E. coli. D, effect of rPpS1V on PO activity of preactivated P. rapae hemolymph. Hemolymph was activated by incubation with M. luteus for 30 min before adding recombinant proteins. All values are presented as the mean (n = 3). Error bars represent S.E. Asterisks indicate significant differences from PBS as the control. ***, p < 0.001.
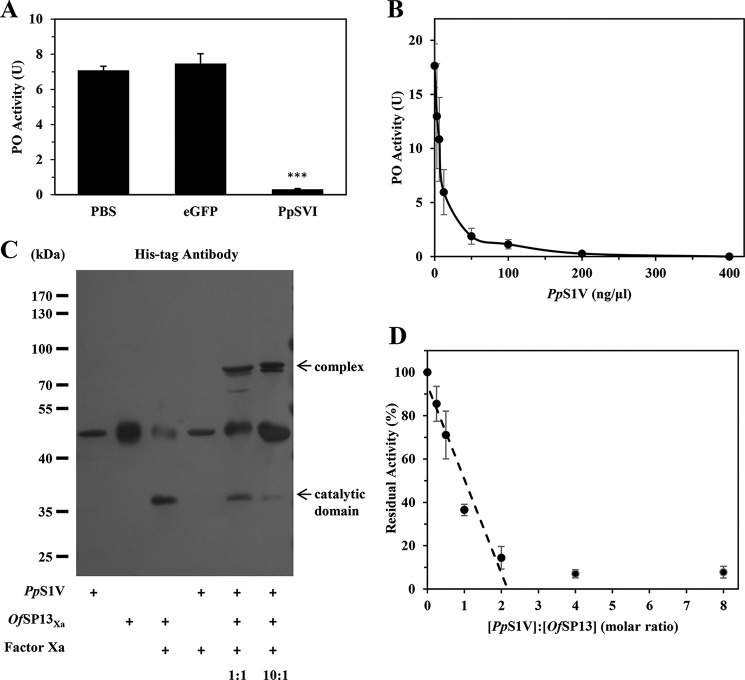
When proteins were incubated with host hemolymph before adding elicitor, rPpS1V (0.5 μg) from both E. coli and Sf9 cells showed activity to significantly suppress the melanization of host hemolymph (Fig. 6B; ANOVA: df = 7, F = 125.63, p < 0.001). rPpS1V purified from E. coli suppresses the melanization of host hemolymph in a dose-dependent manner (Fig. 6C). When proteins were incubated with preactivated host hemolymph, only the PO inhibitor phenylthiourea (PTU) showed significant suppression of PO activity (Fig. 6D; ANOVA: df = 7, F = 125.63, p < 0.001). There were no significant differences among other treatments without PTU (Fig. 6D; ANOVA: df = 6, F = 2.01, p = 0.14; PTU was excluded in this ANOVA). These results show that PpS1V and P. puparum venom inhibit the host hemolymph melanization by suppressing the PPO activation pathway rather than by directly inhibiting activated PO.
Identification of Complexes with Host Hemolymph Proteinases
Serpin inhibits serine proteases by forming covalent complexes with the target protease (13). To identify potential target proteases of PpS1V in host hemolymph, pulldown assays were conducted followed by LC-MS/MS identification.
When PpS1V was incubated with preactivated hemolymph, there were two nonspecific bands of 37 and 78 kDa detected by fluorescence staining and three specific bands recognized by the His tag antibody (Fig. 7A). Two of these three specific bands were present with minor size differences at ∼78 kDa, and the third band was below intact PpS1V with an apparent mass of 41 kDa. After PpS1V was incubated with naïve hemolymph 30 min before adding elicitor, one more specific band (64 kDa) was detected by fluorescence staining and recognized by the His tag antibody (Fig. 7A).
FIGURE 7.
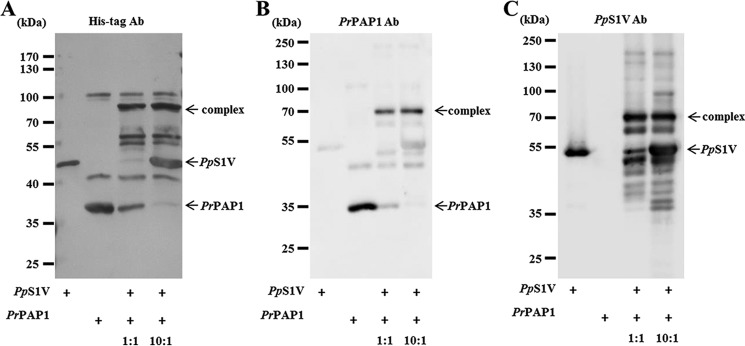
Identification of PrHP8 and PrPAP1 in the complexes of PpS1V formed with P. rapae hemolymph proteins. A, SDS-PAGE analysis of PpS1V pulldown samples followed by fluorescence staining and immunoblotting using antibody against His tag. Lane M, marker; lanes 1, hemolymph plus M. luteus followed by rPpS1V 30 min later; lanes 2, hemolymph plus rPpS1V followed by M. luteus 30 min later. B and C, amino acid sequences of mature PrHP8 (B) and PrPAP1 (C) with peptides identified by MS (highlighted residues). Trypsin cleavage sites are demarcated in blue. The sequence used for antibody production against PrPAP1 is underlined. D, phylogenetic tree of PrPAP and PrHP8. BmPPAE, B. mori prophenoloxidase-activating enzyme, NP_001036832.1; BmBAEEase, B. mori BAEEase (BAEEase is named based on its ability to hydrolyze the synthetic substrate N-benzoyl-l-arginine ethyl ester (BAEE)), BAE73254.1; DmEaster, D. melanogaster Easter, NP_524362.2; DmSPE, D. melanogaster Spätzle processing enzyme, NP_651168.1; DpHP8, D. plexippus hemolymph proteinase 8, EHJ73832.1; HaPPAF1, H. armigera prophenoloxidase-activating factor 1, ABU98654.1; MsHP8, AAV91006.1; MsPAP1, AAX18636.1; MsPAP2, AAL76085.1; MsPAP3, AAO74570.1; TtPCE, Tachypleus tridentatus proclotting enzyme, P21902.1. Red indicates identified PrPAP1 and PrHP8. Green indicates OfSP13, an ortholog to PrPAP1. E, specificity of PrPAP1 antibody against P. rapae hemolymph. F, SDS-PAGE analysis of PpS1V pulldown sample followed by immunoblotting using antibody (Ab) against PrPAP1.
The specific band at 41 kDa below the intact PpS1V mainly contained peptides of PpS1V (supplemental Table S1) and appeared to be cleaved PpS1V. The band at ∼78 kDa contained peptides of PpS1V and two host hemolymph proteinases (supplemental Table S1), comp47849_c0 (Fig. 7B) and comp44977_c0 (Fig. 7C), present in the translated P. rapae transcriptome. The best blastx match for comp47849_c0 in the NCBI Protein Database was Manduca sexta hemolymph proteinase 8 (AAV91006.1) with 59% identities and 72% positives, and the best match for comp44977_c0 was Danaus plexippus PPO-activating enzyme precursor (EHJ64711.1) with 60% identities and 72% positives. We therefore named comp47849_c0 and comp44977_c0 P. rapae hemolymph proteinase 8 (PrHP8) (Fig. 7, B and D) and P. rapae PPO-activating proteinase 1 (PrPAP1) (Fig. 7, C and D), respectively. Peptides of PrHP8 and PpS1V were also identified in a specific band at 64 kDa (supplemental Table S1).
According to our phylogenetic analysis (Fig. 7D), PrPAP1 clusters with M. sexta PAPs (39–41), Bombyx mori prophenoloxidase-activating enzyme (42), Helicoverpa armigera prophenoloxidase-activating factor 1 (43), and O. furnacalis serine protease 13 (OfSP13) (44), which activate PPO into PO. This indicates that PrPAP1 is likely to be involved in PPO activation in P. rapae. PrHP8 is clustered with Drosophila melanogaster Easter, D. melanogaster Spätzle processing enzyme (45), and M. sexta hemolymph proteinase 8 (MsHP8) (46), which activate pro-Spätzle in the Toll pathway.
We hypothesized that PpS1V inhibits PPO activation of P. rapae hemolymph by forming a complex with PrPAP1. The complex was detected in the PpS1V pulldown sample (Fig. 7, E and F) but was not present in the control sample.
Interaction between rPpS1V and rPrPAP1
To confirm the function of PrPAP1 in the PPO activation cascade and its interaction with PpS1V, rPrPAP1Xa was expressed in the baculovirus system. Activated PAPs, but not pro-PAPs, hydrolyze a colorimetric peptide substrate, acetyl-Ile-Glu-Ala-Arg-p-nitroanilide (IEARpNA) (39–41). The culture medium containing rPrPAP1Xa showed a high IEARase activity (data not shown), suggesting that rPrPAP1Xa had already been activated for an unknown reason. Compared with culture medium harvested from eGFP-expressing recombinant baculoviruses, culture medium containing activated rPrPAP1Xa strongly enhanced the melanization of P. rapae hemolymph with or without M. luteus (Fig. 8; for hemolymph without M. luteus, Student's t test: t = 19.4598, df = 4, p < 0.001; for hemolymph with M. luteus, ANOVA: df = 2, F = 746.663, p < 0.001). These results indicate that PrPAP1 acts in the PPO activation cascade, which can be abolished by excessive PpS1V (1 μg) (Fig. 8).
FIGURE 8.
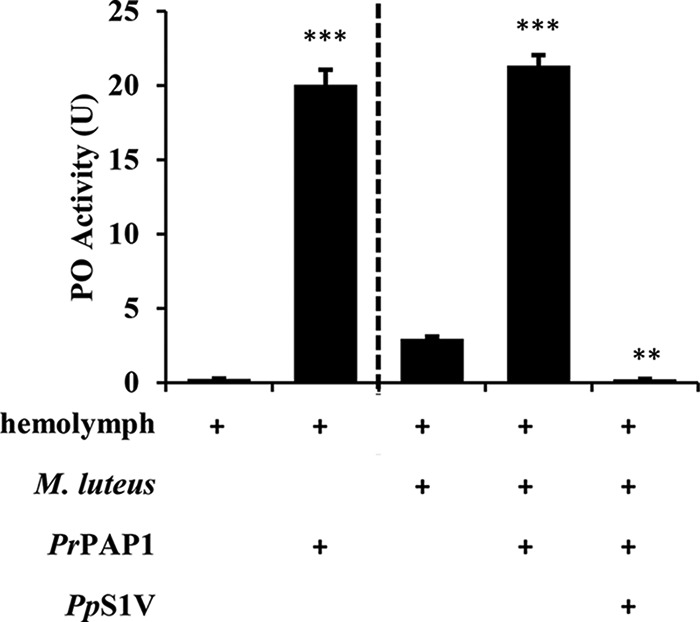
Effect of PrPAP1 on melanization of P. rapae hemolymph. Ten microliters of diluted P. rapae hemolymph was mixed with M. luteus (0.5 μg), purified PpS1V, or 10 μl of culture medium containing activated PrPAP1 (cultured Sf-900 II medium harvested from eGFP-expressing recombinant baculoviruses was used as control). After incubation at room temperature for 5–15 min, PO activity was assayed as described under “Experimental Procedures.” All values are presented as the mean (n = 3). Error bars represent S.E. Asterisks indicate significant differences from the control. **, p < 0.01; ***, p < 0.001.
A characteristic feature of serpin-proteinase interactions is the formation of a covalent complex of a serpin with its target proteinase (44, 47). Immunoblotting band intensities of serially diluted rPpS1V and rPrPAP1Xa indicate that the concentration of rPrPAP1Xa in culture medium was ∼5 ng/μl. After incubating rPpS1V with culture medium containing activated rPrPAP1Xa, complexes above 70 kDa were detected by Western blotting using antibodies against His tag, PrPAP1, and PpS1V (Fig. 9, A–C), indicating that PpS1V forms a covalent complex with PrPAP1.
FIGURE 9.
SDS-stable complex formation between rPpS1V and PrPAP1. Purified PpS1V was incubated with culture medium containing PrPAP1 at room temperature for 10 min. SDS-PAGE analyses were performed followed by immunoblotting by using antibody (Ab) against His tag (A), PrPAP1 (B), and PpS1V (C), respectively.
To further investigate the inhibition of PrPAP1 by PpS1V, we tested the hydrolysis inhibition of a colorimetric peptide substrate, IEARpNA, by PrPAP1. PrPAP1 activity decreased linearly as the PpS1V concentration increased (Fig. 10). The stoichiometry of inhibition is 2.3, indicating that PpS1V is an efficient inhibitor of PrPAP1.
FIGURE 10.
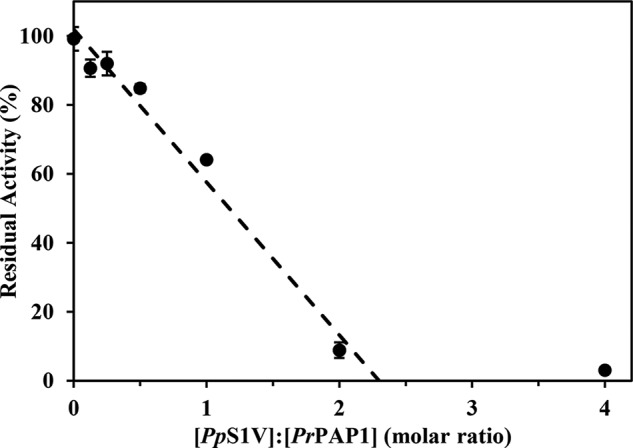
Stoichiometry of inhibition of PrPAP1 by PpS1V. rPpS1V was incubated with culture medium containing PrPAP1 at different molar ratios for 10 min at room temperature. The residual amidase activity was measured using IEARpNA as substrate. The intersection of a line generated by linear regression extrapolated to the x axis occurred at a molar ratio of 2.3. All values are presented as the mean (n = 3). Error bars represent S.E.
PpS1V inhibits O. furnacalis PPO Activation via a Complex with OfSP13, an Ortholog to PrPAP1
We also tested the effect of PpS1V on the hemolymph melanization of a non-natural host, the Asian corn borer, O. furnacalis. rPpS1V showed strong inhibition of the melanization of O. furnacalis hemolymph (Fig. 11A; ANOVA: df = 2, F = 133.818, p < 0.001) in a dose-dependent manner (Fig. 11B).
FIGURE 11.
PpS1V inhibits PPO activation of O. furnacalis hemolymph by forming complex with OfSP13, a homolog to PrPAP1. A, effect of PpS1V on PO activity of O. furnacalis hemolymph. B, dose-dependent suppression of O. furnacalis hemolymph PO activity by PpS1V. C, SDS-stable complex formation between PpS1V and OfSP13 detected by antibody against His tag. D, stoichiometry of inhibition of OfSP13 by PpS1V. OfSP13Xa was incubated with factor Xa at 37 °C for 6 h to produce active OfSP13. The intersection of a line generated by linear regression extrapolated to the x axis occurred at a molar ratio of 2.2. All values are presented as the mean (n = 3). Error bars represent S.E.
OfSP13 is the best reciprocal hit to PrPAP1 when reciprocal blastn was performed between O. furnacalis (50) and P. rapae transcriptome (33). OfSP13 has already been demonstrated to cleave PPO into PO and play an important role in PPO activation (44, 51). To test whether PpS1V forms a complex with OfSP13, we mixed rPpS1V with factor Xa-activated OfSP13Xa and recorded a higher molecular weight complex by immunoblotting (Fig. 11C). Activity of activated OfSP13Xa decreased linearly as PpS1V concentration increased (Fig. 11D). The stoichiometry of inhibition is 2.2 (Fig. 11D), similar to that of PrPAP1 by PpS1V. These results demonstrate that PpS1V inhibits O. furnacalis hemolymph melanization through forming a complex with OfSP13, an ortholog to PrPAP1.
Discussion
Inhibiting host melanization is an immunosuppression strategy in parasitoid wasps. However, quite diverse proteins could be recruited in different parasitoid systems for this convergent function. Virulence proteins include serine proteinase homolog (24, 49) and unrelated peptide Vn 4.6 (22) from Cotesia rubecula venom, extracellular superoxide dismutase (25) and serpin (23) from Leptopilina boulardi venom, and epidermal growth factor-like proteins (26, 48, 52) from Microplitis demolitor PDV. With an estimated 600,000 species, parasitoid wasps are one of the most abundant and diverse insect groups on earth (53). Such high diversity may present an untapped source of bioactive compounds with potential in pest control and drug discovery.
By assay-guided fractionation, a splicing isoform of serpin, PpS1V, was identified from P. puparum venom that inhibits host melanization. This serpin isoform was also detected in previous venom proteomic research of P. puparum (37). However, the previous study only used gene loci to calculate the expression level. Splicing isoforms were not considered, mainly because of the limitation of isoform expression estimation without a reference genome. Thus, PpS1V was not included in the reported 70-venom protein set.
In insects, serpin genes have evolved alternative splicing from a single gene to produce multiple functional serpins, which differ in RCL. Alternate splice forms were first reported in M. sexta serpin-1 with 12 isoforms (54, 55) and identified in other insects such as B. mori (56), Choristoneura fumiferana (57), and Anopheles gambiae (58). Based on our phylogenetic analysis, alternative splicing seems to be common in insect serpins. It is also reasonable to propose that the venom isoform of PpSerpin-1 evolved after the invention of the venom apparatus in Hymenoptera.
For the PpSerpin-1 gene, 16 splicing isoforms were predicted by bioinformatics approaches. Except for the venom isoform PpS1V, the tissue distributions and functions of the other 15 splicing isoforms remain unknown.
It is thought that venom proteins are mainly recruited through gene duplication from non-venom proteins followed by natural selections for adaptive traits (59). Several cases of gene duplications have been described in parasitoid venoms, including Ci-48a-like proteins and reprolysin-like proteins from M. demolitor (60), RhoGAP domain-containing proteins from Leptopilina (61), and γ-glutamyl transpeptidases from Aphidius ervi (62). However, gene duplication is not the only way venom has become diverse. The origins of venom proteins also include co-option of single copy genes (63), de novo synthesis of novel venom proteins from non-coding DNA, alternative splicing (64, 65), and lateral gene transfer from microorganisms (66). P. puparum serpin-1 provides an example of venom recruitment through alternative splicing. Another example is the sarco/endoplasmic reticulum calcium ATPase from Ganaspis sp. 1 that has a venom isoform and a non-venom isoform (67).
Pulldown results indicate that possible targets of PpS1V are hemolymph proteinase PrHP8 and PrPAP1. This is similar to serpin-1J in M. sexta (40, 47, 54, 55, 68). Manduca serpin-1J regulates PPO activation through inhibition of M. sexta PAP3 (MsPAP3) (40, 54) and the Toll pathway through inhibition of MsHP8 (47). PrPAP1 is clustered with proteinases involved in the PPO activation cascade. The role of PrPAP1 in P. rapae melanization and its interaction with PpS1V were also confirmed. In contrast, PrHP8 is clustered with proteinases involved in Toll pathway activation. Thus, PpS1V may also have a role in regulation of the Toll pathway.
In summary, we identified a splicing isoform of serpin in P. puparum venom. This serpin isoform shows activity to inhibit the host's PPO activation through forming a complex with PrPAP1. This study provides a case of parasitoid venom evolution through alternative splicing and sheds light on the mechanisms by which parasitoid venom suppresses host immunity.
Experimental Procedures
Insect Rearing
Laboratory cultures of P. puparum and its host P. rapae were maintained at 25 °C with a photoperiod of 14:10 h (light:dark) as described previously (27, 28). Once emerged, the wasps were collected and held in glass containers without hosts and fed ad libitum on 20% (v/v) honey solution to lengthen life span. P. rapae was reared in a greenhouse on cabbages grown throughout the year. Larvae of O. furnacalis were maintained on an artificial diet at 28 °C under a relative humidity of 70–90% and a photoperiod of 16:8 h (light:dark) as described previously (44, 51).
Venom Protein Extraction
Mated female wasps aged 3–7 days after eclosion were anesthetized on ice and then dissected in PBS on an ice plate under a stereoscope (Olympus). The venom reservoirs were washed several times using PBS and then transferred to an Eppendorf tube on ice. After centrifugation at 16,000 × g and 4 °C for 10 min, the supernatant was filtered with a 0.22-μm Millipore (Billerica, MA) filter and stored at −80 °C until use.
Hemolymph Collection
The plasma was collected from naïve P. rapae pupae or larvae on ice and diluted four times in anticoagulant (4 mm sodium chloride, 40 mm potassium chloride, 8 mm EDTA, 9.5 mm citric acid, 27 mm sodium citrate, 5% sucrose, 0.1% polyvinylpyrrolidone, and 1.7 mm PIPES, pH 6.8). After removing hemocytes by centrifugation at 3000 × g and 4 °C for 10 min, hemolymph samples were collected and stored at −80 °C for further assays.
Phenoloxidase Activity Assay
For the PO activity assay of P. rapae hemolymph, 15 μl of diluted hemolymph was mixed with 5 μl of inhibitor (venom protein or saturated PTU) and 5 μl of elicitor (0.1 μg/μl M. luteus). For the PO activity assay of O. furnacalis hemolymph, 2 μl of larval hemolymph was mixed with 8 μl of recombinant proteins. Then samples were placed on a rotary mixer at 25 °C for around 10 min. After adding 800 μl of substrate solution (20 mm dopa in PBS, pH 6.5), samples (200 μl) were measured at A470 in 96-well plates for 20 min (Varioskan Flash multimode reader, Thermo Scientific). One unit of PO activity was defined as 0.001 ΔA470/min. To increase the throughput of the PO assay, the assay was further modified and conducted in 384-well plates. Ten microliters of diluted P. rapae hemolymph was mixed with 5 μl of proteins, 5 μl of elicitor (0.1 μg/μl M. luteus), and 5 μl of substrate solution (50 mm l-dopa in PBS, pH 7.5). The plates were measured at A470 at 25 °C every 5 min for 2 h using a Varioskan Flash multimode reader. Statistical analyses were performed using Data Processing System (DPS) v13.5 (69). Comparisons of two samples were conducted using Student's t test. Comparisons of multiple samples were performed using ANOVA followed by Tukey's multiple comparison test.
Fractionation of Venom Proteins
Venom protein was diluted with buffer A (25 mm Tris-HCl, pH 7.5) to 2 ml and loaded onto a Bio SAX column (5 μm, 4.6 × 50 mm; Agilent Technologies, Wilmington, DE) using a Biologic Duo-Flow high performance chromatography system (Bio-Rad). Proteins were eluted at a flow rate of 0.5 ml/min with a gradient of buffer B (25 mm Tris-HCl, 1 m NaCl, pH 7.5) against buffer A. Proteins were monitored by absorbance at 280 nm. Fractions of 200 μl were collected in a deep well plate (Eppendorf) using a BioLogic BioFrac fraction collector (Bio-Rad) and then desalted using a Zeba spin desalting plate (Thermo Scientific) according to the manufacturers' instructions. The desalted fractions were stored at −80 °C until use.
Protein Identification of Isolated Venom Protein
To identify potential candidates with PO inhibitory activity, a pooled fraction of 200 μl was digested by trypsin using the filter-aided sample preparation method (70). Then mass spectrometric analysis was performed using an Easy nLC HPLC system (Thermo Scientific) followed by Q-Exactive (Thermo Finnigan). In this study, samples were first loaded on a Thermo Scientific EASY column (5 μm, 2 cm × 100 μm, C18) and then separated on another Thermo Scientific EASY column (3 μm, 75 μm × 100 mm, C18) with a flow rate of 250 nl/min. Buffer A was water with 0.1% formic acid, buffer B was 84% acetonitrile with 0.1% formic acid, and the gradient was from 0–50% buffer B in 50 min and then 50–100% buffer B in 4 min. The charge-to-mass ratios of peptides and fractions of peptides were collected 10 times after every full scan. The resulting MS/MS spectra were searched against the translated P. puparum transcriptomic database (37) using Mascot software (71). The maximum number of missed cleavages was set as 2. Carbamidomethyl of cysteine and oxidation of methionine were set as fixed and variable modifications, respectively. Peptide confidence ≤0.01 was used to filter the peptide identification. This part of the work was done by Shanghai Applied Protein Technology Co., Ltd. (Shanghai, China).
Specific RT-PCR
Female wasps were dissected in Ringer's saline (182 mm KCl, 46 mm NaCl, 3 mm CaCl2, 10 mm Tris-HCl) with 1 unit/μl RNase inhibitor (TOYOBO, Osaka, Japan) on an ice plate under a stereoscope (Olympus). The total RNA was extracted from different tissues of female wasps using TRIzol reagent according to the manufacturer's protocol and then was reverse transcribed using TransScript One-step gDNA Removal and cDNA Synthesis SuperMix (TransGen, China) with random primers. The isoform-specific primers were designed to span exon 7 and exon 8 using PerlPrimer V1.1.21 (72) and are listed in Table 1. The isoform-specific sequence of PpS1V was amplified by PCR using TransTaq HiFi DNA polymerase and confirmed by sequencing.
TABLE 1.
Primers used for construction of recombinant plasmids or specific RT-PCR
| Insert DNA or linear vector | Forward primer | Reverse primer |
|---|---|---|
| Linear pFast-Bac-HTB | 5′-GGTCGTTGGGATATCGTAATCGTGATG-3′ | 5′-CTCGAGGCATGCGGTACCAAGCTTG-3′ |
| pET-28a-PpS1V | 5′-AGCAAATGGGTCGCGGATCCGAGGACAAGGCAGTGGAGGC-3′ | 5′-GTGGTGGTGCTCGAGTTATTCAGCGATCGGATCAG-3′ |
| pFast-Bac-HTB-PpS1V | 5′-GATATCCCAACGACCGAGGACAAGGCAGTGGAGGC-3′ | 5′-ACCGCATGCCTCGAGTTATTCAGCGATCGGATCAG-3′ |
| pET-28a-eGFP | 5′-AGCAAATGGGTCGCGGATCCATGGTGAGCAAGGGCGAGGAG-3′ | 5′-GTGGTGGTGCTCGAGTTACTTGTACAGCTCGTCCATG-3′ |
| pFast-Bac1-PrPAP1 | 5′-CCACCATCGGGCGCGGATCCATGAAGTGTTTCATTGTG-3′ | 5′-TTCTCGACAAGCTTGGTACCTTAGTGATGGTGATGGTGATGGCCTCCGCCAACATTAGTAGATTGTAG-3′ |
| pFast-Bac1-PrPAP1Xa | 5′-AGATCCGCCCTTCTATTTCAATACCACATTTGCC-3′ | 5′-TGAAATAGAAGGGCGGATCTTTGGAGGCATAATTG-3′ |
| Isoform O (specific RT-PCR) | 5′-AAGCAAAGTTACGCAGAAGG-3′ | 5′-GTTGTCATCACCGAATCTAATGTC-3′ |
| 18S rRNA (specific RT-PCR) | 5′-CGAGCGATGAACCGACAG-3′ | 5′-CGGGGAGGTAGTGACGAA-3′ |
Sequence Analysis
Signal peptides were predicted using SignalP 4.1 (73). Gene structure was predicted by Splign (74) using transcript variants of N. vitripennis gene LOC100122505 and transcripts from the P. puparum transcriptome. Multiple amino acid sequence alignments were performed using MUSCLE v3.8 (75). Regions with low similarities were manually removed. Phylogenetic analysis was conducted by PhyML version 20131022 using default settings (76). For phylogenetic tree construction of PpS1V, the variable region at the C terminus was excluded. The structure of PpS1V was modeled by SWISS-MODEL (77, 78) using Protein Data Bank code 2H4R as the template and visualized using PyMOL v1.7.0.0.
Recombinant Protein Expression and Purification
Recombinant plasmids were generated using the ClonExpress One Step Cloning kit (Vazyme, China). To perform recombination cloning, both insert DNA fragments and linear pFast-Bac-HTB vector were amplified by PCR. The primers used are listed in Table 1. Linear pET-28a vector was generated by digestion with BamHI and XhoI (TaKaRa, Dalian, China), and linear pFast-Bac1 vector was generated by digestion with BamHI and KpnI (TaKaRa). In PrPAP1, the predicted activation site RSDR122 was mutated into IEGR122 using the Mut Express II Fast Mutagenesis kit (Vazyme, China) and plasmid pFast-Bac1-PrPAP1 as the template. IEGR is a preferred cleavage site for commercially available factor Xa protease, which theoretically cleaves PrPAP1Xa at IEAR122 and produces active PrPAP1. For protein expression in E. coli, recombinant pET-28a plasmids were transferred into BL21(DE3) and confirmed by sequencing. After growth in autoinduction medium containing 100 μg/μl kanamycin at 20 °C for 48 h, E. coli cells were harvested by centrifugation at 10,000 × g and 4 °C for 10 min. For recombinant protein expression using the baculovirus system, recombinant pFast-Bac plasmids were used to generate recombinant baculoviruses according to the manufacturer's instructions. Sf9 cells (2 × 106 cells/ml) in 100 ml of Sf-900 II serum-free medium (Invitrogen) were infected with the recombinant baculovirus and incubated at 28 °C with shaking at 90 rpm in 250-ml flasks. The Sf9 cells were harvested 72 h after infection by centrifugation at 1000 × g for 20 min at 4 °C. For secreted protein expression, culture medium containing PrPAP1Xa was harvested 96 h after infection. The recombinant proteins in both E. coli and Sf9 cells were purified using the His-Bind Purification kit (Novagen) according to the manufacturer's instruction. The concentration of protein was determined using method of Bradford (79).
Pulldown Assay
One milliliter of diluted P. rapae hemolymph containing 50 μl of saturated PTU was incubated with 100 μl of M. luteus (1 μg/μl) and 5 μl of recombinant protein (2 μg/μl) at 4 °C overnight. After centrifugation at 12,000 × g at 4 °C for 20 min, the supernatant was incubated with 25 μl of cOmplete His tag purification resins (Roche Applied Science) at 4 °C for 2 h. The hemolymph with resins was then loaded onto spin columns and spun at 1000 × g for 2 min followed by reloading and spinning several times until all the sample had been loaded. The resins were washed three times with 300 μl of washing buffer (1 m NaCl, 120 mm imidazole, 40 mm Tris-HCl, pH 7.9) before eluting with 50 μl of elution buffer (1 m imidazole, 0.5 m NaCl, 20 mm Tris-HCl, pH 7.9). Eluted proteins were then analyzed by SDS-PAGE followed by Lumitein (Biotium, Hayward, CA) protein gel staining and immunoblotting.
Protein Identification in Pulldown Complexes
To identify potential target proteins in pulldown complexes, the excised gel slices were in gel-digested by trypsin and lyophilized separately followed by mass spectrometry on a 1DLC-LTQ-Velos instrument (Thermo Finnigan, San Jose, CA) as described previously (37). Briefly, samples were desalted on Zorbax 300 SB-C18 (Agilent Technologies) and then separated on an RP-C18 column (150-μm inner diameter, 150-mm length) (Column Technology Inc., Fremont, CA). Buffer A, buffer B, and the gradient were the same as mentioned above. The charge-to-mass ratios of peptides and fractions of peptides were collected 20 times after every full scan. The resulting MS/MS spectra were searched using BioworksBrowser 3.3 (Thermo Electron, Bremen, Germany) against the translated P. rapae transcriptome with the manually added bait sequence (e.g. PpS1V). Carbamidomethylation of cysteine and oxidation of methionine were set as fixed and variable modifications, respectively. The number of maximum missed cleavages was set to 2. ΔCN (≥0.1) and cross-correlation scores (Xcorr; one charge ≥1.9, two charges ≥2.2, and three charges ≥3.75) were used to filter the peptide identification. The criterion unique identified peptides ≥2 was used to filter the protein identification. This part was done by Shanghai Applied Protein Technology Co., Ltd.
Inhibition of Amidase Activity by PpS1V
For inhibition assays, PpS1V was incubated with factor Xa-activated OfSP13Xa or culture medium containing activated PrPAP1 at different molar ratios. After incubation at room temperature for 10 min, residual amidase activity was measured with 200 μl of 50 μm IEARpNA in 100 mm Tris-HCl, pH 8.0, 100 mm NaCl, 5 mm CaCl2 as colorimetric substrate and monitored at A405 in a microplate reader (BioTek Instruments, Winooski, VT). One unit of amidase activity was defined as 0.001 ΔA405/min. OfSP13Xa was activated by treatment at 37 °C for 6 h with bovine factor Xa protease (New England BioLabs, Ipswich, MA).
Antibody Production
Antibodies against PrPAP1 were generated using PolyExpress Custom Polyclonal Antibody Production Service (GenScript, Nanjin, China). Briefly, peptide CHSGFSDSDNSRPDD was synthesized and conjugated with keyhole limpet hemocyanin for generating rabbit polyclonal antibody (Fig. 7C). Antibodies against PrPAP1 were further subjected to antigen affinity purification using a synthesized peptide coupling column. Rabbit polyclonal antibodies against PpS1V were generated using purified rPpS1V from E. coli and purified using the Montage Antibody Purification kit with PROSEP-A media (Millipore).
Western Blotting
Proteins were separated by 12% SDS-PAGE and then transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad) using a Mini-ProTEAN Tetra system (Bio-Rad) at 16 V for 16 h. The PVDF membrane was blocked and washed. For detection of PrPAP1 and PpS1V, antibodies against PrPAP1 and PpS1V (diluted 1:1000) were used as primary antibodies followed by goat anti-rabbit IgG-horseradish peroxidase (HRP) conjugate (Sigma-Aldrich; diluted 1:5000) as the secondary antibody. For detection of recombinant proteins with His tag, the primary antibody was THETM His tag mouse antibody (GenScript, Nanjin, China; diluted 1:2000). The secondary antibody was HRP- or alkaline phosphatase-conjugated goat anti-mouse IgG antibody (GenScript; diluted 1:2000). When HRP-conjugated secondary antibody was used, protein bands on membranes were detected using ECL Western blotting substrate (Promega, Madison, WI) and imaged using the Chemi Doc-It 600 Imaging System (UVP, Cambridge, UK). When alkaline phosphatase-conjugated secondary antibody was used, protein bands on membranes were detected using 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium staining buffer containing 165 μg/ml 5-bromo-4-chloro-3-indolyl phosphate and 330 μg/ml nitro blue tetrazolium in 100 mm Tris, pH 9.5, 150 mm NaCl, 5 mm MgCl2.
Author Contributions
Z. Y. conducted most of the experiments, analyzed the results, and wrote most of the paper. Q. F. and Y. L. conducted experiments on gene cloning and specific RT-PCR. S. X. and L. Y. conducted experiments for recombinant protein expression and purification. F. W. performed sequence analysis. G. Y. and C. A. conceived and designed the research. J. H. W. and C. A. provided input on data interpretation. G. Y. and J. H. W. wrote the paper with Z. Y.
Supplementary Material
Acknowledgments
We thank Chuan-Xi Zhang, working at the Institute of Insect Sciences, Zhejiang University, for generously providing recombinant pFast-HTE vector. We also thank Sha-sha Zhang, working at the College of Agriculture and Biotechnology, China Agricultural University, for generously providing purified recombinant OfSP13Xa protein. We thank David Stanley, working at the United States Department of Agriculture, for carefully revising and polishing the manuscript.
This work was supported by National Natural Science Foundation of China Grants 31272098 and 31472038, Major International (Regional) Joint Research Project of National Natural Science Foundation Grant 31620103915, and China National Science Fund for Distinguished Young Scholars Grant 31025021 and by National Institutes of Health Grant RO1GM098667 (to J. H. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) KX268468

This article contains supplemental Table S1.
Z. Yan, Q. Fang, S. Xiao, L. Yang, F. Wang, J. H. Werren, and G. Ye, unpublished data.
- PPO
- prophenoloxidase
- PO
- phenoloxidase
- PAP
- prophenoloxidase-activating proteinase
- HP
- hemolymph proteinase
- RCL
- reactive center loop
- PDV
- polydnavirus
- PTU
- phenylthiourea
- VRE
- venom reservoir equivalent
- eGFP
- enhanced green fluorescent protein
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- ANOVA
- analysis of variance
- PpS1V
- P. puparum serpin-1, venom isoform
- rPpS1V
- recombinant PpS1V
- Pr
- P. rapae
- OfSP13
- O. furnacalis serine protease 13
- Dopa
- 3,4-dihydroxyphenylalanine
- Ms
- M. sexta
- IEARpNA
- acetyl-Ile-Glu-Ala-Arg-p-nitroanilide
- POI
- phenoloxidase inhibitor.
References
- 1. Gillespie J. P., Kanost M. R., and Trenczek T. (1997) Biological mediators of insect immunity. Annu. Rev. Entomol. 42, 611–643 [DOI] [PubMed] [Google Scholar]
- 2. Bangham J., Jiggins F., and Lemaitre B. (2006) Insect immunity: the post-genomic era. Immunity 25, 1–5 [DOI] [PubMed] [Google Scholar]
- 3. Vavricka C. J., Christensen B. M., and Li J. (2010) Melanization in living organisms: a perspective of species evolution. Protein Cell 1, 830–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Takahashi D., Garcia B. L., and Kanost M. R. (2015) Initiating protease with modular domains interacts with β-glucan recognition protein to trigger innate immune response in insects. Proc. Natl. Acad. Sci. U.S.A. 112, 13856–13861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kanost M. R., and Jiang H. (2015) Clip-domain serine proteases as immune factors in insect hemolymph. Curr. Opin. Insect Sci. 11, 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carton Y., Poirié M., and Nappi A. J. (2008) Insect immune resistance to parasitoids. Insect Sci. 15, 67–87 [Google Scholar]
- 7. Nappi A., Poirié M., and Carton Y. (2009) The role of melanization and cytotoxic by-products in the cellular immune responses of Drosophila against parasitic wasps. Adv. Parasitol. 70, 99–121 [DOI] [PubMed] [Google Scholar]
- 8. Kanost M. R. (1999) Serine proteinase inhibitors in arthropod immunity. Dev. Comp. Immunol. 23, 291–301 [DOI] [PubMed] [Google Scholar]
- 9. Kanost M. R., and Jiang H. B. (1997) Serpins from an insect, Manduca sexta, in Chemistry and Biology of Serpins (Church F. C., Cunningham D. D., Ginsburg D., Hoffman M., Stone S. R., and Tollefsen D. M., eds) pp. 155–161, Springer, New York: [DOI] [PubMed] [Google Scholar]
- 10. Nappi A. J., Frey F., and Carton Y. (2005) Drosophila serpin 27A is a likely target for immune suppression of the blood cell-mediated melanotic encapsulation response. J. Insect Physiol. 51, 197–205 [DOI] [PubMed] [Google Scholar]
- 11. Gulley M. M., Zhang X., and Michel K. (2013) The roles of serpins in mosquito immunology and physiology. J. Insect Physiol. 59, 138–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Law R. H., Zhang Q., McGowan S., Buckle A. M., Silverman G. A., Wong W., Rosado C. J., Langendorf C. G., Pike R. N., Bird P. I., and Whisstock J. C. (2006) An overview of the serpin superfamily. Genome Biol. 7, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gettins P. G. (2002) Serpin structure, mechanism, and function. Chem. Rev. 102, 4751–4804 [DOI] [PubMed] [Google Scholar]
- 14. Whisstock J. C., and Bottomley S. P. (2006) Molecular gymnastics: serpin structure, folding and misfolding. Curr. Opin. Struct. Biol. 16, 761–768 [DOI] [PubMed] [Google Scholar]
- 15. Gubb D., Sanz-Parra A., Barcena L., Troxler L., and Fullaondo A. (2010) Protease inhibitors and proteolytic signalling cascades in insects. Biochimie 92, 1749–1759 [DOI] [PubMed] [Google Scholar]
- 16. Celli J., and Finlay B. B. (2002) Bacterial avoidance of phagocytosis. Trends Microbiol. 10, 232–237 [DOI] [PubMed] [Google Scholar]
- 17. Nielsen-LeRoux C., Gaudriault S., Ramarao N., Lereclus D., and Givaudan A. (2012) How the insect pathogen bacteria Bacillus thuringiensis and Xenorhabdus/Photorhabdus occupy their hosts. Curr. Opin. Microbiol. 15, 220–231 [DOI] [PubMed] [Google Scholar]
- 18. Dillman A. R., Chaston J. M., Adams B. J., Ciche T. A., Goodrich-Blair H., Stock S. P., and Sternberg P. W. (2012) An entomopathogenic nematode by any other name. PLoS Pathog. 8, e1002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Asgari S. (2006) Venom proteins from polydnavirus-producing endoparasitoids: their role in host-parasite interactions. Arch. Insect Biochem. Physiol. 61, 146–156 [DOI] [PubMed] [Google Scholar]
- 20. Schmidt O., Theopold U., and Strand M. (2001) Innate immunity and its evasion and suppression by hymenopteran endoparasitoids. Bioessays 23, 344–351 [DOI] [PubMed] [Google Scholar]
- 21. Poirié M., Carton Y., and Dubuffet A. (2009) Virulence strategies in parasitoid Hymenoptera as an example of adaptive diversity. C. R. Biol. 332, 311–320 [DOI] [PubMed] [Google Scholar]
- 22. Asgari S., Zareie R., Zhang G., and Schmidt O. (2003) Isolation and characterization of a novel venom protein from an endoparasitoid, Cotesia rubecula (Hym: Braconidae). Arch. Insect Biochem. Physiol. 53, 92–100 [DOI] [PubMed] [Google Scholar]
- 23. Colinet D., Dubuffet A., Cazes D., Moreau S., Drezen J. M., and Poirié M. (2009) A serpin from the parasitoid wasp Leptopilina boulardi targets the Drosophila phenoloxidase cascade. Dev. Comp. Immunol. 33, 681–689 [DOI] [PubMed] [Google Scholar]
- 24. Asgari S., Zhang G., Zareie R., and Schmidt O. (2003) A serine proteinase homolog venom protein from an endoparasitoid wasp inhibits melanization of the host hemolymph. Insect Biochem. Mol. Biol. 33, 1017–1024 [DOI] [PubMed] [Google Scholar]
- 25. Colinet D., Cazes D., Belghazi M., Gatti J.-L., and Poirié M. (2011) Extracellular superoxide dismutase in insects: characterization, function, and interspecific variation in parasitoid wasp venom. J. Biol. Chem. 286, 40110–40121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beck M. H., and Strand M. R. (2007) A novel polydnavirus protein inhibits the insect prophenoloxidase activation pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 19267–19272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cai J., Ye G. Y., and Hu C. (2004) Parasitism of Pieris rapae (Lepidoptera: Pieridae) by a pupal endoparasitoid, Pteromalus puparum (Hymenoptera: Pteromalidae): effects of parasitization and venom on host hemocytes. J. Insect Physiol. 50, 315–322 [DOI] [PubMed] [Google Scholar]
- 28. Zhang Z., Ye G. Y., Cai J., and Hu C. (2005) Comparative venom toxicity between Pteromalus puparum and Nasonia vitripennis (Hymenoptera: Pteromalidae) toward the hemocytes of their natural hosts, non-target insects and cultured insect cells. Toxicon 46, 337–349 [DOI] [PubMed] [Google Scholar]
- 29. Zhu J. Y., Ye G. Y., and Hu C. (2008) Morphology and ultrastructure of the venom apparatus in the endoparasitic wasp Pteromalus puparum (Hymenoptera: Pteromalidae). Micron 39, 926–933 [DOI] [PubMed] [Google Scholar]
- 30. Zhu J. Y., Ye G. Y., Dong S. Z., Fang Q., and Hu C. (2009) Venom of Pteromalus puparum (Hymenoptera: Pteromalidae) induced endocrine changes in the hemolymph of its host, Pieris rapae (Lepidoptera: Pieridae). Arch. Insect Biochem. Physiol. 71, 45–53 [DOI] [PubMed] [Google Scholar]
- 31. Fang Q., Wang L., Zhu Y., Stanley D. W., Chen X., Hu C., and Ye G. (2011) Pteromalus puparum venom impairs host cellular immune responses by decreasing expression of its scavenger receptor gene. Insect Biochem. Mol. Biol. 41, 852–862 [DOI] [PubMed] [Google Scholar]
- 32. Fang Q., Wang L., Zhu J., Li Y., Song Q., Stanley D. W., Akhtar Z. R., and Ye G. (2010) Expression of immune-response genes in Lepidopteran host is suppressed by venom from an endoparasitoid, Pteromalus puparum. BMC Genomics 11, 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu Y., Fang Q., Liu Y., Gao L. F., Yan Z. C., and Ye G. Y. (2015) The endoparasitoid Pteromalus puparum influences host gene expression within first hour of parasitization. Arch. Insect Biochem. Physiol. 90, 140–153 [DOI] [PubMed] [Google Scholar]
- 34. Fang Q., Wang F., Gatehouse J. A., Gatehouse A. M., Chen X. X., Hu C., and Ye G. Y. (2011) Venom of parasitoid, Pteromalus puparum, suppresses host, Pieris rapae, immune promotion by decreasing host C-type lectin gene expression. PLoS One 6, e26888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Söderhäll I., Wu C., Novotny M., Lee B. L., and Söderhäll K. (2009) A novel protein acts as a negative regulator of prophenoloxidase activation and melanization in the freshwater crayfish Pacifastacus leniusculus. J. Biol. Chem. 284, 6301–6310 [DOI] [PubMed] [Google Scholar]
- 36. Asgari S., and Rivers D. B. (2011) Venom proteins from endoparasitoid wasps and their role in host-parasite interactions. Annu. Rev. Entomol. 56, 313–335 [DOI] [PubMed] [Google Scholar]
- 37. Yan Z., Fang Q., Wang L., Liu J., Zhu Y., Wang F., Li F., Werren J. H., and Ye G. (2016) Insights into the venom composition and evolution of an endoparasitoid wasp by combining proteomic and transcriptomic analyses. Sci. Rep. 6, 19604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Benarafa C., and Remold-O'Donnell E. (2005) The ovalbumin serpins revisited: Perspective from the chicken genome of clade B serpin evolution in vertebrates. Proc. Natl. Acad. Sci. U.S.A. 102, 11367–11372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jiang H., Wang Y., Yu X. Q., and Kanost M. R. (2003) Prophenoloxidase-activating proteinase-2 from hemolymph of Manduca sexta. J. Biol. Chem. 278, 3552–3561 [DOI] [PubMed] [Google Scholar]
- 40. Jiang H., Wang Y., Yu X. Q., Zhu Y., and Kanost M. (2003) Prophenoloxidase-activating proteinase-3 (PAP-3) from Manduca sexta hemolymph: a clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem. Mol. Biol. 33, 1049–1060 [DOI] [PubMed] [Google Scholar]
- 41. Jiang H., Wang Y., and Kanost M. R. (1998) Pro-phenol oxidase activating proteinase from an insect, Manduca sexta: a bacteria-inducible protein similar to Drosophila easter. Proc. Natl. Acad. Sci. U.S.A. 95, 12220–12225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Satoh D., Horii A., Ochiai M., and Ashida M. (1999) Prophenoloxidase-activating enzyme of the silkworm, Bombyx mori. Purification, characterization, and cDNA cloning. J. Biol. Chem. 274, 7441–7453 [DOI] [PubMed] [Google Scholar]
- 43. Lee S. Y., Cho M. Y., Hyun J. H., Lee K. M., Homma K. I., Natori S., Kawabata S. I., Iwanaga S., and Lee B. L. (1998) Molecular cloning of cDNA for pro-phenol-oxidase-activating factor I, a serine protease is induced by lipopolysaccharide or 1,3-β-glucan in Coleopteran insect, Holotrichia diomphalia larvae. Eur. J. Biochem. 257, 615–621 [DOI] [PubMed] [Google Scholar]
- 44. Chu Y., Liu Y., Shen D., Hong F., Wang G., and An C. (2015) Serine proteases SP1 and SP13 mediate the melanization response of Asian corn borer, Ostrinia furnacalis, against entomopathogenic fungus Beauveria bassiana. J. Invertebr. Pathol. 128, 64–72 [DOI] [PubMed] [Google Scholar]
- 45. Jang I. H., Chosa N., Kim S. H., Nam H. J., Lemaitre B., Ochiai M., Kambris Z., Brun S., Hashimoto C., Ashida M., Brey P. T., and Lee W. J. (2006) A spätzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev. Cell 10, 45–55 [DOI] [PubMed] [Google Scholar]
- 46. An C., Ishibashi J., Ragan E. J., Jiang H., and Kanost M. R. (2009) Functions of Manduca sexta hemolymph proteinases HP6 and HP8 in two innate immune pathways. J. Biol. Chem. 284, 19716–19726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. An C., Ragan E. J., and Kanost M. R. (2011) Serpin-1 splicing isoform J inhibits the proSpätzle-activating proteinase HP8 to regulate expression of antimicrobial hemolymph proteins in Manduca sexta. Dev. Comp. Immunol. 35, 135–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lu Z., Beck M. H., and Strand M. R. (2010) Egf1.5 is a second phenoloxidase cascade inhibitor encoded by Microplitis demolitor bracovirus. Insect Biochem. Mol. Biol. 40, 497–505 [DOI] [PubMed] [Google Scholar]
- 49. Zhang G., Lu Z. Q., Jiang H., and Asgari S. (2004) Negative regulation of prophenoloxidase (proPO) activation by a clip-domain serine proteinase homolog (SPH) from endoparasitoid venom. Insect Biochem. Mol. Biol. 34, 477–483 [DOI] [PubMed] [Google Scholar]
- 50. Liu Y., Shen D., Zhou F., Wang G., and An C. (2014) Identification of immunity-related genes in Ostrinia furnacalis against entomopathogenic fungi by RNA-seq analysis. PLoS One 9, e86436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chu Y., Zhou F., Liu Y., Hong F., Wang G., and An C. (2015) Ostrinia furnacalis serpin-3 regulates melanization cascade by inhibiting a prophenoloxidase-activating protease. Insect Biochem. Mol. Biol. 61, 53–61 [DOI] [PubMed] [Google Scholar]
- 52. Lu Z., Beck M. H., Wang Y., Jiang H., and Strand M. R. (2008) The viral protein Egf1.0 is a dual activity inhibitor of prophenoloxidase-activating proteinases 1 and 3 from Manduca sexta. J. Biol. Chem. 283, 21325–21333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Heraty J. (2009) Parasitoid diversity and insect pest management, in Insect Biodiversity: Science and Society (Adler P. H., and Foottit R. G., eds) pp. 445–462, Wiley-Blackwell, Hoboken, NJ [Google Scholar]
- 54. Jiang H., and Kanost M. R. (1997) Characterization and functional analysis of 12 naturally occurring reactive site variants of serpin-1 from Manduca sexta. J. Biol. Chem. 272, 1082–1087 [DOI] [PubMed] [Google Scholar]
- 55. Jiang H., Wang Y., Huang Y., Mulnix A. B., Kadel J., Cole K., and Kanost M. R. (1996) Organization of serpin gene-1 from Manduca sexta. Evolution of a family of alternate exons encoding the reactive site loop. J. Biol. Chem. 271, 28017–28023 [DOI] [PubMed] [Google Scholar]
- 56. Zou Z., Picheng Z., Weng H., Mita K., and Jiang H. (2009) A comparative analysis of serpin genes in the silkworm genome. Genomics 93, 367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zheng Y. P., He W. Y., Béliveau C., Nisole A., Stewart D., Zheng S. C., Doucet D., Cusson M., and Feng Q. L. (2009) Cloning, expression and characterization of four serpin-1 cDNA variants from the spruce budworm, Choristoneura fumiferana. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 154, 165–173 [DOI] [PubMed] [Google Scholar]
- 58. Suwanchaichinda C., and Kanost M. R. (2009) The serpin gene family in Anopheles gambiae. Gene 442, 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wong E. S., and Belov K. (2012) Venom evolution through gene duplications. Gene 496, 1–7 [DOI] [PubMed] [Google Scholar]
- 60. Burke G. R., and Strand M. R. (2014) Systematic analysis of a wasp parasitism arsenal. Mol. Ecol. 23, 890–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Goecks J., Mortimer N. T., Mobley J. A., Bowersock G. J., Taylor J., and Schlenke T. A. (2013) Integrative approach reveals composition of endoparasitoid wasp venoms. PLoS One 8, e64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Colinet D., Anselme C., Deleury E., Mancini D., Poulain J., Azéma-Dossat C., Belghazi M., Tares S., Pennacchio F., Poirié M., and Gatti J. L. (2014) Identification of the main venom protein components of Aphidius ervi, a parasitoid wasp of the aphid model Acyrthosiphon pisum. BMC Genomics 15, 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mrinalini and Werren J. H. (2016) Parasitoid wasps and their venoms, in Evolution of Venomous Animals and Their Toxins (Gopalakrishnakone P., and Malhotra A., eds) pp. 1–26, Springer, Rotterdam, Netherlands [Google Scholar]
- 64. Siigur E., Aaspõllu A., and Siigur J. (2001) Sequence diversity of Vipera lebetina snake venom gland serine proteinase homologs—result of alternative-splicing or genome alteration. Gene 263, 199–203 [DOI] [PubMed] [Google Scholar]
- 65. Vaiyapuri S., Wagstaff S. C., Harrison R. A., Gibbins J. M., and Hutchinson E. G. (2011) Evolutionary analysis of novel serine proteases in the venom gland transcriptome of Bitis gabonica rhinoceros. PLoS One 6, e21532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Martinson E. O., Martinson V. G., Edwards R., Mrinalini, and Werren J. H. (2016) Laterally transferred gene recruited as a venom in parasitoid wasps. Mol. Biol. Evol. 33, 1042–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mortimer N. T., Goecks J., Kacsoh B. Z., Mobley J. A., Bowersock G. J., Taylor J., and Schlenke T. A. (2013) Parasitoid wasp venom SERCA regulates Drosophila calcium levels and inhibits cellular immunity. Proc. Natl. Acad. Sci. U.S.A. 110, 9427–9432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ragan E. J., An C., Yang C. T., and Kanost M. R. (2010) Analysis of mutually exclusive alternatively spliced Serpin-1 isoforms and identification of Serpin-1 proteinase complexes in Manduca sexta hemolymph. J. Biol. Chem. 285, 29642–29650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tang Q. Y., and Zhang C. X. (2013) Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 20, 254–260 [DOI] [PubMed] [Google Scholar]
- 70. Wiśniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 71. Perkins D. N., Pappin D. J., Creasy D. M., and Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 72. Marshall O. J. (2004) PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 20, 2471–2472 [DOI] [PubMed] [Google Scholar]
- 73. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 74. Kapustin Y., Souvorov A., Tatusova T., and Lipman D. (2008) Splign: algorithms for computing spliced alignments with identification of paralogs. Biol. Direct 3, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Edgar R. C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., and Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 [DOI] [PubMed] [Google Scholar]
- 77. Arnold K., Bordoli L., Kopp J., and Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 78. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.